Abstract
Retroviruses assemble new virus particles that are released by budding from the plasma membranes of infected cells. Gag proteins, encoded by retroviruses, orchestrate the assembly of virus particles in close collaboration with host cell machinery. The earliest steps in retrovirus assembly—those immediately following synthesis of Gag on cytosolic ribosomes—are poorly understood. Rous sarcoma virus (RSV) offers a unique model system for dissecting these early steps because the RSV Gag protein undergoes transient nuclear trafficking prior to plasma membrane transport. Other Gag proteins, including those of human immunodeficiency virus (HIV), murine leukemia virus (MLV), foamy virus and retrotransposons in Schizosaccharomyces pombe and Drosophila, have also been detected in the nucleus, suggesting that nuclear trafficking of Gag proteins is a common property of retroviruses and retrotransposons. In addition to retroviruses, many structural proteins of unrelated viruses, including influenza M1, NEP and NP proteins,38 Borna disease virus N and P proteins28,56 and coronavirus N protein,23,57 undergo nuclear localization and bind viral RNAs to form viral ribonuclear protein (RNP) complexes that are exported from the nucleus for packaging into virus particles. Similarly, nuclear trafficking of the RSV Gag protein is required for efficient encapsidation of the viral genomic RNA (gRNA) into assembling virus particles.19 Recently, we reported that the viral RNA itself appears to be a key factor in controlling the nucleus/cytosol distribution of RSV Gag.22 Our data demonstrate that binding of RSV RNA to the Gag protein promotes Gag-CRM1-RanGTP binding, resulting in export of the retroviral RNP from the nucleus. We propose that association of the viral RNA induces a conformational change in Gag that reveals its nuclear export signal (NES) and prepares that complex for its journey to the plasma membrane for budding. This work challenges existing dogmas regarding the molecular basis of Gag-mediated selection of gRNA for packaging and may lead to novel paradigms for the mechanism of retroviral genome encapsidation.
Key words: gag proteins, retroviral assembly, nucleocytoplasmic trafficking, viral RNA packaging
Absolute Requirement for Genomic RNA Packaging
Retroviruses cause severe immunodeficiency diseases and cancer in humans and animals. The avian pathogen Rous sarcoma virus (RSV) was the first retrovirus identified and the first virus linked to cancer. RSV remains a fundamentally informative model for studying retrovirus replication, and it serves as the prototype for the oncoretrovirus family. Basic steps in the retroviral replication cycle that were discovered by studying avian retroviruses have been directly applicable to understanding and treating the human retroviruses human immunodeficiency virus (HIV) and human T cell lymphotropic viruses (HTLV), which infect tens of millions of people worldwide.
A key step in the transmission of retroviruses within and between hosts is the assembly and release of new virions from infected cells. To make an infectious virus, the retroviral RNA genome must be encapsidated by the Gag protein. Despite the absolute requirement for genome packaging, no antiviral drugs that target packaging or assembly have been developed yet for clinical use. Therefore, establishing a deeper understanding of Gag-mediated viral RNA packaging and virus release is an important step toward developing potential new antiviral agents. In addition, the design of optimized retroviral vectors would be enhanced by a clearer understanding of genome encapsidation. However, impeding progress are many basic questions regarding Gag-viral gRNA interactions that remain unanswered: (1) Where in the cell does the Gag protein initially select gRNA for packaging? (2) Do cellular factors contribute to packaging specificity or efficiency? (3) How does nuclear trafficking of Gag influence genome incorporation? (4) How are viral RNAs destined for translation or packaging “marked” by host or viral factors?
Selection of the Genomic RNA for Packaging by Gag
Retroviruses convert their RNA genomes into a DNA provirus that is integrated into the host chromosome. Postintegration, RNA polymerase II transcribes a full-length viral RNA that is processed like an mRNA to have a 5′ cap and a 3′ poly(A) tail. This full-length viral RNA has three potential fates: (1) it can be spliced to form subgenomic mRNAs (env and src in RSV); (2) it can remain as full-length gRNA, becoming encapsidated into virions and released from the cell; or (3) it can be maintained as unspliced mRNA and undergo nuclear export and localization to cytosolic ribosomes, serving as the template for translation of viral structural proteins Gag and Gag-Pol. Both populations of unspliced viral RNAs must circumvent cellular mechanisms that normally retain unspliced mRNAs in the nucleus.53 A key question that remains in retrovirology is whether the gRNA and unspliced viral mRNAs undergo different nuclear export pathways that determine whether they are encapsidated (gRNA) or translated (mRNA).
