Abstract
The inner nuclear membrane (INM) accommodates a specific set of integral membrane proteins many of which interact with chromatin and/or in metazoan cells with the lamina network. The localization of these proteins characterizes this membrane area of the nuclear envelope (NE) despite the fact that the INM forms a membrane continuum with the outer nuclear membrane (ONM) and the remaining endoplasmic reticulum (ER). In fact, the INM can be regarded as a highly specialized membrane subdomain of the ER. How the specific protein composition of the INM is established and maintained and whether this is achieved via a single unifying mechanism is by and large unclear. Recent experiments shed light on some aspects of the process.
Key words: nuclear pore complex, Nup188, inner nuclear membrane, membrane targeting, nuclear localization signal, SUN2, nuclear envelope
Two Principle Modes for INM Targeting
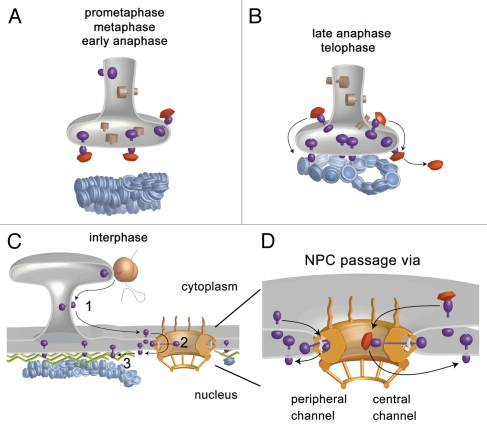
Two fundamentally different mechanisms of INM protein targeting can be envisioned; the first functioning in all eukaryotic cells during interphase, and another that is additionally employed in cells undergoing open mitosis. During this process, characterized by NE breakdown, the membranes of the NE are reabsorbed into and distributed throughout the mitotic ER, which serves as a reservoir for INM proteins1,2 (Fig. 1A). In telophase, the NE structure reforms around the decondensing chromatin. But how are INM components sorted out from the mitotic ER? The emerging picture is that the membranes giving rise to the INM segregate initially by binding to chromatin (Fig. 1B). In vitro experiments have shown that certain protein-chromatin interactions can target membranes to chromatin.3 Interestingly, most nucleoplasmic domains of INM proteins posses an overall basic amino acid composition which might render them competent for DNA binding.4 Indeed, live cell imaging of GFP-tagged INM proteins such as lamin B receptor (LBR), MAN1, LAP2beta, as well as the transmembrane nucleoporins NDC1 and POM121 suggest that the binding of these proteins to chromatin feeds membranes from the mitotic ER into the emerging NE.5 This chromatin binding could not only be important for the establishment of the first membrane contacts to chromatin, but also crucial for the re-localization and enrichment of integral nuclear membrane proteins, that probably freely diffuse in the membrane plane of the ER, on chromatin and/or the nuclear lamina by a capturing mechanism.
Figure 1.
Model for the targeting of transmembrane proteins to the INM at the end of mitosis (A and B) and in interphase (C and D). (A) During mitosis INM proteins (violet) are dispersed throughout the ER and their basic domains are shielded by nuclear import receptors (red). (B) At the end of mitosis, binding of RanGTP to the transport receptors releases them from INM proteins in the vicinity of chromatin. This probably together with dephosphorylation (not shown) allows INM proteins to interact with chromatin and segregates them from bulk ER proteins (brown). (C) In interphase, integral INM proteins are synthesized at the rough ER, translocate in the plane of the ER and ONM membrane to the NPC (1). After passage through the NPC (2), INM proteins reach their site of destination (3). (D) The nucleoplasmic domains of integral INM proteins could pass the NPC either via the peripheral channels in proximity to the pore membrane or via the central channel, possibly by the action of transport receptors (red).
