Abstract
Developmental programs that govern the embryonic diversification of distinct kinds of muscles in vertebrates remain obscure. For instance, the most widely recognized attribute of early diversity among skeletal myoblasts is their ability to differentiate exclusively into fibers with slow or fast contractile properties. However, we know little about the developmental basis and genetic regulation of this seminal event in vertebrate myogenesis. Here we show that in the zebrafish, the u-boot gene acts as a myogenic switch that regulates the choice of myoblasts to adopt slow versus fast fiber developmental pathways. In u-boot mutant embryos, slow muscle precursors abort their developmental program, failing to activate expression of the homeobox gene prox1 and transfating into muscle cells with fast fiber properties. Using oligonucleotide-mediated translational inhibition, we have investigated the role of prox1 in this program. We find that it functions in the terminal step of the u-boot controlled slow fiber developmental pathway in the regulation of slow myofibril assembly. Our findings provide new insight into the genetic control of slow versus fast fiber specification and differentiation and indicate that dedicated developmental pathways exist in vertebrates for the elaboration of distinct elements of embryonic muscle pattern.
Keywords: zebrafish, Hedgehog, u-boot, prox1, slow myoblast, myofibril
Locomotion is an essential element of the behavioral repertoire of most animals and is mediated by skeletal muscle fibers that have unique properties. For example, their shapes and sizes, orientations and attachments, profiles of gene expression, and patterns of innervation can uniquely identify many muscles in different regions of the vertebrate body. Although our understanding of the molecular mechanisms that specify myogenic fate has been considerably furthered over the recent years (for review, see Arnold and Braun 2000), relatively little is known about the subsequent developmental pathways that distinguish muscle fibers from one another during embryogenesis. Although it is conceivable that autonomous properties of myoblasts, as well as inductive influences from interacting tissues, could be instrumental in dictating the attributes of individual muscles, the genetic pathways involved in mediating these processes have remained largely unappreciated. The likelihood that such dedicated pathways can indeed exist is supported by advances in our understanding of muscle pattern diversification in insects; in this case, it has been possible to analyze the development of single muscle cells and to dissect the genetic programs that are devoted to build them (for review, see Frasch 1999; Roy and VijayRaghavan 1999).
The process of myogenesis in vertebrate embryos is best studied in the somites. In amniotes, myogenesis is intimately coupled to somitogenesis and occurs in a defined somitic compartment called the myotome (for review, see Arnold and Braun 2000). It is clear that the general myogenic program within the myotome is influenced by a number of intersecting signals, most notably those emanating from axial structures like the notochord and neural tube and those from the dorsal epidermis. Whether the interplay of these inductive signals and autonomous properties of myotomal cells results in the emergence of distinct classes of muscle cell types with specific developmental and cellular properties is, however, unclear. Perhaps the most widely recognized property used to distinguish different kinds of muscle fibers in vertebrates is the expression pattern of myosin heavy chain (MyHC) isoforms (for review, see Hughes and Salinas 1999). Thus, there are slow versus fast muscle cells that preferentially express slow versus fast isoforms of MyHC, respectively. Although it is well established that physiological cues play profound roles in modulating MyHC isoform expression in mature skeletal muscles, there is growing evidence indicating that vertebrate myoblasts become fated to differentiate as slow or fast fibers early in embryogenesis (for review, see Currie and Ingham 1998; Stockdale 1992; Hughes and Salinas 1999). The details of the kinds of developmental decisions that govern this early diversity of muscle cell types remain to be elucidated.
