Abstract
In eukaryotes, many nuclear processes are spatially compartmentalized. Previously, we have shown that in Trypanosoma cruzi, an early-divergent eukaryote, DNA replication occurs at the nuclear periphery where chromosomes remain constrained during the S phase of the cell cycle. We followed Orc1/Cdc6, a pre-replication machinery component and the proliferating cell nuclear antigen (PCNA), a component of replication machinery, during the cell cycle of this protozoon. We found that, at the G1 stage, TcOrc1/Cdc6 and TcPCNA are dispersed throughout the nuclear space. During the G1/S transition, TcOrc1/Cdc6 migrates to a region close to nuclear periphery. At the onset of S phase, TcPCNA is loaded onto the DNA and remains constrained close to nuclear periphery. Finally, in G2, mitosis and cytokinesis, TcOrc1/Cdc6 and TcPCNA are dispersed throughout the nuclear space. Based on these findings, we propose that DNA replication in T. cruzi is accomplished by the organization of functional machineries in a spatial-temporal manner.
Key words: nuclear organization, replication sites, PCNA, Orc1/Cdc6, Trypanosoma cruzi
Introduction
In all organisms, DNA replication is a complex event involving multiple proteins and enzymes to ensure accurate and timely duplication of the genetic material. The initial step that occurs at the G1 phase of the cell cycle is the assembly of pre-replication machinery into the chromatin at replication origins. In unicellular eukaryotes such as yeast and metazoans, the replication origin is recognized by the heterohexamer origin recognition complex (ORC), which contains six proteins, Orc1–Orc6. Once bound to DNA, ORC recruits Cdc6 and Cdt1, and together, these molecules recruit the helicase component, mini-chromosome maintenance (MCM), which is fundamental for the initiation of DNA replication. During the G1/S transition, the binding of regulatory factors and components of the replication fork to the DNA allows unwinding at the origin, the recruitment of replicative DNA polymerases and finally, the establishment of replication forks.1,2 The high processivity of DNA polymerases is warranted by the proliferating cell nuclear antigen (PCNA), which associates with double-stranded DNA, forming a ring-like structure.3 At the replication fork, PCNA not only acts as a linker between DNA polymerase and the DNA backbone, it also ensures continuous leading strand synthesis, completion of Okazaki fragment synthesis,4 removal of RNA primers,5,6 and ligation of two adjacent fragments, thus concluding DNA replication.7,8
We have previously shown that in the proliferative stage of the life cycle of Trypanosoma cruzi, an early-divergent eukaryote and the agent of Chagas' disease, the duplication of genetic material, is constrained at the nuclear periphery. At the nuclear periphery, chromosomes are concentrated during S phase of the cell cycle.9
To better understand the organization of DNA replication in this organism, we have examined whether the pre-replication and replication machineries remain constrained at the nuclear periphery during the entire cell cycle or if these replication molecules are recruited to the replication sites in a spatial-temporal manner. As a marker for the pre-replication machinery, we used a molecule named Orc1/Cdc6, which, in trypanosomes, replaces the six Orcs and the Cdc6 normally found in other eukaryotes.10 As a marker for the replication machinery, we used PCNA. PCNA was the first protein identified at the replication foci during the S phase11,12 and since then, it has been widely used as a marker for replication foci.13 In trypanosomatids, it has been recently demonstrated that PCNA co-localizes with the replication foci in Leishmania major.14 TcOrc1/Cdc6 and TcPCNA were found forming the following two distinct distribution patterns: a peripheral one, in which TcOrc1/Cdc6 and TcPCNA are constrained close to nuclear periphery and a dispersed pattern, in which Orc1/Cdc6 and PCNA are dispersed throughout the nuclear space. These patterns are related to the cell cycle. The peripheral pattern is found during the S phase, whereas the dispersed distribution is found in cells at G1, G2, mitosis and cytokinesis.
Results
Distribution of TcOrc1/Cdc6 and TcPCNA in the nuclei of T. cruzi.
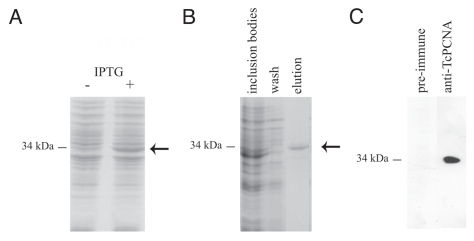
The localization of Orc1/Cdc6 and PCNA in the nuclei of the epimastigote form of T. cruzi, which corresponds to a parasite proliferative stage, was investigated using an anti-TcOrc1/Cdc6 antibody10 and an anti-serum of rabbits and mice, which were immunized with rTcPCNA. Figure 1A shows the expression of recombinant TcPCNA in IPTG-induced cultures of E. coli BL21DE3, and the 34 kDa protein detected in urea solubilized inclusion bodies. rTcPCNA purified by Ni-columns (Fig. 1B) was used to immunize animals. The antibodies were found to be specific to PCNA in whole epimastigote cell extracts (Fig. 1C).
Figure 1.
Preparation and specificity of anti-TcPCNA antibodies. Coomassie Blue stained SDS-PAGE of E. coli BL21 total extracts (A) without (−) or after IPTG induction (+) or (B) after Ni-agarose purification. Arrows indicate rTcPCNA migration. (C) T. cruzi epimastigotes were boiled in the SDS-PAGE loading buffer, and the supernatants were analyzed by SDS-PAGE, followed by western blot using either anti-rTcPCNA immune or pre-immune serum rabbit. The numbers on the left of each gel correspond to the molecular weight standards.
