Abstract
The conserved heterochromatic location of centromeres in higher eukaryotes suggests that intrinsic properties of heterochromatin are important for chromosome inheritance. Based on this hypothesis, mutations in Drosophila melanogaster that alter heterochromatin-induced gene silencing were tested for effects on chromosome inheritance. Here we describe the characterization of the Su(var)2-10 locus, initially identified as a Suppressor of Position-Effect Variegation. Su(var)2-10 is required for viability, and mutations cause both minichromosome and endogenous chromosome inheritance defects. Mitotic chromosomes are improperly condensed in mutants, and polytene chromosomes are structurally abnormal and disorganized in the nucleus. Su(var)2-10 encodes a member of the PIAS protein family, a group of highly conserved proteins that control diverse functions. SU(VAR)2-10 proteins colocalize with nuclear lamin in interphase, and little to no SU(VAR)2-10 is found on condensed mitotic chromosomes. SU(VAR)2-10 is present at some polytene chromosome telomeres, and FISH analyses in mutant polytene nuclei revealed defects in telomere clustering and telomere–nuclear-lamina associations. We propose that Su(var2-10 controls multiple aspects of chromosome structure and function by establishing/maintaining chromosome organization in interphase nuclei.
Keywords: Chromosome, condensation, inheritance, nuclear organization, position-effect variegation
Heterochromatin is an enigmatic component of higher eukaryotic genomes. The paucity of genes and the abundance of repetitive sequences in heterochromatin contribute to it being described as functionally inert. However, heterochromatin houses essential single copy genes (Liu et al. 1998) and the rDNA loci, the most highly transcribed genes in the genome (for review, see Williams and Robbins 1992). In addition, the centromere—the site of kinetochore formation, spindle attachment, and checkpoint control during mitosis and meiosis—is usually buried deep within heterochromatin. Elegant studies from a variety of organisms (Goday and Pimpinelli 1989; Bass et al. 1997) indicate that heterochromatin plays other important roles in chromosome inheritance. For example, heterochromatic homology is required for faithful homolog pairing and chromosome segregation during male and female meiosis in Drosophila (McKee and Karpen 1990; Dernburg et al. 1996b; Karpen et al. 1996).
Heterochromatin can silence the expression of genes that are normally found in euchromatin, resulting in a phenomenon known as position-effect variegation (PEV). PEV manifests as the mosaic or variegated expression of an affected locus owing to its abnormal juxtaposition near centric heterochromatin or telomeres (for review, see Wallrath 1998). Changes in the chromatin structure surrounding a variegating gene motivated the hypothesis that PEV is caused by spreading of the heterochromatic state into neighboring regions of euchromatin (for review, see Wakimoto 1998). However, PEV also occurs when heterochromatic associations produce large-scale alterations in the nuclear organization of chromosomes. For example, a large block of heterochromatin inserted into the Drosophila brown (bw) gene induces the mislocalization of the homologously paired normal copy of the bw locus to a nuclear region that contains centric heterochromatin (Csink and Henikoff 1996; Dernburg et al. 1996a). Similarly, the silencing of lymphoid-associated genes in mammalian B lymphocytes correlates with their association with Ikaros proteins in a region of interphase nuclei containing centric heterochromatin (Brown et al. 1997). These observations challenge the notion that variegated gene expression results simply from cis-spreading of the heterochromatic state, and demonstrate that nuclear organization also contributes to PEV (Weiler and Wakimoto 1995).
Loci that alter heterochromatin-induced gene silencing likely encode chromosomal proteins that control normal heterochromatic functions. One hundred or more Drosophila genes are predicted to modify PEV in trans (Suppressors and Enhancers of PEV, or Su(var)s and E(var)s (Reuter and Wolff 1981; Sinclair et al. 1989). Do modifiers of PEV encode proteins that also contribute to chromosome inheritance? The strongest data linking silencing loci to chromosome inheritance come from the fission yeast Schizosaccharomyces pombe. Three genes (Swi6, Clr4, and Rik1) originally identified from mutations that alleviated silencing were shown to encode centromere-associated proteins necessary for chromosome inheritance (Allshire et al. 1995). A small number of Drosophila PEV modifiers have been linked to chromosome inheritance functions (Wines and Henikoff 1992; Kellum and Alberts 1995; Fanti et al. 1998), including Heterochromatin Protein 1, which promotes accurate chromosome segregation in embryos and is required to prevent telomere fusions in diploid cells (Kellum and Alberts 1995; Fanti et al. 1998). Interestingly, SUV39H1, a human homolog of Drosophila SU(VAR)3-9 and S. pombe CLR4 proteins, binds centromeric regions on human metaphase chromosomes. SUV39H1 overexpression analyses and histone methyltransferase activity imply a role in assembling heterochromatic structures (Melcher et al. 2000; Rea et al. 2000). Nonetheless, the link between PEV modifiers and heterochromatin-mediated inheritance functions remains largely unexplored; to date, no comprehensive investigation of the relationship between PEV modifiers and inheritance has been reported for any multicellular eukaryote.
We have devised a method to screen Drosophila PEV modifiers systematically for roles in chromosome inheritance. J21A, one of a large collection of Dp(1;f)1187 (Dp1187) minichromosome deletion derivatives, is greatly reduced in overall size and contains only two-thirds of the DNA sequences required for normal centromere function (Murphy and Karpen 1995b; Sun et al. 1997). Previous studies demonstrated that J21A transmission is dominantly affected by mutations in many genes required for inheritance, whereas the inheritance of normal chromosomes is unaffected. J21A inheritance is sensitized to the reduced dosage of genes involved in different aspects of inheritance, including spindle components, antipoleward forces, condensation, replication, sister chromatid cohesion, and overall chromosome architecture (Murphy and Karpen 1995a; Cook et al. 1997; Lopez et al. 2000; Dobie et al. 2001). J21A has been used to screen for dominant effects of new mutations on transmission, bypassing the aneuploidy-induced lethality that can result when screening for recessive mutations that impact endogenous chromosome inheritance (Dobie et al. 2001). We have recently demonstrated that nearly half of Su(var) and E(var) mutations dominantly alter the transmission of J21A, suggesting that a large proportion of PEV modifier loci influence chromosome inheritance (H. Le, K. Donaldson, K. Cook, and G. Karpen, in prep.). Here we describe the cloning and characterization of one locus identified in this screen, Su(var)2-10. We demonstrate that Su(var)2-10 is required for proper chromosome structure and chromosome inheritance. Furthermore, we present protein localization data and cytological analyses that suggest that Su(var)2-10 controls chromosome structure and function, and other cellular tasks, by establishing and/or maintaining interphase chromosome organization within the nucleus.
Results
Su(var)2-10 mutations cause dominant reductions in sensitized minichromosome transmission
Su(var)2-10 was identified as a candidate chromosome inheritance locus in a pilot screen for dominant effects of PEV modifiers on the transmission of a sensitized Dp1187 minichromosome derivative, J21A (H. Le, K. Donaldson, K. Cook, and G. Karpen, in prep.). J21A shows reduced monosome transmission from parents to progeny (27%), in comparison to the normal 50% monosome transmission of full-length minichromosomes (Murphy and Karpen 1995b). Su(var)2-101 and Su(var)2-102 caused dominant zygotic and maternal reductions in J21A transmission. As shown in Table 1, Su(var)2-10 mutations exhibited weak but reproducible decreases in J21A transmission when the Su(var)2-10 mutation was inherited from the father (Table 1, left column, without transgene, rows 3 and 5). When Su(var)2-101 or Su(var)2-102 was inherited from heterozygous mothers, zygotic and maternal defects combined to reduce J21A transmission levels to 7% and 8%, respectively (Table 1, left column, without transgene, rows 2 and 4). Previous statistical analyses determined that J21A transmission rates of <22% or >37% differed significantly (p < 0.05) from the normal rate of 27% (Dobie et al. 2001). These data indicate that Su(var)2-10 is required early in development when maternal RNAs are being utilized, and also later in development when zygotic transcription is required. Surprisingly, raising the dose of Su(var)2-10+ to three copies using a transgene (Table 1, right column, with transgene, row 1) also reduced J21A transmission, indicating that cells are also sensitive to increases in Su(var)2-10 product.
Table 1.
J21A minichromosome transmission rates in Su(var)2-10 heterozygous females
| Genotype
|
− transgenea (% ± SD)
|
+ transgenea (% ± SD)
|
|---|---|---|
| +/+ | 27 ± 12 | 14 ± 9 |
| Su(var)2-101/+ | 7 ± 5 | 10 ± 7 |
| Su(var)2-101/+ b | 20 ± 10 | 30 ± 13 |
| Su(var)2-102/+ | 8 ± 4 | 23 ± 8 |
| Su(var)2-102/+ b | 16 ± 9 | 25 ± 14 |
Monosome transmission for the J21A minichromosome from individual females of the indicated genotype was measured as described in Materials and Methods, and the average percent transmission ±SD is shown.
The transgene construct is described and illustrated in Supplemental Material at www.genesdev.org.
Mutation chromosome inherited from the father, revealing only the zygotic defects caused by the mutation.
