Abstract
Linear plasmids and chromosomes of the bacterial genus Streptomyces have proteins of unknown characteristics and function linked covalently to their 5′ DNA termini. We purified protein attached to the end of the pSLA2 linear plasmid of Streptomyces rochei, determined the N-terminal amino acid sequence, and used this information to clone corresponding genes from a S. rochei cosmid library. Three separate terminal protein genes (here designated as tpgR1, tpgR2, and tpgR3), which map to the S. rochei chromosome and to 100-kb and 206-kb linear plasmids contained in S. rochei, were isolated and found to encode a family of similar but distinct 21-kD proteins. Using tpgR1 to probe a genomic DNA library of Streptomyces lividans ZX7, whose linear chromosome can undergo transition to a circular form, we isolated a S. lividans chromosomal gene (tpgL) that we found specifies a protein closely related to, and functionally interchangeable with, TpgR proteins for pSLA2 maintenance in S. lividans. Mutation of tpgL precluded propagation of the pSLA2 plasmid in a linear form and also prevented propagation of S. lividans cells that contain linear, but not circular, chromosomes, indicating a specific and essential role for tpg genes in linear DNA replication. Surprisingly, Tpg proteins were observed to contain a reverse transcriptase-like domain rather than sequences in common with proteins that attach covalently to the termini of linear DNA replicons.
Keywords: Telomere, terminal protein, linear DNA replication, chromosome circularization, tpgR, tpgL
Streptomycetes are a group of eubacteria that are not only the source of more than 60% of the world's antibiotics, but also are believed to be the origin of many of the genes that make bacteria resistant to antibiotics (Davies 1997). Among the vehicles proposed to be involved in the transfer of antibiotic-resistance genes among, and from, Streptomyces species are linear extrachromosomal replicons (Davies 1994). How linear plasmids—and also the linear chromosomes found in streptomycetes—replicate their DNA and consequently reproduce the genes they carry has in recent years become a subject of increasing interest (Shiffman and Cohen 1992; Lin et al. 1993; Chang and Cohen 1994; Chen 1996; Chang et al. 1996; Lin and Chen 1997; Huang et al. 1998; Qin and Cohen 1998; Redenbach et al. 1999; Hiratsu et al. 2000; Qin and Cohen 2000; Volff and Altenbuchner 2000; Yamasaki et al. 2000).
Bacillus subtilis phage φ29 DNA has been an extensively studied model for elucidation of the mechanism of linear DNA replication in prokaryotic cells (for review, see Salas 1991; Salas et al. 1995). A terminal protein (TP) covalently bound to the 5′ end of the phage DNA forms a heterodimer with the phage-encoded DNA polymerase, catalyzing covalent linkage of the TP to the first nucleotide of the nascent DNA strand and priming strand-displacing synthesis of full-length φ29 DNA (Salas 1991; Salas et al. 1995; Mendez et al. 1997; Gonzales-Huici et al. 2000). In eukaryotic cells, replication of adenoviruses, which also have protein bound covalently to 5′ DNA termini, occurs by a fundamentally similar protein-primed strand-displacement mechanism (van der Vliet 1995; Hay 1996; King et al. 1997; de Jong and van der Vliet 1999; Liu et al. 2000).
Like the φ29 and adenovirus linear replicons, Streptomyces linear plasmids and chromosomes have covalently attached protein at their 5′ DNA ends (Hirochika and Sakaguchi 1982; Kinashi et al. 1987; Sakaguchi 1990; Lin et al. 1993). However, in contrast to adenovirus and φ29, Streptomyces linear DNAs replicate divergently from a site located near the center of the molecule, generating 3′ leading-strand overhangs at the telomeres (Chang and Cohen 1994). The recessed 5′ ends of the lagging strands produced by the joining together of Okazaki fragments (Kurosawa et al. 1975) are then extended (patched) to produce full-length duplex DNA molecules (Chang and Cohen 1994). Because Streptomyces chromosomes and plasmids have similar termini (Huang et al. 1998; Qin and Cohen 1998) and Streptomyces chromosomes also replicate bidirectionally from internal origins of replication (Musialowski et al. 1994), replication of chromosomes of streptomycetes is presumed also to generate telomeric 3′ overhangs that require patching. Sequence analysis of Streptomyces telomeres indicates that they have similarities to, and differences with, the telomeres of eukaryotic chromosomes. The 3′ overhangs of both types of replicons contain multiple short repeats (Huang et al. 1998; Qin and Cohen 1998; for review of eukaryotes, see Greider 1996; Lingner and Cech 1998; McEachern et al. 2000). However, whereas Streptomyces telomeres contain inverted repeats, eukaryote telomeres consist of a long series of tandem direct repeats. Moreover, the proteins that bind to eukaryote telomeres are not covalently attached to the terminus (Greider 1996; Bryan and Cech 1999).
To understand the biological role of the terminal proteins of Streptomyces linear replicons, we set out to: (1) isolate and characterize proteins bound covalently to the DNA ends of pSLA2, a 17-kb linear plasmid of Streptomyces rochei that has been a paradigm in recent investigations of the structure and function of Streptomyces telomeres (Chang and Cohen 1994; Chang et al. 1996; Qin and Cohen 1998; 2000); (2) identify and map the genes that encode terminal proteins in Streptomyces; and (3) investigate the role of terminal proteins in the replication of Streptomyces chromosomes and plasmids as linear DNA molecules. Our results indicate the existence of a family of structurally distinct, but related and functionally interchangeable, telomere-binding terminal proteins in Streptomyces; show that these proteins are encoded by genes carried by chromosomes and plasmids of different Streptomyces species; and demonstrate that terminal proteins are essential for the propagation of Streptomyces replicons in a linear, but not circular, form.
Results
Identification and characterization of proteins bound covalently to the termini of pSLA2 DNA
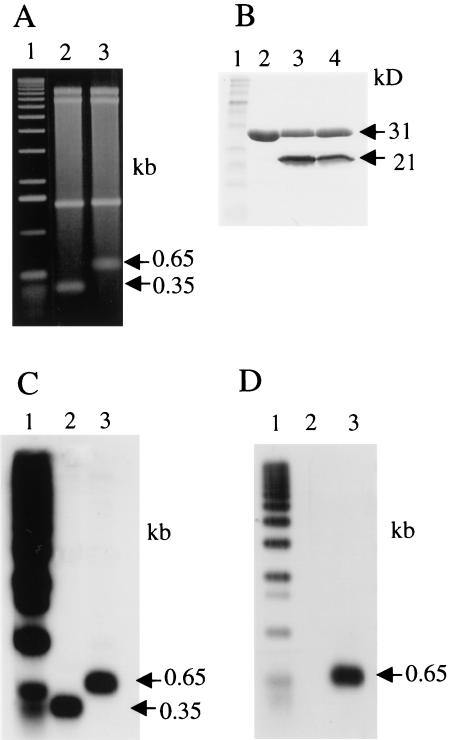
Covalent linkage of proteins with the termini of Streptomyces linear replicons was first indicated by evidence that pSLA2 linear plasmid DNA is sensitive to 3′, but not 5′, exonucleases and that DNA mobility in gels is altered by treatment with pronase but not with SDS (Hirochika and Sakaguchi 1982). We found that the protein component of the pSLA2 DNA–terminal protein complex (i.e., pSLA2–TP) was resistant also to treatment with a phenol/chloroform mixture, as are proteins bound covalently to the termini of other linear replicons (Rekosh et al. 1977; Salas et al. 1978; Yehle 1978), and used this property for terminal protein isolation. The pSLA2 DNA–TP complex from gently lysed cells was electroeluted from agarose gels and purified further by phenol/chloroform extraction in the presence of 1% SDS, followed by ethanol precipitation. Digestion of this complex with BglII endonuclease yielded bands migrating in SDS-agarose gels as 8.2-kb, 6.6-kb, 1.4-kb, and 0.65-kb DNA fragments (Fig. 1A, lane 3). Treatment of the BglII-digested complex by proteinase K prior to electrophoresis led to replacement of the 0.65-kb band by one migrating at the 0.35-kb position (Fig. 1A, lanes 2,3), which is the size expected for both of the terminal fragments of pSLA2 DNA that has been treated with BglII (Hirochika et al. 1984; Qin and Cohen 1998). Migration of the putative telomeric band was not affected by 8 M urea, 2% SDS, or 4 M guanidine hydrochloride; however, treatment with 0.1 N NaOH and renaturation or treatment with piperidine reproduced the effect of proteinase K treatment (data not shown).