With the end of mitosis and the reestablishment of the NE, the problem of INM targeting demands an alternative solution. During interphase, the ER and the ONM are continuous and the ONM is connected to the INM at the pore membrane, the membrane area facing nuclear pore complexes (NPCs). Integral membrane proteins could in principle still laterally diffuse within the plane of this membrane continuum and reach the INM from the ER, the site of integral membrane protein synthesis (Fig. 1C). Base on early work on the localization of the INM protein LAP1, it was long assumed that the capture of the proteins at the INM is the main mechanism of their targeting. Indeed, LAP1 localization requires the expression of its binding partner lamin A.6 Enrichment of other INM proteins has also been found to be supported by chromatin and/or lamina interactions.7–9 Thus, this capture mechanism appears to be important for enriching INM protein at their site of action. But is diffusion in the ER membrane system sufficient to explain how transmembrane proteins initially reach the INM? And, is free diffusion across the pore membrane with the necessity of passing nucleoplasmic domains of INM proteins through the NPC a realistic scenario? Such skepticism was nourished by a study which demonstrated that proper targeting of an INM reporter in HeLa cells is energy-dependent.10 This finding provided the first persuasive evidence that diffusion was probably not the only mechanism for targeting proteins to the INM and opened way to further investigations. As the passage through the pore membrane and the NPC might restrict free diffusion of integral membrane proteins, the NPC is likely the bottleneck for INM proteins on their way to the nucleus-similarly as for soluble proteins.
What are the Targeting Signals?
Transport of soluble proteins through the NPC has been intensively studied and in most cases is signal-dependent. Among the best investigated nuclear targeting signals are the mono- and bipartite classical nuclear localization signals (NLSs) (reviewed in ref. 11). Despite a wealth of information, it is still not straightforward to predict an NLS from the primary amino acid sequence of a protein as basic sequence patches are present in many proteins without rendering them nuclear. The best possible evidence to prove NLS function is still to redirect an otherwise non-nuclear protein to the nucleus by fusing it with the putative NLS.
It has been suggested that basic stretches found in the nucleoplasmic domains of many INM proteins serve as NLSs for receptor-mediated translocation through the NPC.12 However, the prevalence of these sequences within INM proteins could also be an adaption to their chromatin binding function. Complicating the issue, chromatin-binding motifs could in principle support both the capture of INM proteins by chromatin-binding and nuclear import. It is therefore difficult to distinguish the function of a putative NLS in nuclear uptake of an INM protein from a role in retention by chromatin binding.
Presently, only a few INM protein targeting sequences are characterized in detail, the classical example being LBR. Here, both the nucleoplasmic amino-terminal domain13 and the first transmembrane region with some flanking amino acids14 have shown to be sufficient for INM targeting. Although both elements can act independently of each other, it is still an open question as to whether both are necessary in context of the full-length protein. Interestingly, the amino-terminal domain of LBR is targeted to the nucleus as a truncated soluble domain13 and it contains basic sequence patches which resemble NLSs. However, a chimeric protein consisting of an isolated classical NLS derived from a soluble nucleoplasmic protein fused to a membrane-bound reporter was unable to localize to the INM.15 This could indicate that an isolated NLS is unable to target a protein to the INM or that the distance of the NLS to the transmembrane domain is critical to NLS functionality. Also the nucleoplasmic domains of other INM proteins localize to the nucleoplasm as truncated soluble fragments, suggesting that they also contain NLSs that might trigger receptor-mediated transport.16–18 However, this should not be taken as final proof and the importance of these putative NLSs for INM targeting still remains to be clarified.
In a recent study, we have found that targeting of human SUN2 protein to the INM relies on multiple sequence elements.19 One such element is indeed a classical NLS, which is able to bind the transport receptors importin alpha and beta in vitro. The second is a basic arginine cluster that does not function as an NLS but rather serves to recruit COPI components and to retrieve SUN2 from the Golgi to the ER in case it escapes along the secretory pathway. Potential COPI binding motifs can be identified in a number of INM proteins and such motifs might emerge as a general feature that promote INM localization. The third element is the SUN domain which is located within the luminal space between ONM and INM and which mediates interaction with KASH (Klarsicht, ANC-1 and Syne/Nesprin homology) proteins in the ONM.20 All three elements together determine proper localization of SUN2 to the INM. It remains to be seen whether other INM proteins require a combination of different targeting signals for efficient localization.
