Abstract
In order to survive and propagate, RNA viruses must achieve a balance between the capacity for adaptation to new environmental conditions or host cells with the need to maintain an intact and replication competent genome. Several virus families in the order Nidovirales, such as the coronaviruses (CoVs) must achieve these objectives with the largest and most complex replicating RNA genomes known, up to 32 kb of positive-sense RNA. The CoVs encode sixteen nonstructural proteins (nsp 1–16) with known or predicted RNA synthesis and modification activities, and it has been proposed that they are also responsible for the evolution of large genomes. The CoVs, including murine hepatitis virus (MHV) and SARS-CoV, encode a 3′-to-5′ exoribonuclease activity (ExoN) in nsp14. Genetic inactivation of ExoN activity in engineered SARS-CoV and MHV genomes by alanine substitution at conserved DE-D-D active site residues results in viable mutants that demonstrate 15- to 20-fold increases in mutation rates, up to 18 times greater than those tolerated for fidelity mutants of other RNA viruses. Thus nsp14-ExoN is essential for replication fidelity, and likely serves either as a direct mediator or regulator of a more complex RNA proofreading machine, a process previously unprecedented in RNA virus biology. Elucidation of the mechanisms of nsp14-mediated proofreading will have major implications for our understanding of the evolution of RNA viruses, and also will provide a robust model to investigate the balance between fidelity, diversity and pathogenesis. The discovery of a protein distinct from a viral RdRp that regulates replication fidelity also raises the possibility that RNA genome replication fidelity may be adaptable to differing replication environments and selective pressures, rather than being a fixed determinant.
Key words: coronavirus, replication, proofreading, quasispecies, fidelity, mutator phenotype, nidovirales, recombination, SARS, mutagenesis, vaccine
Introduction
Coronaviruses (CoVs) are a family of RNA viruses that cause significant diseases in humans such as severe acute respiratory syndrome (SARS) and other respiratory infections, as well as a variety of respiratory, gastrointestinal and other infections in an increasingly large variety of mammals and birds. The capacity CoVs possess for trans-species movement and adaptation, long recognized in the laboratory,1 was confirmed in nature by the recent emergence of several animal coronavirus pathogens of domesticated animals and SARS-CoV. Data on the latter emergence event indicated that SARS likely resulted from human infection and adaptation by a Bat SARS-like CoV.2,3 Finally, the “post-SARS” identification and analysis of a vast diversity of newly identified coronaviruses across bat species,4 along with the successful synthetic resurrection of a Bat SARS-like CoV,5 suggests that many, if not all, mammalian CoVs may originate from bats.6 However, the mechanisms of CoV host species movement and adaptation for replication and pathogenesis are poorly understood. This review will discuss the role of RNA replication fidelity in RNA virus replication and pathogenesis, and will focus on a novel exoribonuclease universally encoded within CoV genomes that likely mediates RNA-dependent RNA proofreading during virus replication.7,8
Impact of Replication Fidelity and Diversity on RNA Virus Adaptation and Pathogenesis
Due to high error rates of RNA replication, RNA viruses exist as quasispecies, defined as “ensembles of related genotypes”.9,10 There is evidence that evolutionary selection targets RNA virus quasispecies populations rather than individual variants,11,12 and that cooperative interactions between variants influences RNA virus pathogenesis. Cell culture passage of a mumps vaccine strain associated with meningitis resulted in reduced neurovirulence that correlated with heterogeneity at specific positions in multiple viral genes.13 For West Nile virus, increased genetic heterogeneity after mosquito cell passage correlated with growth fitness in those cells.14 Another study provided evidence suggesting that innate immunity can limit poliovirus pathogenesis by restricting viral diversity during transit to vulnerable tissues such as brain.15 Despite the clear linkage between replication fidelity and pathogenesis, and although numerous studies support high mutation rates of RNA viruses, the range of genetic variability tolerated by specific viruses is not well understood. A four-fold increase in mutation frequency of poliovirus through chemical mutagenesis reduced infectivity by 95%.16 In contrast, a 2-to-3-fold decrease in mutation frequency (3Dpol-G64S) reduced the capacity of the mutant poliovirus to compete with WT virus in direct competition assays in culture and in mice and resulted in highly attenuated viruses with restricted tissue tropism in mice.12,15,17 Vesicular stomatitis virus (VSV) likely has a narrow tolerance for alteration in replication fidelity, based on the finding that mutations from chemical mutagenesis at two defined single-nucleotide positions in VSV genomes could be only moderately increased (2–3-fold) without abolishing infectivity.18 Underscoring the contribution of diversity to pathogenesis, modulation of replication fidelity is a new and promising approach for engineering live-attenuated RNA virus vaccines. Poliovirus RdRp mutants with restricted genetic diversity elicit a protective immune response in mice comparable to the Sabin type 1 vaccine strain but have superior genetic stability since increased replication fidelity minimizes reversion to virulence.17 Although constitutively low replication fidelity (m = 10−4 to 10−5 substitutions per nt per round of replication) is thought to be a key determinant of RNA virus quasispecies diversity, adaptation and virulence, the tolerated range of increased or decreased fidelity has appeared to be constrained to less than 4-fold. This is in sharp contrast to DNA genomes of bacteria such as E. coli that may have profoundly greater fidelity (10−9 to 10−11 substitutions per gene per round of replication) but may tolerate up to 1,000 fold differences in replication fidelity in viable mutator phenotype bacteria.
Coronavirus Transcription, Replication and Recombination
CoVs are enveloped viruses that have positive-sense, non-segmented RNA genomes 27–32 kb in length. The basic gene organization and replication is similar for all CoVs and is illustrated for SARS-CoV (Fig. 1). Gene 1 encodes all predicted replicase/transcriptase proteins, which are translated from input genome RNA (RNA1). Genes 2–9 encode structural and accessory proteins, which are translated from separate subgenomic (sg) mRNAs.19–22 CoVs, as members of the Nidovirales order, generate not only new genome RNA, but also a 3′-nested set (Nido = nest) of subgenomic mRNAs (sgRNAs). Along with a portion of the 3′ genome sequence, each CoV sgRNA also contains the first approximately 70 nt of the 5′ leader sequence (Fig. 2). Coronavirus RNA synthesis can be conceptualized as involving two stages: genome replication and subgenomic RNA transcription. In genome replication, the plus-strand genome RNA is transcribed into a full-length minus-strand template RNA, and then significantly more plus-strand genome RNAs are synthesized from that minus-strand template. In subgenomic RNA transcription, 3′-nested subgenomic RNAs are transcribed to serve as templates for translation of the viral structural and accessory proteins. This stage of viral RNA synthesis involves a discontinuous RNA transcription model termed transcription attenuation during negative strand RNA synthesis.21–25 During negative strand synthesis, the viral RdRp recognizes virus-specific conserved sequences termed transcriptional regulatory sequences (TRSs), located just upstream of each subgenomic ORF. At these points, the polymerase either reads through to the next TRS or dissociates from the template strand, then re-associates with the leader TRS, located in the 5′ UTR, and completes synthesis of a set of subgenomic length negative strand RNA containing an antileader RNA sequence and equivalent in size to each viral mRNA. These subgenomic negative strand RNAs then function as the principal templates for the production of subgenomic mRNAs that are 3′-coterminal, and that all possess an ∼70 nt 5′ leader sequence. Because of this transcription mechanism, alterations in TRS sequences can influence viral replication efficiency.26,27 Importantly, the primary TRS sequence seems less important, as recombinant viruses engineered to encode completely new TRS networks are viable, suggesting that regulatory sequences flanking the TRS elements are critical regulators of subgenomic transcription.27 Recently, Wu and Brian have shown that artificial, marked positive-sense subgenomic mRNAs can function as templates for minus strand synthesis and likely contribute to amplifying the amounts of plus-strand sg mRNAs synthesized during infection. Moreover, mRNAs also can serve as templates for the synthesis of smaller sg mRNAs by recognition of internal TRS elements as well.28 The specific mechanism that confers the capacity of the polymerase to dissociate and reassociate is not well understood, but is thought to be mediated by complementary binding of the anti-TRS on the nascent minus strand with the leader template TRS on the plus strand.29–31 As rare misaligned leader-body junctions are occasionally seen during transcription,21,22 it is possible that one or more unique RNA modifying activities, like nsp14 ExoN which encodes a 3′–5′ exonuclease activity (see below), may process the ends of incomplete negative strand RNAs to promote base-pairing and the priming of antileader RNA synthesis.