Figure 1.
Analysis of components of the pLSA2–TP Complex. (A) Agarose gel electrophoresis of the BglII-digested TP complex. A 1-kb DNA ladder was end-labeled with 32PO4 by T4 kinase after dephosphorylation with calf intestinal alkaline phosphatase (CIP): (lane 1) 32PO4 end-labeled 1-kb DNA ladder; (lane 2) pSLA2 complex digested with BglII and then with proteinase K; (lane 3) pSLA2 complex digested with BglII. Two sets of this gel were identically prepared for Southern blotting and Western blotting. (B) SDS-PAGE of the DNase I-digested complex. The 12% polyacrylamide gel was stained with Coommassie Brilliant Blue R-250. (Lane 1) Protein size marker (BIO-RAD); (lane 2) DNase I alone shown as control; (lanes 3,4) pSLA2 DNA complex isolated from filter-binding assay (see Materials and Methods) and treated with DNase I. The samples shown are duplicates from independent experiments. (C) Southern blot of lanes shown in A using pSLA2 terminal BglII fragments as probe. (D) Western blot of lanes shown in A using anti-TpgR1 antibody.
Preparations of the pSLA2–TP complex isolated and purified as described in Materials and Methods were treated with pancreatic DNase I and subjected to polyacrylamide gel electrophoresis, yielding a single band migrating as a 21-kD protein (Fig. 1B, lanes 3,4). N-Terminal microsequencing (Fig. 2) indicated that this band contains at least two closely related proteins differing only at three positions (positions 4, 7, and 14), where similar but not identical amino acids were present (Micro1 and Micro2, Fig. 2). Oligonucleotide primers containing degenerate sequences corresponding to the 40 N-terminal amino acids of Micro1 and Micro2 were designed and used to generate PCR products employing S. rochei total DNA as template. This amplification yielded two distinct DNA species (PCR2 and PCR1, Fig. 2) whose nucleotide sequences matched exactly the amino acid sequences of Micro1 and Micro2, respectively, except that PCR1 contains an isoleucine, rather than a proline, at position 20 (Fig. 2).
Figure 2.
Alignment of the amino acid sequences of proteins isolated from pSLA2–TP complexes and deduced from the nucleotide sequences of PCR products and tpg genes. Micro1 and Micro2 indicate two N-terminal amino acid sequences from microsequencing of the 21-kD protein shown in Figure 1. PCR1 and PCR2 indicate the two amino acid sequences deduced from sequence of PCR products amplified using total DNA of Streptomyces rochei and degenerate primers corresponding to Micro1 and Micro2. TpgR1: Protein sequence corresponding to the S. rochei tpgR1 gene on cosmid pBC67. TpgR2: Protein sequence corresponding to the S. rochei tpgR2 gene on cosmid pBC66. TpgR3: Protein sequence corresponding to the S. rochei tpgR3 gene on cosmid pBC65. TpgL: Protein sequence corresponding to the tpgL gene of S. lividans ZX7.
A family of terminal protein genes carried by plasmids and chromosome of S. rochei
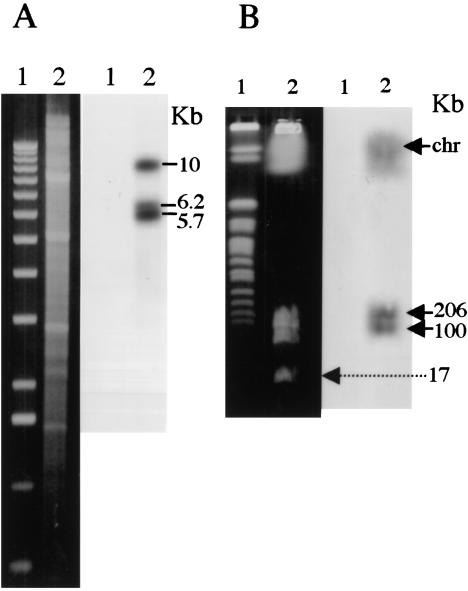
Southern blot hybridization of BamHI-digested total DNA from S. rochei using as probe a mixture of PCR1 and PCR2 detected three bands migrating at 5.7-kb, 6.1-kb, and 9.0-kb positions (Fig. 3A). To isolate the DNA sequences represented by these bands, we constructed and cloned a cosmid library of S. rochei total DNA in Escherichia coli (see Materials and Methods) and analyzed it by colony hybridization using the PCR products as probes. We identified 24 positive clones among 1920 cosmid-bearing colonies. BamHI and KpnI restriction enzyme site mapping of the cosmids plus Southern blot hybridization analysis revealed three distinct groupings, each containing a different BamHI fragment that corresponded in size to one of the bands detected by the PCR probe in BamHI-digested S. rochei total DNA.
Figure 3.
Southern blot hybridization analysis of Streptomyces rochei total DNA probed with a mixture of PCR products corresponding to Micro1 and Micro2. (A) Agarose gel electrophoresis. Left panel shows ethidium-bromide-stained gel. Right panel shows the Southern blot. (Lane 1) 1-kb DNA ladder; (lane 2) BamHI-digested total DNA. The positions of bands about 5.7 kb, 6.1 kb, and 9.0 kb in length are indicated relative to a 1-kb DNA ladder. (B) Pulsed-field gel electrophoresis analysis of S. rochei DNA. Left panel shows ethidium-bromide-stained gel where a DNA size marker (yeast chromosomes, lane 1) and S. rochei total DNA (lane 2) were separated. Right panel shows Southern blot of the gel.
DNA sequencing of restriction-enzyme-generated fragments of randomly chosen members of the three groups of cosmids confirmed the existence in S. rochei of ORFs encoded by a family of three distinct but closely related genes. The N-terminal region specified by these ORFs corresponds exactly to the N-terminal regions of proteins encoded by PCR products we synthesized using S. rochei total DNA as template (see Fig. 2). Analysis of the sequence by the Frame program (Bibb et al. 1984) indicated that codons in all three ORFs contain a G or a C in the third position, as is typical of Streptomyces species, whose DNA contains about 73% (G + C) (Enquist and Bradley 1971). The genes corresponding to the ORFs we identified were assigned the names tpgR1 (terminal protein gene 1 of S. rochei; cosmid pBC67), tpgR2 (cosmid pBC66), and tpgR3 (cosmid pBC65). Southern blot hybridization analysis and sequencing of PCR-amplified products corresponding to fractions of S. rochei total DNA that had been separated by pulsed-field gel electrophoresis showed that tpgR1 is contained on a band migrating in the position of chromosomal DNA, whereas tpgR2 and tpgR3 are carried, respectively, by 206-kb and 100-kb linear plasmids that were previously identified in this S. rochei strain (Fig. 3B; Kinashi et al. 1994). The third plasmid present in this S. rochei strain (i.e., the 17-kb SLA2 plasmid) lacks detectable homology with the probe, and no PCR product was generated using tpg-specific primers and pSLA2 DNA as template. Analysis of the tpgR genes further indicated that the Micro1 and Micro2 proteins attached to pSLA2 telomeres consist of a mix of products encoded by tpgR1 and the terminal protein gene of at least one of the two large linear plasmids resident in S. rochei; the two plasmid-specified proteins are not distinguishable from the N-terminal sequence. The tpgR1 gene was overexpressed in E. coli and confirmed to encode a 21-kD protein (data not shown).
