Abstract
The continued expansion of microbial sequence data has allowed for the detection of an increasing number of conserved RNA motifs by using comparative sequence analysis. Recently, we reported the discovery of two structured non-coding RNA motifs, called glnA and downstream-peptide, that have similarity in sequence and secondary structure. In this report, we describe data demonstrating that representatives of both RNA motifs selectively bind the amino acid l-glutamine. These glutamine aptamers are found exclusively in cyanobacteria and marine metagenomic sequences, wherein several glnA RNA representatives reside upstream of genes involved in nitrogen metabolism. These motifs have genomic distributions that are consistent with a gene regulation function, suggesting they are components of glutamine-responsive riboswitches. Thus, our findings implicate glutamine as a regulator of cyanobacterial nitrogen metabolism pathways. Furthermore, our findings expand the collection of natural aptamer classes that bind amino acids to include glycine, lysine and glutamine.
Key words: cooperative, E-loop, glutamate, pseudoknot, riboswitch
Introduction
Riboswitches are genetic regulatory elements composed solely of RNA that bind metabolites and control gene expression commonly without the involvement of protein factors.1 Most simple riboswitches are composed of an aptamer domain and an expression platform, where the aptamer functions as a receptor for a specific metabolite and the expression platform modulates the expression of one or more genes in a ligand-dependent fashion.2,3 Riboswitches are usually found in the 5′ untranslated regions (UTRs) of bacterial mRNAs and often control gene expression in cis either at the level of transcription or translation, although other regulatory mechanisms are also known.4 In most cases, metabolite binding triggers a structural rearrangement that affects the formation of either a terminator stem or a base-paired element that occludes the ribosome binding site. In addition, there is a known example of a trans-acting riboswitch5 as well as eukaryotic riboswitches6 that modulate expression by controlling alternative mRNA spicing in algae,7 plants8 and fungi.9
Comparative sequence analysis methods have been developed for novel riboswitch class discovery.10–12 These techniques involve computational searches through genomic and metagenomic databases for sequences that are conserved both in their primary and secondary structures.13,14 Through one of these searches, the glnA motif and the Downstream-peptide motif (Fig. 1) were discovered in cyanobacteria and marine metagenomic sequences.15 These motifs are both approximately 60 nucleotides in length, comprise three base-paired regions, and share highly similar portions of conserved sequence and structure. However, there are some minor structural differences that distinguish the two motifs. The glnA motif includes a predicted E-loop connecting the junction of stems P2 and P3 (J2/3) with J3/1. In contrast, the Downstream-peptide motif lacks an E-loop and P3 stem, but instead forms a pseudoknot. An additional difference between the two RNAs is their genetic placement. While the Downstream-peptide motif is found exclusively in the 5′ UTRs of short polypeptides of unknown function that are typically 17 to 100 amino acids in length, the glnA motif is frequently positioned upstream of a variety of genes involved in nitrogen metabolism including ammonium transporters, glutamine and glutamate synthetases, and nitrogen regulatory protein PII.15
Figure 1.
Consensus sequence and secondary structure models for two candidate riboswitch aptamer families. (A) The glnA motif is a 3-stem junction (stems are named P1, P2 and P3) that carries an E-loop and a possible single long-distance base pair (dashed line). (B) The Downstream-peptide motif is formed by three extended base-paired substructures wherein P1 and P2 are nearly identical to those of the glnA motif. The motif lacks P3 and E-loop features, but nucleotides in this region form a pseudoknot. Like glnA RNAs, the Downstream-peptide motif can potentially form a single long-range base pair. The two motifs also carry identical nucleotides at the base of P1 and in the junction. These models are derived using methods and data reported previously in reference 15.
The two motifs share various qualities with previously characterized riboswitches, including size, complexity, degree of sequence conservation, and genetic context. Considering the nature of the genes downstream of the glnA motif, we speculated that l-glutamine could be the ligand for this candidate riboswitch aptamer. Glycine and lysine are known ligands for riboswitches,16,17 this establishes a precedent for riboswitch aptamers recognizing amino acids. Furthermore, glutamine is an attractive riboswitch ligand candidate given its involvement in nitrogen regulation processes.
Nitrogen is often a limiting nutrient in marine environments which makes accurately monitoring internal nitrogen levels particularly important for aquatic bacteria.18 Although glutamine is known to be a key indicator of the state of nitrogen metabolism in proteobacteria and firmicutes,19,20 other compounds are thought control this set of pathways in cyanobacteria.21–23 In this study, we demonstrate that the glnA and Downstream-peptide motifs are structural variants of a novel aptamer class responsive to glutamine, providing the first evidence that this amino acid is an important signaling molecule in the regulation of nitrogen metabolism in cyanobacteria.
