Abstract
The Saccharomyces cerevisiae CDC9 gene encodes a DNA ligase protein that is targeted to both the nucleus and the mitochondria. While nuclear Cdc9p is known to play an essential role in nuclear DNA replication and repair, its role in mitochondrial DNA dynamics has not been defined. It is also unclear whether additional DNA ligase proteins are present in yeast mitochondria. To address these issues, mitochondrial DNA ligase function in S.cerevisiae was analyzed. Biochemical analysis of mitochondrial protein extracts supported the conclusion that Cdc9p was the sole DNA ligase protein present in this organelle. Inactivation of mitochondrial Cdc9p function led to a rapid decline in cellular mitochondrial DNA content in both dividing and stationary yeast cultures. In contrast, there was no apparent defect in mitochondrial DNA dynamics in a yeast strain deficient in Dnl4p (Δdnl4). The Escherichia coli EcoRI endonuclease was targeted to yeast mitochondria. Transient expression of this recombinant EcoRI endonuclease led to the formation of mitochondrial DNA double-strand breaks. While wild-type and Δdnl4 yeast were able to rapidly recover from this mitochondrial DNA damage, clones deficient in mitochondrial Cdc9p were not. These results support the conclusion that yeast rely upon a single DNA ligase, Cdc9p, to carry out mitochondrial DNA replication and recovery from both spontaneous and induced mitochondrial DNA damage.
INTRODUCTION
The yeast Saccharomyces cerevisiae relies on two known DNA ligase proteins, Cdc9p and Dnl4p, to carry out nuclear DNA replication, repair and recombination. Cdc9p, the yeast homolog of mammalian DNA ligase I, is an essential protein that is responsible for joining Okazaki fragments during yeast lagging-strand DNA synthesis (1,2). A role for CDC9 in repair of nuclear DNA has also been demonstrated. Clones lacking a functional CDC9 gene are hypersensitive to the DNA-damaging agent methyl methane sulfonate (3,4). In addition, cell-free extracts prepared from cdc9 temperature-sensitive mutants lack both nucleotide excision repair and base excision repair activity when preincubated at the non-permissive temperature (5). Consistent with a role in DNA repair, CDC9 gene expression is induced following exposure to DNA-damaging agents such as UV irradiation, methyl methane sulfonate and γ-irradiation (6,7).
The other known yeast DNA ligase gene, DNL4, is a homolog of mammalian DNA ligase IV (8). Studies reveal that this gene participates in non-homologous end-joining of yeast nuclear DNA (9–11). These studies further demonstrated that, in contrast to strains deficient in Cdc9p, Δdnl4 mutants do not display DNA replication or recombination defects. DNL4 is not essential for yeast viability.
While nuclear DNA ligase function has become fairly well understood, mitochondrial DNA ligase function is less well understood. Recently, Willer and colleagues demonstrated that Cdc9p localizes to the mitochondria as well as to the nucleus and is required for mitochondrial function (12). The CDC9 gene was found to encode two distinct forms of Cdc9p. Translation initiation from an ‘upstream’ AUG codon gives rise to a mitochondrial form of Cdc9p (M-Cdc9p) while initiation from a ‘downstream’ codon results in a nuclear Cdc9p (N-Cdc9p). M-Cdc9p contains at its N-terminus a targeting sequence that directs the protein into the mitochondria. Interestingly, mammalian cells rely upon a similar mechanism to target DNA ligase III protein to the mitochondria (13).
However, whereas mitochondrial DNA ligase III has recently been shown to play an essential role in mammalian mitochondrial DNA repair and replication (14), a similar function for M-Cdc9p has yet to be formally established. The experiments of Willer and colleagues definitively demonstrated mitochondrial localization of Cdc9p and showed that strains lacking M-Cdc9p fail to grow on the non-fermentable carbon source glycerol (12). However, their experiments did not address whether M-Cdc9p was required for replication or repair of the mitochondrial genome, as one would predict if M-Cdc9p functions as a mitochondrial DNA ligase, or conversely, was required for some other area of mitochondrial metabolism necessary for growth on glycerol. Additionally, it is not known if Dnl4p or other unidentified DNA ligases also function in the yeast mitochondria.
The results detailed below indicate that Cdc9p is the major, if not the sole, DNA ligase in S.cerevisiae mitochondria. Furthermore, we provide evidence that Cdc9p is involved in mitochondrial replication and repair.
MATERIALS AND METHODS
Yeast strains
2780-49B (MATα cdc9-1, his7, leu2, can1, ura3, hom3), referred to hereafter as cdc9-1ts (a generous gift from Barbara Garvik and Lee Hartwell, Fred Hutchinson Cancer Research Center, Seattle, WA). 2780-49BWT (MATα his7, leu2, can1, ura3, hom3), derived from cdc9-1ts by replacing (15) the cdc9-1 allele with a wild-type allele derived from strain FF18734 (see below). PRSY003,1 (MATα leu2-3, trp1-289, ura3-52, his7, lys1-1, dnl4::KANMX4), referred to hereafter as Δdnl4. FF18734 (MATα leu2-3, trp1-289, ura3-52, his7-2, lys1-1), the wild-type strain from which Δdnl4 was derived (9). Both PRSY003,1 and FF18734 were generous gifts from Tomas Lindahl (Imperial Cancer Research Fund, Clare Hall Laboratories, South Mimms, UK).
Plasmid constructs
Mitochondrial targeted GFP construct. The first 141 bp of the CDC9 gene encode an apparent mitochondrial targeting sequence (12). This DNA was amplified by PCR and cloned into the multiple cloning site of the pYES2 vector (downstream of the GAL1 promoter) using the primers 5′-AAGCTTATGCGCAGATTACTGACCGGT-3′ and 5′-GAATTCGGAAGTGAAGAATCTAGCCAAAG-3′. The coding region of a yeast codon-optimized green fluorescent protein (GFP) gene (16; a generous gift from Brendan Cormack, Johns Hopkins University) was amplified by PCR using the primers 5′-TATTAAGAATTCTAAAGGTG-3′ and 5′-CTCGAGCTGCAGTTATT-3′. The GFP gene was then cloned into the aforementioned plasmid downstream of and in-frame with the CDC9 mitochondrial targeting coding sequence to yield plasmid pYES2-MGFP.