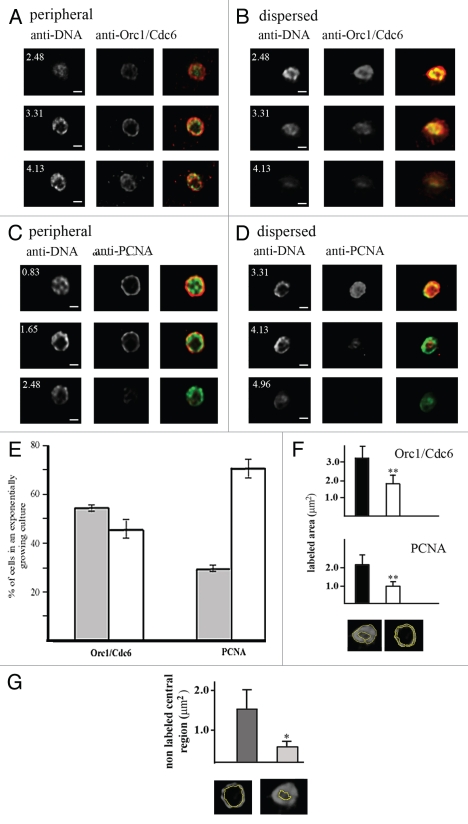
Trypanosoma cruzi, as well as other kinetoplastids presents a structure named kinetoplast, which contains the mitochondrial genetic material. In epimastigote cells, analyzed in this study, the kinetoplast is localized close to nucleus as represented in the supplementary material (Fig. S1a) and DAPI staining labels both nucleus and kinetoplast (Fig. S1b). Since kinetoplast is close to nucleus, which sometimes impairs the nuclear visualization, we decided to label T. cruzi DNA using an anti-DNA monoclonal antibody (mAbB17). This antibody was obtained from NZB/NZW mice, which spontaneously develop a systemic autoimmune disease that closely resembles human systemic lupus. Due to unknown reasons, this antibody is not able to recognize kinetoplast DNA. Immunofluorescence assays using anti-TcOrc1/Cdc6 or anti-TcPCNA showed that TcOrc1/Cdc6 and TcPCNA labels just the nucleus. It is expected once the replication origins, as well as replication machineries working in kinetoplast DNA replication are quite different from that working on nuclear DNA replication. TcOrc1/Cdc6 and TcPCNA present two patterns of nuclear distribution in an exponentially growing culture. Analysis of different Z-sections, obtained by confocal microscopy, illustrate the following two main patterns: a peripheral pattern, in which molecules are constrained close to nuclear periphery (Fig. 2A and C), and a dispersed pattern, in which TcOrc1/Cdc6 or TcPCNA is dispersed throughout the nuclear space (Fig. 2B and D). The TcOrc1/Cdc6 and TcPCNA labeled area of dispersed and peripheral patterns were measured (as showed in the bottom of Fig. 2F). We found that in fact the labeled area of peripheral pattern is smaller than the dispersed pattern (Fig. 2F). Comparing the anti-TcOrc1/Cdc6 labeling with anti-DNA labeling we found that when TcOrc1/Cdc6 is constrained close to nuclear periphery, DNA is also constrained at this region (Fig. 2A). However, when TcOrc1/Cdc6 is dispersed through the nuclear space, DNA is also dispersed (Fig. 2B). These data are not unexpected once we have shown that TcOrc1/Cdc6 binds DNA.10 But it is interesting to note that the entire DNA (and not only the replication origins) can also be found constrained close to nuclear periphery (Fig. 2A). Also, even when DNA is close to nuclear periphery, TcOrc1/Cdc6 is outsider (Fig. 2A), strongly suggesting that chromatin structures in loops putting replication origin closer to nuclear periphery.
Figure 2.
There are two patterns of TcOrc1/Cdc6 and TcPCNA distribution in the nuclei of T. cruzi epimastigote cells. Epimastigote cells were fixed with 2% paraformaldehyde, permeabilized with Triton X-100 and incubated with (A and B) anti-TcOrc1/Cdc6 (red) or (C and D) anti-TcPCNA antibodies (red). All cells were labeled with mAbB17 (green), a monoclonal anti-DNA antibody. Images shown for each pattern were acquired at different Z-sections by a confocal microscope. The white numbers indicate the distance between the section and the top limit of each nucleus. N indicates nuclei, k indicates kinetoplasts and bars represent 1 µm. (E) Graph shows average ± standard deviation of three independent experiments (n = 100), indicating the proportion of cells in an exponentially growing culture presenting TcOrc/Cdc6 or TcPCNA constrained at the nuclear periphery (gray box) or dispersed throughout the entire nucleus (white box). (F) Graphs show labeled area by anti-TcOrc1/Cdc6 or anti-TcPCNA in dispersed (black) or peripheral (white) patterns (n = 50). To perform this analysis the labeled areas were circled as exemplified in the bottom of (F) and these areas were quantified using the Image J program. **p < 0.01. (G) Graph shows the central non-labeled area by anti-TcOrc1/Cdc6 (dark gray) or anti-TcPCNA (light gray) antibodies. To perform this analysis the central non-labeled areas were circled as exemplified in the bottom of (G) and these areas were quantified by Image J. *p < 0.05.
Images suggest that the peripheral pattern of TcOrc1/Cdc6 is more constrained close to nuclear periphery than the TcPCNA peripheral pattern. To confirm that, we measured the central non-labeled area (as represented in the bottom of Fig. 2G) of nuclei presenting peripheral patterns. We found that central nonlabeled area from nuclei labeled with anti-TcPCNA is smaller than the central non-labeled area from nuclei labeled with anti-TcOrc1/Cdc6 (Fig. 2G). Quantitative analyses of the distribution of these patterns in exponentially growing cells (n = 100 in three experiments) showed that TcOrc1/Cdc6 is constrained at the nuclear periphery in 54% of cells, whereas TcPCNA is localized at the nuclear periphery in 24% of cells (Fig. 2E).
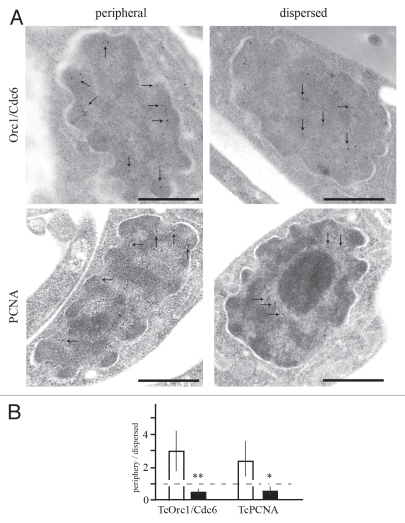
To further explore the nuclear localization of these molecules, ultrathin sections were labeled with each antibody and sections observed by transmission electron microscope. While in some cells TcOrc1/Cdc6 and TcPCNA are constrained in a more peripheral nuclear region (between nuclear membrane and the eletrodense chromatin), these molecules are not juxtaposed with nuclear membrane in 90% of the cells (Fig. 3A and left parts), suggesting that the physical contact between nuclear membrane proteins and TcOrc1/Cdc6 or TcPCNA is not necessary for the localization of these proteins close to nuclear periphery. The right parts in Figure 3A show the dispersed patterns of both molecules visualized by immunoelectronic assay. Figure 3B is the quantification of data present in Figure 3A, showing that the difference between the nuclear localization of TcOrc1/Cdc6 or TcPCNA in the peripheral or dispersed pattern is statically significant.
Figure 3.
