Abstract
The aerial architecture of flowering plants is determined to a large extent by shoot growth and shoot branching arising from the initiation and growth of axillary meristems. We have identified an Arabidopsis mutant, supershoot (sps), which is characterized by a massive overproliferation of shoots, such that a single plant can generate 500 or more inflorescences. Analysis of the mutant plants shows that the primary defect is because of an increase in the number of meristems formed in leaf axils, together with release of bud arrest, resulting in reiterative branch formation from rosette and cauline leaves. The SPS gene is shown here to encode a cytochrome P450, and together with a 3- to 9-fold increase in levels of Z-type cytokinins in sps mutant plants, indicate a role for SPS in modulating hormone levels. The expression pattern of SPS, with strong expression at the leaf axils, correlates well with the phenotypic defects. Our results indicate that control of shoot branching in Arabidopsis may be accomplished in part by suppression of axillary meristem initiation and growth through the localized attenuation of cytokinin levels at sites of bud initiation.
Keywords: Axillary meristem, bud initiation, branching, cytochrome P450, cytokinins, apical dominance, Arabidopsis
The shoot system plays an important role in generating the immense diversity of plant forms. The overall architecture of the shoot system is derived from the activity of the primary shoot apical meristem (SAM), arising during embryogenesis, together with the activity of the additional meristems subsequently formed after seed germination. The SAM provides the main axis of the plant body. The plant architecture is then further modified by shoot branching, performed by the activity of the postembryonic meristems. The complexity of the branching pattern depends on when and where these branches develop. These characteristics, although plastic in their response to environmental cues, are genetically determined (for review, see Halle 1999). The developmental program that specifies branching patterns in different plant species is of fundamental importance for generating species-specific plant forms.
Axillary branching is the most common process by which lateral branching occurs in angiosperms (Sussex and Kerk 2001). The axillary branching process generally involves two developmental stages: the formation of axillary meristems in the leaf axils and subsequent axillary bud growth. In many plant species, including Arabidopsis, the growth of axillary meristems is inhibited by the primary inflorescence, a phenomenon generally known as apical dominance. Most studies of shoot branching have emphasized the mechanisms controlling this bud outgrowth. The plant hormones auxin and cytokinin appear to play a major role in controlling this process, with cytokinin as a key factor in promoting bud growth, whereas auxin has an inhibitory effect (for review, see Tamas 1995; Li and Bangert 1992). Therefore, the outcome appears to depend on the ratio of the two hormones. However, the underlying regulatory mechanisms and genetic pathways have not yet been established.
Experimental procedures applied to this problem have progressed from classic studies with exogenous application of hormones, to more recent studies of transgenic plants transformed with bacterial hormone biosynthesis genes (Morris 1986; Tamas 1995; Chatfield et al. 2000; Hedden and Phillips 2000). Recently, the isolation and characterization of mutants with altered shoot branching patterns has been used to study this phenomenon. The defects in these mutants fall into two classes. In the first class, mutations do not effect early stages of axillary meristem development in terms of the timing of axillary meristem formation or the number of the axillary meristems produced. Therefore, the increased number of branches in the mutants is caused only by the release of inhibited axillary buds. An Arabidopsis auxin-insensitive (axr1-12) mutant (Stirnberg et al. 1999) and a maize teosinte branched 1 (tb1) mutant (Doebley et al. 1997) confer this type of mutation. In the second class, mutations cause a defect in axillary meristem initiation. In tomato, these include the blind, torosa, and lateral suppressor (ls) mutants (Schmitz and Theres 1999). In Arabidopsis, a loss of function mutation in the REVOLUTA gene, which is necessary for apical meristem development, also inhibits axillary meristem formation (Talbert et al. 1995).
Even though molecular and genetic analysis of the previously isolated mutants defective in the branching pattern has been invaluable, the information obtained is still fragmentary, and no clear mechanism has been defined. It is obvious that additional informative mutants are required for analysis before the mechanisms underlying this process can be unraveled. In this paper we describe an Arabidopsis mutant with an abnormal shoot branching pattern, designated supershoot (sps). The mutant plants show a massive proliferation of shoots because of an increase in the number of bud initials. The SPS gene has been further investigated by molecular cloning, as well as by study of the temporal and spatial expression pattern. In addition, possible involvement of the SPS gene in modulating hormone levels has been addressed.
Results
sps mutants show extreme shoot proliferation and branching
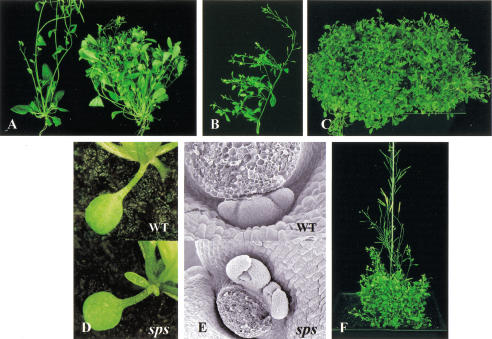
The sps mutants were isolated from a collection of Ds insertion lines, generated by transpositions of a Ds gene trap element from a single T-DNA locus (Sundaresan et al. 1995; see also Materials and Methods). The mutant plants were identified by overproliferation of shoots, resulting in several hundred inflorescences per plant after 4–5 mo (Fig. 1A–C). The developmental phenotypes of sps mutants are not detectable in young seedlings, but become noticeable after the plants produce a few leaves. The sps mutant has the same rate of leaf initiation as wild type during vegetative growth, and the timing of the transition from vegetative to reproductive phase is not affected. The phenotypic defects in the sps mutants become more pronounced after the plants start to flower, at which stage the mutant plants produce primary inflorescences with reduced internode elongation, and secondary inflorescences begin to emerge.
Figure 1.
Shoot branching patterns of sps mutants. (A) Comparison of wild-type (left) and sps mutant (right) showing axillary inflorescences from the rosette leaves. (B) Multiple axillary inflorescences have developed from each single cauline leaf of sps mutant. (C) A single sps mutant plant at 4 mo. (D) Axillary meristem emerging from the axil of a sps cotyledon (bottom), wild-type plant (top) for comparison. (E) Cauline leaf axil from wild-type plant (top) with single arrested axillary meristem, and from sps mutant (bottom) with multiple axillary meristems. The first inflorescences formed in the cauline leaf axils have been excised for convenience in observation. (F) sps mutant plant carrying Ac transposase showing a revertant wild-type sector because of excision of the Ds element.
The number of secondary inflorescences developing from the rosette leaves in wild-type plants varies, but are usually fewer than five. In wild-type plants almost all leaf axils, but not cotyledon axils, have the developmental potential to form an axillary meristem, with a lower frequency in the first pair of leaves. However, the growth of these axillary meristems in wild-type plants is suppressed to various degrees. In contrast, sps mutant plants continue to produce axillary inflorescences from the axils of both rosette leaves (up to 45) and cauline leaves (up to 29 branches per rosette inflorescence), resulting in as many as 500–600 inflorescences per plant after 4–5 mo growth (Fig. 1A–C). Axillary inflorescences in the mutant plants can grow out from every leaf axil and in some extreme cases, from the axils of cotyledons (Fig. 1D). Importantly, in contrast to wild-type plants, multiple axillary meristems are commonly formed in sps plants in the axils of both rosette and cauline leaves (Fig. 1E). Therefore, the drastic increase in the number of axillary branches in the sps mutant is not simply because of loss of apical dominance resulting in the release of suppressed meristems as commonly found in other altered branching mutants, but is also attributable to multiple meristem initiation at the leaf axils.