Subcellular Targeting Signals and Assembly Domains in Gag Overlap
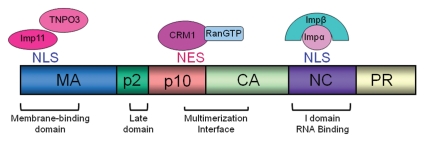
Initiation of particle formation begins with the binding of Gag to viral gRNA.3,25,45,53 The Gag NC domain binds selectively to the viral genome through its association with the cis-acting packaging element ψ, located in the 5′UTR of the retroviral genome.10,53,59 In the absence of other viral elements, Gag alone is necessary and sufficient for the formation and release of virus-like particles (VLPs) from the plasma membrane, and cellular RNAs can substitute for viral gRNA. Three assembly domains were identified in RSV Gag, and functional homologues exist in HIV and other Gag proteins.50,55 In RSV, the “M” (membrane-binding) domain is in the N-terminal 86 residues of MA and directs Gag to the plasma membrane52 (Fig. 1). The “I” (interaction) domain within NC mediates Gag-RNA and Gag-Gag interactions.5,50 Release of virus particles is controlled by the “L” (late) domain by recruiting host machinery, including endosomal sorting factors.12,16,42
Figure 1.
Overlapping assembly and nuclear trafficking signals in the RSV Gag polyprotein. Schematic diagram showing the RSV Gag cleavage products, assembly domains, nuclear targeting sequences and host nuclear transport factors that bind to Gag. MA, matrix; CA, capsid; NC, nucleocapsid; PR, protease; imp11, importin-11; TNPO3, transportin 3; imp-α, importin-α; impβ, importin-β.
In addition to the essential assembly domains, RSV Gag contains targeting motifs that control its intracellular trafficking.7,46 It was previously thought that Gag proteins were targeted directly to the plasma membrane after translation on cytosolic ribosomes; however, we discovered that RSV Gag enters the nucleus via an active transport pathway mediated by nuclear targeting signals in MA and NC (Fig. 1). The NC domain contains a classical nuclear localization signal (NLS), with basic residues that interact directly with importin-α, which in turn binds to importin-β.22 Overexpression of importin-β promotes nuclear localization of Gag in virus-infected cells. The NLS in NC coincides with the I domain, and the same basic residues required for viral RNA packaging are also involved in the binding to importin-α.7,22
The MA domain contains a complex, noncanonical NLS that interacts with two nonclassical importins, importin-11 and transportin-SR (also called transportin-3, TNPO3).7 Nuclear import of Gag is enhanced by importin-11 overexpression, suggesting that importin-11 is sufficient to drive import through the MA NLS. However, the contribution of TNPO3 in nuclear entry or subnuclear localization of Gag has not been thoroughly investigated. It is not clear whether both import factors are utilized during Gag import or whether these are redundant nuclear import pathways. Moreover, the structural basis for the binding of the atypical NLS in MA to the noncanonical import factors importin-11 and TNPO3 is of interest because the molecular interactions of these unusual import receptors with their cargoes remain unknown.
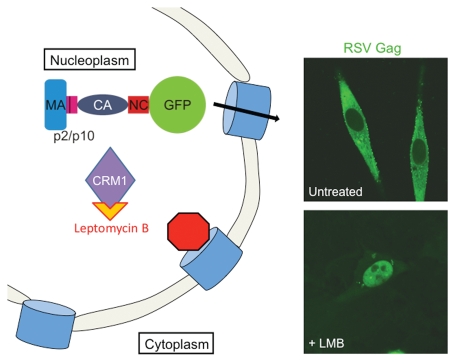
The nuclear localization of RSV Gag is transient, and egress through the nuclear pore occurs when the nuclear export signal (NES) in the p10 domain binds to the host export factor CRM1.46,48 This unexpected trafficking step was discovered when cells expressing RSV Gag were treated with the drug leptomycin B (LMB), which covalently binds to CRM1, preventing its interaction with hydrophobic NESs18,46 (Fig. 2). The NES in p10 consists of an 11-amino acid sequence: 219LTD WAR VRE EL229 with the hydrophobic residues L219, W222, V225 and L229 required for export.48
Figure 2.
The RSV Gag protein undergoes CRM1-dependent nucleocytoplasmic trafficking. PR domain of the RSV protein was replaced by GFP (green fluorescent protein). Nuclear export of the Gag protein is inhibited by treatment of cells expressing Gag-GFP with leptomycin B (LMB). LMB binds covalently to CRM1 to interfere with Gag transit through the nuclear pore complex (blue barrels) in the nuclear envelope. The Gag-GFP protein localizes primarily to the cytoplasm and at discrete foci along the plasma membrane in untreated quail fibroblasts. Treatment of cells with LMB results in the accumulation of Gag-GFP in the nucleus.