Ultrastructural immunocytochemistry showing the localization of TcOrc1/Cdc6 and TcPCNA. (A) Exponentially growing epimastigotes were fixed and processed for immune electron microscopy and labeled with anti-TcOrc1/Cdc6 (top parts) or with antiTcPCNA (bottom parts), followed by anti-rabbit IgG conjugated to 10 nm gold particles. Arrows indicate the gold particles seen in each micrograph. Bars = 0.5 µm. (B) Statistical analysis of the patterns observed in (A). First, we defined “close to nuclear periphery” as the region between nuclear membrane and eletrodense chromatin. Then, we analyzed different images and determined the number of gold particles “close to nuclear periphery” or “dispersed through the entire nucleus”. The ratio between “close to nuclear periphery“ and “dispersed through the entire nucleus” was plotted for peripheral pattern (white box) or dispersed pattern (black box). Interrupted line indicates the ratio 1 that means the same number of gold particles close to nuclear periphery and dispersed through the entire nucleus. **p < 0.01 and *p < 0.05.
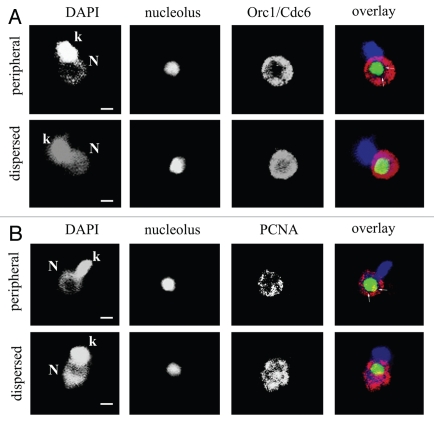
Next, we localized TcOrc1/Cdc6 and TcPCNA in parasites expressing green fluorescent protein (GFP) fused with the first 33 amino acids of the T. cruzi histone, H2B. This hybrid protein is found exclusively as a large dot in the nucleolus of epimastigotes.15 As shown in Figure 4, when anti-TcOrc1/Cdc6 is found mainly close to nuclear periphery, the central area, where anti-TcOrc1/Cdc6 is absent is larger than the nucleolar region (arrows in Fig. 4A, upper part). In the dispersed pattern, anti-TcOrc1/Cdc6 is juxtaposed with minimal co-localization with GFP (Fig. 4A and lower part). Similarly, in spite of TcPCNA being juxtaposed to the nucleolus in some regions of the nuclei with peripheral patterns, it is clear that the area in the central region that anti-TcPCNA is absent from is larger than the nucleolar region (arrows in Fig. 4B, upper part). In cells with the dispersed pattern, TcPCNA labeling is juxtaposed to the nucleolus (Fig. 4B and lower part). These data show that the peripheral pattern is not a consequence of the nucleolus extension, but instead it represents compartmentalization of TcOrc1/Cdc6 and TcPCNA in precise nuclear regions.
Figure 4.
Localization of TcOrc1/Cdc6 and TcPCNA according to the position of the nucleolus. Epimastigote cells expressing GFP in the nucleolus (green) were labeled with (A) anti-TcOrc1/Cdc6 (red) or with (B) anti-TcPCNA (red) by immunofluorescence assay. All cells were stained with DAPI (blue). Single optical sections were acquired using confocal microscopy. N indicates nuclei, k indicates kinetoplasts and bars represent 1 µm.
Different patterns of TcOrc1/Cdc6 and TcPCNA localization are related to the cell cycle.
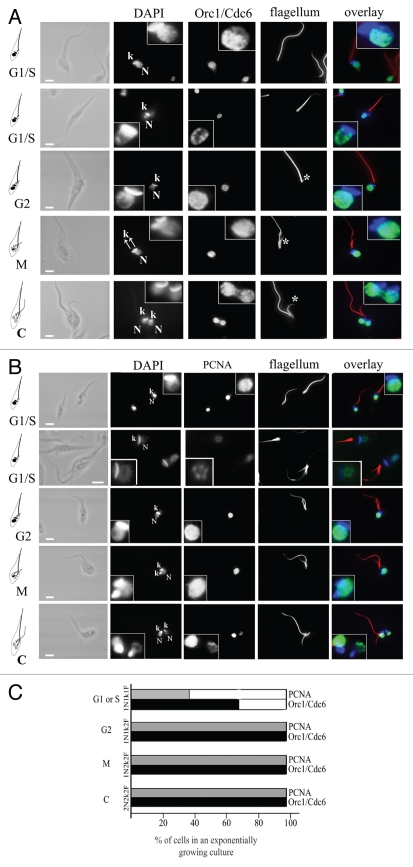
The cell cycle stage of an epimastigote cell can be inferred by the number of flagella, nuclei and kinetoplasts. Kinetoplasts correspond to the genetic material of the unique mitochondrion in this organism. As schematically represented in Figure 5, cells with one nucleus, one kinetoplast and one flagellum (1N1k1F) are in the G1 or S stage of the cell cycle. Cells with one nucleus, one kinetoplast and two flagella (1N1k2F) are in the G2 phase. Cells with one nucleus, two kinetoplasts and two flagella (1N2k2F) are close to mitosis, and cells with two nuclei, two kinetoplasts and two flagella (2N2k2F) are in cytokinesis.16 The cells were stained with mAb25, which labels flagella, DAPI and anti-TcOrc1/Cdc6 (Fig. 5A) or anti-TcPCNA (Fig. 5B). As shown in the selected images, the peripheral pattern was observed in cells containing one nucleus, one kinetoplast and one flagellum, which are cells in the G1 or S phase of the cell cycle. The dispersed pattern was found predominantly in the remaining stages of the cell cycle when stained with anti-TcOrc1/Cdc6 and anti-TcPCNA. Quantitative analyses showed that 100% of cells in G2, mitosis and cytokinesis cells contain TcOrc1/Cdc6 and TcPCNA dispersed throughout the entire nucleus, 67% of cells at the G1/S phase contain TcOrc1/Cdc6 constrained close to nuclear periphery and only 29% of cells at the G1/S phase contain TcPCNA constrained close to nuclear periphery (Fig. 5C).
Figure 5.
TcOrc1/Cdc6 and TcPCNA localization varies according to the stage of the cell cycle. Exponentially growing cultures of T. cruzi epimastigote forms were harvested by centrifugation, immobilized on glass slides, fixed in 2% formaldehyde and permeabilized with 0.1% Triton-X100. Cells were incubated with (A) anti-TcOrc1/Cdc6 (green) or with (B) anti-TcPCNA antibodies (green). All cells were incubated with mAbAC to label the flagellum (red). Cells were also stained with DAPI (blue). N indicates nuclei, k indicates kinetoplasts and * indicates the new flagellum. The inserted box shows a zoomed image. Bars represent 2 µm. (C) Graph shows average of three independent experiments (n = 100), indicating the proportion of cells presenting TcOrc/Cdc6 (black columns) or TcPCNA (gray columns) constrained at the nuclear periphery (empty box) or dispersed throughout the entire nucleus (filled box) in each stage of the cell cycle.