Branching in sps plants is reiterative and partially acropetal
Reiterative branch initiation in sps mutants usually appears first from the axils of the rosette leaves and later in development, from the axils of the cauline leaves. In wild-type Arabidopsis, the axillary meristems developing from the axils of rosette and cauline leaves produce a few leaf primodia before changing to initiate flower buds, with the concomitant internode elongation resulting in lateral branches. In the axils of wild-type rosette leaves, the first one or two leaves produced by axillary meristems usually lack internode elongation and thus stay at the base of the branch. These axillary meristem-derived rosette leaves can also initiate additional meristems. However, these meristems in the wild-type plants do not develop further and remain as very small arrested axillary buds. In sps mutant plants, these axillary meristems continue to develop into inflorescence branches that produce additional leaves at the base of the stems. Many higher order branches continue to form that reiterate the same developmental pattern and generate the extreme branching phenotype.
In wild-type Arabidopsis, after the transition from shoot apical meristem to inflorescence meristem, axillary meristems arise in a basipetal pattern, first in the youngest leaves, then spread toward the older leaves (Hempel and Feldman 1994; Grbic and Bleecker 2000). However, this pattern of axillary meristem formation is perturbed in the rosette leaves of sps mutants. The mutant plants appear to have an acropetal pattern of axillary meristem formation superimposed on the normal basipetal pattern found in wild-type plants. That is, in sps mutants, axillary meristems that are more developmentally advanced are found in the upper rosette leaves as well as in the leaves at the base of the plants, and the buds that lie in between are relatively less developed (Fig. 2).
Figure 2.
Patterns of axillary inflorescence development in the rosette leaves after bolting. R1–R7 represent rosette leaves 1–7. R1 is the oldest rosette leaf. Cot represents a cotyledon. (A–C) Wild-type axillary buds in the axils of rosette leaves: R7 (A), R5 (B), and R3 (C). (D–F) sps axillary buds in the axils of rosette leaves: R7 (D), R3 (E), and R1 (F). The photographs were taken at the same magnification for the comparison. Arrows indicate the developing axillary inflorescence. (G) Diagrammatic representation of the axillary buds developing in each leaf axil and shown as dotted bars. Relative sizes of the axillary buds are shown as relative sizes of bars. Eight plants from wild type and an sps mutant allele (see text) were used for the analysis. As all of the wild-type plants show a similar pattern of axillary inflorescence development, a representative wild-type plant is shown here (wt), whereas eight individual sps plants are shown (sps: 1–8).
Besides the drastic changes in branching pattern, the sps mutation also causes other developmental defects. However, all these defects are confined to the aerial part of the plant, and we have not been able to detect any changes in the development of the root system. In sps mutants, newly emerging leaves are usually curled, leaf shape is more serrated, and the veins of the adult leaves become strikingly prominent and appear dark green compared to wild type (Fig. 3A). The mutant plants also contain higher levels of chlorophyll in mature leaves (data not shown). The pattern of the vasculature in the rosette leaves is less complex than that in wild-type leaves. In sps mutants, the primary, secondary, and tertiary veins are normal; however, the quaternary veins are usually missing (Fig. 3B). The sps mutants are also defective in floral development. Anthers do not usually release pollen grains and stigmas remain underdeveloped (Fig. 3D). Some flowers show more extreme phenotypes, such as the reduction or absence of petals and stamens. Consequently, the mutants have extremely reduced fertility. Finally, the mutant plants show delayed senescence, so that they can be maintained in a green and healthy state for > 4–5 mo (Fig. 1C).
Figure 3.
Other developmental defects in the sps mutants. (A,B) Details of the leaf veins in wild-type and sps plants. (A) Prominent dark green veins on the leaf surface of the sps adult leaf (right); veins are not apparent in the wild-type leaf (left). (B) Pattern of leaf vasculature in wild-type and sps plants observed after clearing. The sps mutant (right) has less complexity in vascular patterning compared to the wild type (left). (C,D) Floral morphology and development: sps flower with a reduced number of organs and is not fully developed (D), compared to wild type (C).
The SPS gene encodes a cytochrome P450
Genetic analysis indicates that sps mutation cosegregates with a transposed Ds element and is recessive to wild type. To further investigate the molecular nature of the defect, we cloned and characterized the SPS gene. A fragment of genomic DNA flanking the Ds element from a mutant plant was amplified by thermal asymmetric interlaced-PCR (TAIL-PCR) for detailed analysis. A search of the Arabidopsis thaliana genomic database revealed that the Ds element has transposed into the coding region of a putative cytochrome P450, assigned previously as CYP79F1 by the P450 Nomenclature Committee, located on chromosome 1 (BAC clone accession number AC006341). In addition, there is another closely related sequence immediately upstream of the SPS gene that has been designated CYP79F2. The two genes show 89% sequence identity at both nucleotide and amino acid levels, indicating a recent duplication event. This high sequence identity between the two genes indicates that both members are involved in the same metabolic step. Nevertheless, the extreme phenotypic defects caused by a mutation in the CYP79F1 gene alone indicate at least a degree of nonredundancy, perhaps because of differences in expression (see below). The donor T-DNA locus used in the transposon tagging experiment was also found to be within the same BAC clone, 7 kb away from the CYP79F1 gene. Locations of the two Cytochrome P450s as well as the donor T-DNA are shown in Figure 4.
Figure 4.
Positions of the donor T-DNA and insertion sites of Ds gene trap elements in different sps alleles. Positions of the SPS/CYP79F1 gene and the duplicated gene, CYP79F2, on the BAC clone accession number AC006341, are shown. Boxes represent exons. Insertion sites of Ds elements in the SPS gene are indicated as small arrowheads; position of the donor T-DNA is indicated as big arrowhead. Insertion sites of the Ds element in sps-1 to sps-5 are in the exons, whereas in sps-6, it is in the 5′ untranslated leader sequence.
