Abstract
We have studied four Caenorhabditis elegans homologs of the Rad21/Scc1/Rec8 sister-chromatid cohesion protein family. Based on the RNAi phenotype and protein localization, it is concluded that one of them, W02A2.6p, is the likely worm ortholog of yeast Rec8p. The depletion of C. elegans W02A2.6p (called REC-8) by RNAi, induced univalent formation and splitting of chromosomes into sister chromatids at diakinesis. Chromosome synapsis at pachytene was defective, but primary homology recognition seemed unaffected, as a closer-than-random association of homologous fluorescence in situ hybridization (FISH) signals at leptotene/zygotene was observed. Depletion of REC-8 also induced chromosome fragmentation at diakinesis. We interpret these fragments as products of unrepaired meiotic double-stranded DNA breaks (DSBs), because fragmentation was suppressed in a spo-11 background. Thus, REC-8 seems to be required for successful repair of DSBs. The occurrence of DSBs in REC-8-depleted meiocytes suggests that DSB formation does not depend on homologous synapsis. Anti-REC-8 immunostaining decorated synaptonemal complexes (SCs) at pachytene and chromosomal axes in bivalents and univalents at diakinesis. Between metaphase I and metaphase II, REC-8 is partially lost from the chromosomes. The partial loss of REC-8 from chromosomes between metaphase I and metaphase II suggests that worm REC-8 might function similarly to yeast Rec8p. The loss of yeast Rec8p from chromosome arms at meiosis I and centromeres at meiosis II coordinates the disjunction of homologs and sister chromatids at the two meiotic divisions.
Keywords: Meiosis, sister chromatid cohesion, meiotic pairing, C. elegans, synaptonemal complex, recombination
The orderly disjunction of chromosomes during mitosis requires the coordinated separation of sister chromatids at the onset of anaphase. Precocious separation of sister chromatids can cause their missegregation and the formation of aneuploid daughter cells, because the spindle apparatus can regularly disjoin only chromatids, which it recognizes as pairs by their physical connection. Therefore, the cohesion of sister chromatids after chromosome replication at S-phase is an essential function. Only at the moment when all chromosomes are aligned at the equator of the dividing cell, is association of sister chromatids released, which permits their movement to opposite poles. To obtain proper centromeric orientation and disjunction, the best place to tether sister chromatids would be the centromeric region. However, Rattner et al. (1988) detected a presumptive sister chromatid linking protein all along muntjak chromosome arms. Conversely, metaphase chromosomes of cells treated with spindle poisons often show split sister chromatids that are only connected at their centromeric regions (C-mitosis — Rieder and Cole 1999; Nasmyth et al. 2000). This led Rieder and Cole (1999) to propose that chromatids are tethered at their centromeres and along their arms by different structures. It is likely that the primary role of centromeric cohesion is in proper chromosome segregation, whereas arm cohesion serves in facilitating mitotic recombinational repair of DNA lesions via the sister molecule, but has acquired an additional function in meiosis (see below). Several proteins mediating the cohesion of sister chromatids are now known from a variety of organisms (Guacci et al. 1997; Michaelis et al. 1997; Furuya et al. 1998; Losada et al. 1998; Skibbens et al. 1999; Tóth et al. 1999). Four of these proteins (Smc1, Smc3, Scc1/Mcd1/Rad21, and Scc3) are part of a multisubunit complex called cohesin in vertebrates (Losada et al. 1998) and in yeast (Tóth et al. 1999).
Whereas in mitosis sister chromatids are separated during a single anaphase movement, in meiosis first homologous chromosomes and then sister chromatids are separated in the course of two subsequent division steps. It has long been realized that the maintenance of bivalents by chiasmata might be based on sister-chromatid cohesion. If sister-chromatid cohesion is lost from chromosome arms distal to a chiasma, the chiasma would lose its capacity to hold homologs together (Maguire 1982, 1990; Buonomo et al. 2000). Therefore, loss of sister-chromatid cohesion along the chromosome arms at anaphase I would abolish the association of homologs. Loss of cohesion at or around the centromeres would allow sisters to separate at anaphase II. Thus, as in mitosis, one might expect two cohesion apparatuses to be present in meiosis, one along the chromosome arms and the other at the centromeres (Maguire et al. 1993). In Drosophila, the protein MEI-S332 was found to be associated with centromeres until sister chromatids separate, which made it a likely candidate for a centromere-specific cohesion protein both in mitosis and meiosis (Moore et al. 1998). On the other hand, it was found by Moens and Spyropoulos (1995) in the mouse that Cor1, a protein of the axes of meiotic chromosomes, is lost from chromosome arms at the onset of chromosomal disjunction in anaphase I, whereas it is retained at the centromeres up to anaphase II. Thus, the two meiotic divisions could be coordinated by the sequential destabilization of elements of one and the same cohesion apparatus in the arms and centromeres.
Recently, this hypothesis was confirmed in the budding yeast by the characterization of Rec8p, which is a meiosis-specific version of the cohesion protein Scc1/Rad21. Rec8p is lost from chromosome arms and centromeric regions in the predicted order (Klein et al. 1999; Buonomo et al. 2000). The absence of Rec8p caused chaotic segregation of chromosomes. A second cohesion protein, Smc3, which is not exclusively meiotic, showed similar behavior. Both proteins colocalize with chromosome cores at pachytene and seem to be components of the lateral elements of the synaptonemal complex (SC) with an additional role in recombination (Klein et al. 1999). Members of the Rec8/Scc1/Rad21 protein family with exclusive or predominant meiotic expression have been identified and characterized in the fission yeast, in Arabidopsis thaliana, and in man (Bhatt et al. 1999; Parisi et al. 1999; Watanabe and Nurse 1999).
Whereas cytological studies in budding yeast have shown that Rec8p is directly involved in chiasma maintenance, and suggest its dual role in the stabilization of bivalents and the cohesion of sister chromatids (Klein et al. 1999; Buonomo et al. 2000), direct evidence for a similar role of Rec8p homologs in higher organisms is lacking. Yeast is a highly derived eukaryotic microorganism with some meiotic particularities (Loidl 2000), thus, the existence of homologous proteins in plants and animals does not imply that their functions have been conserved in evolution. Therefore, we have set out to study the meiotic function of Rec8p homologs in the animal model system Caenorhabditis elegans. The nematode worm C. elegans offers the advantage of the easy identification of meiotic stages attributable to the linear array of meiotic stages along the hermaphrodite gonad. Chromosomes in diplotene/diakinesis can be studied easily, whereas this crucial stage is not amenable to cytological investigation in yeast. Moreover, chromosomal nondisjunction can be detected by a noncytological assay in the worm (Albertson et al. 1997). The chromosomes of C. elegans are holocentric during mitosis, whereas during the two meiotic divisions they are functionally monocentric. The sites of meiotic centromeres are not predetermined, as it was found that either end of a chromosome can adopt the role of the centromere during meiosis I (Albertson and Thomson 1993; Wicky and Rose 1996). This facultative centromere formation and the independence of chromosome synapsis from the initiation of recombination in the worm (Dernburg et al. 1998) make its meiosis notably different from yeast meiosis. The wide phylogenetic distance between Saccharomyces cerevisiae and C. elegans and the differences in meiosis would suggest that any mechanism of sister chromatid cohesion they have in common is highly conserved among eukaryotes. Therefore, we searched for C. elegans homologs of the Scc1p/Rec8p family of cohesion proteins. We tested their cellular localization by immunocytology, and studied the consequences of their loss using double-stranded RNA mediated interference (RNAi).