SUN2 is not the only vertebrate NE protein that employs a classical NLS to promote its targeting. Also POM121, a transmembrane NPC protein, contains classical NLSs that bind importin alpha and beta and are required for proper NPC localization.21,22 Given that at least some membrane proteins employ NLSs as targeting sequences, the question of how these are recognized within the cell becomes important. Nuclear transport receptors functioning in the import of soluble cargos to the nucleus seem to be prime candidates. The best evidence for an involvement of transport receptors in INM targeting comes from work in Saccharomyces cerevisiae. Here, it was shown that two INM proteins, Heh1 and Heh2 that are related to vertebrate MAN1 and LEM2, bind the yeast homologues of importin alpha via a basic sequence motif.23 INM targeting of these proteins is indeed dependent on importin alpha and beta as well as on the GTPase Ran. Although presently it cannot be excluded that the requirement of importins and Ran is indirect (e.g., ensuring nuclear localization of Heh1/Heh2 interacting proteins), this data suggests that passage of membrane proteins to the INM could be mediated by mechanisms similar to the transport of soluble cargo to the nucleoplasm.
In metazoans, the presence of basic, importin-binding patches in the nucleoplasmic domains of INM proteins could also arise from the necessity to shield these domains from undesired interactions during mitosis. At the onset of mitosis, interactions of INM proteins with chromatin and the lamina are broken, likely driven by various phosphorylations on INM proteins, nuclear lamins and chromatin. Chaperoning nuclear transport receptors may then bind and burrow the released basic regions of INM proteins while they reside in the mitotic ER (Fig. 1A). Although the idea is currently purely speculative, in telophase, RanGTP could help to unmask the chromatin binding sites of these proteins by dissociating bound transport receptors in the vicinity of chromatin. This unshielding along with other changes like dephosphorylation could render these proteins competent for chromatin re-binding (Fig. 1B), similar to what has been proposed for nucleoporin binding.24
The Road to the Inner Nuclear Membrane
If indeed the picture emerges that transport receptors are required for INM targeting of at least some proteins during interphase, it remains to be seen where in the process they are involved. The process can be dissected into three steps (Fig. 1C); first, newly synthesized INM proteins inserted into the rough ER need to reach and perhaps bind the cytoplasmic site of the NPC. Whether this initial journey is by free diffusion in the plane of the ER and ONM or whether there exists a directed transport mechanism is currently unclear. It is hard to imagine how import receptors could help transport along the ER and ONM, but they might be important for establishing initial contacts to the cytoplasmic site of the NPC.
The second step is the crossing of the NE. This includes the passage of the transmembrane region(s) of a future INM protein through the plane of the pore membrane. The nucleoplasmic domain needs to pass through the NPC, whereas the luminal domain moves along in the perinuclear space of the NE (Fig. 1D). Soluble cargos traverse the NPC bound to transport receptors presumably within a hydrophobic meshwork of FG repeat containing NPC proteins25 localized in the central channel of the pore. To use this central passageway, the nucleoplasmic domains of INM proteins would need to considerably extend from the membrane surface into the central channel. However, unlike soluble nuclear transport cargo, the nucleoplasmic domains of INM proteins possess a size limitation of ∼60 kDa,10,15 suggesting that each type of cargo might take a distinct route for NPC passage. Hence, INM proteins could take a different route leading through the peripheral, 9 nm wide channels of the NPC.6,26 Although these channels might be wide enough to allow passage of the nucleoplasmic domains of INM proteins, it is difficult to envision these domains passing in a rather voluminous complex with transport receptors like importin alpha (and probably beta).