The PCR fragment flanking the Ds element was used as a probe to screen an Arabidopsis leaf and stem cDNA library. The longest SPS cDNA clone isolated from the screen is 1.38 kb long. This cDNA is probably not a full-length clone but is truncated at the 5′ end. This assumption is based on the fact that the predicted start codon of the open reading frame from the available genomic and EST sequences corresponding to the SPS gene, ATTS5112, locates 442 bp before the beginning of the isolated cDNA. This prediction is supported by the information derived from the CYP79F1 closely related gene, CYP79F2. The longest CYP79F2 cDNA clone isolated is 1.8 kb long, consistent with its being full length and confirming the predicted size of the gene products for both genes. The SPS gene is predicted to encode 538 amino acids, with a calculated molecular mass of 61.5 kD. The Cytochrome P450 family is known to be involved in several important biochemical pathways, such as the biosynthesis of brassinosteroids, flavonoids, Gibberellins, and lignin (for review, see Chapple 1998). From the available Arabidopsis genomic database, six predicted P450 proteins have been assigned as members of the CYP79 family. SPS/CYP79F1 shows 46% sequence identity to CYP79C1, and less to other CYP79 members. However, the function of CYP79C1 is not known. In Arabidopsis, CYP79B2 and CYP79B3 can convert tryptohane to indole-3-acetaldoxime, a precursor to IAA and indole glucosinolates (Hull et al. 2000). In other plant species, CYP79A1 from Sorghum bicolor (Koch et al. 1995) and CYP79B1 from Sinapis alba (Bak et al. 1998) have been shown to catalyze the conversion of tyrosine to p-hydroxyphenylacetaldoxime in the biosynthesis of a plant secondary metabolite, the cyanogenic glucoside dhurrin.
To confirm that Ds disruption is indeed the cause of the mutant phenotype, we characterized additional independently isolated sps alleles. A total of 10 sps alleles have been isolated so far, and six alleles have been characterized in more detail. In all six cases, the mutants carried different Ds insertions within the SPS gene (Fig. 4). Mutants sps-1 to sps-5 have Ds elements inserted into the exons of the gene, whereas the insertion site of the Ds element in sps-6 is in the 5′ untranslated leader sequence. All sps alleles show overall phenotypes very similar to one another, consistent with the prediction that they are all null alleles as expected for Ds gene trap insertions within a transcribed region.
Furthermore, reversion experiments show that in sps mutants carrying Ac, excision of the Ds element from the CYP79F1 gene can restore wild-type phenotype. Figure 1F shows a somatic excision resulting in a mosaic plant and shows a sector with a wild-type inflorescence arising from the mutant background. Seeds collected from the wild-type inflorescence in this mosaic plant segregate for revertant wild-type progeny as well as for kanamycin sensitivity, confirming that excision of the Ds element results in reversion.
Taken together, these results show that the developmental defects observed in the sps mutant are caused by disruption of the CYP79F1 gene. Additional confirmation was derived from overexpression studies using transgenic plants. Arabidopsis plants of the WS ecotype were generated in which the cDNA sequence for the closely related duplicate gene, CYP79F2, was placed under the control of CaMV 35S promoter. Whereas many transformants showed seedling lethality, indicating that constitutive overexpression may be deleterious, a few plants could be recovered in which cosuppression resulted in phenotypic changes similar to those observed in loss of function sps mutants (not shown).
SPS expression pattern monitored by gene trap Ds element
Three of the sps alleles confer GUS reporter gene expression because of Ds gene trap element insertions in the correct orientation (Sundaresan et al. 1995). The three sps alleles show similar GUS staining pattern but differ in their staining intensity. To study the pattern of SPS gene expression, the allele with the strongest GUS reporter gene expression, designated sps-2, was selected for more detailed observations. In seedlings, GUS expression is first detected 4–5 d after germination at the branching zone of the vascular tissue that lies at the base of the shoot apical meristem (Fig. 5A). A few days later, the expression level is more intense and found in the vascular tissue of both the hypocotyl and the cotyledons (Fig. 5B).
Figure 5.
SPS expression patterns monitored by GUS expression from the Ds gene trap insertion in sps-2. (A–D) GUS staining patterns of heterozygous sps plants. (A) Staining detected at the vascular junction of a 7-d-old seedling. (B) Extensive staining in the vascular tissue of a 10-d-old seedling. (C) Strong SPS expression at the cauline leaf axil. (D) Staining at the receptacle of flowers. (E) GUS staining patterns of heterozygous (left) and homozygous sps (right) plants.
The expression of the SPS gene is restricted solely to the aerial part of the plant, continuing through the vegetative phase to reproductive phase, and no expression is detected in the roots. Within the inflorescence, SPS expresses in the vascular tissue of the stem and cauline leaves, with the strongest intensity at the base of the cauline leaves where axillary meristems will be initiated (Fig. 5C). SPS is also expressed at the receptacle of the flower and silique (Fig. 5D). The expression pattern in the leaf vasculature appears to be developmentally regulated. It is first observed only in the mid vein in the young leaves and then, when the leaves are more mature, in the finer veins. In addition, SPS-GUS expression levels observed in homozygous mutant plants appear several-fold more intense than those observed in heterozygous plants (Fig. 5E), indicating that expression of SPS may be controlled by a feedback regulatory mechanism that enhances expression level on disruption of the SPS gene.
The expression pattern of SPS as shown by the GUS reporter correlates spatially with the morphological abnormalities observed in the sps mutants. These results indicate that SPS may act locally to control the development of the aerial plant parts. The absence of SPS expression in the roots (Fig. 5B) is consistent with our failure to detect any abnormalities in the root systems of sps mutants. We have recently recovered Ds gene trap insertions in the CYP79F2 gene and showed that this closely related gene is expressed primarily in the roots, and that disruption of this gene has no effect on shoot branching (T. Tantikanjana and V. Sundaresan, unpubl.). The non-overlapping expression patterns of these two genes may explain why mutation of SPS produces a severe shoot phenotype, despite the presence of the CYP79F2 gene. We are currently investigating CYP79F2 gene disruption for possible defects in root development.
Elevated levels of cytokinins in sps mutants
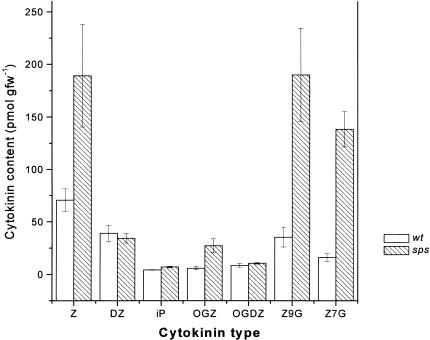
Several of the physiological changes in the aerial part of the plant are strikingly similar to the effects of exogenous application of cytokinin and transgenic plants showing cytokinin overproduction (Hobbie et al. 1994). These physiological changes include the release of lateral buds from apical dominance and an increase in bud initiation as well as the delay of senescence. In addition, one effect of cytokinin that is known to enhance chloroplast development is apparent, particularly in sps leaf veins that are dark green. The serrated leaf phenotype observed in sps mutants is also a characteristic of transgenic plants expressing bacterial cytokinin biosynthesis genes (Rupp et al. 1999). Therefore, we determined the levels of cytokinins in this mutant (Fig. 6): zeatin (Z), dihydrozeatin (DZ), N6-(2-isopentenyl) adenine (iP), cytokinin O-glucosides (Z-O-glucoside [OGZ] and DZ-O-glucoside [OGDZ]), and cytokinin N-glucosides (Z-7-glucoside [Z-7-G] and Z-9-glucoside [Z-9-G]). For the analysis, we used a simplified method for quantifying cytokinins in potentially active forms. Cytokinin ribosides and nucleotides were converted to bases during the process of anhydrous methanolysis, whereas the O-, 7-, and 9-glucoside linkages were not cleaved appreciably (Wang et al. 1995; Yong et al. 2000). Thus, Z, DZ, iP, OGZ, and OGDZ levels presented in this paper refer to the total content of each compound (base) in free, riboside and nucleotide forms. We find that sps mutants have higher levels of Z (2.7×) when compared with wild-type siblings. The levels of OGZ, Z-7-G, and Z-9-G are also increased (five- to ninefold) in the mutant plants. However, the levels of iP, DZ, and OGDZ (minor cytokinins) are not significantly different from wild-type levels (Fig. 6).