Su(var)2-10 is essential for the inheritance of endogenous chromosomes
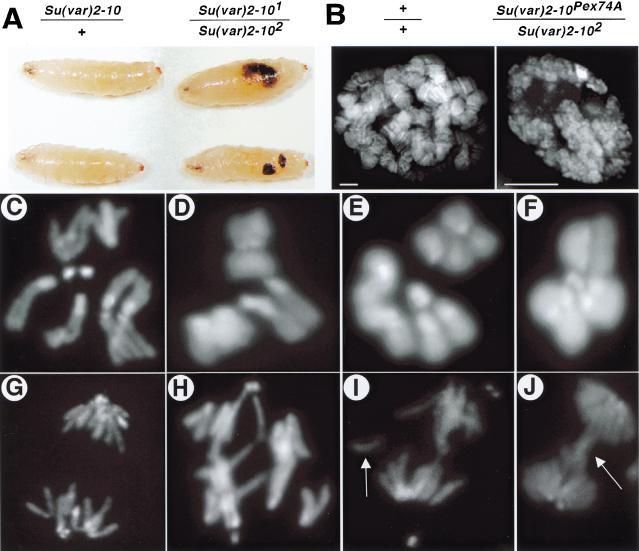
Data from deficiency mapping and lethality rescue experiments indicated that multiple lethal mutations exist along the Su(var)2-101 and Su(var)2-102 chromosomes. To determine if Su(var)2-10 is required for viability, we examined the development of Su(var)2-101/Su(var)2-102 and Su(var)2-102/Su(var)2-10Pex74a trans-heterozygous animals. [Su(var)2-10Pex74a is a deletion of the coding region; see Supplemental Material at www.genesdev.org.] These mutant animals died as late larvae and early pupae, and 3%–15% of Su(var)2-101/Su(var)2-102 larvae had melanotic tumors (Fig. 1A).
Figure 1.
Chromosome structure and function defects in Su(var)2-10 homozygotes. (A) Melanotic tumors are visible in homozygous third-instar larvae (right side), but not heterozygous larvae (left side). (B) Mutant salivary gland nuclei (right panel) are significantly smaller and contain disorganized polytene chromosomes in comparison to wild-type nuclei (left panel). Note the absence of banding in the mutant nuclei. Bar, 5 μm. (C–J) DAPI-stained mitotic chromosome preparations from larval neuroblasts. Top panels are metaphase (C–F), bottom panels are in anaphase (G–J). C and G are controls, the rest are Su(var)2-10 trans-heterozygotes (see Materials and Methods). Note the abnormal chromosome condensation in D—F, and the aberrant segregation (H), chromosome fragments (I, arrow), and anaphase bridges (J, arrow) in mutant cells. The magnification for all mitotic figures is 1250×.
Is Su(var)2-10 required for the inheritance of all chromosomes, or is the effect minichromosome-specific? The late lethal phase of Su(var)2-10 trans-heterozygotes allowed us to examine mitotically active neuroblast tissue from mutant third-instar larvae. Balancer chromosomes expressing Green Fluorescent Protein were used to distinguish Su(var)2-101/Su(var)2-102 larvae from their siblings. The structure and function of chromosomes in Su(var)2-10 mutants were grossly abnormal in both males and females. Two major types of chromosome defects were observed: abnormally condensed chromosomes in metaphase (Fig. 1D–F) and aberrantly segregating chromosomes in anaphase (Fig. 1G–J). Telomere fusions, characteristic of mutants for another PEV modifier, Su(var)2-5 (Fanti et al. 1998), were not observed in Su(var)2-10 mutants. Anaphase segregation defects included chromosome fragmentation (arrow, Fig. 1I) and bridging (arrow, Fig. 1J). Penetrance of the mitotic defects was temperature-sensitive (Table 2), and the percentage of mutant larvae with melanotic tumors also increased with temperature (from 15% at 25°C to 63% at 28°C). Judging normal condensation as the appearance of distinct sister chromatids in mitotic spreads, we found a slight but statistically significant increase in improperly condensed metaphases in control versus mutant neuroblasts at 25°C (22% to 35%, respectively; p < 0.002), but a very large increase at 28°C (18% versus 61%; p < 0.0001). Anaphase segregation defects were significantly elevated in Su(var)2-10 mutants (p < 0.0001 for both conditions), but did not show temperature sensitivity (18% at 25°C and 19% at 28°C; Table 2). The significantly elevated frequencies of mitotic defects and the recessive lethality displayed by homozygous mutants demonstrate that Su(var)2-10 is essential for the normal inheritance of all chromosomes, not just sensitized minichromosomes.
Table 2.
Endogenous chromosome defects in Su(var)2-10 mutants
|
|
+/+
|
Su(var)2-101/Su(var)2-102
|
||
|---|---|---|---|---|
| 25°C
|
28°C
|
25°C
|
28°C
|
|
| Mitotic index | 1.04 | 1.01 | 1.39 | 1.26 |
| Abnormally condensed metaphases (%) | 22 | 18 | 35 | 61 |
| Aberrant anaphases (%) | 2 | 2 | 18 | 19 |
The mitotic index, percentage of undercondensed metaphases, and percentage of aberrant anaphases are shown for the indicated genotypes at 25°C and 28°C. The number of brains scored for each data set was the following: +/+ at 25°C = 9; +/+ at 28°C = 5; Su(var)2-101/Su(var)2-102at 25°C = 13; Su(var)2-102/Su(var)2-102at 28°C = 8.
Su(var)2-10 mutations affect polytene chromosome structure
Many Drosophila larval tissues contain polytene cells. These interphase nuclei undergo multiple rounds of DNA replication without mitotic division, yielding large chromosomes that are amenable to high-resolution cytological analyses. To determine whether Su(var)2-10 controls general aspects of chromosome architecture, we analyzed polytene chromosome structure in whole-mount salivary gland nuclei from control and Su(var)2-102/Su(var)2-10Pex74a third instar larvae. Similar results were obtained for Su(var)2-101/Su(var)2-102 larvae, although the phenotypes were less severe. Mutant salivary glands were reduced in size, and the consistent banding pattern of normal polytene chromosomes was severely disrupted. (Fig. 1B). Examination of individual 0.2-μm optical sections from whole-mount salivary gland nuclei indicated that polytene chromosomes were generally disorganized and abnormally condensed (right panel, Fig. 1B). These chromosome structure defects are not simply a result of reduced nuclear size because comparably sized nuclei from younger wild-type larvae exhibited normal polytene chromosome structure (data not shown). In sum, Su(var)2-10 promotes proper chromosome structure in both polytene and diploid cells, and influences the organization of polytene chromosomes.
Molecular characterization of Su(var)2-10
Previous studies mapped the variegation phenotype of Su(var)2-10 to chromosome 2R (Wustmann et al. 1989). We deficiency-mapped the lethal phenotype to polytene region 45A, and complementation-tested a collection of P-element-induced lethal mutations localized to this area by the Berkeley Drosophila Genome Project (BDGP; Spradling et al. 1999). One insertion, P{PZ}l(2)0369703697, failed to complement Su(var)2-101 and Su(var)2-102 for lethality. Precise excisions of P{PZ}l(2)0369703697 reverted the lethal phenotype of the P insertion and complemented both Su(var)2-10 EMS alleles for lethality. These data demonstrate that P{PZ}l(2)0369703697 is a bona fide allele of Su(var)2-10, which we call Su(var)2-1003697. P{PZ}l(2)0369703697 is inserted into the zimp (zinc finger-containing, Miz1, PIAS3-like) transcription unit (Mohr and Boswell 1999), named for its similarity to known proteins (see below) and without knowledge of allelism to Su(var)2-10. The genetically based Su(var)2-10 nomenclature will be used here because of historical precedence (Reuter and Wolff 1981). We confirmed that Su(var)2-10 encodes this transcription unit by examining the structures and complementation behaviors of imprecise P-element excisions, by identifying single base mutations in the coding region for Su(var)2-101 and Su(var)2-102, and by transgene-mediated rescue of the lethality, variegation, and minichromosome transmission defects (see Table 1, Materials and Methods, and Supplemental Material at www.genesdev.org).
SU(VAR)2-10 is a member of the PIAS/Miz1/ARIP3 protein family
Analysis of cDNAs and ESTs demonstrated that Su(var) 2-10 is alternatively spliced to produce at least six different transcripts, whose presence in animals was confirmed by Northern analysis (data not shown). Conceptual translation of Su(var)2-10 transcripts yields at least four predicted protein products (522 aa, 537 aa, 554 aa, and 593 aa). SU(VAR)2-10 isoforms are identical across a 514-amino-acid domain, but differ in their COOH termini. Future studies on alternative splicing at the 5′ end of the transcripts may identify additional isoforms. Protein motif searches revealed two domains present in all of the SU(VAR)2-10 isoforms: a SAP domain at the extreme NH2 termini from aa 2 to 36, which is a putative DNA-binding domain found in diverse nuclear proteins (Aravind and Koonin 2000), and a zinc finger from aaa 325 to 367. SU(VAR)2-10 proteins are highly homologous to a diverse group of proteins that includes members of the Protein Inhibitor of Activated STAT (PIAS) family, which appear to be involved in transcriptional regulation (see below; Chung et al. 1997; Liu et al. 1998).
SU(VAR)2-10 proteins are localized in the nucleoplasm and are not concentrated along condensed mitotic chromosomes
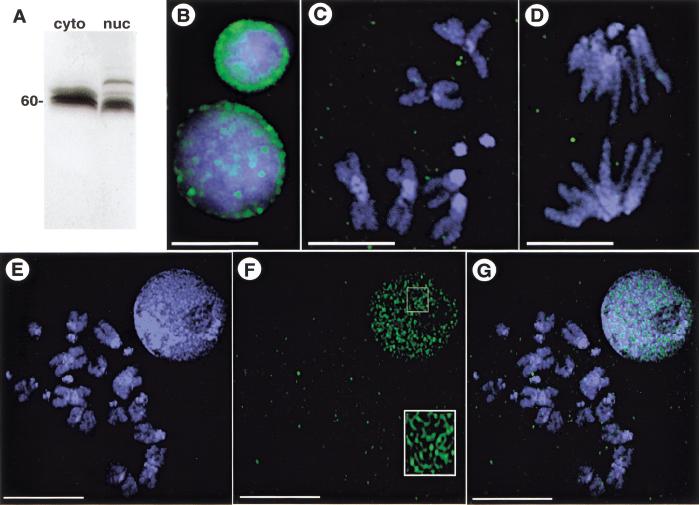
To test whether SU(VAR)2-10 proteins localize to chromosomes, antibodies that recognize all SU(VAR)2-10 isoforms were generated. Affinity-purified antibodies recognize a triplet at ∼60 kD in 0–12-h cytoplasmic extracts from wild-type embryos, and at least four bands in a nuclear extract from the same developmental stage, whose molecular weights are close to the sizes of the predicted isoforms (Fig. 2A).