An unexpected solution to this problem might arise from the suggestion that smaller importin alpha variants are utilized for targeting of INM proteins. A sequence motif distinct from NLSs and close to the transmembrane domain of a baculovirus-derived INM protein was found to interact with importin alpha-16 contemporaneously with its insertion into the rough ER in SF9 cells.27 Shorter importin alpha isoforms have more recently been detected in human cells and S. cerevisiae, but at lower abundance than the canonical importin alpha.28,29 Whether these importin alpha isoforms are indeed required for the passage through the NPC is still an open question. Of note, the binding site in yeast Heh2 for the proposed shorter importin alpha isoform is distinct from the basic sequence required for full-length importin alpha binding. Thus, the question of whether importin alpha (and probably beta) binding is necessary for and even possible during NPC passage of INM proteins persists.
The third step is the release of the INM protein from the NPC and probably an initial free diffusion in the plane of the INM. But how is directionality of transport ensured? If importins were indeed utilized for the translocation process, the dissociation of the imported protein from transport receptors by RanGTP would contribute to directionality. For the shorter importin alpha isoforms it is presently unclear what would control their dissociation from INM proteins before or after translocation through the pore. In any case, whether NPC translocation is assisted or not, the interaction with chromatin, lamins or other binding partners of INM proteins is surely important for retaining INM proteins in the nuclear compartment and currently the best explanation for their enrichment in the nucleus.
Traversing the NPC
Despite the fact that there is no high resolution structure of the NPC, significant progress has been made in assigning the localization and function of individual proteins within this huge macromolecular assembly. From a wealth of studies it has become apparent that NPC proteins might be categorized into components, which are localized close to the pore membrane and are important for the overall structure of the NPC, and nucleoporins, which contain large unstructured regions mostly consisting of FG repeats, and are localized in the center and periphery of the NPC. These FG-containing nucleoporins are important for both setting the diffusion limit of the NPC and the selective transport of soluble cargos into the nucleus.
Consistent with the concept that passage of the nucleoplasmic domains of INM proteins is not mediated by the central channel, only nucleoporins localized in proximity to the pore membrane have been implicated in INM targeting so far (with the exception of yeast Nup2p which is localized to the nucleoplasmic site of the NPC). In human cells, RNAi against Nup155 affects INM targeting of LAP2, LEM2 and LBR.30 Similarly, in S. cerevisiae, deletion of Nup170, which is homologous to metazoan Nup155, causes aberrant INM targeting of Heh1/Heh2.23 Further, depletion of yeast Nup188 or the integral membrane nucleoporin Pom152 reduces INM targeting of the ubiquitin ligase Doa10.31
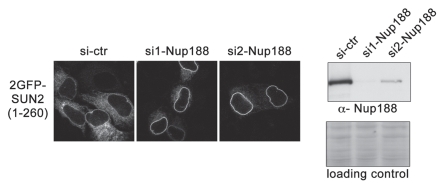
We have recently shown that in vitro assembled Xenopus nuclei lacking Nup188 show an increased passage of integral membrane proteins through the NPC.32 Consistently, depletion of human Nup188 by RNAi in HeLa cells promotes INM targeting of a 2GFP-SUN2(1-260) fusion protein, which is otherwise inefficiently targeted to the INM due to the size of its nucleoplasmic domain that is larger than 60 kDa (Fig. 2 and reviewed in ref. 19). Interestingly, depletion of Nup188 in in vitro assembled nuclei does not affect the kinetics of nuclear import of soluble cargos. Moreover, NPCs lacking Nup188 are still functional in excluding large, soluble dextran molecules. Also the Nup62 complex, which most likely forms a significant part of the hydrophobic meshwork within the central pore, is still present in NPCs lacking Nup188. Together, these data show that the central channel of the NPC is not affected by depletion of Nup188. The different effects of Nup188 depletion on NPC passage of soluble and transmembrane proteins suggest that these cargos do indeed possess different molecular requirements within the NPC. Hence, it appears likely that membrane-bound and soluble cargos follow different pathways through the NPC and probably pass the pore by employing different mechanisms.