Results
Given its characteristics revealed by bioinformatics, we hypothesized that glnA motif RNAs might be representatives of a newfound riboswitch aptamer class. Because these RNAs are encoded upstream of several genes involved in nitrogen metabolism, we tested a 67 nucleotide glnA representative from Synechococcus elongatus, termed 67 glnA (Fig. 2A) with l-glutamine (Fig. 2B) and a collection of potential ligands related to this set of metabolic pathways (Fig. 3). In-line probing assays revealed that 67 glnA binds most tightly to L-glutamine with an apparent dissociation constant (KD) of approximately 575 µM (Fig. 2B and C). The shape of the binding curve matches that expected for a one-to-one interaction between the RNA and its ligand.
Figure 2.
The 67 glnA RNA binds l-glutamine. (A) Sequence and secondary structural model for the 67 glnA RNA from S. elongatus. Circled positions indicate areas of the RNA where internucleotide linkages undergo reduced (red) or constant (yellow) scission as ligand concentrations are increased when subjected to in-line probing (data from B). Nucleotides depicted in lowercase identify guanosine residues added to the construct to facilitate efficient in vitro transcription. Asterisks indicate the boundaries of the annotations for in-line probing results that could be clearly resolved by PAGE. (B) In-line probing analysis of 5′ 32P-labeled 67 glnA RNA. Precursor RNAs (Pre) were loaded onto gel lanes after treatment as follows: NR, no reaction; T1, partial digest with RNase T1 (cleaves after G residues); −OH, partial alkaline-mediated degradation. Additional lanes were loaded with precursor RNAs subjected to in-line probing conditions without ligand (-), or were subjected to inline probing conditions in the presence of various concentrations of l-glutamine ranging from 1 µM to 10 mM. Vertical lines designate areas where band intensities decrease as the RNA is exposed to higher concentrations of ligand. Band intensities of numbered regions were quantified and used to assess the extent of ligand binding. (C) Plot of the normalized fraction of band modulation (interpreted as fraction of RNAs bound to ligand) versus the logarithm of the concentration of ligand. Regions are as depicted in (B). The line represents the curve expected for a 1-to-1 RNA-ligand interaction with a KD of 575 µM.
Figure 3.
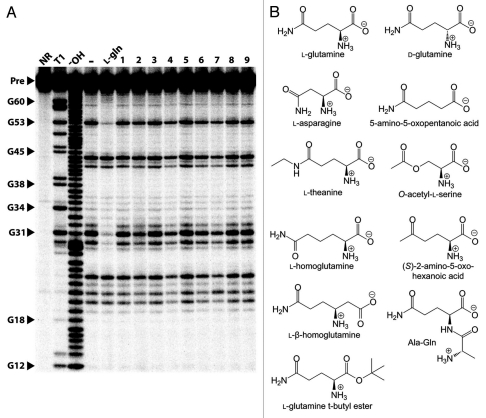
The 67 glnA RNA binds L-glutamine but rejects a variety of other compounds. (A) In-line probing analysis of the 67 glnA RNA with several glutamine analogs. Precursor RNAs were subjected to in-line probing conditions in the presence of 10 mM L-glutamine (L-gln) or 10 mM of the various compounds as follows: Ala-Gln (1), L-glutamine t-butyl ester (2), L-theanine (3), O-acetyl-L-serine (4), L-homoglutamine (5), L-β-homoglutamine (6), (S)-2-amino-5-oxo-hexonic acid (7), 5-amino-oxopentanoic acid (8) and asparagine (9). Other annotations are as described for Figure 2B. (B) Chemical structures of compounds tested with both the 67 glnA and 83 DP RNAs.
With the exception of d-glutamine, which is bound by the 67 glnA aptamer with approximately 1/10th the affinity of its more common isomer (Sup. Fig. S1), all other compounds tested were rejected by the aptamer even at concentrations as high as 10 mM (Fig. 3A). The compounds tested include the natural amino acid l-asparagine, the glutamine analogues l-glutamine t-butyl ester, l-theanine, O-acetyl-l-serine, l-homoglutamine, l-β-homoglutamine, (S)-2-amino-5-oxo-hexanoic acid, 5-amino-5-oxopentanoic acid, and the dipeptide Ala-Gln (Fig. 3B). Ligand-binding specificity was also assessed for a second representative of the glnA motif from a marine metagenomic sequence. This RNA binds to l-glutamine with a KD of approximately 150 µM, whereas putrescine, l-lysine, 2-oxoglutarate, γ-aminobutyric acid (GABA), glutaric acid, succinate, succinic semialdehyde, agmatine, pyridoxal phosphate and glutamate are all rejected at 1 mM (data not shown). Despite the lower KD value of this RNA, further experiments were conducted with the 67 glnA RNA due to more pronounced positions of modulating cleavage intensity in our in-line probing assasy.