Figure 2.
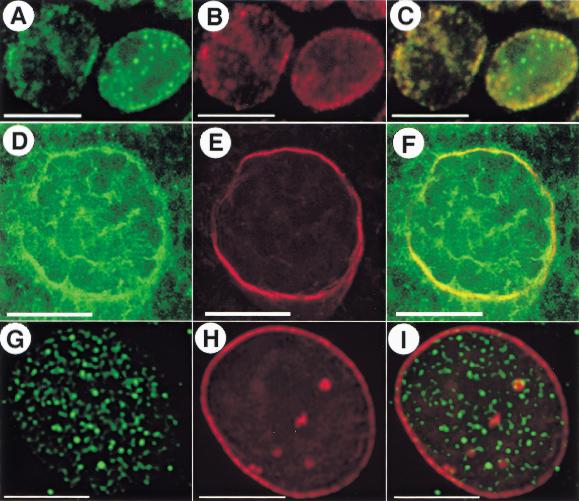
SU(VAR)2-10 localization in third-instar larval neuroblasts and S2 cells. (A) Western analysis of affinity-purified anti-SU(VAR)2-10. The antibodies recognize three distinct bands in a cytoplasmic extract (cyto) from 0–12-h embryos, and at least four bands in a nuclear extract (nuc) from the same developmental stage. (B–G) Indirect immunofluorescence showing SU(VAR)2-10 localization in mitotic cells. Green represents SU(VAR)2-10 protein, and blue DAPI-stained DNA. (B–D) In larval neuroblasts, SU(VAR)2-10 does not localize to chromosomes during metaphase (C) or anaphase (D), and is only prominent in interphase nuclei (B), where it is organized around the nuclear periphery and in numerous intranuclear spots. Bar, 5 μm. (E–G) Volume view of interphase and metaphase nuclei from S2 cells. (E) DNA alone, (F) SU(VAR)2-10 alone, and (G) the merged image. SU(VAR)2-10 antibodies stain interphase nuclei, but are not concentrated on metaphase chromosomes (F,G). Note the threadlike localization pattern in interphase cells (inset in F). Bar, 10 μm.
We examined SU(VAR)2-10 distribution in diverse cell types including early embryos, larval neuroblasts, S2 tissue culture cells, and polytenized larval salivary glands. Subcellular localization varied according to the phase of the cell cycle. During interphase in all cell types, SU(VAR)2-10 proteins are both cytoplasmic and nuclear, with concentrated staining near the nuclear periphery and in the nucleoplasm. For example, SU(VAR)2-10 proteins localized to the nuclear periphery in interphase neuroblast cells, and distinct nucleoplasmic foci were also observed (Fig. 2B). Surprisingly, SU(VAR)2-10 proteins did not localize to condensed metaphase or anaphase chromosomes in diploid nuclei from early embryos (data not shown) or larval neuroblast cells (Fig. 2C,D). Similarly, staining in S2 cells occurred in a punctate pattern in interphase, and metaphase chromosomes showed no distinct concentration of SU(VAR)2-10 (Fig. 2E–G). Interestingly, at higher magnifications the punctate interphase localization pattern of SU(VAR)2-10 appeared as a threadlike network (inset, Fig. 2F), but whether the network either overlapped with or contacted the DNA could not be determined. The absence of SU(VAR)2-10 antibody staining from mitotic chromosomes is not likely to be an artifact of the staining procedure because two different fixation protocols yielded the same results for larval neuroblast tissue (see Materials and Methods).
Given the striking hypocondensation and anaphase segregation defects in homozygous mutants, it is surprising that SU(VAR)2-10 proteins are predominantly found in interphase cells and not at specific sites along mitotic chromosomes. It is possible that our antibodies do not recognize chromosome-bound isoforms of SU(VAR)2-10, that an undetectable quantity of SU(VAR)2-10 is bound to chromosomes, or that SU(VAR)2-10 proteins are only transiently associated with chromosomes in the tissues we have analyzed. Regardless, the nucleoplasmic staining and concentration near the nuclear periphery in interphase nuclei suggest that SU(VAR)2-10 proteins perform essential cellular functions during interphase.
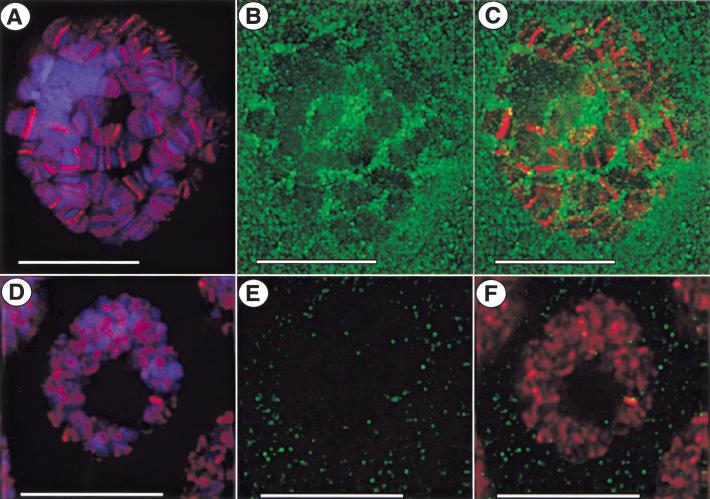
To further investigate the interphase distribution of SU(VAR)2-10 proteins, we examined localization in larval salivary glands, where polytenization allows for better visualization of interphase chromosome morphology than in diploid cells. Antibodies to the GAGA transcriptional activator, which binds SU(VAR)2-10 in two-hybrid assays (E. Andrulis and J. Lis, pers. comm.), were included as controls for the staining procedure. Figure 3 shows single, deconvolved optical sections from individual whole-mount nuclei. As expected, anti-GAGA factor antibodies bound distinct sites along polytene chromosomes (Fig. 3A). In these same nuclei, SU(VAR)2-10 staining was found in the cytoplasm, with more concentrated localization in the nucleoplasm (Fig. 3B). SU(VAR)2-10 staining was generally not found at specific chromosomal regions (Fig. 3C); however, weak chromosomal staining was observed. SU(VAR)2-10 proteins predominantly occupied interchromosomal spaces, and staining intensity was higher along the edges of the chromosomes.
Figure 3.
SU(VAR)2-10 does not overlap with GAGA factor in whole-mount larval salivary gland nuclei. Indirect immunofluorescence showing SU(VAR)2-10 and GAGA-factor localization in whole-mount salivary gland nuclei from third-instar larvae. In all panels, green is anti-SU(VAR)2-10 staining, red anti-GAGA staining, and blue DAPI-stained DNA. (A–C) Images from a single 0.2-μm optical section from the z-series stack of the deconvolved image. GAGA factor binds to discrete sites along polytene chromosomes (A), and does not overlap with anti-SU(VAR)2-10 staining (C). SU(VAR)2-10 is predominantly present in the interchromosomal spaces of the nucleoplasm and near the nuclear periphery, and weakly stains chromosomes (B). Bar, 15 μm. (D–F) SU(VAR)2-10 and GAGA-factor localization in whole-mount salivary gland nuclei from Su(var)2-101/Su(var)2-102 mutant larvae. (D) Nuclei are reduced in size and chromosome morphology is abnormal in mutant nuclei, yet GAGA factor is still able to associate with the DNA in a banded pattern. (E) SU(VAR)2-10 staining is decreased both in the cytoplasm and the nucleus. Bar, 10 μm.
In whole-mount salivary gland nuclei from Su(var)2-101/Su(var)2-102 mutant larvae (Fig. 3D–F), a dramatic decrease in the cytoplasmic levels of SU(VAR)2-10 was observed, and little if any anti-SU(VAR)2-10 staining was found in the nucleus (Fig. 3E). Although chromosome morphology and organization were disrupted in these nuclei, GAGA factor continued to associate with DNA in a banded pattern (Fig. 3D). Therefore, although SU(VAR)2-10 and GAGA factor may associate at some stage(s) in the cell cycle, these results suggest that proper Su(var)2-10 function is not absolutely required for GAGA factor to bind DNA in interphase.
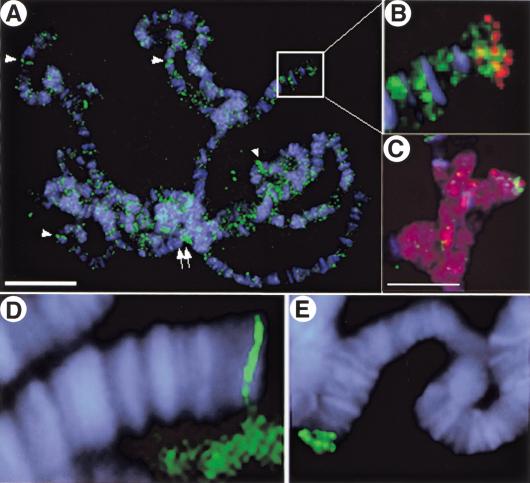
SU(VAR)2-10 proteins displayed a weak, punctate association with squashed polytene chromosomes (Fig. 4). The chromosomal staining patterns were variable; nonetheless, a small number of euchromatic bands were labeled (arrowheads, Fig. 4A–C), and the telomeric regions of chromosomes 2L (arrow, Fig. 4A,B) and 4 (Fig. 4C) were stained consistently. The heterochromatic chromocenter was also stained in these squashed preparations (double arrows, Fig. 4A). These staining patterns likely represent local areas of SU(VAR)2-10 binding that become apparent only after the high levels of cytoplasmic and nucleoplasmic SU(VAR)2-10 are removed by extraction and squashing. A second fixation protocol that removed the majority of the nucleoplasmic pool confirmed the striking telomeric localization for SU(VAR)2-10 proteins (Fig. 4D,E). SU(VAR)2-10 did not colocalize with Heterochromatin Protein 1 (HP1) at the telomere (Fig. 4B,C); the proteins occupied distinct subdomains at the end of chromosomes 2L and 4, with HP1 located more distally along the chromosome.
