Abstract
Glycogen synthase kinase 3β (GSK3β) can regulate a broad range of cellular processes in a variety of cell types and tissues through its ability to phosphorylate its substrates in a cell- and time-specific manner. Although it is known that Axin and presenilin help to recruit β-catenin/Smad3 and tau protein to GSK3β, respectively, it is not clear how many of the other GSK3β substrates are recruited to it. Here, we have established the binding of GSK3β with a novel scaffold protein, STRAP, through its WD40 domains. In a new finding, we have observed that STRAP, GSK3β and Axin form a ternary complex together. We show for the first time that intracellular fragment of Notch3 (ICN3) binds with GSK3β through the ankyrin repeat domain. This binding between STRAP and GSK3β is reduced by small-molecule inhibitors of GSK3β. Further studies revealed that STRAP also binds ICN3 through the ankyrin repeat region, and this binding is enhanced in a proteasomal inhibition-dependent manner. In vivo ubiquitination studies indicate that STRAP reduces ubiquitination of ICN3, suggesting a role of STRAP in stabilizing ICN3. This is supported by the fact that STRAP and Notch3 are co-upregulated and co-localized in 59% of non-small cell lung cancers, as observed in an immunohistochemical staining of tissue microarrays. These results provide a potential mechanism by which STRAP regulates GSK3β function and Notch3 stabilization and further support the oncogenic functions of STRAP.
Key words: STRAP, GSK3β, Notch3, axin, lung cancer, ubiquitination
Introduction
The serine/threonine protein kinase glycogen synthase kinase 3β (GSK3β) is a highly conserved protein kinase. In mammals, GSK-3 is encoded by two genes, termed GSK-3α and GSK-3β.1 It has been shown that a variety of signaling pathways acting on cells can result in a reversible inhibition of its enzymatic activity. Interestingly, most of the substrates of GSK3β are functionally inhibited after phosphorylation. Almost all of the GSK3β substrates are required to have a priming phosphate at n + 4 (where n is the site of phosphorylation of a serine or threonine residue) to be phosphorylated in turn by GSK3β. GSK3β phosphorylates and thereby regulates the functions of many metabolic, signaling and structural proteins. One of the most important roles of GSK3β is the regulation by phosphorylation of numerous transcription factors including AP-1, c-Myc, Notch, c-Jun, nuclear factor kappa B (NFκB), nuclear factor of activating T cells (NFAT) and heat shock factor-1 (HSF-1), and thereby the regulation of genes involved in cell proliferation, cell death, immunity, apoptosis, development, metabolism regulation, neuronal growth and differentiation, cell polarity and cell fate. Since GSK3β is involved in such a variety of signaling pathways and cellular functions, it is thought that agents that target specific functions of GSK3β may be needed to selectively interfere with GSK3β signaling. Towards this end, it is necessary to understand how GSK3β regulates its many roles in the cell. Since GSK3β has a predominant role in the control of several intracellular pathways, its activity needs to be carefully regulated. GSK3β is phosphorylated at Serine9 by Protein Kinase B (PKB) in response to insulin signaling. This phosphorylation creates a primed pseudosubstrate that occupies the catalytic groove of GSK3β and prevents phosphorylation of exogenous substrates that leads to inhibition of GSK3β activity toward its substrates.2
On the other hand, the above mechanism appears to play no role in the regulation of GSK3β by Wnt signaling pathway where recruiting of β-catenin to GSK3β is mediated through scaffolds such as Axin and adenomatous polyposis coli (APC) proteins, and Dishevelled disrupts this complex formation. It is certainly a very critical property of GSK3β to avoid any cross-talk of the various pathways regulated by it, as this can lead to ectopic or inappropriate stabilization of key GSK3β substrates, such as β-catenin and c-Myc, and drive cells towards oncogenesis. It seems that protein complex formation with specific scaffold proteins and intracellular localization are effective ways to regulate this enzyme. Apart from β-catenin, GSK3β is shown to phosphorylate c-Myc, c-Jun, Cyclin E, Notch and Cyclin D1 and target them for proteolytic degradation through ubiquitin-proteasome system.3
Intracellular domains of Notch1 and Notch2 have been validated as GSK3β substrates. Notch signaling, conserved from flies to mammals, regulates cell fate decisions through direct cell-cell interactions.4 Notch signaling is known to regulate a wide variety of developmental processes such as hematopoiesis, neurogenesis, myogenesis, wing formation and somite segregation.5 Notch signaling pathway relies only on a few key components, where binding of ligands from one cell to Notch receptors in neighboring cells triggers two serial proteolytic cleavages of Notch receptor, which results in a release of the intracellular domain of Notch (ICN) from the plasma membrane. The intracellular domain of Notch has four main sub-domains: the RAM, ankyrin repeat, RE/AC and C-terminal region. ICN then travels to the nucleus and associates with a CSL (CBF-1/Suppressor of Hairless/Lag-1) DNA binding protein to activate transcription of target genes. Hes and Hey have been well established to be primary downstream targets following Notch activation.6,7
Patients suffering from T-cell lymphoblastic leukemia (T-ALL) provided the first evidence for an oncogenic function of Notch. About 1% of the cases possess a specific chromosomal translocation, t(7;9), that produces a truncated Notch 1 receptor that corresponds to ICN1, which behaves in a constitutively active fashion. More recently, two types of activating mutations within Notch 1 were found in 55–60% of human T-ALL cases.8 Tissue microarray studies have shown that high expression levels of Jagged1 and/or Notch 1 in human breast cancer are associated with a more aggressive disease course.9 Ligand-driven activation of Notch pathway seems to play a role during the development of T-ALL and some other solid tumors by inducing increased proliferation, protection from apoptosis and maintenance of cancer initiating cells.10 Notch and Kras have been proposed to synergize during pancreatic cancer development.11 Notch signaling has also been suggested to be required in the hypoxia-induced EMT and cell migration in tumor cells.
Just as other Notches, Notch3 plays a role in development indicated by its ability to alter cell fate in animals expressing gain-of-function mutants of Notch3.12,13 Most studies relating to the role of Notch in cancer focus on Notch1, and little is known about the role of Notch3 in epithelial tumors, such as lung carcinomas. Recently, Notch3 was shown to be upregulated in non-small cell lung cancers (NSCLC).14 This is significant as Notch3 expression in normal adult lungs is restricted only to the smooth muscle cells of blood vessels.15 Furthermore, inhibition of the Notch3 pathway using a dominant-negative receptor dramatically decreased the malignant potential of lung cancer cells, as evidenced by reduced growth in soft agar and increase in growth factor dependence. Treatment of lung cancer cells with a γ-secretase inhibitor inhibited Notch3 signaling, reduced tumor cell proliferation and induced apoptosis.16 Recent reports have shown increased expression of Notch3 in T-cell leukemias and epithelial malignancies arising from pancreas, ovary, breast and lung.17–21 How Notch3 is upregulated in these cancers and what is the mechanism for the possible Notch3-mediated carcinogenesis is not known. Here we show that STRAP binds with GSK3β and forms a ternary complex together with Axin. We, for the first time, show that GSK3β and STRAP bind with ICN3 through the ankyrin repeat region. Finally, we show that STRAP decreases ubiquitination of ICN3 and may help to stabilize it. STRAP is already known to be upregulated in 78% lung cancers and we found that STRAP, and ICN3 are co-upregulated in 59% of lung cancers. These reports suggest that STRAP may stabilize Notch3 in non-small cell lung cancers.
Results
STRAP binds to GSK3β through its WD40 domain region.
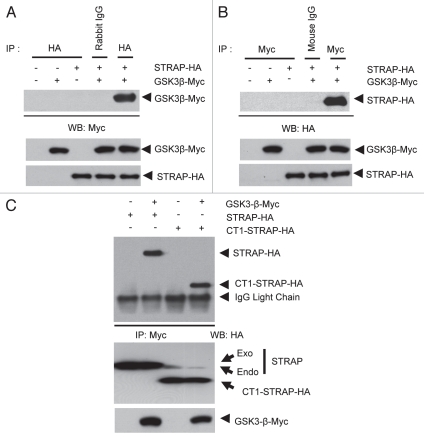
Ewing et al. reported the possibility of an interaction between the scaffold protein STRAP and GSK3β, the classic enzyme in the Wnt and insulin signaling pathways. During a large-scale analysis of human protein-protein interactions using mass spectroscopy, they predicted that STRAP binds with GSK3β with a probability of only 0.5. Since they had not validated the binding between STRAP and GSK3β, we decided to determine the interaction between STRAP and GSK3β and the functional outcome of this binding. 293T cells were transfected with myc-tagged GSK3β and HA-tagged STRAP. Cells were lysed, and the lysates were incubated with either anti-HA, anti-Myc or appropriate control pre-immune IgG antibody followed by incubation with protein G-sepharose beads. Figure 1A and B show that GSK3β-Myc was co-immunoprecipitated with STRAP-HA and vice versa. Corresponding negative controls with either transfection of single plasmid (second and third lane) or immunoprecipitation with a pre-immune Rabbit or Mouse IgG (fourth lane of both western blots) did not show any co-immunoprecipitated proteins indicating that the binding between STRAP and GSK3β was a specific one.
Figure 1.
GSK3β and STRAP physically interact with each other. (A) STRAP-HA and GSK3β-myc constructs were transiently transfected into 293T cells. Cells were subjected to lysis 48 hours after tranfection, immunoprecipitation using 1 µg of pre-immuneanti-rabbit IgG or 1 µg anti-HA antibody and immunoblotted with anti-myc antibody as indicated. Bottom parts show comparable expression of GSK3β-myc and STRAP-HA in the lysates. (B) Same as above except immunoprecipitations were done with anti-mouse IgG and anti-myc antibodies and immunoblotting was done with anti-HA antibody. All antibodies are from Santa Cruz Biotechnology. Bottom parts show comparable expression of GSK3β and STRAP in the lysates. (C) GSK3β interacts with the WD40-domain region of STRAP. STRAP-HA, CT1-STRAP-HA and GSK3β-myc constructs were transiently transfected into 293T cells. Immunoprecipitation was done with anti-myc and immunoblotting with anti-HA antibody as indicated. Light chain of the myc antibody used for immunoprecipitation is visible just below the CT1-STRAP-HA band. Bottom parts show comparable expression of STRAP-HA, CT1-STRAP-HA and GSK3β-myc in the lysates.
