Abstract
Previous studies suggest that the amino-terminal ubiquitin-like (ubl) domain of Rad23 protein can recruit the proteasome for a stimulatory role during nucleotide excision repair in the yeast Saccharomyces cerevisiae. In this report, we show that the 19S regulatory complex of the yeast proteasome can affect nucleotide excision repair independently of Rad23 protein. Strains with mutations in 19S regulatory subunits (but not 20S subunits) of the proteasome promote partial recovery of nucleotide excision repair in vivo in rad23 deletion mutants, but not in other nucleotide excision repair-defective strains tested. In addition, a strain that expresses a temperature-degradable ATPase subunit of the 19S regulatory complex manifests a dramatically increased rate of nucleotide excision repair in vivo. These data indicate that the 19S regulatory complex of the 26S proteasome can negatively regulate the rate of nucleotide excision repair in yeast and suggest that Rad23 protein not only recruits the 19S regulatory complex, but also can mediate functional interactions between the 19S regulatory complex and the nucleotide excision repair machinery. The 19S regulatory complex of the yeast proteasome functions in nucleotide excision repair independent of proteolysis.
Keywords: Rad23 protein, DNA repair, Saccharomyces cerevisiae, proteasome
Nucleotide excision repair (NER) promotes the removal of various types of bulky base damage from DNA by a multistage process involving ∼30 different proteins (for review, see Friedberg et al. 1995). The majority of these proteins are highly conserved from yeast to man (for review, see Friedberg et al. 1995). Mutations in NER genes lead to hypersensitivity to killing as well as hypermutability following exposure to DNA-damaging agents such as ultraviolet (UV) radiation. Defective NER in humans predisposes to skin cancer following sunlight exposure, as exemplified by the hereditary NER-defective disease xeroderma pigmentosum (XP). The regulation of NER in living cells is therefore a question of both fundamental and clinical interest. In this study, we demonstrate novel functional relationships between NER and the proteasome in the yeast Saccharomyces cerevisiae.
Rad23 protein is one of the multiple proteins involved in NER in S. cerevisiae (Friedberg et al. 1995). The precise function of this protein in this process is not clear. Extracts from cells deleted of the RAD23 gene do not support detectable NER in vitro (Wang et al. 1997; Russell et al. 1999). Such mutants, however, display a level of UV radiation sensitivity that is intermediate between that of wild-type strains and strains deleted for other RAD genes that are indispensable for NER, such as RAD1, RAD2, RAD3, etc. (Watkins et al. 1993; Mueller and Smerdon 1996). In some studies, this intermediate UV radiation sensitivity has been correlated with a partial decrease in NER in vivo (Mueller and Smerdon 1996).
Human cells possess two homologs of the yeast RAD23 gene, designated HHR23A and HHR23B (Masutani et al. 1994). HHR23B protein binds tightly to human XPC protein and stimulates the rate of NER in vitro (Masutani et al. 1994, 1997; Li et al. 1997). On the other hand, deletion of the mouse HHRAD23A or HHRAD23B genes does not result in increased sensitivity to UV radiation in mouse embryo fibroblasts (Friedberg and Meira 2000). Remarkably, mice deleted of the HHRAD23B gene show defective post-natal growth and HHRAD23A HHRAD23B double deletion mutants are inviable (Friedberg and Meira 2000). These observations suggest the existence of as yet unidentified essential functions of HHRAD23 protein, which is partially redundant between the A and B forms.
Levels of human HHRAD23A protein are regulated in a cell cycle-dependent manner (Kumar et al. 1999). In addition, the yeast RAD23 gene is suggested to have a role in cell cycle progression, which is redundant with the ubiquitin-like (ubl) protein DSK2, as loss of both genes results in a temperature-dependent block in spindle pole body duplication (Biggins et al. 1996).
No catalytic function has been associated with yeast Rad23 protein. The protein forms a tight complex with Rad4 protein (a protein with limited amino-acid sequence homology to human XPC protein) through its carboxy-terminal region (Guzder et al. 1995; Wang et al. 1997; Schauber et al. 1998) and the Rad23/Rad4 complex has been shown to preferentially bind UV-irradiated DNA (Guzder et al. 1998, 1999; Jansen et al. 1998). The amino-terminal region of Rad23 protein contains a ubl domain that is required for optimal levels of NER in vivo. Deletion of this domain results in a UV radiation-sensitive phenotype that is intermediate between that of wild-type and rad23 deletion strains (Watkins et al. 1993). Recent studies have shown that the ubl domain is required for a physical interaction between Rad23 protein and the 26S proteasome in yeast (Schauber et al. 1998; Russell et al. 1999).
The 26S proteasome is a large protein complex involved in the degradation of proteins targeted by the ubiquitin pathway. The complex is a heterotrimer, comprised of a 20S core particle and two copies of a 19S regulatory complex (19S-RC) (for review, see Baumeister et al. 1998; Coux et al. 1996). The 20S complex confers the proteolytic activities of the proteasome, whereas the 19S-RC confers ATP-dependence and specificity for ubiquitin protein conjugates. The 19S-RC is comprised of at least 18 subunits, including six ATPases (Rpt1–Rpt6) belonging to the AAA (ATPases associated with a variety of cellular activities) family, as well as the non-ATPase subunits Rpn1, Rpn2 and Rpn10 (Glickman et al. 1999).
The AAA ATPases are believed to facilitate unwinding of protein substrates to permit their passage through the proteolytic chamber of the proteasome (Rubin and Finley 1995; Weissman et al. 1995). It has been suggested that this activity may also be adapted for the disassembly or rearrangement of protein complexes without proteolysis (Neuwald et al. 1999; Russell et al. 1999). Escherichia coli contains an orthologous protein designated ClpX, which is a member of the AAA superfamily, and has both a regulatory role in proteolysis and a non-proteolytic role in the disassembly of a protein–DNA complex during lysogeny of bacteriophage Mu (Mhammedi-Alaoui et al. 1994; Levchenko et al. 1995; Jones et al. 1998). Recently, the AAA domain of the yeast Yme1 protein was shown to possess a chaperone-like activity, further supporting a non-proteolytic function of the 19S-RC (Leonhard et al. 1999).
Previous studies from our laboratories show that antibodies specific for the 19S-RC AAA ATPase subunit Sug1 can inhibit NER in vitro. In addition, we showed that a physical interaction between the ubl domain of Rad23 protein and the 26S proteasome is required for optimal NER in vitro (Russell et al. 1999) and that this requirement does not involve the proteolytic functions of the 26S proteasome, but is rather dependent on a novel, as yet unidentified function of the 19S-RC (Russell et al. 1999). These observations prompted speculation of the involvement of the 19S-RC during NER in yeast by a chaperone-like or protein complex remodeling activity that may modulate the rate or efficiency of NER in vivo (Russell et al. 1999).
In this report, we provide new evidence for involvement of 19S-RC ATPases in regulating the dynamics of NER in living cells in a manner that has no requirement for proteolysis. Specifically, we show that alleles carrying point mutations in the conserved ATPase domain of several subunits of the 19S-RC alter the rate of NER in vivo. These mutant alleles also result in partial suppression of the UV radiation-sensitive and NER-defective phenotype of rad23 deletion mutants, demonstrating a Rad23-independent effect of the proteasome on NER. Finally, we show that alleles completely suppress the NER defect associated with deletion of the ubl domain of rad23, but have no effect on the NER defect of other deletion mutant strains tested that are defective in NER. Collectively, these observations indicate that in addition to the stimulatory role in NER described previously, the 19S-RC has a negative role in regulating NER that is mediated by the ubl domain of Rad23 protein.