Figure 4.
SU(VAR)2-10 is generally associated with squashed polytene chromosomes and does not colocalize with HP1 at telomeres. Indirect immunofluorescence showing SU(VAR)2-10 (green) on squashed polytene chromosomes (DAPI-stained DNA is blue in all panels). (A) SU(VAR)2-10 associates with polytene chromosomes in a general punctate pattern. The chromocenter is labeled (arrow), as are some euchromatic bands (arrowheads), and the tips of chromosomes 2L (B) and 4 (C), Bar in A, 15 μm. (B) Magnified view of area boxed in A showing that SU(VAR)2-10 (green) does not colocalize with HP1 (red) at the tip of chromosome 2L. (C) HP1 (red) densely stains the chromocenter and chromosome 4, and SU(VAR)2-10 (green) is found near the tip of chromosome 4. Bar, 10 μm. (D,E) SU(VAR)2-10 proteins localize distinctly to telomeres.
SU(VAR)2-10 staining is closely associated with lamin around the nuclear periphery during interphase
SU(VAR)2-10 staining was closely associated with lamin staining around the nuclear periphery of interphase cells from neuroblasts and salivary glands (Fig. 5). In neuroblast nuclei, the proteins tightly colocalized around the nuclear periphery, and distinct spots of SU(VAR)2-10 staining were present in the nuclear lumen that did not colocalize with lamin (Fig. 5C). In salivary gland nuclei, portions of SU(VAR)2-10 colocalized with nuclear lamin, but the colocalization was sometimes variable (cf. Figs. 3B and 5F), most likely owing to differences in antibody penetration of the sheath that surrounds the whole-mount salivary glands. In S2 cells, where SU(VAR)2-10 localization near the nuclear periphery was less pronounced, SU(VAR)2-10 staining overlapped with lamin at only a few discrete foci (Fig. 5I). These differences may reflect tissue-specific distributions for the various SU(VAR)2-10 isoforms, and further experiments are needed to determine whether SU(VAR)2-10 and lamin proteins bind, and whether the association of these proteins is necessary for normal cellular function.
Figure 5.
SU(VAR)2-10 and lamin colocalize around the nuclear periphery in interphase neuroblasts and salivary gland cells. Indirect immunofluorescence showing simultaneous localization of SU(VAR)2-10 and nuclear lamin in interphase cells from third-instar larval neuroblasts (A–C), whole-mount salivary gland nuclei (D–F), and interphase S2 cells (G–I). In all panels, green is anti-SU(VAR)2-10 staining and red is anti-lamin staining. SU(VAR)2-10 and lamin are tightly colocalized in neuroblasts (A–C) and salivary gland cells (D–F), but the two do not colocalize in S2 cells (G–I). Note the SU(VAR)2-10-specific staining in the interior of neuroblast nuclei (A,C). (A–C,G–I) Bar, 5 μm. (D–F) Bar, 15 μm.
SU(VAR)2-10 isoforms influence chromosome condensation independently of transcription
The homology with PIAS proteins suggests that SU(VAR)2-10 isoforms may influence chromosome structure and function indirectly, perhaps by regulating the transcription of other genes that control chromosome structure and function. To address the potential role in transcription, SU(VAR)2-10 inhibition in syncytial Drosophila embryos was compared to a general block in transcription. Chromosome divisions were examined with real-time deconvolution microscopy, using embryos expressing a Histone–GFP fusion protein (Clarkson and Saint 1999).
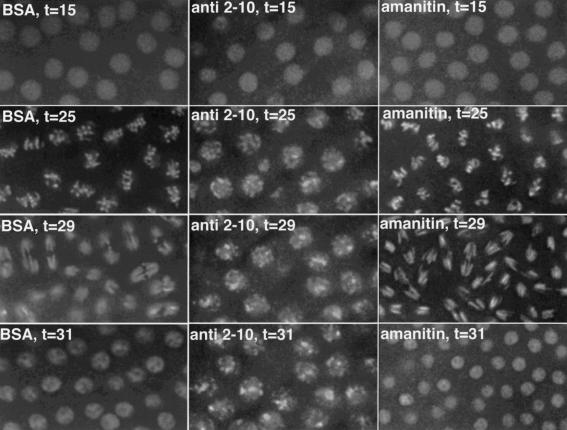
Affinity-purified, anti-SU(VAR)2-10 antibodies were injected into pre-cycle 13 embryos, and two divisions were filmed post-injection. Embryos were exquisitely sensitive to the concentration of the SU(VAR)2-10 antibodies. Injecting anti-SU(VAR)2-10 antibodies at a concentration of 10 mg/mL yielded massive chromosome condensation defects that eventually resulted in a prophase/prometaphase arrest (middle panels, Fig. 6). Embryos injected with anti-SU(VAR)2-10 antibodies at a concentration of 4 mg/mL showed major chromosome segregation defects, but no defects in condensation (data not shown). Importantly, the defects produced by disrupting SU(VAR)2-10 activity in early embryos precisely mimic the mitotic phenotypes generated by Su(var)2-10 mutations in third instar larvae (see above). Condensation defects were never observed in uninjected control embryos, or in control embryos injected with the BSA-supplemented filtrate from the antibody concentration procedure (left panels, Fig. 6; see Materials and Methods).
Figure 6.
SU(VAR)2-10 antibodies disrupt chromosome condensation and segregation in early embryos. Chromosome condensation and segregation are disrupted in embryos injected with anti-SU(VAR)2-10 antibodies (middle panels), but not in embryos injected with BSA (left panels) or α-amanitin (right panels), an RNA polymerase inhibitor. Embryos shown are at equivalent time points during the second mitotic cycle after injection. Time in minutes (t) postinjection is indicated for each panel. Note that at t = 25 metaphase chromosomes in the BSA- and α-amanitin-injected embryos are properly condensed and aligned at the metaphase plate. These nuclei divide and continue to cycle. During the second postinjection cycle in anti-SU(VAR)2-10-injected embryos, condensed metaphase chromosomes are never formed, and the mitotic cycles arrest prior to metaphase.
The transcription profile of early Drosophila development suggests that these defects are not caused by altering SU(VAR)2-10-mediated transcriptional activation. Zygotic transcription is dispensable until cycle 14 (Merrill et al. 1988; Wieschaus and Sweeton 1988), and although some zygotic RNAs can be visualized prior to cycle 13 (Zalokar 1976), the rapidity of these cycles likely precludes the production of functional transcripts (O'Farrell et al. 1989). Because our antibody injection experiments were performed on pre-cycle-13 embryos, it is unlikely that inhibition of zygotic transcription is responsible for the observed phenotypes. To address the role of zygotic transcription in generating the Su(var)2-10 embryonic defects, we examined embryos injected with α-amanitin, an effective RNA polymerase II inhibitor in Drosophila embryos (Edgar and Schubiger 1986). If SU(VAR)2-10 antibody injections caused the condensation and segregation defects via failed transcriptional activation, we would expect that a general inhibition of transcription would yield similar defects. However, amanitin injections yielded no defects in chromosome condensation or anaphase segregation (right panels, Fig. 6). We conclude that inhibiting SU(VAR)2-10 activity in early embryos directly results in chromosome structure and function defects that are not caused by the inability to express other genes. Although these data do not exclude a potential role for SU(VAR)2-10 in transcriptional repression of a gene whose misexpression causes the observed phenotypes, it is unlikely given the absence of transcription in early embryos.
Interphase chromosome organization is altered in Su(var)2-10 mutant nuclei
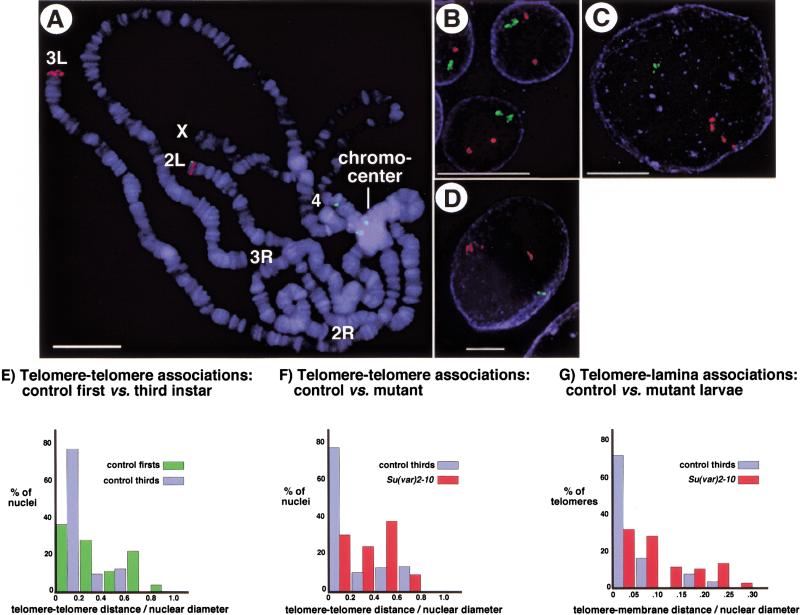
SU(VAR)2-10 protein localization and the pleiotropic mutant phenotypes suggest that this locus may play a global role in chromosome and nuclear function. To examine the role of SU(VAR)2-10 in nuclear organization directly, we monitored telomere–telomere and telomere–lamina associations in whole-mount salivary gland nuclei using Fluorescence In Situ Hybridization (FISH). Interphase chromosomes are often organized in a Rabl configuration where centromeres and telomeres lie at opposite ends of the nucleus (Dernburg 1995). Fluorescently labeled DNA probes specifically hybridized to centric and telomeric regions (Fig. 7A), and three-dimensional analyses showed that chromosomes in whole-mount nuclei from control, third instar larvae assume a Rabl configuration (Fig. 7C). The average distance between telomeric hybridization signals in wild-type third instar larvae was 5.5 μm, and nearly 80% of nuclei yielded a telomere distance : nuclear diameter ratio of <0.2 (blue bars, Fig. 7F). By contrast, nuclei from first instar larval glands showed a dramatic separation of telomeric signals, with 63% of nuclei exhibiting a telomere distance : nuclear diameter ratio >0.25 (Fig. 7B; green bars, Fig. 7F, p < 0.01). These results suggest that telomeres are not as tightly clustered during first instar developmental stages, and imply that telomere clustering in salivary gland nuclei occurs during later stages of larval development.