After validating the specific interaction between STRAP and GSK3β, we decided to do a preliminary mapping of the region of STRAP that mediates this interaction. We tested whether STRAP binds GSK3β through its WD domain region or the C-terminal low complexity region. We used a STRAP deletion construct that has only the WD40 region, i.e., the N-terminal 294 amino acids, but lacks the C-terminal 57 amino acids (CT1-STRAP). When a co-immunoprecipitation assay was performed in a similar way as above, CT1-STRAP-HA was co-precipitated equally well with GSK3β as the wild-type STRAP-HA, indicating that GSK3β binds STRAP through the WD40 domain region (Fig. 1C, lanes 2 and 4). Further search to find the exact STRAP region that binds to GSK3β was prohibited by the fact that any deletions in the WD40 region have a tendency to make STRAP unstable. Together, these results indicate that STRAP specifically associates with GSK3β though its WD40 domain region. It is possible that STRAP may either recruit an upstream signaling kinase to bind with GSK3β, or it can recruit a substrate to GSK3β.
Effect of GSK3β inhibitors on STRAP/GSK3β binding.
To understand whether STRAP has any preference towards binding activated or inhibited state of GSK3β, we repeated the co-immunoprecipitation assays between GSK3β and STRAP in the presence of the upstream inhibitor of GSK3β, lithium chloride and three other small-molecule inhibitors, namely AR-A01441, SB415286 and SB216763. LiCl is an ATP noncompetitive inhibitor of GSK3β activity (Ki 2 mM) that has been used extensively in studies investigating the functional role of GSK3β.23–25 The lithium ion competes with the Mg++ ion that is necessary for GSK3β activity. LiCl has also been reported to acutely elevate phosphatidylinositol-3-phosphate levels in some cell types, thereby activating PKB.26 Activated PKB phosphorylates and inhibits GSK3β, suggesting that LiCl has the potential to inhibit this kinase both directly and indirectly in cells. AR-A01441, SB415286 and SB216763 are among the new potent, highly selective and cell-permeable small-molecule inhibitors of GSK3β. These compounds inhibit their target protein kinase in an ATP competitive manner.27 These small-molecule inhibitors bind to the Val135 and Asp133 residues in the catalytic domain of GSK3β and inhibit GSK3β in vitro at 0.01 mM ATP with IC50s less than 100 nM. In the case of some other small-molecule inhibitors, it is speculated that small-molecule inhibitors can cross react with other structurally similar enzymes and result in a range of cellular alterations not related to the inhibition of enzyme of concern. But a review of literature shows that the effect of these small-molecule inhibitors in very similar to knocking down GSK3β using shRNA or siRNA. Hongisto et al. showed that inhibition of GSK3β using small-molecule inhibitors like SB216763 and LY294002 had a very similar effect to using GSK3β-specific shRNA or lithium chloride, resulting in protection of cerebellar granule neurons from trophic-deprivation-induced death, probably by blocking the stress-induced Bim protein elevation.28 Rinnab et al. showed that in androgen receptor (AR)-positive prostate cancer 22Rv1 PCa cells, inhibition of GSK3β using both shRNA or small-molecule inhibitors like SB216763 and maleimide decreased AR-dependent transcriptional activity by decreasing the intracellular AR protein levels.29 In a report by Tighe et al., both SB-415286 and AR-A014418 led to an aligning defect of chromosomes during mitosis, resulting in non-disjunction and knockdown of GSK3β using siRNA had a very similar effect.30 Finally, inhibition of GSK3β by small molecular inhibitors or shRNA lead to increased β-catenin in the cells.31,32 These reports suggest that using small-molecule inhibitors is a practical and valid approach to study the effect of GSK3β inhibition in the cells.
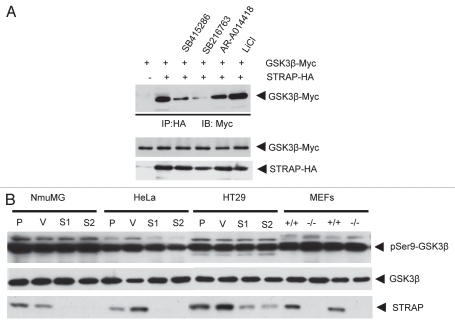
Comparison between lane 2 and lane 6 of Figure 2A indicates that LiCl did not have any effect on binding between STRAP and GSK3β, suggesting that STRAP may not have any dependence on the active or inactive state of GSK3β to bind with it. In contrast to this, STRAP binding with GSK3β was reduced significantly in presence of AR-A014418 (lane 5) and reduced considerably in presence of the other two small-molecule inhibitors, SB216763 and SB415286 (Fig. 2A, lane 3 and 4). Since these inhibitors directly bind with the catalytic domain of GSK3β, this domain or, more likely, the region surrounding the catalytic site seems to play a role in the binding of GSK3β with STRAP.
Figure 2.
STRAP and GSK3β phosphorylation status. (A) Effect of lithium chloride and small-molecule inhibitors of GSK3β on STRAP and GSK3β binding. STRAP-HA and GSK3β-myc constructs were transiently transfected into 293T cells. 35 hours after transfection, cells were treated with SB415286 (20 µM), SB216763 (25 µM) and AR-A014418 (20 µM) as shown in figure. 48 hours after tranfection, cells were subjected to lysis, immunoprecipitation using 1 µg anti-HA antibody and immunoblotted with anti-myc antibody as indicated. Bottom parts show comparable expression of GSK3β-myc and STRAP-HA in the lysates. (B) STRAP has no effect of the phosphorylation/activation status of GSK3β in a part of cell lines. MEFs from wild-type and STRAP-null mice were used, and STRAP was also knocked down in NmuMG, HeLa and HT29 cells using a lentiviral shRNA construct (Open Biosystems). Lysates were prepared, and total proteins (30 µg) were analyzed for phospho-Ser9-GSK3β, total GSK3β (Cell Signaling) and also β-actin as a loading control. (P: parental cells; V: vector control cells; S1 and S2: two STRAP knockdown clones; +/+: wild-type MEFs and −/−: STRAP-null MEFs).
STRAP does not alter phosphorylation/activation status of GSK3β.
STRAP may regulate signaling upstream to GSK3β by acting as a scaffold protein to recruit an upstream inhibitory regulator of GSK3β, such as PKB (Akt) to GSK3β. This will result in phosphorylation of GSK3β at Ser9 residue in the N-terminal region. We decided to test whether STRAP affects activation status of GSK3β in a range of human and mouse cell lines. We predicted that a significant change in the total pool of intracellular STRAP may alter the activation status of GSK3β if STRAP was crucial for mediating signaling upstream of GSK3β. We used the wild-type and STRAP-null MEFs and STRAP-knockdown clones derived from HeLa, HT29 and NMuMG cells. Lysates from these cell lines were analyzed, as seen in Figure 2B. Western analyses using the phospho-Ser9 specific antibody showed no difference in Ser9 phosphorylation of GSK3β in these cell lines (Fig. 2B). These results rule out the possibility that STRAP may affect signaling upstream of GSK3β or activation status of GSK3β.
STRAP forms a ternary complex with GSK3β and axin.
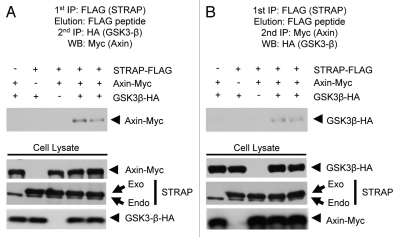
Scaffolding proteins like Axin and APC mediate the recruitment of β-catenin to GSK3β. Considering that STRAP may play a similar role in recruiting substrates into a complex with GSK3β, we tried to determine whether STRAP is present in a complex together with Axin. 293T cells were transfected as described above, either with all Myc-tagged Axin, HA-tagged GSK3β and FLAG-tagged STRAP together or in combinations of two of them together. Lysates were prepared similarly, and STRAP was immunoprecipitated with anti-FLAG antibody. After a final wash, the protein complexes bound with the beads were eluted with FLAG peptide. The eluates were then subjected to a second immunoprecipitation with either anti-HA (Fig. 3A) or anti-Myc antibodies (Fig. 3B) to pull down GSK3β or Axin, respectively. The proteins eluted after second immunoprecipitation were subjected to western analysis for the third protein. The results indicate that STARP, GSK3β and Axin formed a ternary complex with each other. The role of Axin to recruit β-catenin to GSK3β has been extensively studied. Only recently has Axin been shown to aid recruitment of substrates other than β-catenin, such as Smad3 to GSK3β.33 APC is the other scaffold protein that helps Axin to recruit β-catenin to GSK3β. It is possible that STRAP may help Axin in recruiting substrates like β-catenin, Smad3 or some yet unknown substrate to GSK3β. Our experiments failed to show any binding of β-catenin with STRAP (data not shown) indicating that STRAP is unlikely to play any role in recruiting β-catenin to GSK3β.
Figure 3.
STRAP and GSK3β form a ternary complex together with Axin. (A) 293T cells were co-transfected with STRAP-FLAG, GSK3β-HA and Axin-myc in different combinations as indicated. Cell lysates were prepared and co-immunoprecipitated with 2.5 µg of anti-Flag antibody. After five washes with the wash buffer, bound proteins were eluted using the FLAG peptide (Sigma). The eluate was diluted in the lysis buffer and subjected to a second immunoprecipitation with the anti-HA antibody. After washes, the bound proteins were eluted and analyzed by western blotting with anti-myc antibody. Bottom parts show comparable expression of STRAP-FLAG, GSK3β-HA and Axin-myc in the lysates. (B) Same as above except the second immunoprecipitation was done using anti-myc antibody and western analysis was done using anti-HA antibody.
GSK3β binds with intracellular fragment of Notch3.
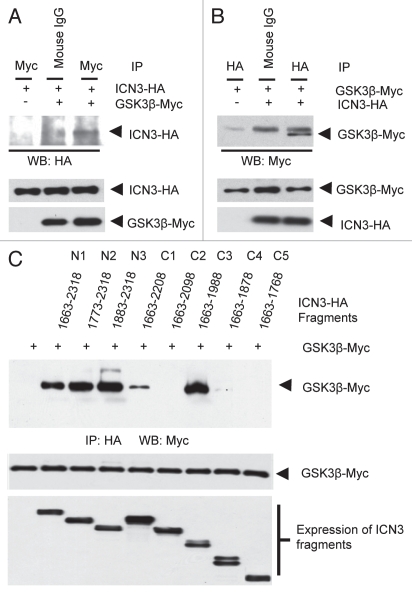
Earlier reports have shown that GSK3β binds and phosphorylates intracellular fragments of Notch1 and Notch2.34–36 There is no report yet of GSK3β interacting or phosphorylating Notch3. Though Notch3 shares an overall good homology with Notch1 and Notch2, they differ in certain regions like the transactivating domain (TAD). We used co-immunoprecipitation assays in 293T cells after transient transfection to assess interaction of HA-tagged ICN3 with GSK3β-Myc. We successfully showed for the first time that GSK3β interacts with ICN3. This is evident from the third lanes of parts A (where ICN3 was coprecipitated with GSK3β) and B (where GSK3β was coprecipitated with ICN3) of Figure 4. Binding of Notch3 with GSK3β predicts that Notch3 is also a possible substrate for GSK3β. Recent reports by Espinosa et al. showed that GSK3β binds and phosphorylates ICN1 and ICN2. This phosphorylation inhibited the activity of ICN1 and ICN2. Interestingly, Notch3 stabilization has been reported to occur and contribute to the progression of lung cancer. Recent studies also suggest that increased Ser9 phosphorylation that inhibits GSK3β predicts a good prognosis for lung cancer patients.37 It is possible to hypothesize that GSK3β phosphorylation may lead to destabilization of Notch3. It will need further work to find out the exact role of GSK3β in Notch3 mediated signaling pathway.