Results
Mutations in 19S proteasome subunits suppress UV radiation sensitivity and defective NER in a rad23 deletion mutant
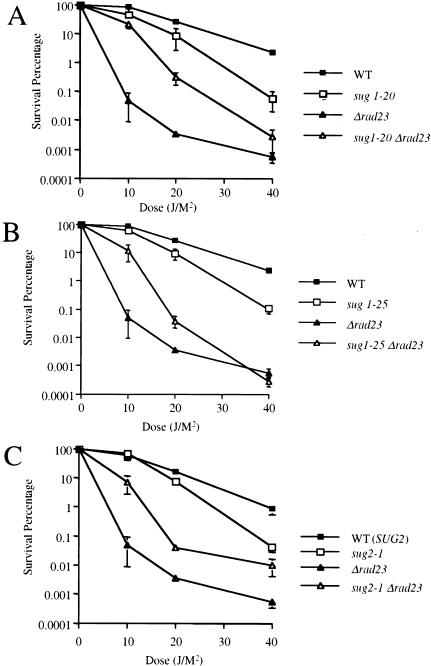
To investigate relationships between the SUG1 RPT6 and RAD23 genes during NER in yeast, we constructed a rad23 deletion mutant (rad23Δ) in the genetic background of several different sug1 rpt6 and sug2 rpt4 mutant strains. Previous studies show that sug mutants are slightly more UV radiation-sensitive than congenic wild-type strains (Russell et al. 1999). Consistent with these results, we again observed increased UV radiation sensitivity of the sug1-20, sug1-25, and sug2-1 mutant strains in multiple independent experiments (Fig. 1A–C). In addition, we confirmed the UV radiation sensitivity of the rad23Δ mutant (Fig. 1A–C). Surprisingly, we observed enhanced survival of sug1-20 rad23Δ, sug1-25 rad23Δ, and sug2-1 rad23Δ double mutant strains compared to otherwise isogenic rad23Δ single mutants (Fig. 1A–C). Analysis of diploid strains showed that this phenotype of the mutant sug alleles is recessive (data not shown). These data suggest that mutations in sug rpt genes can partially suppress defective NER associated with the complete absence of Rad23 protein.
Figure 1.
UV radiation sensitivity in sug1-2 and rad23 mutant strains. (▪) Wild-type strain Sc507 (A,B); Sc342 (C). (□) sug mutant strains sug1-20 (A); sug1-25 (B); sug2-1 (C). (▴) rad23 null strain. (▵) sug rad23 strain, in the same order as sug single mutant strains.
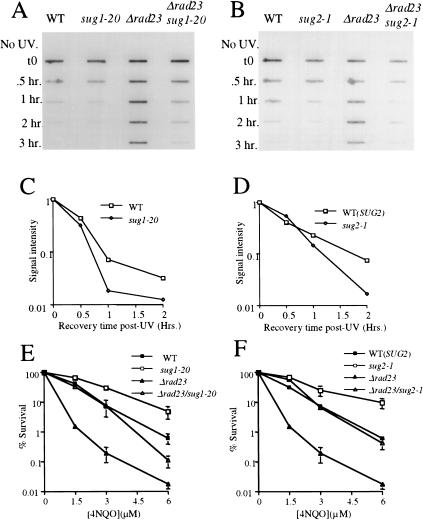
To determine whether the suppression of UV radiation sensitivity specifically reflects enhanced NER, we measured directly the loss of photoproducts from the DNA of living cells using an immunoslotblot assay (see Materials and Methods). In a wild-type strain, removal of (6-4) photoproducts following exposure of cells to UV radiation was rapid and efficient, with most of the lesions removed within 1 h after exposure to UV radiation (Fig. 2A,B; left lanes). As anticipated, loss of (6–4) photoproducts from DNA was defective in the rad23Δ strain, with no loss of lesions observed during a 3-h incubation period (Fig. 2A,B; second right lanes). Consistent with the results shown in Figure 1, both the sug1-20 rad23Δ (Fig. 2A, right lane) and sug2-1 rad23Δ (Fig. 2B, right lane) double mutant strains showed an enhanced rate of loss of (6–4) photoproducts from DNA compared with the rad23Δ single mutant strain. Similar results were obtained when measuring the loss of cyclobutane pyrimidine dimers (CPD) from DNA using the same assay with CPD-specific antibodies (data not shown).
Figure 2.
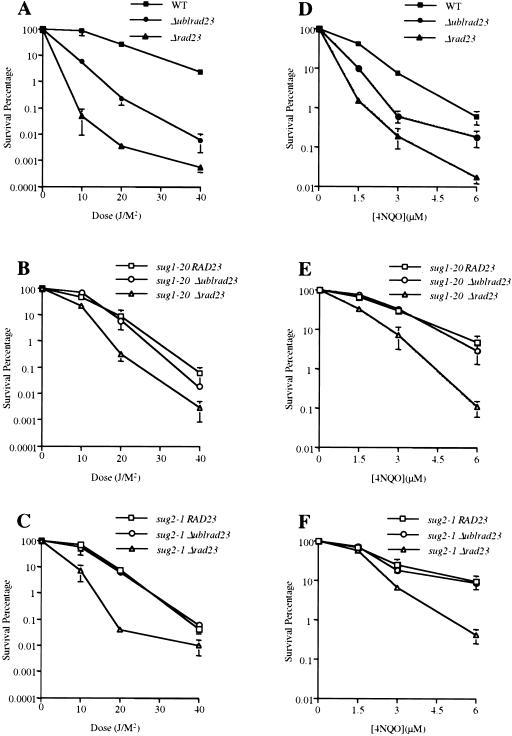
4NQO sensitivity and removal of photoproducts in vivo in sug1-2 and rad23 mutant strains. (A,B) Time course of (6-4) photoproduct lesion removal as detected by antibody. No UV indicates DNA from cells immediately before UV treatment and t0 immediately post UV treatment. All other points are time points of recovery at 30°C post-treatment. (WT) Parental strain for sug1-20 (A) and sug2-1 (B) strains. (C,D) Quantitation of slot blot. Intensity of signal measured using NIH Image1.62. Data was normalized with the measurement at t0=1. (E,F) 4NQO survival: (▪) wild-type strain Sc507 (E) and Sc342 (F); (□) sug mutant strains sug1-20 (E) and sug2-1 (F); (▴) rad23 null strain; (▵) sug rad23 strain, in the same order as sug single mutant strains.
Collectively, the results presented in Figures 1 and 2, A and B, indicate that mutations in the AAA ATPase subunits of the proteasome 19S-RC can partially suppress the NER defect of a rad23 null mutant. In addition, these results confirm our previous demonstration of functional relationships between this complex and the NER machinery (Russell et al. 1999).
The consistently decreased survival of sug mutants in the presence of a wild-type RAD23 gene following exposure to UV radiation notwithstanding (Fig. 1), these mutants reproducibly manifested an enhanced rate of removal of (6–4) photoproducts. This is shown both qualitatively in Figure 2, A and B, (cf. left and adjacent lanes) and quantitatively in Figure 2, C and D. In order To investigate this apparent paradox further, we examined survival of the sug RAD23+ mutants after exposure to the UV radiation-mimetic chemical 4-nitroquinoline-1-oxide (4-NQO), a well-characterized compound that generates lesions in DNA that are removed exclusively by NER in wild-type cells (Friedberg et al. 1995).