Figure 1.
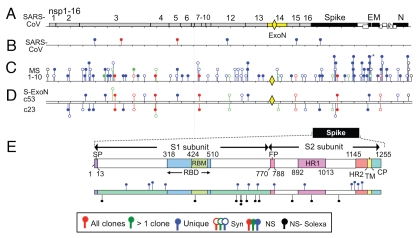
SARS-CoV genome organization, reverse genetics and nsp14-ExoN motifs and mutations. (A) SARS-CoV genome. ∼29.7 kb single-strand (+) RNA. ORF1ab replicase gene is shown in gray with nonstructural protein domains (nsp) 1–16 indicated. Putative RNA synthesis proteins are indicated as in text. Nsp14/ExoN is yellow. (B) Schematic shows nsp14 with three ExoN motifs in red and zinc finger motif hatched. (C) Alignment and MHV and SARS-CoV nsp14 motifs, with conserved active site residues in red. Location of alanine substitutions are indicated, with resulting mutant.
Figure 2.
Coronavirus subgenomic (sg) mRNA synthesis. See text for discussion. Red arrows show direction of RNA synthesis.
Coronaviruses and Recombination
Recombination has shaped the population genetic structure of coronaviruses, promoting cross-species transmission and pathogenesis while complicating vaccine design.5,27,32 Coronaviruses are quite capable of mediating homologous RNA recombination, with rates approaching 20% during mixed infection of closely related strains in the same group.33–35 This high recombination frequency is likely due to the large size of the genome, paired with replication machinery that is already equipped to dissociate and reassociate from the template RNA (site-assisted copy choice recombination), as well as the availability of full-length and subgenomic-length strands for template switching.36 This view is supported by studies showing targeted recombination between specially designed mutant subgenomic mRNAs and genome length templates.37,38 Viable mutant CoVs can be recovered with artificial TRS sequences, as long as the leader and intergenic TRS sequences match. Further, these artificial TRS sequence viruses prevent recombination with virus containing the native TRS sequences.27 These factors combine to allow for the predicted rapid evolution of structural genes, especially within the Spike gene, which undergoes high positive selective pressure during emergence and host-shift events.39–42
Coronavirus Replicase Protein Expression and Functions
The viral proteins responsible for CoV replication, transcription and recombination are encoded as protein domains of the largest known RNA virus polyproteins (Fig. 1). The CoV ORF1a and fusion ORF1ab replicase polyproteins are expressed from the ∼20 kb gene 1 on the input genome RNA and subsequently are processed by viral proteinases to yield 16 mature nonstructural proteins (nsps 1–16), as well as several intermediate precursors, as recently reviewed in reference 43. Nsps 4–16 are significantly conserved in all known CoVs and have been experimentally demonstrated or predicted to be critical enzymes in CoV RNA synthesis and modification, including: nsp7–nsp8, hexadecamer with putative processivity activities; nsp8, primase;44 nsp12, RNA-dependent RNA polymerase (RdRp);45 nsp13, RNA helicase-ATPase (Hel);46,47 nsp14, exoribonuclease (ExoN),48 and methyltransferase;49 nsp15 endoribonuclease (EndoU);50–54 and nsp16, RNA 2′-O-methyltransferase (MT).55,56 In addition, multiple nsps may be required for certain functions in RNA synthesis, as it has recently been shown that interactions of nsps 10, 14 and 16 are required for methyl transferase activity.57 Remarkably, viruses with mutations that ablate the 2′-O-methyl transferase activity encoded in nsp16 are highly sensitive to the activity of the interferon stimulated gene IFIT-2, which likely inhibits the translation of RNA molecules lacking 2′-O-methyl modifications.58
Coronavirus nsp14: A Multifunctional Protein with Exoribonuclease and Methyltransferase Activity
At 27 to 32 kb, the positive-sense RNA genomes of CoVs (Fig. 1) are the largest known RNA genomes59,60 at up to twice the length in nucleotides as those of the next-largest non-segmented RNA virus.59 The bioinformatics prediction of a putative exoribonuclease (ExoN) encoded in replicase nonstructural protein 14 (nsp14) of all CoVs led to the speculation that ExoN functions in proofreading during replication and that acquisition of ExoN by a precursor virus was critical for expansion and maintenance of the large RNA genomes of CoVs.55 The amino-terminal half of the 59-kDa nsp14 includes 3′-to-5′ ExoN motifs I (DE), II (D) and III (D), which were originally identified in cellular enzymes of the DEDD superfamily, including those that catalyze DNA proofreading.55,61,62 Bacterially expressed SARS-CoV nsp14 has been shown to have 3′-to-5′ ExoN activity in vitro, and alanine substitution of the DE-D-D residues profoundly impairs or abolishes this activity.48 The intracellular RNA targets for ExoN activity likely include viral RNA intermediates; importantly, however, they may also include cellular mRNAs, noncoding RNAs and/or microRNAs, which encode or regulate critical antiviral activities during infection.63 Studies from our laboratories also indicated that the carboxy-terminal half of nsp14 has independent functions in RNA synthesis and virulence in animals,64,65 a conclusion which was confirmed by the demonstration that the carboxy-terminal half of nsp14 encodes a novel cap N7-methyltransferase function.49 Proteomic studies, while controversial, indicate robust two-way interactions between nsp14, nsp8 and ORF9b or nsp14 and nsp10; the latter study also hinting at less robust interactions with nsp5 and ORF3b. Clearly, additional biochemical and genetic studies are needed to identify the nsp14 viral and cellular protein interactome.66,67
Nsp14-ExoN is Required for Replication Fidelity
Substitution of alanine at DE Motif I of either MHV (M-ExoN) or SARS-CoV (S-ExoN) allows recovery of viable mutants with modest replication defects of less than 1 log in peak titer compared to wild type, indicating that functional ExoN activity is not required for CoV replication in culture. Sequencing revealed 12- to 20-fold increases in mutation frequency and up to 14-fold increase in mutation rate than comparably isolated and sequenced wildtype MHV or SARS-CoV (Fig. 3).7,8 Thus, both M-ExoN and S-ExoN have similar high-level mutator phenotypes, from 5 to 20 times greater than that seen with other RNA viruses with described fidelity defects, representing the highest-level mutator phenotype of any RNA virus. The mutator phenotype of M-ExoN was maintained over at least 17 passages, as was the stable replication defect. The mutations accumulated in a linear manner over passage consistent with a stable mutator phenotype. Interestingly, M-ExoN has an estimated mutation rate similar to other known wt RNA viruses, while wt MHV in fact had a mutation rate 15-fold less than other RNA viruses, suggesting that ExoN confers very high replication fidelity on CoV genome replication. This result was consistent with predictions that ExoN may be required for the stability of large RNA genomes.55,59 To test whether these findings could be applied across divergent CoVs, the studies also were performed with S-ExoN in Vero cells with very limited replication cycles and complete genome sequencing (Sanger) of 10 wt and 10 S-ExoN plaque isolates from a single round of infection. The results showed a 21-fold increase in mutational frequency and 14-fold increase in mutation rate across the S-ExoN genomes compared with wt SARS-CoV. For both S-ExoN and M-ExoN, mutations were distributed across the genomes with no statistical bias for regions of the genome, for type of mutation (codon position, transversion, transition) or for synonymous, non-synonymous or non-coding mutations. The analysis was only performed on viable replication-competent populations or plaques, and therefore excluded lethal or profoundly deleterious individual or combination mutations. Thus, the results likely highly under-represent the numbers, types and locations of mutations that would be detected in analysis of total viral genomes from a round of replication that would include both non-viable or minor mutations and defective interfering genomes. When the mutation patterns in the genomes from each of the 10 plaque isolates of S-ExoN were compared, 100 mutations were identified, and both the individual mutations and the “mutation sets” for each genome were unique. Finally, the growth of multiple plaque isolates showed that all plaque isolates had growth patterns with defects compared to wt SARS-CoV but indistinguishable from the initial recovered S-ExoN population or from each other.