Figure 4.
GSK3β binds specifically with ICN3. (A) ICN3-HA and GSK3β-myc constructs were transiently transfected into 293T cells. Cells were subjected to lysis 48 hours after tranfection, immunoprecipitation using 1 µg pre-immune mouse IgG or 1 µg anti-myc antibody and immunoblotted with anti-HA antibody as indicated. Bottom parts show comparable expression of GSK3β-myc and ICN3-HA in the lysates. (B) Same as above except immunoprecipitations were done with pre-immune rabbit IgG and anti-HA antibodies, and immunoblotting was done using anti-myc antibody. The band above the GSK3β band is the heavy chain. All antibodies are from Santa Cruz Biotechnology. Bottom parts show comparable expression of GSK3β-myc and ICN3-HA in the lysates. (C) The ANK domain 1,863–2,000 aa region of Notch3-IC physically interacts with GSK3β. HEK-293T cells transfected with 1 µg of HA-GSK3β and various deletion constructs of ICN3 as indicated. The lysates from these cells were incubated with anti-HA antibodies for 3 hours and then with G-sepharose beads for 1 hour. Complexes were precipitated anti-HA antibody and analyzed by western blot with anti-myc antibody to detect GSK3β-myc. Bottom parts show equal expression of the ICN3 fragments and GSK3β.
1,880–2,000 aa region of Notch3 is important for GSK3β binding.
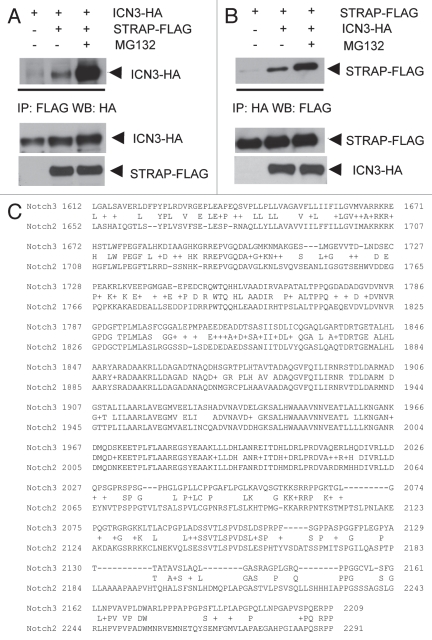
GSK3β binds to ICN2 through the ankyrin repeat domain, ANK.34 The ANK domain of ICN2 has six ankyrin repeats, and ankyrin repeat 6 is most crucial for this interaction. Notch3 has a high homology with Notch2 in the ANK domain region. The ANK domain of ICN2 extends from 1,824 to 2,064 aa, and the ANK domain of ICN3 extends from 1,790 to 2,000 aa. The high homology in this region is evident from Figure 5C. We decided to study the region of Notch3 that is necessary for binding with GSK3β. For this, we decided to generate serial deletion constructs of the intracellular portion of Notch3 using the pCDNA3 mICN3-HA.
Figure 5.
STRAP binds ICN3, and this binding is significantly upregulated in presence of MG132. (A) 1 µg of STRAP-FLAG and ICN3-HA constructs were transiently transfected into HEK-293T cells. Where indicated, cells were treated with 40 µM of the proteasomal inhibitor MG132 (Sigma Biotechnology) for 5 hours before lysis. Forty-eight hours after transfection, cells were subjected to lysis, immunoprecipitation using 1 µg anti-FLAG antibody and immunoblotted with anti-HA antibody. (B) Same as above except immunoprecipitation was done using anti-HA antibody and western analysis with anti-FLAG antibody. For both (A and B), bottom parts show comparable expression of STRAP-FLAG and ICN3 in the lystaes. (C) Homology between mouse ICN3 and ICN2. Protein sequences of mouse ICN3 and ICN2 were compared, and homology is indicated by the common sequence placed between the Notch2 and Notch3 sequences. (D) STRAP binds weakly with ICN1. One µg of STRAP-HA, GSK3β-HA and ICN1-myc constructs were transiently transfected into HEK-293T cells as indicated. Forty-eight hours after transfection, cells were subjected to lysis, immunoprecipitation using 1 µg anti-HA antibody and immunoblotted with anti-myc antibody. Heavy chain band is visible just above the GSK3β-myc band. Bottom parts show comparable expression of STRAP-HA, GSK3β-HA and ICN3-myc in the lysates.
The fragments were amplified by PCR, gel purified and digested with restriction enzymes XbaI and XhoI. The fragments were ligated in pCDNA3.1 digested with the same enzymes. All constructs have a C-terminal HA tag. The expression was verified by western analysis. The fragments N1 (1,663–2,318 same as WT ICN3), N2 (1,773 –2,318), N3 (1,883–2,318), C1 (1,663–2,208), C2 (1,663–2,098), C3 (1,663–1,988), C4 (1,663–1,878) and C5 (1,663–1,768) showed comparable expressions (Fig. 4C, bottom part). These fragments were overexpressed in 293T cells with GSK3β-Myc. Cell lysates were used for immunoprecipitation with anti-HA antibody and the immune complexes were subjected to western blot analysis with anti-Myc antibody. Results from the co-immunoprecipitation assays suggest that the region of ICN3 from 1,880 to 2,000 aa is vital for binding with GSK3β (Fig. 4C). Looking at the high homology between ICN2 and ICN3 in the ANK domain, it is possible to predict that the ankyrin repeat 6 of ICN3 (1,972–2,001 aa) might be crucial for the interaction. At the same time, the 1,990–2,100 aa region seems to have an inhibitory effect on ICN3 and GSK3β binding (Fig. 4C). This inhibitory effect seems to be amplified when this region is freely mobile as C-terminal tail in the C2 construct.
STRAP binding to ICN3 is enhanced in a proteasome inhibition-dependent manner.
Apart from β-catenin, c-Myc, c-Jun, Notch family proteins and Cyclin E are targeted for ubiquitination and proteolysis after GSK3β-mediated phosphorylation. Axin acts a as a docking protein that allows substrates like β-catenin, Smad3 and even some priming kinases like CK1 to be in a complex with GSK3β. Although STRAP was not involved in β-catenin processing, we decided to determine whether STRAP was involved in the GSK3β-mediated processing of the other GSK3β substrates. We have already found that ICN3 binds with GSK3β and is a putative new substrate for it. So we decided to determine if STRAP could functionally interact with ICN3.
Co-immunoprecipitation assays were done in 293 T cells transfected with FLAG-tagged STRAP and HA-tagged ICN3 as described above. Cells were treated with MG132 5 hours before lysis, and the lysates were subjected to immunoprecipitation and western blot analysis (Fig. 5A and B). It is evident from the second lanes of both parts A and B of Figure 5 that STRAP effectively binds to ICN3. Interestingly, short treatment with MG132 significantly enhanced the interaction between STRAP and ICN3 (Fig. 5, lane 3 of parts A and B). As the amount of ICN3 or STRAP present in lysates used for the immunoprecipitation are comparable to each other, this finding indicates that the form of ICN3 that binds with STRAP might be unstable or rapidly degraded in absence of proteasomal inhibition.
STRAP interacts with ICN3 through the same ANK domain region as GSK3β.
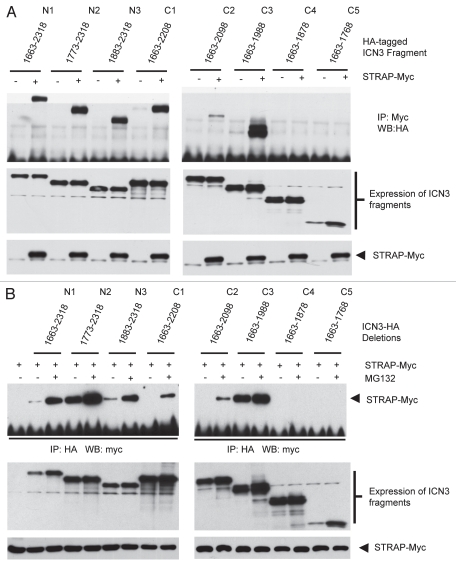
In order to determine the region of ICN3 that binds with STRAP, we performed coimmunoprecipitation experiments using 293T cells as discussed above. HA-tagged ICN3 fragments were expressed either alone or together with STRAP-Myc in presence of MG132. Myc-tagged STRAP was immunoprecipitated, and coprecipitated ICN3 fragments were detected by western blotting with anti-HA antibody (Fig. 6A). We found that ICN3 fragments N1 (1,663–2,318), N2 (1,773–2,318), N3 (1,883–2,318), C1 (1,663–2,208) and C3 (1,663–1,988) demonstrated strong interaction with STRAP in presence of proteasomal inhibitor MG132 as evident from lanes 2, 4, 6, 8 and 12 of Figure 6A. In contrast, the C2 (1,663–2,098) fragment bound STRAP with relatively less affinity (lane 10) whereas fragments C4 (1,663–1,878) and C5 (1,663–1,768) completely failed to bind with STRAP (lane 14 and 16). In a reverse experiment, after treating the cells with or without MG132, consistent results were obtained, where all the ICN3 fragments bound with STRAP except the fragments C4 (1,663–1,878) and C5 (1,663–1,768) (Fig. 6B). These data suggest that STRAP also appears to bind ICN3 through the 1,883–2,000 aa region, which is the same highly conserved ANK domain of Notch3. This is the same region that mediates ICN3 binding with GSK3β. This may be expected, as the ankyrin repeat region is one of the most adapted motifs for protein-protein interactions in Notch3.38 It also appears that the region from 1,990–2,100 aa may have an inhibitory effect on STRAP-ICN3 binding, just as the case with GSK3β. Taken together, this indicates that STRAP binds with ICN3 through a region of 1,880 to 2,000 which is similar to the overall region of ICN3 crucial for its binding with GSK3β.
Figure 6.