TcOrc1/Cdc6 and TcPCNA are localized close to nuclear periphery during the S phase.
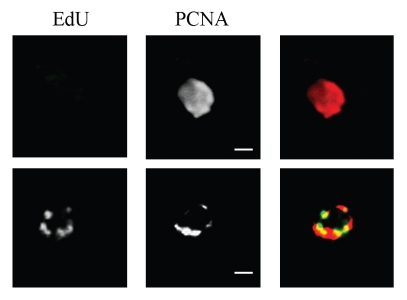
As mentioned, it is not possible to distinguish between cells at the G1 or S phase according to the number of nuclei, kinetoplasts and flagella. We, along with others, have already shown that it is possible to synchronize T. cruzi epimastigote cells with hydroxyurea.9,17,18 Hydroxyurea inhibits the nucleotide reductase enzyme, thereby consuming dNTPs. In the absence of dNTPs, DNA elongation is blocked and most of the cells are arrested very early in S phase. We have also shown that 6 h after hydroxyurea release, cells are in the S phase, replicating their DNA.9 Analysis of cells at 0 or 6 h after release (S phase) showed that most of these cells display the peripheral pattern for both TcOrc1/Cdc6 and TcPCNA (Table 1). These numbers strongly suggest that the peripheral patterns correspond to cells whose DNA is being replicated. To confirm that TcPCNA is close to nuclear periphery of cells at S stage of the cell cycle, we performed a nucleotide pulse labeling. We have previously shown that the shorter pulse to visualize deoxynucleotide incorporation in T. cruzi epimastigote cells is 2 hours.9 Therefore, we incubated cells for 2 h with EdU and these cells were analyzed concerning TcPCNA localization. We found that in EdU non-labeled cells TcPCNA is dispersed through the nuclear space (Fig. 6A and top part), while in 100% of EdU labeled cells TcPCNA is found at nuclear periphery (Fig. 6B and bottom part). These results confirm that TcPCNA is at nuclear periphery during DNA replication at S phase.
Table 1.
Patterns of TcOrc1/Cdc6 and TcPCNA localization after HU release
| Hours after HU release (stage of the cell cycle) | TcOrc1/Cdc6 dispersed | TcOrc1/Cdc6 peripheral | TcPCNA dispersed | TcPCNA peripheral |
| 0 (G1/S transition) | 25.3 ± 2.1 | 74.7 ± 2.1 | 20.7 ± 1.5 | 79.3 ± 1.5 |
| 6 (S) | 11.6 ± 2.1 | 89.5 ± 0.7 | 11 ± 2 | 89 ± 2 |
| 20 (G1) | 55.6 ± 2.5 | 44.3 ± 2.5 | 89.7 ± 1.5 | 10.3 ± 1.5 |
*Numbers are mean of percentage of cells ± standard deviations of each pattern. Numbers are media of three independent experiments (n = 100).
Figure 6.
TcPCNA is found at nuclear periphery of replicating cells. Exponentially growing cells were exposed to EdU (green) for 2 h, fixed in 2% formaldehyde, permeabilized with 0.1% Triton-X100 and incubated with anti-TcPCNA (red). Bars are 1 µm.
To confirm that the dispersed pattern visualized in cells with one nucleus, one kinetoplast and one flagellum corresponds to G1 cells, we followed cells after hydroxyurea release until the cells reached the G1 stage. It has been shown that 18 h after the release, most of the cells are still at G2.18 To examine cells just after mitosis, we took epimastigotes 20 h after hydroxyurea release. At this point, 25% of the cells were in cytokinesis (two nuclei, two kinetoplasts and two flagella) and 75% of cells had concluded the cell cycle (one nucleus, one kinetoplast and one flagellum). Most of these 1N1k1F cells contain TcOrc1/Cdc6 and TcPCNA dispersed throughout the nuclear space (Table 1), confirming that these molecules are dispersed at the G1 stage of the cell cycle. It is important to note that these 1N1k1F cells differ greatly in their TcOrc1/Cdc6 and TcPCNA localization patterns. While 55.6% of cells present TcOrc1/Cdc6 dispersed throughout the nuclear space, 89.7% of these cells present TcPCNA throughout the nuclear space (Table 1). These numbers suggest that TcOrc1/Cdc6 reaches the nuclear periphery before TcPCNA does.
Constraining of TcOrc1/Cdc6 and TcPCNA close to nuclear periphery.
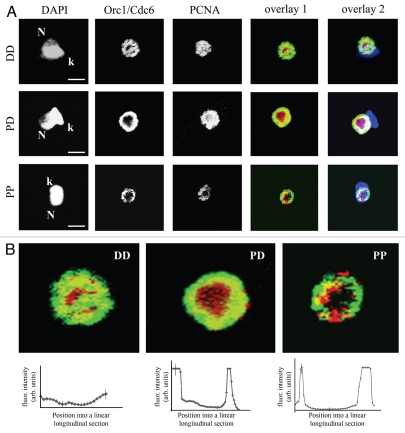
To further verify the possibility that TcOrc1/Cdc6 localizes close to nuclear periphery before TcPCNA does, exponentially growing cells were labeled with both anti-TcOrc1/Cdc6 and anti-TcPCNA. Among the cells with one nucleus, one kinetoplast and one flagellum (G1 and S phase cells), we found the following: 42% of cells had both TcPCNA and TcOrc1/Cdc6 dispersed throughout the nuclear space (DD pattern), 27% of cells had TcOrc1/Cdc6 constrained at the nuclear periphery and TcPCNA dispersed throughout the entire nucleus (DP pattern) and 31% of cells had both TcOrc1/Cdc6 and TcPCNA constrained at the nuclear periphery (PP pattern) (Fig. 7A and Table 2). The fourth possibility, dispersed TcOrc1/Cdc6 with peripheral TcPCNA, was not found. These data clearly demonstrated that TcOrc1/Cdc6 localizes to the nuclear periphery before TcPCNA does during the T. cruzi cell cycle. The three patterns were also observed in different z-stacks by confocal microscope (Fig. S2). Analysis of the double staining at high magnification showed that co-localization of TcOrc1/Cdc6 and TcPCNA was observed only when both molecules were distributed to the nuclear periphery (Fig. 7B). Graphs below each pattern in Figure 7 show the mean intensity of co-localization signals (found by the Image J co-localization finder plug-in) in a longitudinal section of several nuclei. Since the co-localization of these molecules represents replication origins that have just been fired, we propose that replication starts close to nuclear periphery in epimastigote forms of T. cruzi. Note that even when TcOrc1/Cdc6 and TcPCNA are constrained close to nuclear periphery, TcOrc1/Cdc6 is further outside than TcPCNA (statistic presented at Fig. 2G and 7). Considering that the direction of the replication fork is from TcOrc1/Cdc6 (replication origins) to TcPCNA, we concluded that DNA replication occurs from a region close to nuclear periphery to nuclear interior.