Mitochondrial targeted CDC9–EcoRI construct. The CDC9 mitochondrial targeting sequence was amplified by PCR using the primers 5′-GGTACCATGCGCAGATTACTGACC-3′ and 5′-GAGCTCGTGAAGAATCTAGCCAAAG-3′. The mitochondrial targeting sequence was cloned into the pYES2 vector downstream of the GAL1 promoter. The coding region of the Escherichia coli EcoRI gene (17; a generous gift from Jasper Rine, University of California, Berkeley) was PCR amplified using the primers 5′-GAGCTCTAATAAAAAACAGTCAAATAGG-3′ and 5′-CTCGAGTCACTTAGATGTAAGCTGTTC-3′. The resulting EcoRI gene was cloned downstream of and in-frame with the CDC9 mitochondrial targeting coding sequence to yield the pYES2-MiRI construct.
pYES2-CDC9 and pYES2-N-CDC9 constructs. pYES2-CDC9 was created by PCR amplification of the CDC9 gene (including ∼700 bp of sequence upstream of the coding region) from total genomic DNA and cloning it into the pYES2 plasmid using primers 5′-GGTACCCGATATGCCTCCAGTATT-3′ and 5′-CTCGAGGCATATCTAATTTTGCATGTGGGA-3′. pYES2-N-CDC9 was constructed by amplifying the CDC9 gene (as above) with primers that introduced a mutation and novel restriction site at the first ATG of CDC9. The primers used to introduce the mutation were 5′-TTCATCAATTACTCGAGGAGATTACTG-3′ and 5′-CAGTAATCTCCTCGAGTAATTGATGAA-3′. The mutated CDC9 gene was cloned into pYES2. Expression of the CDC9 and N-CDC9 genes was not controlled by the GAL1 promoter.
Yeast transformation
The lithium acetate method (18) was used to transform yeast.
Growth curves
Yeast cultures were grown to mid-logarithmic phase in glucose medium at 28°C. They were sedimented by centrifugation and resuspended in medium containing glucose or glycerol prewarmed to 37°C. The cultures were kept at 37°C for the duration of the experiment. Growth was monitored by measuring the optical density at 600 nm every hour.
Growth curve of CDC9–EcoRI transformed yeast
Yeast cultures were grown to mid-logarithmic phase in glucose medium at 28°C, sedimented by centrifugation and resuspended in galactose medium. Galactose yeast cultures were incubated at 37°C for 0 or 30 min for the strain 2780-49BWT and cdc9-1ts yeast or at 28°C for 0 min or 1 h for Δdnl4 yeast. Subsequently the yeast were sedimented by centrifugation and resuspended in glycerol medium at 28°C. The yeast were kept in glycerol medium at 28°C for the remainder of the experiment. Growth was monitored as above.
Protein and DNA preparations
Total genomic DNA preparations were performed by the small-scale glass bead purification method (19). Nuclear and mitochondrial protein extracts were prepared as described (20,21). Extracts were purified on Percoll gradients and stored at –80°C.
Ligase assays
DNA ligase assays were performed on yeast mitochondrial protein extracts using a nicked DNA:DNA duplex or a nicked DNA:RNA duplex substrate (see below). An aliquot of 5 µg of mitochondrial protein extract was incubated at 25 or 37°C for 5 min subsequent to the reaction. The aliquot of extract was then allowed to react with the substrate for 5–10 min at room temperature before addition of formamide-containing electrophoresis buffer and treatment at 95°C for 5 min. Products formed were separated by electrophoresis on a 22.5% polyacrylamide/7 M urea gel. As a positive control, substrate was incubated with 1 U T4 DNA ligase (New England Biolabs, Beverly, MA).
Nicked DNA:DNA duplex. The substrate was made as previously described (22). Briefly, a 15mer oligodeoxynucleotide (5′-CCCAGTCACGACGTT-3′) and a 32P-labeled 17mer oligonucleotide (5′-GTAAAACGACGGCCAGT-3′) were annealed onto the M13mp18 single-stranded DNA phage vector (New England Biolabs). These hybridize to immediately adjacent regions to form a double-stranded DNA:DNA template resembling a nick. Aliquots of 3 × 104 c.p.m. substrate per reaction were used.
Nicked DNA:RNA duplex. The substrate was made as previously described (23,24). Briefly, 16mer oligomers of deoxythymidylic acid (dT16) (Pharmacia Biotech, Piscataway, NJ) were pretreated with alkaline phosphatase and radioactively labeled with 32P. The ligase substrate was formed by annealing 5′-32P-labeled dT16 to polyriboadenylic acid [poly(rA)] (Pharmacia Biotech). These hybridize to immediately adjacent regions to form a double-stranded DNA:RNA template resembling a nick. Aliquots of 3 × 103 c.p.m. substrate per reaction were used.
Southern blot of spontaneously damaged DNA in logarithmically growing yeast
Logarithmically growing yeast were incubated in glucose medium at 37°C for 0, 1, 2 or 4 h and total cellular DNA isolated. Southern blots were performed on 1 µg genomic DNA that had been cut with SacII and resolved on 0.9% agarose. Southern blot hybridization was performed using a DNA probe obtained by PCR amplification of the mitochondrial genome from position 13986 to 16724. This probe (labeled using the RadPrime DNA labeling system; Gibco BRL, Gathersburg, MD) hybridizes to a 12.5 kb mitochondrial DNA SacII fragment.
Southern blot of spontaneously damaged DNA in stationary phase yeast
Yeast were grown in glucose medium for ∼3 days (optical density at 600 nm ∼5.0) and then shifted to the non-permissive temperature for 0, 1, 2 or 4 h before total cellular DNA was isolated. A Southern blot hybridization was performed on the DNA as above.
Northern blot of CDC9–EcoRI carrying yeast
Cultures of yeast were grown to mid-logarithmic phase in glucose medium then incubated in galactose-containing medium at 28°C to induce MiR1 gene expression. Total cellular RNA was isolated by a previously described method (19). Northern blots were performed on 20 µg total cellular RNA using an [α-32P]dATP-labeled CDC9–EcoRI fusion gene-specific probe.