Figure 2.
Depletion of Nup188 promotes targeting of a reporter protein to the INM in human cells. HeLa cells were transfected with either a control siRNA or two different siRNAs to Nup188 (9 nM siRNA, 2 × 48 hrs). After 72 hrs, cells were transiently transfected with 2GFP-SUN2(1-260) (Turgay et al. 2010). Cells were either fixed in 1% PFA and analyzed by confocal microscopy or cell extracts were analyzed by immunoblotting using an antibody against Nup188. Loading control: Ponceau S staining of nitrocellulose membrane used for antibody detection of Nup188.
With the exception of yeast Nup2, all nucleoporins implicated so far in the passage of INM proteins through the NPC do not contain FG-repeats. Probably, the parts of the NPC facing the pore membrane contain, if at all, only few FG-repeats, since the structural nucleoporins localized in this region are mostly devoid of these. As the proposed function of transport receptors during NPC passage is to interact with the hydrophobic FG-repeat meshwork, the passage of INM proteins through the peripheral channels of the NPC is likely independent of transport factors.
Outlook
Currently, we are far from understanding how NPC passage of INM proteins is accomplished. The ongoing work in the field aiming to elucidate the structural organization of the NPC will be of utmost importance in order to reconcile biochemical data with topological constraints within the pore. It will also be challenging to understand whether there are dynamic rearrangements within the pore that accompany INM protein passage. At present, it is unclear whether all INM proteins follow the same path through the NPC, i.e., whether there exist differences between membrane proteins that solely rely on a diffusion-retention mechanism for INM targeting and those INM proteins carrying transport receptor binding sites. At least in cells undergoing open mitosis, these NLSs might not primarily promote NPC passage, but could also serve as binding sites for chaperoning transport receptors in the mitotic cytosol and as a means to control RanGTP-dependent chromatin binding during mitotic exit. A more systematic analysis of different INM proteins with respect to their targeting signals and nucleoporin requirements in yeast and vertebrate cells will be needed to shed light on the pathway(s) of NPC passage.
Acknowledgements
This work was supported by the Swiss National Science Foundation (grant 3100-133135) to UK.
Extra View to: Turgay Y, Ungricht R, Rothballer A, Kiss A, Csucs G, Horvath P, et al. A classical NLS and the SUN domain contribute to the targeting of SUN2 to the inner nuclear membrane. EMBO J. 2010;189:1129–1142. doi: 10.1038/emboj.2010.119. and Theerthagiri G, Eisenhardt N, Schwarz H, Antonin W. The necleoporin Nup188 controls passage of membrane proteins across the nuclear pore complex. J Cell Biol. 2010;189:1129–1142. doi: 10.1083/jcb.200912045.