Figure 6.
Cytokinin levels in wild-type and sps plants. Cytokinin bases (free and released) and cytokinin glucosides (free and released) were measured using scintillation proximity immnoassay after hydrolysis and purification (including the HPLC step to resolve different cytokinins). Cytokinin bases: zeatin (Z), dihydrozeatin (DZ), and N6-(2-isopentenyl) adenine (iP); cytokinin O-glucosides (zeatin O-glucoside [OGZ] and dihydrozeatin O-glucoside [OGDZ]); cytokinin N-glucosides (zeatin-7-glucoside [Z-7-G] and zeatin-9-glucoside [Z-9-G]). Data were derived from five samples.
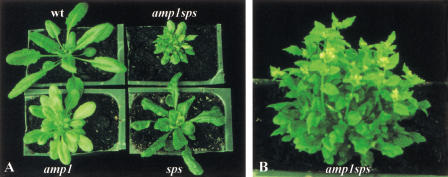
The Arabidopsis mutant, altered meristem program1 (amp1) has also been shown to have elevated levels of endogenous cytokinin (Chaudhury et al. 1993; Chin-Atkins et al. 1996). Even though both sps and amp1 show loss of apical dominance, reduced fertility, and delayed senescence, they differ significantly with respect to other developmental phenotypes. Several amp1 phenotypes, such as polycotyly, faster rate of leaf initiation and abnormal phyllotaxy, are not observed in sps mutants. Conversely, the multiple mersitem in the leaf axils, the reduced number of floral organs, and the dark green leaf veins, characteristic of sps mutants, have not been reported for the amp1 mutant. To date, there is no molecular information on the structure and function of the AMP1 gene. The phenotype of sps and amp1 double mutants is additive (Fig. 7). The double mutants have a faster rate of leaf initiation, abnormal phyllotaxy, dark green veins, abnormal flowers, very short internodes, and extreme shoot branching. Therefore, it is likely that if these two genes regulate cytokinin levels, they function via different pathways and/or have different spatial and temporal expression patterns.
Figure 7.
Double mutant analysis with sps and amp1. (A) Phenotypes of 4-wk-old seedlings of wild type, amp1, sps, and amp1sps mutants. (B) 6-wk-old amp1sps plant.
These differences are underscored by the different phenotypes of dark-grown sps and amp1 mutant seedlings. Exogenous application of cytokinin can mimic some of the light responses in the dark-grown seedlings (Chory et al. 1995). These include inhibition of hypocotyl elongation, relaxed apical hooks, and partially opened cotyledons. Similar responses are also found in the amp1 mutant (Chin-Atkins et al. 1996). However, when sps segregating families are germinated in the dark for 7 d, there are no phenotypic differences from wild-type seedlings (data not shown). This result is consistent with the expression pattern as no GUS activity is detected in the dark-grown seedlings, and indicates that SPS has no function in repression of photomorphogenesis. SPS gene expression in these dark grown seedlings can be induced after 24 h of light exposure and resembles that seen in light-grown seedlings (Fig. 5A).
Auxin is believed to play a central role in controlling branching pattern, and the loss of apical dominance is usually associated with decreased auxin levels. In addition, it has been shown that decreased auxin levels can result in the elevation of cytokinin content (Palni et al. 1988; Bangerth 1994; Zhang et al. 1995). To determine whether higher levels of cytokinins observed in the sps mutant are the consequence of decreased auxin levels, we measured the free IAA content in this mutant compared to the wild-type plants. The free IAA level in sps mutant plants was found to be up to eightfold higher than that in the wild-type plants (223.8 ± 37.67 pg IAA/mg fresh weight in sps plants, n = 3, compared with 28.55 ± 2.67 pg/mg in wild-type plants, n = 4). Because different plant samples were used for the auxin and cytokinin measurements, the relative increases in the levels of the two hormones are not directly comparable. Nevertheless, it is clear that there is a higher level of free auxin in the sps mutants as compared to wild-type plants, so that the increased cytokinin levels and the proliferative shoot branching pattern observed in the sps mutant cannot be explained as a consequence of decreased auxin levels.
Discussion
Proliferative shoot branching in sps mutants is through bud initiation as well as bud growth
Shoot branching in plants is a fundamental process that has been the subject of investigation for many decades. A critical aspect of this process is the multiplication of postembryonically derived meristems, which is controlled in planta by a developmental program that restricts axillary meristem formation and further bud outgrowth. The loss of function mutation in the SUPERSHOOT gene results in an increase in the number of axillary buds initiated as well as their release from apical dominance, indicating that SUPERSHOOT negatively regulates axillary meristem formation and growth in Arabidopsis.
Recent detailed studies of normal axillary meristem development in Arabidopsis have been valuable as a baseline for the study of mutants with altered shoot architecture (Hempel and Feldman 1994; Grbic and Bleecker 2000; Long and Barton 2000). After the transition from the vegetative to the reproductive phase, axillary meristems form in the youngest leaves before they form in the older leaves. As a result, axillary buds with more advanced developmental stages are found near the shoot apex. In sps mutant plants, a higher frequency of branches develop early from the base of the plants, indicating that the SPS gene suppresses axillary meristem formation in an acropetal pattern by exerting a stronger effect on the older leaves. This hypothesis is well supported by the pattern of expression of the SPS gene, which is expressed weakly at the base of newly formed leaves and becomes more intense as the leaves mature. However, it is likely that SPS is not the only regulator involved, as the basipetal developmental gradient is not completely abolished in the sps mutants (Fig. 2). Acropetal formation of the axillary meristems can also be induced in wild-type plants by using conditions that prolong vegetative growth (Long and Barton 2000). Yet even under these conditions, axillary meristem growth switches to a basipetal pattern after the transition to the reproductive phase. It is not known at this time why the pattern of bud formation changes after the transition to flowering, but it has been postulated that axillary meristem activation during vegetative and reproductive development are independently controlled (Schmitz and Theres 1999; Schumacher et al. 1999; Grbic and Bleecker 2000). Our results indicate that SPS might have a major role as a suppressor of bud formation acropetally during the reproductive phase. The only other mutant affecting shoot branching that has been investigated in detail is the axr1-12 mutant involved in the auxin response pathway, which was found to retain the normal basipetal pattern of bud initiation (Stirnberg et al. 1999).