Induction of the CDC9–EcoRI endonuclease
A culture of yeast harboring the pYES2-MiRI construct was grown to mid-logarithmic phase in glucose medium then incubated for 4 h in galactose-containing medium at 28°C to induce MiR1 gene expression. Yeast were then sedimented by centrifugation, resuspended in glycerol medium and grown at 28°C for 4 h. Total cellular DNA was isolated from the yeast before galactose incubation, after galactose incubation and after glycerol incubation. Southern blots were performed on 1 µg newly prepared genomic DNA that had been digested with SacII and electrophoretically resolved on a 0.9% agarose gel.
Confocal microscopy of yeast
Yeast cultures were grown to mid-logarithmic phase in glucose medium. Yeast transformed with pYES2-MGFP were sedimented by centrifugation and resuspended in galactose medium at an optical density at 600 nm of ∼0.3. This culture was then allowed to grow for 4–6 h at 28°C. A portion (2.5 µl) of this culture was placed on a glass slide and immobilized with 4 µl of 1% low melting point agarose (Gibco BRL) that had been prewarmed to 45°C. A glass coverslip was immediately placed on top of the cell/agar mixture, sealed with nail polish and examined. To stain the mitochondria, 2.5 µl of this same yeast culture was treated for 30 min at room temperature with a 200 nM solution of MitoTracker Red in 1% DMSO, 99% glucose medium (CM-H2XRos; Molecular Probes, Eugene, OR) in the dark (25). The mixture was immobilized on a glass coverslip and immediately examined. A Bio-Rad MRC-1024 confocal system was used to examine the yeast. A 100× oil immersion objective (Olympus) was used and the samples were excited at 488 nm, with emission analyzed at 522 and 605 nm. Digitized fluorescent images were analyzed with Adobe Photoshop.
RESULTS
In vitro analysis of S.cerevisiae mitochondrial DNA ligase function
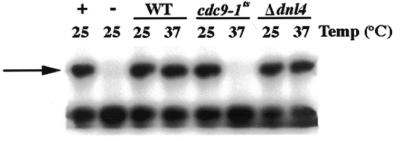
Percoll-purified mitochondrial protein extracts were prepared from yeast strains harboring mutations in the CDC9 and DNL4 genes. Since the CDC9 gene is essential for yeast viability, a strain harboring a temperature-sensitive allele of this gene (referred to as cdc9-1ts; see Materials and Methods) was used. The DNL4 gene is not essential and thus a strain harboring a null allele (referred to as Δdnl4; see Materials and Methods) of this gene was used. In addition, a mitochondrial protein extract was prepared from strain 2780-49BWT, an isogenic derivative of cdc9-1ts that harbors a wild-type allele of CDC9 (see Materials and Methods). Two different DNA ligase assays were performed on the purified mitochondrial protein extracts. The first assay measured the ability of mitochondrial protein extracts to ligate a plasmid engineered to mimic a nick in duplex DNA (see DNA:DNA duplex in Materials and Methods). This ligase substrate was prepared by annealing two synthetic oligodeoxynucleotides (one of which was a radiolabeled 17mer) side-by-side onto M13 single-stranded virion DNA (22). Ligation of the two oligodeoxynucleotides resulted in a 32mer product that was detected by electrophoresis on a denaturing acrylamide gel. As Figure 1 indicates, incubation of the nicked DNA duplex substrate with mitochondrial protein extracts from strain 2780-49BWT and Δdnl4 yeast yielded product (indicated by the arrow) regardless of the preincubation temperature. Product was also formed using a mitochondrial protein extract prepared from strain FF18734, the wild-type parental strain from which Δdnl4 was derived (data not shown). In contrast, mitochondrial protein extracts prepared from the strain harboring the cdc9-1ts allele were capable of producing product when preincubated at 25°C, but not when preincubated at 37°C (Fig. 1). It has been previously shown (22) that Cdc9p is functional at 25°C, but is irreversibly inactivated by incubation at 37°C. This indicates that Cdc9p is present in S.cerevisiae mitochondria, confirming a previous report (12). More significantly, the absence of residual DNA ligase activity in heat-treated mitochondrial protein extracts from cdc9-1ts yeast suggests that Cdc9p is the only mitochondrial DNA ligase capable of ligating a nicked DNA:DNA duplex.
Figure 1.
Yeast mitochondrial protein extracts ligate nicked DNA:DNA substrates. Mitochondrial protein extracts were prepared from strain 2780-49BWT, cdc9-1ts and Δdnl4 yeast and their ability to seal a nick in a DNA:DNA duplex substrate was assayed. Reactions were carried out as described in Materials and Methods, with extracts preincubated at the temperatures indicated. The lower band indicates unligated substrate DNA, while the upper band (arrow) indicates the ligated DNA product. The positive control (+) is substrate plus T4 DNA ligase; the negative control (–) is substrate alone.
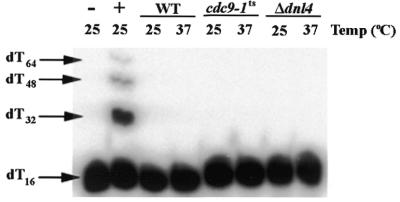
It has been shown that recombinant yeast Dnl4p is incapable of sealing nicks present on DNA:DNA duplex molecules, although it is capable of ligating oligodeoxynucleotides annealed on a complementary oligoribonucleotide molecule (9). Thus, the results shown in Figure 1 provide no information about whether yeast mitochondria contain Dnl4p. To test this hypothesis, mitochondrial protein extracts were tested for their ability to ligate 16mer oligodeoxythymidylic acid monomers (dT16) annealed side-by-side onto a poly(rA) backbone (see DNA:RNA duplex in Materials and Methods). Since Cdc9p is incapable of ligating this substrate (9,23), product formation under these conditions would reveal the presence of Dnl4p. As Figure 2 reveals, while T4 DNA ligase efficiently multimerized dT16 (arrows indicate ligation products in Fig. 2), no product was formed following incubation of this substrate with mitochondrial extracts from any of the strains. In contrast, Dnl4p ligase activity was detected in a nuclear protein extract prepared from the wild-type yeast strain FF18734 (data not shown). This suggests that Dnl4p is not present in yeast mitochondria, although we cannot rule out the presence of a very small amount of Dnl4p in this compartment.