To ensure that the binding observed was not the result of non-specific interactions between the RNA and glutamine, 67 glnA mutant RNA constructs M1 and M2 (Fig. 2A) containing two consecutive disruptive mutations in either the P2 or P3 stems, respectively, were prepared. As expected, we did not detect any modulation of these structurally disrupted RNAs upon addition of glutamine to in-line probing reactions (Sup. Fig. S2A and B), indicating that the banding pattern changes seen with the wildtype RNA with glutamine are caused by selective interactions. We then tested constructs M3 and M4 (Fig. 2A) in which the mismatched base pairs were restored with compensatory mutations. These RNAs regain structural modulation in response to glutamine addition, and exhibit KD values similar to that of the 67 glnA RNA (Sup. Fig. S2C–F).
We considered several reasons why glutamate, rather than or in addition to glutamine, could be the natural metabolite ligand for glnA motif aptamers. First, the chemical structures of glutamine and glutamate differ only by a side chain amino or hydroxyl group, respectively. Both these groups at a minimum could serve as a single hydrogen bond donor source. Second, some representatives of the glnA motif are found upstream of genes directly involved in glutamate synthesis. Third, the concentrations of both glutamine and glutamate are exceptionally high in bacteria. In Escherichia coli, glutamine is present in the bacteria at a concentration of approximately 4 mM, whereas the intracellular level of glutamate is approximately 100 mM.24 For a riboswitch to be selective for glutamine, it would need to discriminate against glutamate by more than 20-fold, given the exceptionally high concentration of this competing amino acid.
As noted above, we assessed binding by glutamate without observing binding at 1 mM amino acid concentration. However, to investigate possible glutamate binding at physiologically relevant concentrations, we conducted in-line probing assays using the 67 glnA RNA and 100 mM glutamate as the primary buffering agent. Although no structural modulation was observed in the presence of this high concentration of glutamate, the addition of 1 mM glutamine to an in-line probing assay containing 100 mM glutamate produced the pattern of RNA cleavage products expected for glutamine binding (Sup. Fig. S3).
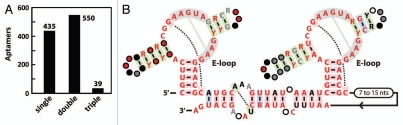
Interestingly, over half of the glnA RNAs are arranged in tandem orientations, where two or sometimes three aptamers are found grouped together with only small segments of intervening sequence in between (Fig. 4A). Of the double glnA aptamer arrangements, over half share a similar intervening sequence and an additional portion of conserved nucleotides 3′ of the aptamers that has the potential to base-pair with the intervening sequence (Fig. 4B). Multiple aptamers arrangements have been observed previously and can serve various biological purposes. For example, two aptamers sensitive to different ligands can influence the expression of the same gene, functioning similarly to two-input Boolean logic gates.25 Multiple aptamers that recognize the same compound can be utilized to achieve sharper, more digital responses to changing ligand concentrations either by using multiple terminator stems26 or cooperativity.17,27 In the case of glnA RNAs, the amount of intervening sequence between the aptamers is often apparently too small to allow for the use of multiple expression platforms, so we speculate that they might function in a cooperative fashion. To test this, several tandem glnA constructs were made and tested via in-line probing. These constructs included a three aptamer arrangement and a representative the group of RNAs shown in Figure 4B, both with and without the conserved portion of 3′ sequence. Our in-line probing analysis of these tandem constructs did not reveal any evidence of cooperative binding (data not shown), such as a steeper dose-response curve than that exhibited by non-cooperative riboswitches. Our data does not rule out cooperative function, since various factors such as inadequate construct length or non-physiological assay conditions could confound our assays.
Figure 4.
Tandem glutamine aptamers. (A) Distribution of glutamine aptamers among single, double and triple arrangements. Aptamers were grouped together if the amount of intervening sequence was less than 100 nucleotides. (B) Consensus sequence and structure of the most common tandem arrangement of glutamine aptamers. Annotations are as described in Figure 1A.