STRAP also binds ICN3 through the ANK domain region. (A) ICN3 deletion fragments C4 and C5 do not bind with STRAP. HEK-293 cells were transiently transfected with 1 µg of STRAP-myc and the HA-tagged ICN3 deletion constructs in different combinations as indicated. All cells were treated with the proteasomal inhibitor MG132 (40 µM) for 5 hours before cell lysis. Cells were lysed 48 hours after transfection, subjected to immunoprecipitation with 1.5 µg of anti-myc antibody and immunoblotted with anti-HA antibody to detect co-immunoprecipitated ICN3 deletion fragments. The middle part indicates the same western blot as in the top part, after stripping and immunoblotting with anti-myc antibody to reveal equal immunoprecipitation of STRAP-myc. The bottom two parts indicate comparable expressions of the ICN3 deletion constructs and STRAP-myc in the lysates. (B) This is a reverse of the experiment in part (A). HEK-293 cells were transiently transfected with 1 µg of STRAP-myc and HA-tagged ICN3 deletion constructs. Lane 1 is a negative control transfected only with STRAP-myc. Cell in lanes 3, 5, 7, 9, 11, 13, 15, 17 and 19 were treated with 40 µM of MG132 for 5 hours. Cells were lysed 48 after transfection and subjected to immunoprecipitation with 1.5 µg of anti-HA antibody. Western analysis of the bound proteins was done using anti-myc antibody. Lower part indicates comparable expressions of STRAP-myc and ICN3 deletion constructs in the lysates.
STRAP decreases ICN3 ubiquitination.
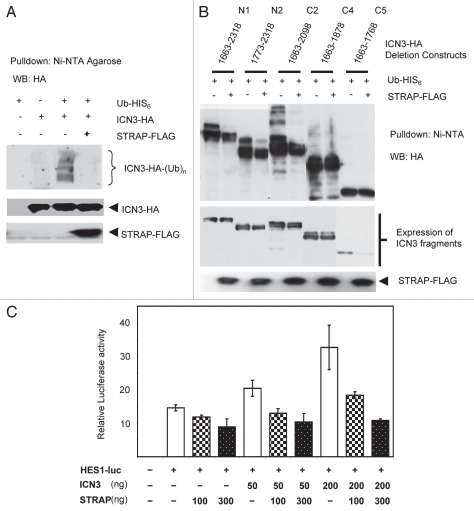
STRAP binds with ICN3, and this binding appears to be enhanced in the presence of proteasomal inhibition. This suggested that STRAP might preferably bind with the form of ICN3 that tends to accumulate when 26S proteasomes are inhibited. This in turn may indicate two possibilities. First is that STRAP can bind with post GSK3β phosphorylated and/or ubiquitinated form of ICN3 and targets it to proteasomes. The other possibility is that STRAP may bind the ubiquitinated form of ICN3 and help it to be docked to some deubiquitinating proteins that can take off the ubiquitin residues of ICN3 to return it to the total cellular pool of ICN3. In order to test these possibilities, plasmids expressing FLAG-tagged STRAP, HA-tagged ICN3 and hexa-histidine-tagged ubiquitin were expressed in 293T cells in combinations as indicated in Figure 7A, and ubiquitinated proteins in the cell lysates were pulled down with Ni-NTA (Nickel-nitrilo triacetic acid) agarose beads. The beads were then washed and eluted proteins were analyzed by western blotting with anti-HA antibody. The western analysis shows only ubiquitinated forms of ICN3 as only ubiquitinated proteins were pulled down (Fig. 7A). Lane 3 shows ICN3 was poly-ubiquitinated in absence of STRAP. When STRAP was co-expressed, this ubiquitination of ICN3 was significantly inhibited. The total expression of exogenous ICN3 remained comparable in the cells as can be seen from the bottom part indicating that the decrease in the ubiquitinated form of ICN3 was not due to a decrease in the total level of overexpressed ICN3. Decrease in ubiquitination can have several effects on the functional aspects of a protein, but most commonly it will lead to stabilization of the protein in the cell.
Figure 7.
Effect of STRAP on ICN3 activity. (A) STRAP decreases ubiquitination of ICN3. HEK-293 cells were transfected with 0.8 µg of ICN3 and His6-tagged ubiquitin and 1 µg of STRAP-FLAG in combinations as indicated. The cells were lysed in a modified lysis buffer as detailed the Materials and Methods. Proteins tagged with His6-ubiquitin molecules were pulled down with Nickel-Nitrilo Tri-Acetic Acid (Ni-NTA) agarose beads. Eluted proteins were subjected to electrophoresis and immunoblotting with anti-HA antibody to specifically detect ubiquitinated species of ICN3. Lower parts indicate equal expression of ICN3 in the lysates. (B) STRAP does not alter ubiquitination of ICN3 fragments C4 (1,663–1,878) andC5 (1,663–1,768). HEK-293 cells were transfected with 0.8 µg of ICN3 deletion constructs and His6-tagged ubiquitin and 1 µg of STRAP-FLAG in combinations as indicated. The rest of the procedure was as described above. Top part shows ubiquitination pattern of the Notch3 fragments in the absence and presence of STRAP-FLAG and lower part shows the expression of STRAP-FLAG and Notch3 deletion constructs in the lysates. (C) STRAP inhibits Notch3 mediated transactivation. HEK-293 cells were plated in 12-well plates, transfected with 0.5 µg of the HES1-promoter luciferase reporter construct and different combinations of ICN3-HA and STRAP-FLAG. All wells were also transfected with 20 ng of beta-galactosidase construct. Cells were lysed, luciferase activity was normalized using beta-galactosidase activity and averaged for triplicates before representing here. The experiment was replicated three times.
When a similar experiment was repeated with a few select fragments of ICN3, it was observed that STRAP expression decreased ubiquitination of the WT ICN3, i.e., N1 (1,663–2,318), fragment N2 (1,773–2,318) and fragment C2 (1,663–2,098), but did not have much effect on the ubiquitination of fragments C4 (1,663–1,878) and C5 (1,663–1,768) (Fig. 7B). This further supports the specificity of STRAP in decreasing the ubiquitination of ICN3, as it does not decrease ubiquitination of ICN3 fragments it does not bind to. Taken together, these data suggest that STRAP may play a role in stabilization of ICN3 in the cells and can possibly lead to a decreased turnover or longer half-life of ICN3.
STRAP has an inhibitory effect on Notch3-mediated transcriptional activity.
STRAP decreases ubiquitination of ICN3 and may stabilize it. An increase in the half-life of the ICN3 protein by STRAP will lead to a larger intracellular pool of ICN3. This can possibly lead to an increase in the transcriptional activity by ICN3. We used the Hes1-luciferase reporter construct to study the effect of STRAP on ICN3 induced transcriptional activity. ICN3 was able to induce the reporter activity in HeLa cells, and STRAP inhibited this induction in a dose-dependent manner when co-expressed with ICN3 (Fig. 7C). This is a paradoxical effect compared to our initial expectations. However, there are increasing reports indicating that ubiquitination of some transcription factors helps them for certain protein-protein interactions with other transcriptional activators. Ubiquitination of Notch IC may facilitate formation of such a transcriptional activation complex and may explain why STRAP can decrease ICN3 ubiquitination and at the same time reduce the transcriptional activity of ICN3.
STRAP and ICN3 show significant co-upregulation in non-small cell lung cancer (NSCLC).
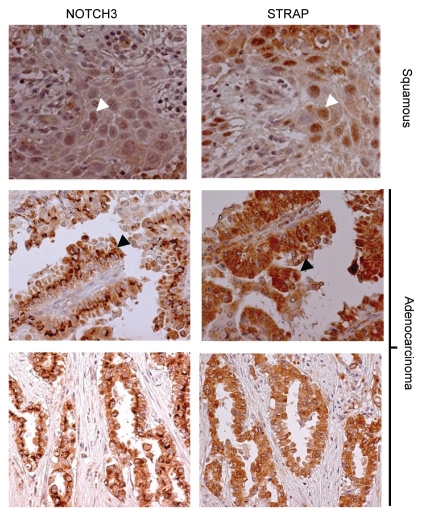
Previous reports by Haruki et al. have shown that ICN3 is upregulated to a significant degree in lung cancers. STRAP is also overexpressed in 78% of lung cancers.14 Here we have shown that STRAP binds with Notch in a proteasomal inhibition-dependent manner, and furthermore, STRAP seems to decrease ubiquitination of ICN3. These facts taken together suggested that STRAP might be one of the factors that help stabilization of ICN3 in lung cancer. To test this concept, we stained serial sections of a lung tissue microarray (TMA) with anti-Notch3 and anti-STRAP antibodies. The TMA consisted of duplicate samples from 42 lung cancer patients. Figure 8 shows sample staining patterns for both STRAP and Notch3 in a pair-wise manner. Each sample on the TMA was scored for the percentage of tumor cells showing staining (N) and for the intensity of staining (I), scored as 0 (no expression) to 3 (highest expression). These two numbers were then multiplied (N × I) to get the staining score for each spot. The score for the duplicate spots was averaged and then compared pair wise between STRAP and Notch3 staining. Using the Pearson's pairwise comparison ratio we obtained an overall correlation of 59% for STRAP and Notch3 in the lung cancer TMA. This indicates a highly significant correlation between STRAP and ICN3 levels in lung cancer and strengthens the idea that STRAP may help to stabilize Notch3 in lung cancer. Interestingly, both STRAP and ICN3 showed a nuclear localization in the two samples form patients suffering from squamous cell carcinomas. Conversely, all other samples, mostly adenocarcinomas, showed an intracytoplasmic localization for both STRAP and ICN3. This favors a cell type-specific role for the STRAP-ICN3 interaction. This may also result in a differential effect of STRAP on ICN3 function in a cell type-dependent manner as far as induction of transcriptional activity is concerned.
Figure 8.
Immunohistochemical analysis of Notch3 and STRAP expression in lung cancer TMA. Left hand columns show expression of Notch3 in lung cancer TMA (Novus anti-Notch3 antibody). Right hand column shows expression of STRAP in the serial section of the same lung cancer samples. All of the positively stained pulmonary adenocarcinomas show a predominantly cytoplasmic localization of Notch3 and STRAP (bottom two parts), whereas the squamous carcinomas exhibit a predominantly nuclear localization of both Notch3 and STRAP (top part).
Discussion
In addition to the roles of STRAP in TGFβ signaling, mRNA splicing and transport, PDK1 signaling, EWS signaling, MAPK pathway regulation, maintenance of mesenchymal morphology, regulation of GSK3 signaling and Notch signaling are newly identified functions of STRAP that are discussed here. It is likely the WD40 domain's based rigid scaffold platform that allows STRAP to mediate such diverse protein functions.