Figure 7.
Defects in telomere–telomere and telomere–lamina associations occur in Su(var)2-10 mutant cells. FISH analyses using centric (green) and telomeric (red) probes to analyze chromosome organization in interphase polytene nuclei are shown. (A) Squashed polytene chromosomes from larval salivary glands were hybridized with the DNA probes and counterstained with DAPI (blue) to visualize DNA. Centric probes bind the heterochromatic chromocenter as expected, and telomeric probes bind the tips of chromosomes 2L and 3L Bar, 15 μm. Telomeric hybridization signals are separated in whole-mount nuclei from first-instar larval salivary glands (B), but are more tightly associated in nuclei from third-instar larvae (C). Telomere–telomere and telomere–lamina associations are disrupted in Su(var)2-10 mutant nuclei (D). Blue represents nuclear lamin staining in B–D Bar, 10 μm. (E–G) Bar graphs representing telomere–telomere and telomere–lamina distances relative to nuclear diameter for wild-type first-instar (green bars) and third-instar (blue bars) larvae, and for Su(var)2-10 mutant larvae (red bars). Two-sample t-tests confirmed that the differences between control and mutant ratios were statistically significant (see text for p-values).
Telomere clustering was aberrant in Su(var)2-101/Su(var)2-102 third instar larvae (Fig. 7D). Two separated telomere hybridization signals were observed in 73% of mutant nuclei, compared to only 9% for wild-type controls. The average distance between telomeric hybridization signals in Su(var)2-10 mutants was 11.6 μm, and 70% of nuclei yielded a telomere distance : nuclear diameter ratio >0.25 (red bars, Fig. 7G, p < 0.001). Telomere clustering defects are not the result of arrested development at first instar stages because the average size of the Su(var)2-10 mutant nuclei analyzed in these experiments (30.96 μm) is similar to third instar control nuclei (29.84 μm; also cf. sizes in Fig. 7C,D). This analysis also revealed Su(var)2-10 mutant nuclei have aberrant associations between telomeres and the nuclear lamina (cf. red and blue bars, Fig. 7H). On average, telomeric hybridization signals in mutant nuclei were positioned 3.0 μm from the nuclear lamina with 26% of telomeres within 1 μm of the lamina (red bars, Fig. 7H). By contrast, the average telomere-to-lamina distance in wild-type controls was only 1.7 μm, and 72% of telomeres were within 1 μm of the nuclear lamina (blue bars, Fig. 7H, p < 0.01). We conclude that telomere–telomere and telomere–lamina associations are severely disrupted in Su(var)2-10 mutant polytene nuclei, demonstrating that SU(VAR)2-10 plays a role in the organization of chromosomes in the interphase nucleus.
Donaldson and Karpen have proposed that variegated gene expression from terminally deleted minichromosomes (e.g., the yellow+ gene in γ878) is linked to the juxtaposition of telomeric chromatin, which in turn influences nuclear organization and gene expression (Donaldson and Karpen 1997). Su(var)2-10 mutations dominantly suppress this terminal-deficiency-associated PEV (TDA-PEV; data not shown), which correlates with the telomere localization of the protein and mutant effects on telomere clustering and lamina interactions.
Discussion
The diverse phenotypes displayed by Su(var)2-10 mutants indicate that the gene plays multiple roles in the cell. Decreased minichromosome transmission and defective anaphase movements of endogenous chromosomes in embryos and neuroblasts demonstrate a role in chromosome inheritance. Its involvement in chromosome structure is demonstrated by condensation defects during metaphase in embryos and neuroblasts and in interphase polytene cells. The suppression of variegation phenotype indicates a role in gene expression, and the homology to the PIAS protein family and the presence of melanotic tumors in mutant larvae suggest that SU(VAR)2-10 functions in the JAK-STAT signal transduction pathway. In fact, studies performed recently by the Darnell lab have demonstrated genetic and biochemical interactions between Drosophila PIAS Su(var)2-10 and components of the JAK-STAT pathway (A. Betz and J.E. Darnell, pers. comm.). Surprisingly, SU(VAR)2-10 proteins are not localized to mitotic chromosomes and in interphase cells are localized to telomeres, the nuclear lamina, and the nuclear lumen. Lastly, the defects in telomere clustering and telomere–lamina associations and the mutant effects on TDA-PEV demonstrate a role for SU(VAR)2-10 in telomere function and nuclear organization. Lethality is most likely caused by a combination of these defects, and the various phenotypes may have a common underlying cause, namely, aberrant chromosome structure and nuclear organization.
Relating the SU(VAR)2-10 homologs to SU(VAR)2-10 function
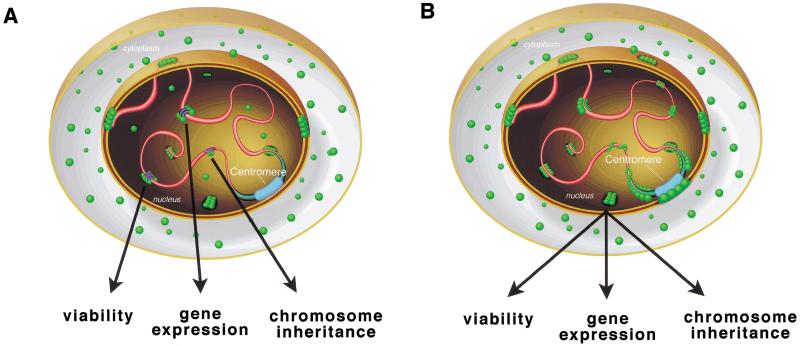
Data from these Drosophila studies may clarify the mode of action for mammalian PIAS proteins. Although PIAS1 was initially characterized as an inhibitor of STAT1-mediated gene activation (Liu et al. 1998), recent work indicates that PIAS proteins are not acting exclusively in the JAK-STAT pathway; PIAS1 also acts as a nuclear receptor coregulator for the androgen, glucocorticoid, and progesterone receptors (Tan et al. 2000). How the protein coordinates interactions with multiple binding partners is unclear. Similarly, SU(VAR)2-10 isoforms could act in concert with multiple transcriptional regulation complexes (Fig. 8A); however, a role in transcription does not exclude additional, more direct roles in chromosome inheritance. In Saccharomyces cerevisiae, CPF1 acts as a centromere-binding protein and a transcriptional regulator (Mellor et al. 1990). In addition, human Centromere Protein C (CENP-C) interacts with two nucleolar transcription factors, UBF1 and UBF2 (Pluta and Earnshaw 1996). These observations indicate that transcriptional regulation and chromosome inheritance can utilize common factors, and suggest that either the factors have dual function, or that the processes are linked by a common mechanism (e.g., the nuclear organization of chromosomes as discussed below).
Figure 8.
Models for SU(VAR)2-10 function. In a wild-type interphase cell, SU(VAR)2-10 proteins (green balls) are present in the cytoplasm and the nucleus, where the predominant localization observed is near the nuclear periphery and in interchromosomal spaces (pink tubes, chromosomes; blue tubes, heterochromatin; wide blue tube, centromere). (A) SU(VAR)2-10 isoforms may function in a variety of transcriptional regulation complexes, associating with a number of chromosome-bound transcription factors (dark blue and purple ovals), and each association may control a different cellular response (e.g., viability, gene expression, or chromosome inheritance). (B) SU(VAR)2-10 coordinates chromosome organization in interphase nuclei, ensuring normal viability, gene expression, and chromosome inheritance. Importantly, the two models presented here are not mutually exclusive.
Several observations argue against models for Su(var) 2-10 function based solely on transcriptional regulation. First, we have observed direct, transcription-independent effects on chromosome condensation and segregation in our antibody microinjection experiments. Similarly, one SU(VAR)2-10 ortholog, KChAP, binds voltage-gated potassium channel subunits and modulates channel currents; however, its ability to up-regulate currents does not require transcription (Kuryshev et al. 2000). Second, SU(VAR)2-10 proteins do not associate with polytene chromosomes in a banded pattern, which substantially differs from the localization of known transcription factors and cofactors, for example, GAGA factor, a transcriptional activator; and SMRTER (SMRT-related ecdysone receptor-interacting factor), a nuclear receptor coregulator (Tsai et al. 1999). Finally, our in vivo functional analyses of Su(var)2-10 suggest that PIAS homologs function in cellular processes beyond transcription, including chromosome structure, inheritance, and the organization of chromosomes in the nucleus. Regardless of whether these effects are direct or indirect, the exciting possibility that SU(VAR)2-10 and other PIAS proteins link signal transduction to chromosome behavior warrants further analysis.