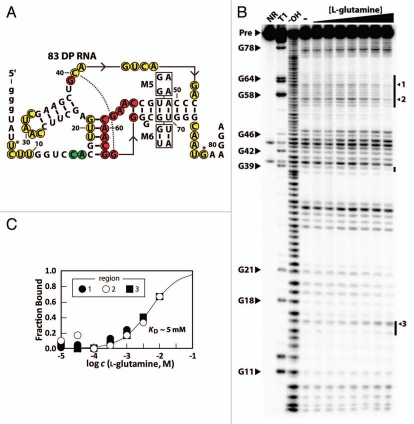
Despite the fact that the genetic contexts of glnA motif and Downstream-peptide motif RNAs are distinct, we speculated that the Downsteam-peptide motif might also bind glutamine due to sequence and structural similarities of the aptamer families. Again we used in-line probing on a representative member of the Downstream-peptide motif from Synechococcus sp. CC9902 (83 DP RNA) and determined that the RNA binds glutamine with an apparent KD of approximately 5 mM (Fig. 5). Unlike with the 67 glnA RNA, we did not detect any binding between the 83 DP RNA and D-glutamine at concentrations up to 10 mM (Sup. Fig. S4). Additionally, the RNA does not appreciably bind to any of the other compounds tested with the 67 glnA RNA at 10 mM concentrations (Sup. Fig. S4).
Figure 5.
The 83 DP RNA is an aptamer for glutamine. (A) Sequence and predicted secondary structure of the 83 DP RNA. Nucleotides circled in green indicate internucleotide linkages that undergo greater scission when the RNA is exposed to ligand. Other annotations are as described for Figure 2A. (B) In-line probing analysis of 5′ 32P-labeled 83 DP RNA with various concentrations of L-glutamine ranging from 10 µM to 10 mM. Annotations are as described for Figure 2B, with the exception that arrows indicate specific bands which were used to make the KD plot in (C). (C) Plot representing ligand binding as described for Figure 2C. The line represents the curve expected for a 1-to-1 interaction using a KD value 5 mM.
The predicted pseudoknot was validated by examining the ligand-binding functions of disruptive and compensatory mutations. Specifically, construct M5 was designed as a mutant 83 DP RNA with two disruptive mutations in the pseudoknot. As with the 67 glnA disruption mutation constructs, M5 RNA does not bind to glutamine (Sup. Fig. S5A). By contrast, a construct with compensatory mutations that restore base pairing within the pseudoknot (M6 RNA) binds to glutamine with a KD value similar to that of the DP RNA (Sup. Fig. S5B and C).
Discussion
The glnA motif and the Downstream-peptide motif share a variety of structural features and are able to bind glutamine while strongly discriminating against a variety of structurally related analogues. Mutational studies indicated the interactions between glutamine and the RNAs examined in this study are specific, because small disruptions of various predicted base-paired regions affect the ability of the RNAs to bind ligand. Additionally, these experiments support the accuracy of our secondary structural models because the compensatory mutations used in constructs M3, M4 and M6 indicate that the structure rather than the precise sequence in these putative stems is important. All of our findings are consistent with the hypothesis that these RNAs are subtypes of a novel glutamine riboswitch class, although we have not experimentally assessed gene regulation function.
Highly conserved nucleotides in loops and bulges are often indicative of positions essential for forming riboswitch aptamer binding pockets. We suspect that several residues in the three stem junction of the glnA aptamer and analogous positions on the Downstream-peptide aptamer are involved in the formation of the aptamer binding pocket. Additionally, the high degree of conservation of the nucleotides in the P1 stem leads us to believe that some of these residues directly participate in ligand binding as well.
Neither of the glutamine aptamer subtypes bind to any other compounds tested in our in-line probing assays. This observation, in conjunction with the high sequence similarity between the two related RNAs, implies that the binding pockets of both subtypes are similar. The fact that these RNAs reject glutamate, l-homoglutamine and (S)-2-amino-5-oxo-hexanoic acid indicates that the aptamers are highly sensitive to the length and composition of the amino acid side chain. This is an important characteristic for a receptor that must be responsive to a single amino acid, given that there are many other natural amino acids in all cells. Because the aptamers tested were both sensitive to the removal of the amino group in the side chain, the RNA likely makes one or more hydrogen bonds with the ligand at this position. Similarly, removal of the amino group attached to the α-carbon also causes a loss of binding, as is evident by our in-line probing results with the compound 5-amino-5-oxopentanoic acid. The loss of hydrogen bonds and/or ionic interactions is likely responsible for this compound's inability to serve as an aptamer ligand.
Large chemical groups on either the N or C termini of the amino acid are also not tolerated because neither Ala-Gln nor l-glutamine t-butyl ester are bound by the aptamers. The addition of bulky chemical groups to glutamine likely results in steric clashes with the RNA, which may indicate that the aptamers form a highly-enclosed pocket as do many other riboswitch aptamers. In the case of l-glutamine t-butyl ester, it is also possible that the removal of the negative charge from the C terminal oxygen causes a loss of a favorable ionic interaction.
The 67 glnA RNA can bind D-glutamine, albeit with a reduced affinity when compared to the more biologically prevalent l-isomer. This might indicate that the reversal of two groups bonded to the α-carbon of the ligand causes the loss of only weak contacts between the compound and the RNA. Alternatively, if important bonds are broken, this could be partially mitigated by the formation of other fortuitous interactions when the chirality of the ligand is reversed.