In the zebrafish embryo, myogenesis is initiated precociously in the presomitic mesoderm in a special group of adaxial cells that develop close to the notochord (for review, see Currie and Ingham 1998). One of the earliest events in this myogenic episode is the induction of myoD and patched1 (ptc1) in these cells around the embryonic shield at the end of gastrulation (Concordet et al. 1996; Weinberg et al. 1996). Elegant cell–labeling experiments have shown that the adaxial cells mature into a subset of muscle cells in the fish myotome—the slow muscle cells (Devoto et al. 1996). After their specification in the adaxial mesoderm, the slow myoblasts progressively migrate out through the somite and differentiate to form a layer of slow muscles on the surface of the myotome. A small population of slow fibers continue to reside medially next to the notochord and develop into Engrailed (Eng)-expressing slow fibers called muscle pioneers (MPs; Patel et al. 1989; Hatta et al. 1991; Halpern et al. 1993). The rest of the myotome below the slow fibers and around the MPs consists of fast muscles that are derived from nonadaxial mesodermal cells in the somites (Devoto et al. 1996). There is now compelling evidence to indicate that the specification of the slow myoblasts is directed by secreted Hedgehog (Hh) proteins from axial structures like the notochord and the neural tube (Blagden et al. 1997; Du et al. 1997; Lewis et al. 1999). For example, loss-of-function mutations in components of the Hh signaling pathway reduce or eliminate adaxial myoD and ptc1 expression and compromise the specification of slow muscle fibers (van Eeden et al. 1996; Schauerte et al. 1998; Lewis et al. 1999; Barresi et al. 2000). Conversely, ectopic expression of Hh paralogs is able to convert the entire myotome into slow muscle (and MP) identity at the expense of fast muscle (Currie and Ingham 1996; Hammerschmidt et al. 1996; Blagden et al. 1997; Du et al. 1997). Myogenesis in the precursors of the fast muscles is initiated later, in conjunction with the onset of somitogenesis and is believed to be regulated in these cells by mechanisms independent of Hh signaling (Blagden et al. 1997).
These observations suggest that two distinct developmental programs drive the pattern of myogenic differentiation in the zebrafish somite that are spatially and temporally discrete. By default, the majority of the mesodermal cells are fated to develop as fast muscles, whereas Hh activity singles out a subpopulation of somitic cells (the adaxial cells) and directs them to mature into slow muscle fibers. Here, using mutations in the u-boot (ubo) gene, we provide evidence that this Hh-dependent fiber-type specification operates through the activation of a myogenic switch that selectively propels naïve myoblasts to the slow muscle differentiation pathway. In ubo mutant embryos, slow myoblasts fail to express the slow muscle-specific homeobox gene prox1 and transfate into fast MyHC-expressing fibers resembling fast muscles. We show that loss of Prox1 activity accounts for one, but not all, of the effects of the ubo mutation, indicating that ubo is the pivotal regulator of the developmental pattern of the slow fibers. Thus, the making of slow muscles is driven not only by the activity of a core myogenic program but, in addition, through the superimposition on this core element, of a specialized pathway that is unique to this muscle type. Taken together, our findings provide a striking example of a genetic pathway that connects the activity of an inductive signal with the specification and terminal differentiation of a distinct cell type during vertebrate embryogenesis.
Results
Two distinct populations of muscle cell types in the zebrafish myotome: Single-celled slow fibers and syncytial fast fibers
At 24 h postfertilization (hpf), the myotome of the zebrafish embryo consists of a chevron-shaped block of muscles. A few recent studies have shown that at least two distinct populations of muscles can be identified using slow and fast MyHC antibodies raised against MyHC isoforms of other vertebrates (Devoto et al. 1996; Blagden et al. 1997). In addition, two regulatory proteins, Eng and Prox1, have also been reported to be expressed in subsets of muscles in the myotome at this stage (Patel et al. 1989; Hatta et al. 1991; Glasgow and Tomarev 1998). However, comparatively little is known about the cellular features of the muscles, and the expression patterns of the above markers in the different cell-types also have not been investigated in detail.