Figure 2.
Yeast mitochondrial protein extracts do not ligate nicked DNA:RNA substrates. DNA ligase reactions were carried out on mitochondrial protein extracts in a manner similar to Figure 1, however, the substrate was a DNA:RNA duplex. The lower band (dT16) indicates unligated substrate, while the upper bands (dT32, dT48 and dT64) indicate products. The positive control (+) is substrate plus T4 DNA ligase; the negative control (–) is substrate alone.
Cdc9p is required for growth in glycerol, while Dnl4p is not
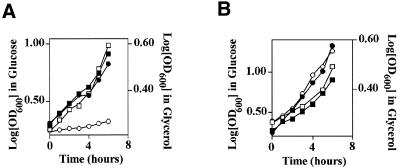
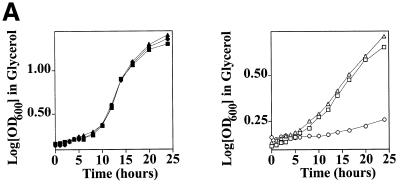
Translation of mitochondrial Cdc9p begins from the first initiation codon while transcription from a second, downstream initiation codon produces nuclear Cdc9p. Inactivation of the upstream codon results in a CDC9 gene that produces only nuclear Cdc9p (12). Therefore, we introduced plasmids encoding either the wild-type CDC9 gene (pYES2-CDC9) or a mutated CDC9 gene that had an altered upstream initiation codon (pYES2-N-CDC9) into cdc9-1ts yeast. As Figure 3A indicates, the cdc9-1ts yeast strain harboring the pYES2-CDC9 gene grew at the non-permissive temperature in both glucose and glycerol media (the parental cdc9-1ts strain could not grow in either medium at this temperature). In contrast, the cdc9-1ts yeast strain harboring the pYES2-N-CDC9 plasmid grew in glucose but not in glycerol medium. Δdnl4 yeast displayed no apparent defect in the ability to grow in glycerol medium (Fig. 3B). These data provide independent confirmation that Cdc9p is essential for mitochondrial function and further indicate that Dnl4p is not.
Figure 3.
Growth of DNA ligase mutant yeast strains in glucose and glycerol. Yeast strains were grown to mid-log phase in glucose, sedimented by centrifugation and resuspended in either glucose or glycerol medium at 37°C. The growth rate was monitored by determining the optical density at 600 nm. Filled symbols depict growth in glucose, while open symbols depict growth in glycerol. (A) cdc9-1ts pYES2-CDC9 strain (squares); cdc9-1ts pYES2-N-CDC9 strain (circles). (B) 2780-49BWT strain (squares); Δdnl4 strain (circles).
In vivo characterization of mitochondrial DNA ligase activity
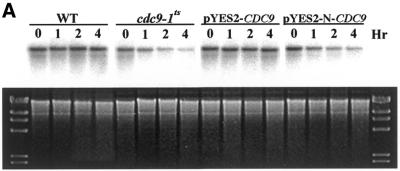
The results shown in Figure 3 indicate that mitochondrial Cdc9p has an essential role in one or more mitochondrial functions, but do not address whether mitochondrial Cdc9p is specifically required for mitochondrial DNA stability. To gain more insight into the fate of the mitochondrial genome in yeast lacking functional Cdc9p, 2780-49BWT, cdc9-1ts and cdc9-1ts strains harboring either the pYES2-CDC9 or the pYES2-N-CDC9 plasmid were grown in glucose medium to mid-log phase at the permissive temperature and then shifted to the non-permissive temperature. Total cellular DNA was then isolated from these cultures immediately and after 1, 2 and 4 h. This DNA was cut with SacII, resolved by agarose gel electrophoresis, transferred to nitrocellulose and hybridized with a mitochondrial DNA probe. Figure 4 shows the results of this Southern blot analysis. Both strain 2780-49BWT yeast and the cdc9-1ts strain harboring the pYES2-CDC9 plasmid maintained a constant mitochondrial DNA copy number throughout the time course (quantitation of the Southern blot revealed that mitochondrial DNA content changed by <15% during the time course). In contrast, the mitochondrial DNA content of the cdc9-1ts strain and the cdc9-1ts strain harboring the pYES2-N-CDC9 plasmid was reduced by 62 and 52%, respectively. As the image of the ethidium bromide stained agarose gel indicates, this is not the result of unequal loading of total cellular DNA. Analogous experiments performed on Δdnl4 strains revealed that their mitochondrial DNA content was indistinguishable from that seen in strain 2780-49BWT yeast, whether they were grown in glucose or glycerol (data not shown).
Figure 4.
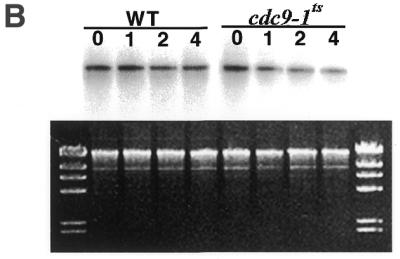
Cdc9p is required for mitochondrial DNA maintenance. Yeast strains were grown in glucose to mid-log (A) or to stationary (B) phase at the permissive temperature and subsequently shifted to the non-permissive temperature. At the times indicated genomic DNA was harvested and mitochondrial DNA was examined. Strain 2780-49BWT, cdc9-1ts yeast, cdc9-1ts yeast harboring pYES2-CDC9 and cdc9-1ts yeast harboring pYES2-N-CDC9 were analyzed. The upper panels depict Southern blot hybridizations to a mitochondrial DNA probe, the lower panels depict the corresponding ethidium bromide stained agarose gels. The molecular weight marker is HindIII-digested phage λ DNA.
These results clearly indicate that mitochondrial Cdc9p is required for maintenance of mitochondrial DNA during growth. We also examined whether Cdc9p was required for the maintenance of mitochondrial DNA in yeast that were in stationary phase. Strain 2780-49BWT and cdc9-1ts yeast were grown in glucose medium at 25°C to stationary phase (see Materials and Methods), placed at 37°C and then analyzed as above. We observed that the mitochondrial DNA content of the 2780-49BWT strain was reduced by <15% during a 4 h incubation at 37°C, while under these same conditions the mitochondrial DNA content of the cdc9-1ts strain was reduced by 54% (Fig. 4B). These data indicate that Cdc9p is required for maintenance of mitochondrial DNA during logarithmic growth and in stationary phase.