Southern blot analysis (Fig. 1C) of the gel shown in Figure 1A using the 0.35-kb BglII fragment of pSLA2 as probe confirmed that this DNA fragment, whose mobility is altered by proteinase K digestion, is the terminal fragment of the pSLA2 linear replicon. To show that the tpgR gene products can bind to this fragment and consequently slow its migration, we subjected an identically prepared gel to Western blot analysis using as probe a polyclonal antibody generated against the TpgR1 protein that we had expressed in E. coli (Fig. 1D). As seen, anti-TpgR1 antibody detects a protein that, when attached in S. rochei to the 0.35-kb terminal fragments of pSLA2, caused these fragments to migrate at the 0.65-kb position.
The terminal protein gene of Streptomyces lividans ZX7
Unlike S. rochei, the Streptomyces species Streptomyces lividans can be readily transformed with plasmid DNA (Bibb et al. 1978, 1980), and accordingly has been used widely in studies of plasmid biology (Kieser et al. 1982; Kieser 1984; Hopwood et al. 1987; Deng et al. 1988; Kendall and Cohen 1988; Stein et al. 1989; Stein and Cohen 1990; Qin and Cohen 1998). While certain strains of S. lividans have linear chromosomes, others have undergone DNA rearrangements that result in chromosome circularization and the loss of telomeres (Lin and Chen 1997; Volff et al. 1997; Volff and Altenbuchner 2000). Like the telomeres of S. rochei linear plasmids, the 5′ ends of S. lividans linear chromosomal DNA are protected by covalently attached protein (Lin et al. 1993). We hypothesized that this protein may function in the replication in S. lividans of linear DNA of plasmid pSLA2 (Chang and Cohen 1994), which has no detectable tpg gene of its own (see above), and thus that the putative S. lividans terminal protein may have domain similarities with the Tpg proteins of S. rochei. To isolate the postulated S. lividans tpg gene, we constructed a cosmid library containing S. lividans ZX7 genomic DNA, as described in Materials and Methods, and probed this library with S. rochei tpgR1 DNA. Four positive cosmid clones were detected among 1920 isolates; BamHI restriction mapping, Southern blot hybridization analysis, and DNA sequencing of cosmids present in these clones showed that all of the cosmids include an ORF (here designated as tpgL) encoding a 21-kD protein that shows 61% identity and 77% similarity to TpgR1 of S. rochei. The amino acid sequence of TpgL is compared with the sequences of TpgR proteins in Figure 2. Analysis of the genome database of Streptomyces coelicolor (http://www.sanger.ac.uk/Projects/S_coelicolor), which is closely related to S. lividans, indicates that an ORF identical to tpgL and present on cosmid 8D11 of a S. coelicolor A3(2) cosmid DNA library (Redenbach et al. 1996) is located ∼120 kb from the end of the recently sequenced chromosome of S. coelicolor strain M145. There are only two base-pair differences between the tpg genes of ZX7 and M145, and no amino acid differences. Southern blot hybridization analysis of BamHI-, BglII-, KpnI-, and PstI-digested total DNA of S. coelicolor strains A3(2) and M145 and of S. lividans strains 1326 and ZX7 using the ZX7 tpgL gene as probe detected one band for each enzyme digest (data not shown), suggesting that S. lividans and S. coelicolor each contains only one tpg locus.
Tpg protein is required for the propagation of linear plasmids and bacteria containing linear chromosomes
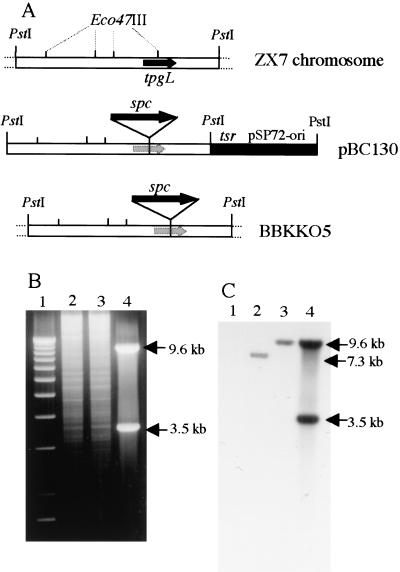
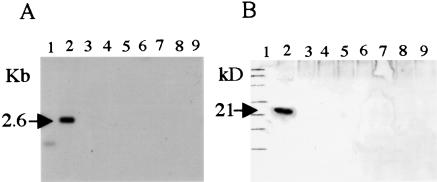
We could not readily inactivate the chromosomal tpgL gene in ZX7 cells by a single-gene-replacement strategy using a pBC130 plasmid-borne tpgL copy containing an inserted spectinomycin-resistance gene (spcr; Fig. 4A). However, during these experiments, we observed rare spcr tsrs survivors (frequency ∼ 10−3 versus our normal gene replacement frequency of 10−2–3 × 10−2); examination of several of these survivors by Southern blot analysis confirmed disruption of the tpgL gene, as shown in Figure 4, B and C, for BKKO5. When telomeres are deleted from Streptomyces linear replicons, the internal replication origin allows their propagation as circular DNA molecules (Shiffman and Cohen 1992; Lin et al. 1993; Chang and Cohen 1994; Chang et al. 1996; Lin and Chen 1997; Volff et al. 1997; Qin and Cohen 1998; Volff and Altenbuchner 2000). Southern blot analysis of chromosomal DNA isolated from two individual survivors (BKKO5 and BKKO6) indicated that these survivors had, in fact, undergone the telomere loss (Fig. 5A) characteristic of recombinational chromosome circularization in Streptomyces (Lin and Chen 1997; Volff et al. 1997; Qin and Cohen 1998; Volff and Altenbuchner 2000). Further analysis of BKK05 after culture for one life cycle in the absence of spectinomycin revealed that spontaneous loss of the spcr gene, which had been inserted into the chromosomal tpgL gene, had occurred in 70% of cells. Because tpgL is located about 120 kb from one end of the chromosome, the finding suggested that the nonlinear chromosomal replicons formed in the absence of a functional tpgL gene undergo further sequence deletion; this conclusion is consistent with previous evidence that the subtelomere regions of S. lividans chromosomes that have undergone telomere removal and circularization are structurally unstable (Lin and Chen 1997; Volff et al. 1997; Volff and Altenbuchner 2000). Additionally, Western blot analysis of DNase I-treated preparations obtained from filter-binding of S. lividans chromosomal DNA detected a TP only in the preparation derived from ZX7 and not in the preparations from BKKO5, BKKO6, or from five separate spcs BKK05 derivatives (BKC1–BKC5), all of which had lost their telomeres (Fig. 5B).
Figure 4.