In contrast to the results observed with UV radiation, the 19S-RC mutant alleles tested showed enhanced survival following 4NQO treatment (Fig. 2E,F), correlating with the enhanced ability to remove NER-specific lesions induced by UV radiation (Fig. 2C,D). Furthermore, as was observed with UV radiation exposure, we noted enhanced survival following 4NQO treatment of sug1-20 rad23Δ and sug2-1 rad23Δ double mutant strains compared with otherwise isogenic rad23Δ single mutants (Fig. 2C,D). Why then are these sug mutants more sensitive to UV radiation? It is well established that survival of colony-forming ability after exposure to UV radiation can reflect multiple responses to DNA damage and is therefore a relatively nonspecific indicator of defects in individual DNA repair pathways (for review, see Friedberg et al. 1995). Therefore, the observation that the sug RAD23+ mutants are more sensitive to killing by UV radiation than wild-type controls suggests an effect of 19S-RC functions on a cellular responses to UV radiation other than NER. Consistent with this interpretation, it has been reported recently that mutations in selected 19S-RC components (including RPT6 SUG1) can inhibit the activation of Gcn4p target genes induced by UV radiation or the alkylating agent methylmethanesulfonate (MMS) (Stitzel et al. 2001). In addition, we have shown that double mutants containing the sug alleles tested here and deleted of either the RAD23 or RAD4 genes required for NER, manifest increased sensitivity to killing following exposure to MMS (T.G. Gillette and E.C. Friedberg, unpubl.).
Suppression of defective NER is specific for deletion of the RAD23 gene
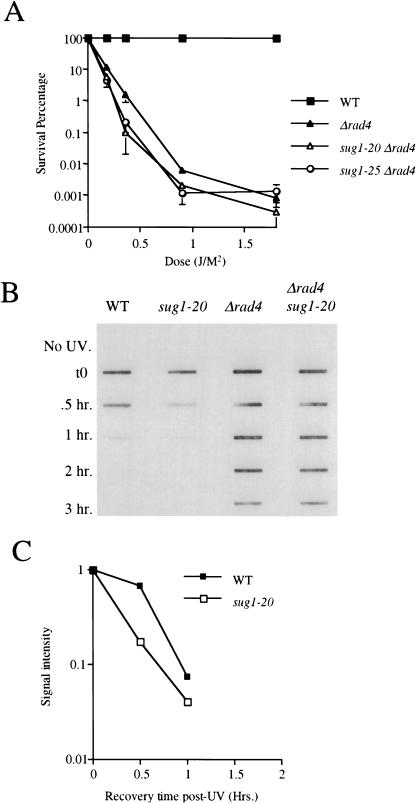
As indicated above, several studies have shown that Rad23 protein forms a stable complex with Rad4 protein. Rad4 protein has also been shown to interact with the Rad7 subunit of the Rad7 Rad16 Abf1 complex in vitro (Guzder et al. 1995; Wang et al. 1997; Reed et al. 1999). To determine whether sug1 mutants also suppress defective NER in rad4 and/or rad7 mutants, we deleted the RAD4 and RAD7 genes in independent experiments in both sug1-20 and sug1-25 mutant strains. No suppression of UV radiation sensitivity was observed in either sug mutant combination with a rad4Δ mutant (Fig. 3A). In addition, examination of the removal of both (6-4) photoproducts (Fig. 3B) and of CPD from DNA (data not shown) showed no detectable rescue of NER in a rad4Δ sug1-20 double mutant. It is instructive to note that consistent with the results described above, in the independent experiment shown in Figure 3, B and C, the rate of removal of (6–4) photoproducts in the sug1-20 mutant is once again increased compared with the wild-type strain.
Figure 3.
UV radiation sensitivity and NER in the sug1 rad4 double mutant. (A) UV survival curves: (▪) Sc507 wild-type strain; (▴) rad4 null mutant; (▵) sug1-20 rad4 double mutant; (○) sug1-25 rad4 strain. (B) Removal of UV lesions in vivo. Time course of (6-4) photoproduct lesion removal as detected by antibody. (C) Quantitation of slot blot (in A). Intensity of signal measured using NIH Image1.62. Data was normalized with the measurement at t0=1.
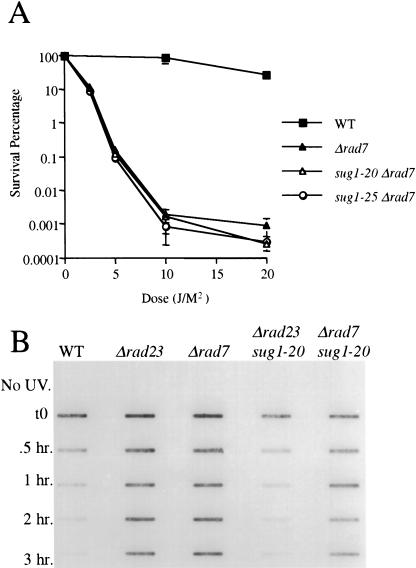
Unlike the extreme UV radiation sensitive phenotype of a rad4Δ mutant, rad7Δ mutants strains manifest a moderate level of sensitivity similar to that observed in rad23Δ strains. The molecular phenotypes of rad7 and rad23 mutants, however, are distinct. Rad7 protein is required for overall (global) genome NER, and its absence has no effect on transcription-coupled repair (TCR) in vivo (Verhage et al. 1994). In contrast, loss of Rad23 protein impairs both global genome NER and TCR in vivo (Mueller and Smerdon 1996). As was the case for the rad4Δ mutant, rad7Δ sug1-20 and rad7Δ sug1-25 double mutant strains showed no suppression of UV radiation sensitivity (Fig. 4A). Similarly, no rescue of NER was observed by measuring the loss of photoproducts from the DNA of cells in vivo (Fig. 4B). We therefore conclude that the partial recovery of NER in sug mutant strains is specifically associated with loss of the RAD23 gene.
Figure 4.
UV radiation sensitivity and NER in the sug1 rad7 double mutant. (A): (▪) Sc507 wild-type strain; (▴) rad7 null mutant; (▵) sug1-20 rad7 double mutant; (○) sug1-25 rad7 strain. (B) Removal of UV lesions in vivo. Time course of (6-4) photoproduct lesion removal as detected by antibody.
Defective proteolysis by the proteasome does not suppress defective NER in the rad23Δ mutant
Previous studies from our laboratories showed that the roles of the proteasome in NER is not dependent on proteolysis (Russell et al. 1999). To determine whether partial suppression of the NER defect in rad23Δ mutants in the presence of sug mutations is mimicked by defective proteolytic function of the proteasome, we constructed a rad23Δ mutant in the genetic background of a strain carrying double mutations in the 20S proteolytic subunit genes PRE1 and either PRE4 or PRE2 (Heinemeyer et al. 1993; Hilt et al. 1993). The pre1-1 pre4-1 and pre1-1 pre2-2 mutant strains have a severe defect in proteolysis mediated by the 26S proteasome (Heinemeyer et al. 1993; Hilt et al. 1993; Ferdous et al. 2001). The in vivo phenotype of these mutants includes defective growth in the presence of canavanine, an accumulation of ubiquitin protein conjugates when stressed (e.g., by starvation) and a severe sensitivity to growth at elevated temperatures (Heinemeyer et al. 1993; Hilt et al. 1993). In vitro, extracts from the pre1-1 pre2-2 strain contain <95% of the chymotrypsin-like activity observed in the isogenic wild-type strain and the pre1-1 pre4-1 strain has <75% of the peptidase activity of a wild-type strain (Heinemeyer et al. 1993; Hilt et al. 1993; Ferdous et al. 2001). This is in stark contrast to the sug mutant alleles used in these studies, which manifest no significant defect in proteolysis (Ferdous et al. 2001; Russell and Johnston 2001)
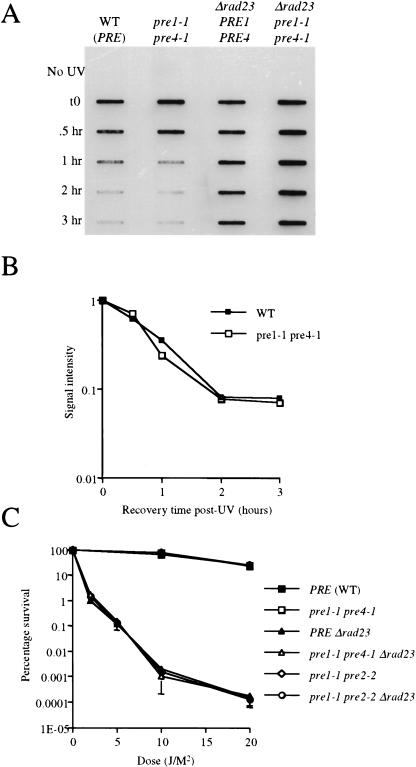
We measured NER in pre mutant strains using the immunoslotblot assay described above, except that the strains were grown overnight at 30°C and switched to 37°C for 90 min before UV irradiation and then returned to 37°C for the 3-hr repair interval. Both the parental wild-type and pre1-1 pre4-1 mutant strains showed similar rates of removal of (6-4) photoproducts from DNA (Fig. 5A,B). In contrast, the rad23Δ and the rad23Δ pre1-1 pre4-1 triple mutant strains showed no detectable loss of lesions from DNA (Fig. 5A). A similar result was observed with the rad23Δ pre1-1 pre2-2 strain (data not shown). Furthermore, as shown previously (Russell et al. 1999), the UV radiation sensitivity of the pre1-1 pre2-2 and pre1-1 pre4-1 mutant strains is indistinguishable from that of wild-type strains (Fig. 5C), and no suppression of this phenotype was observed in rad23Δ pre1-1 pre4-1 or rad23Δ pre1-1 pre2-2 triple mutant (Fig. 5C). These data reinforce the lack of any correlation between defective proteolysis and suppression of the rad23 mutant phenotype. Therefore, our results provide further support for functional interactions of the 19S subunit in NER, which are independent of proteolysis.