The additional meristems observed in sps mutants initiate only from the leaf axils. In this respect, they differ from transgenic plants ectopically expressing Knotted1 (Kn1) homologs, which show ectopic shoot meristems on vegetative leaves (Sinha et al 1993; Chuck et al. 1996). The mutant axillary buds are always on the adaxial side of the leaves, as in the case of normal axillary meristem formation in wild-type plants (e.g., Talbert et al. 1995; McConnell and Barton 1998). These results indicate that the role of SPS in restricting meristem initiation is confined to the adaxial cells at the leaf axils, reflecting a position-dependent competence for meristem formation. The formation of axillary meristems in the cotyledonary axils in sps mutants is of particular interest (Fig. 1D). This finding indicates that even the cotyledonary axils are competent for axillary bud formation, although in wild-type plants, the process usually occurs only in the leaf axils.
SPS action on shoot proliferation is short range
The mode of gene action in controlling branching pattern has been explored in a few plant species by grafting experiments. The rms2 and rms3 mutants in pea and dad1-1 mutant in Petunia are shown to be defective in graft-transmissible substances that inhibit branching, indicating that the wild-type gene products act as diffusible suppressor of axillary development (Beveridge et al. 1994, 1996; Napoli 1996). To date there is no molecular information on the nature of these two genes. On the other hand, the lateral suppressor (ls) mutant, which has no side shoots, except for the sympodial shoot and the lateral shoot immediately below, is defective in a graft-nontransmissible gene product (Tucker 1979). The homology between the LS gene and the GAI gene involved in the negative regulation of GA signal transduction raises the possibility that the GA response pathway may play a role in controlling the formation of axillary meristems (Schumacher et al. 1999). Despite this sequence similarity, however, more evidence is required to support this hypothesis.
The expression pattern of the SPS gene, particularly its strong expression pattern at the leaf axils, indicates that SPS acts locally to suppress axillary bud growth and de novo bud formation. This hypothesis is further supported by somatic reversion data from mosaic plants. Somatic excision of the Ds element from the sps gene caused complete restoration of the wild-type inflorescence within a definable region of a mutant plant, with no alleviation of the mutant phenotype in adjacent inflorescences (Fig. 1F).
On the other hand, there are indications of a limited nonlocal or longer range action of the SPS gene in some developmental processes, perhaps as a consequence of perturbations in hormone levels. For example, even though SPS expression is detected in the receptacle of the flower buds, this cannot explain the defects in flower development as the expression is detectable only after the floral organs are already formed.
The SPS gene may act as a modulator of cytokinin metabolism
The phenotypic changes in the sps mutants, together with the changes in hormone levels, indicate that SPS may be involved in hormone homeostasis. Extensive physiological studies, as well as more recent transgenic studies (e.g., Klee et al. 1987), indicate that auxin plays a central role in maintaining apical dominance. Detailed analysis of an auxin insensitive mutant, axr1-12, showed that the timing of axillary meristem formation and the number of axillary meristems produced are unaffected, and that the increased number of branches in this mutant is because of the release of inhibited axillary buds (Stirnberg et al. 1999). Therefore, it is likely that the role of auxin in controlling branching pattern arises only after the axillary meristems have been initiated.
Unlike the known hormone mutants characterized thus far, supershoot mutants show an increase in the number of axillary meristems in each axil. It has been generally accepted that cytokinin can promote shoot formation in tissue culture as well as promote release from apical dominance. The striking similarity between the aerial structure of the sps mutants and the effects of cytokinin overproduction indicated the possible involvement of the sps gene in regulating cytokinin levels. Our results show that the sps mutant has higher levels of cytokinins as compared to wild type, implicating SPS as a negative regulator in modulating cytokinin levels.
There are several factors that influence the active cytokinin levels in planta: de novo synthesis, oxidative degradation, formation and hydrolysis of inactive conjugates, transport, and subcellular compartmentalisation (Brzobohaty et al. 1994; Kaminek et al. 1997). The high levels of deactivated cytokinins (Z-7- and Z-9-glucosides) in the sps mutant indicate that this gene is not likely to be involved in either the formation or the hydrolysis of these conjugates. It is also unlikely that the SPS gene directly influences cytokinin biosynthesis in the roots, because the expression pattern and the phenotypic changes observed in the mutants are confined to the aerial part without any detectable expression or phenotypic changes in the root system, which is the main source of cytokinin biosynthesis. It appears more likely that SPS may act by modulating oxidative breakdown of cytokinins. It has been shown that a reduction in cytokinin oxidase activity results in higher levels of active cytokinins as well as cytokinin conjugates (Frank et al. 2000). An increase in the cytokinin conjugates may result from a feedback mechanism that enhances the cytokinin conjugative pathway in an attempt to down-regulate the higher levels of cytokinins in planta.
While this paper was under revision, two groups have independently reported that CYP79F1 can catalyze the conversion of chain-elongated methionines to their aldoximes, the initial step in biosynthesis of short-chain aliphatic glucosinolates (Hansen et al. 2001; Reintanz et al. 2001). Glucosinolates are secondary metabolites that have no known effect on plant growth and development, and it is very unlikely that they exert a direct effect on shoot branching. Moreover, a connection between glucosinolates and cytokinins has not been reported previously. Therefore, it is not clear how a lesion in glucosinolate synthesis could affect cytokinin metabolism. However, it is likely that the loss of CYP79F1 function interferes with methionine metabolism, resulting in changes in the pool sizes of methionine and methionine metabolites. It is interesting to note that there are structural similarities between the methionine metabolite, S-adenosyl-methionine (SAM) and its derivatives, and cytokinin. It has been shown that S-adenosyl-L-homocystein (SAH) hydrolase can bind to cytokinin, indicating that adenine analogs such as cytokinins could modulate SAH hydrolase activity in plants by acting as inhibitors (Mitsui et al. 1996). The reverse scenario might also be possible, that is, an accumulation of adenine analogs from SAM metabolism in sps mutants might act to inhibit cytokinin oxidase. In this regard, we note that transgenic plants in which SAM metabolism has been altered have been found to show phenotypic changes reminiscent of the effect of cytokinin, including highly branched stems and an enhanced ability to form shoots in tissue culture (Noh and Minocha 1994; Kumar et al. 1996). The nature of this crosstalk between primary and secondary metabolism needs to be investigated further.