References
- 1.Ellenberg J, Siggia ED, Moreira JE, Smith CL, Presley JF, Worman HJ, et al. Nuclear membrane dynamics and reassembly in living cells: targeting of an inner nuclear membrane protein in interphase and mitosis. J Cell Biol. 1997;138:1193–1206. doi: 10.1083/jcb.138.6.1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Yang L, Guan T, Gerace L. Integral membrane proteins of the nuclear envelope are dispersed throughout the endoplasmic reticulum during mitosis. J Cell Biol. 1997;137:1199–1210. doi: 10.1083/jcb.137.6.1199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pyrpasopoulou A, Meier J, Maison C, Simos G, Georgatos SD. The lamin B receptor (LBR) provides essential chromatin docking sites at the nuclear envelope. EMBO J. 1996;15:7108–7119. [PMC free article] [PubMed] [Google Scholar]
- 4.Ulbert S, Platani M, Boue S, Mattaj IW. Direct membrane protein-DNA interactions required early in nuclear envelope assembly. J Cell Biol. 2006;173:469–476. doi: 10.1083/jcb.200512078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Anderson DJ, Vargas JD, Hsiao JP, Hetzer MW. Recruitment of functionally distinct membrane proteins to chromatin mediates nuclear envelope formation in vivo. J Cell Biol. 2009;186:183–191. doi: 10.1083/jcb.200901106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Powell L, Burke B. Internuclear exchange of an inner nuclear membrane protein (p55) in heterokaryons: in vivo evidence for the interaction of p55 with the nuclear lamina. J Cell Biol. 1990;111:2225–2234. doi: 10.1083/jcb.111.6.2225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gruenbaum Y, Lee KK, Liu J, Cohen M, Wilson KL. The expression, lamin-dependent localization and RNAi depletion phenotype for emerin in C. elegans. J Cell Sci. 2002;115:923–929. doi: 10.1242/jcs.115.5.923. [DOI] [PubMed] [Google Scholar]
- 8.Ostlund C, Sullivan T, Stewart CL, Worman HJ. Dependence of diffusional mobility of integral inner nuclear membrane proteins on A-type lamins. Biochemistry. 2006;45:1374–1382. doi: 10.1021/bi052156n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sullivan T, Escalante-Alcalde D, Bhatt H, Anver M, Bhat N, Nagashima K, et al. Loss of A-type lamin expression compromises nuclear envelope integrity leading to muscular dystrophy. J Cell Biol. 1999;147:913–920. doi: 10.1083/jcb.147.5.913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ohba T, Schirmer EC, Nishimoto T, Gerace L. Energy- and temperature-dependent transport of integral proteins to the inner nuclear membrane via the nuclear pore. J Cell Biol. 2004;167:1051–1062. doi: 10.1083/jcb.200409149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Marfori M, Mynott A, Ellis JJ, Mehdi AM, Saunders NF, Curmi PM, et al. Molecular basis for specificity of nuclear import and prediction of nuclear localization. Biochim Biophys Acta. 2010 doi: 10.1016/j.bbamcr.2010.10.013. In press. [DOI] [PubMed] [Google Scholar]
- 12.Lusk CP, Blobel G, King MC. Highway to the inner nuclear membrane. Rules for the road. Nat Rev Mol Cell Biol. 2007;8:414–420. doi: 10.1038/nrm2165. [DOI] [PubMed] [Google Scholar]
- 13.Soullam B, Worman HJ. The amino-terminal domain of the lamin B receptor is a nuclear envelope targeting signal. J Cell Biol. 1993;120:1093–1100. doi: 10.1083/jcb.120.5.1093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Smith S, Blobel G. The first membrane spanning region of the lamin B receptor is sufficient for sorting to the inner nuclear membrane. J Cell Biol. 1993;120:631–637. doi: 10.1083/jcb.120.3.631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Soullam B, Worman HJ. Signals and structural features involved in integral membrane protein targeting to the inner nuclear membrane. J Cell Biol. 1995;130:15–27. doi: 10.1083/jcb.130.1.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Brachner A, Reipert S, Foisner R, Gotzmann J. LEM2 is a novel MAN1-related inner nuclear membrane protein associated with A-type lamins. J Cell Sci. 2005;118:5797–5810. doi: 10.1242/jcs.02701. [DOI] [PubMed] [Google Scholar]