The p10 NES is unusual because it contains hydrophobic residues other than leucines, and the spacing of the intervening nonhydrophobic residues is atypical compared to other NESs.48 Therefore, we did not rely solely on LMB sensitivity and mutagenesis of the hydrophobic residues to demonstrate that this sequence was required for Gag nuclear export. Two additional gain-of-function experiments demonstrated that the p10 NES sequence was functional when (a) moved to a different position within Gag and (b) inserted into fibrillarin, a nucleolar protein, conferring nuclear export in an LMB-sensitive manner. These experiments revealed that the p10 NES motif is a positionally independent and transferrable signaling module. Subsequent experiments demonstrated that the export signal in the C-terminal region of p10 overlaps with a sequence that comprises the dimer interface involved in Gag-Gag interactions.36,47 Thus, the distinctive composition of the p10 NES might be explained by the strict structural requirements for this region to adopt specific conformations that are critical for immature Gag assembly, Gag-viral RNA interactions and morphogenesis of the mature virion core. Because of the overlap of assembly domains and subcellular localization signals in Gag, it is challenging to ascertain how these competing signals are regulated to act in a concerted manner.
Directionality of Gag Nuclear Transport
To understand the factors that influence Gag nucleocytoplasmic exchange, it was essential to identify the host partners involved. Bidirectional nuclear transport is mediated by the nuclear pore complex (NPC) and soluble nuclear transport receptors called importins or karyopherins.31,51 Components of the NPC (nucleoporins or Nups) are highly conserved from yeast to mammals. Many Nups have GLPG, FG or FXFG motifs collectively called FG-repeats13 that serve as “docking sites” for soluble nuclear transport factors. Overexpression of FG repeats inhibits specific Nup activity. Using this approach, we identified Nup214 and Nup98 as nucleoporins that interact with Gag during its egress from the nucleus.7
In addition to Nups, nuclear transport requires soluble receptors (importins and exportins) belonging to the importin β superfamily. Directionality is provided by the Ran-GTPase system by producing a high RanGTP:RanGDP ratio in the nucleus compared to the cytoplasm, ensuring that importins bind to cargo in the cytoplasm and release it in the nucleus.31 Importins have separate domains responsible for binding to cargo, Ran-GTP and the NPC. Some NLSs interact with multiple importins, although the reason for using more than a single import factor is unclear.1,17,39 The NLS in RSV MA utilizes importin-11 and TNPO3, but it is unclear whether these importins bind simultaneously to Gag or whether their binding is mutually exclusive. Dissecting the roles of importin-11 and TNPO3 in nuclear import of Gag may enhance our understanding of how different import pathways can influence subsequent events in the nucleus.
Nuclear export of proteins and RNAs is mediated by exportins.8,9,33 RSV Gag binds directly to CRM1 (exportin-1), which also transports proteins and cellular RNAs, including U snRNAs, large rRNAs and 5S rRNA out of the nucleus.9 RanGTP binds to CRM1 cooperatively, and formation of the heterodimer significantly enhances the binding of Gag22 (Fig. 3). However, the association of Gag to CRM1:RanGTP is much more efficient when Gag is bound to the viral gRNA ψ element. The facilitated binding of Gag to the export complex is dependent on the sequence and structural integrity of ψ, as both a ψ mutant and a nonviral RNA are markedly less effective in promoting Gag:CRM1:RanGTP association. Therefore, the viral gRNA ψ packaging sequence serves as a specific and potent signal to stimulate nuclear export of Gag and promote virus assembly. This result provides evidence that the RSV gRNA plays a role in controlling the directionality of protein translocation across the NPC. Although RSV is genetically a very simple virus, it takes advantage of the host's nuclear export machinery by using both viral proteins and RNAs to ensure that it replicates efficiently.
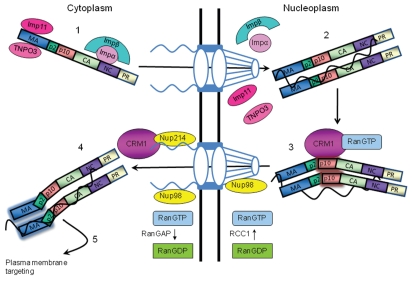
Figure 3.
Current model of RSV Gag nucleocytoplasmic trafficking, as described in detail in the text. RCC1, Regulator of chromosome condensation 1; RanGAP, Ran GTPase activating protein. Shadowing of the p10 domain3 and the MA domain5 indicates that these regions are predicted to undergo a conformational change at this step. The viral RNA is shown as a black curved line. The NPC is illustrated in blue with fibrils extending into the cytoplasm and the basket pointed toward the nucleus. Nup, nucleoporin.