Figure 3.
Mutations detected in SARS-CoV and S-ExoN Mutants. (A) SARS-CoV genome with nsp14-ExoN in yellow and diamond indication location of ExoN mutant. (B) Mutations identified in 10 SARS-CoV plaque clones (∼320 knt). Legend above indicates type of mutations. (C) Mutations identified in 10 S-ExoN plaque clones (∼320 knt). (D) Representative S-ExoN plaque clones showing unique and shared mutations and patterns. (E) Spike glycoprotein showing the schematic of spike subunits and domains. Lower schematic shows unique mutations across spike from 10 S-ExoN plaque clones (upright-blue) and from population Solexa Sequencing (inverted-black).
ExoN: An RNA-dependent RNA Proofreading Machine?
The results from both M-ExoN and S-ExoN mutants, as well as the in vitro ssRNA exonuclease activity of SARS-CoV nsp14, all argue strongly that nsp14-ExoN directly mediates or participates in the prevention or repair of mutations, which would constitute RNA proofreading, an enzymatic activity not previously reported during the replication of RNA viruses. A nuclease activity of influenza virions in removing non-cognate residues from the 3′ termini of capped primers was reported as evidence of proofreading, but this was not tested during replication and has not been further investigated or confirmed by other labs.68 Rather, others have tested for and failed to find evidence for 3′ to 5′ exonuclease activity or proofreading in RNA viruses,69 and have generally concluded that the energy and fitness cost exceeded the need for error recognition or repair mechanisms. Thus it may be that the incorporation of nsp14 ExoN in the coronaviruses both allowed expansion of the genome and then was required for maintenance of the large and complex genome integrity. In this case the central unanswered research question remains: by what mechanism does nsp14 increase fidelity? There are several possible models, each of which would be unprecedented among RNA viruses, and which could occur alone or in combination as functions of nsp14 or with other replicase proteins. (1) Nsp14 ExoN could directly mediate RNA proofreading. This would be analogous to DNA proofreading where the exonuclease activity is oftentimes provided by a subunit distinct from the polymerase activity. In this regard it is notable that DNA proofreading exonucleases also belong to the DE-D-D superfamily. (2) Nsp14 could stimulate an intrinsic putative 3′-to-5′ ExoN activity of the RdRp. This would be similar to RNA proofreading during cellular transcription by eukaryotic RNA pol II and bacterial RNAP.70 (3) Nsp14 could increase RdRp fidelity through an allosteric effect, transmitting conformational changes through the RdRp. Although a precedent of this situation in DNA or RNA proofreading is not obvious, there are numerous examples of cofactor-enzyme allostery. (4) Nsp14 ExoN could participate in repair by RNA recombination. Considering that high-frequency homologous recombination in the form of discontinuous transcription is central to nidovirus gene expression, and that ExoN mutants of human CoV-229E have altered subgenomic mRNA ratios and aberrant mobilities,48 it is reasonable to propose that subgenomic transcriptional and recombination repair may have evolved in CoVs and that nsp14-ExoN is required for this process. A most likely model is that nsp14 cooperates with other CoV enzymes to form a complex that is involved in error recognition and repair. Although nsp14 alone has been shown so far only to hydrolyze nucleotides at the 3′ terminus of RNA, sequential action of nsp15-EndoU and nsp14-ExoN would theoretically allow removal of internal, mismatched nucleotides. Thus, whereas nsp14 can facilitate correction of residues at only the growing end of a nascent RNA chain, cooperative interaction of nsp15 and nsp14 could facilitate correction of residues at other sites, and conceivably in full-length molecules that are no longer nascent. Finally, CoVs encode methyltransferase (nsp16) that interacts specifically with nsp14 for cap methylation and which could also be participating in fidelity or other RNA modifications that allow expansion of the CoV genome.57
Limits of Replication Fidelity and Mutational Diversity in RNA Viruses
The potential for genetic variability of RNA viruses has long been considered to be fundamental to their evolution, adaptation and escape from host responses. However, the effects of changes in replication fidelity, susceptibility to accumulation of deleterious mutations and lethal mutagenesis are not well studied for many viruses. Genetic determinants including size of genome and presence of repair mechanisms such as proofreading, replicase fidelity and recombination, as well as other as yet undetermined factors may have evolved quite differently in distinct virus families. The high mutation rates of RNA viruses also render them particularly susceptible to repeated genetic bottleneck events during replication, transmission between hosts or spread within a host, resulting in progressive deviation from the consensus sequence associated with decreased viral fitness and sometimes extinction.15,71,72 The process by which populations of asexual organisms tend to accumulate deleterious mutations in the absence of recombination is referred to as Muller's ratchet.73 Muller's ratchet has been shown to be applicable to multiple RNA viruses during plaque-to-plaque passage74–78 and to result in accumulation of mutations and lethal mutagenesis and extinction of plaque-passaged viruses. For example, some FMDV clones are susceptible to genetic bottleneck-mediated extinction, while others are resistant.77 Mutagenesis has also been proposed to work as an antiviral strategy.9,79 A major mechanism of action of ribavirin and other RNA mutagens is lethal mutagenesis, as demonstrated with poliovirus,16,80,81 and other RNA viruses, including HIV, FMDV, LCMV and Hantaan virus.82–85 In contrast, SARS-CoV is highly resistant to ribavirin in vitro and in vivo;86,87 in fact, in some instances, the drug exacerbates in vivo disease.88
Impact of ExoN Mutator Phenotype on CoV Genome Diversity and Stability
The susceptibility to varying degrees of genetic bottleneck has been addressed for the M-ExoN and S-ExoN mutator viruses in comparison with wt parental cloned strains. SARS-CoV and S-ExoN were passaged as populations (three independent passages each) at fixed intervals (18 h) and with titer determination and equivalent MOI for passage 20 times (Fig. 4). Interestingly, both SARS-CoV and S-ExoN had similar responses, with selection for increased titer (1–2 log) by passage 10, after which adaptation reached a plateau. Analysis of sequence showed retention of the ExoN mutations at passage 10, indicating that a primary revertant was not the cause of increased growth. Further, up to passage 10 the total mutational diversity of the S-ExoN populations dramatically exceeded that of wt SARS-CoV. These results demonstrated that in population passage with selection limited only to growth at 18 h, massive diversity in the population was tolerated and still allowed adaptation for increased growth. The effect of more stringent genetic bottleneck (Muller's Ratchet) was tested using MHV-ExoN, in which plaque-to-plaque passage of 10 plaques each of wt MHV and M-ExoN was performed in parallel (Fig. 4). Ten clones each of WT-MHV-A59 and MHV-ExoN were subjected to ten serial plaque-to-plaque transfers in murine DBT cells. Two M-ExoN1 clones became non-recoverable during this passage series (one at p4 and one at p8), whereas all MHV clones were recoverable throughout. Moreover, titers from M-ExoN plaque passage showed a trend of decreasing average titer over passage, whereas the titer from WT-MHV remained constant. The results suggest that ExoN mutator viruses may be more susceptible to accumulation of deleterious mutations driven by repeated population bottlenecks. Conversely, it was surprising that the plaque passage revealed no rapid extinction of the mutant, suggesting that other mechanisms have evolved to stabilize populations and prevent lethal mutagenesis, and/or that CoVs tolerate the accumulation of massive mutational loads across the expanded genomes. Comparison of complete genome sequences of extended plaque and population passages will be required to test these possibilities.
Figure 4.
Population and plaque passage of S-ExoN and M-ExoN. (A) Population passage of WT SARS-CoV and S-ExoN (Adapted from Eckerle, et al.7). (B) Parallel plaque passages of 10 plaques each of WT-MHV and M-ExoN. *indicates passage where loss of a plaque lineage occurred.