We did not detect any binding between the DP RNA and d-glutamine. This finding might seemingly contradict our claim that the binding pockets of the glnA and the Downstream-peptide motif are likely similar. However, because the 67 glnA RNA binds to l-glutamine 10-fold more tightly compared to d-glutamine, it is possible that concentrations higher than 10 mM (the highest concentration we tested) would be necessary to detect an interaction between the 83 DP RNA and the d-isomer.
The high frequency of tandem glnA RNA arrangements could mean that these aptamers are often employed to achieve a form of riboswitch-mediated gene control not feasible with a single aptamer. For example, cooperative binding by two aptamers can yield a more digital gene control element.17,26 Although we did not observe evidence of cooperativity in our in-line probing assays, it is possible that the tandem glnA RNAs can function in this manner, but they may require additional portions of sequence elements not included in our constructs or they may require other cellular factors or conditions for proper function.
The KD values for the different glutamine aptamers tested are all relatively high in comparison to those of characterized riboswitch aptamer classes, which range from about 10 pM for the cyclic di-GMP-I aptamer28 to more than 100 µM for glmS ribozymes.29,30 Considering the high concentrations of glutamine in E. coli and likely other bacterial species, it isn't surprising that members of this aptamer class exhibit poor affinity. Regardless, the correlation of an aptamer's KD with the concentrations of ligand needed to trigger riboswitch is not possible with many riboswitches because kinetically-driven riboswitches do not reach thermodynamic equilibrium for ligand binding.31–33
As mentioned previously, riboswitches not only require an aptamer but an expression platform to translate ligand binding of the aptamer into a change in gene expression. There are cases where the glnA and downstream-peptide aptamers are positioned close to predicted terminator stems and ribosomal binding sites, which might be part of riboswitch expression platforms. However, mechanisms for how the aptamers interact with these portions of sequence are not obvious. It is not uncommon for expression platforms to be hard to identify, and we nevertheless suspect the glnA and Downstream-peptide RNAs are components of full riboswitches.
In this study, we have shown that the glnA and Downstream-peptide motifs are naturally occurring aptamers that selectively bind l-glutamine. These elements are often positioned 5′ of several genes involved with nitrogen metabolism in cyanobacteria, which implicates glutamine as an important signaling molecule in the pathways of these organisms. The presence of glutamine-responsive riboswitches might explain why the glutamine-sensing regulatory proteins responsible for nitrogen regulation in other bacterial taxa are absent in cyanobacteria.20 The discovery of these RNAs expands the scope of metabolites that are recognized by natural aptamers, introducing glutamine as the third amino acid to be sensed by natural RNAs along with glycine and lysine. As the amount of available genomic sequence data continues to expand, we expect that many additional metabolite-sensing aptamers will be discovered, including a greater diversity of classes that sense amino acids.
Materials and Methods
Chemicals and DNA oligonucleotides.
The compounds l-glutamine, d-glutamine, Ala-Gln, l-glutamine t-butyl ester, l-theanine, O-acetyl-l-serine, asparagine, putrescine, lysine, 2-oxoglutarate, glutaric acid, succinate, succinic semialdehyde, GABA, agmatine, pyridoxal phosphate, and glutamate were all obtained from Sigma-Aldrich. L-homoglutamine and (S)-2-amino-5-oxo-hexanoic acid were ordered from Toronto Research Chemicals Inc., L-β-homoglutamine was obtained from the PepTech Corporation and 5-amino-5-oxopentanoic acid was purchased from ChemBridge.
The following DNA oligonucleotides were ordered from Sigma-Genosys: primer 1, 5′-TAA TAC GAC TCA CTA TAG GGT AAT CGT TGG CCC AGT TTA TCT GGG TGG AA; primer 2, 5′-TGA GAG GCG CGT TGC TTC AGG CCA AAG ACC TTA CTT CCA CCC AGA TAA A; primer 3, 5−-TAA TAC GAC TCA CTA TAG GGT AAT CGT TGG CCC AGT TTA TCA AGG TGG AA; primer 4, 5′-TAA TAC GAC TCA CTA TAG GGT AAT CGT TGG CCT TGT TTA TCA AGG TGG AA; primer 5, 5′-TGA GAG GCG CGT TGC TTC AGG CCA AAG ACC TTA CTT CCA CCT TGA TAA A; primer 6, 5′-TGA GAG GCG CGT TGC TTC AGC CCA AAG TCC TTA CTT CCA CCC AGA TAA A; primer 7, 5′-TGA GAG GCG CGT TGC TTC AGC TCA AAG TGC TTA CTT CCA CCC AGA TAA A; primer 8, 5′-TAA TAC GAC TCA CTA TAG GGT ATT CTT GGT CCA CGT TGA GCT TCC AAT CGA AGC TGC A; primer 9, 5′-TCC TTC ATT GCC CAC GCC CCC GTT GCT TGG CAT GGG TCT GAC TGC AGC TTC GAT TGG A; primer 10, 5′-TCC TTC ATT GCC CTC GCC CCC GTT GCT TGG CCT GGG TCT GAC TGC AGC TTC GAT TGG AAG CT; primer 11, 5′-TCC TTC ATT GCC CTA GCC CCC GTT GCT TGG CCA GGG TCT GAC TGC AGC TTC GAT TGG AAG CT.