Figure 7.
TcOrc1/Cdc6 localizes to nuclear periphery before TcPCNA. (A) Exponentially growing cultures of T. cruzi epimastigote forms were harvested by centrifugation, immobilized on glass slides, fixed in 2% formaldehyde and permeabilized with 0.1% Triton-X100. Cells were incubated with anti-TcOrc1/Cdc6 (red) and with anti-TcPCNA antibodies (green). Cells were also stained with DAPI (blue). Bars represent 2 µm. N indicates nuclei and k indicates kinetoplasts. Overlay 1 shows the merged images without DAPI and overlay 2 includes DAPI staining. (B) The overlaid images have been zoomed. The patterns observed are TcOrc1/Cdc6 and TcPCNA dispersed (DD), TcOrc1/Cdc6 peripheral and TcPCNA dispersed (PD) and TcOrc1/Cdc6 and TcPCNA peripheral (PP). Graphs below each image show the mean fluorescence intensity of co-localized signal versus the position in a linear longitudinal section that was traced over the image of each pattern (n = 10). The co-localization signal was identified using the Image J “colocalization finder” plug in.
Table 2.
Patterns of TcOrc1/Cdc6 and TcPCNA localization in G1 and S cells
| % of the pattern in the 1N1k1F cells (n = 100) | |
| DD | 42 |
| PD | 27 |
| PP | 31 |
*Numbers are media of three independent experiments.
TcPCNA remains constrained at the nuclear periphery when bound to DNA.
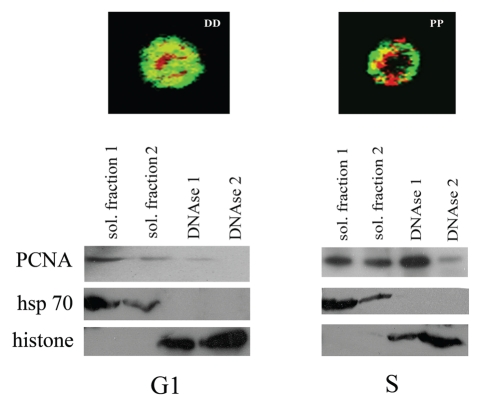
We have previously shown that TcOrc1/Cdc6 remains bound to DNA during the entire cell cycle.10 It is well-established in other models that, at the S phase, PCNA is recruited to the DNA replication fork after extension of the primers by DNA-polymerase α-primase.19 Therefore, we hypothesized that the localization of TcPCNA close to the nuclear periphery during the S phase is a consequence of the recruitment of this molecule by DNA. To check this possibility, we used an in vitro differential extraction assay that distinguishes among cytoplasm/nucleoplasm soluble proteins and proteins bound to DNA.10 Cells were then synchronized with hydroxyurea. Twenty hours after hydroxyurea release (G1 phase), or immediately after release (early S phase, when the DNA replication fork is established), cells were treated with detergent-containing buffer to extract soluble proteins. The obtained pellets were then digested with DNAse to fragment DNA, releasing DNA-bound proteins. Samples were then analyzed by western blotting using an anti-TcPCNA antibody. As expected, in G1 cells (20 h after HU release), when TcPCNA is dispersed throughout the nuclear space, most of the TcPCNA is obtained after detergent extraction. Moreover, 0 h after HU release, when 55% of cells present TcPCNA constrained close to the nuclear periphery, most of the TcPCNA is obtained after DNAse digestion (Fig. 8). This data demonstrates that TcPCNA is located close to the nuclear periphery when it is bound to DNA. Heat shock protein 70 was also analyzed as a control for soluble proteins and histone H4 was analyzed as a control for DNA-binding proteins.
Figure 8.
TcPCNA is bound to DNA when it is localized to the nuclear periphery. Cells were synchronized with hydroxyurea and analyzed 6 h (right part) and 20 h (left part) after release. Pellets were extracted using detergent-containing buffer (soluble fraction 1). The pellets were then extracted again using the same detergent-containing buffer (soluble fraction 2). Pellets were then digested with DNAse, samples were centrifuged again, and the supernatants were saved as DNAse-released proteins (DNAse 1). The pellets were digested with DNAse again, samples were centrifuged and the supernatants were saved as DNAse released proteins (DNAse 2). Samples were analyzed by SDS-PAGE, followed by western blotting using anti-TcPCNA serum. For control, samples were also analyzed by SDS-PAGE, followed by western blotting, using antiheat shock protein 70 (anti-hsp 70) and anti-histone H4 (histone).
Discussion
Based on previous results regarding morphological patterns of each cell cycle stage of T. cruzi replicative forms,16 we established the dynamics of TcOrc1/Cdc6 and TcPCNA, pre-replication and replication machinery components, respectively. After division, cells enter into the G1 phase and TcOrc1/Cdc6 and TcPCNA are dispersed throughout the nuclear space, excluding the nucleolus region. Since TcOrc1/Cdc6 is bound to DNA during the entire cell cycle, TcOrc1/Cdc6 labeling might be reflecting replication origins localization.10 TcOrc1/Cdc6 is located close to nuclear periphery in 56% of the cells from an asynchronous culture (Fig. 2). We have previously shown that 14% of an asynchronous culture corresponds to S phase cells; therefore, we conclude that as cells go through G1, TcOrc1/Cdc6 localizes close to the nuclear periphery, while TcPCNA remains dispersed. Previously, we have shown that chromatin is also dispersed during the G1 phase and becomes peripheral during replication.9 Furthermore, we have shown that TcOrc1/Cdc6 is bound to DNA during the entire cell cycle.10 In this way, it is possible that TcOrc1/Cdc6 localizes close to nuclear periphery region as a consequence of chromosome movement. However, it is also possible that chromosomes move to the nuclear periphery as a result of TcOrc1/Cdc6 localizing close to nuclear periphery. Twenty four percent of the cells in an asynchronous culture present TcPCNA localized close to nuclear periphery. This number is very close to the percentage of cells in S phase (14%), and this small discrepancy might be due to the sensibility of different methodologies. Therefore, we conclude that upon reaching early S phase, the DNA replication fork is established, TcPCNA is loaded onto DNA clustering close to the nuclear periphery.