Figure 5.
UV radiation sensitivity and NER in 20S proteasome mutants. (A) Removal of UV lesions in vivo. Time course of (6-4) photoproduct lesion removal as detected by antibody. Method the same as in Fig. 2, except cells moved to 37°C for 90 min before UV treatment and subsequent recovery at 37°C post-UV treatment. (B) Quantitation of slot blot (in A). Intensity of signal measured using NIH Image1.62. Data was normalized with the measurement at t0=1. (C): (▪) Wild-type strain; (□) pre1-1 pre4-1 mutant; (⋄) pre1-1 pre2-2 mutant; (▴) rad23 null mutant; (▵) rad23 pre1-1 pre4-1 mutant strain; (○) rad23 pre1-1 pre2-2 strain.
19S mutants rescue the NER defect associated with loss of the Ubl domain of Rad23 protein
Previous studies showed that a physical interaction between Rad23 protein and the 19S subunit of the proteasome is mediated through the ubl domain located at the amino-terminus of Rad23 protein (Schauber et al. 1998; Russell et al. 1999). To investigate the effect of sug mutations on strains expressing Rad23 protein deleted of this domain, we transformed rad23Δ or rad23Δ sug1-20 strains with a centromeric plasmid vector (pJW160) carrying a construct (Δublrad23) that expresses Rad23 protein missing the amino-terminal 60 amino acids (Watkins et al. 1993). This truncated Rad23 protein fails to co-immunoprecipitate components of the 19S-RC (S. Reed and E.C. Friedberg, unpubl.). As anticipated, loss of the ubl domain resulted in a level of UV radiation sensitivity intermediate between wild-type strains and strains deleted of the entire Rad23 protein (Fig. 6A). Significantly, in the background of the sug1-20 mutation, the loss of this domain had no further effect on the level of UV radiation sensitivity (Fig. 6B). Essentially identical results were observed with the sug2-1 allele (Fig. 6C). These observations indicate that mutations in these subunits of the 19S-RC are epistatic to loss of the ubl domain of Rad23 protein.
Figure 6.
Sensitivity to UV radiation and 4NQO of strains carrying rad23 lacking the ubl domain. UV/4NQO survival curves. (A,D): (▪) Sc507 wild-type strain; (▴) rad23 null mutant; (●) rad23 null mutant carrying a centromeric plasmid expressing Δubl rad23. (B,E): (□) sug1-20 strain; (▵) sug1-20 rad23 mutant; (○) sug1-20 rad23 mutant carrying a centromeric plasmid expressing Δubl rad23. (C,F): (□) sug2-1 strain; (▵) sug2-1 rad23 mutant; (○) sug2-1 rad23 mutant carrying a centromeric plasmid expressing Δubl rad23.
We also examined survival of the Δubl rad23 double mutant strain in response to 4NQO treatment. Unlike the case with UV radiation exposure, the sug mutant strains showed enhanced resistance to this NER-specific DNA- damaging agent that was unaffected by additional loss of the ubl domain (Fig. 6D–F). Therefore, we show with two different DNA-damaging agents that 19S mutations are epistatic to the loss of the ubl domain of Rad23 protein. These results suggest that the increased sensitivity to DNA-damaging agents in Δrad23 sug double mutants, that is, mutants that are completely deleted for the RAD23 gene, actually reflects the loss of the C-terminal domain of Rad23 protein.
A temperature-degradable sug mutant manifests enhanced NER
As discussed above, careful examination of the kinetics of the loss of (6–4) photoproducts form the DNA of UV-irradiated cells reproducibly showed that the sug mutants that are wild-type for RAD23, have a slightly enhanced rate of NER compared with the congenic wild-type strain (Figs. 2A–-D; 3B,C). To investigate this further, we examined NER in a strain that expresses temperature-degradable Sug2 protein (YHM11.2). This strain was constructed by the N-degron strategy originally described by Varshavsky and his colleagues (Dohmen et al. 1994; McDonald and Byers 1997). This strain has been characterized independently and shown to grow at 30°C, but arrests in the G2 phase of the cell cycle at 37°C (McDonald and Byers 1997).
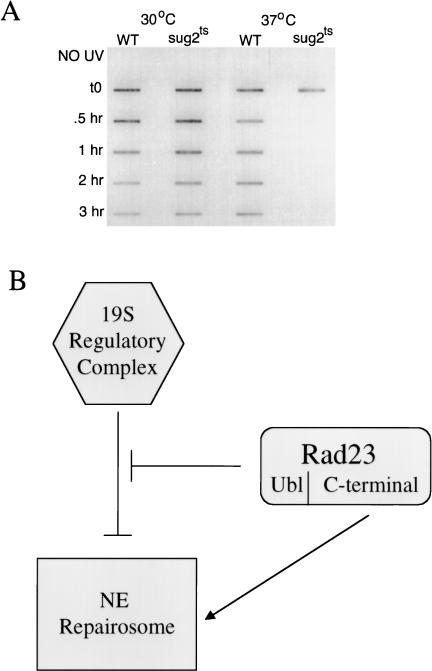
The mutant strain and its congenic parent were grown overnight at 30°C. The culture was divided and one half was switched to 37°C, while the other half was maintained at the permissive temperature. After 5 h, both cultures were exposed to UV radiation and incubation was continued at the respective temperatures for a further 3 h. Cultures maintained at 30°C showed a normal rate of loss of (6–4) photoproducts (Fig. 7, lanes 1 and 2). Those maintained at the non-permissive temperature at which Sug2 protein is degraded, however, showed a markedly enhanced rate of loss of (6-4) photoproducts from DNA (Fig. 7A, cf. lanes 3 and 4). Similar results were obtained when measuring the loss of CPD from DNA using the slot blot assay with CPD-specific antibodies (data not shown).
Figure 7.
(A) Removal of photoproducts in vivo after degradation of Sug2 protein. Removal of UV lesions in vivo. Time course of (6-4) photoproduct lesion removal as detected by antibody. Strains where grown to mid-log phase at 30°C and then switched to the indicated temperature for 5 h before UV treatment as well as the post-treatment recovery period. (WT) Isogenic parent of the sug2ts strain. (B) Model of Rad23 and 19S genetic interactions in NER. In this model, the 19S regulatory complex of the proteasome is shown to negatively modulate the rate of NER in vivo. This effect is attenuated by interaction of the amino-terminal ubl domain of Rad23 protein. The carboxyl terminus of Rad23 enhances NER through direct interaction with other subunits of the nucleotide excision repairosome.