Analysis of Streptomyces lividans ZX7 chromosomal DNA showing disruption of tpgL gene. (A) Schematic representation of tpgL gene disruption. tsr, thiostrepton-resistance gene; spc, spectinomycin-resistance gene. (B) ethidium-bromide-stained agarose gel. (Lane 1) 1-kb DNA ladder; (lane 2) ZX7 total DNA; (lane 3) BKKO5 total DNA; (lane 4) pBC130 plasmid DNA. (C) Southern blot of same gel probed with 32P-labeled pBC130. All DNAs were digested with PstI.
Figure 5.
Analysis of telomeres and terminal protein in S. lividans ZX7 and its derivatives lacking the tpgL gene. (A) Southern blot of PstI-digested total DNA of ZX7 (lane 2), BKKO5 (lane 3), BKKO6 (lane 4), and BKC1–BKC5 (lanes 5–9) was hybridized with the 2.6-kb PstI chromosome end fragment of ZX7 (Lin et al. 1993). (Lane 1) The 1-kb DNA ladder. (B) Western blot of DNase I-digested filter-binding assay samples of ZX7 (lane 2), BKKO5 (lane 3), BKKO6 (lane 4), and BKC1–BKC5 (lanes 5–9) against TpgR1 antibody. (Lane 1) Protein size marker (BIO-RAD).
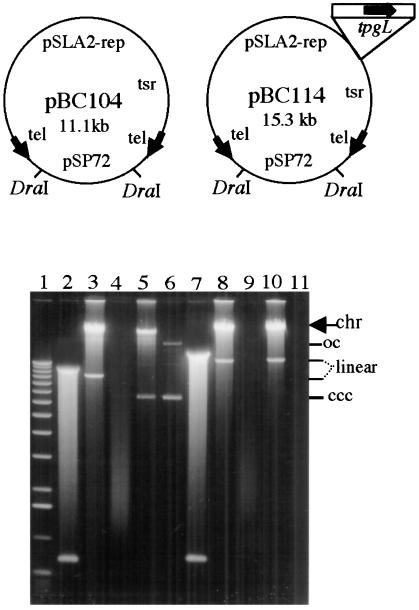
pSLA2-derived plasmids that included an E. coli plasmid replicon and either lacked or contained the tpgL gene (pBC104 and pBC114, respectively, Fig. 6) were constructed as described in Materials and Methods and cloned in E. coli; as seen in Figure 6, each plasmid is cleaved at two locations by DraI endonuclease. Previous work has shown that the terminal protein recognizes precisely, and links covalently to, native telomere ends present on plasmid DNA molecules introduced into S. lividans by transformation (Shiffman and Cohen 1992; Qin and Cohen 1998, 2000). Whereas DNA of both the pBC104 and pBC114 plasmids yielded transformants at a frequency of 8 × 103/μg DNA when treated with DraI and introduced into protoplasts of ZX7, only the tpgL-containing plasmid pBC114 yielded transformants after DraI cleavage and introduction into a host (BKKO5) that lacks a chromosomal tpgL gene. Nonlinear DNA from both plasmids yielded transformants (frequency of 2 × 106 transformants/μg DNA) in bacteria mutated in tpgL. Six randomly selected transformants of ZX7, which contains an intact tgpL gene, all contained linear plasmids after receiving DraI-treated pBC104 or pBC114 DNA (Fig. 6, shown for two transformants in lanes 3 and 10), as did six transformants of BKK05 that had received DraI-cleaved pBC114 (Fig. 6, lane 8). Additionally, 12 randomly selected BKKO5 clones transformed with uncut pBC104 all contained circular plasmids (shown for one transformant in lane 5), as did ZX7 transformed with uncut DNA (data not shown; Qin and Cohen 1998). The linear form of plasmids was confirmed by treatment with NaOH, which causes loss of the double-strand linear DNA band (Fig. 6, lanes 4,9,11; Qin and Cohen 1998, 2000), but does not affect the circular DNA band (lane 6). Collectively, the experiments described in this section indicate that tpgL is essential for replication of both chromosomal and plasmid DNA in a linear form in S. lividans.
Figure 6.
Plasmid replicons in Streptomyces lividans following tpgL gene disruption. (Top) Schematic map of pSLA2 derivatives pBC104 and pBC114. The difference between these two plasmids is that pBC114 contains a functional tpgL gene; the site of insertion is shown. tsr, thiostrepton-resistance gene; tel, 365-bp pSLA2 telomere end indicated by arrowheads pointing away from the centrally located replication origin of pSLA2. The locations of DraI-cleavage sites are indicated. (Bottom) Analysis of plasmid DNA replication of pSLA2 derivatives in S. lividans ZX7 and in the tpgL disrupted circular chromosomal derivative BKKO5. (chr) Chromosomal DNA; ccc (complete closed circular), linear, and oc (open circular) designate different forms of plasmid DNAs. (Lane 1) The 1-kb DNA-size ladder (Life Technologies); (lane 2) DraI-digested pBC104 plasmid DNA isolated from Escherichia coli; (lane 3) DNA isolated from ZX7 transformants receiving DraI-cleaved pBC104; (lane 4) same DNA as in lane 3, but following treatment with 0.2 N NaOH (30 min at 37°C); (lane 5) DNA isolated from BKKO5 transformants receiving uncleaved pBC104; (lane 6) same DNA as in lane 5 following treatment with 0.2 N NaOH; (lane 7) DraI-digested pBC114; (lane 8) DNA isolated from S. lividans BKKO5 transformants receiving DraI-cleaved pBC114 plasmid DNA isolated from E. coli; (lane 9) same as lane 8 following NaOH treatment; (lane 10) same as lane 8 except that host is ZX7; (lane 11) same as lane 10 following NaOH treatment. Streptomyces linear plasmid DNA was isolated by treatment with proteinase K and SDS (Qin and Cohen 1998, 2000) from BKKO5 and ZX7 transformants and electrophoresed for 12 hr at 30 V in 0.6% agarose gel.
Discussion
Because the 5′ ends of Streptomyces linear plasmids are known to be attached covalently to terminal protein, these linear DNA molecules were thought until recently to replicate by the protein-primed full-length strand-displacement mechanism demonstrated for adenovirus and φ29 (Salas 1991, 1995) and therefore thought not to require telomere patching. However, because Streptomyces linear replicons are now known to replicate bidirectionally from an internal site at the center of the molecule (Shiffman and Cohen 1992; Chang and Cohen 1994; Musialowski et al. 1994), producing replicative intermediates that contain a 3′ overhang of leading-strand DNA (Chang and Cohen 1994), their telomeres must undergo patching in order to generate blunt-ended postreplicative DNA molecules. Deletion of widely spaced palindromic DNA sequences in the telomeres of Streptomyces linear plasmids precludes plasmid DNA replication in a linear form (Qin and Cohen 1998, 2000), leading to the proposal that pairing of sequences at the terminus with internal sequences near the base of the overhang provides a recognition site for DNA-binding proteins involved in telomere patching. However, the specific proteins that accomplish this task have not been known. Streptomyces linear chromosomes (Lin et al. 1993), which contain telomeres similar to those present on Streptomyces linear plasmids and also replicate bidirectionally, are presumed to undergo replication and telomere patching by a similar process (Chen 1996; Huang et al. 1998; Qin and Cohen 1998).
The S. rochei linear replicon pSLA2, which is 17 kb in length, can use interchangeably the tpgR gene products specified by the S. rochei chromosome and two large linear plasmids present in this bacterial host. S. lividans ZX7, which contains the chromosomal tpg gene tpgL, related to the tpgR genes, requires tpgL to propagate its chromosome as a linear replicon and also to propagate pSLA2 derivatives as linear plasmids. S. lividans cells that survive after disruption of tpgL undergo the telomere loss characteristic of Streptomyces chromosomes that have undergone recombinational circularization. Additionally, pSLA2 derivatives, which can replicate as linear DNA in the presence of a functional tpgL gene, can be propagated only as circular replicons in S. lividans cells lacking tpgL. Therefore, tpg function is normally required for the replication of either Streptomyces chromosomes or plasmids as linear molecules.