It has been shown that exogenous application of auxin can stimulate oxidative breakdown of active cytokinin (Palni et al. 1988; Zhang et al. 1995). In addition, removal of the endogenous source of auxin by decapitation leads to an increase in the cytokinin content of xylem exudate (Bangerth 1994), indicating that auxin can influence apical dominance via inhibition of cytokinin synthesis or export from the roots. However, the higher levels of auxin in sps mutants indicate that the increase in cytokinins in these mutants is not because of a reduction in auxin levels. Rather, the changes in the auxin level in sps plants may be part of the feedback mechanisms involved in hormone interactions. Several lines of evidence show that the manipulation of cytokinin levels in plants by transformation with the bacterial cytokinin biosynthesis gene results in more auxin (Binns et al. 1987; Makarova et al. 1996). Finally, it is important to note that the cytokinin and auxin measurements on the entire aerial parts of the plants may not reflect the changes in levels of these hormones at specific developmental sites. Because the SPS gene is most strongly expressed at specific zones such as leaf axils, it is quite likely that the increases in cytokinin levels in sps mutants are not uniform but are most severe at these sites. As discussed above, our observations of the revertant sectors on mosaic plants also support the idea that the action of SPS is local rather than long range. In conclusion, the results from this study lead us to suggest that Arabidopsis plants control shoot branching and the development of aerial structures in part by attenuating cytokinins at the sites of bud initiation to levels that inhibit de novo bud formation as well as the release of bud growth.
Materials and methods
Plant materials and growth conditions
All lines were derived from Arabidopsis thaliana WS ecotype. Plants were grown on soil at 20°C and 60% humidity under a 16 h light/8 h dark cycle. The sps mutants were maintained by transferring to new soil once every 6–8 wk.
Generation of Ds insertion lines
The targeted transposon tagging system used was a modified system based on the inducible system previously reported (Balcells et al. 1994) and the gene trap system (Sundaresan et al. 1995). Both the autonomous element (Ac) and a nonautonomous element (a modified Ds) (Sundaresan et al. 1995) were placed on the same T-DNA to simplify the generation of the independent insertion lines. Expression of the transposase was driven by the induction of a heat-shock promoter. The transactivition of the Ds elements was performed by subjecting the starter line during flowering stage to high temperature (40°C–42°C) as described (Balcells et al. 1994). A detailed description of the transposon mutagenesis system will be published elsewhere. Reversion analysis was performed by using heat treatment with the sps heterozygous plants. The heat-shocked seeds were then germinated and observed for the restoration of the wild-type sectors.
Molecular cloning of the SPS gene
Genomic sequences flanking the Ds element were obtained with TAIL-PCR, according to Parinov et al. (1999). Sequencing of the flanking sequences was performed with an ABI sequencer. The flanking sequences were used as probe to screen the SPS cDNA clones from Arabidopsis thaliana 5′-STRETCH cDNA library (Clontech) prepared from 4.5 wk-old Columbia leaves and stem-grown under light. The screening of the SPS cDNA was performed according to the manufacturing protocol. All cloning procedures were performed according to standard methods (Sambrook et al. 1989)
Staining for GUS expression
Plant materials were stained in GUS staining solution (100 mM Na Phosphate at pH 7.0, 10 mM EDTA, 0.1% Triton X-100, 1 mg/mL of X-Gluc [Biosynth AG]). Plant materials were generally stained in the GUS solution containing 1 mM potassium ferricyanide and 1 mM potassium ferrocyanide; the specificity of localization was confirmed by increasing the potassium ferricyanide and potassium ferrocyanide to 10mM. The samples were incubated at 37°C overnight after being placed under vacuum for 10 min in a dessicator. The stain solution was removed and the tissues were cleared by incubating with several changes of 70% ethanol.
Scanning electron microscopy
Samples were fixed in 2% glutaldehyde in 25 mM sodium phosphate buffer (pH 6.8), infiltrated with 0.5% osmium tetroxide, dehydrated through an ethanol series, and critical-point dried using liquid carbon dioxide. After coating with gold, samples were viewed using a scanning electron microscope (model JSM 5310-LV; JEOL). For the analysis of flower organs, samples were placed on a double-sided sticky tape and frozen in liquid nitrogen before viewing.
Quantification of cytokinins
Aerial parts, excluding the primary inflorescences of the plants, were collected after bolting for the analysis. Data were derived from five samples of the sps mutants and wild-type siblings. Cytokinins were extracted and the extract subjected to acid-catalyzed methanolysis in the presence of the water-scavenging reagent, 2,2-dimethoxypropane. This procedure (Chaudhury et al. 1993; Wang et al. 1995) converts 9-ribosides and nucleotides to bases, whereas O-, 7-, and 9-glucoside linkages are not cleaved appreciably. After three-step column purification (Chaudhury et al. 1993), the bases (free plus bound and glucosides) were further purified by HPLC for quantification by scintillation proximity immunoassay (Wang et al. 1995; Yong et al. 2000). Cytokinin bases (Z, DZ, and iP) and Z-9-glucoside were measured directly, but the O-glucosides were first hydrolyzed to bases with β-glucosidase (Yong et al. 2000). Z-7-glucoside was converted to Z by periodate oxidation and cleavage with freshly distilled cyclohexyl-amine (Letham and Gollnow 1985) before immunoassay.
Quantification of free IAA
Plant material weighing 0.5 g was homogenized and extracted in 4 mL cold methanol containing 0.02% (w/v) DEDTCA (diethyl dithiocarbamic acid) as an antioxidant at 4°C for 2 h with stirring. The extracts were then filtered and evaporated to dryness. Before homogenization, 50 ng 13C6-IAA (internal standard, Cambridge Isotope Laboratories) was added to each sample. The samples were then dissolved in 1 mL methanol and transferred to 1.5 mL Eppendorf tubes and evaporated to dryness. They were then dissolved in 500 μL Na-phosphate buffer (0.05 M at pH 7.0 containing 0.02% [w/v] DEDTCA) and purified and analyzed by gas chromatography-selected reaction monitoring-masspectrometry (GC-SRM-MS) according to Edlund et al. (1995).
Generation of sps amp1 double mutants
The amp1 mutant was derived from Columbia ecotype. The sps heterozygous plants were crossed with amp1 homozygous plants. The double amp1sps mutants were selected from the segregating F2 seedlings for the investigation. The amp1 homozygotes were confirmed by using primers for a polymorphism closely linked to the amp1 allele (A.M. Chaudhury, pers. comm.), and the sps homozygotes were confirmed by using a combination of SPS gene-specific primers and a Ds specific primer.
Analysis of leaf vasculature
Leaves were cleared by incubating at 70°C for 30 min in 95% ethanol followed by incubating at 90°C for 30 min in lactic acid : phenol : glycerol:water (1 : 1 : 1 : 1) (Telfer and Poethig 1994).
Acknowledgments
We thank Drs. Nam Hai Chua and Jaideep Mathur for helpful advice, Dr. A. Chaudhury for providing amp1 seeds, Ms. Chan Yang Sun for SEM analysis. This research was supported by grants from the National Science and Technology Board of Singapore. G.S. and K.L. acknowledge financial support from the Swedish Research Council.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL director@ima.org.sg; FAX (65) 872-7012.
Article and publication are at http://www.genesdev.org/cgi/doi/10.1101/gad.887301.