- 17.Furukawa K, Pante N, Aebi U, Gerace L. Cloning of a cDNA for lamina-associated polypeptide 2 (LAP2) and identification of regions that specify targeting to the nuclear envelope. EMBO J. 1995;14:1626–1636. doi: 10.1002/j.1460-2075.1995.tb07151.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ostlund C, Ellenberg J, Hallberg E, Lippincott-Schwartz J, Worman HJ. Intracellular trafficking of emerin, the Emery-Dreifuss muscular dystrophy protein. J Cell Sci. 1999;112:1709–1719. doi: 10.1242/jcs.112.11.1709. [DOI] [PubMed] [Google Scholar]
- 19.Turgay Y, Ungricht R, Rothballer A, Kiss A, Csucs G, Horvath P, et al. A classical NLS and the SUN domain contribute to the targeting of SUN2 to the inner nuclear membrane. EMBO J. 2010;29:2262–2275. doi: 10.1038/emboj.2010.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Starr DA, Fridolfsson HN. Interactions between nuclei and the cytoskeleton are mediated by SUNKASH nuclear-envelope bridges. Annu Rev Cell Dev Biol. 2010;26:421–444. doi: 10.1146/annurev-cellbio-100109-104037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Doucet CM, Talamas JA, Hetzer MW. Cell cycle-dependent differences in nuclear pore complex assembly in metazoa. Cell. 2010;141:1030–1041. doi: 10.1016/j.cell.2010.04.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yavuz S, Santarella-Mellwig R, Koch B, Jaedicke A, Mattaj IW, Antonin W. NLS-mediated NPC functions of the nucleoporin Pom121. FEBS Lett. 2010;584:3292–3298. doi: 10.1016/j.febslet.2010.07.008. [DOI] [PubMed] [Google Scholar]
- 23.King MC, Lusk CP, Blobel G. Karyopherin-mediated import of integral inner nuclear membrane proteins. Nature. 2006;442:1003–1007. doi: 10.1038/nature05075. [DOI] [PubMed] [Google Scholar]
- 24.Walther TC, Askjaer P, Gentzel M, Habermann A, Griffiths G, Wilm M, et al. RanGTP mediates nuclear pore complex assembly. Nature. 2003;424:689–694. doi: 10.1038/nature01898. [DOI] [PubMed] [Google Scholar]
- 25.Frey S, Richter RP, Gorlich D. FG-rich repeats of nuclear pore proteins form a three-dimensional meshwork with hydrogel-like properties. Science. 2006;314:815–817. doi: 10.1126/science.1132516. [DOI] [PubMed] [Google Scholar]
- 26.Beck M, Lucic V, Forster F, Baumeister W, Medalia O. Snapshots of nuclear pore complexes in action captured by cryo-electron tomography. Nature. 2007;449:611–615. doi: 10.1038/nature06170. [DOI] [PubMed] [Google Scholar]
- 27.Saksena S, Summers MD, Burks JK, Johnson AE, Braunagel SC. Importin-alpha-16 is a translocon-associated protein involved in sorting membrane proteins to the nuclear envelope. Nat Struct Mol Biol. 2006;13:500–508. doi: 10.1038/nsmb1098. [DOI] [PubMed] [Google Scholar]
- 28.Braunagel SC, Williamson ST, Ding Q, Wu X, Summers MD. Early sorting of inner nuclear membrane proteins is conserved. Proc Natl Acad Sci USA. 2007;104:9307–9312. doi: 10.1073/pnas.0703186104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liu D, Wu X, Summers MD, Lee A, Ryan KJ, Braunagel SC. Truncated isoforms of Kap60 facilitate trafficking of Heh2 to the nuclear envelope. Traffic. 2010;11:1506–1518. doi: 10.1111/j.1600-0854.2010.01119.x. [DOI] [PubMed] [Google Scholar]
- 30.Mitchell JM, Mansfeld J, Capitanio J, Kutay U, Wozniak RW. Pom121 links two essential subcomplexes of the nuclear pore complex core to the membrane. J Cell Biol. 2010;191:505–521. doi: 10.1083/jcb.201007098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Deng M, Hochstrasser M. Spatially regulated ubiquitin ligation by an ER/nuclear membrane ligase. Nature. 2006;443:827–831. doi: 10.1038/nature05170. [DOI] [PubMed] [Google Scholar]
- 32.Theerthagiri G, Eisenhardt N, Schwarz H, Antonin W. The nucleoporin Nup188 controls passage of membrane proteins across the nuclear pore complex. J Cell Biol. 2010;189:1129–1142. doi: 10.1083/jcb.200912045. [DOI] [PMC free article] [PubMed] [Google Scholar]