A search of GenBank databases for sequence relationships to the family of tpg genes and proteins we identified revealed no similarity to terminal proteins linked covalently to 5′ DNA termini of previously studied linear replicons. However, the Tpg proteins of Streptomyces contain a domain (in TpgL, amino acids 24–83) that includes a predicted Helix–Turn–Helix motif characteristic of DNA-binding proteins (amino acids 37–58, score 9.3 versus threshold of 4.0; SeqWeb version 1.2, HTH Scan). This domain shows 27% identity and 54% similarity to the reverse transcriptase domain of human immunodeficiency virus (HIV; Vergne et al. 2000). The reverse transcriptases (RTs) of hepatitis B and other hepadnaviruses initiate protein-primed synthesis of minus strands of viral DNA on an RNA template by covalent attachment of RT to the first deoxynucleotide of the nascent DNA chain (Wang and Seeger 1992; Beck and Nassal 1997; Lanford et al. 1997). Interestingly, the viral DNA strand remains bound to the polymerase, paralleling the covalent linkage of 5′ ends of Streptomyces linear plasmid lagging-strand DNA to Tpg proteins. Although the Tpgs we identified are much smaller than known RT proteins, they may participate in protein-primed DNA synthesis at Streptomyces linear plasmid telomeres using an RNA template that corresponds in sequence to the 3′ leading-strand DNA overhangs generated as replication intermediates (see Chang and Cohen 1994). The resulting terminal segment of lagging-strand DNA—still carrying its covalently attached Tpg—may then be joined to the 5′ end of the most distal Okazaki fragment of that strand. If sequence similarity between Tpg proteins and RT does, in fact, have functional significance, protein-primed telomere patching in Streptomyces linear replicons may be mediated by RNA rather than by the modified rolling-hairpin parvoviruslike mechanism considered previously (Huang et al. 1998; Qin and Cohen 1998). For either mechanism, palindrome-mediated fold back of 3′ leading-strand overhangs may provide a recognition site, an anchor, for Tpg protein binding, as suggested previously (Qin and Cohen 1998).
Although S. rochei and S. lividans have widely different phenotypic characteristics (Williams et al. 1983), and the amino acid sequences of the Tpg proteins of these two species show no more than 61% identity (similarity ∼77%), nevertheless, pSLA2 can use the tpg gene products of either species. This functional interchangeability of chromosomally encoded Tpg proteins of S. lividans and S. rochei potentially allows the plasmid to continue to replicate in a linear form when transferred in nature between these species. Additionally, the functional interchangeability of Tpg proteins from disparate Streptomyces species suggests that the mechanism for telomere patching is similar among different streptomycetes. Finally, we note that the ability of plasmids to acquire tpg genes enables such plasmids to replicate in, and thus potentially recombine with, the genomes of streptomycetes whose chromosomes lack tpg genes as a result of telomere deletion and circularization.
Materials and methods
Bacterial strains, plasmids, and general methods
Streptomyces rochei 7434-AN4 (Hirochika and Sakaguchi 1982) and Streptomyces lividans ZX7 (Zhou et al. 1988) were kindly provided by K. Sakaguchi and X. Zhou, respectively. The S. coelicolor strains A3(2) and M145 and the S. lividans strain 1326 (Hopwood et al. 1983) were kindly provided by D.A. Hopwood. The E. coli host was E. coli DH5α (Life Technologies), and pSP72 (Promega) was used as the cloning vector. Standard methods were used for culturing cells, DNA cloning, and so on in E. coli (Sambrook et al. 1989) and Streptomyces (Kieser et al. 2000). Restriction-enzyme-digested DNA fragments were extracted from agarose gel by Qiaquick Gel Extraction Kit (Qiagen). Southern blot hybridization followed the procedure of Church and Gilbert (1984). DNA sequencing was performed using an Applied Biosystems (ABI) Prism 310 Genetic Analyzer and ABI dye terminator sequencing kit. DNA sequencing was carried out for both strands. The degenerate oligonucleotides 5′-GTSGACAGCATCGGSGACGG-3′ and 5′-CGSGTSCCCTTSAGCTGCTT-3′ (S: G or C) were made (Life Technologies) for amplifying the N-terminal DNA sequences of tpgR genes according to the preference for G or C in the third position of codons in Streptomyces (see Bibb et al. 1984). Total DNA libraries of S. rochei and S. lividans ZX7 were constructed by using E. coli–Streptomyces shuttle cosmid vectors pHZ1358 (K. Bao and Z. Deng, pers. comm.) and pHZ132 (Hu et al. 1994), respectively. The 30–40-kb inserts for the library were made by Sau3AI partial digestion of chromosome DNA and were purified by sucrose gradient fractionating. The packaging ligation mixture for library construction used the Gigapack III Gold Packaging Extract kit (Stratagene). Pulsed-field gel electrophoresis (PFGE) of S. rochei DNA was performed as described by Redenbach et al. (1996). Linear plasmid DNA was isolated as described by Qin and Cohen (1998).
Purification of terminal protein of pSLA2–TP complex
Isolation of the pSLA2 DNA–TP complex followed a Streptomyces total-DNA isolation procedure (Kieser et al. 2000), except that the lysozyme solution was changed to 10% sucrose, 2 mg/mL lysozyme, 50 μg/mL RNase A, 50 mM Tris at pH 8.0, 25 mM EDTA at pH 8.0, and 0.1 mM PMSF. The pSLA2 DNA–TP complex was separated from total DNA by agarose gel electrophoresis and rescued from the gel by electroelution. The pSLA2 DNA–TP complex was further purified by phenol/chloroform extraction and ethanol precipitation. Under optimal extraction conditions the pSLA2 complex represented about 10% of the total DNA of S. rochei. Purified pSLA2 DNA–TP complex (in 1 M NaCl, 0.1 mM PMSF) was sonicated to fragment the DNA into small pieces (0.3–5 kb) with a Sonicator W-375 at high output for 10 sec × 6, and the product was applied to Whatman glass microfiber filter for filter-binding assays (Coombs and Pearson 1978). The filter was washed with 5 volumes of 1 M NaCl, and the DNA–TP complex was eluted from the filter with 0.1 M NaCl containing 0.5% SDS. The eluted DNA–TP complex was precipitated with 0.1 volume of 3 M sodium acetate and 2.5 volumes of ethanol. The pellet was dissolved in TE buffer (containing 0.1 mM PMSF) and treated with protease-free DNase I (Worthington, DPRFS grade) at 37°C for 2 h. The material was concentrated by ultrafiltration with Centricon-10 (Millipore), and TP was separated from the DNase I-digested pSLA2 complex by SDS-PAGE. The gel was transferred onto a PVDF membrane, and the protein band (21 kD) was cut out for amino acid microsequencing of the N terminus (Proseq Microsequencing). For Western blotting the eluates from filters (see above) were digested with DNase I, applied to 12% SDS-PAGE gels and electrophoresed for 2 h at 80 V.