References
- Bak S, Nielsen HL, Halkier BA. The presence of CYP79 homologues in glucosinolate-producing plants shows evolutinary conservation of the enzymes in the conversion of amino acid to aldoxime in the biosynthesis of cyanogenic glucosides and glucosinolates. Plant Mol Biol. 1998;38:725–734. doi: 10.1023/a:1006064202774. [DOI] [PubMed] [Google Scholar]
- Balcells L, Sundberg E, Coupland G. A heat-shock promoter fusion to the Ac transposase gene drives inducible transposition of a Ds element during Arabidopsis embryo development. Plant J. 1994;5:755–764. [Google Scholar]
- Bangerth F. Response of cytokinin concentration in the xylem exudate of bean (Phaseolus vulgaris L.) plants to decapitation and auxin treatment and relationship to apical dominance. Planta. 1994;194:439–442. [Google Scholar]
- Beveridge CA, Ross JJ, Murfet IC. Branching mutant rms-2 in Pisum sativum. Grafting studies and endogenous indole-3-acetic acid levels. Plant Physiol. 1994;104:953–959. doi: 10.1104/pp.104.3.953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beveridge CA, Ross JJ, Murfet IC. Branching in Pisum; action of genes Rms3 and Rms4. Plant Physiol. 1996;110:859–865. doi: 10.1104/pp.110.3.859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Binns AN, Labriola J, Black RC. Initiation of auxin autonomy in Nicotiana glutinosa cells be the cytokinin-biosynthesis gene from Agrobacterium tumefaciens. Planta. 1987;171:539–548. doi: 10.1007/BF00392304. [DOI] [PubMed] [Google Scholar]
- Brzobohaty B, Moore I, Plame K. Cytokinin metabolism: Implications for regulation of plant growth and development. Plant Mol Biol. 1994;26:1483–1497. doi: 10.1007/BF00016486. [DOI] [PubMed] [Google Scholar]
- Chapple C. Molecular-genetic analysis of plant cytochrome P450-dependent monooxygenases. Annu Rev Plant Physiol Plant Mol Biol. 1998;49:311–343. doi: 10.1146/annurev.arplant.49.1.311. [DOI] [PubMed] [Google Scholar]
- Chatfield PS, Stirnberg P, Forde BG, Leyser O. The hormonal regulation of axillary bud growth in Arabidopsis. Plant J. 2000;24:159–169. doi: 10.1046/j.1365-313x.2000.00862.x. [DOI] [PubMed] [Google Scholar]
- Chaudhury AM, Letham S, Craig S, E.S. D. amp1-a mutant with high cytokinin levels and altered embryonic pattern, faster vegetative growth, constitutive photomorphogenesis and precocious flowering. Plant J. 1993;4:907–916. [Google Scholar]
- Chin-Atkins AN, Craig S, Hocart CH, Dennis ES, Chaudhury AM. Increased endogenous cytokinin in the Arabidopsis amp1 mutant corresponds with de-etiolation responses. Planta. 1996;198:549–556. doi: 10.1007/BF00262641. [DOI] [PubMed] [Google Scholar]
- Chory J, Cook RK, Dixon R, Elich T, Li HM, Lopez E, Mochizuki N, Nagpal P, Pepper A, Poole D, et al. Signal-transduction pathways controlling light-regulated development in Arabidopsis. Philos Trans R Soc Lond B Biol Sci. 1995;350:59–65. doi: 10.1098/rstb.1995.0138. [DOI] [PubMed] [Google Scholar]
- Chuck G, Lincoln C, Hake S. KNAT1 induces lobed leaves with ectopic meristems when overexpressed in Arabidopsis. Plant Cell. 1996;8:1277–1289. doi: 10.1105/tpc.8.8.1277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doebley J, Stec A, Hubbard L. The evolution of apical dominance in maize. Nature. 1997;386:485–488. doi: 10.1038/386485a0. [DOI] [PubMed] [Google Scholar]
- Edlund A, Eklöf S, Sundberg B, Moritz T, Sandberg G. A microscale technique for gas chromatography-mass spectrometry measurements of picogram amounts of Indole-3-acetic acid in plant tissues. Plant Physiol. 1995;108:1043–1047. doi: 10.1104/pp.108.3.1043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank M, Rupp H-M, Prinsen E, Motyka V, Van Onckelen H, Schmülling T. Hormone autotrophic growth and differentiation identifies mutant lines of Arabidopsis with altered cytokinin and auxin content or signaling. Plant Physiol. 2000;122:721–730. doi: 10.1104/pp.122.3.721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grbic V, Bleecker AB. Axillary meristem development in Arabidopsis thaliana. Plant J. 2000;21:215–223. doi: 10.1046/j.1365-313x.2000.00670.x. [DOI] [PubMed] [Google Scholar]
- Halle F. Ecology of reiteration in tropical trees. In: Kurmann MH, Hemsley AR, editors. The evolution of plant architecture. Kew: Royal Botanic Gardens; 1999. pp. 93–107. [Google Scholar]
- Hansen CH, Wittstock U, Olsen CE, Hick AJ, Pickett JA, Halkier BA. Cytochrome P450 CYP79F1 from Arabidopsis catalyzes the conversion of dihomomethionine and trihomomethionine to the corresponding aldoximes in the biosynthesis of aliphatic glucosinolates. J Biol Chem. 2001;276:11078–11085. doi: 10.1074/jbc.M010123200. [DOI] [PubMed] [Google Scholar]
- Hedden P, Phillips AL. Manipulation of hormone biosynthetic genes in transgenic plants. Curr Opin Biotech. 2000;11:130–137. doi: 10.1016/s0958-1669(00)00071-9. [DOI] [PubMed] [Google Scholar]
- Hempel FD, Feldman LJ. Bi-directional inflorescence development in Arabidopsis thaliana: Acropetal initiation of flowers and basipetal initiation of paraclades. Planta. 1994;192:276–286. [Google Scholar]
- Hobbie L, Timpte C, Estelle M. Molecular genetics of auxin and cytokinin. Plant Mol Biol. 1994;26:1499–1519. doi: 10.1007/BF00016487. [DOI] [PubMed] [Google Scholar]
- Hull AK, Vij R, Celenza JL. Arabidopsis cytochrome P450s that catalyze the first step of tryptophan-dependent indole-3-acetic acid biosynthesis. Proc Natl Acad Sci. 2000;97:2379–2384. doi: 10.1073/pnas.040569997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaminek M, Motyka V, Vackova R. Regulation of cytokinin content in plant cells. Physiol Plantarum. 1997;101:689–700. [Google Scholar]
- Klee HJ, Horsch RB, Hinchee MA, Hein MB, Hoffmann NL. The effects of overproduction of two Agrobacterium tumefaciens T-DNA auxin biosynthetic gene products in transgenic petunia plants. Genes & Dev. 1987;1:86–96. [Google Scholar]
- Koch BM, Sibbesen O, Halkier BA, Svendsen I, Moller BL. The primary sequence of cytochrome P450tyr, the multifunctional N-hydroxylase catalyzing the conversion of L-tyrosine to p-hydroxyphenylacetaldehyde oxime in the biosynthesis of the cyanogenic glucoside dhurrin in Sorghum bicolor (L.) Arch Biochem Biophys. 1995;323:177–186. doi: 10.1006/abbi.1995.0024. [DOI] [PubMed] [Google Scholar]