The enhanced NER observed under these experimental conditions is unlikely to result from the block in G2 that is a phenotype of the N-degron sug2 mutant, as increased NER is not observed in G2 phase of the cell cycle in yeast (Friedberg et al. 1995). Furthermore, inducible NER in yeast is confined to the G1 phase of the cell cycle (Scott and Waters 1997). These data suggest that Sug2 protein, acting in the 19S-RC, normally functions to modulate the rate of NER in yeast. Collectively, our data also suggest that this phenotype, as well as the suppressive effects of these mutations on rad23 deletion mutants, is mediated by the loss of some functions of the 19S-RC.
Discussion
The present studies demonstrate a role for the 19S-RC of the yeast proteasome in modulating NER. Most dramatically, temperature-regulatable degradation of the 19S regulatory subunit Sug2 results in a marked increase in the rate in NER of photoproducts in vivo, suggesting that at least one function of the proteasome in NER is to negatively modulate the rate of this process. This is supported by the observations of an increased rate of NER in vivo in strains with point mutations in SUG genes. In addition, we observed that mutations in the 19S ATPases Sug1 and Sug2 partially suppress UV radiation sensitivity and defective NER in the absence of Rad23 protein, providing evidence of a Rad23-independent relationship between the proteasome and NER. These mutations also suppress the NER defect associated with loss of the ubl domain of Rad23 protein, suggesting that the ubl domain modulates the effect of the 19S complex on NER.
Lommel et al. have also reported recently that strains carrying point mutations in 19S-RS genes have a slightly increased rate of NER (Lommel et al. 2000; Fig 2A,B). They interpreted this result as indicative of a role for the 26S proteasome in the regulation of NER, which is dependent on proteolysis. Only strains with mutations in 19S regulatory subunit genes, however, were tested in this study (Lommel et al. 2000). In our experiments, strains with mutations in 20S subunits that are severely defective in proteolysis had no observable effect on NER, consistent with previous studies (Russell et al. 1999). Therefore, although we cannot exclude the possibility that proteolysis may yet be shown to have some role in a cellular response to UV radiation-induced DNA damage in yeast, we have not observed any correlation between defective proteolysis and NER.
Previous studies showed a physical interaction between the 26S proteasome and the ubl domain of Rad23 protein and suggested that this interaction serves to recruit the 26S proteasome for some function in NER (Schauber et al. 1998; Russell et al. 1999). The observation that mutations in the 19S ATPases Sug1 and Sug2 can partially suppress UV radiation sensitivity and defective NER in the absence of Rad23 protein, indicates that modulation of NER can also arise from an interaction between the proteasome and the NER machinery that is independent of Rad23 protein. A number of experimental observations suggest that a likely target of this interaction is the NER–RNA polymerase II basal transcription factor TFIIH. First, a complex that includes the ubl domain of Rad23 protein, the 19S-RC of the proteasome and TFIIH, but is exclusive of the 20S proteolytic core has been detected (L. Sun and T. Kodadek, pers. comm.). Second, interactions between subunits of the 19S-RC and TFIIH have been reported in human cells (Weeda et al. 1997). Finally, a direct interaction between the 19S-RC subunit Sug1 and the TFIIH subunit Ssl2 has been shown in yeast (W. Huang and E.C. Friedberg, unpubl.).
Parallels between phenotypes of 19S-RC mutant strains with respect to NER and transcription in vivo suggest an inhibitory role for the 19S complex in both processes. For example, sug1-20 has been shown to suppress the mutant phenotype of the cdc68 allele (whose gene product has now been identified as a transcription elongation factor) independently of proteolysis (Xu et al. 1995; Russell and Johnston 2001). Furthermore, the sug2-1 mutant was originally isolated as a recessive suppressor of a transcriptional defect in GAL4 (Russell et al. 1996; Orphanides et al. 1999). Here, we show that these sug alleles can also suppress defective NER resulting from the loss of the ubl domain of Rad23 protein. It is instructive to note that the sug mutations examined are recessive, indicating that the phenotypes are attributable to the loss of some wild-type inhibitory function of the 19S-RC on both transcription and NER in vivo.
Parallels between the roles of the 19S-RC in NER and transcription are also observed in vitro. The in vitro observations, however, point to a stimulatory role for the 19S complex in both processes. For example, recent findings (Ferdous et al. 2001), show that transcriptional elongation in vitro is inhibited by an antiserum against Sug1 protein. Furthermore, heat treatment of an extract from a temperature-sensitive sug1 mutant results in loss of transcriptional activity that can be restored by the addition of purified 19S-RC (Ferdous et al. 2001). Therefore, it would appear that the 19S-RC has both positive and negative modulating roles in transcription. This duality is mirrored for NER. Specifically, antiserum against Sug1 protein, or specific alkylation (by N-ethylmaleimide treatment) of a cysteine residue in Sug1 protein partially inhibits NER in vitro (Russell et al. 1999).
A simple explanation for the apparently contradictory results observed in experiments in vitro and in vivo may relate to the different end point measured. The in vitro NER assay measures repair synthesis of DNA and may be sensitive to only one aspect of 19S-RC function. In contrast, the in vivo assays measure DNA damage removal and cellular survival and may more reflect the summation of multiple modes of regulation by the 19S complex. These differences are also reflected with respect to the RAD23 gene, the loss of which completely eliminates repair synthesis in vitro, but has only an intermediate effect on survival compared with the loss of other NER genes. We cannot rule out, however, that the partial inhibition of NER activity in vitro by both the Sug1 antibody and the N-ethylmaleimide treatment reflects interference with the interaction between the 19S regulatory complex and the ubl domain of Rad23 protein.
The data presented here also suggest that the primary, if not the exclusive role of the ubl domain of Rad23 in NER is to modulate regulation of NER by the 19S complex (Fig. 7B). Therefore, the level of UV radiation sensitivity and defective NER observed in yeast strains deleted for just the ubl domain of Rad23 protein (which is intermediate between that resulting from complete absence of Rad23 protein and a wild-type strain) reflects the loss of modulation of NER by the 19S complex rather than a direct role of the ubl domain of Rad23 protein in the biochemical mechanism of NER. This conclusion is supported by the observation that loss of the ubl domain does not result in a further increase in UV radiation sensitivity or sensitivity to 4NQO in sug1 and sug2 mutants, reflecting the epistatic relationship between sug mutations and loss of the ubl domain of Rad23 protein.
Complementation of rad23Δ sug1 or rad23Δ sug2 double mutants with the carboxy-terminal fragment of RAD23 deleted of just the ubl domain restores NER to levels observed in sug mutants alone. Therefore, the carboxy-terminal domain of Rad23 protein is clearly required for a direct biochemical role in NER (Fig. 7B). Rad23 protein has been shown to interact stably with Rad4 protein, an interaction that does not require the amino-terminal ubl domain (Guzder et al. 1995; Schauber et al. 1998). Therefore, Rad23 protein apparently has two distinct roles in NER, which require different domains. Studies are in progress to investigate the phenotype of Rad23 point mutants defective in one, but not the other of these functions.
The ubl domain of Rad23 is apparently critical for interactions with the proteasome that is required for regulation of NER. This domain, however, apparently does not signal the degradation of rad23 protein. Rad23 protein is very stable with a half-life of over 6 hr (Watkins et al. 1993). The principal signal for protein degradation by the ubiquitin pathway is a cis-acting Lys48-linked polyubiquitin chain, as opposed to a single or several ubiquitin monomers (Thrower et al. 2000). A ubiquitin-conjugating enzyme variant has been shown to assemble novel polyubiquitin chains for processing through the RAD6-dependent post-replicative repair pathway (Spence et al. 1995). These polyubiquitin chains are unique in that they are linked at Lys63, and when conjugated to specific yeast proteins, do not destabilize them, suggesting that the proteasome does not recognize these modified proteins as degradation signals (Spence et al. 1995, 2000). Therefore, the ubl domain of Rad23, like monomers of ubiquitin and Lys63-linked polyubiquitin chains, may lack critical interactions required for targeted degradation by the proteasome pathway.