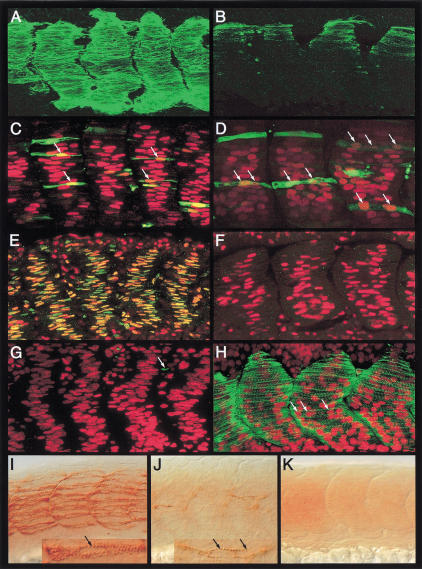
We have used confocal microscopy to visualize the different kinds of muscles and their morphologies. Double labeling with antibodies to slow MyHC and Prox1 revealed a layer of thin fibers decorating the surface of the myotome—the slow-twitch fibers (Fig. 1A). The expression of the nuclear protein Prox1 clearly revealed that the slow fibers consist of individual muscle cells that stretch across the myotomal boundaries and are not syncytial myotubes as is characteristic of mature skeletal muscles (Fig. 1A). A special subset of the slow fibers (about two to six fibers per somite), the MPs, remain bundled together in the medial region of the myotome (Hatta et al. 1991). We observed that like the surface slow muscles, the MPs are also mononucleated (Fig. 1B,C).
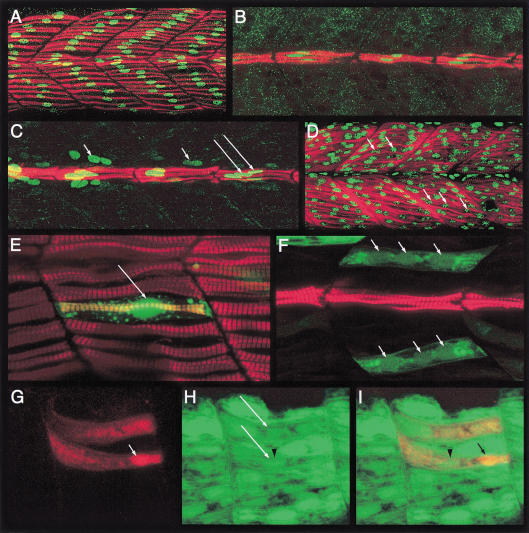
Figure 1.
The zebrafish myotome consists of mononucleate slow fibers and multinucleate fast fibers. (A) Myotomal segments of an embryo, stained with mAb F59 (red) and anti-Prox1 (green) showing the surface layer of mononucleate slow fibers. (B) Medial view of the same embryo showing the mononucleate muscle pioneers (MPs). (C) Similar view of an embryo stained with mAb F59 (red) and mAb 4D9 (anti- Engrailed [Eng], green). Note the high levels of Eng in the MP cells (long arrows) and a lower level in the surrounding multinucleate fast muscle cells that are devoid of mAb F59 labeling (small arrows). (D) Fast MyHC (red) expression in the fast muscle cells. The nuclei have been highlighted with propidium iodide (green; arrows). Note the multinucleate nature of these fibers. (E) Myotomal segments of an α-actin-GFP plasmid–injected wild-type embryo, counterstained with mAb F59 (red), showing a single GFP-labeled (green) surface slow fiber. The accumulation of GFP in the nucleus clearly reveals its mononucleate morphology (arrow). (F) A similar image of another embryo showing GFP-labeled (green) fast muscle fibers. Note their multinucleate morphology (arrows). The MPs, labeled with mAb F59, are also visible. (G) A pair of fast muscle fibers in a host embryo that have been derived from donor myoblasts labeled with rhodamine-dextran (red). A donor nucleus is indicated by the arrow. (H) GFP expression (green) in the α-actin-GFP transgenic host embryo. The two fast fibers shown in G, which have received contributions from donor myoblasts, are indicated by long arrows. The arrow head indicates a host nucleus associated with high levels of GFP expression. (I) Merged view of panels G and H, showing the pair of composite fast fibers. The arrow indicates the donor nucleus as in G and the arrow head the host nucleus as in H. All embryos shown here and in subsequent figures are at 24 h postfertilization and oriented anterior to the left, dorsal to the top, unless otherwise mentioned.