Results
Four SCC1/REC8 homologs
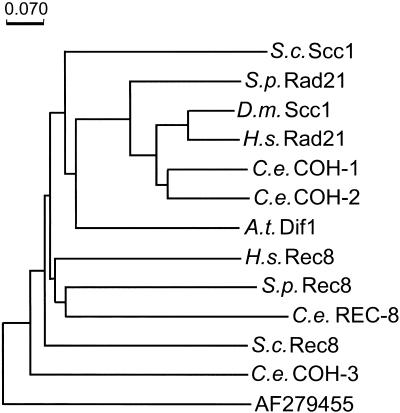
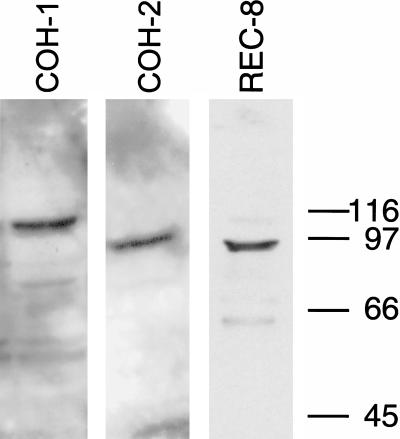
In a database screen, K08A8.3, F10G7.4, F08H9.1, and W02A2.6 were identified as four putative C. elegans homologs of the genes for the Rad21/Scc1/Rec8 sister-chromatid cohesion family of proteins (Fig. 1). RNAi-depletion of the products of three of them resulted in a Him (high incidence of males) phenotype and caused reduction of embryo viability, consistent with meiotic defects. (A Him phenotype is generated as a consequence of missegregation of a sex chromosome in the germ line of XX hermaphrodites to produce X0 males.) Portions of the three open reading frames (ORFs) were amplified by PCR and cloned to allow their expression as GST-fusion proteins. The fusion proteins were purified and used to immunize rabbits. The resulting antisera were tested on Western blots and antisera that recognized a single band of proper molecular weight in N2 worms were used for further analysis (Fig. 2). On the basis of RNAi-depletionphenotypes and immunolocalization, W02A2.6p was identified as the most likely ortholog of the Schizosaccharomyces pombe and S. cerevisiae meiotic cohesion protein Rec8p, and hence was named REC-8. Assigning a clear mitotic or meiotic role to K08A8.3p, F10G7.4p, and F08H9.1p proved to be more difficult, and they were designated cohesion protein homologs COH-1, COH-2, and COH-3 (Fig. 1).
Figure 1.
Phylogenetic tree of Rad21/Scc1/Rec8 family members, calculated by the Phylo_Win program using the Neighbor Joining method (Galtier et al. 1996). The tree is based on a Clustal W-alignment with minor modifications (Thompson et al. 1994) of the amino terminus of Scc1/Rec8 proteins. S.c. = Saccharomyces cerevisiae, S.p. = Schizosaccharomyces pombe, D.m. = Drosophila melanogaster, H.s. = Homo sapiens, C.e. = Caenorhabditis elegans, A.t. = Arabidopsis thaliana. For protein sequences used see Parisi et al. (1999), Bhatt et al. (1999), and Klein et al. (1999). The next noncohesion protein hit (EMBL/GenBank accession no. AF279455) in a PSI-BLAST search with H. sapiens Scc1p as the query, served as the outgroup. The scale bar denotes the number of substitutions per site.
Figure 2.
Expression of K08A8.3p (COH-1), F10G7.4p (COH-2), and W02A2.6p (REC-8) in Caenorhabditis elegans N2. Antisera against the three proteins detected single prominent bands. The minor bands most likely correspond to breakdown products. COH-1 and COH-2 have predicted molecular weights of 75 kD, whereas the bands detected by the respective antisera indicate a slightly aberrant migration. The band detected by the REC-8 antiserum corresponds exactly to the predicted 89 kD of the protein. All antisera could be blocked efficiently by recombinant protein, indicating that the observed signals are specific (not shown).
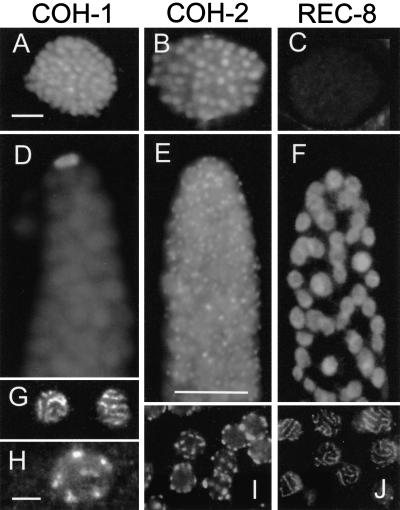
The product of coh-1 was detected by immunocytology in somatic and embryonic nuclei (Fig. 3A) but was not found to be localized to the condensed chromosomes of cells in mitosis. Within the gonad, the strongest immunostaining of COH-1 was found in the nucleus of the distal tip cell (which is not part of the germ line) (Fig. 3D). COH-1 was strikingly absent from the mitotic and the transition zone of the gonads but reappeared on pachytene and diplotene/diakinesis chromosomes (Fig. 3 G,H). RNAi-depletion eliminated COH-1-immunostaining and caused both reduced embryo viability to ∼20% of the brood and a slightly increased incidence (∼2%) of males in F1 worms around 36 h after the delivery of double-stranded RNA (dsRNA). The meiotic pairing of chromosomes appeared normal.
Figure 3.
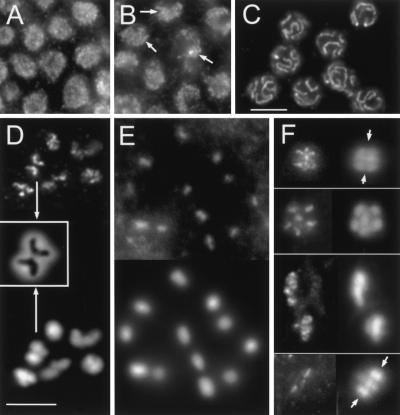
Immunolocalization of Rec8p homologs. COH-1 is present in the nuclei of embryos (A) and of the distal tip cell (D). It also localizes to chromosomes in pachytene (G) to diakinesis (H). COH-2 is localized in embryonic nuclei (B) and forms foci from the mitotic up to the pachytene zone of the gonad (E, I). In pachytene nuclei the foci are exclusively peripheral (I). REC-8 can not be detected in embryos (C). It is homogeneously distributed in early nuclei of the gonad (F) and is condensed into threads in pachytene nuclei (J). Bars: 5 μm in A for A–C, 10 μm in E for D–F, 5 μm in H for G–J.
coh-2 was expressed primarily in germline cells as seen by immunofluorescence. Its product was present as dots, mainly at the periphery but also inside nuclei of the mitotic zone (Fig. 3E). In later stages, COH-2 assumed an exclusively peripheral distribution (Fig. 3I). In pachytene nuclei, ∼10 (10.1 ± 1.5 [n = 25 nuclei]) distinct spots were counted but by diplotene and later COH-2 was not detectable. COH-2 was present in the nuclei of embryos but was not associated with condensed mitotic chromosomes. RNAi with coh-2 dsRNA depleted the protein from germ cells as judged by immunostaining. RNAi caused complete embryonic lethality up to 36 h after delivery of the dsRNA. At later time points, presumably when the effect of RNAi ceased, some survivors appeared, of which ∼3% were males. SC formation in COH-2 RNAi-depleted cells appeared normal. At diakinesis, bivalents were present but nondisjunction seemed to occur at anaphase I, as chromosome number was irregular in the rare metaphase II nuclei amenable to analysis.
RNAi-depletion of COH-3 failed to produce reduced embryonic viability or a Him phenotype, nor did it cause aberrant meiotic chromosome behavior, although coh-3 has been shown to be expressed above somatic level in the germline (see http://cmgm.stanford.edu/∼kimlab/germline cited in Reinke et al. 2000).
Immunolocalization of REC-8 to chromosomes in the gonad
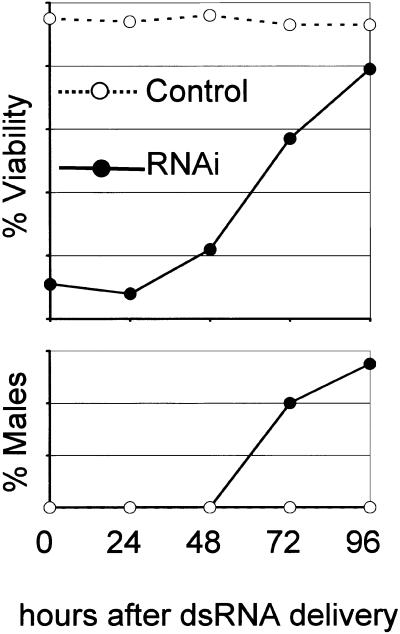
RNAi depletion of REC-8, the fourth Rad21/Scc1/Rec8p homolog, caused high embryonic lethality, a stronger Him phenotype (11% males, see Fig. 4) and cytological defects.
Figure 4.