Transcription and purification of RNAs.
Pairs of oligonucleotides were used in primer extension reactions to make full-length double-stranded DNA (dsDNA) products to use as templates for in vitro transcription reactions. Primers 1 and 2 were used for 67 glnA, 2 and 3 for M1, 4 and 5 for M3, 1 and 6 for M2, 1 and 7 for M4, 8 and 9 for 83 DP, 8 and 10 for M5, and 8 and 11 for M6. A 100 µl solution containing 300 pmoles of each primer, 50 mM Tris-HCl (pH 8.3 at 23°C), 75 mM KCl, 3 mM MgCl2, 10 µM dithiothreitol (DTT), 1 mM of each of the four deoxynucleoside triphosphates (dNTPs) and 8 U/µl of SuperScript II Reverse Transcriptase (Invitrogen) was heated for 2 hours at 42°C. The full length dsDNA was then purified using the QIAquick PCR Purification Kit (QIAgen) following the manufacturer's protocol and was eluted in a volume of 50 µl.
Twenty microliters of dsDNA was used as a template in a 100 µl in vitro transcription reaction containing 80 mM HEPES (pH 7.5 at 23°C), 40 mM DTT, 24 mM MgCl2, 2 mM spermidine, 2.5 mM each of the four ribonucleoside 5′ triphosphates (NTPs), and 10 units/µl bacteriophage T7 RNA polymerase. Samples were heated for 2 hours at 37°C and purified via denaturing (8 M urea) 6% polyacrylamide gel electrophoresis (PAGE). A band containing the RNA was cut from the gel and soaked in a solution of 10 mM Tris-HCl (pH 7.5 at 23°C), 200 mM NaCl and 1 mM EDTA (pH 8.0 at 23°C). The RNAs were then concentrated by adding 2.5 volumes of cold (−20°C) ethanol, centrifuging for 20 minutes at 17,900 g. The resulting pellet was dried, resuspended in water, and stored at −20°C until use.
In-line probing assays.
RNAs were 5′-32P radiolabeled and subjected to in-line probing analyses, which have been described in detail previously in reference 34 and 35. Briefly, 5′ triphosphates were removed from the RNAs using alkaline phosphatase (Roche) according to the manufacturer's protocol. The dephosphorylated RNAs were 5′-32P radiolabeled by incubation with [γ-32P] ATP and T4 polynucleotide kinase (New England Biolabs), following the manufacturer's directions. Denaturing 6% PAGE was subsequently employed to purify the RNAs as described above.
The radiolabeled RNAs were then incubated at 23°C in a solution containing 75 mM Tris-HCl (pH 8.3 at 23°C), 20 mM MgCl2, 100 mM KCl, and various different potential ligands (see results section for full list) at concentrations ranging from 1 µM to 10 mM in most cases. When RNAs were subjected to in-line probing with 100 mM l-glutamate, the amino acid replaced the Tris-HCl as the buffering agent. NaOH were used to adjust this modified in-line probing solution to pH 8.3.
After incubating for approximately 40 hours, the products of in-line probing reactions were separated by denaturing 10% PAGE. The gels were then dried and imaged using a Storm 820 PhosphorImager (GE Healthcare). The relative intensities of the various degradation products were quantified using SAFA v1.1 software.36 The bands which modulated the most intensely were used to make KD estimates. Note that in Figure 5A, circled positions before nucleotide 34 may actually be shifted due to a gel compression issue, but by one or two positions at most. In the cases where the concentrations of ligand used were insufficiently high to saturate RNA binding, the data was normalized to KD values that best explained the data assuming a standard one-to-one interaction.
Acknowledgements
We thank Thalyana Smith-Vikos for performing preliminary in-line probing binding assays on representatives of the Downstream-peptide motif. This work was supported by funding to R.R.B. from the NIH (GM022778) and from the Howard Hughes Medical Institute.