We, for the first time, have established that STRAP interacts with GSK3β, a kinase that regulates phosphorylation of a great variety of proteins, including but not limited to enzymes and transcription factors. Looking at the list of its validated substrates, GSK3β appears to be at the crossroads of diverse cellular signaling pathways. Considering the limited pool of GSK3β that would be available in a cell at a given time, it is not well-understood how GSK3β would regulate certain pathways selectively in a cell- and time-dependent manner and be able to insulate other pathways at the same time. The emerging theme is that the abundance of various scaffold or docking proteins in a cell will decide what substrates and subsequently pathways are being regulated by GSK3β in that particular cell. Axin/ APC mediated docking of β-catenin and presenilin-mediated docking of tau protein are very well known examples of such mechanisms.39,40 The fact that STRAP is a novel scaffold protein that binds GSK3β through WD40 domains raises a possibility that STRAP may have a similar role in processing of one or a few of the GSK3β substrates. The ability of STRAP to form homo-oligomers can further enhance the possibility of such a role for STRAP. Known scaffold proteins in GSK3β signaling pathway, namely, Axin, APC and presenilin are all phosphorylated by GSK3β. Possibility of GSK3β mediated phosphorylation of STRAP needs to be tested by careful kinase assays.
In another completely novel finding, we observed that STRAP, GSK3β and Axin can form a triple complex together (Fig. 3). Though this supports a role for STRAP in β-catenin processing, our assays indicate that STRAP failed to interact with β-catenin in absence or presence of GSK3β or the proteasomal inhibitor MG132 (data not shown). Recently, however, the role of Axin in GSK3β signaling has been shown to be more versatile, as it also recruits Smad3 to GSK3β.33 Smad3 is phosphorylated and degraded as an effect of GSK3β phosphorylation. This finding can imply that Axin may play a similar role for other known and yet-unknown substrates of GSK3β. Just as APC acts as an additional scaffold to recruit β-catenin to GSK3β, STRAP may play a similar role in recruiting or processing of Smad3 or other yet unknown substrates to GSK3β together with Axin. Consistent with this, previous studies have already shown that STRAP interacts with Smad3 and MAP1B, another known substrate of GSK3β.41,42
Notch1 and Notch2 are among the validated substrates of GSK3β. Though Notch3 shares a high homology with Notch1 and Notch2 in some regions, the N-terminal and C-terminal regions of ICN3 (intracellular domain of Notch3) are considerably different from ICN1 and ICN2. We show for the first time that ICN3 also interacts strongly with GSK3β, raising a possibility that it could be another substrate of GSK3β (Fig. 4). Using serial deletion constructs of ICN3, we have mapped the 1,880 to 2,000 aa region of ICN3 to be crucial for mediating the interaction with GSK3β (Fig. 4C). This region falls within the ANK domain of ICN3 that is made of six ankyrin repeats. Ankyrin repeat is one of the most widely existing protein motifs in nature.38 It consists of 30–34 amino acid residues and exclusively functions to mediate protein-protein interactions, further validating the region we mapped in our studies. The Notch family members also exhibit a very high homology with each other in the ANK domain compared to other regions. Our finding is consistent with a previous study showing that the ankyrin repeat number 6 in the ANK domain of ICN2 mediates the interaction of ICN2 with GSK3β.34 Further studies with targeted deletions in the ANK domain of ICN3 will be needed to see if the same ankyrin repeat is involved in GSK3β and ICN3 binding.
A previous study by Foltz et al. found that GSK3β-mediated phosphorylation destabilizes ICN1, while a study by Jin et al. found that the same phosphorylation has a stabilizing effect on ICN1, inducing its transcriptional activity.35,36 In light of this conflicting data about the outcome of GSK3β-mediated phosphorylation of Notch1 protein, the exact effect of this interaction of ICN3 with GSK3β and the probable outcome of GSK3β-mediated phosphorylation may be complicated to predict. But additional data shows that GSK3β also downregulates Notch2 activity possibly by helping its degradation.34 Again, considering that the GSK3β-mediated phosphorylation of other transcription factors including β-catenin, c-Myc, c-Jun, Snail, Notch2, HIF-1α and also Notch1, destabilizes and inhibits them, it may be predicted that GSK3β may have a similar inhibitory effect on ICN3.43 In other words, GSK3β-mediated phosphorylation would probably lead to ubiquitination and proteasomal degradation of ICN3, thus reducing its transcriptional ability.
After finding that STRAP did not seem to bind β-catenin, we looked at any possible interaction of STRAP with ICN3, as it was one of the new probable substrates of GSK3β we had found. Our results suggest that STRAP interacts with ICN3 (Fig. 5). This might add STRAP to the list of known WD40-domain proteins like β-TRCP and Fbw7 that help to process substrates of GSK3β. These WD40 proteins bind these substrates only after they are phosphorylated by GSK3β. This is achieved through the WD40 domains that are considered to be very efficient for recognizing post-translationally modified substrates, especially phosphorylated ones. On a similar note, it is conceivable that binding of STRAP with ICN3 may be enhanced after specific residues have been phosphorylated by GSK3β. Further understanding may come after confirming the phosphorylation sites for GSK3β and performing co-immunoprecipitation experiments with the specific ICN3 point mutants.
But in an interesting observation, we noted that the interaction between STRAP and ICN3 was significantly enhanced when the cells were pretreated with the proteasomal inhibitor MG132 for a short period of time (Fig. 5A and B). Treatment with MG132 is usually employed when a particular form of the protein, which may be more relevant to investigation than the total protein pool, is being degraded rapidly. The half life of Notch3 was calculated to be 0.7 days, or approximately 17 hours.44 So a pretreatment of the cells for 5 hours may not lead to a significant elevation in the total ICN3 protein, but it can possibly lead to a relatively higher accumulation of the fraction of the total level of ICN3 protein that is being rapidly degraded by proteasomes after ubiquitination. This data may just indicate that the phosphorylated form of ICN3 is degraded rapidly after phosphorylation. Accumulation of this phosphorylated form of ICN3 in an ubiquitinated or non-ubiquitinated form seems to have significantly enhanced ability for binding with STRAP. There is still a chance that this data may suggest that STRAP can also preferentially bind to a form of ICN3 that was ubiquitinated. As in case of GSK3β, the binding between STRAP and ICN3 appears to be through the 1,883–2,000 aa region that is known as the ANK domain (Fig. 6). This is expected since the ankyrin repeats in this domain are one of the most ideal modules for protein-protein interactions. Data from our experiments does not indicate any competition between STRAP and ICN3 to bind with GSK3β. Though this does suggest that STRAP, ICN3 and GSK3β might from a ternary complex together, a triple-binding assay did not show a ternary complex formation among these proteins. This appears to be mostly due to the relatively weak and transient binding between GSK3β and its possible substrate, ICN3; and the relatively insensitive method used to for detection.
STRAP seems to interact with a form of ICN3 that may be rapidly degraded. So when we looked at the effect of STRAP on the ubiquitination of ICN3, STRAP consistently decreased Notch3 ubiquitination (Fig. 7A). We hypothesize that STRAP can either carry a phosphorylated ICN3 to a phosphatase to remove the phosphate group and avoid ubiquitination or, later in the process, dock it to an ubiquitin-specific protease after ICN3 has already been ubiquitinated. This has been found to be the case for β-catenin. It was shown that PR55α, the regulatory subunit of protein phosphatase 2A (PP2A), controls β-catenin dephosphorylation and degradation.45 Very surprisingly, PR55α is a WD40 domain protein with seven WD40 repeats, just like STRAP. The same report also showed that PR55α interacted with both β-catenin and Axin. This strengthens the chance of a similar role for STRAP and raises the possibility that STRAP may act as a regulatory subunit for a phosphatase like PP2A. A more detailed knowledge of Nocth3 phosphorylation sites and availability of phospho-specific antibodies will be necessary for additional experiments in this regard. If STRAP helps to target the phosphorylated ICN3 to a specific phosphatase, it can inhibit the ubiquitination and proteasomal degradation of ICN3.
Another plausible explanation appears to be that STRAP can recruit the ubiquitinated ICN3 to a deubiquitinating enzyme (DUB). Ubiquitination of target proteins is reversible process and the removal of Ub can rescue proteins from degradation or re-modulate their activity. The deconjugation of ubiquitin involves removing the covalently linked ubiquitin molecules and is accomplished by the deubiquitinating enzymes (DUBs). The majority of the approximately 100 DUBs in the human genome belong to the ubiquitin-specific protease (USP) subclass of DUBs and others belong to the ubiquitin C-terminal hydrolase (UCH) subclass. Though such a deubiquitinating protein has not been identified for Notch proteins yet, there are several reports demonstrating such a role for USPs. An example for such case is c-Myc, a vastly studied substrate of GSK3β. c-Myc is phosphorylated by GSK3β on Thr-58 and carried by Fbw7 to the E3 ubiquitin ligase complex SCF (Skp, Cullin, F-box containing complex) to be ubiquitinated.45 But the same Fbw7 can also dock ubiquitinated c-Myc to an ubiquitin specific protease, specifically USP-28.47 This leads to de-ubiquitination of c-Myc and it returns back to the normal cellular pool. STRAP might play a similar role for Notch3, though STRAP may not regulate Notch3 in a dual manner, as Fbw7 regulates c-Myc.
Yet another possibility is that STRAP may act just by the mechanism of steric hindrance. In this scenario, binding of STRAP with ICN3 will not allow ICN3 to bind with its two known E3 ubiquitin ligases, namely, Fbw7, a component of an SCF-class ubiquitin ligase (E3) complex, and Itch, a Hect-type E3 ubiquitin ligase.48 Consistent with this idea, in the case of c-Myc, it was shown that when only the WD40 domain region of Fbw7 was overexpressed without the remaining c-terminal 350 aa, it acted in a dominant negative fashion to inhibit Fbw7 activity and stabilized c-Myc.46 STRAP has 7 WD40 domains just as Fbw7 but naturally lacks the large C-terminal region observed in Fbw7 and thus may act in a dominant-negative fashion to inhibit ubiquitination and subsequent degradation of ICN3 by Fbw7. To find the effect of STRAP on ICN3 level, we used two lung cancer cell lines (A549 and H460) that are known to have stabilized ICN3.19 Using lentiviral shRNA, we had a partial success in knocking down STRAP to about 60% in these cell lines. Contrary to our expectations, we did not observe any change in ICN3 in these cell lines. This could possibly be due to the fact that the level of STRAP in these cells is considerably high, and even a 50 to 70% knockdown possibly leaves behind adequate amount of STRAP protein to interact with and stabilize the relatively smaller amount of ICN3 present in these cells. The second possibility is that STRAP may stabilize ICN3 in a cell type-specific manner. Future experiments with better knockdown of STRAP in a range of different cell lines might be needed to resolve this problem.