To visualize the fast muscle population, we used fast MyHC antibodies and counter-stained the preparations with propidium iodide to reveal their nuclei. In contrast to the atypical single-celled slow muscles, the fast muscles consist of conventional syncytial myofibers that are arranged in diagonal arrays on the dorsal and ventral halves of the myotome, with their nuclei displayed along the length of the myotubes (Fig. 1D). This contrasts with a previous report that the majority of muscle cells in the zebrafish embryo, up to 30 hpf, are mononucleate (Kimmel and Warga 1987), but it is in agreement with a detailed ultrastructural analysis that clearly indicated the presence of superficial mononucleate and deep polynucleate fibers in the myotome as early as 24 hpf (Waterman 1969). To resolve this issue definitively and to confirm the association of mononucleate versus the polynucleate phenotype with distinct fiber types, we randomly labeled muscle fibers by injecting fertilized eggs with a skeletal muscle α-actin-GFP construct that expresses in all differentiated muscles of both lineages (Higashijima et al. 1997; S. Roy, C. Wolff, and P.W. Ingham, unpubl.) and counter-stained these embryos with antibodies to slow MyHC. In all cases examined, we observed an absolute concordance of the mononucleate phenotype with differentiated slow muscle cells, whereas all GFP-expressing differentiated fast fibers were syncytial (Fig. 1E,F).
To further establish whether the syncytial nature of the fast muscle cells arises by fusion of myoblasts, we transplanted rhodamine-dextran–labeled wild-type donor cells into embryos stably transgenic for the same α-actin-GFP construct (Higashijima et al. 1997). We reasoned that if the transplanted donor cells were fated to form fast myoblasts and if fast muscles are made by myoblast fusion, then donor-host chimeric myotubes should be traceable through the presence of the rhodamine label in the GFP-expressing host fast muscle syncytia. Indeed, such mosaic fast fibers were observed in abundance in the chimeric embryos, confirming that fast muscles are derived by fusion events (Fig. 1G–I). On the other hand and as expected, donor cells fated to be slow myoblasts always differentiated as GFP-negative, single-celled slow fibers (see later section). These results clearly show that mononucleate morphology is in fact an exclusive attribute of one class of muscle cells—the slow-twitch fibers, which coexist in the myotome with fusion-derived multinucleate fast muscles.
Sequential activation of gene expression during maturation defines a distinct developmental pattern for the slow muscle cells
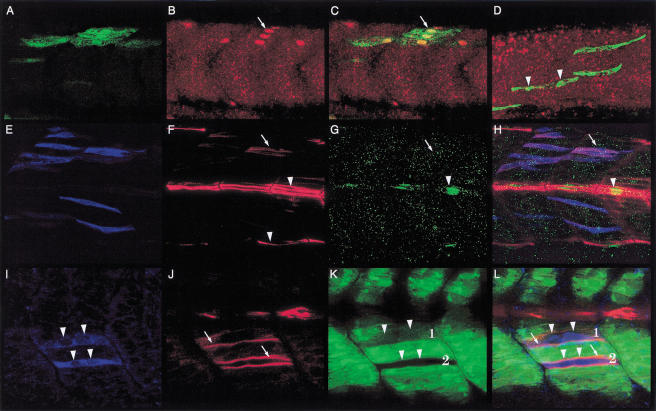
Although myoD and ptc1 are among the earliest markers to be expressed in the adaxial cells, the first indication that they are slow muscle precursors is revealed by the expression of slow MyHC, beginning around the eight to ten somite stage (Devoto et al. 1996; Blagden et al. 1997; S. Roy, C. Wolff, and P.W. Ingham, unpubl.). Examination of the expression profiles of Eng and slow MyHC in 12 somite stage embryos showed that the expression of Eng in a subset of the slow myoblasts, the presumptive MPs, occurs after the initiation of contractile protein gene expression, but before the migration of the slow myoblasts (Fig. 2A–C). Prox1 expression is absent at these stages (Glasgow and Tomarev 1998; data not shown), being first detectable in all slow myoblasts coincident with the onset of their radial migration through the somite (Fig. 2D,E). The pattern of Eng expression clearly indicates that unlike slow MyHC and Prox1, it is not a general property of the differentiation pattern of the slow fibers (Devoto et al. 1996). In addition, it also appears not to be an exclusive attribute of the MP cells (Hatta et al. 1991; Devoto et al. 1996). Eng is induced in all muscle cells irrespective of their lineage (at high levels in slow MPs and subsequently at low levels in surrounding fast fibers; see Fig. 1C) that remain in close proximity to the midline, reflecting its dependence on high or continued levels of Hh signaling (C. Wolff, S. Roy, P.W. Ingham, in prep.). Thus, the progressive transition of the adaxial cells to mature slow fibers proceeds through a sequence of discrete steps revealed by distinct patterns of gene expression.