Applications
The genomes of positive-strand RNA viruses have considerable capacity to evolve quickly in response to changing ecologic conditions and/or host environments. Mutation rate is a critical parameter for understanding virus evolution, and restriction in genetic diversity within a population of viruses leads to lower adaptability and pathogenicity. Moreover, a general trend toward an inverse correlation between genome size and replication fidelity has been demonstrated by high variations in RdRp error rates that range from about 10−4 to 10−6.89 Based primarily on studies with enteroviruses, RNA viruses with smaller genome sizes seem to regulate replication fidelity by a long distance network of dynamic interactions throughout the 3Dpol RdRp that function to regulate RNA binding and recognition, ligand recognition and binding, protein conformation and RNA synthesis.90 Fidelity can be further modified by virus-host interactions that regulate RNA replicase fidelity or RNA recombination and repair. In contrast to other positive strand RNA viruses, CoVs appear to have expanded replicase fidelity by acquiring and evolving a unique enzymatic activity, encoded with the ExoN nsp14 replicase protein.7,8 Clearly, the existing paradigm of a lumbering error-prone RdRp will be replaced with one that recognizes a more complex, highly tuned and regulated protein machine that was likely essential for the expansion of the CoV genome. There is a critical need to elucidate the molecular mechanisms governing ExoN fidelity regulation, and we are in the process of designing in vitro ExoN mutant assays. Nonetheless, the existence of a non-essential exogenous activity which appears to modify CoV RdRp fidelity provides novel opportunities for experimental testing of the fundamental relationships between fidelity and replication, recombination, adaptation, cross species transmission, genome evolution and pathogenesis while simultaneously providing new avenues for therapeutic enhancement of lethal mutagenesis and the rational design of live attenuated vaccines.91,92 Historically, the extent of genetic diversity in RNA virus populations has most often been analyzed by sequencing a small number of genomes at low coverage or a small region at high coverage. The former approach lacks resolution while the latter is narrowly focused, so extrapolation to the entire genome can be misleading. Deep sequencing approaches like mRNA-seq provide new opportunities for high resolution mapping of mutation distributions across genomes.7
Genome Evolution
The availability of mutator phenotype mutants of both MHV and SARS that can tolerate up to 20-fold increase in mutation rate, accumulate massive mutational diversity at the population level and survive extended population and plaque passage, represents a powerful tool to directly test long-standing questions concerning the role of diversity and fidelity in virus replication, pathogenesis and evolution. How do CoVs maintain a large and complex genome over time while allowing sufficient mutation rates for adaptation and trans-species movement? Is the fidelity of wt CoV replication fixed by highly selected interactions between the RdRp, ExoN and possibly other viral and cellular proteins or is it flexible in response to altered environmental conditions as has been shown for bacteria such as E. coli? What are the limitations to CoV genome diversity in vitro and in vivo? Does the ExoN mutator phenotype result in more rapid adaptation or attenuation associated with lethal mutational load and rapid extinction during infection in vivo? Does the mutator phenotype increase susceptibility to mutagens for lethal mutagenesis? Clearly, CoVs provide a rich empirical platform to address these interesting and unique questions in virus evolution and adaptation.
Replication and Recombination
CoVs use a unique discontinuous mechanism to transcribe a series of progressively larger subgenomic mRNAs, and each contains a leader RNA sequence that is derived from the 5′ end of the genome. RNA recombination and coronavirus subgenomic transcription occur by template switching mechanisms, which can occur either by sequence-assisted base pairing and hydrogen bonding networks or sequence independent processes in mismatched regions. Given the potential need to appropriately process and match the 3′ ends of nascent RNAs and their templates, the ExoN mutator phenotype provides a novel approach to investigate the role of fidelity in regulating both full length and subgenomic length RNA synthesis, transcription attenuation during negative strand RNA synthesis and as potential regulators of RNA recombination distributions and frequencies across the genome. The possibility that ExoN interacts with other RNA modifying enzymes such as nsp15 EndoU and nsp16 2′-O-MTase or interact with putative RNA processivity components encoded in nsp7 and 8 to modify RNA repair rates or recombination frequencies may serve as a rich environment for future research.
Experimental Evolution
Of the 335 new emerging infectious diseases that were identified between 1940–2004, 60.3 percent were zoonoses, 71.8 percent of which originated in wildlife.93 As pathogen emergence has also been increasing overtime, coupled with greater rates of global dissemination,93 the threat to global health and economies is profound. Strategies that can identify viral threats that emerge as a consequence of advantageous mutations in response to select evolutionary pressure(s) would provide profound advances in the ability of the Global Health Response Network to control emerging diseases. The existence of a defined ExoN-mediated mutator phenotype may allow for mechanistic insights and modeling of the mutation repertoires that govern: (a) the rapid selection of host range expanded mutant viruses which represent precursors to future epidemic emergences; (b) pathways of escape from therapeutic human monoclonal antibodies and drugs; (c) limits of genome variation and stability; and (d) replicase mutations and interacting networks which restore fidelity in passaged ExoN mutant viruses. For example, CoV phylogeny is punctuated by numerous shifts between host species and cross-species transmission is readily achieved in co-cultures and during persistent infections in vitro.1,94,95 In nature, human coronavirus (HCoV) °C43 emerged around 1,900 from closely related bovine CoVs, whereas HCoV 229E likely emerged from closely related group 1 bat coronaviruses around 1,800 in Africa,96,97 leading some to propose that nearly all human and animal CoVs originated from a vast reservoir of strains circulating in bat species.6,98 SARS-CoV is also a zoonotic virus that crossed the species barrier, most likely originating from bats, following amplification in other species (civet cats, raccoon dogs), prior to transmission to humans.2,3,99 Our group has used synthetic biology to reconstruct full-length genomes of SARS-like bat CoV precursors to the 2002–2003 epidemic strains.5,100,101 Although these strains replicate but do not spread because they lack the appropriate receptor-binding domain, recombinant bat coronaviruses harboring the SARS-CoV receptor binding domain (RBD) replicate efficiently and use angiotensin 1 converting enzyme 2 (ACE2) as a receptor for docking and entry. These data suggest that the trimeric spike glycoprotein of CoVs may be plastic and modular in design, readily allowing for protein domains to be exchanged between divergent S glycoproteins from different strains.32 We propose that blending the ExoN mutator phenotype into synthetically reconstructed zoonotic viruses provides a strategy to rapidly identify pathway components and mutation sets that govern trans-species movement in cell or organ cultures and in vivo. We hypothesize that the CoV ExoN mutator phenotype constitutes a robust investigative platform to predict mutations and possibly recombinants in advance of their occurrence by identifying advantageous mutations governing host range and virus cross-species transmission.
Pathogenesis
Genetic diversity within a quasispecies has been proposed to contribute to pathogenesis by cooperative interactions among engineered variant viruses within a population.12,17 However, the impact of reduced replication fidelity on pathogenesis remains largely untested for RNA viruses. The CoV ExoN mutator phenotype represents a unique property with unclear consequences for adaptation and pathogenesis in animals. Although increased fidelity attenuates virulence of poliovirus and restricts tissue tropism, the ExoN mutator activity might increase virus virulence and tissue tropism because of the increased population diversity and spread into novel tissues. Alternatively, ExoN decreased fidelity might attenuate virulence because the mutation frequency may approach error catastrophe and drive self-annihilation of S-ExoN in vivo. The growth defects observed in culture seem to support the prediction that S-ExoN will be attenuated in animals, but these impairments could be trumped by potential enhanced adaptability of S-ExoN. To assess these possibilities, pathogenesis studies in animal models are currently underway. Of note, a low-fidelity exonuclease mutant of cytomegalovirus showed accelerated evolution of drug resistance in cell culture.102 Increased mutation rates in the GII.4 noroviruses have been proposed to account for their epochal evolution, increased diversity and the striking increase in the frequency of human epidemics in winter.103,104 Thus, fidelity regulation is a broadly relevant topic with far-ranging appeal in RNA virus evolution and pathogenesis. Importantly, ExoN represents an important and unique high impact target for understanding CoV replication fidelity, quasispecies diversity and pathogenesis and is strongly coupled with the potential of developing universal strategies to build safe live attenuated CoV vaccines.