Supplementary Material
References
- 1.Breaker RR. Riboswitches: from ancient gene-control systems to modern drug targets. Future Microbiol. 2009;4:771–773. doi: 10.2217/fmb.09.46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Barrick JE, Breaker RR. The distributions, mechanisms and structures of metabolite-binding riboswitches. Genome Biol. 2007;8:239. doi: 10.1186/gb-2007-8-11-r239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dambach MD, Winkler WC. Expanding roles for metabolite-sensing regulatory RNAs. Curr Opin Microbiol. 2009;12:161–169. doi: 10.1016/j.mib.2009.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Roth A, Breaker RR. The structural and functional diversity of metabolite-binding riboswitches. Annu Rev Biochem. 2009;78:305–334. doi: 10.1146/annurev.biochem.78.070507.135656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Loh E, Dussurget O, Gripenland J, Vaitkevicius K, Tiensuu T, Mandin P, et al. A trans-acting riboswitch controls expression of the virulence regulator PrfA in listeria monocytogenes. Cell. 2009;139:770–779. doi: 10.1016/j.cell.2009.08.046. [DOI] [PubMed] [Google Scholar]
- 6.Wachter A. Riboswitch-mediated control of gene expression in eukaryotes. RNA Biol. 2010;7:67–76. doi: 10.4161/rna.7.1.10489. [DOI] [PubMed] [Google Scholar]
- 7.Croft MT, Moulin M, Webb ME, Smith AG. Thiamine biosynthesis in algae is regulated by riboswitches. Proc Natl Acad Sci USA. 2007;104:20770–20775. doi: 10.1073/pnas.0705786105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wachter A, Tunc-Ozdemir M, Grove BC, Green PJ, Shintani DK, Breaker RR. Riboswitch control of gene expression in plants by splicing and alternative 3' end processing of mRNAs. Plant Cell. 2007;19:3437–3450. doi: 10.1105/tpc.107.053645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cheah MT, Wachter A, Sudarsan N, Breaker RR. Control of alternative RNA splicing and gene expression by eukaryotic riboswitches. Nature. 2007;447:497–500. doi: 10.1038/nature05769. [DOI] [PubMed] [Google Scholar]
- 10.Rodionov DA, Vitreschak AG, Mironov AA, Gelfand MS. Regulation of lysine biosynthesis and transport genes in bacteria: yet another RNA riboswitch? Nucleic Acids Res. 2003;31:6748–6757. doi: 10.1093/nar/gkg900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Barrick JE, Corbino KA, Winkler WC, Nahvi A, Mandal M, Collins J, et al. New RNA motifs suggest an expanded scope for riboswitches in bacterial genetic control. Proc Natl Acad Sci USA. 2004;101:6421–6426. doi: 10.1073/pnas.0308014101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Weinberg Z, Barrick JE, Yao Z, Roth A, Kim JN, Gore J, et al. Identification of 22 candidate structured RNAs in bacteria using the CMfinder comparative genomics pipeline. Nucleic Acids Res. 2007;35:4809–4819. doi: 10.1093/nar/gkm487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yao Z, Barrick J, Weinberg Z, Neph S, Breaker RR, Tompa M, et al. A computational pipeline for highthroughput discovery of cis-regulatory noncoding RNA in prokaryotes. PLoS Comput Biol. 2007;3:126. doi: 10.1371/journal.pcbi.0030126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tseng H, Weinberg Z, Gore J, Breaker RR, Ruzzo WL. Finding non-coding RNAs through genome-scale clustering. J Bioinform Comput Biol. 2009;7:373–388. doi: 10.1142/s0219720009004126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Weinberg Z, Wang J, Bogue J, Yang J, Corbino K, Moy R, et al. Comparative genomics reveals 104 candidate structured RNAs from bacteria, archaea and their metagenomes. Genome Biol. 2010;11:31. doi: 10.1186/gb-2010-11-3-r31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sudarsan N, Wickiser JK, Nakamura S, Ebert MS, Breaker RR. An mRNA structure in bacteria that controls gene expression by binding lysine. Genes Dev. 2003;17:2688–2697. doi: 10.1101/gad.1140003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mandal M, Lee M, Barrick JE, Weinberg Z, Emilsson GM, Ruzzo WL, et al. A glycine-dependent riboswitch that uses cooperative binding to control gene expression. Science. 2004;306:275–279. doi: 10.1126/science.1100829. [DOI] [PubMed] [Google Scholar]
- 18.Goldman JC. Identification of nitrogen as a growth-limiting nutrient in wastewaters and coastal marine waters through continuous culture algal assays. Water Res. 1976;10:97–104. [Google Scholar]