SU(VAR)2-10 and the nuclear organization of chromosomes
We propose a parsimonious model that accounts for the behaviors of SU(VAR)2-10 and possibly other PIAS proteins. The primary role for SU(VAR)2-10 may be to promote normal chromosome organization in the interphase nucleus and generate/maintain the chromatin structures required for normal gene expression and chromosome inheritance (Fig. 8B). The organization of chromosomes in the interphase nucleus is a coordinated, nonrandom process. Three broad levels of nuclear architecture are generally consistent in diverse cell types (e.g., diploid versus polytene cells). First, chromosomes can be organized in a Rabl configuration with centromeres at one end of the nucleus and telomeres at the other (for review, see Dernburg 1995). Second, chromosomes are not intertwined within the nucleus; rather, they occupy distinct domains (Hochstrasser et al. 1986; Bridger and Bickmore 1998). Third, discrete chromosome regions are in close contact with the nuclear lamina (Marshall et al. 1996). Various chromosomal functions are associated with the nuclear lamina. The perinuclear localization of genes in S. cerevisiae can induce transcriptional silencing (Andrulis et al. 1998), and in Drosophila embryos, focal points for the initiation of chromosome condensation occur near the nuclear periphery (Hiraoka et al. 1989). Furthermore, interphase centromeres in mammalian cells and Drosophila embryos tend to cluster at the nuclear periphery and around nucleoli (for review, see Pluta et al. 1995). Therefore, nuclear organization defects in interphase could result in multiple chromosome abnormalities, including altered gene expression, defective chromosome condensation at prophase, and aberrant segregation in anaphase. All are phenotypes exhibited by Su(var)2-10 mutants. The strongest direct support for a nuclear organization role for Su(var)2-10 comes from the defective telomere–telomere and telomere–lamina interactions in Su(var)2-10 mutant nuclei. Consistent with a role in these processes, SU(VAR)2-10 proteins localize to telomeres and are concentrated at the nuclear periphery during interphase. The recent description of the SAP domain, an amino acid motif present in the human scaffold attachment factors SAF-A and SAF-B, Ku70 DNA repair proteins, and all SU(VAR)2-10 proteins, further implicates Su(var)2-10 in determining interphase chromosome organization (Aravind and Koonin 2000). The SAP domain of SU(VAR)2-10 isoforms may link specific chromosomal regions to the nuclear lamina; moreover, the transcriptional regulation properties assigned to PIAS family members may be the secondary consequence of a primary defect in interphase chromosome organization.
Here we have progressed from identifying a mutation that causes a chromosome transmission phenotype to a molecular entry into heterochromatin biology and the organization of chromosomes within the nucleus. Future studies of how Su(var)2-10 regulates chromosome structure and function will include identifying its binding partners and resolving the functions of the different isoforms. Categorizing the complexes in which SU(VAR)2-10 proteins are found will help determine its modes of action. More broadly, data generated from these studies will expand current views on the biological functions of the PIAS protein family, and how interphase chromosome architecture contributes to transcriptional regulation and chromosome inheritance.
Materials and methods
Genetic techniques
All genetic crosses were carried out at 25°C, and minichromosome transmission assays were performed as described previously (Cook et al. 1997). The two Su(var)2-10 EMS alleles were kindly provided by Gunter Reuter (Wustmann et al. 1989), and fly lines were obtained from the Bloomington and Umea Drosophila Stock Centers. Precise and imprecise excisions of the Su(var)2-1003697 P element were generated by genetically introducing a source of transposase to fly lines bearing the P element and selecting for loss of the rosy+ eye color marker. In all analyses presented, the control is our standard y1 ; ry506 strain.
Mitotic defects
Mitotic chromosome squashes were prepared from third-instar larval brain tissue as described in protocol 2 of Gatti et al. (1994), except neither hypotonic incubation nor colchicine treatment was used. Mitotic figures from mutants were compared to those from y1 ; ry506 control larvae. At least 50 fields per brain were scored for mitotic figures, where a field was defined as the region visible at 100× magnification with 1.25× optivar on a Zeiss Axiophot fluorescence microscope. Rare aneuploid metaphase figures in mutant brains were not included in quantitating the defects. The mitotic index (M.I.) was calculated as the total of metaphase plus anaphase figures divided by the total number of fields scored.
cDNA mapping and transformation rescue experiments
Plasmid rescue of genomic DNA flanking the Su(var)2-1003697 insertion yielded a 5-kb XhoI fragment that was used to probe an embryonic cDNA library (a gift from Carl Thummel). Six Su(var)2-10 cDNAs were isolated, and additional ESTs from the locus were identified from the BDGP database. The 5-kb XhoI fragment was also used to screen the BDGP P1 clones from polytene region 45A. A 15-kb EcoRI fragment from P1300 that contained the locus was subcloned into the pYES transformation vector (a gift from Pamela Geyer; Patton et al. 1992). Germ line transformation into y1 ; ry506 produced an X-chromosome transgene that was used in the lethality and minichromosome transmission rescue experiments. The element was remobilized to generate a third chromosome insertion used for analyzing rescue of the PEV suppression phenotype.
Analyzing the Su(var)2-10 alleles
To sequence Su(var)2-101 and Su(var)2-102, single-embryo PCR was performed using primers that span the coding region. DNA sequences for purified PCR products were determined by the Salk Institute DNA Sequencing Facility, using an ABI377 automated sequencer (Perkin Elmer), and sequences were analyzed using the Sequencher software package (Gene Codes). Sequences from Su(var) chromosomes that did not alter minichromosome transmission (H. Le, K. Donaldson, K. Cook, and G. Karpen, in prep.) and y1 ; ry506 were used as controls. A T → A mutation in Su(var)2-101 yields a leucine-to-methionine change in amino acid 327, and a G → A mutation in Su(var)2-102 yields a tryptophan-to-STOP change in amino acid 260. Su(var)2-1003697 excisions were defined as precise or imprecise based on complementation, Southern blot, and PCR analyses. Of 107 excisions, 31 were precise and 76 were imprecise. Four imprecise excisions, including Su(var)2-10Pex74a, failed to complement Su(var)2-101 for lethality and deleted 3′ into the coding region. Su(var)2-10Pex74a removes all of the Su(var)2-10 coding sequences (see Supplemental Material at www.genesdev.org).
Generating SU(VAR)2-10 antibodies
PCR was used to generate a 690-bp fragment (representing amino acids 124–354) from the middle portion of Su(var)2-10 that was cloned into a 6xHis-tagging vector (pQE-30, Qiagen). The tag was added to the N terminus of this 230-aa fragment. The fusion protein was expressed in Escherichia coli and purified over a Ni-NTA column. Three extra bands coeluted from the Ni-NTA column with the fusion protein; fractions containing the fusion protein were pooled and further purified over Mono-Q and Mono-S columns. All four bands coeluted from both columns. Western blotting with anti-RGS-HIS antibodies (Qiagen), which recognize the 6xHis tag bound to all four bands, indicated that these were degradation products from the carboxyl end of the 6xHis/SU(VAR)2-10 fusion protein. Preparative gels were run, and the full-length fusion protein was excised from these gels for use in raising Guinea Pig antibodies (Covance).
Western analysis
SU(VAR)2-10 antibodies were affinity-purified from Western blots using the 6xHis/SU(VAR)2-10 fusion protein. 10 μL of purified cytoplasmic and nuclear extracts from 0–12-h embryos (gifts from the lab of J. Kadonaga, UC San Diego) were electrophoresed through a 12.5% SDS-PAGE gel and transferred to PVDF membrane using a Bio-Rad electrophoretic transfer cell. The blot was probed with a 1 : 100 dilution of affinity-purified anti-SU(VAR)2-10 antibody, and a 1 : 5000 dilution of a horseradish peroxidase-conjugated Goat anti-Guinea Pig secondary antibody (Chemicon). Bands were visualized using ECL Western blotting detection reagents (Amersham Pharmacia Biotech).
Cytological analyses
Mitotic figures were analyzed using a Zeiss Axiophot fluorescence microscope equipped with a Princeton Instruments cooled CCD camera. Images were captured using IP Lab Spectrum Imaging Software (Scanalytics) and arranged using Adobe Photoshop. For antibody staining experiments, 0–4-h embryos were prepared and stained as described in Theurkauf (1994). Two protocols were used to prepare and stain squashed neuroblast tissue (Andrew and Scott 1994; Fanti et al. 1998). S2 cell (Invitrogen) maintenance and staining procedures are described in Blower and Karpen (2001). Whole-mount salivary glands were stained as in Goldberg et al. (1998), except that DNA was stained 5 min in 1-μg/mL DAPI. Squashed polytene chromosomes were prepared and stained as described in Platero et al. (1995) or Andrew and Scott (1994). Primary antibody dilutions were as follows: affinity-purified guinea pig anti-SU(VAR)2-10, 1 : 10; monoclonal anti-lamin , 1 : 10; monoclonal anti-HP1, 1 : 10; and affinity-purified rabbit anti-GAGA factor, 1 : 100. Goat anti-guinea pig-FITC, goat anti-mouse-Cy3 (Chemicon), and donkey anti-rabbit-Cy5 (Jackson ImmunoResearch Laboratories) secondaries were used at a 1 : 100 dilution. The monoclonal anti-lamin consisted of T47 (provided by S. Wasserman, UC San Diego) and T40 (provided by J. Sedat, UC San Francisco). The monoclonal anti-HP1 was C1A9, provided by B. Wakimoto (U. of Washington). The affinity-purified rabbit anti-GAGA factor came from the lab of J. Lis (Cornell University). Salivary gland defect and protein localization images were captured using a DeltaVision Optical Sectioning Microscope and were deconvolved and modeled using the DeltaVision softWoRx software package (Applied Precision).