RNAi-induced reduction in viability and induction of Him phenotype for rec-8. Worms were soaked in dsRNA for 12 h and then transferred to fresh plates every 24 h. The percentage of viability and sex of the laid offspring was determined for each 24 h interval. Viability was strongly reduced for the first two intervals. At later times, when a sufficiently high number of viable embryos appeared because of fading of the RNAi-effect, increased incidence of male offspring was observed. No data could be collected for time points later than 96 h as worms ran out of sperm and produced unfertilized eggs. In total, viability was determined for ∼3000 embryos.
Indirect immunostaining with rabbit or rat antiserum against a 350 amino acid fragment of REC-8 showed that the protein was absent from somatic cells (Fig. 3C). In the gonad, REC-8 was localized as small grains all over the nuclei of the mitotic zone (Figs. 3F, 5A), whereas in the transition zone and in pachytene nuclei, it assembled into short and long threads, respectively (Figs. 3J, 5B,C). In diakinesis, REC-8 was localized along the axes of chromosomes (Fig. 5D). This pattern of localization in leptotene/zygotene to diakinesis nuclei is the same as the one described for HIM-3, which is a component of the axes of meiotic chromosomes with some homology to yeast Hop1p (Zetka et al. 1999).
Figure 5.
REC-8 immunostaining pattern throughout gonadal mitosis and meiosis. (A) Grains in nuclei of the mitotic zone. (B) Formation of short threads (arrows) in the transition zone (leptotene/zygotene). (C) REC-8 delineates synaptonemal complexes (SCs) in pachytene. In D and E immunostaining is shown on top and DAPI at the bottom, in F immunostaining is on the left and DAPI on the right. Diakinesis: Labeling of chromosomal axes in bivalents (D) and spo-11 univalents (E). Insert in D shows an enlarged DAPI-stained bivalent (arrows) with the inverted image of the REC-8 axes superimposed. (F) From top to bottom: Metaphase I (side view, two bivalents in focus, arrows denote equator), early anaphase I (top view of half-nucleus; the other half is in a different focal plane underneath), late anaphase I (separating nuclei out of alignment due to preparation), metaphase II (side view, three chromosomes in the focal plane; arrows denote equator, polar body outside the area shown). Bar in C represents 10 μm in A–C; bar in D, 5 μm in D–F.
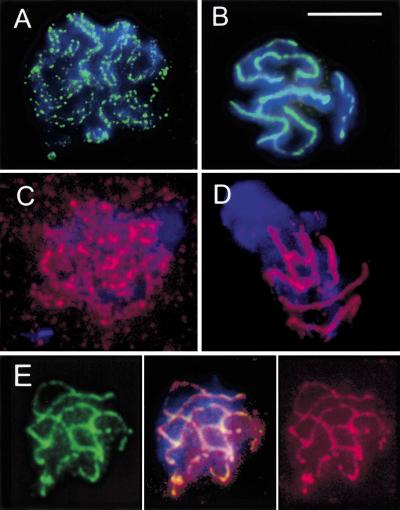
To better visualize chromosome pairing and the spatial relationship of HIM-3 and REC-8, a nuclear spreading technique for high resolution of SCs was performed. Preparations were immunostained with rat anti-REC-8 and rabbit anti-HIM-3 (kindly provided by Monique C. Zetka, University of California, Davis) either separately or simultaneously. In leptotene/zygotene nuclei, both HIM-3 and REC-8 were present as dots along unsynapsed chromosomal axes (Fig. 6A,C), which sometimes could be seen to be associated loosely in pairs (Fig. 6A). The side-by-side arrangement of chromosomes prior to synapsis is reminiscent of the presynaptic alignment of homologs known from a variety of organisms. In pachytene nuclei, both HIM-3 and REC-8 antibodies decorated the six SCs (Fig. 6B,D). HIM-3/REC-8 double immunostaining produced an almost complete overlap (Fig. 6E), suggesting that REC-8 is a component of or associated with axial and lateral elements at leptotene/zygotene and pachytene, respectively, which persists after the decomposition of the SC.
Figure 6.
Immunostaining of HIM-3 (A,B,E left: green) and REC-8 (C,D,E right: red) of spread nuclei at leptotene/zygotene (A,C) and pachytene (B,D,E). Note the parallel association of forming axial elements (presynaptic alignment) in A. Both proteins delineate the six synaptonemal complexes (SCs) at pachytene (B,D) and show nearly perfect colocalization by double immunostaining (E, center). Counterstaining of chromatin by DAPI (blue). Bar: 10 μm.
To test if REC-8 localization or maintenance is dependent on meiotic double-stranded DNA breaks (DSBs), we performed immunostaining in spo-11 mutant animals that do not form DSBs and chiasmata (Dernburg et al. 1998). REC-8 was present along the SC and the axes of diplotene/diakinesis univalents (where it colocalized with HIM-3), which indicated that the protein is loaded independently of DSB induction and recombination, and confirmed that it is a component of individual chromosomes rather than of bivalents (Fig. 5E).
From metaphase I through to metaphase II REC-8 signals appeared to become progressively less intense or smaller (Fig. 5F). In metaphase I, both the poleward and equatorial axes of bivalents were decorated with REC-8 (Fig. 5F, top) whereas in early anaphase I chromosomes, the length of REC-8 immunosignals seemed to be restricted to the original poleward portion of metaphase I chromosomes. In late anaphase I, individual chromosomes could no longer be discriminated and REC-8 spots were visible within the highly condensed DAPI-positive masses of the two half-nuclei. In metaphase II short tracts of REC-8 were observed in the equatorial plane (Fig. 5G).
No immunofluorescence was detected after rec-8 RNAi, i.e., in the gonads of F1 worms from embryos laid by animals 24 h or later after dsRNA injection. Also the staining with preimmune serum was negative.
The cytological phenotype of REC-8-depleted worms
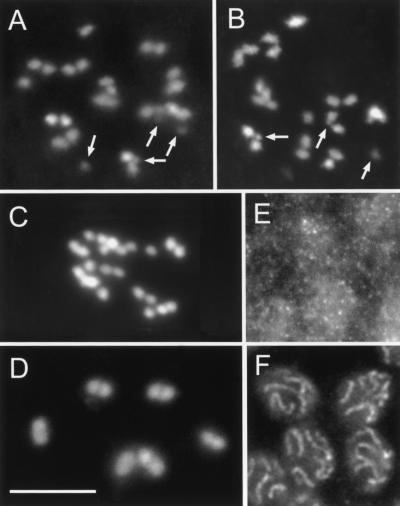
Rec-8-RNAi both by soaking and injection produced a variety of effects on meiotic chromosomes. In injection experiments, effects were manifested in F1 worms grown from embryos that were laid 24- to 84 h after dsRNA administration. The most striking consequence of REC-8-depletion was the presence of ∼24 DAPI-positive linear structures, as well as several additional dots in diakinesis (Fig. 7A,B), whereas untreated controls showed six bivalents (Fig. 7D). This indicates that homologous chromosomes were not connected by chiasmata and, moreover, that they split into their sister chromatids. The separation of homologs was always complete, whereas the sister chromatids were seen as separate units which were either lying side-by-side or completely detached. Double RNAi with either coh-1 or coh-2 and rec-8 dsRNA did not result in a stronger separation of sisters (data not shown), making the possibility that COH-1 or COH-2 partially substitutes for REC-8 rather unlikely. We consider it likely that residual associations between sister chromatids are caused by the absence of a repulsive force rather than by the existence of nonREC-8-mediated cohesion. However, we cannot rule out the possibility that another, as yet unidentified, cohesion protein provides a weak connection.
Figure 7.
Cytological effects of REC-8 depletion. (A,B) Separated sister chromatids and chromosomal fragments (arrows; some fragments slightly out of focus) at diakinesis (DAPI staining). (C) Sister chromatids are separated, but no fragments occur after REC-8 depletion in spo-11 deletion animals. (D) Wild-type (untreated) diakinesis showing six bivalents (DAPI). (E) Dispersed HIM-3 immunostaining in pachytene zone after RNAi. (F) Immunolocalization of HIM-3 in wild-type pachytene. Bar: 10 μm.