Vaccines and Therapeutics
Live attenuated vaccines elicit strong protective immune responses with low risk of disease, leading to robust tools that protect the public health against pathogens like measles, poliovirus, mumps, smallpox, herpesviruses and rubella. Safety concerns clearly exist as evidenced by vaccine reversion to virulence and the development of serious and lethal disease in a low percentage of vaccinees. The present regulatory environment in the US is now limiting live attenuated virus vaccine use because of safety concerns, attesting to the need for rational approaches that prevent reversion to virulence. The high conservation of nsp14 ExoN sequences among CoVs and lack of close orthologs in cells suggests that nsp14 ExoN may be a promising target for live attenuated virus design or antiviral therapeutics. Clearly, studying the ExoN mutator phenotype in pathogenesis and as a rational approach to develop reversion-resistant live attenuated vaccines provides a potential rapid response strategy to control future emerging CoV diseases in human and domesticated animals. Current treatment regimens for SARS-CoV include ribavirin, a nucleoside analog that induces lethal mutagenesis of other RNA viruses such as poliovirus, foot and mouth disease virus, hepatitis C virus among others.16,81,105,106 However, its precise mechanism of action against CoVs has not been determined and the high replication fidelity of WT MHV and SARS-CoV in cell culture suggests that drug-induced viral extinction therapies employed against other RNA viruses might not be as effective against CoVs.7,8 A possible recalcitrance of CoVs to RNA mutagens is also suggested by our demonstration that at least in cell culture SARS-CoV tolerates a 16.5-fold increase in substitution frequency, whereas a two- to six-fold increase in mutation frequency was sufficient to cause lethal mutagenesis of poliovirus in cell culture.7,8,16 Moreover, ribavirin is clearly ineffective against mouse-adapted SARS-CoV and appears to exacerbate disease, suggesting that the ExoN activity in wildtype viruses may reduce the efficacy of this important antiviral.88 The high conservation of nsp14 ExoN sequences among CoVs and lack of close orthologs in cells suggests that nsp14 ExoN might represent a promising target for design and development of antiviral drugs and raises the possibility that a single drug targeting ExoN might be effective against multiple coronaviruses, including potential zoonotic SARS-CoV-like viruses from bats that emerge in the future. However, given numerous examples of viruses evolving drug resistance, an ExoN-targeted companion drug in a combination therapy would not only attenuate pathogenesis by altering error rates, but also prevent reversion from other compounds in the cocktail. Investigating the potential of ExoN targeted mutations as a universal strategy to construct live attenuated, reversion proof CoV vaccines and antivirals seems broadly relevant. Studies investigating the pathogenesis of ExoN mutants in animal models, along with their tenability as vaccine candidates, are currently in progress.
Summary
The identification of a stable mutator phenotype and possible dedicated RNA-dependent RNA-proofreading complex is significant for several reasons. The availability of the M-ExoN and S-ExoN mutant viruses constitutes a unique system to directly study the impact of profoundly decreased replication fidelity and increased diversity on replication and pathogenesis of RNA viruses. The use of both traditional di-deoxy (Sanger) and ultradeep (Solexa) sequencing in conjunction with virus passage and persistent infection will allow establishment of new models for understanding the range and limitations on diversity and mutational load and will aid in developing tool sets for evaluating, comparing and annotating sequence diversity across RNA virus genomes. Massive sequence annotation and analysis during different stages of acute infection (intracellular, virion), as well as over time (passage, persistent infection) will allow mapping of genetic regions highly tolerant or resistant to change and to define potential epistatic interaction networks not predictable by other approaches as well as conserved across the Coronaviridae. Passage and sequencing of ExoN mutator viruses also will provide a system to rapidly generate extensive libraries of individual mutations and complete mutation sets across the genome that can be tested in individual coding or regulatory domains for effects on immune response, host range and virulence. Such studies of ExoN-generated decreased fidelity and increased diversity also represent two potentially broadly applicable strategies for live vaccine design that simultaneously attenuate and prevent reversion to virulence. Finally, the mutator viruses and studies of ExoN mutant revertants will result in important insights into the evolutionary mechanisms by which the Nidovirales acquired (or lost) the capacity for RNA proofreading, and will allow testing of the limitations of size and complexity of RNA as a replicating molecule. As such, ExoN should be considered as a high impact research target for protecting the public health against future emerging CoV diseases.
Acknowledgements
This work was supported by grants from the National Institutes of Health: SERCEB-U54-AI057157 (M.R.D., R.S.B., E.F.D.), R01-AI26603 (M.R.D., L.D.E.) and contract HHSN272200800060C. Additionally, R.L.G. received funding from F32-AI080148 and L.D.E. received funding from T32-AI049824. The funders had no role in study design, data collection and analysis, decision to publish or preparation of the manuscript.
Abbreviations
- SARS
severe acute respiratory syndrome
- CoV
coronavirus
- MHV
murine hepatitis virus
- nsp
nonstructural protein
- ExoN
exonuclease
- RdRp
RNA-dependent RNA polymerase
- DE-D-D
Asp-Glu-Asp-Asp active site residues
- ORF
open reading frame
References
- 1.Baric RS, Yount B, Hensley L, Peel SA, Chen W. Episodic evolution mediates interspecies transfer of a murine coronavirus. J Virol. 1997;71:1946–1955. doi: 10.1128/jvi.71.3.1946-1955.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lau SK, Woo PC, Li KS, Huang Y, Tsoi HW, Wong BH, et al. Severe acute respiratory syndrome coronavirus-like virus in Chinese horseshoe bats. Proc Natl Acad Sci USA. 2005;102:14040–14045. doi: 10.1073/pnas.0506735102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Li W, Shi Z, Yu M, Ren W, Smith C, Epstein JH, et al. Bats are natural reservoirs of SARS-like coronaviruses. Science. 2005;310:676–679. doi: 10.1126/science.1118391. [DOI] [PubMed] [Google Scholar]
- 4.Donaldson EF, Haskew AN, Gates JE, Huynh J, Moore CJ, Frieman MB. Metagenomic Analysis of the Virome of three North American Bat Species: Viral Diversity Between Different Bat Species that Share a Common Habitat. J Virol. 2010 doi: 10.1128/JVI.01255-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Becker MM, Graham RL, Donaldson EF, Rockx B, Sims AC, Sheahan T, et al. Synthetic recombinant bat SARS-like coronavirus is infectious in cultured cells and in mice. Proc Natl Acad Sci USA. 2008;105:19944–19949. doi: 10.1073/pnas.0808116105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Vijaykrishna D, Smith GJ, Zhang JX, Peiris JS, Chen H, Guan Y. Evolutionary insights into the ecology of coronaviruses. J Virol. 2007;81:4012–4020. doi: 10.1128/JVI.02605-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Eckerle LD, Becker MM, Halpin RA, Li K, Venter E, Lu X, et al. Infidelity of SARS-CoV Nsp14-exonuclease mutant virus replication is revealed by complete genome sequencing. PLoS Pathog. 2010;6:1000896. doi: 10.1371/journal.ppat.1000896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Eckerle LD, Lu X, Sperry SM, Choi L, Denison MR. High fidelity of murine hepatitis virus replication is decreased in nsp14 exoribonuclease mutants. J Virol. 2007;81:12135–12144. doi: 10.1128/JVI.01296-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bull JJ, Meyers LA, Lachmann M. Quasispecies made simple. PLoS Comput Biol. 2005;1:61. doi: 10.1371/journal.pcbi.0010061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Eigen M. The origin of genetic information: viruses as models. Gene. 1993;135:37–47. doi: 10.1016/0378-1119(93)90047-7. [DOI] [PubMed] [Google Scholar]