FISH analyses for nuclear organization
Telomere probes for chromosomes 2L and 3L were generated by genomic PCR from the 457-bp subtelomeric repeat within the Taq minisatellite from Chromosome 2L (Walter et al. 1995). Single-stranded AACAC 40-mer oligonucleotides were used as probes for chromosome 2R-specific centric heterochromatin (Dernburg et al. 1996a). Telomeric and centric probes were fluorescently labeled as described in Dernburg et al. (1996a) using Cy3-dUTP (Amersham Pharmacia Biotech) and FITC-dUTP (Enzo Diagnostics), respectively. Larval salivary glands were isolated and fixed as above for whole-mount glands, and FISH was performed as described in Dernburg (1999), using 100 ng of each probe. Glands were stained with anti-lamin antibodies as above following the FISH procedure.
Antibody injections into early embryos
Microinjections were performed as described (Francis-Lang et al. 1999; Sharp et al. 2000), except that divisions were filmed using DeltaVision Optical Sectioning Microscopy (Applied Precision). Histone–GFP transgenic flies were developed by R. Saint (Clarkson and Saint 1999) and were provided by B. Sullivan (UC Santa Cruz). Affinity-purified anti-SU(VAR)2-10 antibodies in 1× PBS/50% glycerol were concentrated using Microcon YM-10 centrifugal filter devices (Millipore Corporation). BSA was added to the filtrate from this concentration procedure to a final concentration of 10 mg/mL, and this solution was injected as a control. Purity of the antibody injection solutions and BSA-supplemented control was verified by Coommassie gel. α-Amanitin was injected at 500 μg/mL in 1× PBS/50% glycerol.
Statistical analyses
Chi square tests were used to compare the percentages of mitotic defects in control versus mutant neuroblasts. Two-sample t-tests were performed to determine the statistical significance of observed differences between control and mutant telomere–telomere and telomere–lamina interactions. Means were compared assuming equal and unequal variances. Both methods produced the same p-value for all cases examined.
Acknowledgments
We thank Mike Blower, Cameron Kennedy, and Mike Wood for technical assistance; Jim Cordeiro and Bill Sullivan for assistance with embryo microinjections; Steve Wasserman and members of the Karpen lab for helpful comments on the manuscript; and Richard Gardner, Hiep Le, and Vivienne Velasco for essential guidance. We are grateful to Gunter Reuter and Barbara Wakimoto for providing the Su(var)2-10 EMS alleles; and to Erik Andrulis, Jim Darnell, Sue Parkhurst, James Darnell, and Judith Lengyel for discussing data prior to publication. Finally, we thank members from the labs of Pamela Geyer, Jim Kadonaga, John Lis, John Sedat, Carl Thummel, and Steve Wasserman for providing reagents used in these studies. This work was supported by NIH grant R01-GM54549 to G.H.K., an NIH Training Grant CA64041 and a Chappman Fellowship to K.L.H., and an American Cancer Society Postdoctoral Fellowship to K.R.C.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
Article and publication are at www.genesdev.org/cgi/doi/10.1101/gad.877901.
References
- Allshire RC, Nimmo ER, Ekwall K, Javerzat JP, Cranston G. Mutations derepressing silent centromeric domains in fission yeast disrupt chromosome segregation. Genes & Dev. 1995;9:218–233. doi: 10.1101/gad.9.2.218. [DOI] [PubMed] [Google Scholar]
- Andrew DJ, Scott MP. Immunological methods for mapping protein distributions on polytene chromosomes. Methods Cell Biol. 1994;44:353–370. doi: 10.1016/s0091-679x(08)60923-1. [DOI] [PubMed] [Google Scholar]
- Andrulis ED, Neiman AM, Zappulla DC, Sternglanz R. Perinuclear localization of chromatin facilitates transcriptional silencing. Nature. 1998;394:592–595. doi: 10.1038/29100. [DOI] [PubMed] [Google Scholar]
- Aravind L, Koonin EV. SAP—A putative DNA-binding motif involved in chromosomal organization. Trends Biochem Sci. 2000;25:112–114. doi: 10.1016/s0968-0004(99)01537-6. [DOI] [PubMed] [Google Scholar]
- Bass HW, Marshall WF, Sedat JW, Agard DA, Cande WZ. Telomeres cluster de novo before the initiation of synapsis: A three-dimensional spatial analysis of telomere positions before and during meiotic prophase. J Cell Biol. 1997;137:5–18. doi: 10.1083/jcb.137.1.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blower, M.D. and Karpen, G.H. 2001. The role of Drosophila CENP-A/CID in kinetochore formation, cell-cycle progression and interactions with heterochromatin. Nature Genetics (in press). [DOI] [PMC free article] [PubMed]
- Bridger JM, Bickmore WA. Putting the genome on the map. Trends Genet. 1998;14:403–409. doi: 10.1016/s0168-9525(98)01572-8. [DOI] [PubMed] [Google Scholar]
- Brown KE, Guest SS, Smale ST, Hahm K, Merkenschlager M, Fisher AG. Association of transcriptionally silent genes with Ikaros complexes at centromeric heterochromatin. Cell. 1997;91:845–854. doi: 10.1016/s0092-8674(00)80472-9. [DOI] [PubMed] [Google Scholar]
- Chung CD, Liao J, Liu B, Rao X, Jay P, Berta P, Shuai K. Specific inhibition of Stat3 signal transduction by PIAS3. Science. 1997;278:1803–1805. doi: 10.1126/science.278.5344.1803. [DOI] [PubMed] [Google Scholar]
- Clarkson M, Saint R. A His2AvDGFP fusion gene complements a lethal His2AvD mutant allele and provides an in vivo marker for Drosophila chromosome behavior. DNA Cell Biol. 1999;18:457–462. doi: 10.1089/104454999315178. [DOI] [PubMed] [Google Scholar]
- Cook KR, Murphy TD, Nguyen TC, Karpen GH. Identification of trans-acting genes necessary for centromere function in Drosophila melanogaster using centromere-defective minichromosomes. Genetics. 1997;145:737–747. doi: 10.1093/genetics/145.3.737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Csink AK, Henikoff S. Genetic modification of heterochromatic association and nuclear organization in Drosophila. Nature. 1996;381:529–531. doi: 10.1038/381529a0. [DOI] [PubMed] [Google Scholar]
- Dernburg AF. Fluorescence in situ hybridization in whole-mount tissues. In: Bickmore WA, editor. Chromosome structural analysis: A practical approach. New York: Oxford University Press; 1999. pp. 125–145. [Google Scholar]
- Dernburg AF, Sedat JW, Cande WZ, Bass HW. Cytology of telomeres. In: Blackburn EH, Greider CW, editors. Telomeres. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1995. pp. 295–338. [Google Scholar]
- Dernburg AF, Broman KW, Fung JC, Marshall WF, Philips J, Agard DA, Sedat JW. Perturbation of nuclear architecture by long-distance chromosome interactions. Cell. 1996a;85:745–759. doi: 10.1016/s0092-8674(00)81240-4. [DOI] [PubMed] [Google Scholar]
- Dernburg AF, Sedat JW, Hawley RS. Direct evidence of a role for the heterochromatin in meiotic chromosome segregation. Cell. 1996b;86:135–146. doi: 10.1016/s0092-8674(00)80084-7. [DOI] [PubMed] [Google Scholar]
- Dobie KW, Kennedy CK, Velasco VM, McGrath TL, Weko J, Patterson RW, Karpen GH. Identification of chromosome inheritance modifiers in Drosophila melanogaster. Genetics. 2001;157:1623–1637. doi: 10.1093/genetics/157.4.1623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donaldson KM, Karpen GH. Trans-suppression of terminal deficiency-associated position effect variegation in a Drosophila minichromosome. Genetics. 1997;145:325–337. doi: 10.1093/genetics/145.2.325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar BA, Schubiger G. Parameters controlling transcriptional activation during early Drosophila development. Cell. 1986;44:871–877. doi: 10.1016/0092-8674(86)90009-7. [DOI] [PubMed] [Google Scholar]
- Fanti L, Giovinazzo G, Berloco M, Pimpinelli S. The heterochromatin protein 1 prevents telomere fusions in Drosophila. Mol Cell. 1998;2:527–538. doi: 10.1016/s1097-2765(00)80152-5. [DOI] [PubMed] [Google Scholar]
- Francis-Lang H, Minden J, Sullivan W, Oegema K. Live confocal analysis with fluorescently labeled proteins. Methods Mol Biol. 1999;122:223–239. doi: 10.1385/1-59259-722-x:223. [DOI] [PubMed] [Google Scholar]
- Gatti M, Bonaccorsi S, Pimpinelli S. Looking at Drosophila mitotic chromosomes. Methods Cell Biol. 1994;44:371–391. doi: 10.1016/s0091-679x(08)60924-3. [DOI] [PubMed] [Google Scholar]
- Goday C, Pimpinelli S. Centromere organization in meiotic chromosomes of Parascaris univalens. Chromosoma. 1989;98:160–166. doi: 10.1007/BF00329679. [DOI] [PubMed] [Google Scholar]
- Goldberg M, Lu H, Stuurman N, Ashery-Padan R, Weiss AM, Yu J, Bhattacharyya D, Fisher PA, Gruenbaum Y, Wolfner MF. Interactions among Drosophila nuclear envelope proteins lamin, otefin, and YA. Mol Cell Biol. 1998;18:4315–4323. doi: 10.1128/mcb.18.7.4315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiraoka Y, Minden JS, Swedlow JR, Sedat JW, Agard DA. Focal points for chromosome condensation and decondensation revealed by three-dimensional in vivo time-lapse microscopy. Nature. 1989;342:293–296. doi: 10.1038/342293a0. [DOI] [PubMed] [Google Scholar]