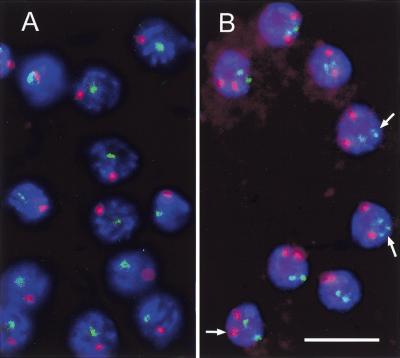
Univalent formation at diakinesis can be caused by a failure of homology recognition and/or synapsis, or to the inability to undergo crossing-over or to transform crossovers into stable chiasmata. To discriminate between these possibilities, we analyzed chromosome pairing at leptotene/zygotene (in nuclei of the transition zone) and pachytene by fluorescence in situ hybridization (FISH). Two regions on chromosomes I and V were differentially labeled and the frequencies of associated signal pairs were recorded and compared to untreated controls (Fig. 8A,B). Association of like signals was interpreted as homologous pairing, whereas the association of signals of different colors is a measure of the frequency of random collisions (Loidl et al. 1994). In controls, homologous pairing was frequent both in leptotene/zygotene and pachytene nuclei (Table 1). In REC-8-depleted animals, homologous pairing was completely disrupted in pachytene (Table 1). In leptotene/zygotene nuclei, pairing was not or only slightly reduced compared to the untreated control (Table 1). To verify that leptotene/zygotene pairing had indeed taken place in cells depleted of REC-8, evaluation was performed in F1 individuals laid by injected worms within a certain time window after injection. After 24 h, when the corresponding leptotene/zygotene cell population in worms of the same batch had most likely proceeded to pachytene, pairing was absent in such nuclei, which confirmed that RNAi had been effective at the early meiotic stages. Thus, we think it likely that primary homology recognition and alignment (see also Fig. 6A) are not dependent on REC-8, although we cannot rule out the possibility that a small amount of the protein that may be produced even after RNAi is sufficient for presynaptic alignment, but not synapsis.
Figure 8.
Detection of homologous pairing at pachytene in wild-type (A) and REC-8-depleted (B) gonads, as determined by two color FISH (green: chromosome I locus; red: chromosome V locus; blue: DNA staining by DAPI). Pairing is deficient in nuclei lacking the protein. Arrows indicate split sister chromatids. Bar: 10 μm.
Table 1.
Homologous pairing evaluated by the association of FISH signals
|
|
REC-8-depleted
|
Untreated controls
|
||||
|---|---|---|---|---|---|---|
| mitotic zone (n = 198)
|
leptotene/zygotenea (n = 155)
|
pachytene (n = 159)
|
mitotic zone (n = 125)
|
leptotene/zygotenea (n = 141)
|
pachytene (n = 135)
|
|
| Homologous pairingb at both locic | 0 | 93% | 2% | n.d.e | 94% | 100% |
| Homologous pairing at one locus | 7% | 4% | 3% | 15% | 2% | 0 |
| No pairing | 85% | 3% | 91% | 85% | 4% | 0 |
| Pairing of nonhomologous locid | 8% | 1% | 4% | n.d.e | 6% | 11% |
Nuclei in the transition zone of gonads were defined as corresponding to leptotene/zygotene. These nuclei are characterized by the crested appearance of chromatin after DAPI staining.
Loci were classified as paired if the corresponding FISH signals merged into a single signal or were separated by ≤ the width of a signal.
Two loci on chromosomes I and V were tested (see Materials and methods).
Pairing categories can sum up to >100% because nuclei showing nonhomologously paired loci may at the same time have homologous loci paired. The category “Pairing of nonhomologous loci” is shown to demonstrate the relative rareness of these events. The frequency of associations of nonhomologous loci is a measure for random associations and has to be subtracted from the frequency of homologous associations to determine the frequency of nonrandom, specific homologous pairing (see Loidl et al. 1994).
n.d. = not determined; for the mitotic zone of the control, pairing was determined only for the locus on chromosome V.
In leptotene/zygotene and pachytene nuclei, signals corresponding to a locus on a single chromosome sometimes appeared as doublets (Fig. 8B), which indicates that the separation of sister chromatids, which is almost complete in diakinesis, starts at these early stages. However, extensive splitting of sister chromatids at pachytene was rare. It is possible that the sister chromatids of zygotene and pachytene chromosomes remain linked even on the reduction of REC-8 to a cytologically undetectable level, as they are likely to be coiled around each other. Only when chromosomes become shorter and more rigid at the transition to diakinesis, the sites where they are intertwined become fewer, and finally the sisters fall apart.
To determine the structure of pachytene chromosomes after depletion of REC-8, we performed immunostaining with antibodies against the putative SC-component HIM-3 in RNAi-treated animals. In wild-type pachytene, HIM-3 was organized in long threads (Fig. 7F); whereas after RNAi treatment, HIM-3 granules were distributed all over the nuclei (Fig. 7E). Also in spreads, no HIM-3-positive axial structures were detected, except for the occasional occurrence of very weak dots that we interpret as being caused by variable response of worms to RNAi (data not shown). This supports a role of REC-8 as a structural component of axial and lateral elements. Although these elements serve as a scaffold with a role in the structural organization of chromatids, their lack or insufficiency had no obvious consequence for chromatin condensation at later stages, as this appeared normal in DAPI-stained diplotene/diakinesis nuclei of REC-8-depleted animals.
Chromosome fragments occur at diakinesis in REC-8-depleted cells
In addition to separate sister chromatids, 0–10 small DAPI-positive structures were present in diakinesis after RNAi with rec-8 dsRNA (Fig. 7A,B). To test the possibility that these structures are chromosome fragments that originate from unrepaired meiotic DSBs, we performed RNAi with rec-8 RNA in spo-11 mutant animals. SPO-11 is the enzyme that produces DSBs in meiosis and is essential for the initiation of meiotic recombination (Dernburg et al. 1998). No fragments were found in diakineses of treated spo-11 worms (Fig. 7C), which suggests that they originate from DSBs.
In REC-8 depleted wild-type cells, chromatid fragmentation was observed in F1 animals arising from embryos laid 24 h or more after dsRNA injection. This is also the time when chromatid splitting first appeared. Fragments were also found after dsRNA administration by soaking. The average number of fragments was less than the expected number of DSBs. It is possible that some fragments escaped detection because of their small size or because they adhered to their intact sister chromatids, just like sisters did not always completely separate after depletion of REC-8 (see above). It cannot, however, be excluded that in the complete absence of REC-8, DSBs would not occur at all and that the observed fragments are caused by the leakiness of the RNAi phenotype.
Discussion
Functions of three C. elegans cohesion proteins
We have identified four C. elegans proteins with homology to the Rad21/Scc1/Rec8 family of cohesin components (Fig. 1) (for review, see van Heemst and Heyting 2000). Three of them, namely coh-1, coh-2, and rec-8, showing reduced viability and a Him phenotype on RNAi-depletion, were characterized further.
The predominant occurrence of COH-1 in somatic nuclei and the failure of RNAi to exert an effect on chromosome pairing at pachytene or later, suggests that COH-1 has its main function in somatic nuclei. However, the observed reduction of embryo viability and the slight increase in males by RNAi are reconcilable both with the disturbance of embryonic mitoses or meiosis. A function of COH-1 in meiosis can, therefore, not be ruled out.
COH-2 was detected by immunostaining in nondividing embryonic and germline nuclei. Correspondingly, F10G7.4 (coh-2) expression in the germline has been found to be only slightly, if at all increased above somatic expression (see http://cmgm.stanford.edu/∼kimlab/germline cited in Reinke et al. 2000). The low viability and slight Him phenotype caused by RNAi might be attributable to nondisjunction at meiosis I as we observed aberrant chromosome numbers in interkinesis/metaphase II nuclei. Pachytene pairing, SC formation and diakinesis bivalent formation appeared undisturbed in RNAi individuals. Because no signals were detectable by immunostaining in RNAi-treated worms, we believe that RNAi produces a strong loss-of-function phenotype, and truly reflects the relatively mild effect of a coh-2 defect on meiosis.
REC-8 is a component of meiotic chromosome axes and has a function in sister chromatid cohesion
C. elegans REC-8 shows 17.3% identity and 41.6% similarity to S. cerevisiae Rec8p. The depletion of REC-8 caused defective chromosome pairing and the separation of sister chromatids prior to the first meiotic division, leading to chromosomal nondisjunction. This together with the localization of REC-8 at chromosomal axes strongly suggests that it is the ortholog of Rec8p, a yeast meiotic sister chromatid cohesion protein (Molnar et al. 1995; Klein et al. 1999).