Expression of the EcoRI endonuclease in the mitochondria of yeast
It has been previously shown that inducible expression of the E.coli EcoRI endonuclease gene in yeast led to nuclear DNA damage (17,26). We therefore reasoned that it might be possible to create a CDC9–EcoRI fusion gene that would target the bacterial endonuclease to the yeast mitochondrial compartment. Such an inducible system would permit us to examine whether Cdc9p plays a role in the response of mitochondrial DNA to induced damage.
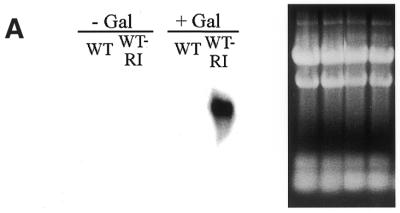
We examined the inducible system used to regulate expression of a mitochondrially targeted EcoRI endonuclease. A CDC9–EcoRI fusion gene, under the control of the GAL1 promoter, was introduced into strain 2780-49BWT yeast (see Materials and Methods). Total cellular RNA from these transformed yeast (and control untransformed strain 2780-49BWT yeast) was subjected to northern blot analysis after 4 h in galactose-containing medium. Figure 5A (right) shows an ethidium bromide stained agarose gel run under native conditions indicating that this result is not due to unequal loading of RNA. The left panel of Figure 5A shows that there is no expression of the CDC9–EcoRI fusion gene prior to galactose induction (–Gal) but that the CDC9–EcoRI fusion gene is expressed after induction in galactose medium (+Gal). As expected, the untransformed strain 2780-49BWT yeast did not express the CDC9–EcoRI transcript before or after galactose induction. Similar results were obtained when the CDC9–EcoRI construct was introduced into both Δdnl4 and cdc9-1ts yeast (data not shown).
Figure 5.
Expression of the mitochondrially targeted EcoRI endonuclease is controlled by galactose induction. (A) Transcription of the CDC9–EcoRI plasmid is tightly controlled by the GAL1 promoter. (Left) Northern blot analysis of untransformed strain 2780-49BWT yeast and strain 2780-49BWT yeast harboring the CDC9–EcoRI construct before and after galactose induction. (Right) Ethidium bromide stained agarose gel of the RNA used in the northern blot. (B) A CDC9–GFP fusion protein is targeted to yeast mitochondria and detected by confocal fluorescence microscopy. (Top left) Yeast expressing the CDC9–GFP fusion; (top middle) yeast stained with the mitochondria-specific dye MitoTracker Red; (top right) yeast expressing the CDC9–GFP fusion protein stained with MitoTracker Red. The bottom set of panels depict GFP green and red fluorescence and the MitoTracker Red red and green fluorescence.
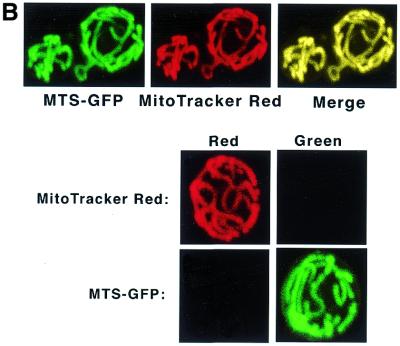
To confirm that the mitochondrial targeting sequence of the CDC9 gene would deliver a heterologous protein to the yeast mitochondrion, a fusion gene under the control of the GAL1 promoter was constructed using the CDC9 mitochondrial targeting sequence fused to the 5′-end of the green fluorescent protein (GFP) gene (see Materials and Methods). This gene was introduced into strain 2780-49BWT yeast. Figure 5B (top left) depicts a confocal fluorescence microscopic image of yeast expressing the Cdc9–GFP protein. The top middle panel shows an image of yeast stained with the mitochondria-specific fluorochrome MitoTracker Red. The top right-hand panel of Figure 5B shows a confocal fluorescence image of yeast expressing the Cdc9–GFP fusion protein that had also been stained with MitoTracker Red. It is clear that there is complete overlap between the dye and GFP expression, indicating that the Cdc9–GFP protein is correctly targeted to the mitochondrion. Additionally, GFP fluorescence is only detected at 522 nm and MitoTracker Red fluorescence is only detected at 605 nm (Fig. 5B, bottom). Finally, strain 2780-49BWT yeast carrying the CDC9–GFP construct that were not induced with galactose did not express Cdc9–GFP protein.
Analyses of yeast following induced mitochondrial DNA damage
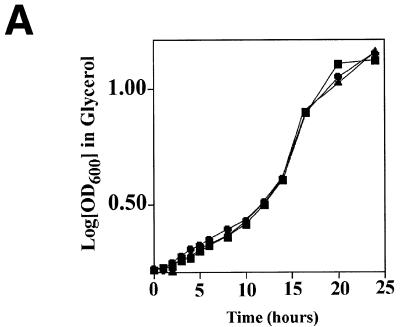
The CDC9–EcoRI fusion gene was introduced into both strain 2780-49BWT and cdc9-1ts yeast (see Materials and Methods). Both strains were incubated in galactose-containing medium at the non-permissive temperature for 30 min. Immediately following this period of induction the yeast were isolated and resuspended in glycerol-containing medium and their growth rate monitored. Transient expression of the fusion protein in strain 2780-49BWT yeast resulted in a transient growth lag of ∼2 h, from which yeast quickly regained normal growth kinetics (Fig. 6A). In contrast, transient expression of the mitochondrial targeted EcoRI protein in cdc9-1ts yeast led to a growth lag lasting ∼12 h. This result suggests that expression of EcoRI endonuclease in yeast mitochondria damages that organelle’s DNA, thereby inhibiting the yeast’s ability to utilize glycerol as a carbon source. The data presented in Figure 6A further suggest that it is possible for mitochondria to recover from this DNA damage and that this process is critically dependent upon Cdc9p.
Figure 6.