Polyubiquitination and ubiquitin-dependent degradation of HHR23A protein has been observed during the S phase of the cell cycle in human cells infected with human papilloma virus (Kumar et al. 1999). There is no evidence, however, that this degradation is directly related to the role of HHR23A in NER. Recent genetic studies by Ortolan et al. (2000) have shown that deletion of the genes encoding the ubiquitin-conjugating enzymes Ubc4 and Ufd2 can also partially suppress the UV sensitivity of a rad23 deletion. This evidence has been proposed to support a role for proteolysis in down-regulating NER (Ortolan et al. 2000). This study did not identify a specific target for ubiquitination, and no evidence was provided for increased degradation of NER proteins in the absence of Rad23 protein. In this study we provide evidence that there is no correlation between defective proteolysis and NER. This suggests that the role of ubiquitination in NER observed by Ortolan et al. (2000) may be non-proteolytic. It has been reported that Cdc34–SCFMet30-mediated ubiquitination regulates the function of the transcription factor Met4 in a non-proteolytic manner (Kaiser et al. 2000). Furthermore, it has been observed that ubiquitin chain formation has a non-proteolytic regulatory role in translation (Spence et al. 2000). Therefore, ubiquitination of proteins does not necessarily lead to proteolysis.
The precise biological roles of proteasome interactions with the NER machinery is not clear. The AAA ATPase superfamily of proteins (which includes the Rpt subunits) are involved in numerous aspects of cell metabolism and perform chaperone-like functions that aid the assembly or disassembly of protein complexes (Neuwald et al. 1999). It has been shown recently that the base of the 19S complex, containing the Rpt subunits, has an intrinsic chaperone-like activity that is biochemically separable from both the ubiquitin recognition function of the 19S lid and the proteolytic function of the 20S core complex (Braun et al. 1999). Such a chaperone-like activity may be involved in assembly or disassembly of the NER machinery. The recent observation that the 19S complex can also function in transcription elongation by RNA polymerase II independently of proteolysis (Ferdous et al. 2001) suggests that the 19S complex has a similar role in both transcription and NER.
Materials and methods
Strains
The yeast S. cerevisiae strains used in this study are listed in Table 1. Sc507 was derived from Sc342 and was constructed by inserting a bacteriophage T7 epitope S10 at the amino terminus of the wild-type sug1 gene. All of the sug1 mutant strains were congenic to Sc507. The sug2-1 mutant strain was derived directly from Sc342. To construct the double mutants of sug1 sug2 or pre1-1 pre4-1 and rad genes, the mutant strains were transformed with the rad23, rad4 or rad7 knockout plasmids and screened in the selective media. The surviving strains were then subjected to the Southern hybridization to confirm the disruption of the rad gene of interest. Plasmid pJW160 (Watkins et al. 1993), a centromeric vector expressing Rad23 with the ubl domain deleted (residues 2–60), was introduced to strains Sc507R23, WH120R23 and WH201R23 and maintained in URA dropout media. All other yeast strains were maintained and grown in the yeast extract-peptone-dextrose (YPD) medium.
Table 1.
Saccharomyces cerevisiae strains used in this study
| Strain
|
Genotype
|
Source
|
|---|---|---|
| Sc342 (same as W3031A) | MATa ade2–1 trp1–1 leu2–3, 112 his3–11, 15 ura3–1 can1–100 | Russell et al. 1999 |
| Sc507 | W303–1A S10–SUG1 | Russell et al. 1999 |
| Sc654 | W303–1A S10–sug1–1 | Russell et al. 1999 |
| Sc657 | W303–1A S10–sug1–3 | Russell et al. 1999 |
| Sc658 | W303–1A S10–sug1–20 | Russell et al. 1999 |
| Sc660 | W303–1A S10–sug1–25 | Russell et al. 1999 |
| Sc671 | W303–1A sug2–1 | Russell 2001 |
| Sc507R23 | W303–1A S10–SUG1 rad23∷HIS3 | This study |
| WH101R23 | W303–1A S10–sug1–1 rad23∷HIS3 | This study |
| WH103R23 | W303–1A S10–sug1–3 rad23∷HIS3 | This study |
| WH120R23 | W303–1A S10–sug1–20 rad23∷HIS3 | This study |
| WH125R23 | W303–1A S10–sug1–25 rad23∷HIS3 | This study |
| WH201R23 | W303–1A sug2–1 rad23∷HIS3 | This study |
| WH120R4 | W303–1A S10–sug1–20 Δrad4∷HIS3 | This study |
| WH125R4 | W303–1A S10–sug1–25 Δrad4∷HIS3 | This study |
| WH120R7 | W303–1A S10–sug1–20 Δrad7∷LEU2 | This study |
| WH125R7 | W303–1A S10–sug1–25 Δrad7∷LEU2 | This study |
| WCG4a | MATa ura3 leu2–3, 112 his3–11, 15 CanS Gal+ | Hilt et al. 1993 |
| YH129/14 | WCG4a pre1–1 pre4–1 | Hilt et al. 1993 |
| TG1141R23 | WCG4a pre1–1 pre4–1 rad23∷HIS3 | This study |
| WGC4 11/22a | WCG4a pre1–1 pre2–2 | Heinemeyer et al. 1993 |
| TG1122R23 | WCG4a pre1–1 pre2–2 rad23∷HIS3 | This study |
| TGR23 | WCG4a rad23∷HIS3 | This study |
| Wx257–5c | MATa ura3–52 leu2–3, 112 trp1Δ his3Δ | Mcdonald et al. 1997 |
| YHM11.2 | Wx257–5c pcs1td | Mcdonald et al. 1997 |
Measurement of UV radiation sensitivity
Protocols for measuring UV sensitivities in yeast strains were modified from Huang et al. (1998). Cells were grown in YPD medium to mid-log phase (around 107 cells/mL). Serial 10-fold dilutions of each culture were plated on the YPD plate, then irradiated with the germicidal UV lamp at 1 J/sec/m2. For the experiments involving the rad4 mutant strains, the rate of irradiation was reduced to 0.18 J/sec/m2 by increasing the distance between the plates and the UV lamp. Following irradiation, the plates were wrapped with aluminum foil and incubated at 30°C until colonies appeared.
Measurement of 4-NQO sensitivity
Cells were grown in YPD medium to mid-log phase, harvested by centrifugation, washed twice in sterile water, then resuspended in sterile water at a final cell density of approximately 0.5×107 cells/mL. Ten milliliters of the suspension was treated with 4-NQO at the indicated dose for 1 h at 30°C. The cells are harvested by centrifugation and washed twice in sterile water. Serial 10-fold dilutions of each culture were plated on YPD plates and incubated at 30°C until colonies appeared.