Our working model depicting how host and viral factors contribute to the nucleo-cytoplasmic trafficking of RSV Gag is shown in Figure 3. RSV Gag is imported into the nucleus as a monomer in association with importin-11 or TNPO3 bound to the NLS in MA (step 1). In addition, importin-α binds directly to the NLS in NC and recruits importin-β to import Gag into the nucleus. The importins disassociate from Gag due to the high intranuclear concentration of Ran-GTP (step 2). Gag then binds to the gRNA ψ packaging sequence, stimulating Gag-Gag dimerization. gRNA binding induces a conformational change in Gag, exposing the p10 NES, which binds to the CRM1:RanGTP heterodimer (step 3). The Gag:gRNA:CRM1:RanGTP complex translocates through the NPC via interactions with Nup214 and Nup98. Once outside the nuclear envelope, Gag:gRNA disassociates from CRM1 (step 4). We speculate that Gag undergoes another conformational change to expose the membrane-targeting signal in MA to direct the viral RNP to the plasma membrane (step 5). Features of this model are being tested in ongoing experiments in our laboratory.
Nuclear Localization of Gag Proteins of Retroviruses and Retrotransposons
In addition to RSV, other retroviral Gag proteins have been detected in the nucleus. HIV Gag was reported to contain an NLS and an NES within the MA domain, and mutation of the NES interferes with gRNA packaging;14 however this result has not been further explored in subsequent studies. Moreover, a mutant protein containing deletion of the gRNA-binding region of the HIV Gag NC domain localizes to the nucleus, although the functional significance of this finding is unclear.21 Finally, HIV Gag:gRNA complexes were observed near centrioles just outside of the nuclear envelope in one study, but not in another.26,43 Thus, whether HIV Gag gRNA packaging involves nuclear events has not been resolved and remains an intriguing question.
Feline leukemia virus (FIV), another lentivirus, forms Gag:gRNA complexes at the nuclear rim.26 In addition, a subset of Mason-Pfizer monkey virus (MPMV) Gag protein is associated with the NPC, and deletion of a putative NLS in Gag results in a gRNA packaging defect.4,54 Although it is possible that genome packaging may occur near the nucleus in these retroviruses, none of these studies has yet determined where the initial site of Gag:gRNA contact occurs.
In murine leukemia virus (MLV) infected cells, approximately 18% of the Gag precursor (Pr65) is present in nuclei.37 The authors hypothesized that Gag could potentially play two roles in the nucleus: regulation of splicing or initiation of viral RNA dimerization, as a requirement for genome packaging. A recent report suggested that cellular RNAs incorporated into MLV virions are nuclear-localized, suggesting that MLV Gag might encounter these RNAs in the nucleus.20 Other nuclear RNAs (e.g., 7SK RNA and snRNA U6) are enriched in MLV particles,40 supporting the idea that MLV Gag protein might traffic through the nucleus to recruit RNAs there. Foamy virus Gag proteins localize predominantly to the nucleus, and the NLS in Gag coincides with the nucleic acid binding domain in the C-terminal region.49,58 The role of Gag nuclear targeting in foamy viruses is not known, although a mutant with impaired nuclear targeting (but persistent localization to subnuclear structures) had reduced infectivity.58 Further experiments are needed to determine whether localization of these retroviral Gag proteins to the nucleus is related to RNA incorporation or other functions.
Several retrotransposon Gag proteins localize to the nucleus. Drosophila retrotransposons HeT-Amel and TARTvir play a role in maintenance of telomeres, and their Gag proteins are specifically targeted to chromosomal ends during interphase.41,44 Schizosaccharomyces pombe retrotransposon Tf1 Gag protein localizes to both nuclei and nucleoli,2,11 and alterations in Tf1 Gag nuclear import are associated with defective particle assembly.27 Recently, it was reported that a mutant of Saccharomyces cerevisiae retrotransposon Ty3 Gag is nuclear-localized and binds Ty3 RNA, suggesting a possible mechanism for RNA packaging.29 Thus, nuclear transport of Gag is prevalent among retroviruses and retrotransposons, emphasizing that probing the mechanisms underlying Gag nuclear trafficking, identification of Gag-interacting host factors and strategies to reveal the relevance of nuclear trafficking to virus replication will have broad implications.