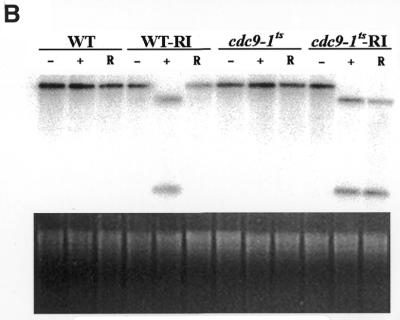
Expression of a mitochondrial-targeted EcoRI endonuclease in yeast. (A) The effect of EcoRI expression on growth in glycerol. Cultures were grown to mid-log phase in glucose. Yeast were sedimented by centrifugation, incubated for 30 min at the non-permissive temperature in galactose and their growth rate in glycerol subsequently determined. (Left) Wild-type incubated in galactose (filled squares); wild-type harboring the CDC9–EcoRI fusion gene not induced with galactose (filled triangles); wild-type harboring the CDC9–EcoRI gene induced with galactose (filled circles). (Right) cdc9-1ts incubated in galactose (open squares); cdc9-1ts harboring the CDC9–EcoRI fusion gene not induced with galactose (open triangles); cdc9-1ts harboring the CDC9–EcoRI fusion gene induced with galactose (open circles). (B) Analysis of mitochondrial DNA. Genomic DNA was isolated from yeast clones treated as described above (except that the incubation in galactose was performed at the non-permissive temperature; see text) and mitochondrial DNA analyzed. (Upper) Southern blot hybridization; (lower) ethidium bromide stained agarose gel. Wild-type yeast (WT), wild-type yeast harboring the CDC9–EcoRI fusion gene (WT-RI), cdc9-1ts yeast and cdc9-1ts yeast harboring the CDC9–EcoRI fusion gene (cdc9-1ts-RI) were examined. Three conditions were analyzed: yeast not exposed to galactose (–); yeast exposed to galactose (+); yeast exposed to galactose and permitted to recover in glycerol medium (R).
Mitochondrial targeted EcoRI creates DNA double-strand breaks in the mitochondrial genome
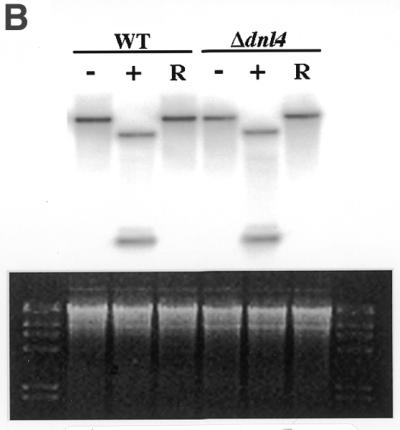
To test this hypothesis, Southern blot analysis was performed on strain 2780-49BWT and cdc9-1ts yeast before and after expression of the recombinant EcoRI endonuclease to look for the presence of mitochondrial DNA double-strand breaks. We were concerned that it might prove difficult to detect these lesions in cdc9-1ts yeast that had been incubated at the non-permissive temperature, since incubation of this strain at this temperature leads to a rapid loss of mitochondrial DNA (see Fig. 4). Since it has been shown (3) that even at the permissive temperature cdc9 yeast have a partial defect in DNA ligase function, we examined whether expression of the recombinant EcoRI endonuclease at the permissive temperature would lead to mitochondrial DNA double-strand breaks. Strain 2780-49BWT and cdc9-1ts yeast were grown in glucose medium to mid-logarithmic phase and subsequently shifted to galactose medium at the permissive temperature for 0 or 4 h. These yeast were then sedimented by centrifugation, resuspended in glycerol and incubated at the permissive temperature for 4 h. At this point genomic DNA was isolated, cut with SacII and Southern blot analysis performed using a mitochondrial DNA probe. This probe hybridizes to a 12.5 kb SacII fragment of intact mitochondrial DNA. However, if the recombinant EcoRI enzyme has cut the mitochondrial genome in vivo, the probe will hybridize to two fragments of 1.0 and 4.3 kb. Figure 6B shows that after induction of EcoRI endonuclease both the strain 2780-49BWT and cdc9-1ts mitochondrial genomes were fragmented. However, after incubation in glycerol medium at the permissive temperature, mitochondrial DNA levels were restored to normal levels in strain 2780-49BWT yeast while no recovery was detected in the cdc9-1ts yeast.
The effects of expression of mitochondrial targeted EcoRI in Δdnl4 yeast was also assessed. Interestingly, as Figure 7A indicates, Δdnl4 yeast displayed no increased sensitivity to the growth inhibitory effects of expression of EcoRI endonuclease, as compared to strain 2780-49BWT yeast. Southern blot analysis revealed that Δdnl4 yeast were fully able to restore their mitochondrial DNA content to normal levels (Fig. 7B). Thus, we can conclude that Dnl4p is not necessary for recovery following the introduction of DNA double-strand breaks within the mitochondrial genome.
Figure 7.
Expression of a mitochondrial-targeted EcoRI endonuclease in yeast. An analysis similar to that depicted in Figure 6 was performed using Δdnl4 yeast. (A) Yeast cultures: Δdnl4 incubated in galactose (squares); Δdnl4 strains harboring the CDC9–EcoRI fusion gene not incubated in galactose (triangles); Δdnl4 strains harboring the CDC9–EcoRI fusion gene incubated in galactose (circles). (B) Analysis of mitochondrial DNA as in Figure 6B.
DISCUSSION
The results presented herein clearly establish that the CDC9 gene is essential for maintenance of mitochondrial DNA in yeast. These results further indicate that the other known DNA ligase gene of yeast, DNL4, is not required for mitochondrial DNA maintenance. Biochemical analysis revealed the presence of Cdc9p in yeast mitochondrial protein extracts. However, the inability to detect residual DNA ligase activity in heat-treated mitochondrial extracts from cdc9-1ts yeast suggests that Cdc9p is the sole DNA ligase present in yeast mitochondria.