- 19.Jiang P, Peliska JA, Ninfa AJ. Enzymological characterization of the signal-transducing uridylyltransferase/uridylyl-removing enzyme (EC 2.7.7.59) of Escherichia coli and its interaction with the PII protein. Biochemistry. 1998;37:12782–12794. doi: 10.1021/bi980667m. [DOI] [PubMed] [Google Scholar]
- 20.Forchhamme K. Glutamine signalling in bacteria. Front Biosci. 2007;12:358–370. doi: 10.2741/2069. [DOI] [PubMed] [Google Scholar]
- 21.Muro-Pastor MI, Reyes JC, Florencio FJ. Cyanobacteria perceive nitrogen status by sensing intracellular 2-oxoglutarate levels. J Biol Chem. 2001;276:38320–38328. doi: 10.1074/jbc.M105297200. [DOI] [PubMed] [Google Scholar]
- 22.Vázquez-Bermúdez MF, Herrero A, Flores E. Carbon supply and 2-oxoglutarate effects on expression of nitrate reductase and nitrogen-regulated genes in Synechococcus sp. strain PCC 7942. FEMS Microbiol Lett. 2003;221:155–159. doi: 10.1016/S0378-1097(03)00208-8. [DOI] [PubMed] [Google Scholar]
- 23.Forchhammer K. Global carbon/nitrogen control by PII signal transduction in cyanobacteria: from signals to targets. FEMS Microbiol Rev. 2004;28:319–333. doi: 10.1016/j.femsre.2003.11.001. [DOI] [PubMed] [Google Scholar]
- 24.Bennett BD, Kimball EH, Gao M, Osterhout R, Van Dien SJ, Rabinowitz JD. Absolute metabolite concentrations and implied enzyme active site occupancy in Escherichia coli. Nat Chem Biol. 2009;5:593–599. doi: 10.1038/nchembio.186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sudarsan N, Hammond MC, Block KF, Welz R, Barrick JE, Roth A, et al. Tandem riboswitch architectures exhibit complex gene control functions. Science. 2006;314:300–304. doi: 10.1126/science.1130716. [DOI] [PubMed] [Google Scholar]
- 26.Welz R, Breaker RR. Ligand binding and gene control characteristics of tandem riboswitches in Bacillus anthracis. RNA. 2007;13:573–582. doi: 10.1261/rna.407707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kwon M, Strobel SA. Chemical basis of glycine riboswitch cooperativity. RNA. 2008;14:25–34. doi: 10.1261/rna.771608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Smith KD, Lipchock SV, Ames TD, Wang J, Breaker RR, Strobel SA. Structural basis of ligand binding by a c-di-GMP riboswitch. Nat Struct Mol Biol. 2009;16:1218–1223. doi: 10.1038/nsmb.1702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Winkler WC, Nahvi A, Roth A, Collins JA, Breaker RR. Control of gene expression by a natural metabolite-responsive ribozyme. Nature. 2004;428:281–286. doi: 10.1038/nature02362. [DOI] [PubMed] [Google Scholar]
- 30.Cochrane JC, Lipchock SV, Smith KD, Strobel SA. Structural and chemical basis for glucosamine 6-phosphate binding and activation of the glmS ribozyme. Biochemistry. 2009;48:3239–3246. doi: 10.1021/bi802069p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wickiser JK, Cheah MT, Breaker RR, Crothers DM. The kinetics of ligand binding by an adenine-sensing riboswitch. Biochemistry. 2005;44:13404–13414. doi: 10.1021/bi051008u. [DOI] [PubMed] [Google Scholar]
- 32.Wickiser JK, Winkler WC, Breaker RR, Crothers DM. The speed of RNA transcription and metabolite binding kinetics operate an FMN riboswitch. Mol Cell. 2005;18:49–60. doi: 10.1016/j.molcel.2005.02.032. [DOI] [PubMed] [Google Scholar]
- 33.Gilbert SD, Stoddard CD, Wise SJ, Batey RT. Thermodynamic and kinetic characterization of ligand binding to the purine riboswitch aptamer domain. J Mol Biol. 2006;359:754–768. doi: 10.1016/j.jmb.2006.04.003. [DOI] [PubMed] [Google Scholar]
- 34.Soukup GA, Breaker RR. Relationship between internucleotide linkage geometry and the stability of RNA. RNA. 1999;5:1308–1325. doi: 10.1017/s1355838299990891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Regulski EE, Breaker RR. In-line probing analysis of riboswitches. Methods Mol Biol. 2008;419:53–67. doi: 10.1007/978-1-59745-033-1_4. [DOI] [PubMed] [Google Scholar]
- 36.Das R, Laederach A, Pearlman SM, Herschlag D, Altman RB. SAFA: Semi-automated footprinting analysis software for high-throughput quantification of nucleic acid footprinting experiments. RNA. 2005;11:344–354. doi: 10.1261/rna.7214405. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.