- 11.Lauring AS, Andino R. Quasispecies theory and the behavior of RNA viruses. PLoS Pathog. 2010;6:1001005. doi: 10.1371/journal.ppat.1001005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Vignuzzi M, Stone JK, Arnold JJ, Cameron CE, Andino R. Quasispecies diversity determines pathogenesis through cooperative interactions in a viral population. Nature. 2006;439:344–348. doi: 10.1038/nature04388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sauder CJ, Vandenburgh KM, Iskow RC, Malik T, Carbone KM, Rubin SA. Changes in mumps virus neurovirulence phenotype associated with quasispecies heterogeneity. Virology. 2006;350:48–57. doi: 10.1016/j.virol.2006.01.035. [DOI] [PubMed] [Google Scholar]
- 14.Ciota AT, Ngo KA, Lovelace AO, Payne AF, Zhou Y, Shi PY, et al. Role of the mutant spectrum in adaptation and replication of West Nile virus. J Gen Virol. 2007;88:865–874. doi: 10.1099/vir.0.82606-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pfeiffer JK, Kirkegaard K. Increased fidelity reduces poliovirus fitness and virulence under selective pressure in mice. PLoS Pathog. 2005;1:11. doi: 10.1371/journal.ppat.0010011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Crotty S, Cameron CE, Andino R. RNA virus error catastrophe: direct molecular test by using ribavirin. Proc Natl Acad Sci USA. 2001;98:6895–6900. doi: 10.1073/pnas.111085598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Vignuzzi M, Wendt E, Andino R. Engineering attenuated virus vaccines by controlling replication fidelity. Nat Med. 2008;14:154–161. doi: 10.1038/nm1726. [DOI] [PubMed] [Google Scholar]
- 18.Holland JJ, Domingo E, de la Torre JC, Steinhauer DA. Mutation frequencies at defined single codon sites in vesicular stomatitis virus and poliovirus can be increased only slightly by chemical mutagenesis. J Virol. 1990;64:3960–3962. doi: 10.1128/jvi.64.8.3960-3962.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sawicki SG, Sawicki DL. Coronaviruses use discontinuous extension for synthesis of subgenome-length negative strands. Adv Exp Med Biol. 1995;380:499–506. doi: 10.1007/978-1-4615-1899-0_79. [DOI] [PubMed] [Google Scholar]
- 20.Sawicki SG, Sawicki DL. Coronavirus transcription: a perspective. Curr Top Microbiol Immunol. 2005;287:31–55. doi: 10.1007/3-540-26765-4_2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schaad MC, Chen W, Peel SA, Baric RS. Studies into the mechanism for MHV transcription. Adv Exp Med Biol. 1993;342:85–90. doi: 10.1007/978-1-4615-2996-5_14. [DOI] [PubMed] [Google Scholar]
- 22.Baric RS, Yount B. Subgenomic negative-strand RNA function during mouse hepatitis virus infection. J Virol. 2000;74:4039–4046. doi: 10.1128/jvi.74.9.4039-4046.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sawicki SG, Sawicki DL. A new model for coronavirus transcription. Adv Exp Med Biol. 1998;440:215–219. doi: 10.1007/978-1-4615-5331-1_26. [DOI] [PubMed] [Google Scholar]
- 24.Sawicki SG, Sawicki DL. Coronavirus transcription: subgenomic mouse hepatitis virus replicative intermediates function in RNA synthesis. J Virol. 1990;64:1050–1056. doi: 10.1128/jvi.64.3.1050-1056.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pasternak AO, Spaan WJ, Snijder EJ. Nidovirus transcription: how to make sense…? J Gen Virol. 2006;87:1403–1421. doi: 10.1099/vir.0.81611-0. [DOI] [PubMed] [Google Scholar]
- 26.van Marle G, Dobbe JC, Gultyaev AP, Luytjes W, Spaan WJ, Snijder EJ. Arterivirus discontinuous mRNA transcription is guided by base pairing between sense and antisense transcription-regulating sequences. Proc Natl Acad Sci USA. 1999;96:12056–12061. doi: 10.1073/pnas.96.21.12056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yount B, Roberts RS, Lindesmith L, Baric RS. Rewiring the severe acute respiratory syndrome coronavirus (SARS-CoV) transcription circuit: engineering a recombination-resistant genome. Proc Natl Acad Sci USA. 2006;103:12546–12551. doi: 10.1073/pnas.0605438103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wu HY, Brian DA. Subgenomic messenger RNA amplification in coronaviruses. Proc Natl Acad Sci USA. 2010;107:12257–12262. doi: 10.1073/pnas.1000378107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sola I, Moreno JL, Zuniga S, Alonso S, Enjuanes L. Role of nucleotides immediately flanking the transcription-regulating sequence core in coronavirus subgenomic mRNA synthesis. J Virol. 2005;79:2506–2516. doi: 10.1128/JVI.79.4.2506-2516.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Curtis KM, Yount B, Sims AC, Baric RS. Reverse genetic analysis of the transcription regulatory sequence of the coronavirus transmissible gastroenteritis virus. J Virol. 2004;78:6061–6066. doi: 10.1128/JVI.78.11.6061-6066.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zuniga S, Sola I, Alonso S, Enjuanes L. Sequence motifs involved in the regulation of discontinuous coronavirus subgenomic RNA synthesis. J Virol. 2004;78:980–994. doi: 10.1128/JVI.78.2.980-994.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Graham RL, Baric RS. Recombination, reservoirs and the modular spike: mechanisms of coronavirus cross-species transmission. J Virol. 2010;84:3134–3146. doi: 10.1128/JVI.01394-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Baric RS, Fu K, Chen W, Yount B. High recombination and mutation rates in mouse hepatitis virus suggest that coronaviruses may be potentially important emerging viruses. Adv Exp Med Biol. 1995;380:571–576. doi: 10.1007/978-1-4615-1899-0_91. [DOI] [PubMed] [Google Scholar]
- 34.Baric RS, Fu K, Schaad MC, Stohlman SA. Establishing a genetic recombination map for murine coronavirus strain A59 complementation groups. Virology. 1990;177:646–656. doi: 10.1016/0042-6822(90)90530-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Makino S, Keck JG, Stohlman SA, Lai MM. High-frequency RNA recombination of murine coronaviruses. J Virol. 1986;57:729–737. doi: 10.1128/jvi.57.3.729-737.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fu K, Baric RS. Evidence for variable rates of recombination in the MHV genome. Virology. 1992;189:88–102. doi: 10.1016/0042-6822(92)90684-H. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Koetzner CA, Parker MM, Ricard CS, Sturman LS, Masters PS. Repair and mutagenesis of the genome of a deletion mutant of the coronavirus mouse hepatitis virus by targeted RNA recombination. J Virol. 1992;66:1841–1848. doi: 10.1128/jvi.66.4.1841-1848.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Masters PS, Rottier PJ. Coronavirus reverse genetics by targeted RNA recombination. Curr Top Microbiol Immunol. 2005;287:133–159. doi: 10.1007/3-540-26765-4_5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Stavrinides J, Guttman DS. Mosaic evolution of the severe acute respiratory syndrome coronavirus. J Virol. 2004;78:76–82. doi: 10.1128/JVI.78.1.76-82.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hu J, Wang J, Xu J, Li W, Han Y, Li Y, et al. Evolution and variation of the SARS-CoV genome. Genomics Proteomics Bioinformatics. 2003;1:216–225. doi: 10.1016/S1672-0229(03)01027-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lau SK, Woo PC, Li KS, Huang Y, Wang M, Lam CS, et al. Complete genome sequence of bat coronavirus HKU2 from Chinese horseshoe bats revealed a much smaller spike gene with a different evolutionary lineage from the rest of the genome. Virology. 2007;367:428–439. doi: 10.1016/j.virol.2007.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yeh SH, Wang HY, Tsai CY, Kao CL, Yang JY, Liu HW, et al. Characterization of severe acute respiratory syndrome coronavirus genomes in Taiwan: molecular epidemiology and genome evolution. Proc Natl Acad Sci USA. 2004;101:2542–2547. doi: 10.1073/pnas.0307904100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Perlman S, Netland J. Coronaviruses post-SARS: update on replication and pathogenesis. Nat Rev Microbiol. 