The requirement for Cdc9p for maintenance of yeast mitochondrial DNA in rapidly dividing cultures implies an involvement of this molecule in DNA replication. This view is supported by the finding that Cdc9p is essential for nuclear DNA replication and by the lack of evidence that any other mitochondrial DNA ligase molecules exist. However, as the results presented above indicate, mitochondrial Cdc9p is essential for mitochondrial stability in stationary phase cultures of yeast. Indeed, the rate of loss of mitochondrial DNA observed in stationary phase cultures is indistinguishable from that seen in cultures undergoing logarithmic growth. While it is conceivable that some mitochondrial DNA replication occurs in stationary phase yeast, it seems unlikely that mitochondrial DNA replication occurs to a similar extent in both logarithmically dividing and stationary phase yeast. Thus, we conclude that Cdc9p is also required to stabilize non-replicating yeast mitochondrial DNA.
It is known that mitochondrial DNA is subject to extremely high levels of oxidative DNA damage. This type of damage occurs in yeast that are growing logarithmically and recent data has shown that oxidative DNA damage also accumulates when yeast are in stationary phase (27,28). Therefore, the observed loss of mitochondrial DNA upon Cdc9p inactivation may be due to a defect in base excision repair. A number of findings support this hypothesis. It is known that Cdc9p is essential for base excision repair in the yeast nucleus (5). Additionally, a number of other yeast base excision repair enzymes participate in both nuclear and mitochondrial repair processes (29,30). Furthermore, it has been shown that in higher eukaryotes the same DNA ligase molecule catalyzes both nuclear and mitochondrial base excision repair (14,31). Therefore, a role for Cdc9p in base excision repair can be proposed. It is possible that the Cdc9p ligase is able to bind to DNA ends formed during base excision repair in order to protect them from immediate degradation by mitochondrial DNA endonucleases. There is a precedent for a DNA ligase having this function, as in vitro studies have suggested that human DNA ligase III is able to protect DNA ends in this manner (32). This explanation may account for the loss of mitochondrial DNA observed in CDC9-1ts yeast incubated at the non-permissive temperature.
We also observed that Cdcp9 was required for recovery following induced mitochondrial DNA double-strand breaks created by a recombinant EcoRI endonuclease. The results presented in Figures 6 and 7 clearly indicate that strain 2780-49BWT and Δdnl4 yeast were able to rapidly recover from induced mitochondrial DNA double-strand breaks, while cdc9-1ts clones were not. However, the mechanism by which this recovery occurs is not clear. One can imagine a number of possible explanations for this result. First, the recovery of strain 2780-49BWT and Δdnl4 yeast may be due to double-strand break repair. While it is not known whether DNA end-joining occurs in the yeast mitochondria, it has long been known that mitochondrial DNA homologous recombination occurs in yeast during mating (33) and that DNA biolistically introduced into this organelle can homologously recombine with the mitochondrial genome in mitotic yeast (34). Since there are 19 EcoRI restriction sites within the yeast mitochondrial genome, one might expect that if either homologous recombination or DNA end-joining were responsible for mitochondrial DNA recovery there would be transient formation of multimer products as these fragments were rejoined. However, despite repeated efforts, we have failed to detect any (data not shown). Thus, we conclude that if mitochondrial DNA double-strand break repair occurs, the repair intermediates are too short lived to be detected. Alternatively, it is possible that the Cdc9p-dependent recovery of normal levels of mitochondrial DNA occurs through compensatory re-replication of a small pool of uncut molecules. It is noteworthy that overexposed images of the experiments presented in Figures 6 and 7 reveal the existence of low levels of uncut mitochondrial DNA.
Cdc9p comprises a core catalytic domain that shows a high degree of similarity to other DNA ligase proteins as well as additional N- and C-terminal domains that presumably interact with other DNA repair and replication proteins. By altering regions of Cdc9p outside the conserved core region it may be possible to functionally separate the mitochondrial DNA repair and replication functions of Cdc9p, thereby gaining additional insight into the various roles this protein plays in yeast mitochondrial genome dynamics. Experiments of this nature are currently in progress.
Acknowledgments
ACKNOWLEDGEMENTS
We thank Drs Barbara Garvik and Lee Hartwell as well as Tomas Lindahl for kindly providing the yeast strains used herein. The GFP gene was provided by Dr Brendan Cormack. This work was supported in part by grants from the NIH (CA61906 and AG16678) and the American Heart Association (AHA MN 9951198Z). L.B. was supported by a fellowship from the American Heart Association.
References
- 1.Johnston L.H. and Nasmyth,K.A. (1978) Saccharomyces cerevisiae cell cycle mutant cdc9 is defective in DNA ligase. Nature, 274, 891–893. [DOI] [PubMed] [Google Scholar]
- 2.Johnston L.H. (1983) The cdc9 ligase joins completed replicons in baker’s yeast. Mol. Gen. Genet., 190, 315–317. [DOI] [PubMed] [Google Scholar]
- 3.Johnston L.H. (1979) The DNA repair capability of cdc9, the Saccharomyces cerevisiae mutant defective in DNA ligase. Mol. Gen. Genet., 170, 89–92. [DOI] [PubMed] [Google Scholar]
- 4.Montelone B.A., Prakash,S. and Prakash,L. (1981) Spontaneous mitotic recombination in mms8-1, an allele of the CDC9 gene of Saccharomyces cerevisiae. J. Bacteriol ., 147, 517–525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wu X., Braithwaite,E. and Wang,Z. (1999) DNA ligation during excision repair in yeast cell-free extracts is specifically catalyzed by the CDC9 gene product. Biochemistry, 38, 2628–2635. [DOI] [PubMed] [Google Scholar]