Dang et al.19 first showed that level of ICN3 was significantly upregulated in human lung carcinomas when compared to the surrounding normal lung tissue from same patients. Overexpression of Notch3 later was shown to contribute to the tumorigenic behavior of the lung cancer cell lines.14,16 The exact mechanism behind this Notch3 upregulation is not known. We have shown before that STRAP is upregulated in 78% of lung cancers. Since our current work indicates that STRAP may stabilize ICN3, we hypothesized that STRAP upregulation may be one of the possible causes behind the stabilization and subsequent upregulation of Notch3 observed in lung cancer patients. Immunohistochemical staining of tissue microarray (TMA) with duplicate serial sections of lung tumors showed that STRAP and ICN3 are coupregulated in about 59% of all (Fig. 8). The localization pattern for both STRAP and ICN3 is mostly cytoplasmic and only slightly nuclear in the majority of the lung adenocarcinomas. In an interesting observation, we found that only in the squamous carcinomas, both ICN3 and STRAP had a distinct nuclear localization. The reasons behind this are unclear at this time but emphasize a cell type-dependent role for STRAP in Notch3 signaling. A previous report has shown that squamous carcinomas of the cervix had a much higher nuclear distribution of ICN3 when compared with cervical adenocarcinomas.49 Nuclear ICN3 was shown to be associated with adverse clinical outcome. Another study reported that immunohistological staining of pancreatic adenocarcinomas showed both cytoplasmic and nuclear staining patterns. Again, statistical analysis suggested that nuclear localization of ICN3 was associated with a more aggressive tumor phenotype and a poorer prognosis.50 These results suggest a potential role of STRAP in the upregulation of ICN3 in NSCLC.
The possibility of ICN3 stabilization by STRAP suggested that STRAP might be able to affect Notch3-mediated transcriptional activity. We expected that STRAP would be able to induce the transactivating ability of ICN3. Surprisingly, we found that STRAP was able to inhibit Notch3-induced activation of the HES-1 promoter in a dose-dependent manner (Fig. 7C). Apart from novel functions, like membrane trafficking, endocytosis, cell cycle control, protein kinase activation and DNA repair, ubiquitination of some transcriptional factors is now being reported to be crucial for their transactivating ability.51 For example, during tumorigenesis, Lys-63-linked polyubiquitination of the transcription factor Myc by the E3 ubiquitin ligase HectH9 is required for the transactivation of multiple Myc target genes.52 Salghetti et al. showed the studies on the activity of a transcription factor containing the VP16 transactivating domain (TAD) is regulated through ubiquitination. More recently, ubiquitination of Gal4 protein was shown to be essential for its binding to a promoter in vivo during transcriptional assays.54 These interactions are being achieved through the ubiquitin-binding domains (UBDs) present in the interacting proteins. It is proposed that Lys-48-linked polyubiquitin chains may be important for protein degradation, while Lys-63-linked polyubiquitin chains may play a role in signal transduction and transcriptional activation.55 Ubiquitination of Notch IC through Lys-63-linked chains may facilitate formation of such a transcriptional activation complex. STRAP can possibly help deubiquitination of both Lys-48 and Lys-63 linked polyubiquitin chains and thus may prolong the half-life but yet reduce the transcriptional activity of ICN3. Future experiments will include elaborate studies to find ubiquitination sites on ICN3 and the type of polyubiquitin chains on these residues.
As mentioned earlier, STRAP knockout mice had multiple defects including angiogenesis, cardiogenesis, somitogenesis, neural tube closure and embryonic turning. Though Notch3 mice were viable, fertile and developed normally, they have some defects in vasculogenesis.56,57 Reports have indicated that Notch-Dll4 signaling is essential for vascular development in the embryo as well as during tumor angiogenesis.58–60 Notch signaling is also supposed to play a major role during somitogenesis.61 Interestingly, Notch signaling in Drosophila melanogaster was implicated in tubulogenesis of the tracheal tree during development, whereas the Drosophila homolog of STRAP, named pterodactyl, was also shown to be crucial for tubulogenesis.62,63 Additional experiments will be needed to know whether Notch plays any role in the phenotype observed in STRAP-knockout mice or the tubulogenesis defect observed in pterodactyl knockout in Drosophila. This may be a challenging task considering the very diverse range of functions of STRAP.
Materials and Methods
Cell culture and plasmids.
Wild-type and STRAP-null mouse embryonic fibroblasts (MEFs), HEK-293, HT29, NmuMG and HeLa were maintained in DMEM supplemented with 10% fetal bovine serum (FBS), antibiotics and glutamine (GIBCO BRL). Axin-myc (in pCDNA3.1) was a gift from Dr. Michele Kimple (Duke University). HA-tagged GSK3β (in pCDNA3) and myc-tagged GSK3β (in pJ3M vector) were gifts from Dr. Gordon Mills (MD Anderson Cancer Center) and Dr. Alan Diehl (University of Pennsylvania Cancer Center) respectively. Murine STRAP and CT-1-STRAP constructed using the pCDNA3 vector have been described previously (Datta et al. 1998). HA-tagged mICN3 and myc-tagged mICN1 (both in pCDNA3) were a gift from Dr. Jon Aster (Brigham and Women's Hospital, Harvard University). HA-tagged β-catenin was a gift from Dr. Stephen Byers (Georgetown University School of Medicine, WA). GSK3β inhibitors SB216763 and SB416286 were purchased from Sigma, and AR-A01441 was purchased from Calbiochem. To generate serial deletion constructs of ICN3, we generated DNA fragments coding for ICN3 deletion constructs using PCR. We added XhoI and XbaI endonuclease restriction sites at their ends and subcloned these fragments into the pCDNA3.1 vector after digesting with XhoI and XbaI. All primers were carefully designed to add an HA tag in frame to the C terminus of the ICN3 fragments. Primer sequences are available upon request.
Western blot analysis.
For immunoblotting, whole-cell lysates were prepared in a cold lysis buffer with 0.01 M Tris-HCl (pH 7.4), 0.01 M NaCl, 1 mM EDTA, sodium ortho-vanadate, 0.1% SDS and protease inhibitors (Aprotinin, Leupeptin and PMSF) and sonicated before centrifugation at 14,000 rpm for 15 min. The proteins were separated by 10% SDS/PAGE, transferred to nitrocellulose membrane (Biorad) and probed with primary antibodies from the following sources: Santa Cruz Biotechnologies (HA and Myc), BD Biosciences (STRAP) and Sigma (FLAG). Primary antibodies were incubated for 3 hr at room temperature, followed by incubation with species-specific secondary antibodies for 1 hr at room temperature. The signal was visualized by enhanced chemiluminescence assay (Amersham Pharmacia Biotech, Pittsburgh, PA).
Co-immunoprecipitation.
HEK-293T cells were plated in 60 mm dish and transfected next day at 40% confluency with appropriate combination of plasmids using Lipofectamine reagent (Invitrogen) using 1:3 ratio in serum-free media. The serum-free media was changed with serum-containing media 3 hours after transfection. Where needed, cells were treated with proteasomal inhibitor MG132 (4 hr) or GSK3β inhibitors (12 hr) as indicated in respective figures. Cells were solubilized in 1 ml of lysis buffer (50 mM Tris, 150 mM NaCl, 10 mM EDTA, 0.02% NaN3, 50 mM NaF, 1 mM Na3VO4, 0.7% NP-40, 0.5 mM dithiothreitol, 0.02% SDS and protease inhibitors aprotinin, PMSF and leupeptin). An equal amount of each protein lysate was incubated with the appropriate antibodies as indicated in the figures, for 3 hours at 4°C, followed by incubation with 20 µl of protein G-Sepharose beads (dry volume) equilibrated with lysis buffer (Sigma Biochemicals, St. Louis, MO) for 1 hour. The immune complexes were washed with the lysis buffer five times. The beads were finally boiled in 50 µl of 2x SDS sample buffer (125 mM Tris-HCL pH 6.8, 20% glycerol, 4% SDS, 2% β-mercaptoethanol, 0.001% bromophenol blue), and the samples then were separated on 10% SDS-PAGE and transferred to PVDF membranes (Biorad). Bound proteins were analyzed by western blot analysis using appropriate antibodies. Protein lysates used for immunoprecipitation were also analyzed by western blot analysis with other antibodies to check for comparable expression of proteins across all transfections.
Reporter assays.
For studying effect of STRAP on ICN3-mediated HES1 promoter induction, HeLa cells were plated in 12-well plates. After 30 h, HES1-luciferase construct (0.5 µg/ well) along with expression plasmids for ICN3-HA (two doses of 50 and 200 ng) and/or STRAP (two doses of 100 and 300 ng) were transfected into the cells using Lipofectamine and Plus reagent following the manufacturers protocol. After approximately 48 hours, cells were lysed, and luciferase assays were performed using a luminometer (BD bioscience) according to the manufacturer's protocol. Transfection of each construct was performed in triplicate in each assay, and a total of three assays were performed on three separate days. All wells were transfected with 25 ng of β-galactosidase to serve as a control for the transfection efficiency. Ratios of luciferase readings to β-gal readings were taken for each experiment and triplicates were averaged. Bars represent the averages of the normalized values, with error bars indicating the standard deviation.
Production of STRAP shRNA lentivirus.
Second-generation VSV-G pseudotyped high titers lentivirus was generated by transient co-transfection of 293T cells with a three-plasmid combination as follows:
One 15 cm dish containing 1 × 107 293T cells was transfected using Lipofectamine2000 (Invitrogen) with 5 µg STRAP shRNA lentiviral vector (pGIPZ, Open Biosystems), 3.75 µg pCMV Δ8.91 and 1.25 µg pMD VSV-G. For vector control lentivirus, empty lentiviral vector was used instead of STRAP shRNA lentiviral vector. A shRNA construct targeting human and mouse STRAP was obtained from Open Biosystems. The 21 bp sequence was 5′-GCT CAT GTA CTC TCA GGA CAT-3′. Supernatants were collected every 12 hr between 36 to 96 hr after transfection, pulled together and frozen at −70°C.
Lentiviral transduction.
For lentiviral transduction, 1 × 105 cells were seeded in 6-well tissue culture plates and infected the following day with STRAP or vector control lentivirus. The cells were then selected for 7 days with puromycin, and when cultures reached near confluency, cells were trypsinized and processed by FACS analysis to separate cells with highest GFP expression. To generate stable STRAP-knockdown clones, these cells were plated at high dilutions in 10 cm petridishes and colonies obtained from single cells were screened for STRAP expression by western blot analysis.
Immunohistochemical analysis.
Tissue Microarray (TMA) slides containing 42 duplicate samples of different lung carcinomas were obtained from the Lung SPORE project at Vanderbilt University. The slides were placed in the sodium citrate solution, microwaved for 45 sec at full power and heated in a prewarmed steamer for 25 min. After cooling at room temperature for 15 min, the slides were washed three times with PBS. After antigen retrieval, the specimens were treated with 3% H2O2 (DAKO) for 5 min to quench endogenous peroxidase activity, and a protein block treatment (Dako, Inc.,) was performed prior to primary antibody addition. Tissues were incubated with anti-STRAP antibody (BD Biosciences) at 1:400, anti-Notch3 antibody (Novus Biological) at 1:200. After primary antibody incubation, the slides were washed three times with PBS. The specimens were then incubated for 10 min at room temperature with biotin-labeled goat anti-mouse immunoglobulin (DAKO). Slides were lightly counterstained with Mayer's hematoxylin (Mayer's, VWR) for nuclear staining. Afterwards, the slides were dehydrated by sequential incubation in 95% ethanol, 100% ethanol and 100% ethanol for 5 min each before transferring to xylene.