PCR and DNA sequencing analysis of tpg gene distribution in S. rochei
DNA bands representing the S. rochei chromosome and the three linear plasmids (206 kb, 100 kb, and 17 kb) of this species were cut from PFGE low-melting-point agarose gels, and the DNA was extracted by melting the gel for 10 min at 65°C. PstI was added to cut the large DNA molecules after the agarose gel had melted. The rescued DNA was purified by phenol/chloroform extraction and ethanol precipitation. PCR reactions were carried out with Platinum Taq HIFI DNA polymerase (Life Technologies), PCR primers 5′-AGCCATATGGTGGACTCG ATCGGAGAC-3′ and 5′-CCCTCGTAGAACTGGGCCGCTA CAG-3′, and the above four templates. PCR products rescued from agarose gel were directly sequenced. The sequences of PCR products were compared with the cloned tpgR gene sequences from library cosmids.
Overexpression of TpgR1 protein in E. coli
A plasmid supplying tRNA genes corresponding to the codons for arginine (two consecutive AGG, AGA, and CGG), proline (CCC), and isoleucine (ATA), all of which are rare in E. coli (Kane 1995; Ivanov et al. 1997), was used to overexpress the tpgR1 gene in E. coli. A pACYC-derived plasmid (pPIRL) that encodes tRNAs corresponding to these rare codons was constructed with three tRNA genes of isoleucine, arginine, and leucine from Stratagene and a proline tRNA gene from Dr. H. Zhong. The chromosomal tpgR1 gene sequence was amplified from pBC67 by a PCR using a Platinum Taq HIFI DNA polymerase (Life Technologies) (PCR primers: 5′-AGCCATATGGT GGACTCGATCGGAGAC-3′ and 5′-CCCTCGTAGAACTGG GCCGCTACAG-3′) and cloned into TA cloning vector pCR2.1 (Invitrogen) as plasmid pBC98. The correctness of the construct was confirmed by DNA sequencing analysis. The tpgR1 gene was further cloned into NdeI/BamHI-digested pET28(a) from pBC98 (as plasmid pBC116), which enabled production of a fusion protein containing a histidine tag at the N-terminal end. After IPTG induction, E. coli BL21(DE3) containing pBC116 and pPIRL overexpressed the TpgR1 fusion protein, which was further purified by Ni2+-column chromatography. The fusion protein was confirmed by Western blot analysis using His-tag antibody and used for the production of rabbit anti-TpgR1 polyclonal antibody.
Southern–Western blot analysis of telomere DNA–TP complex
The purified pSLA2 DNA–TP complex was digested with BglII, and half the mixture was further digested with proteinase K at 50°C in the presence of 0.5% SDS. A 1-kb DNA ladder (Life Technologies) was end-labeled with 32PO4 by T4 kinase after dephosphorylation with CIAP. Samples of a labeled 1-kb DNA ladder, the BglII-digested pSLA2 complex, and the complex digested with both BglII and proteinase K were loaded in duplicate onto an electrophoresis gel in 1× TAE buffer containing 0.05% SDS. One section of the gel was processed for Southern blotting using the telomere BglII fragment as probe while the TP was removed from the telomere fragment during the denaturation step of Southern blotting. Western blotting was performed with the adjacent duplicate part of the agarose gel. Double-strand DNA fragments (including terminal DNA–TP complex) in agarose gel were electrophoretically transferred onto Nylon membrane (Hybond-N, Amersham) in 1× TAE buffer containing 0.05% SDS, using a voltage of 100 V for 1 h at 4°C. After the transferred DNA was fixed onto Nylon membrane by UV cross-linking, the membrane was analyzed by Western blotting with anti-TpgR1 antibody.
Terminal protein gene disruption and complementation
A tpgL gene disruption plasmid (pBC130; Fig. 4) was constructed by inserting a spectinomycin-resistance gene (spcr) at the Eco47III site of the tpgL gene following partial digestion of the tpgL gene cloned in E. coli. To mutate the chromosomal tpgL gene of ZX7, protoplasts were transformed with pBC130, which lacks the ability to replicate in Streptomyces and contains a gene (tsr) encoding thiostrepton resistance; transformants (spcr tsrr) were selected on R5 containing spectinomycin. Clones in which a double crossover leading to replacement of chromosomal tpgL by the insertionally mutated gene were isolated by the testing of spores for sensitivity to thiostrepton by replica-plating colonies selected in the presence of spectinomycin and in the absence of thiostrepton (spcr tsrs). Replacement of wild-type tpgL by the insertionally mutated gene in spcr tsrs bacteria was confirmed by Southern blot hybridization. For tpgL gene complementation, a 4.2-kb BglII fragment containing the entire tpgL gene was cloned into the pQC18-derived replicon pBC104 to generate pBC114 (Fig. 6; Qin and Cohen 1998). ZX7 and its derivative BKK05 (see above) were transformed with the DNA of these two plasmids following digestion with DraI.
Acknowledgments
These investigations were supported by NIH grant AI08619 to S.N.C. K.B. thanks Chris Miller and other coworkers for helpful discussions, Zixin Deng and Xiufen Zhou (Shanghai Jiao Tong University), and Yunsheng Fu and his group at Hubei University (People's Republic of China) for their encouragement.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL sncohen@stanford.edu; FAX (650) 725-1536.
Article and publication are at http://www.genesdev.org/cgi/doi/10.1101/gad.896201.
References
- Beck J, Nassal M. Sequence- and structure-specific determinants in the interaction between the RNA encapsidation signal and reverse transcriptase of avian hepatitis B virus. J Virol. 1997;71:4971–4980. doi: 10.1128/jvi.71.7.4971-4980.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bibb MJ, Ward JM, Hopwood DA. Transformation of plasmid DNA into Streptomyces at high frequency. Nature. 1978;274:398–400. doi: 10.1038/274398a0. [DOI] [PubMed] [Google Scholar]
- Bibb MJ, Schottel JL, Cohen SN. A DNA cloning system for interspecies gene transfer in antibiotic-producing Streptomyces. Nature. 1980;284:526–531. doi: 10.1038/284526a0. [DOI] [PubMed] [Google Scholar]
- Bibb MJ, Findlay PR, Johnson MW. The relationship between base composition and codon usage in bacterial genes and its use for the simple and reliable identification of protein-coding sequences. Gene. 1984;30:157–166. doi: 10.1016/0378-1119(84)90116-1. [DOI] [PubMed] [Google Scholar]
- Bryan TM, Cech TR. Telomerase and the maintenance of chromosome ends. Curr Opin Cell Biol. 1999;11:318–324. doi: 10.1016/S0955-0674(99)80043-X. [DOI] [PubMed] [Google Scholar]
- Chang PC, Cohen SN. Bidirectional replication from an internal origin in a linear Streptomyces plasmid. Science. 1994;265:952–954. doi: 10.1126/science.8052852. [DOI] [PubMed] [Google Scholar]
- Chang PC, Kim ES, Cohen SN. Streptomyces linear plasmids that contain a phage-like, centrally located, replication origin. Mol Microbiol. 1996;22:789–800. doi: 10.1046/j.1365-2958.1996.01526.x. [DOI] [PubMed] [Google Scholar]