Immunoslotblot assay
In vivo DNA repair was measured by growing the mutant and isogenic parental strains at the indicated temperature in YPD to mid-log phase before harvesting. Cells were harvested by centrifugation, washed in sterile water, then resuspended in 0.1 m PBS (pH 7.4) at a final cell density of approximately 2×107 cells/ml. The strains were irradiated in dishes such that the depth of the cell suspension was <3 mm. Each strain was exposed to UV light at a dose of 20 J from a germicidal lamp at a fluence of at 1 J/sec/m2 with constant agitation. The strains were returned to their respective temperature in the dark to allow for a period of repair. Aliquots were taken before and immediately after UV treatment and at times indicated. All manipulations were carried out in a safe light to avoid photoreactivation. Genomic DNA was prepared using standard techniques. Equal amounts of DNA (5 μg) from each time point were applied to Hybond N+ transfer membrane (Amersham) using the S&S minifold I slot blot (Schliecher & Schuell Inc.). DNA was denatured and fixed on the membrane by adding NaOH to .0.4 m to each sample before binding. Removal of UV radiation-induced photoproducts from the genomic DNA was examined by Western blotting with monoclonal CPD or (6-4) photoproduct antibodies (25) followed by a secondary antibody/horseradish peroxidase conjugate. This secondary antibody is detected with Renaissance chemilumenescence reagent (NEN) and exposure to autoradiography film. Examining lesion removal in short time periods and carrying out the post-UV recovery in PBS removes the potential loss of signal from replication. Data for Figures 2 and 3 were quantitated using National Institutes of Health Image1.62.
Acknowledgments
We thank our laboratory colleagues for valuable discussions and critical review of the manuscript. We also thank Toshio Mori for the generous gift of CPD and (6–4) photoproduct antibodies and Heather McDonald for the yeast strains YHM11.2 and Wx257-5c. These studies are supported by research grant CA-12424 from the United States Public Health Service (E.C.F.).
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL friedberg.errol@pathology.swmed.edu; FAX (214) 648-4067.
Article and publication are at http://www.genesdev.org/cgi/doi/10.1101/gad.869601.
References
- Baumeister W, Walz J, Zuhl F, Seemuller E. The proteasome: Paradigm of a self-compartmentalizing protease. Cell. 1998;92:367–380. doi: 10.1016/s0092-8674(00)80929-0. [DOI] [PubMed] [Google Scholar]
- Biggins S, Ivanovska I, Rose MD. Yeast ubiquitin-like genes are involved in duplication of the microtubule organizing center. J Cell Biol. 1996;133:1331–1346. doi: 10.1083/jcb.133.6.1331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braun BC, Glickman M, Kraft R, Dahlmann B, Kloetzel PM, Finley D, Schmidt M. The base of the proteasome regulatory particle exhibits chaperone-like activity. Nat Cell Biol. 1999;1:221–226. doi: 10.1038/12043. [DOI] [PubMed] [Google Scholar]
- Coux O, Tanaka K, Goldberg AL. Structure and functions of the 20S and 26S proteasomes. Annu Rev Biochem. 1996;65:801–847. doi: 10.1146/annurev.bi.65.070196.004101. [DOI] [PubMed] [Google Scholar]
- Dohmen RJ, Wu P, Varshavsky A. Heat-inducible degron: A method for constructing temperature-sensitive mutants. Science. 1994;263:1273–1276. doi: 10.1126/science.8122109. [DOI] [PubMed] [Google Scholar]
- Ferdous, A., Gonzalez, F., Sun, L., Kodadek, T., and Johnston, S.A. 2001. The 19S regulatory particle of the proteasome is required for efficient transcription elongation by RNA polymerase II. Mol. Cell (in press). [DOI] [PubMed]
- Friedberg EC, Meira LB. Database of mouse strains carrying targeted mutations in genes affecting cellular responses to DNA damage. Version 4. Mutat Res. 2000;459:243–274. doi: 10.1016/s0921-8777(00)00006-9. [DOI] [PubMed] [Google Scholar]
- Friedberg EC, Walker GC, Siede W. DNA repair and mutagenesis. Washington DC.: ASM Press; 1995. [Google Scholar]
- Glickman MH, Rubin DM, Fu H, Larsen CN, Coux O, Wefes I, Pfeifer G, Cjeka Z, Vierstra R, Baumeister W, Fried V, Finley D. Functional analysis of the proteasome regulatory particle. Mol Biol Rep. 1999;26:21–28. doi: 10.1023/a:1006928316738. [DOI] [PubMed] [Google Scholar]
- Guzder SN, Habraken Y, Sung P, Prakash L, Prakash S. Reconstitution of yeast nucleotide excision repair with purified Rad proteins, replication protein A, and transcription factor TFIIH. J Biol Chem. 1995;270:12973–12976. doi: 10.1074/jbc.270.22.12973. [DOI] [PubMed] [Google Scholar]
- Guzder SN, Sung P, Prakash L, Prakash S. Affinity of yeast nucleotide excision repair factor 2, consisting of the Rad4 and Rad23 proteins, for ultraviolet damaged DNA. J Biol Chem. 1998;273:31541–31546. doi: 10.1074/jbc.273.47.31541. [DOI] [PubMed] [Google Scholar]
- ————— Synergistic interaction between yeast nucleotide excision repair factors NEF2 and NEF4 in the binding of ultraviolet-damaged DNA. J Biol Chem. 1999;274:24257–24262. doi: 10.1074/jbc.274.34.24257. [DOI] [PubMed] [Google Scholar]
- Heinemeyer W, Gruhler A, Mohrle V, Mahe Y, Wolf DH. PRE2, highly homologous to the human major histocompatibility complex-linked RING10 gene, codes for a yeast proteasome subunit necessary for chrymotryptic activity and degradation of ubiquitinated proteins. J Biol Chem. 1993;268:5115–5120. [PubMed] [Google Scholar]
- Hilt W, Enenkel C, Gruhler A, Singer T, Wolf DH. The PRE4 gene codes for a subunit of the yeast proteasome necessary for peptidylglutamyl-peptide-hydrolyzing activity. Mutations link the proteasome to stress- and ubiquitin-dependent proteolysis. J Biol Chem. 1993;268:3479–3486. [PubMed] [Google Scholar]
- Huang W, Feaver WJ, Tomkinson AE, Friedberg EC. The N-degron protein degradation strategy for investigating the function of essential genes: Requirement for replication protein A and proliferating cell nuclear antigen proteins for nucleotide excision repair in yeast extracts. Mutat Res. 1998;408:183–194. doi: 10.1016/s0921-8777(98)00031-7. [DOI] [PubMed] [Google Scholar]
- Jansen LE, Verhage RA, Brouwer J. Preferential binding of yeast Rad4.Rad23 complex to damaged DNA. J Biol Chem. 1998;273:33111–33114. doi: 10.1074/jbc.273.50.33111. [DOI] [PubMed] [Google Scholar]
- Jones JM, Welty DJ, Nakai H. Versatile action of Escherichia coli ClpXP as protease or molecular chaperone for bacteriophage Mu transposition. J Biol Chem. 1998;273:459–465. doi: 10.1074/jbc.273.1.459. [DOI] [PubMed] [Google Scholar]
- Kaiser P, Flick K, Wittenberg C, Reed SI. Regulation of transcription by ubiquitination without proteolysis: Cdc34/SCF(Met30)-mediated inactivation of the transcription factor Met4 [In Process Citation] Cell. 2000;102:303–314. doi: 10.1016/s0092-8674(00)00036-2. [DOI] [PubMed] [Google Scholar]
- Kumar S, Talis AL, Howley PM. Identification of HHR23A as a substrate for E6-associated protein-mediated ubiquitination. J Biol Chem. 1999;274:18785–18792. doi: 10.1074/jbc.274.26.18785. [DOI] [PubMed] [Google Scholar]
- Leonhard K, Stiegler A, Neupert W, Langer T. Chaperone-like activity of the AAA domain of the yeast Yme1 AAA protease. Nature. 1999;398:348–351. doi: 10.1038/18704. [DOI] [PubMed] [Google Scholar]