In vivo ubiquitination assay.
HEK-293T cells were transfected with appropriate combinations of plasmids expressing his6-tagged ubiquitin, STRAP-FLAG and HA-ICN3 as indicated. Forty-three hours after transfection, the cells were treated or not with 50 µM of MG132 for 5 hours. The cells were then lysed in highly denaturing conditions using 1 ml lysis buffer with 8 M urea at pH 8. This inhibits the otherwise rapid deubiquitination of proteins by deubiquinases after using regular cell lysis conditions. Cell lysates were then sonicated and centrifuged at 13,000 rpm for 20 min. A fraction was saved for western analysis and the rest was incubated with 50 µl of 50% slurry of Nickel-Nitrilo triacetic acid (Ni-NTA) agarose beads slurry for 4 hours on a rocker at room temperature. Ni-NTA beads bind to histidine residues of proteins effectively and strongly pull down hexa-histidine tagged proteins, his-ubiquitinated proteins in this case. The beads were washed five times in a buffer with 8 M urea, 20 mM Imidazole kept at a pH of 6.3. This helps to remove background of endogenous proteins binding within NTA beads. His-ICN3 bound, with the beads were finally eluted with a buffer containing 8 M urea, 250 mM β-mercaptoethanol (BME) and 200 mM Imidazole at a pH of 4.5. The proteins were eluted in two batches first with 50 µl and then with 30 µl of elution buffer. The elauates were then boiled at 95°C for 6 min with 1x SDS sample buffer and stored at −80°C until further analysis. 15 µl of this eluate was analyzed by SDS-PAGE followed by western blotting with anti-HA antibody to detect ICN3 species that were ubiquitinated with HA-tagged ubiquitin.
Acknowledgements
This work was supported by R01 CA95195, CA113519, NCI SPORE grant in lung cancer (5P50CA90949, project #4) and by the Department of Veterans Affairs Merit Review Award (to P.K.D.).
Abbreviations
- STRAP
serine threonine receptor-associated protein
- TGFβ
transforming growth factor-β
- MEF
mouse embryonic fibroblast
- TβRI and TβRII
TGFβ receptors type I and type II
- PDK1
pyruvate dehydrogenase kinase, isozyme 1
- ERK
extracellular signal-regulated kinase
- MAP1B
microtubule-associated protein 1B
- GFP
green fluorescent protein
- PDGF
platelet-derived growth factor
References
- 1.Woodgett JR. Molecular cloning and expression of glycogen synthase kinase-3/factor A. EMBO J. 2000;9:2431–2438. doi: 10.1002/j.1460-2075.1990.tb07419.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cross DA, Alessi DR, Cohen P, Andjelkovich M, Hemmings BA. Inhibition of glycogen synthase kinase-3 by insulin mediated by protein kinase B. Nature. 1995;378:785–789. doi: 10.1038/378785a0. [DOI] [PubMed] [Google Scholar]
- 3.Xu C, Kim NG, Gumbiner BM. Regulation of protein stability by GSK3 mediated phosphorylation. Cell Cycle. 2009;8:4032–4039. doi: 10.4161/cc.8.24.10111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lai EC. Notch signaling: control of cell communication and cell fate. Development. 2004;131:965–973. doi: 10.1242/dev.01074. [DOI] [PubMed] [Google Scholar]
- 5.Lewis J. Notch signalling and the control of cell fate choices in vertebrates. Semin Cell Dev Biol. 1998;9:583–589. doi: 10.1006/scdb.1998.0266. [DOI] [PubMed] [Google Scholar]
- 6.Sasai Y, Kageyama R, Tagawa Y, Shigemoto R, Nakanishi S. Two mammalian helix-loop-helix factors structurally related to Drosophila hairy and Enhancer of split. Genes Dev. 1992;6:2620–2634. doi: 10.1101/gad.6.12b.2620. [DOI] [PubMed] [Google Scholar]
- 7.Zhong TP, Rosenberg M, Mohideen MA, Weinstein B, Fishman MC. gridlock, an HLH gene required for assembly of the aorta in zebrafish. Science. 2000;287:1820–1824. doi: 10.1126/science.287.5459.1820. [DOI] [PubMed] [Google Scholar]
- 8.Weng AP, Ferrando AA, Lee W, Morris JP, 4th, Silverman LB, Sanchez-Irizarry C, et al. Activating mutations of NOTCH1 in human T cell acute lymphoblastic leukemia. Science. 2004;306:269–271. doi: 10.1126/science.1102160. [DOI] [PubMed] [Google Scholar]
- 9.Bismar TA, Demichelis F, Riva A, Kim R, Varambally S, He L, et al. Defining aggressive prostate cancer using a 12-gene model. Neoplasia. 2006;8:59–68. doi: 10.1593/neo.05664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Indraccolo S, Minuzzo S, Masiero M, Amadori A. Ligand-driven activation of the notch pathway in T-ALL and solid tumors: why Not(ch)? Cell Cycle. 2010;9:80–85. doi: 10.4161/cc.9.1.10346. [DOI] [PubMed] [Google Scholar]
- 11.De La O JP, Murtaugh L. Notch and Kras in pancreatic cancer: at the crossroads of mutation, differentiation and signaling. Cell Cycle. 2009;8:1860–1864. doi: 10.4161/cc.8.12.8744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dang TP, Eichenberger S, Gonzalez A, Olson S, Carbone DP. Constitutive activation of Notch3 inhibits terminal epithelial differentiation in lungs of transgenic mice. Oncogene. 2003;22:1988–1999. doi: 10.1038/sj.onc.1206230. [DOI] [PubMed] [Google Scholar]
- 13.Apelqvist A, Li H, Sommer L, Beatus P, Anderson DJ, Honjo T, et al. Notch signalling controls pancreatic cell differentiation. Nature. 1999;400:877–881. doi: 10.1038/23716. [DOI] [PubMed] [Google Scholar]
- 14.Haruki N, Kawaguchi KS, Eichenberger S, Massion PP, Olson S, Gonzalez A, et al. Dominant-negative Notch3 receptor inhibits mitogen-activated protein kinase pathway and the growth of human lung cancers. Cancer Res. 2005;65:3555–3561. doi: 10.1158/0008-5472.CAN-04-3132. [DOI] [PubMed] [Google Scholar]
- 15.Villa N, Walker L, Lindsell CE, Gasson J, Iruela-Arispe ML, Weinmaster G. Vascular expression of Notch pathway receptors and ligands is restricted to arterial vessels. Mech Dev. 2001;108:161–164. doi: 10.1016/s0925-4773(01)00469-5. [DOI] [PubMed] [Google Scholar]
- 16.Konishi J, Kawaguchi KS, Vo H, Haruki N, Gonzalez A, Carbone DP, Dang TP. Gamma-secretase inhibitor prevents Notch3 activation and reduces proliferation in human lung cancers. Cancer Res. 2007;67:8051–8057. doi: 10.1158/0008-5472.CAN-07-1022. [DOI] [PubMed] [Google Scholar]
- 17.Talora C, Campese AF, Bellavia D, Pascucci M, Checquolo S, Groppioni M, et al. Pre-TCR-triggered ERK signalling-dependent downregulation of E2A activity in Notch3-induced T-cell lymphoma. EMBO Rep. 2003;4:1067–1072. doi: 10.1038/sj.embor.7400013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Miyamoto Y, Maitra A, Ghosh B, Zechner U, Argani P, Iacobuzio-Donahue CA, et al. Notch mediates TGF alpha-induced changes in epithelial differentiation during pancreatic tumorigenesis. Cancer Cell. 2003;3:565–576. doi: 10.1016/s1535-6108(03)00140-5. [DOI] [PubMed] [Google Scholar]
- 19.Dang TP, Gazdar AF, Virmani AK, Sepetavec T, Hande KR, Minna JD, et al. Chromosome 19 translocation, overexpression of Notch3 and human lung cancer. J Natl Cancer Inst. 2000;92:1355–1357. doi: 10.1093/jnci/92.16.1355. [DOI] [PubMed] [Google Scholar]
- 20.Lu KH, Patterson AP, Wang L, Marquez RT, Atkinson EN, Baggerly KA, et al. Selection of potential markers for epithelial ovarian cancer with gene expression arrays and recursive descent partition analysis. Clin Cancer Res. 2004;10:3291–3300. doi: 10.1158/1078-0432.CCR-03-0409. [DOI] [PubMed] [Google Scholar]
- 21.Yamaguchi N, Oyama T, Ito E, Satoh H, Azuma S, Hayashi M, et al. NOTCH3 signaling pathway plays crucial roles in the proliferation of ErbB2-negative human breast cancer cells. Cancer Res. 2008;68:1881–1888. doi: 10.1158/0008-5472.CAN-07-1597. [DOI] [PubMed] [Google Scholar]
- 22.Ewing RM, Chu P, Elisma F, Li H, Taylor P, Climie S, et al. Large-scale mapping of human protein-protein interactions by mass spectrometry. Mol Syst Biol. 2007:3. doi: 10.1038/msb4100134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hong M, Chen DC, Klein PS, Lee VM. Lithium reduces tau phosphorylation by inhibition of glycogen synthase kinase-3. J Biol Chem. 1997;272:25326–25332. doi: 10.1074/jbc.272.40.25326. [DOI] [PubMed] [Google Scholar]
- 24.Klein PS, Melton DA. A molecular mechanism for the effect of lithium on development. Proc Natl Acad Sci USA. 1997;93:8455–8459. doi: 10.1073/pnas.93.16.8455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Stambolic V, Ruel L, Woodgett JR. Lithium inhibits glycogen synthase kinase-3 activity and mimics wingless signaling in intact cells. Curr Biol. 1996;6:1664–1668. doi: 10.1016/s0960-9822(02)70790-2. [DOI] [PubMed] [Google Scholar]
- 26.Chalecka-Franaszek E, Chuang DM. Lithium activates the serine/threonine kinase Akt-1 and suppresses glutamate-induced inhibition of Akt-1 activity in neurons. Proc Natl Acad Sci USA. 1999;96:8745–8750. doi: 10.1073/pnas.96.15.8745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Coghlan MP, Culbert AA, Cross DA, Corcoran SL, Yates JW, Pearce NJ, et al. Selective small-molecule inhibitors of glycogen synthase kinase-3 modulate glycogen metabolism and gene transcription. Chem Biol. 2000;7:793–803. doi: 10.1016/s1074-5521(00)00025-9. [DOI] [PubMed] [Google Scholar]