Separation of sister chromatids in REC-8-depleted animals seems to start as early as in leptotene/zygotene as judged from the occasional presence of more than two FISH signals in nuclei of the transition zone. In diakinesis most sisters are completely disconnected although they may still lie side-by-side. The presence of single sister chromatids is expected to cause considerable disturbances during the following anaphase. The few nuclei we observed that had gone beyond diakinesis, showed irregular chromosome numbers. In S. pombe carrying a rec8 deletion, sister chromatids separate at meiosis I with high regularity. This equational first division is followed by a random second division (Watanabe and Nurse 1999). In the case of S. pombe, this led to a reduction in spore viability of the rec8Δ strain to 21%. Given that this organism has only three chromosomes in the haploid, this still high spore viability can be explained by the fairly high probability for each spore to receive at least one copy of each chromosome by a division in which three pairs of chromatids are randomly segregated. Also in REC-8-depleted C. elegans there might be a slight preference for chromosomes to divide equationally at meiosis I, as the residual side-by-side alignment of sisters would tend to promote their coorientation and subsequent disjunction. We do not know whether a second division is attempted or if restitution nuclei are formed after a single division.
Homologous alignment is independent of synapsis
The application of a spreading method to leptotene/zygotene nuclei of C. elegans clearly revealed parallel alignment of homologous axes at a distance (Fig. 6A), resembling the presynaptic alignment found in many other organisms, including yeast (Scherthan et al. 1992; for review, see Loidl 2000). In wild-type meiosis, however, it is difficult to tell whether homologous alignment in transition-zone nuclei precedes SC formation or is the consequence of distinct SC initiation sites along the homologs.
In animals depleted of REC-8, primary homology recognition and alignment in the nuclei of the transition zone (corresponding to leptotene/zygotene), as revealed by FISH, appeared unaffected (Fig. 8; Table 1). Conversely, FISH analysis and immunostaining of axial elements using HIM-3 as a marker revealed that pachytene pairing was eliminated and SCs did not form. This observation suggests that the homologous associations in nuclei of the transition zone reflect SC-independent alignment in the course of primary homology recognition.
Chromosome fragments in diakinesis nuclei of REC-8-depleted animals suggest meiotic DSB formation in the absence of synapsis
Evidence from the budding yeast, S. cerevisiae, that initiation of meiotic recombination precedes the connection of homologous chromosomes by the SC led to a revision of the classical view of meiotic pairing (Hawley and Arbel 1993). The traditional view had held that the SC provides the physical link between pairing chromosomes and the framework within which the molecular process of recombination is initiated and completed (for review, see Loidl 1990). In yeast it was found that homologous synapsis is preceded by early steps in the meiotic recombination process (Padmore et al. 1991) and requires them to take place (Alani et al. 1990). The initial event in meiotic recombination is the induction of DSBs that are catalyzed by Spo11p, a type II topoisomerase (Bergerat et al. 1997; Keeney et al. 1997). DSBs are extended to gaps and repaired by association with the homologous chromosome whose sequence at the site of the gap serves as the template for repair synthesis. This repair process is recombinogenic and synapsis seems to initiate at the sites of interchromosomal recombination (Chua and Roeder 1998).
Contrary to the situation in yeast, both in Drosophila (McKim et al. 1998) and in C. elegans (Dernburg et al. 1998), deletion of spo-11 does not prevent SC formation. This could mean that these organisms either conform with the classical theory, namely that synapsis is required for the initiation of molecular recombination, or that DSBs can occur independently of synapsis. Here, we observed DSB-generated chromosomal fragments in diakineses of REC-8-depleted animals. At the same time RNAi-depletion of REC-8 caused a defect in synapsis. Unless short tracts of SC are formed which we were unable to detect, this would mean that DSBs originate independently of synapsis but cannot be repaired properly. It is likely that repair is hampered by the unavailability of a homologous template in the absence of pairing. Although the sister chromatid is also a potential source of homology for recombinational repair, repair from the sister is actively suppressed in yeast meiosis (Arbel et al. 1999). It can substitute for repair by the homolog if this is not available, as is the case in haploid meiosis (de Massy et al. 1994; Gilbertson and Stahl 1994). In the absence of REC-8, not only is synapsis disturbed, but also sisters are disconnected. Therefore, neither the homolog nor the sister would be available as a template for recombinational repair of DSBs in REC-8-depleted C. elegans meiosis.
A conserved role for Rec8 proteins?
It has been shown in the fission yeast that the meiosis-specific protein Rec8 is involved in the formation of axial elements, chromosome pairing, and sister-chromatid cohesion (Molnar et al. 1995). The budding yeast ortholog was found to be distributed along the axes of paired chromosomes during pachytene, to be maintained only at or near the centromeres during anaphase I and to disappear completely at anaphase II (Klein et al. 1999). In the absence of Rec8p, the segregation of chromatids is random. The observations in budding yeast favored a model (Klein et al. 1999; Nasmyth et al. 2000) according to which the loss of sister chromatid cohesion distal to chiasmata allows the separation of homologs during the first meiotic division while sister centromeres move to the same pole (Moens and Spyropoulos 1995). The separation of sister centromeres is made possible only by the loss of cohesins from the centromeric region prior to the second meiotic division. Thus, in budding yeast the sequential disappearance of Rec8p from the arms and the centromeres ensures the coordination of reductional and equational chromosome separation at meiosis I and II, respectively, and it is reasonable to assume that centromere and arm cohesion are mediated by the same structures. Also the behavior of the mouse protein Cor1 (also known as Scp3), which is a component of SCs and meiotic chromosome cores from zygotene to diakinesis but is retained only at centromeres after metaphase I, was believed to indicate the existence of a common structure for centromere and arm cohesion (Moens and Spyropoulos 1995). It is doubtful if Cor1/Scp3 is a cohesion protein, as centromere cohesion appears unaffected in zygotene nuclei of knockout mice (Yuan et al. 2000). Even if Cor1/Scp3 is not the cohesion protein itself, it might require the latter for its localization, and therefore, mirror its stepwise loss from the chromosomes. Thus, the proposal of a single protein being responsible for arm and centromere cohesion in the mouse could still be valid.
In fission yeast this may be different, as it was shown that in the absence of Rec8p the first meiotic division is equational (Watanabe and Nurse 1999). Thus, there must exist an additional structure which persists at centromeres and promotes the faithful disjunction of sister chromatids. Likewise, in maize there exists a meiotic cohesion mutant, dsy1, which shows a massive failure of chiasma maintenance during diplotene and diakinesis, yet the adhesion of sister centromeres was much less affected (Maguire et al. 1993). This led Maguire et al. (1993) to suggest that centromere cohesion is mediated by different or additional factors to those that function in the arms.
Here, we observed in C. elegans the partial loss of REC-8 between metaphase and anaphase I (Fig. 5F). In early anaphase I, linear tracts of REC-8 were present that appeared to be shorter than in half-bivalents at diakinesis/metaphase I, and in late anaphase the size of REC-8 signals was reduced further. This can be interpreted as a gradual loss, beginning at the noncentromeric chromosome ends, which progresses towards the regions with centromeric activity. Bivalents would separate as soon as REC-8 has disappeared from the regions between the noncentromeric ends and the chiasma whereas the proximal portions of sister chromatids are still connected. The mostly distal position of chiasmata would explain why there is no massive reduction of REC-8 in anaphase I as compared to metaphase I. A low amount of REC-8 was seen in the equatorial plane of metaphase II division plates. This agrees with the experiments of Albertson et al. (1997) in which it was shown that the ends of the chromatids that had kinetic activity in the first division were located at the equator, whereas the opposite ends adopted kinetic function in metaphase II.
Whereas for yeast it has been assumed previously that after metaphase I Rec8p is retained only at or near the centromeres, the limited resolution of yeast cytology leaves room for the interpretation that Rec8p is localized between the centromere and the nearest crossover (Tóth et al. 2000). Also in C. elegans it appears that from anaphase I onward, REC-8 is retained in the region between the facultative centromere and the (mostly single) chiasma rather than at the centromere alone. It is even possible that the centromere occupies the whole region between the telomere and the chiasma. But irrespective of its precise spatial relationship to the centromere, the reduction of the amount of REC-8 from metaphase I to metaphase II is consistent with its exclusive maintenance in chromosomal regions that retain cohesion until anaphase II. This is similar to S. cerevisiae Rec8p, which is involved in the timing of meiotic divisions by mediating both centromere-proximal and distal arm cohesion.