Formation of Viral Nucleoprotein Complexes in the Nucleus
Nuclear entry is a critical step for replication of many viruses, including herpes simplex virus, adenovirus and influenza, and there may be important parallels with RSV Gag nuclear trafficking.35 Influenza uses the nuclear compartment to assemble new viral RNPs for encapsidation. Early in infection, incoming viral RNPs are imported into the nucleus using redundant NLSs. Transcription and nuclear export of viral mRNAs occurs, and then newly synthesized viral proteins are imported into the nucleus where they bind viral RNA to form new RNP complexes. The matrix (M1) protein mediates nuclear export of viral RNPs, which occurs in a CRM1-dependent manner.15,30,32 NS2 and NP proteins are also involved, possibly through an interaction with M1-viral RNP.15,24 Once exported into the cytoplasm, M1-viral RNP complexes are targeted to the plasma membrane.6,32 Association with M1 prevents viral RNPs from reentering the nucleus.32 The similarities between influenza M1 and the Gag protein of RSV are intriguing: both undergo active nuclear transport, bind to viral RNAs to form RNPs for packaging, mediate viral RNP nuclear egress, and finally are targeted to the plasma membrane for budding.24
Opportunities and Challenges
It remains to be seen whether RSV is unique among retroviruses in the requirement for Gag nuclear trafficking to efficiently encapsidate its genome. However, there is evidence suggesting that the HIV, MPMV and FIV Gag proteins bind to RNAs either at the nuclear rim or just outside the nucleus.4,26,43,54 Why would retroviral Gag proteins bind to their unspliced gRNAs in or near the nucleus rather than finding them in the cytoplasm? One possibility is that selecting the genome in the nucleus would facilitate selective encapsidation of unspliced rather than spliced viral RNAs. For example, selective packaging of gRNA would occur if spliced viral mRNAs in the nucleus were not available for encapsidation by Gag because they were “marked” co-transcriptionally with splicing machinery. Deposition of exon junction complex factors would lead spliced viral mRNAs to exit the nucleus and enter into the translation pathway.34,35 In contrast, full-length viral genomic RNAs do not undergo splicing and instead would be “marked” for encapsidation by binding to Gag. The need to differentiate spliced from unspliced viral RNAs is especially problematic for RSV because the ψ sequence is present on both RNA species. Identification of the host factors bound to spliced and unspliced viral RNAs will be an important step in uncovering the mechanisms that control specific packaging of the retroviral gRNA by Gag.
Acknowledgements
I gratefully acknowledge the members of my laboratory and collaborators who contributed to the work described in this review, including Lisa Z. Scheifele, Rachel Garbitt-Hirst, Scott Kenney, Eileen Ryan, Timothy L. Lochmann, Kristen Butterfield-Gerson, Nicole Gudleski, John Flanagan, Maria Bewley and Anita K. Hopper. I give special thanks to Andrea Beyer for critical review of the manuscript. The Penn State College of Medicine Sequencing Core and Imaging Core were used in this research. This work was supported by a grant from the National Institutes of Health R01 CA76534 (L.J.P.), funds from the Penn State College of Medicine (L.J.P. and A.K.H.), a Graduate Research Fellowship from the National Science Foundation (L.Z.S.), and a Graduate Research Supplement Award from Penn State College of Medicine through the Pennsylvania State Department of Health (L.J.P., T.L.L., and N.G.). This project was funded, in part, under a grant with the Pennsylvania Department of Health using Tobacco Settlement Funds. The Department of Health specifically disclaims responsibility for any analyses, interpretations or conclusions.
Extra View to: Gudleski N, Flanagan JM, Ryan EP, Bewley MC, Parent LJ. Directionality of nucleocytoplasmic transport of the retroviral gag protein depends on sequential binding of karyopherins and viral RNA. Proc Natl Acad Sci USA. 2010;107:9358–9363. doi: 10.1073/pnas.1000304107.
References
- 1.Arnold M, Nath A, Hauber J, Kehlenbach RH. Multiple importins function as nuclear transport receptors for the rev protein of human immunodeficiency virus type 1. J Biol Chem. 2006;281:20883–20890. doi: 10.1074/jbc.M602189200. [DOI] [PubMed] [Google Scholar]