TcPCNA remains at the replication fork and moves away from the replication origin as DNA elongates. Therefore, the co-localization of TcOrc1/Cdc6 with TcPCNA is expected to occur only during the firing of replication origins. The most extensive co-localization of the two proteins is close to nuclear periphery. Therefore, we concluded that T. cruzi DNA replication occurs mainly close to nuclear periphery, whereas mammalian early-replicating genes are replicated in the nucleus interior and mid and late-replicating genes are replicated at the nuclear periphery.20 It is important to note here that in mammalian cells the chromatin is present in the same location throughout the cell cycle and replication spreads to the relevant compartment at a defined time of S phase. As a quite different system, we showed that in T. cruzi entire chromatin is relocated from nuclear interior to a more peripheral zone (compare Fig. 2A with B) and reviewed in reference 9. These dynamics of TcOrc1/Cdc6 and TcPCNA, together with chromosome movement are schematically represented in Figure 9.
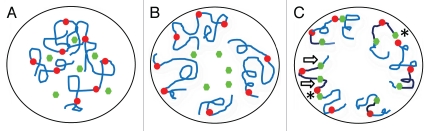
Figure 9.
Schematic representation of TcOrc1/Cdc6 and TcPCNA dynamics during the cell cycle of T. cruzi. When a new cycle starts, chromosomes (blue), TcOrc1/Cdc6 (red) and TcPCNA (green) are dispersed throughout the nuclear space (A). As cells go through G1, chromosomes and TcOrc1/Cdc6 localize close to nuclear periphery, while TcPCNA is dispersed throughout nuclear space (B). At the onset of the S phase, TcPCNA is loaded onto replication origins (c-asterisks). Once DNA replication begins, TcPCNA follows the replication fork that goes from replication origin (located closed to nuclear periphery) into nuclear interior direction (direction is indicated by arrows on C). Chromosomes regions between TcOrc1/Cdc6 and TcPCNA has already duplicated and are represented in dark blue. Chromosomes regions that are not replicated are in ligth blue.
The molecular bases that warrant the occurrence of DNA replication close to nuclear periphery in T. cruzi remain unknown. One possibility is that the import of nucleotides from the cytoplasm establishes this organization. Another hypothesis is that since T. cruzi does not control transcription, it should physically separate the mechanisms of transcription and replication. In fact, T. cruzi transcription occurs throughout the nuclear space and has a main transcription center close to the nucleolus.21
In other eukaryotes, the ORC complex is additionally involved in a wide range of cellular processes, such as transcriptional silencing and heterochromatin formation, cell cycle checkpoints, mitotic chromosome assembly, sister chromatid cohesion (as reviewed in ref. 22) and establishments of centriole and centrosome copy numbers.23 In the same way, PCNA is known to interact with many proteins related to DNA replication, DNA repair, translation DNA synthesis, DNA methylation, cell cycle regulation, chromatin metabolism, chromatin cohesion and apoptosis.24–26 Therefore, at this point, we cannot infer that the dynamics of TcOrc1/Cdc6 and TcPCNA observed in T. cruzi are exclusively due to DNA replication. It is important to note, however, that TcPCNA co-localizes with sites of active DNA replication (Fig. 6). Nonetheless, the recruitment of molecules to specific sites in the T. cruzi nucleus illustrates the compartmentalization of specific machineries of this simpler organism, contributing to the understanding of the spatial-temporal process involved in nuclear organization among eukaryotes.
Materials and Methods
Cell culture.
The experiments were performed with exponentially growing T. cruzi epimastigotes (Y strain) that were cultured in liver infusion tryptose medium supplemented with 10% fetal bovine serum at 28°C.27 The transfected lineage, named pTexΔ, contains a sequence coding for green fluorescence protein (GFP) fused with amino acids 4–28 of the histone H2B from the T. cruzi gene.15 The transfected cells were cultured in the same conditions as above in the presence of 0.5 mg/ml of Geneticin G418. Cells were synchronized with hydroxyurea as described in reference 9.
Construction, protein purification, polyclonal antibody and western blot analyses.
T. cruzi PCNA (TcPCNA) sequence was amplified from genomic DNA of epimastigotes CL-Brener strain by PCR, with oligonucleotides PCNAF (5′-GCT AGC CTG GAG GCT CAA GTT C) and PCNAR (5′-CGA TCG TCA CAT CAC GTC ATC C). The PCR product was inserted into a TOPO vector (Invitrogen) and then into the Nhe I site of pET28a (Novagen). The plasmids containing the correct fragment orientation were transferred to Escherichia coli BL21DE3, and the recombinant protein was purified after induction of expression with 0.5 mM isopropylthio-β-D-galactoside (IPTG) for 3 h at 37°C. For protein purification, bacteria were re-suspended in 50 mM Tris-HCl (pH 8), and 300 mM NaCl supplemented with EDTA-free complete protease inhibitors (Roche). The bacteria were also treated with 10 µg/ml of lysozyme for 30 min on ice. Bacteria were lysed by sonication (7 pulses of 15 sec each), and the insoluble recombinant TcPCNA (rTcPCNA) was washed and re-suspended in 8 M urea containing 500 mM NaCl and 20 mM sodium phosphate at a pH of 8.0. Solubilized proteins were applied to a Ni-Agarose column and, after washes in 8 M urea, 500 mM NaCl and 20 mM sodium phosphate at a pH of 6.0, purified rTcPCNA was obtained by elution with the same buffer adjusted to a pH of 4.0. Eluted rTcPCNA was dialyzed against PBS and used to immunize rabbits and mice. Immunoblots were performed using sodium dodecyl sulfate-(SDS) boiled extracts of 1 × 107 cells per lane, with serum from rabbit diluted 1:200. Detection was done by ECL (Amersham Biosciences) using standard protocols.
Immunofluorescence.