- Hochstrasser M, Mathog D, Gruenbaum Y, Saumweber H, Sedat JW. Spatial organization of chromosomes in the salivary gland nuclei of Drosophila melanogaster. J Cell Biol. 1986;102:112–123. doi: 10.1083/jcb.102.1.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karpen GH, Le MH, Le H. Centric heterochromatin and the efficiency of achiasmate disjunction in Drosophila female meiosis. Science. 1996;273:118–122. doi: 10.1126/science.273.5271.118. [DOI] [PubMed] [Google Scholar]
- Kellum R, Alberts BM. Heterochromatin protein 1 is required for correct chromosome segregation in Drosophila embryos. J Cell Sci. 1995;108:1419–1431. doi: 10.1242/jcs.108.4.1419. [DOI] [PubMed] [Google Scholar]
- Kuryshev YA, Gudz TI, Brown AM, Wible BA. KChAP as a chaperone for specific K(+) channels. Am J Physiol Cell Physiol. 2000;278:C931–C941. doi: 10.1152/ajpcell.2000.278.5.C931. [DOI] [PubMed] [Google Scholar]
- Liu B, Liao J, Rao X, Kushner SA, Chung CD, Chang DD, Shuai K. Inhibition of Stat1-mediated gene activation by PIAS1. Proc Natl Acad Sci. 1998;95:10626–10631. doi: 10.1073/pnas.95.18.10626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez JM, Karpen GH, Orr-Weaver TL. Sister-chromatid cohesion via MEI-S332 and kinetochore assembly are separable functions of the Drosophila centromere. Curr Biol. 2000;10:997–1000. doi: 10.1016/s0960-9822(00)00650-3. [DOI] [PubMed] [Google Scholar]
- Marshall WF, Dernburg AF, Harmon B, Agard DA, Sedat JW. Specific interactions of chromatin with the nuclear envelope: Positional determination within the nucleus in Drosophila melanogaster. Mol Biol Cell. 1996;7:825–842. doi: 10.1091/mbc.7.5.825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKee BD, Karpen GH. Drosophila ribosomal RNA genes function as an x-y pairing site during male meiosis. Cell. 1990;61:61–72. doi: 10.1016/0092-8674(90)90215-z. [DOI] [PubMed] [Google Scholar]
- Melcher M, Schmid M, Aagaard L, Selenko P, Laible G, Jenuwein T. Structure–function analysis of SUV39H1 reveals a dominant role in heterochromatin organization, chromosome segregation, and mitotic progression. Mol Cell Biol. 2000;20:3728–3741. doi: 10.1128/mcb.20.10.3728-3741.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mellor J, Jiang W, Funk M, Rathjen J, Barnes CA, Hinz T, Hegemann JH, Philippsen P. CPF1, a yeast protein which functions in centromeres and promoters. EMBO J. 1990;9:4017–4026. doi: 10.1002/j.1460-2075.1990.tb07623.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merrill PT, Sweeton D, Wieschaus E. Requirements for autosomal gene activity during precellular stages of Drosophila melanogaster. Development. 1988;104:495–509. doi: 10.1242/dev.104.3.495. [DOI] [PubMed] [Google Scholar]
- Mohr SE, Boswell RE. Zimp encodes a homologue of mouse miz1 and PIAS3 and is an essential gene in Drosophila melanogaster. Gene. 1999;229:109–116. doi: 10.1016/s0378-1119(99)00033-5. [DOI] [PubMed] [Google Scholar]
- Murphy TD, Karpen GH. Interactions between the nod+ kinesin-like gene and extracentromeric sequences are required for transmission of a Drosophila minichromosome. Cell. 1995a;81:139–148. doi: 10.1016/0092-8674(95)90378-x. [DOI] [PubMed] [Google Scholar]
- ————— Localization of centromere function in a Drosophila minichromosome. Cell. 1995b;82:599–609. doi: 10.1016/0092-8674(95)90032-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Farrell PH, Edgar BA, Lakich D, Lehner CF. Directing cell division during development. Science. 1989;246:635–640. doi: 10.1126/science.2683080. [DOI] [PubMed] [Google Scholar]
- Patton JS, Gomes XV, Geyer PK. Position-independent germline transformation in Drosophila using a cuticle pigmentation gene as a selectable marker. Nucleic Acids Res. 1992;20:5859–5860. doi: 10.1093/nar/20.21.5859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Platero JS, Hartnett T, Eissenberg JC. Functional analysis of the chromo domain of HP1. EMBO J. 1995;14:3977–3986. doi: 10.1002/j.1460-2075.1995.tb00069.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pluta AF, Earnshaw WC. Specific interaction between human kinetochore protein CENP-C and a nucleolar transcriptional regulator. J Biol Chem. 1996;271:18767–18774. doi: 10.1074/jbc.271.31.18767. [DOI] [PubMed] [Google Scholar]
- Pluta AF, Mackay AM, Ainsztein AM, Goldberg IG, Earnshaw WC. The centromere: Hub of chromosomal activities. Science. 1995;270:1591–1594. doi: 10.1126/science.270.5242.1591. [DOI] [PubMed] [Google Scholar]
- Rea S, Eisenhaber F, O'Carroll D, Strahl BD, Sun ZW, Schmid M, Opravil S, Mechtler K, Ponting CP, Allis CD, et al. Regulation of chromatin structure by site-specific histone H3 methyltransferases. Nature. 2000;406:593–599. doi: 10.1038/35020506. [DOI] [PubMed] [Google Scholar]
- Reuter G, Wolff I. Isolation of dominant suppressor mutations for position-effect variegation in Drosophila melanogaster. Mol Gen Genet. 1981;182:516–519. doi: 10.1007/BF00293947. [DOI] [PubMed] [Google Scholar]
- Sharp DJ, Brown HM, Kwon M, Rogers GC, Holland G, Scholey JM. Functional coordination of three mitotic motors in Drosophila embryos. Mol Biol Cell. 2000;11:241–253. doi: 10.1091/mbc.11.1.241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinclair DA, Lloyd VK, Grigliatti TA. Characterization of mutations that enhance position-effect variegation in Drosophila melanogaster. Mol Gen Genet. 1989;216:328–333. doi: 10.1007/BF00334372. [DOI] [PubMed] [Google Scholar]
- Spradling AC, Stern D, Beaton A, Rhem EJ, Laverty T, Mozden N, Misra S, Rubin GM. The Berkeley Drosophila Genome Project gene disruption project. Single P-element insertions mutating 25% of vital Drosophila genes. Genetics. 1999;153:135–177. doi: 10.1093/genetics/153.1.135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun X, Wahlstrom J, Karpen G. Molecular structure of a functional Drosophila centromere. Cell. 1997;91:1007–1019. doi: 10.1016/s0092-8674(00)80491-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan J, Hall SH, Hamil KG, Grossman G, Petrusz P, Liao J, Shuai K, French FS. Protein inhibitor of activated STAT-1 (signal transducer and activator of transcription-1) is a nuclear receptor coregulator expressed in human testis. Mol Endocrinol. 2000;14:14–26. doi: 10.1210/mend.14.1.0408. [DOI] [PubMed] [Google Scholar]
- Theurkauf WE. Immunofluorescence analysis of the cytoskeleton during oogenesis and early embryogenesis. Methods Cell Biol. 1994;44:489–505. doi: 10.1016/s0091-679x(08)60928-0. [DOI] [PubMed] [Google Scholar]
- Tsai CC, Kao HY, Yao TP, McKeown M, Evans RM. SMRTER, a Drosophila nuclear receptor coregulator, reveals that EcR-mediated repression is critical for development. Mol Cell. 1999;4:175–186. doi: 10.1016/s1097-2765(00)80365-2. [DOI] [PubMed] [Google Scholar]
- Wakimoto BT. Beyond the nucleosome: Epigenetic aspects of position-effect variegation in Drosophila. Cell. 1998;93:321–324. doi: 10.1016/s0092-8674(00)81159-9. [DOI] [PubMed] [Google Scholar]
- Wallrath LL. Unfolding the mysteries of heterochromatin. Curr Opin Genet Dev. 1998;8:147–153. doi: 10.1016/s0959-437x(98)80135-4. [DOI] [PubMed] [Google Scholar]
- Walter MF, Jang C, Kasravi B, Donath J, Mechler BM, Mason JM, Biessmann H. DNA organization and polymorphism of a wild-type Drosophila telomere region. Chromosoma. 1995;104:229–241. doi: 10.1007/BF00352254. [DOI] [PubMed] [Google Scholar]
- Weiler KS, Wakimoto BT. Heterochromatin and gene expression in Drosophila. Annu Rev Genet. 1995;29:577–605. doi: 10.1146/annurev.ge.29.120195.003045. [DOI] [PubMed] [Google Scholar]
- Wieschaus E, Sweeton D. Requirements for X-linked zygotic gene activity during cellularization of early Drosophila embryos. Development. 1988;104:483–493. doi: 10.1242/dev.104.3.483. [DOI] [PubMed] [Google Scholar]
- Williams SM, Robbins LG. Molecular genetic analysis of Drosophila rDNA arrays. Trends Genet. 1992;8:335–340. doi: 10.1016/0168-9525(92)90277-b. [DOI] [PubMed] [Google Scholar]
- Wines DR, Henikoff S. Somatic instability of a Drosophila chromosome. Genetics. 1992;131:683–691. doi: 10.1093/genetics/131.3.683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wustmann G, Szidonya J, Taubert H, Reuter G. The genetics of position-effect variegation modifying loci in Drosophila melanogaster. Mol Gen Genet. 1989;217:520–527. doi: 10.1007/BF02464926. [DOI] [PubMed] [Google Scholar]
- Zalokar M. Autoradiographic study of protein and RNA formation during early development of Drosophila eggs. Dev Biol. 1976;49:425–437. doi: 10.1016/0012-1606(76)90185-8. [DOI] [PubMed] [Google Scholar]