- 2.Balasundaram D, Benedik MJ, Morphew M, Dang VD, Levin HL. Nup124p Is a nuclear pore factor of schizosaccharomyces pombe that is important for nuclear import and activity of retrotransposon Tf1. Mol Cell Biol. 1999;19:5768–5784. doi: 10.1128/mcb.19.8.5768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Berkowitz R, Fisher J, Goff SP. RNA packaging. Curr Top Microbiol Immunol. 1996;214:177–218. doi: 10.1007/978-3-642-80145-7_6. [DOI] [PubMed] [Google Scholar]
- 4.Bohl CR, Brown SM, Weldon RA., Jr The Pp24 phosphoprotein of mason-pfizer monkey virus contributes to viral genome packaging. Retrovirology. 2005;2:68. doi: 10.1186/1742-4690-2-68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bowzard JB, Bennett RP, Krishna NK, Ernst SM, Rein A, Wills JW. Importance of basic residues in the nucleocapsid sequence for retrovirus Gag assembly and complementation rescue. J Virol. 1998;72:9034–9044. doi: 10.1128/jvi.72.11.9034-9044.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bui M, Wills EG, Helenius A, Whittaker G. Role of the infeluenza virus M1 protein in nuclear export of viral ribonucleoproteins. J Virol. 2000;74:1781–1786. doi: 10.1128/jvi.74.4.1781-1786.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Butterfield-Gerson KL, Scheifele LZ, Ryan EP, Hopper AK, Parent LJ. Importin beta family members mediate alpharetrovirus Gag nuclear entry via interactions with MA and NC. J Virol. 2006;80:1798–1806. doi: 10.1128/JVI.80.4.1798-1806.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Carmody SR, Wente SR. Mrna nuclear export at a glance. J Cell Sci. 2009;122:1933–1937. doi: 10.1242/jcs.041236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cullen BR. Nuclear RNA export pathways. Mol Cell Biol. 2000;20:4181–4187. doi: 10.1128/mcb.20.12.4181-4187.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.D'Souza V, Summers MF. How retroviruses select their genomes. Nat Rev Microbiol. 2005;3:643–655. doi: 10.1038/nrmicro1210. [DOI] [PubMed] [Google Scholar]
- 11.Dang VD, Levin HL. Nuclear import of the retrotransposon Tf1 is governed by a nuclear localization signal that possesses a unique requirement for the FXFG nuclear pore factor Nup124p. Mol Cell Biol. 2000;20:7798–7812. doi: 10.1128/mcb.20.20.7798-7812.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Demirov DG, Freed EO. Retrovirus budding. Virus Res. 2004;106:87–102. doi: 10.1016/j.virusres.2004.08.007. [DOI] [PubMed] [Google Scholar]
- 13.Doye V, Hurt E. From nucleoporins to nuclear pore complexes. Curr Opin Cell Biol. 1997;9:401–411. doi: 10.1016/s0955-0674(97)80014-2. [DOI] [PubMed] [Google Scholar]
- 14.Dupont S, Sharova N, DeHoratius C, Virbasius CM, Zhu X, Bukrinskaya AG, et al. A novel nuclear export activity in HIV-1 matrix protein required for viral replication. Nature. 1999;402:681–685. doi: 10.1038/45272. [DOI] [PubMed] [Google Scholar]
- 15.Elton D, Simpson-Holley M, Archer K, Medcalf L, Hallam R, McCauley J, et al. Interaction of the influenza virus nucleoprotein with the cellular Crm1-mediated nuclear export pathway. J Virol. 2001;75:408–419. doi: 10.1128/JVI.75.1.408-419.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Freed EO. Viral late domains. J Virol. 2002;76:4679–4687. doi: 10.1128/JVI.76.10.4679-4687.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fries T, Betz C, Sohn K, Caesar S, Schlenstedt G, Bailer SM. A novel conserved nuclear localization signal is recognized by a group of yeast importins. J Biol Chem. 2007;282:19292–19301. doi: 10.1074/jbc.M700217200. [DOI] [PubMed] [Google Scholar]
- 18.Fukuda M, Asano S, Nakamura T, Adachi M, Yoshida M, Yanagida M, et al. Crm1 is responsible for intracellular transport mediated by the nuclear export signal. Nature. 1997;390:308–311. doi: 10.1038/36894. [DOI] [PubMed] [Google Scholar]
- 19.Garbitt-Hirst R, Kenney SP, Parent LJ. Genetic evidence for a connection between rous sarcoma virus Gag nuclear trafficking and genomic RNA packaging. J Virol. 2009;83:6790–6797. doi: 10.1128/JVI.00101-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Garcia EL, Onafuwa-Nuga A, Sim S, King SR, Wolin SL, Telesnitsky A. Packaging of host My RNAs by Murine leukemia virus may occur early in Y RNA biogenesis. J Virol. 2009;83:12526–12534. doi: 10.1128/JVI.01219-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Grigorov B, Decimo D, Smagulova F, Pechoux C, Mougel M, Muriaux D, et al. Intracellular HIV-1 Gag localization is impaired by mutations in the nucleocapsid zinc fingers. Retrovirology. 