Given the similarity of the meiotic cohesion apparatus in budding yeast and the worm, both provide themselves as systems to study the coordination of reductional and equational meiotic division. In particular this will be of use when searching for factors that affect the retarded loss of centromeric (proximal) Rec8p. It will also be interesting to explore whether the meiotic cohesion apparatus that is common to budding yeast, C. elegans (and probably also the mouse), is shared with other species or if centromeric and arm cohesion can be mediated by different proteins.
Materials and methods
Worm strains and culture conditions
The wild-type (N2 Bristol) strain was obtained from the Caenorhabditis Genetics Center (University of Minnesota, St. Paul, MN). The spo-11 mutant strain with a deletion in ORF T05E11.4 (Dernburg et al. 1998) was kindly provided by Anne Villeneuve (Stanford University). Strains were cultured at 20°C on NGM-plates with E. coli OP50 following the methods described by Brenner (1974).
dsRNA interference (RNAi)
Target sequences were PCR-amplified from C. elegans first strand cDNA and cloned into the pGEM-T Easy vector (Promega, Madison, WI). Plasmids containing the insert were linearized with either PstI or ApaI and were used for in vitro transcription with T7 and SP6 RNA polymerases, respectively. After removing the template by DNase treatment, RNA was phenol-extracted and precipitated over night at −20°C. Single-stranded RNA (ssRNA) was used for making dsRNA, which was then precipitated. The typical yield of a single preparation was about 10 μg of dsRNA.
dsRNA was administered by soaking or injection. For soaking, about 10 μg of dsRNA was dissolved in 40 μl of 1× M9 buffer (0.3% KH2PO4, 0.6% Na2HPO4, 0.5% NaCl, 1 mM MgS04). L4 larvae were put on an NGM plate without OP50 for 3 h and then 40–60 worms were transferred to the dsRNA solution in an Eppendorf tube and were soaked for 12 h at 20°C. Alternatively, dsRNA solution was injected into the gonads of young adult hermaphrodites according to Fire et al. (1998). Injected animals (P) were transferred to NGM agar plates with E. coli OP50, singled after 24 h, and then transferred to fresh NGM plates every 12 h. Efficient RNAi was confirmed by testing F2 viability and (in the case of REC-8) the DAPI phenotype of a few F1 worms from each individual batch, and siblings were used for further experiments.
Production and testing of antisera
For antibody production, regions of the K08A8.3 (coh-1), F10G7.4 (coh-2), and W02A2.6 (rec-8) ORFs were amplified by PCR from C. elegans first strand cDNA prepared from polyA+ RNA. The amplified fragments were chosen to encode regions with little homology between the three ORFs to reduce the possibility of crossreactivity. The regions chosen were: K08A8.3 aa146–513, F10G7.4 aa161–440, and W02A2.6 aa511–781. The amplified fragments were first cloned into pGEM-T Easy vector and verified by sequencing. Subsequently, the fragments were transferred to a pGEX vector (Pharmacia, Uppsala, Sweden) using suitable restriction sites. After transformation into E. coli BL21 the fragments were expressed as glutathione-S-transferase (GST) fusions and purified from crude E. coli lysates on glutathione Sepharose (Pharmacia). The purified protein was lyophilized and used for immunization of rabbits and, in the case of W02A2.6p, both rabbits and rats (Eurogentech, Seraing, Belgium).
Antisera were tested on immunoblots for their ability to recognize a dilution of the antigen used for immunization but also to recognize the endogenous protein in Bristol N2 worms (Fig. 2). Worm lysates were made by pelleting worms in 1× M9 buffer. The worm pellet was resuspended in SDS sample buffer and worms were first mechanically crushed with a pestle and subsequently sonicated to break the DNA. Proteins were separated on 8% SDS-polyacrylamide gels and blotted onto a PVDF membrane (Millipore, Bedford, MA). To detect the protein, primary antisera were used at a dilution of 1:500 to 1:1000. Primary antibodies were detected by chemi-luminescence following the manufacturer's instructions (Pierce SuperSignal, Pierce, Rockford, IL). Where necessary, primary antisera were blocked with acetone powder prepared from E. coli OP50 to eliminate background bands emanating from contaminating E. coli proteins (Harlow and Lane 1988). Specificity of antisera was also tested by in situ staining in comparison with preimmune sera.
Conventional preparation
To prepare whole gonads for light microscopy and FISH, gravid hermaphrodites were transferred to 10 μl of M9 buffer (as above) on a poly-L-lysine-coated microscope slide. (For coating, 3 μl of a 0.1% solution of poly-L-lysine were spread out on a clean slide and left to dry.) The gonads were released by cutting worms with a fine injection needle behind the pharynx and in the tail region. An equal volume of 7.4% formaldehyde was added, and a coverslip was placed on the tissue and gently squashed to remove excess fixative. Slides were then immediately frozen in liquid nitrogen for 10 min, the coverslip was cracked off, and slides were transferred into 96% ethanol at −20°C for 5 min. Slides were then allowed to warm up in ethanol to above 0°C and transferred to 1× PBS. Nuclei and chromosomes were stained by mounting the preparations with 2 μg/ml 4‘,6-diamidino-2-phenylindole (DAPI) in anti-fading agent (Vectashild, Vector Laboratories, Burlingame, CA). For storage or for subsequent application of the FISH procedure, slides were dehydrated in an ethanol series (2× 70%, 1× 96%, 2 min each) and air dried. For indirect immunostaining worms were prepared as above and squashed under a coverslip without the addition of paraformaldehyde. After removal of coverslips on ice, preparations were fixed in a series of methanol/methanol:acetone (1:1)/acetone for 5 min each at−2°C, and immediately transferred to 1× PBS without drying.
SC spreading procedure
The spreading procedure applied is a modification of a protocol for the production of whole mount SCs in plants by Albini and Jones (1984). Ten to 15 young adults were transferred to a vessel with 1× PBS to rid them of adhering bacteria. Worms were opened under the dissecting microscope and gonads were cut open. The worms with their gonads cut open were transferred to 10 μl 1× M9 buffer on a poly-L-lysine-coated slide. Worms were squashed with the flat end of a brass rod to crush the tissues. Ten microliters of 1% Lipsol (L.I.P. Ltd., Shipley, UK) were added to the cell suspension and spreading of cells and nuclei was monitored in phase contrast. When meiocytes had swelled and poured out from the gonadal tubes, the process was terminated by the addition of 30 μl fixative (4% paraformaldehyde and 3.4% sucrose in distilled water). Care was taken to keep the cells concentrated in a small area of the slide. The slides were stored horizontally and left to dry.
Immunostaining
Conventional or spread preparations were washed three times 5 min in 1× PBS and blocked with 3% BSA in 1× PBS under a coverslip for 30 min at 37°C in a humid chamber. The coverslip was rinsed off and the specimen was incubated with primary antibody (Ab) diluted in Ab buffer (1% BSA, 0.5% Triton X-100, 0.05% sodium azide, and 1% acetone powdered E. coli OP50 in 1× PBS) (Harlow and Lane 1988). Dilutions were 1:500 for anti HIM-3, 1:100 for anti COH-1 and anti COH-2, and 1:50 for anti REC-8 antibodies. As a positive control of immunostaining, mouse monoclonal antibody directed against histone H1 (1:30; MAB1276, Chemicon International Inc., Temecula, CA) was used. Slides were incubated with the primary Ab under a coverslip for 90 min at room temperature. The coverslip was then rinsed off and slides were washed three times 5 min in 1× PBS. Incubation with the secondary Ab (anti-rabbit Cy3 [1:250], anti-rabbit FITC [1:500] or anti-rat FITC [1:500]) in Ab buffer was done for 30 min at room temperature. For double immunostaining (rabbit anti-HIM-3/rat anti-REC-8), primary Ab’s were applied simultaneously, and secondary Ab's were applied consecutively. Finally, slides were washed three times in 1× PBS containing 0.1% Tween 20, and mounted in Vectashield supplemented with 2μg/ml DAPI.
FISH
Pooled cosmids C53D5 and R119 (obtained from Alan Coulson, Nematode Functional Genomics Group, Sanger Centre) were used as a probe for the left end chromosome I. Cosmid DNA was isolated following standard procedures. To create a probe for a distal region on the right arm of chromosome V, a 133-bp sequence from the coding region of the 5S rDNA repeat was amplified from genomic DNA by PCR using the oligonucleotides GGCACTTGTCCATAGACGC and CTTACAACATC CAGGATTCC as primers. The cosmids and the PCR product were labeled with digoxigenin-11-dUTP or biotin-21-dUTP using the BioNick Labeling System (Life Technologies, Rockville MD) according to the manufacturer's instructions.