- Kumar A, Taylor MA, Arif SAM, Davies HV. Potato plants expressing antisense and sense S-adenosylmethionine decarboxylase (SAMDC) transgenes show altered levels of polyamines and ethylene: Antisense plants display abnormal phenotypes. Plant J. 1996;9:147–158. [Google Scholar]
- Letham DS, Gollnow BI. Regulators of cell division in plant tissues. XXX. Cytokinin metabolism in relation to radish cotyledon expansion and senescence. J Plant Growth Regul. 1985;4:129–145. [Google Scholar]
- Li CJ, Bangerth F. The possible role of cytokinins, ethylene and indoleacetic acid in apical dominance. In: Karssen C, et al., editors. Progress in plant growth and regulation. Netherlands: Kluwer Academic Publishers; 1992. pp. 431–436. [Google Scholar]
- Long J, Barton MK. Initiation of axillary and floral meristems in Arabidopsis. Dev Biol. 2000;218:341–353. doi: 10.1006/dbio.1999.9572. [DOI] [PubMed] [Google Scholar]
- Makarova RV, Borisova TA, Machackova I, Kefeli VI. Effect of alien ipt gene on hormonal concentrations of plants. In: Smith AR, et al., editors. Plant hormone signal perception and transduction. Netherlands: Kluwer Academic Publishers; 1996. pp. 171–173. [Google Scholar]
- McConnell J, Barton M. Leaf polarity and meristem formation in Arabidopsis. Development. 1998;125:2935–2942. doi: 10.1242/dev.125.15.2935. [DOI] [PubMed] [Google Scholar]
- Mitsui S, Wakasugi T, Sugiura M. A cytokinin-binding protein complex from tobacco leaves. J Plant Growth Regul. 1996;18:39–43. [Google Scholar]
- Morris RO. Genes specifying auxin and cytokinin biosynthesis in phytopathogens. Ann Rev Plant Physiol. 1986;37:509–538. [Google Scholar]
- Napoli C. Highly branched phenotype of the Petunia dad1-1 mutant is reverse by grafting. Plant Physiol. 1996;111:27–37. doi: 10.1104/pp.111.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noh E, Minocha SC. Expression of a human S-adenosylmethionine decarboxylase cDNA in transgenic tobacco and its effects on polyamine biosynthesis. Transgenic Res. 1994;3:26–35. doi: 10.1007/BF01976024. [DOI] [PubMed] [Google Scholar]
- Palni LMS, Burch L, Horgan R. The effect of auxin concentration on cytokinin stability and metabolism. Planta. 1988;194:231–234. doi: 10.1007/BF00394775. [DOI] [PubMed] [Google Scholar]
- Parinov S, Sevugan M, Ye D, Yang WC, Kumaran M, Sundaresan V. Analysis of flanking sequences from dissociation insertion lines: A database for reverse genetics in Arabidopsis. Plant Cell. 1999;11:2263–2270. doi: 10.1105/tpc.11.12.2263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reintanz B, Lehnen M, Reichelt M, Gershenzon J, Kowalczyk M, Sandberg G, Godde M, Uhl R, Palme K. bus, a Bushy Arabidopsis CYP79F1 knockout mutant with abolished synthesis of short-chain aliphatic glucosinolates. Plant Cell. 2001;13:351–367. doi: 10.1105/tpc.13.2.351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rupp H-M, Frank M, Werner T, Strnad M, Schmülling T. Increased steady state mRNA levels of the STM and KNAT1 homeobox genes in cytokinin overproducing Arabidopsis thaliana indicate a role for cytokinins in the shoot apical meristem. Plant J. 1999;18:557–563. doi: 10.1046/j.1365-313x.1999.00472.x. [DOI] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T. Molecular Cloning: A Laboratory Manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- Schmitz G, Theres K. Genetic control of branching in Arabidopsis and tomato. Curr Opin Plant Biol. 1999;2:51–55. doi: 10.1016/s1369-5266(99)80010-7. [DOI] [PubMed] [Google Scholar]
- Schumacher K, Schmitt T, Rossberg M, Schmitz GTK. The lateral suppressor (Ls) gene of tomato encodes a new member of the VHIID protein family. Proc Natl Acad Sci. 1999;96:290–295. doi: 10.1073/pnas.96.1.290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinha N, Williams R, Hake S. Overexpression of the maize homeobox gene, Knotted-1, causes a switch from determinate to indeterminate cell fates. Genes & Dev. 1993;7:787–795. doi: 10.1101/gad.7.5.787. [DOI] [PubMed] [Google Scholar]
- Stirnberg P, Chatfield SP, Leyser HMO. AXR1 acts after lateral bud formation to inhibit lateral bud growth in Arabidopsis. Plant Physiol. 1999;121:839–847. doi: 10.1104/pp.121.3.839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sundaresan V, Springer P, Volpe T, Haward S, Jones JDG, Dean C, Ma H, Martienssen R. Patterns of gene action in plant development revealed by enhancer trap and gene trap transposable elements. Genes & Dev. 1995;9:1797–1810. doi: 10.1101/gad.9.14.1797. [DOI] [PubMed] [Google Scholar]
- Sussex IM, Kerk NM. The evolution of plant architecture. Curr Opin Plant Biol. 2001;4:33–37. doi: 10.1016/s1369-5266(00)00132-1. [DOI] [PubMed] [Google Scholar]
- Talbert PB, Adler HT, Parks DW, Comai L. The REVOLUTA gene is necessary for apical meristem development and for limiting cell divisions in the leaves and stems of Arabidopsis thaliana. Development. 1995;121:2723–2735. doi: 10.1242/dev.121.9.2723. [DOI] [PubMed] [Google Scholar]
- Tamas IA. Hormonal regulation of apical dominance. In: Davies PJ, editor. Plant Hormones. The Netherlands: Kluwer Academic Publishers; 1995. pp. 572–579. [Google Scholar]
- Telfer A, Poethig RS. Leaf development in Arabidopsis. In: Meyerowitz EM, Somerville CR, editors. Arabidopsis. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1994. p. 389. [Google Scholar]
- Tucker DJ. Axillary bud development in the tomato. Ann Bot. 1979;43:393–395. [Google Scholar]
- Wang J, Letham DS, Taverner E, Badenoch-Jones J, Hocart CH. A procedure for quantification of cytokinins as free bases involving scintillation proximity immunoassay. Physiol Plantarum. 1995;95:91–98. [Google Scholar]
- Yong JWH, Wong SC, Letham DS, Hocart CH, Farquhar GD. Effects of elevated [CO2] and nitrogen nutrition on cytokinins in the xylem sap and leaves of cotton. Plant Physiol. 2000;124:767–779. doi: 10.1104/pp.124.2.767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang R, Zhang X, Wang J, Letham DS, Mckinney AA, Higgins TJV. The effect of auxin on cytokinin levels and metabolism in transgenic tobacco tissue expressing an ipt gene. Planta. 1995;196:84–94. [Google Scholar]