2007;4:54. doi: 10.1186/1742-4690-4-54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gudleski N, Flanagan JM, Ryan EP, Bewley MC, Parent LJ. Directionality of nucleocytoplasmic transport of the retroviral Gag protein depends on sequential binding of karyopherins and viral RNA. Proc Natl Acad Sci USA. 2010;107:9358–9363. doi: 10.1073/pnas.1000304107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hsieh PK, Chang SC, Huang CC, Lee TT, Hsiao CW, Kou YH, et al. Assembly of severe acute respiratory syndrome coronavirus RNA packaging signal into virus-like particles is nucleocapsid dependent. J Virol. 2005;79:13848–13855. doi: 10.1128/JVI.79.22.13848-13855.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Huang X, Liu T, Muller J, Levandowski RA, Ye Z. Effect of influenza virus matrix protein and viral RNA on ribonucleoprotein formation and nuclear export. Virology. 2001;287:405–416. doi: 10.1006/viro.2001.1067. [DOI] [PubMed] [Google Scholar]
- 25.Jewell NA, Mansky LM. In the beginning: Genome recognition, RNA encapsidation and the initiation of complex retrovirus assembly. J Gen Virol. 2000;81:1889–1899. doi: 10.1099/0022-1317-81-8-1889. [DOI] [PubMed] [Google Scholar]
- 26.Kemler I, Meehan A, Poeschla EM. Live-cell coimaging of the genomic rnas and Gag proteins of two lentiviruses. J Virol. 2010;84:6352–6366. doi: 10.1128/JVI.00363-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kim MK, Claiborn KC, Levin HL. The long terminal repeat-containing retrotransposon Tf1 possesses amino acids in Gag that regulate nuclear localization and particle formation. J Virol. 2005;79:9540–9555. doi: 10.1128/JVI.79.15.9540-9555.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kobayashi T, Kamitani W, Zhang G, Watanabe M, Tomonaga K, Ikuta K. Borna disease virus nucleo-protein requires both nuclear localization and export activities for viral nucleocytoplasmic shuttling. J Virol. 2001;75:3404–3412. doi: 10.1128/JVI.75.7.3404-3412.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Larsen LS, Beliakova-Bethell N, Bilanchone V, Zhang M, Lamsa A, Dasilva R, et al. Ty3 nucleocapsid controls localization of particle assembly. J Virol. 2008;82:2501–2514. doi: 10.1128/JVI.01814-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ma K, Roy AMM, Whittaker GR. Nuclear export of influenza virus ribonucleoproteins: Identfication of an export intermediate at the nuclear periphery. Virology. 2001;282:215–220. doi: 10.1006/viro.2001.0833. [DOI] [PubMed] [Google Scholar]
- 31.Macara IG. Transport into and out of the nucleus. Microbiol Mol Biol Rev. 2001;65:570–594. doi: 10.1128/MMBR.65.4.570-594.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Martin K, Helenius A. Nuclear transport of influenza virus ribonucleoproteins: The viral matrix protein (M1) promotes export and inhibits import. Cell. 1991;67:117–130. doi: 10.1016/0092-8674(91)90576-k. [DOI] [PubMed] [Google Scholar]
- 33.Mattaj IW, Englmeier L. Nucleocytoplasmic transport: The soluble phase. Annu Rev Biochem. 1998;67:265–306. doi: 10.1146/annurev.biochem.67.1.265. [DOI] [PubMed] [Google Scholar]
- 34.Moore MJ. From birth to death: The complex lives of eukaryotic MRNAs. Science. 2005;309:1514–1518. doi: 10.1126/science.1111443. [DOI] [PubMed] [Google Scholar]
- 35.Moore MJ, Schwartzfarb EM, Silver PA, Yu MC. Differential recruitment of the splicing machinery during transcription predicts genome-wide patterns of mRNA splicing. Mol Cell. 2006;24:903–915. doi: 10.1016/j.molcel.2006.12.006. [DOI] [PubMed] [Google Scholar]
- 36.Nandhagopal N, Simpson AA, Johnson MC, Francisco AB, Schatz GW, Rossmann MG, et al. Dimeric Rous Sarcoma virus capsid protein structure relevant to immature Gag assembly. J Mol Biol. 2004;335:275–282. doi: 10.1016/j.jmb.2003.10.034. [DOI] [PubMed] [Google Scholar]
- 37.Nash MA, Meyer MK, Decker GL, Arlinghaus RB. A subset of pr65gag is nucleus associated in Murine leukemia virus-infected cells. J Virol. 1993;67:1350–1356. doi: 10.1128/jvi.67.3.1350-1356.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nayak DP, Hui EKW, Barman S. Assembly and budding of influenza virus. Virus Research. 2004;106:147–165. doi: 10.1016/j.virusres.2004.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nikolaev I, Cochet MF, Felenbok B. Nuclear import of zinc binuclear cluster proteins proceeds through multiple, overlapping transport pathways. Eukaryotic Cell. 2003;2:209–221. doi: 10.1128/EC.2.2.209-221.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Onafuwa-Nuga AA, King SR, Telesnitsky A. non-random packaging of host RNAs in moloney Murine leukemia virus. J Virol. 79:13528–13537. doi: 10.1128/JVI.79.21.13528-13537.