2009;7:439–450. doi: 10.1038/nrmicro2147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Imbert I, Guillemot JC, Bourhis JM, Bussetta C, Coutard B, Egloff MP, et al. A second, non-canonical RNA-dependent RNA polymerase in SARS coronavirus. EMBO J. 2006;25:4933–4942. doi: 10.1038/sj.emboj.7601368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.te Velthuis AJ, Arnold JJ, Cameron CE, van den Worm SH, Snijder EJ. The RNA polymerase activity of SARS-coronavirus nsp12 is primer dependent. Nucleic Acids Res. 2010;38:203–214. doi: 10.1093/nar/gkp904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Seybert A, Hegyi A, Siddell SG, Ziebuhr J. The human coronavirus 229E superfamily 1 helicase has RNA and DNA duplex-unwinding activities with 5′-to-3′ polarity. Rna. 2000;6:1056–1068. doi: 10.1017/s1355838200000728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ivanov KA, Ziebuhr J. Human coronavirus 229E nonstructural protein 13: characterization of duplex-unwinding, nucleoside triphosphatase and RNA 5′-triphosphatase activities. J Virol. 2004;78:7833–7838. doi: 10.1128/JVI.78.14.7833-7838.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Minskaia E, Hertzig T, Gorbalenya AE, Campanacci V, Cambillau C, Canard B, et al. Discovery of an RNA virus 3′→5′ exoribonuclease that is critically involved in coronavirus RNA synthesis. Proc Natl Acad Sci USA. 2006;103:5108–5113. doi: 10.1073/pnas.0508200103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chen Y, Cai H, Pan J, Xiang N, Tien P, Ahola T, et al. Functional screen reveals SARS coronavirus nonstructural protein nsp14 as a novel cap N7 methyltransferase. Proc Natl Acad Sci USA. 2009;106:3484–3489. doi: 10.1073/pnas.0808790106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bhardwaj K, Sun J, Holzenburg A, Guarino LA, Kao CC. RNA recognition and cleavage by the SARS coronavirus endoribonuclease. J Mol Biol. 2006;361:243–256. doi: 10.1016/j.jmb.2006.06.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Guarino LA, Bhardwaj K, Dong W, Sun J, Holzenburg A, Kao C. Mutational analysis of the SARS virus Nsp15 endoribonuclease: identification of residues affecting hexamer formation. J Mol Biol. 2005;353:1106–1117. doi: 10.1016/j.jmb.2005.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kang H, Bhardwaj K, Li Y, Palaninathan S, Sacchettini J, Guarino L, et al. Biochemical and genetic analyses of murine hepatitis virus Nsp15 endoribonuclease. J Virol. 2007;81:13587–13597. doi: 10.1128/JVI.00547-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ivanov KA, Hertzig T, Rozanov M, Bayer S, Thiel V, Gorbalenya AE, et al. Major genetic marker of nidoviruses encodes a replicative endoribonuclease. Proc Natl Acad Sci USA. 2004;101:12694–12699. doi: 10.1073/pnas.0403127101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bhardwaj K, Guarino L, Kao CC. The severe acute respiratory syndrome coronavirus Nsp15 protein is an endoribonuclease that prefers manganese as a cofactor. J Virol. 2004;78:12218–12224. doi: 10.1128/JVI.78.22.12218-12224.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Snijder EJ, Bredenbeek PJ, Dobbe JC, Thiel V, Ziebuhr J, Poon LL, et al. Unique and conserved features of genome and proteome of SARS-coronavirus, an early split-off from the coronavirus group 2 lineage. J Mol Biol. 2003;331:991–1004. doi: 10.1016/S0022-2836(03)00865-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Decroly E, Imbert I, Coutard B, Bouvet M, Selisko B, Alvarez K, et al. Coronavirus nonstructural protein 16 is a cap-0 binding enzyme possessing (nucleoside-2′O)-methyltransferase activity. J Virol. 2008;82:8071–8084. doi: 10.1128/JVI.00407-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Bouvet M, Debarnot C, Imbert I, Selisko B, Snijder EJ, Canard B, et al. In vitro reconstitution of SARS-coronavirus mRNA cap methylation. PLoS Pathog. 2010;6:1000863. doi: 10.1371/journal.ppat.1000863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Daffis S, Szretter KJ, Schriewer J, Li J, Youn S, Errett J, et al. 2′-O methylation of the viral mRNA cap evades host restriction by IFIT family members. Nature. 2010;468:452–456. doi: 10.1038/nature09489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Gorbalenya AE, Enjuanes L, Ziebuhr J, Snijder EJ. Nidovirales: evolving the largest RNA virus genome. Virus Res. 2006;117:17–37. doi: 10.1016/j.virusres.2006.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Fauquet CM, Fargette D. International Committee on Taxonomy of Viruses and the 3,142 unassigned species. Virol J. 2005;2:64. doi: 10.1186/1743-422X-2-64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zuo Y, Deutscher MP. Exoribonuclease superfamilies: structural analysis and phylogenetic distribution. Nucleic Acids Res. 2001;29:1017–1026. doi: 10.1093/nar/29.5.1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Moser MJ, Holley WR, Chatterjee A, Mian IS. The proofreading domain of Escherichia coli DNA polymerase I and other DNA and/or RNA exonuclease domains. Nucleic Acids Res. 1997;25:5110–5118. doi: 10.1093/nar/25.24.5110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Peng X, Gralinski L, Armour CD, Ferris MT, Thomas MJ, Proll S, et al. Unique signatures of long noncoding RNA expression in response to virus infection and altered innate immune signaling. MBio. 2010:1. doi: 10.1128/mBio.00206-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Sperry SM, Kazi L, Graham RL, Baric RS, Weiss SR, Denison MR. Single-amino-acid substitutions in open reading frame (ORF) 1b-nsp14 and ORF 2a proteins of the coronavirus mouse hepatitis virus are attenuating in mice. J Virol. 2005;79:3391–3400. doi: 10.1128/JVI.79.6.3391-3400.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Eckerle LD, Brockway SM, Sperry SM, Lu X, Denison MR. Effects of mutagenesis of murine hepatitis virus nsp1 and nsp14 on replication in culture. Adv Exp Med Biol. 2006;581:55–60. doi: 10.1007/978-0-387-33012-9_8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.von Brunn A, Teepe C, Simpson JC, Pepperkok R, Friedel CC, Zimmer R, et al. Analysis of intraviral protein-protein interactions of the SARS coronavirus ORFeome. PLoS One. 2007;2:459. doi: 10.1371/journal.pone.0000459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Pan J, Peng X, Gao Y, Li Z, Lu X, Chen Y, et al. Genome-wide analysis of protein-protein interactions and involvement of viral proteins in SARS-CoV replication. PLoS One. 2008;3:3299. doi: 10.1371/journal.pone.0003299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Ishihama A, Mizumoto K, Kawakami K, Kato A, Honda A. Proofreading function associated with the RNA-dependent RNA polymerase from influenza virus. J Biol Chem. 1986;261:10417–10421. [PubMed] [Google Scholar]
- 69.Steinhauer DA, Domingo E, Holland JJ. Lack of evidence for proofreading mechanisms associated with an RNA virus polymerase. Gene. 1992;122:281–288. doi: 10.1016/0378-1119(92)90216-c. [DOI] [PubMed] [Google Scholar]
- 70.Poole AM, Logan DT. Modern mRNA proofreading and repair: clues that the last universal common ancestor possessed an RNA genome? Mol Biol Evol. 2005;22:1444–1455. doi: 10.1093/molbev/msi132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Domingo E, Escarmis C, Lazaro E, Manrubia SC. Quasispecies dynamics and RNA virus extinction. Virus Res. 2005;107:129–139. doi: 10.1016/j.virusres.2004.11.003. [DOI] [PubMed] [Google Scholar]
- 72.Carrillo C, Lu Z, Borca MV, Vagnozzi A, Kutish GF, Rock DL. Genetic and phenotypic variation of foot-and-mouth disease virus during serial passages in a natural host. J Virol. 2007;81:11341–11351. doi: 10.1128/JVI.00930-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Muller HJ. The Relation of Recombination to Mutational Advance. Mutat Res. 1964;106:2–9. doi: 10.1016/0027-5107(64)90047-8. [DOI] [PubMed] [Google Scholar]