- Chen CW. Complications and implications of linear bacterial chromosomes. Trends Genet. 1996;12:192–196. doi: 10.1016/0168-9525(96)30014-0. [DOI] [PubMed] [Google Scholar]
- Church GM, Gilbert W. Genomic sequencing. Proc Natl Acad Sci. 1984;81:1991–1995. doi: 10.1073/pnas.81.7.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coombs DH, Pearson GD. Filter-binding assay for covalent DNA–protein complex: Adenovirus DNA–terminal protein complex. Proc Natl Acad Sci. 1978;75:5291–5295. doi: 10.1073/pnas.75.11.5291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies JE. Origins, acquisition and dissemination of antibiotics resistance determinants. Ciba Found Symp. 1997;207:15–35. [PubMed] [Google Scholar]
- ————— Inactivation of antibiotics and the dissemination of resistance genes. Science. 1994;264:375–382. doi: 10.1126/science.8153624. [DOI] [PubMed] [Google Scholar]
- de Jong RN, van der Vliet PC. Mechanism of DNA replication in eukaryotic cells: Cellular host factors stimulating adenovirus DNA replication. Gene. 1999;236:1–12. doi: 10.1016/s0378-1119(99)00249-8. [DOI] [PubMed] [Google Scholar]
- Deng Z, Kieser T, Hopwood DA. “Strong incompatibility” between derivatives of Streptomyces multi-copy plasmid pIJ101. Mol Gen Genet. 1988;214:286–294. doi: 10.1007/BF00337723. [DOI] [PubMed] [Google Scholar]
- Enquist LW, Bradley SG. Characterization of deoxyribonucleic acid from Streptomyces venezuelae species. Dev Ind Microbiol. 1971;12:225–236. [Google Scholar]
- Gonzales-Huici V, Lazaro JM, Salas M, Hermoso JM. Specific recognition of parental terminal protein by DNA polymerase for initiation of protein-primed DNA replication. J Biol Chem. 2000;275:14678–14683. doi: 10.1074/jbc.m910058199. [DOI] [PubMed] [Google Scholar]
- Greider CW. Telomere length regulation. Annu Rev Biochem. 1996;65:337–365. doi: 10.1146/annurev.bi.65.070196.002005. [DOI] [PubMed] [Google Scholar]
- Hay RT. Adenovirus DNA replication. In: DePamphilis ML, editor. DNA replication in eukaryotic cells. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1996. pp. 699–719. [Google Scholar]
- Hiratsu K, Mochizuki S, Kinashi H. Cloning and analysis of the replication origin and the telomeres of the large linear plasmid pSLA2-L in Streptomyces rochei. Mol Gen Genet. 2000;263:1015–1021. doi: 10.1007/pl00008689. [DOI] [PubMed] [Google Scholar]
- Hirochika H, Sakaguchi K. Analysis of linear plasmids isolated from Streptomyces: Association of protein with the ends of the plasmid DNA. Plasmid. 1982;7:59–65. doi: 10.1016/0147-619x(82)90027-0. [DOI] [PubMed] [Google Scholar]
- Hirochika H, Nakamura K, Sakaguchi K. A linear DNA plasmid from Streptomyces rochei with inverted terminal repetition of 614 base pairs. EMBO J. 1984;3:761–766. doi: 10.1002/j.1460-2075.1984.tb01881.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hopwood DA, Kieser T, Wright HM, Bibb MJ. Plasmids, recombination and chromosome mapping in Streptomyces lividans 66. J Gen Microbiol. 1983;129:2257–2269. doi: 10.1099/00221287-129-7-2257. [DOI] [PubMed] [Google Scholar]
- Hopwood DA, Bibb MJ, Chater KF, Kieser T. Plasmid and phage vector for gene cloning and analysis in Streptomyces. Methods Enzymol. 1987;153:116–166. doi: 10.1016/0076-6879(87)53052-x. [DOI] [PubMed] [Google Scholar]
- Hu Z, Bao K, Zhou X, Zhou Q, Hopwood DA, Kieser T, Deng Z. Repeated polyketide synthase modules involved in the biosynthesis of a heptaene macrolide by Streptomyces sp. FR-008. Mol Microbiol. 1994;14:163–172. doi: 10.1111/j.1365-2958.1994.tb01276.x. [DOI] [PubMed] [Google Scholar]
- Huang C, Lin Y, Huang S, Chen CW. The telomeres of Streptomyces chromosomes contain conserved palindromic sequences with potential to form complex secondary structures. Mol Microbiol. 1998;28:905–916. doi: 10.1046/j.1365-2958.1998.00856.x. [DOI] [PubMed] [Google Scholar]
- Ivanov IG, Saraffova AA, Abouhaidar MG. Unusual effect of clusters of rare arginine (AGG) codons on the expression of human interferon α 1 gene in Escherichia coli. Int J Biochem Cell Biol. 1997;29:659–666. doi: 10.1016/s1357-2725(96)00161-6. [DOI] [PubMed] [Google Scholar]
- Kane JF. Effects of rare codon clusters on high-level expression of heterologous proteins in Escherichia coli. Curr Opin Biotechnol. 1995;6:494–500. doi: 10.1016/0958-1669(95)80082-4. [DOI] [PubMed] [Google Scholar]
- Kendall KJ, Cohen SN. Complete nucleotide sequence of Streptomyces lividans plasmid pIJ101 and correlation of the sequence with genetic properties. J Bacteriol. 1988;170:4634–4651. doi: 10.1128/jb.170.10.4634-4651.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kieser T. Factors affecting the isolation of ccc DNA from Streptomyces lividans and Escherichia coli. Plasmid. 1984;12:19–36. doi: 10.1016/0147-619x(84)90063-5. [DOI] [PubMed] [Google Scholar]
- Kieser T, Hopwood DA, Wright HM, Thompson CJ. pIJ101, a multi-copy broad host-range Streptomyces plasmid: Functional analysis and development of DNA cloning vectors. Mol Gen Genet. 1982;185:223–238. doi: 10.1007/BF00330791. [DOI] [PubMed] [Google Scholar]
- Kieser T, Bibb MJ, Chater KF, Butter MJ, Hopwood DA. Practical Streptomyces genetics. Norwich, UK: John Innes Centre; 2000. [Google Scholar]
- Kinashi H, Shimaji M, Sakai M. Giant linear plasmids in Streptomyces which code for antibiotic biosynthesis genes. Nature. 1987;328:454–456. doi: 10.1038/328454a0. [DOI] [PubMed] [Google Scholar]
- Kinashi H, Mori E, Hatani A, Nimi O. Isolation and characterization of linear plasmids from lankacidin-producing Streptomyces species. J Antibiot. 1994;47:1447–1455. doi: 10.7164/antibiotics.47.1447. [DOI] [PubMed] [Google Scholar]
- King AJ, Teertstra WR, van der Vliet PC. Dissociation of the protein primer and DNA polymerase after initiation of adenovirus DNA replication. J Biol Chem. 1997;272:24617–24623. doi: 10.1074/jbc.272.39.24617. [DOI] [PubMed] [Google Scholar]
- Kurosawa Y, Ogawa T, Hirose S, Okazaki T, Okazaki R. Mechanism of DNA chain growth. XV. RNA-linked nascent DNA pieces in Escherichia coli strains assayed with spleen exonuclease. J Mol Biol. 1975;96:653–664. doi: 10.1016/0022-2836(75)90144-8. [DOI] [PubMed] [Google Scholar]
- Lanford RE, Notvall L, Beames B. Transcomplementation of nucleotide priming and reverse transcription between independently expressed TP and RT domains of hepatitis B virus reverse transcriptase. J Virol. 1997;71:2996–3004. doi: 10.1128/jvi.71.4.2996-3004.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Y, Chen CW. Instability of artificially circularized chromosomes of Streptomyces lividans. Mol Microbiol. 1997;26:709–719. doi: 10.1046/j.1365-2958.1997.5991975.x. [DOI] [PubMed] [Google Scholar]