FISH was performed on conventional preparations according to (Dernburg et al. 1998) with some modifications. Briefly, air-dried slides were permeabilized by applying 20 μl of 1 M NaSCN under a coverslip for 4 min at 90°C (Meyer-Ficca et al. 1998). The slides were then washed three times for 5 min in 2× SSC and the specimens were subjected to RNAse treatment (30 μg per slide) at 37°C for 40 min followed by formaldehyde treatment (3.7% in 2× SSC) for 10 min at room temperature to stop enzymatic reactions. Slides were washed three times for 5 min in 2× SSC, dehydrated in an ethanol series (2× 70%, 1× 96%, 2 min each) and air dried.
Labeled probe DNA (60 ng) mixed with salmon sperm DNA (20 μg) was vacuum-dried and resuspended in 6 μl of formamide (100%) and 6 μl of hybridization mix (4× SSC, 20% dextransulfate). The probe was denatured at 95°C for 5 min and placed on ice for 1 min. After spinning down, it was dropped onto the slide and a coverslip was sealed with rubber cement (Fixogum, Marabuwerke GmbH, Tamm, Germany). DNA on slides was then denatured at 80°C for 10 min, and left up to 48 h at 37°C in a humid chamber to hybridize. Coverslips were rinsed off by incubation of slides in 2× SSC containing 50% formamide in a coplin jar at 42°C. The slides were transferred to 1× SSC, 0.2× SSC, and 0.1× SSC for 5 min at 42°C. Excess liquid was drained and blocking solution (3% BSA, 0.1% Tween 20, 4× SSC) was applied for 30 min at 37°C.
Digoxigenin-labeled probes were detected with rhodamine-conjugated anti-digoxigenin and biotinylated probes with FITC-conjugated streptavidin in detection solution (1% BSA, 0.1% Tween 20, 4× SSC) at 37°C for 50 min. Finally, slides were washed three times for 5 min in washing solution (4× SSC, 0.1% Tween 20) and mounted in Vectashield supplemented with DAPI (2μg/ml).
Fluorescence microscopy and evaluation
Chromosomes stained with the DNA-specific fluorescent dye DAPI, and FISH and immunofluorescence patterns were examined with a Zeiss Axioskop equipped with an epifluorescence system. Appropriate filters were used to detect blue (DAPI), green (FITC), and red (Cy3) emitted light. Black and white images were taken for each wavelength with a cooled CCD camera (Photometrics, Tucson, AZ) and assigned false colors reminiscent of the color of fluorescence. For multicolored preparations, each color was recorded separately and images were merged electronically. Image recording and processing was done with the help of IPLab Spectrum 3.1 software (Scanalytics, Fairfax, VA).
Acknowledgments
We are grateful to Monique Zetka for the generous gift of anti HIM-3 antiserum and Arnold Holik and Andrea Neunteufl for technical assistance. We acknowledge the help of Verena Jantsch-Plunger and of Joerg Fuchs with dsRNA injection and FISH, respectively. We also thank Kim Nasmyth, Tim Schedl, and Maria Siomos for their valuable comments on the manuscript. This work was supported by grant S8211 of the Austrian Science Fund.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL josef.loidl@univie.ac.at; FAX +43-1-4277-9541.
Article and publication are at http://www.genesdev.org/cgi/doi/10.1101/gad.192701.
References
- Alani E, Padmore R, Kleckner N. Analysis of wild-type and rad50 mutants of yeast suggests an intimate relationship between meiotic chromosome synapsis and recombination. Cell. 1990;61:419–436. doi: 10.1016/0092-8674(90)90524-i. [DOI] [PubMed] [Google Scholar]
- Albertson DG, Thomson JN. Segregation of holocentric chromosomes at meiosis in the nematode, Caenorhabditis elegans. Chromosome Res. 1993;1:15–26. doi: 10.1007/BF00710603. [DOI] [PubMed] [Google Scholar]
- Albertson DG, Rose AM, Villeneuve AM. Chromosome organization, mitosis, and meiosis. In: Riddle DL, Blumenthal T, Meyer BJ, Priess JR, editors. C. elegans II. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1997. pp. 47–78. [PubMed] [Google Scholar]
- Albini SM, Jones GH. Synaptonemal complex-associated centromeres and recombination nodules in plant meiocytes prepared by an improved surface-spreading technique. Exp Cell Res. 1984;155:588–592. doi: 10.1016/0014-4827(84)90219-2. [DOI] [PubMed] [Google Scholar]
- Arbel A, Zenvirth D, Simchen G. Sister chromatid-based DNA repair is mediated by RAD54, not by DMC1 or TID1. EMBO J. 1999;18:2648–2658. doi: 10.1093/emboj/18.9.2648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergerat A, de Massy B, Gadelle D, Varoutas P-C, Nicolas A, Forterre P. An atypical topoisomerase II from archaea with implications for meiotic recombination. Nature. 1997;386:414–417. doi: 10.1038/386414a0. [DOI] [PubMed] [Google Scholar]
- Bhatt AM, Lister C, Page T, Fransz P, Findlay K, Jones GH, Dickinson HG, Dean C. The DIF1 gene of Arabidopsis is required for meiotic chromosome segregation and belongs to the REC8/RAD21 cohesin gene family. Plant J. 1999;19:463–472. doi: 10.1046/j.1365-313x.1999.00548.x. [DOI] [PubMed] [Google Scholar]
- Brenner S. The genetics of Caenorhabditis elegans. Genetics. 1974;77:71–94. doi: 10.1093/genetics/77.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buonomo SBC, Clyne RK, Fuchs J, Loidl J, Uhlmann F, Nasmyth K. Disjunction of homologous chromosomes in meiosis I depends on proteolytic cleavage of the meiotic cohesin Rec8p by separin. Cell. 2000;103:387–398. doi: 10.1016/s0092-8674(00)00131-8. [DOI] [PubMed] [Google Scholar]
- Chua PR, Roeder GS. Zip2, a meiosis-specific protein required for the initiation of chromosome synapsis. Cell. 1998;93:349–359. doi: 10.1016/s0092-8674(00)81164-2. [DOI] [PubMed] [Google Scholar]
- de Massy B, Baudat F, Nicolas A. Initiation of recombination in Saccharomyces cerevisiae haploid meiosis. Proc Natl Acad Sci. 1994;91:11929–11933. doi: 10.1073/pnas.91.25.11929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dernburg AF, McDonald K, Moulder G, Barstead R, Dresser M, Villeneuve AM. Meiotic recombination in C. elegans initiates by a conserved mechanism and is dispensable for homologous chromosome synapsis. Cell. 1998;94:387–398. doi: 10.1016/s0092-8674(00)81481-6. [DOI] [PubMed] [Google Scholar]
- Fire A, Xu S, Montgomery MK, Kostas SA, Driver SE, Mello CC. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature. 1998;391:806–811. doi: 10.1038/35888. [DOI] [PubMed] [Google Scholar]
- Furuya K, Takahashi K, Yanagida M. Faithful anaphase is ensured by Mis4, a sister chromatid cohesion molecule required in S phase and not destroyed in G1 phase. Genes & Dev. 1998;12:3408–3418. doi: 10.1101/gad.12.21.3408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galtier N, Gouy M, Gautier C. SEAVIEW and PHYLO_WIN: Two graphic tools for sequence alignment and molecular phylogeny. Comput Appl Biosci. 1996;12:543–548. doi: 10.1093/bioinformatics/12.6.543. [DOI] [PubMed] [Google Scholar]
- Gilbertson LA, Stahl FW. Initiation of meiotic recombination is independent of interhomologue interactions. Proc Natl Acad Sci. 1994;91:11934–11937. doi: 10.1073/pnas.91.25.11934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guacci V, Koshland D, Strunnikov A. A direct link between sister chromatid cohesion and chromosome condensation revealed through the analysis of MCD1 in S. cerevisiae. Cell. 1997;91:47–57. doi: 10.1016/s0092-8674(01)80008-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harlow E, Lane D. Antibodies: A laboratory manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1988. [Google Scholar]