- 6.Peterson TA., Prakash,L., Prakash,S., Osley,M.A. and Reed,S.I. (1985) Regulation of CDC9, the Saccharomyces cerevisiae gene that encodes DNA ligase. Mol. Cell. Biol., 5, 226–235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Johnson A.L., Barker,D.G. and Johnston,L.H. (1986) Induction of yeast DNA ligase genes in exponential and stationary phase cultures in response to DNA damaging agents. Curr. Genet., 11, 107–112. [DOI] [PubMed] [Google Scholar]
- 8.Wei Y.F., Robins,P., Carter,K., Caldecott,K., Pappin,D.J., Yu,G.L., Wang,R.P., Shell,B.K., Nash,R.A. and Schar,P. (1995) Molecular cloning and expression of human cDNAs encoding a novel DNA ligase IV and DNA ligase III, an enzyme active in DNA repair and recombination. Mol. Cell. Biol., 15, 3206–3216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Schar P., Herrmann,G., Daly,G. and Lindahl,T. (1997) A newly identified DNA ligase of Saccharomyces cerevisiae involved in RAD52-independent repair of DNA double-strand breaks. Genes Dev., 11, 1912–1924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Teo S.H. and Jackson,S.P. (1997) Identification of Saccharomyces cerevisiae DNA ligase IV: involvement in DNA double-strand break repair. EMBO J., 16, 4788–4795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wilson T.E., Grawunder,U. and Lieber,M.R. (1997) Yeast DNA ligase IV mediates non-homologous DNA end joining. Nature, 388, 495–498. [DOI] [PubMed] [Google Scholar]
- 12.Willer M., Rainey,M., Pullen,T. and Stirling,C.J. (1999) The yeast CDC9 gene encodes both a nuclear and a mitochondrial form of DNA ligase I. Curr. Biol., 9, 1085–1094. [DOI] [PubMed] [Google Scholar]
- 13.Lakshmipathy U. and Campbell,C. (1999) The human DNA ligase III gene encodes nuclear and mitochondrial proteins. Mol. Cell. Biol., 19, 3869–3876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lakshmipathy U. and Campbell,C. (2000) Mitochondrial DNA ligase III function is independent of Xrcc1. Nucleic Acids Res., 28, 3880–3886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rothstein R. (1991) Targeting, disruption, replacement, and allele rescue: integrative DNA transformation in yeast. Methods Enzymol., 194, 281–301. [DOI] [PubMed] [Google Scholar]
- 16.Cormack B.P., Valdivia,R.H. and Falkow,S. (1996) FACS-optimized mutants of the green fluorescent protein (GFP). Gene, 173, 33–38. [DOI] [PubMed] [Google Scholar]
- 17.Barnes G. and Rine,J. (1985) Regulated expression of endonuclease EcoRI in Saccharomyces cerevisiae: nuclear entry and biological consequences. Proc. Natl Acad. Sci. USA, 82, 1354–1358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ito H., Fukunda,Y., Murata,K. and Kimura,A. (1983) Transformation of intact yeast cells treated with alkali cations. J. Bacteriol ., 153, 163–168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Adams A., Gottshling,D.E., Kaiser,C.A. and Stearns,T. (1997) Methods in Yeast Genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 20.Jazwinski S.M. (1990) Preparation of extracts from yeast. Methods Enzymol., 182, 154–175. [DOI] [PubMed] [Google Scholar]
- 21.Glick B.S. and Pon,L.A. (1995) Isolation of highly purified mitochondria from Saccharomyces cerevisiae. Methods Enzymol., 260, 213–223. [DOI] [PubMed] [Google Scholar]
- 22.Barker D.G., Johnson,A.L. and Johnston,L.H. (1985) An improved assay for DNA ligase reveals temperature-sensitive activity in cdc9 mutants of Saccharomyces cerevisiae. Mol. Gen. Genet., 200, 458–462. [DOI] [PubMed] [Google Scholar]
- 23.Arrand J.E., Willis,A.E., Goldsmith,I. and Lindahl,T. (1986) Differential substrate specificities of the two DNA ligases of mammalian cells. J. Biol. Chem., 261, 9079–9082. [PubMed] [Google Scholar]
- 24.Prigent C., Satoh,M.S., Daly,G., Barnes,D.E. and Lindahl,T. (1994) Aberrant DNA repair and DNA replication due to an inherited enzymatic defect in human DNA ligase I. Mol. Cell. Biol., 14, 310–317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Poot M., and Pierce,R.H. (1999) Detection of changes in mitochondrial function during apoptosis by simultaneous staining with multiple fluorescent dyes and correlated multiparameter flow cytometry. Cytometry, 35, 311–317. [DOI] [PubMed] [Google Scholar]
- 26.Barnes G. and Rio,D. (1997) DNA double-strand break sensitivity, DNA replication, and cell cycle arrest phenotypes of Ku-deficient Saccharomyces cerevisiae. Proc. Natl Acad. Sci. USA, 94, 867–872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Longo V.D., Liou,L.-L., Valentine,J.S. and Gralla,E.B. (1999) Mitochondrial superoxide decreases yeast survival in stationary phase. Arch. Biochem. Biophys., 365, 131–142. [DOI] [PubMed] [Google Scholar]
- 28.Srinivasan C., Liba,A. Imlay,J.A., Valentine,J.S. and Gralla,E.B. (2000) Yeast lacking superoxide dismutase(s) show elevated levels of ‘free iron’ as measured by whole cell electron paramagnetic resonance. J. Biol. Chem., 275, 29187–29192. [DOI] [PubMed] [Google Scholar]
- 29.You H.J., Swanson,R.L., Harrington,C., Corbett,A.H., Jinks-Robertson,S., Senturker,S., Wallace,S.S., Boiteux,S., Dizdaroglu,M. and Doetsch,P.W. (1999) Saccharomyces cerevisiae Ntg1p and Ntg2p: broad specificity N-glycosylases for the repair of oxidative DNA damage in the nucleus and mitochondria. Biochemistry, 38, 11298–11306. [DOI] [PubMed] [Google Scholar]
- 30.Ling F., Makishima,F., Morishima,N. and Shibata,T. (1995) A nuclear mutation defect in mitochondrial recombination in yeast. EMBO J., 14, 4090–4101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pinz K.G. and Bogenhagen,D.F. (1998) Efficient repair of abasic sites in DNA by mitochondrial enzymes. Mol. Cell. Biol., 18, 1257–1265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Calecott K.W., Aoufouchi,S., Johnson,P. and Shall,S. (1996) Srcc1 polypeptide interacts with DNA polymerase β and possibly poly(ADP-ribose) polymerase and DNA ligase III is a novel molecular ‘nick-sensor’ in vitro. Nucleic Acids Res., 24, 4387–4394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dujon B. (1981) Molecular Biology of the Yeast Saccharomyces: Life Cycle and Inheritance. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 34.Butow R.A., Henke,R.M., Moran,J.V., Belcher,S.M. and Perlman,P.S. (1996) Transformation of Saccharomyces cerevisiae mitochondria using the biolistic gun. Methods Enzymol., 264, 265–278. [DOI] [PubMed] [Google Scholar]