- 28.Hongisto V, Vainio JC, Thompson R, Courtney MJ, Coffey ET. The Wnt pool of glycogen synthase kinase-3beta is critical for trophic-deprivation-induced neuronal death. Mol Cell Biol. 2008;28:1515–1527. doi: 10.1128/MCB.02227-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rinnab L, Schütz SV, Diesch J, Schmid E, Küfer R, Hautmann RE, et al. Inhibition of glycogen synthase kinase-3 in androgen-responsive prostate cancer cell lines: are GSK inhibitors therapeutically useful? Neoplasia. 2008;10:624–634. doi: 10.1593/neo.08248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tighe A, Ray-Sinha A, Staples OD, Taylor SS. GSK-3 inhibitors induce chromosome instability. BMC Cell Biol. 2007:8. doi: 10.1186/1471-2121-8-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yu JY, Taylor J, DeRuiter SL, Vojtek AB, Turner DL. Simultaneous inhibition of GSK3alpha and GSK3beta using hairpin siRNA expression vectors. Mol Ther. 2003;7:228–236. doi: 10.1016/s1525-0016(02)00037-0. [DOI] [PubMed] [Google Scholar]
- 32.Coghlan MP, Culbert AA, Cross DA, Corcoran SL, Yates JW, Pearce NJ, et al. Selective small-molecule inhibitors of glycogen synthase kinase-3 modulate glycogen metabolism and gene transcription. Chem Biol. 2000;7:793–803. doi: 10.1016/s1074-5521(00)00025-9. [DOI] [PubMed] [Google Scholar]
- 33.Guo X, Ramirez A, Waddell DS, Li Z, Liu X, Wang XF. Axin and GSK3-control Smad3 protein stability and modulate TGF-signaling. Genes Dev. 2008;22:106–120. doi: 10.1101/gad.1590908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Espinosa L, Inglés-Esteve J, Aguilera C, Bigas A. Phosphorylation by glycogen synthase kinase-3beta downregulates Notch activity, a link for Notch and Wnt pathways. J Biol Chem. 2003;278:32227–32235. doi: 10.1074/jbc.M304001200. [DOI] [PubMed] [Google Scholar]
- 35.Foltz DR, Santiago MC, Berechid BE, Nye JS. Glycogen synthase kinase-3β modulates notch signaling and stability. Curr Biol. 2002;12:1006–1011. doi: 10.1016/s0960-9822(02)00888-6. [DOI] [PubMed] [Google Scholar]
- 36.Jin YH, Kim H, Oh M, Ki H, Kim K. Regulation of Notch1/NICD and Hes1 expressions by GSK-3alpha/beta. Mol Cells. 2009;27:15–19. doi: 10.1007/s10059-009-0001-7. [DOI] [PubMed] [Google Scholar]
- 37.Zheng H, Saito H, Masuda S, Yang X, d Takano Y. Phosphorylated GSK3β-ser9 and EGFR are Good Prognostic Factors for Lung Carcinomas. Anticancer Res. 2007;27:3561–3569. [PubMed] [Google Scholar]
- 38.Mosavi LK, Cammett TJ, Desrosiers DC, Peng ZY. The ankyrin repeat as molecular architecture for protein recognition. Protein Sci. 2004;13:1435–1448. doi: 10.1110/ps.03554604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hart MJ, de los Santos R, Albert IN, Rubinfeld B, Polakis P. Downregulation of beta-catenin by human Axin and its association with the APC tumor suppressor, beta-catenin and GSK3beta. Curr Biol. 1998;8:573–581. doi: 10.1016/s0960-9822(98)70226-x. [DOI] [PubMed] [Google Scholar]
- 40.Takashima A, Murayama M, Murayama O, Kohno T, Honda T, Yasutake K, et al. Presenilin 1 associates with glycogen synthase kinase-3beta and its substrate tau. Proc Natl Acad Sci USA. 1998;95:9637–9641. doi: 10.1073/pnas.95.16.9637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Datta PK, Chytil A, Gorska AE, Moses HL. Identification of STRAP, a novel WD domain protein in transforming growth factor-beta signaling. J Biol Chem. 1998;273:34671–34674. doi: 10.1074/jbc.273.52.34671. [DOI] [PubMed] [Google Scholar]
- 42.Tretyakova I, Zolotukhin AS, Tan W, Bear J, Propst F, Ruthel G, et al. Nuclear export factor family protein participates in cytoplasmic mRNA trafficking. J Biol Chem. 2005;280:31981–31990. doi: 10.1074/jbc.M502736200. [DOI] [PubMed] [Google Scholar]
- 43.Xu C, Kim G, Gumbiner M. Regulation of protein stability by GSK3 mediated phosphorylation. Cell Cycle. 2009;8:4032–4039. doi: 10.4161/cc.8.24.10111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Takahashi K, Adachi K, Yoshizaki K, Kunimoto S, Kalaria RN, Watanabe A. Mutations in NOTCH3 cause the formation and retention of aggregates in the endoplasmic reticulum, leading to impaired cell proliferation. Hum Mol Genet. 2010;19:79–89. doi: 10.1093/hmg/ddp468. [DOI] [PubMed] [Google Scholar]
- 45.Zhang W, Yang J, Liu Y, Chen X, Yu T, Jia J, Liu C. PR55 alpha, a regulatory subunit of PP2A, specifically regulates PP2A-mediated beta-catenin dephosphorylation. J Biol Chem. 2009;284:22649–22656. doi: 10.1074/jbc.M109.013698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Welcker M, Orian A, Jin J, Grim JE, Harper JW, Eisenman RN, et al. The Fbw7 tumor suppressor regulates glycogen synthase kinase-3 phosphorylation-dependent c-Myc protein degradation. Proc Natl Acad Sci USA. 2004;101:9085–9090. doi: 10.1073/pnas.0402770101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Popov N, Wanzel M, Madiredjo M, Zhang D, Beijersbergen R, Bernards R, et al. The ubiquitin-specific protease USP28 is required for MYC stability. Nat Cell Biol. 2007;9:765–774. doi: 10.1038/ncb1601. [DOI] [PubMed] [Google Scholar]
- 48.Qiu L, Joazeiro C, Fang N, Wang HY, Elly C, Altman Y, et al. Recognition and ubiquitination of Notch by Itch, a hect-type E3 ubiquitin ligase. J Biol Chem. 2000;275:35734–35737. doi: 10.1074/jbc.M007300200. [DOI] [PubMed] [Google Scholar]
- 49.Yeasmin S, Nakayama K, Rahman MT, Rahman M, Ishikawa M, Iida K, et al. Expression of nuclear Notch3 in cervical squamous cell carcinomas and its association with adverse clinical outcomes. Gynecol Oncol. 2010;117:409–416. doi: 10.1016/j.ygyno.2010.03.004. [DOI] [PubMed] [Google Scholar]
- 50.Doucas H, Mann CD, Sutton CD, Garcea G, Neal CP, Berry DP, et al. Expression of nuclear Notch3 in pancreatic adenocarcinomas is associated with adverse clinical features and correlates with the expression of STAT3 and phosphorylated Akt. J Surg Oncol. 2008;97:63–68. doi: 10.1002/jso.20894. [DOI] [PubMed] [Google Scholar]
- 51.Chen ZJ, Sun LJ. Nonproteolytic functions of ubiquitin in cell signaling. Mol Cell. 2009;13:275–286. doi: 10.1016/j.molcel.2009.01.014. [DOI] [PubMed] [Google Scholar]
- 52.Adhikary S, Marinoni F, Hock A, Hulleman E, Popov N, Beier R, et al. The ubiquitin ligase HectH9 regulates transcriptional activation by Myc and is essential for tumor cell proliferation. Cell. 2005;123:409–421. doi: 10.1016/j.cell.2005.08.016. [DOI] [PubMed] [Google Scholar]
- 53.Salghetti SE, Caudy AA, Chenoweth JG, Tansey WP. Regulation of transcriptional activation domain function by ubiquitin. Science. 2001;293:1651–1653. doi: 10.1126/science.1062079. [DOI] [PubMed] [Google Scholar]
- 54.Archer CT, Delahodde A, Gonzalez F, Johnston SA, Kodadek T. Activation domain-dependent monoubiquitylation of Gal4 protein is essential for promoter binding in vivo. J Biol Chem. 2008;283:12614–12623. doi: 10.1074/jbc.M801050200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hurley JH, Lee S, Prag G. Ubiquitin-binding domains. Biochem J. 2006;399:361–372. doi: 10.1042/BJ20061138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Domenga V, Fardoux P, Lacombe P, Monet M, Maciazek J, Krebs LT, et al. Notch3 is required for arterial identity and maturation of vascular smooth muscle cells. Genes Dev. 2004;18:2730–2735. doi: 10.1101/gad.308904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Krebs LT, Shutter JR, Tanigaki K, Honjo T, Stark KL, Gridley T. Haploinsufficient lethality and formation of arteriovenous malformations in Notch pathway mutants. Genes Dev. 2004;18:2469–2473. doi: 10.1101/gad.1239204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ridgway J, Zhang G, Wu Y, Stawicki S, Liang WC, Chanthery Y, et al. Inhibition of Dll4 signalling inhibits tumour growth by deregulating angiogenesis. Nature. 2006;444:1083–1087. doi: 10.1038/nature05313. [DOI] [PubMed] [Google Scholar]
- 59.Hellström M, Phng LK, Hofmann JJ, Wallgard E, Coultas L, Lindblom P, et al. Dll4 signalling through Notch1 regulates formation of tip cells during angiogenesis. Nature. 2007;445:776–780. doi: 10.1038/nature05571. [DOI] [PubMed] [Google Scholar]
- 60.Duarte A, Hirashima M, Benedito R, Trindade A, Diniz P, Bekman E, Costa, et al. Dosage-sensitive requirement for mouse Dll4 in artery development. Genes Dev. 2004;18:2474. doi: 10.1101/gad.1239004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Weinmaster G, Kintner C. Modulation of notch signaling during somitogenesis. Annu Rev Cell Dev Biol. 2003;19:367–395. doi: 10.1146/annurev.cellbio.19.111301.115434. [DOI] [PubMed] [Google Scholar]
- 62.Ikeya T, Hayashi S. Interplay of Notch and FGF signaling restricts cell fate and MAPK activation in the Drosophila trachea. Development. 1999;126:4455–4463. doi: 10.1242/dev.126.20.4455. [DOI] [PubMed] [Google Scholar]
- 63.Khokhar A, Chen N, Yuan JP, Li Y, Landis GN, Beaulieu G, et al. Conditional switches for extracellular matrix patterning in Drosophila melanogaster. Genetics. 2008;178:1283–1293. doi: 10.1534/genetics.106.065912. [DOI] [PMC free article] [PubMed] [Google Scholar]