- Hawley RS, Arbel T. Yeast genetics and the fall of the classical view of meiosis. Cell. 1993;72:301–303. doi: 10.1016/0092-8674(93)90108-3. [DOI] [PubMed] [Google Scholar]
- Keeney S, Giroux CN, Kleckner N. Meiosis-specific DNA double-strand breaks are catalyzed by Spo11, a member of a widely conserved protein family. Cell. 1997;88:375–384. doi: 10.1016/s0092-8674(00)81876-0. [DOI] [PubMed] [Google Scholar]
- Klein F, Mahr P, Galova M, Buonomo SBC, Michaelis C, Nairz K, Nasmyth K. A central role for cohesins in sister chromatid cohesion, formation of axial elements, and recombination during yeast meiosis. Cell. 1999;98:91–103. doi: 10.1016/S0092-8674(00)80609-1. [DOI] [PubMed] [Google Scholar]
- Loidl J. The initiation of meiotic chromosome pairing: The cytological view. Genome. 1990;33:759–778. doi: 10.1139/g90-115. [DOI] [PubMed] [Google Scholar]
- ————— Meiosis in budding yeast and in multicellular eukaryotes — similarities and differences. Chromosomes Today. 2000;13:123–137. [Google Scholar]
- Loidl J, Klein F, Scherthan H. Homologous pairing is reduced but not abolished in asynaptic mutants of yeast. J Cell Biol. 1994;125:1191–1200. doi: 10.1083/jcb.125.6.1191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Losada A, Hirano M, Hirano T. Identification of Xenopus SMC protein complexes required for sister chromatid cohesion. Genes & Dev. 1998;12:1986–1997. doi: 10.1101/gad.12.13.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maguire MP. The mechanism of chiasma maintenance. A study based upon behaviour of acentric fragments produced by crossovers in heterozygous paracentric inversions. Cytologia. 1982;47:699–711. [Google Scholar]
- ————— Sister chromatid cohesiveness: Vital function, obscure mechanism. Biochem Cell Biol. 1990;68:1231–1242. doi: 10.1139/o90-183. [DOI] [PubMed] [Google Scholar]
- Maguire MP, Riess RW, Paredes AM. Evidence from a maize desynaptic mutant points to a probable role of synaptonemal complex central region components in provision for subsequent chiasma maintenance. Genome. 1993;36:797–807. doi: 10.1139/g93-105. [DOI] [PubMed] [Google Scholar]
- McKim KS, Green-Marroquin BL, Sekelsky JJ, Chin G, Steinberg C, Khodosh R, Hawley RS. Meiotic synapsis in the absence of recombination. Science. 1998;279:876–878. doi: 10.1126/science.279.5352.876. [DOI] [PubMed] [Google Scholar]
- Meyer-Ficca M, Müller-Navia J, Scherthan H. Clustering of pericentromeres initiates in step 9 of spermiogenesis of the rat (Rattus norvegicus) and contributes to a well-defined genome architecture in the sperm nucleus. J Cell Sci. 1998;111:1363–1370. doi: 10.1242/jcs.111.10.1363. [DOI] [PubMed] [Google Scholar]
- Michaelis C, Ciosk R, Nasmyth K. Cohesins: Chromosomal proteins that prevent premature separation of sister chromatids. Cell. 1997;91:35–45. doi: 10.1016/s0092-8674(01)80007-6. [DOI] [PubMed] [Google Scholar]
- Moens PB, Spyropoulos B. Immunocytology of chiasmata and chromosomal disjunction at mouse meiosis. Chromosoma. 1995;104:175–182. doi: 10.1007/BF00352182. [DOI] [PubMed] [Google Scholar]
- Molnar M, Baehler J, Sipiczki M, Kohli M. The rec8 gene of Schizosaccharomyces pombe is involved in linear element formation, chromosome pairing and sister-chromatid cohesion during meiosis. Genetics. 1995;141:61–73. doi: 10.1093/genetics/141.1.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore DP, Page AW, Tang TTL, Kerrebrock AW, Orr-Weaver TL. The cohesion protein MEI-S332 localizes to condensed meiotic and mitotic centromeres until sister chromatids separate. J Cell Biol. 1998;140:1003–1012. doi: 10.1083/jcb.140.5.1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nasmyth K, Peters J-M, Uhlmann F. Splitting the chromosome: Cutting the ties that bind sister chromatids. Science. 2000;288:1379–1384. doi: 10.1126/science.288.5470.1379. [DOI] [PubMed] [Google Scholar]
- Padmore R, Cao L, Kleckner N. Temporal comparison of recombination and synaptonemal complex formation during meiosis in Saccharomyces cerevisiae. Cell. 1991;66:1239–1256. doi: 10.1016/0092-8674(91)90046-2. [DOI] [PubMed] [Google Scholar]
- Parisi S, McKay MJ, Molnar M, Thompson MA, van der Spek PJ, van Drunen-Schoenmaker E, Kanaar R, Lehmann E, Hoeijmakers JHJ, Kohli J. Rec8p, a meiotic recombination and sister chromatid cohesion phosphoprotein of the Rad21p family conserved from fission yeast to humans. Mol Cell Biol. 1999;19:3515–3528. doi: 10.1128/mcb.19.5.3515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rattner JB, Kingwell BG, Fritzler MJ. Detection of distinct structural domains within the primary constriction using autoantibodies. Chromosoma. 1988;96:360–367. doi: 10.1007/BF00330702. [DOI] [PubMed] [Google Scholar]
- Reinke V, Smith HE, Nance J, Wang J, Van Doren C, Begley R, Jones SJM, Davis EB, Scherer S, Ward S, et al. A global profile of germline gene expression in C. elegans. Mol Cell. 2000;6:605–616. doi: 10.1016/s1097-2765(00)00059-9. [DOI] [PubMed] [Google Scholar]
- Rieder CL, Cole R. Chromatid cohesion during mitosis: Lessons from meiosis. J Cell Sci. 1999;112:2607–2613. doi: 10.1242/jcs.112.16.2607. [DOI] [PubMed] [Google Scholar]
- Scherthan H, Loidl J, Schuster T, Schweizer D. Meiotic chromosome condensation and pairing in Saccharomyces cerevisiae studied by chromosome painting. Chromosoma. 1992;101:590–595. doi: 10.1007/BF00360535. [DOI] [PubMed] [Google Scholar]
- Skibbens RV, Corson LB, Koshland D, Hieter P. Ctf7p is essential for sister chromatid cohesion and links mitotic chromosome structure to the DNA replication machinery. Genes & Dev. 1999;13:307–319. doi: 10.1101/gad.13.3.307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson JD, Higgins DJ, Gibson TJ. CLUSTAL W: Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tóth A, Ciosk R, Uhlmann F, Galova M, Schleiffer A, Nasmyth K. Yeast Cohesin complex requires a conserved protein, Eco1p(Ctf7), to establish cohesion between sister chromatids during DNA replication. Genes & Dev. 1999;13:320–333. doi: 10.1101/gad.13.3.320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tóth A, Rabitsch KP, Gálová M, Schleiffer A, Buonomo SBC, Nasmyth K. Functional genomics identifies monopolin: A kinetochore protein required for segregation of homologs during meiosis I. Cell. 2000;103:1155–1168. doi: 10.1016/s0092-8674(00)00217-8. [DOI] [PubMed] [Google Scholar]
- van Heemst D, Heyting C. Sister chromatid cohesion and recombination in meiosis. Chromosoma. 2000;109:10–26. doi: 10.1007/s004120050408. [DOI] [PubMed] [Google Scholar]
- Watanabe Y, Nurse P. Cohesin Rec8 is required for reductional chromosome segregation at meiosis. Nature. 1999;400:461–464. doi: 10.1038/22774. [DOI] [PubMed] [Google Scholar]
- Wicky C, Rose AM. The role of chromosome ends during meiosis in Caenorhabditis elegans. Bioessays. 1996;18:447–452. doi: 10.1002/bies.950180606. [DOI] [PubMed] [Google Scholar]
- Yuan L, Liu J-G, Zhao J, Brundell E, Daneholt B, Höög C. The murine SCP3 gene is required for synaptonemal complex assembly, chromosome synapsis and male fertility. Mol Cell. 2000;5:73–83. doi: 10.1016/s1097-2765(00)80404-9. [DOI] [PubMed] [Google Scholar]
- Zetka MC, Kawasaki I, Strome S, Muller F. Synapsis and chiasma formation in Caenorhabditis elegans require HIM-3, a meiotic chromosome core component that functions in chromosome segregation. Genes & Dev. 1999;13:2258–2270. doi: 10.1101/gad.13.17.2258. [DOI] [PMC free article] [PubMed] [Google Scholar]