- 74.de la Iglesia F, Elena SF. Fitness declines in Tobacco etch virus upon serial bottleneck transfers. J Virol. 2007;81:4941–4947. doi: 10.1128/JVI.02528-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Yuste E, Sanchez-Palomino S, Casado C, Domingo E, Lopez-Galindez C. Drastic fitness loss in human immunodeficiency virus type 1 upon serial bottleneck events. J Virol. 1999;73:2745–2751. doi: 10.1128/jvi.73.4.2745-2751.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Elena SF, Gonzalez-Candelas F, Novella IS, Duarte EA, Clarke DK, Domingo E, et al. Evolution of fitness in experimental populations of vesicular stomatitis virus. Genetics. 1996;142:673–679. doi: 10.1093/genetics/142.3.673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Escarmis C, Davila M, Charpentier N, Bracho A, Moya A, Domingo E. Genetic lesions associated with Muller's ratchet in an RNA virus. J Mol Biol. 1996;264:255–267. doi: 10.1006/jmbi.1996.0639. [DOI] [PubMed] [Google Scholar]
- 78.Duarte E, Clarke D, Moya A, Domingo E, Holland J. Rapid fitness losses in mammalian RNA virus clones due to Muller's ratchet. Proc Natl Acad Sci USA. 1992;89:6015–6019. doi: 10.1073/pnas.89.13.6015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Anderson JP, Daifuku R, Loeb LA. Viral error catastrophe by mutagenic nucleosides. Annu Rev Microbiol. 2004;58:183–205. doi: 10.1146/annurev.micro.58.030603.123649. [DOI] [PubMed] [Google Scholar]
- 80.Graci JD, Harki DA, Korneeva VS, Edathil JP, Too K, Franco D, et al. Lethal mutagenesis of poliovirus mediated by a mutagenic pyrimidine analogue. J Virol. 2007;81:11256–11266. doi: 10.1128/JVI.01028-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Crotty S, Maag D, Arnold JJ, Zhong W, Lau JY, Hong Z, et al. The broad-spectrum antiviral ribonucleoside ribavirin is an RNA virus mutagen. Nat Med. 2000;6:1375–1379. doi: 10.1038/82191. [DOI] [PubMed] [Google Scholar]
- 82.Sierra S, Davila M, Lowenstein PR, Domingo E. Response of foot-and-mouth disease virus to increased mutagenesis: influence of viral load and fitness in loss of infectivity. J Virol. 2000;74:8316–8323. doi: 10.1128/jvi.74.18.8316-8323.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Chung DH, Sun Y, Parker WB, Arterburn JB, Bartolucci A, Jonsson CB. Ribavirin reveals a lethal threshold of allowable mutation frequency for Hantaan virus. J Virol. 2007;81:11722–11729. doi: 10.1128/JVI.00874-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Loeb LA, Essigmann JM, Kazazi F, Zhang J, Rose KD, Mullins JI. Lethal mutagenesis of HIV with mutagenic nucleoside analogs. Proc Natl Acad Sci USA. 1999;96:1492–1497. doi: 10.1073/pnas.96.4.1492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Martin V, Grande-Perez A, Domingo E. No evidence of selection for mutational robustness during lethal mutagenesis of lymphocytic choriomeningitis virus. Virology. 2008;378:185–192. doi: 10.1016/j.virol.2008.05.016. [DOI] [PubMed] [Google Scholar]
- 86.Chiou HE, Liu CL, Buttrey MJ, Kuo HP, Liu HW, Kuo HT, et al. Adverse effects of ribavirin and outcome in severe acute respiratory syndrome: experience in two medical centers. Chest. 2005;128:263–272. doi: 10.1378/chest.128.1.263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Saijo M, Morikawa S, Fukushi S, Mizutani T, Hasegawa H, Nagata N, et al. Inhibitory effect of mizoribine and ribavirin on the replication of severe acute respiratory syndrome (SARS)-associated coronavirus. Antiviral Res. 2005;66:159–163. doi: 10.1016/j.antiviral.2005.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Day CW, Baric R, Cai SX, Frieman M, Kumaki Y, Morrey JD, et al. A new mouse-adapted strain of SARS-CoV as a lethal model for evaluating antiviral agents in vitro and in vivo. Virology. 2009;395:210–222. doi: 10.1016/j.virol.2009.09.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Sanjuan R, Nebot MR, Chirico N, Mansky LM, Belshaw R. Viral mutation rates. J Virol. 2010;84:9733–9748. doi: 10.1128/JVI.00694-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Yang X, Welch JL, Arnold JJ, Boehr DD. Long range interaction networks in the function and fidelity of poliovirus RNA-dependent RNA polymerase studied by nuclear magnetic resonance. Biochemistry. 2010 doi: 10.1021/bi100833r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Graci JD, Cameron CE. Mechanisms of action of ribavirin against distinct viruses. Rev Med Virol. 2006;16:37–48. doi: 10.1002/rmv.483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Lauring AS, Jones JO, Andino R. Rationalizing the development of live attenuated virus vaccines. Nat Biotechnol. 2010;28:573–579. doi: 10.1038/nbt.1635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Jones KE, Patel NG, Levy MA, Storeygard A, Balk D, Gittleman JL, et al. Global trends in emerging infectious diseases. Nature. 2008;451:990–993. doi: 10.1038/nature06536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Chen W, Baric RS. Molecular anatomy of mouse hepatitis virus persistence: coevolution of increased host cell resistance and virus virulence. J Virol. 1996;70:3947–3960. doi: 10.1128/jvi.70.6.3947-3960.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.McRoy WC, Baric RS. Amino acid substitutions in the S2 subunit of mouse hepatitis virus variant V51 encode determinants of host range expansion. J Virol. 2008;82:1414–1424. doi: 10.1128/JVI.01674-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Pfefferle S, Oppong S, Drexler JF, Gloza-Rausch F, Ipsen A, Seebens A, et al. Distant relatives of severe acute respiratory syndrome coronavirus and close relatives of human coronavirus 229E in bats, Ghana. Emerg Infect Dis. 2009;15:1377–1384. doi: 10.3201/eid1509.090224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Vijgen L, Keyaerts E, Moes E, Thoelen I, Wollants E, Lemey P, et al. Complete genomic sequence of human coronavirus JC43: molecular clock analysis suggests a relatively recent zoonotic coronavirus transmission event. J Virol. 2005;79:1595–1604. doi: 10.1128/JVI.79.3.1595-1604.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Shi Z, Hu Z. A review of studies on animal reservoirs of the SARS coronavirus. Virus Res. 2008;133:74–87. doi: 10.1016/j.virusres.2007.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Woo PC, Lau SK, Chu CM, Chan KH, Tsoi HW, Huang Y, et al. Characterization and complete genome sequence of a novel coronavirus, coronavirus HKU1, from patients with pneumonia. J Virol. 2005;79:884–895. doi: 10.1128/JVI.79.2.884-895.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Sheahan T, Rockx B, Donaldson E, Corti D, Baric R. Pathways of cross-species transmission of synthetically reconstructed zoonotic severe acute respiratory syndrome coronavirus. J Virol. 2008;82:8721–8732. doi: 10.1128/JVI.00818-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Rockx B, Sheahan T, Donaldson E, Harkema J, Sims A, Heise M, et al. Synthetic reconstruction of zoonotic and early human severe acute respiratory syndrome coronavirus isolates that produce fatal disease in aged mice. J Virol. 2007;81:7410–7423. doi: 10.1128/JVI.00505-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Chou S, Marousek GI. Accelerated evolution of maribavir resistance in a cytomegalovirus exonuclease domain II mutant. J Virol. 2008;82:246–253. doi: 10.1128/JVI.01787-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Bull RA, Eden JS, Rawlinson WD, White PA. Rapid evolution of pandemic noroviruses of the GII.4 lineage. PLoS Pathog. 2010;6:1000831. doi: 10.1371/journal.ppat.1000831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Lindesmith LC, Donaldson EF, Baric RS. Norovirus GII.4 Strain Antigenic Variation. J Virol. 2010 doi: 10.1128/JVI.01364-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Pfeiffer JK, Kirkegaard K. A single mutation in poliovirus RNA-dependent RNA polymerase confers resistance to mutagenic nucleotide analogs via increased fidelity. Proc Natl Acad Sci USA. 2003;100:7289–7294. doi: 10.1073/pnas.1232294100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Pfeiffer JK, Kirkegaard K. Ribavirin resistance in hepatitis C virus replicon-containing cell lines conferred by changes in the cell line or mutations in the replicon RNA. J Virol. 2005;79:2346–2355. doi: 10.1128/JVI.79.4.2346-2355.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]