Cells were washed with PBS, attached to glass slides, fixed with 2% formaldehyde in PBS for 20 min, and then permeabilized with 0.1% Triton x100 in PBS for 5 min at room temperature. Cells were incubated with affinity-purified rabbit polyclonal anti-TcPCNA antibody diluted 1:5 in PBS containing 1% BSA (PBS-1% BSA). Alternatively, cells were incubated with affinity-purified mouse polyclonal anti-TcPCNA antibody and affinity-purified rabbit polyclonal anti-TcOrc1/Cdc6 antibody10 diluted in PBS-1% BSA. In some assays, cells were also incubated with the monoclonal antibody mAbAC (culture supernatant), which recognizes an unknown flagellar component in T. cruzi,10 diluted 1:2 in PBS-1% BSA or cells were incubated with the monoclonal antibody anti-DNA (culture supernatant). This antibody was obtained from NZB/NZW mouse, which spontaneously develop a systemic autoimmune disease that closely resembles human systemic lupus. Cells were washed three times with PBS and incubated with Alexa-Fluor 488-conjugated anti-rabbit IgG and with Alexa-Fluor 555-conjugated anti-mouse IgG (Invitrogen), diluted 1:300 in PBS-1% BSA. Alternatively, to extract soluble proteins, cells were first treated with 0.1% Triton X100 in PBS for 5 min at room temperature and then fixed with 2% formaldehyde in PBS for 20 min. Slides were washed again and mounted in Vectashield (Vector) in the presence of 10 µg of 4′,6-diamidino-2-phenylindole dihydrochloride (DAPI, Invitrogen) per ml. Images were acquired with a LaserScan LSM 510 META confocal system using a 100x/1.4 Plan-Apochromatic lens of an Axiovert 200 m microscope. In Figures 2, S1 and S2 images were processed by blind deconvolution using Autoquant x2.1.
EdU incorporation and immunofluorescence.
Cells were incubated with 100 µM of EdU (Click-iT Edu Image Kit, Invitrogen) for 2 h at 28°C. Cells were then washed with PBS, fixed with 2% formaldehyde in PBS for 20 min, and then permeabilized with 0.1% Triton X100 in PBS for 5 min at room temperature. Cells were washed three times with PBS-3% BSA and then incubated with Click-iT reaction cocktail for 30 min at room temperature. Cells were then washed twice with PBS-3% BSA and then incubated with with affinity-purified mouse polyclonal anti-TcPCNA antibody diluted 1:10 in PBS-1% BSA. Cells were washed three times with PBS and incubated with Alexa-Fluor 555-conjugated anti-mouse IgG. Slides were washed again and mounted in Vectashield (Vector). Images were acquired with a LaserScan LSM 510 META confocal system using a 100x/1.4 Plan-Apochromatic lens of an Axiovert 200 m microscope and were processed by blind deconvolution using Autoquant x2.1.
Electron microscopy analysis.
For routine preparations, cells were fixed in 2.5% glutaraldehyde diluted in 0.1 M cacodylate buffer pH 7.2. Cells were postfixed in 1% OsO4, 0.8% KFe(CN)6 and 5 mM CaCl2 in the same buffer. Then, cells were washed, dehydrated in acetone and embedded in Epon. Ultrathin sections were stained with uranyl acetate and lead citrate before observation. For immunocytochemistry analysis, cells were fixed in 0.3% glutaraldehyde, 4% formaldehyde and 1% picric acid diluted in 0.1 M cacodylate buffer at pH 7.2 and then dehydrated at −20°C using a graded acetone series (30%, 50%, 70%, 90% and 2x 100%). The material was progressively infiltrated with Unicryl at lower temperatures and the resin polymerization was carried out in BEEM capsules at −20°C for 5 days under ultraviolet light. Ultrathin sections were obtained in a Leica ultramicrotome (Reichert Ultracuts) and the grids containing the sections were incubated with 50 mM NH4Cl for 30 min. Later, the grids were incubated with blocking solution (3% BSA, 0.5% teleostean gelatin, 0.02% Tween 20 diluted in PBS, pH 8.0) for 30 min and then with the goat serum diluted in blocking solution (1:250). Grids were incubated for 1 h with antimodified histone antibodies diluted in blocking solution. Anti-TcOrc1/Cdc6 was diluted 1:10, while anti-TcPCNA was used undiluted. Then, ultrathin sections were incubated with gold-labeled goat antirabbit antibodies diluted 1:250, for 45 min, washed in blocking solution, and stained with uranyl acetate and lead citrate for further observation in a Zeiss 900 transmission electron microscope. In control assays, sections were not incubated with the primary antibody.
Fractionations of cellular proteins.
Cells at zero or twenty hours after HU release were treated with an extraction buffer [0.1% Triton X-100, 10 mM Tris-HCl at a pH of 7.4, 100 mM NaCl, 300 mM sucrose, 3 mM MgCl2, 50 mM NaF, 1 mM Na3VO4, 0.5 mM PMSF and EDTA-free complete protease inhibitor cocktail (Roche)] for 10 min at 4°C under agitation. Samples were pelleted and supernatants were saved as a soluble fraction (soluble fraction 1). Cells were treated with the extraction buffer again and the second round of supernatants were saved again as a soluble fraction (soluble fraction 2). Pellets were then treated with DNAse (5 units for 106 cells) (Fermentas) for 30 min at room temperature. Samples were pelleted and the supernatants were saved as a DNAse-released fraction (DNAse released 1). Cells were treated with DNAse again, and the second round of supernatants were saved again as a DNAse-released fraction (DNAse released 2). Samples were resolved by SDS-PAGE and analyzed by western blot using anti-anti-hsp70 (1:2,000),28 anti-histone H4 (1:100),29 and anti-TcPCNA antibodies as described above.
Acknowledgements
We are grateful to Alexsander Seixas de Souza for helping with confocal analysis and for helpful discussions. This work was supported with a grant from FAPESP and FAPERJ. P.D.M.G. and S.G.C. are fellows from FAPESP.