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pardue ML, Rashkova S, Casacuberta E, DeBaryshe PG, George JA, Traverse KL. Two retrotransposons maintain telomeres in drosophila. Chromosome Res. 2005;13:443–453. doi: 10.1007/s10577-005-0993-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Parent LJ, Bennett RP, Craven RC, Nelle TD, Krishna NK, Bowzard JB, et al. Positionally independent and exchangeable late budding functions of the Rous Sarcoma virus and human immunodeficiency virus Gag proteins. J Virol. 1995;69:5455–5460. doi: 10.1128/jvi.69.9.5455-5460.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Poole E, Strappe P, Mok HP, Hicks R, Lever AM. HIV-1 Gag-RNA interaction occurs at a perinuclear/centrosomal site; analysis by confocal microscopy and fret. Traffic. 2005;6:741–755. doi: 10.1111/j.1600-0854.2005.00312.x. [DOI] [PubMed] [Google Scholar]
- 44.Rashkova S, Karam SE, Kellum R, Pardue ML. Gag proteins of the two drosophila telomeric retrotransposons are targeted to chromosome ends. J Cell Biol. 2002;159:397–402. doi: 10.1083/jcb.200205039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rein A. Retroviral RNA packaging: A review. Arch Virol. 1994;9:513–522. doi: 10.1007/978-3-7091-9326-6_49. [DOI] [PubMed] [Google Scholar]
- 46.Scheifele LZ, Garbitt RA, Rhoads JD, Parent LJ. Nuclear entry and Crm1-dependent nuclear export of the Rous Sarcoma virus Gag polyprotein. Proc Natl Acad Sci USA. 2002;99:3944–3949. doi: 10.1073/pnas.062652199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Scheifele LZ, Kenney SP, Cairns TM, Craven RC, Parent LJ. Overlapping roles of the Rous Sarcoma virus Gag P10 domain in nuclear export and virion core morphology. J Virol. 2007;81:10718–10728. doi: 10.1128/JVI.01061-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Scheifele LZ, Ryan EP, Parent LJ. Detailed mapping of the nuclear export signal in the Rous Sarcoma virus Gag protein. J Virol. 2005;79:8732–8741. doi: 10.1128/JVI.79.14.8732-8741.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Schliephake AW, Rethwilm A. Nuclear localization of foamy virus Gag precursor protein. J Virol. 1994;68:4946–4954. doi: 10.1128/jvi.68.8.4946-4954.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Swanstrom R, Wills JW. In: Synthesis, Assembly and Processing of Viral Proteins. Retroviruses JM, Coffin SH, Hughes, Varmus HE, editors. Cold Spring Harbor Laboratory Press; 1997. pp. 263–334. [PubMed] [Google Scholar]
- 51.Terry LJ, Shows EB, Wente SR. Crossing the nuclear envelope: Hierarchical regulation of nucleocytoplasmic transport. Science. 2007;318:1412–1416. doi: 10.1126/science.1142204. [DOI] [PubMed] [Google Scholar]
- 52.Verderame MF, Nelle TD, Wills JW. The membrane-binding domain of the Rous Sarcoma virus Gag protein. J Virol. 1996;70:2664–2668. doi: 10.1128/jvi.70.4.2664-2668.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Vogt VM. In: Retroviral Virions and Genomes. Retroviruses JM, Coffin SH, Hughes, Varmus HE, editors. Cold Spring Harbor NY: Cold Spring Harbor Press; 1997. pp. 27–69. [PubMed] [Google Scholar]
- 54.Weldon RA, Jr, Sarkar P, Brown SM, Weldon SK. Mason-Pfizer monkey virus Gag proteins interact with the human sumo conjugating enzyme, hUbc9. Virology. 2003;314:62–73. doi: 10.1016/s0042-6822(03)00348-9. [DOI] [PubMed] [Google Scholar]
- 55.Wills JW, Craven RC. Form, function and use of retroviral Gag proteins. AIDS. 1991;5:639–654. doi: 10.1097/00002030-199106000-00002. [DOI] [PubMed] [Google Scholar]
- 56.Yanai H, Kobayashi T, Hayashi Y, Watanabe Y, Ohtaki N, Zhang G, et al. A methionine-rich domain mediates Crm1-dependent nuclear export activity of borna disease virus phosphoprotein. J Virol. 2006;80:1121–1129. doi: 10.1128/JVI.80.3.1121-1129.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.You J, Dove BK, Enjuanes L, DeDiego ML, Alvarez E, Howell G, et al. Subcellular localization of the severe acute respiratory syndrome coronavirus nucleocapsid protein. J Gen Virol. 2005;86:3303–3310. doi: 10.1099/vir.0.81076-0. [DOI] [PubMed] [Google Scholar]
- 58.Yu SF, Edelmann K, Strong RK, Moebes A, Rethwilm A, Linial ML. The Carboxyl Terminus of the Human Foamy Virus Gag Protein Contains Separable Nucleic Acid Binding and Nuclear Transport Domains. J Virol. 1996;70:8255–8262. doi: 10.1128/jvi.70.12.8255-8262.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zhou J, McAllen JK, Tailor Y, Summers MF. High affinity nucleocapsid protein binding to the mupsi RNA packaging signal of Rous Sarcoma virus. J Mol Biol. 2005;349:976–988. doi: 10.1016/j.jmb.2005.04.046. [DOI] [PubMed] [Google Scholar]