- Levchenko I, Luo L, Baker TA. Disassembly of the Mu transposase tetramer by the ClpX chaperone. Genes & Dev. 1995;9:2399–2408. doi: 10.1101/gad.9.19.2399. [DOI] [PubMed] [Google Scholar]
- Li L, Lu X, Peterson C, Legerski R. XPC interacts with both HHR23B and HHR23A in vivo. Mutat Res. 1997;383:197–203. doi: 10.1016/s0921-8777(97)00002-5. [DOI] [PubMed] [Google Scholar]
- Lommel L, Chen L, Madura K, Sweder K. The 26S proteasome negatively regulates the level of overall genomic nucleotide excision repair. Nucleic Acids Res. 2000;28:4839–4845. doi: 10.1093/nar/28.24.4839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masutani C, Sugasawa K, Yanagisawa J, Sonoyama T, Ui M, Enomoto T, Takio K, Tanaka K, van der Spek PJ, Bootsma D, et al. Purification and cloning of a nucleotide excision repair complex involving the xeroderma pigmentosum group C protein and a human homologue of yeast RAD23. EMBO J. 1994;13:1831–1843. doi: 10.1002/j.1460-2075.1994.tb06452.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masutani C, Araki M, Sugasawa K, van der Spek PJ, Yamada A, Uchida A, Maekawa T, Bootsma D, Hoeijmakers JH, Hanaoka F. Identification and characterization of XPC-binding domain of hHR23B. Mol Cell Biol. 1997;17:6915–6923. doi: 10.1128/mcb.17.12.6915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonald H B, Byers B. A proteasome cap subunit required for spindle pole body duplication in yeast. J Cell Biol. 1997;137:539–553. doi: 10.1083/jcb.137.3.539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mhammedi-Alaoui A, Pato M, Gama MJ, Toussaint A. A new component of bacteriophage Mu replicative transposition machinery: The Escherichia coli ClpX protein. Mol Microbiol. 1994;11:1109–1116. doi: 10.1111/j.1365-2958.1994.tb00387.x. [DOI] [PubMed] [Google Scholar]
- Mueller JP, Smerdon MJ. Rad23 is required for transcription-coupled repair and efficient overall repair in Saccharomyces cerevisiae. Mol Cell Biol. 1996;16:2361–2368. doi: 10.1128/mcb.16.5.2361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neuwald AF, Aravind L, Spouge JL, Koonin EV. AAA+: A class of chaperone-like ATPases associated with the assembly, operation, and disassembly of protein complexes. Genome Res. 1999;9:27–43. [PubMed] [Google Scholar]
- Orphanides G, Wu WH, Lane WS, Hampsey M, Reinberg D. The chromatin-specific transcription elongation factor FACT comprises human SPT16 and SSRP1 proteins. Nature. 1999;400:284–288. doi: 10.1038/22350. [DOI] [PubMed] [Google Scholar]
- Ortolan TG, Tongaonkar P, Lambertson D, Chen L, Schauber C, Madura K. The DNA repair protein Rad23 is a negative regulator of multi-ubiquitin chain assembly. Nat Cell Biol. 2000;2:601–608. doi: 10.1038/35023547. [DOI] [PubMed] [Google Scholar]
- Reed SH, Akiyama M, Stillman B, Friedberg EC. Yeast autonomously replicating sequence binding factor is involved in nucleotide excision repair. Genes & Dev. 1999;13:3052–3058. doi: 10.1101/gad.13.23.3052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubin D M, Finley D. Proteolysis. The proteasome: a protein-degrading organelle? Curr. Biol. 1995;5:854–858. doi: 10.1016/s0960-9822(95)00172-2. [DOI] [PubMed] [Google Scholar]
- Russell SJ, Johnston SA. Evidence that proteolysis of gal4 cannot explain the transcriptional effects of proteasome atpase mutations. J Biol Chem. 2001;276:9825–9831. doi: 10.1074/jbc.M010889200. [DOI] [PubMed] [Google Scholar]
- Russell SJ, Sathyanarayana UG, Johnston SA. Isolation and characterization of SUG2. A novel ATPase family component of the yeast 26 S proteasome. J Biol Chem. 1996;271:32810–32817. doi: 10.1074/jbc.271.51.32810. [DOI] [PubMed] [Google Scholar]
- Russell SJ, Reed SH, Huang W, Friedberg EC, Johnston SA. The 19S regulatory complex of the proteasome functions independently of proteolysis in nucleotide excision repair. Mol Cell. 1999;3:687–695. doi: 10.1016/s1097-2765(01)80001-0. [DOI] [PubMed] [Google Scholar]
- Schauber C, Chen L, Tongaonkar P, Vega I, Lambertson D, Potts W, Madura K. Rad23 links DNA repair to the ubiquitin/proteasome pathway. Nature. 1998;391:715–718. doi: 10.1038/35661. [DOI] [PubMed] [Google Scholar]
- Scott AD, Waters R. Inducible nucleotide excision repair (NER) of UV-induced cyclobutane pyrimidine dimers in the cell cycle of the budding yeast Saccharomyces cerevisiae: evidence that inducible NER is confined to the G1 phase of the mitotic cell cycle. Mol Gen Genet. 1997;254:43–53. doi: 10.1007/s004380050389. [DOI] [PubMed] [Google Scholar]
- Spence J, Sadis S, Haas AL, Finley D. A ubiquitin mutant with specific defects in DNA repair and multiubiquitination. Mol Cell Biol. 1995;15:265–273. doi: 10.1128/mcb.15.3.1265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spence J, Gali RR, Dittmar G, Sherman F, Karin M, Finley D. Cell cycle-regulated modification of the ribosome by a variant multiubiquitin chain. Cell. 2000;102:67–76. doi: 10.1016/s0092-8674(00)00011-8. [DOI] [PubMed] [Google Scholar]
- Stitzel ML, Durso R, Reese JC. The proteasome regulates the UV-induced activation of the AP-1-like transcription factor Gcn4. Genes & Dev. 2001;15:128–133. doi: 10.1101/gad.863801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thrower JS, Hoffman L, Rechsteiner M, Pickart CM. Recognition of the polyubiquitin proteolytic signal. EMBO J. 2000;19:94–102. doi: 10.1093/emboj/19.1.94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verhage R, Zeeman AM, de Groot N, Gleig F, Bang DD, van de Putte P, Brouwer J. The RAD7 and RAD16 genes, which are essential for pyrimidine dimer removal from the silent mating type loci, are also required for repair of the nontranscribed strand of an active gene in Saccharomyces cerevisiae. Mol Cell Biol. 1994;14:6135–6142. doi: 10.1128/mcb.14.9.6135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Wei S, Reed SH, Wu X, Svejstrup JQ, Feaver WJ, Kornberg RD, Friedberg EC. The RAD7, RAD16, and RAD23 genes of Saccharomyces cerevisiae: Requirement for transcription-independent nucleotide excision repair in vitro and interactions between the gene products. Mol Cell Biol. 1997;17:635–643. doi: 10.1128/mcb.17.2.635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watkins JF, Sung P, Prakash L, Prakash S. The Saccharomyces cerevisiae DNA repair gene RAD23 encodes a nuclear protein containing a ubiquitin-like domain required for biological function. Mol Cell Biol. 1993;13:7757–7765. doi: 10.1128/mcb.13.12.7757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weeda G, Rossignol M, Fraser RA, Winkler GS, Vermeulen W, van't Veer LJ, Ma L, Hoeijmakers JH, Egly JM. The XPB subunit of repair/transcription factor TFIIH directly interacts with SUG1, a subunit of the 26S proteasome and putative transcription factor. Nucleic Acids Res. 1997;25:2274–2283. doi: 10.1093/nar/25.12.2274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weissman JS, Sigler PB, Horwich AL. From the cradle to the grave: Ring complexes in the life of a protein [Comment] Science. 1995;268:523–524. doi: 10.1126/science.7725096. [DOI] [PubMed] [Google Scholar]
- Xu Q, Singer RA, Johnston GC. Sug1 modulates yeast transcription activation by Cdc68. Mol Cell Biol. 1995;15:6025–6035. doi: 10.1128/mcb.15.11.6025. [DOI] [PMC free article] [PubMed] [Google Scholar]