Supplementary Material
References
- 1.Bell SP, Dutta A. DNA replication in eukaryotic cells. Annu Rev Biochem. 2002;71:333–374. doi: 10.1146/annurev.biochem.71.110601.135425. [DOI] [PubMed] [Google Scholar]
- 2.Stillman B. Origin recognition and the chromosome cycle. FEBS Lett. 2005;579:877–884. doi: 10.1016/j.febslet.2004.12.011. [DOI] [PubMed] [Google Scholar]
- 3.Krishna TS, Kong XP, Gary S, Burgers PM, Kuriyan J. Crystal structure of the eukaryotic DNA polymerase processivity factor PCNA. Cell. 1994;79:1233–1243. doi: 10.1016/0092-8674(94)90014-0. [DOI] [PubMed] [Google Scholar]
- 4.Waga S, Bauer G, Stillman B. Reconstitution of complete SV40 DNA replication with purified replication factors. J Biol Chem. 1994;269:10923–10934. [PubMed] [Google Scholar]
- 5.Jonsson ZO, Hindges R, Hubscher U. Regulation of DNA replication and repair proteins through interaction with the front side of proliferating cell nuclear antigen. EMBO J. 1998;17:2412–2425. doi: 10.1093/emboj/17.8.2412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tom S, Henricksen LA, Bambara RA. Mechanism whereby proliferating cell nuclear antigen stimulates flap endonuclease 1. J Biol Chem. 2000;275:10498–10505. doi: 10.1074/jbc.275.14.10498. [DOI] [PubMed] [Google Scholar]
- 7.Shen B, Singh P, Liu R, Qiu J, Zheng L, Finger LD. Multiple but dissectible functions of FEN-1 nucleases in nucleic acid processing, genome stability and diseases. Bioessays. 2005;27:717–729. doi: 10.1002/bies.20255. [DOI] [PubMed] [Google Scholar]
- 8.Nishino T, Ishino Y, Morikawa K. Structure-specific DNA nucleases: Structural basis for 3D-scissors. Curr Opin Struct Biol. 2006;16:60–67. doi: 10.1016/j.sbi.2006.01.009. [DOI] [PubMed] [Google Scholar]
- 9.Elias MC, Faria M, Mortara RA, Motta MC, de Souza W, Thiry M, et al. Chromosome localization changes in the Trypanosoma cruzi nucleus. Eukaryot Cell. 2002;1:944–953. doi: 10.1128/EC.1.6.944-953.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Godoy PDM, Nogueira-Junior LA, Paes LS, Cornejo A, Martins RM, Silber AM, et al. Trypanosome prereplication machinery contais a single funcional orc1/cdc6 protein, which is typical of archaea. Eukaryot Cell. 2009;8:1592–1603. doi: 10.1128/EC.00161-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Celis JE, Celis A. Cell cycle-dependent variations in the distribution of the nuclear protein cyclin proliferating cell nuclear antigen in cultured cells: Subdivision of S phase. Proc Natl Acad Sci USA. 1985;82:3262–3266. doi: 10.1073/pnas.82.10.3262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bravo R, Macdonald-Bravo H. Existence of two populations of cyclin/proliferating cell nuclear antigen during the cell cycle: association with DNA replication sites. J Cell Biol. 1987;105:1549–1554. doi: 10.1083/jcb.105.4.1549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Leonhardt H, Rahn HP, Weinzierl P, Sporbert A, Cremer T, Zink D, et al. Dynamics of DNA replication factories in living cells. J Cell Biol. 2000;149:271–280. doi: 10.1083/jcb.149.2.271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kumar D, Minocha N, Rajanala K, Saha S. The distribution pattern of prolferating cell nuclear antigen in the nuclei of Leishmania donovani. Microbiol. 2009;155:3748–3757. doi: 10.1099/mic.0.033217-0. [DOI] [PubMed] [Google Scholar]
- 15.Elias MC, Marques-Porto R, Freymuller E, Schenkman S. Transcription rate modulation through the Trypanosoma cruzi life cycle occurs in parallel with changes in nuclear organisation. Mol Biochem Parasitol. 2001;112:79–90. doi: 10.1016/s0166-6851(00)00349-2. [DOI] [PubMed] [Google Scholar]
- 16.Elias MC, da Cunha JP, de Faria FP, Mortara RA, Freymuller E, Schenkman S. Morphological events during the Trypanosoma cruzi cell cycle. Protist. 2007;158:147–157. doi: 10.1016/j.protis.2006.10.002. [DOI] [PubMed] [Google Scholar]
- 17.Galanti N, Dvorak JA, Grenet J, McDaniel JP. Hydroxyurea-induced synchrony of DNA replication in the Kinetoplstida. Exp Cell Res. 1994;214:225–230. doi: 10.1006/excr.1994.1252. [DOI] [PubMed] [Google Scholar]
- 18.Nardelli SC, da Cunha JP, Motta MC, Schenkman S. Distinct acetylation of Trypanosoma cruzi histone H4 during cell cycle, parasite differentiation and after DNA damage. Chromosoma. 2009;118:487–499. doi: 10.1007/s00412-009-0213-9. [DOI] [PubMed] [Google Scholar]
- 19.Majka J, Burgers PM. The PCNA-RFC families of DNA clamps and clamp loaders. Prog Nucleic Acid Res Mol Biol. 2004;78:227–260. doi: 10.1016/S0079-6603(04)78006-X. [DOI] [PubMed] [Google Scholar]
- 20.Sadoni N, Langer S, Fauth C, Bernardi G, Cremer T, Turner BM, et al. Nuclear organization of mammalian genomes: Polar hromosome territories build up functionally distinct higher order compartments. J Cell Biol. 1999;146:1211–1226. doi: 10.1083/jcb.146.6.1211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dossin FM, Schenkman S. Actively transcribing RNA polymerase II concentrates on spliced leader genes in the nucleus of Trypanosoma cruzi. Eukaryot Cell. 2005;4:960–970. doi: 10.1128/EC.4.5.960-970.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sasaki T, Gilbert DM. The many faces of the origin recognition complex. Curr Opin Cell Biol. 2007;19:337–343. doi: 10.1016/j.ceb.2007.04.007. [DOI] [PubMed] [Google Scholar]
- 23.Hemerly AS, Prasanth SG, Siddiqui K, Stillman B. Orc1 controls centriole and centrosome copy number in human cells. Science. 2009;323:789–793. doi: 10.1126/science.1166745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Maga G, Hubscher U. Proliferating cell nuclear antigen (PCNA): A dancer with many partners. J Cell Sci. 2003;116:3051–3060. doi: 10.1242/jcs.00653. [DOI] [PubMed] [Google Scholar]
- 25.Warbrick E. The puzzle of PCNA's many partners. Bioessays. 2000;22:997–1006. doi: 10.1002/1521-1878(200011)22:11<997::AID-BIES6>3.0.CO;2-#. [DOI] [PubMed] [Google Scholar]
- 26.Prosperi E. The fellowship of the rings: Distinct pools of proliferating cell nuclear antigen trimer at work. FASEB J. 2006;20:833–837. doi: 10.1096/fj.05-5469hyp. [DOI] [PubMed] [Google Scholar]
- 27.Camargo EP. Growth and differentiation in Trypanosoma cruzi. I. Origin of Metacyclic Trpanosomes in Liquid Media. Rev Inst Med Trop Sao Paulo. 1964;12:93–100. [PubMed] [Google Scholar]
- 28.Bangs JD, Uyetake L, Brickman MJ, Balber AE, Boothroyd JC. Molecular cloning and cellular localization of a BiP homologue in Trypanosoma brucei: Divergent ER retention signals in a lower eukaryote. J Cell Sci. 1993;105:1101–1113. doi: 10.1242/jcs.105.4.1101. [DOI] [PubMed] [Google Scholar]
- 29.da Cunha JP, Nakayasu ES, de Almeida I, Schenkman S. Post-translational modifications of Trypanosoma cruzi histone H4. Mol Biochem Parasitol. 2006;150:268–277. doi: 10.1016/j.molbiopara.2006.08.012. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.