- Lin Y, Kieser HM, Hopwood DA, Chen CW. The chromosome DNA of Streptomyces lividans 66 is linear. Mol Microbiol. 1993;10:923–933. doi: 10.1111/j.1365-2958.1993.tb00964.x. [DOI] [PubMed] [Google Scholar]
- Lingner J, Cech TR. Telomerase and chromosome end maintenance. Curr Opin Genet Dev. 1998;8:226–232. doi: 10.1016/s0959-437x(98)80145-7. [DOI] [PubMed] [Google Scholar]
- Liu H, Naismith JH, Hay RT. Identification of conserved residues contributing to the activities of adenovirus DNA polymerase. J Virol. 2000;74:11681–11689. doi: 10.1128/jvi.74.24.11681-11689.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McEachern MJ, Krauskopf A, Blackburn EH. Telomeres and their control. Annu Rev Genet. 2000;34:331–358. doi: 10.1146/annurev.genet.34.1.331. [DOI] [PubMed] [Google Scholar]
- Mendez J, Blanco L, Salas M. Protein-primed DNA replication: A transition between two modes of priming by a unique DNA polymerase. EMBO J. 1997;16:2519–2527. doi: 10.1093/emboj/16.9.2519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Musialowski MS, Flett F, Scott GB, Hobbs G, Smith CP, Oliver SG. Functional evidence that the principal DNA replication origin of the Streptomyces coelicolor chromosome is close to the dnaA-gyrB region. J Bacteriol. 1994;176:5123–5125. doi: 10.1128/jb.176.16.5123-5125.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin ZJ, Cohen SN. Replication at the Streptomyces linear plasmid pSLA2. Mol Microbiol. 1998;28:893–903. doi: 10.1046/j.1365-2958.1998.00838.x. [DOI] [PubMed] [Google Scholar]
- ————— Long palindromes formed in Streptomyces by nonrecombinational intra-strand annealing. Genes & Dev. 2000;14:1789–1796. [PMC free article] [PubMed] [Google Scholar]
- Redenbach M, Kieser HM, Denapaite D, Eichner A, Cullum J, Kinashi H, Hopwood DA. A set of ordered cosmids and a detailed genetic and physical map for the 8 Mb Streptomyces coelicolor A3(2) chromosome. Mol Microbiol. 1996;21:77–96. doi: 10.1046/j.1365-2958.1996.6191336.x. [DOI] [PubMed] [Google Scholar]
- Redenbach M, Bibb M, Gust B, Seitz B, Spychaj A. The linear plasmid SCP1 of Streptomyces coelicolor A3(2) possesses a centrally located replication origin and shows significant homology to the transposon Tn4811. Plasmid. 1999;42:174–185. doi: 10.1006/plas.1999.1419. [DOI] [PubMed] [Google Scholar]
- Rekosh DMK, Russell WC, Bellet AJD, Robinson AJ. Identification of a protein linked to the ends of adenovirus DNA. Cell. 1977;11:283–295. doi: 10.1016/0092-8674(77)90045-9. [DOI] [PubMed] [Google Scholar]
- Sakaguchi K. Invertrons, a class of structurally and functionally related genetic elements that includes linear DNA plasmids, transposable elements and genomes of adeno-type viruses. Microbiol Rev. 1990;54:66–74. doi: 10.1128/mr.54.1.66-74.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salas M. Protein-priming of DNA replication. Annu Rev Biochem. 1991;60:39–71. doi: 10.1146/annurev.bi.60.070191.000351. [DOI] [PubMed] [Google Scholar]
- Salas M, Mellado RP, Vinuela E, Sogo JM. Characterization of a protein covalently linked to the 5′ termini of the DNA of Bacillus subtilis phage φ29. J Mol Biol. 1978;119:269–291. doi: 10.1016/0022-2836(78)90438-2. [DOI] [PubMed] [Google Scholar]
- Salas M, Freire R, Soengas MS, Esteban JA, Mendez J, Bravo A, Serrano M, Blasco MA, Lazaro JM, Blanco L. Protein–nucleic acid interactions in bacteriophage φ29 DNA replication. FEMS Microbiol Rev. 1995;17:73–82. doi: 10.1111/j.1574-6976.1995.tb00189.x. [DOI] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T. Molecular cloning: A laboratory manual. 2nd ed. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- Shiffman D, Cohen SN. Reconstruction of a Streptomyces linear replicon from separately cloned DNA fragments: Existence of a cryptic origin of circular replication within the linear plasmid. Proc Natl Acad Sci USA. 1992;898:6129–6133. doi: 10.1073/pnas.89.13.6129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stein DS, Cohen SN. Mutation and functional analysis of the korA and korB gene products of Streptomyces plasmid pIJ101. Mol Gen Genet. 1990;222:237–244. doi: 10.1007/BF00633838. [DOI] [PubMed] [Google Scholar]
- Stein DS, Kendall KJ, Cohen SN. Identification and analysis of transcriptional regulatory signals for the kil and kor loci of Streptomyces plasmid pIJ101. J Bacteriol. 1989;171:5768–5775. doi: 10.1128/jb.171.11.5768-5775.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Vliet PC. Adenovirus DNA replication. Curr Top Microbiol Immunol. 1995;199:1–30. doi: 10.1007/978-3-642-79499-5_1. [DOI] [PubMed] [Google Scholar]
- Vergne L, Peeters M, Mpoudi-Ngole E, Bourgeois A, Liegeois F, Toure-Kane C, Mboup S, Mulanga-Kabeya C, Saman E, Jourdan J, et al. Genetic diversity of protease and reverse transcriptase sequences in non-subtype-B human immunodeficiency virus type 1 strains: Evidence of many minor drug resistance mutations in treatment-naïve patients. J Clin Microbiol. 2000;38:3919–3925. doi: 10.1128/jcm.38.11.3919-3925.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volff JN, Altenbucher J. A new beginning with new ends: Linearisation of circular chromosomes during bacterial evolution. FEMS Microbiol Lett. 2000;186:143–150. doi: 10.1111/j.1574-6968.2000.tb09095.x. [DOI] [PubMed] [Google Scholar]
- Volff JN, Viell P, Altenbuchner J. Artificial circularization of the chromosome with concomitant deletion of its terminal inverted repeats enhances genetic instability and genome rearrangement in Streptomyces lividans. Mol Gen Genet. 1997;253:753–760. doi: 10.1007/s004380050380. [DOI] [PubMed] [Google Scholar]
- Wang GH, Seeger C. The reverse transcriptase of hepatitis B virus acts as a protein primer for viral DNA synthesis. Cell. 1992;71:663–670. doi: 10.1016/0092-8674(92)90599-8. [DOI] [PubMed] [Google Scholar]
- Williams ST, Goodfellow M, Alderson G, Wellington EMH, Sneath PHA, Sakin MJ. Numerical classification of Streptomyces and related genera. J Gen Microbiol. 1983;129:1743–1813. doi: 10.1099/00221287-129-6-1743. [DOI] [PubMed] [Google Scholar]
- Yamasaki M, Miyashita K, Cullum J, Kinashi H. A complex insertion sequence cluster at a point of interaction between the linear plasmid SCP1 and the linear chromosome of Streptomyces coelicolor A3(2) J Bacteriol. 2000;182:3104–3110. doi: 10.1128/jb.182.11.3104-3110.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yehle CO. Genome-linked protein associated with the 5′ termini of Bacteriophage φ29 DNA. J Virol. 1978;27:776–783. doi: 10.1128/jvi.27.3.776-783.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou X, Deng Z, Firmin JL, Hopwood DA, Kieser T. Site-specific degradation of Streptomyces lividans DNA during electrophoresis in buffers contaminated with ferrous iron. Nucleic Acids Res. 1988;16:4341–4352. doi: 10.1093/nar/16.10.4341. [DOI] [PMC free article] [PubMed] [Google Scholar]