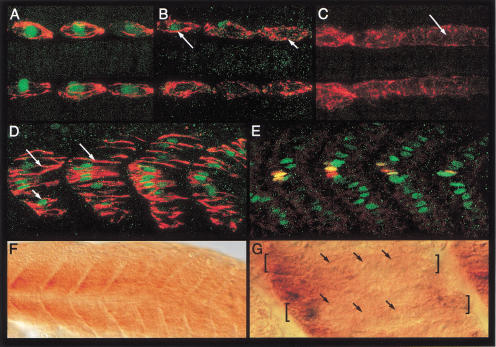
Figure 2.
Temporal profiles of gene expression during development of the slow and fast muscle cells. (A) The anterior-most somites of a 12-somite-stage embryo showing mAb F59 immunoreactivity (red) and Engrailed (Eng) expression (green) in the muscle pioneer (MP) precursors. (B) The mid body somites of the same embryo, showing the transition of Eng-expressing anterior (long arrow) and nonexpressing posterior (short arrow) somites. (C) The caudal end of the same embryo, showing mAb F59 staining extending into the adaxial cells of the unsegmented presomitic mesoderm (arrow), which is devoid of all Eng expression at this stage. Note that the slow MyHC protein at this stage appears randomly distributed in the cytoplasm of the adaxial cells. (D) At the 18 somite stage, anti-Prox1 (green; short arrow) and mAb F59 labeling (red) is observed in all the slow muscle precursors. Note that at this stage, the slow myoblasts have elongated and the MyHC protein appears as long filaments (long arrows). (E) At the same stage, Eng expression (red) is observed to colocalize with Prox1 (green) in a subset of medial cells, the prospective MPs (yellow). (F) Expression of fast MyHC is first observed around the 18 to 20 somite stage. Expression is initiated in the anterior somites and then progresses caudally as the somites mature. (G) High magnification DIC image of the anterior somites of the embryo displayed in F, showing that fast MyHC expression is already restricted to multinucleate myotubes at this stage. Two such myotubes are indicated by the brackets and their nuclei by the arrows. Preparations shown in panels A–C represent dorsal views with anterior to the left.
In contrast to the slow myoblasts and in keeping with the delayed myogenic commitment of the lateral somitic cells as fast muscle precursors, we observed that the expression of fast MyHC in these cells first occurs at the 18 somite stage and progresses in a rostrocaudal wave with the maturation of the somites (Fig. 2F; Bladgen et al. 1997). Furthermore, and again unlike the slow muscles, this expression appears to be restricted from the outset to syncytial myotubes (Fig. 2G).
The ubo mutation uncovers a developmental choice-point in the specification of slow versus fast muscle cells
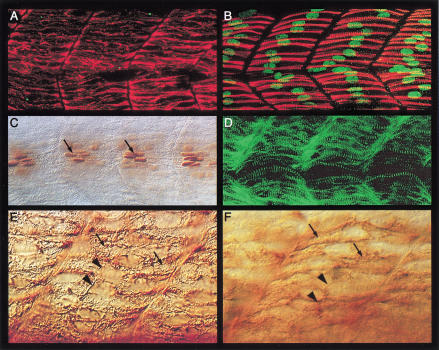
The foregoing analysis indicates that a significant amount of diversity exists among muscle cells in the zebrafish myotome, manifested not only in their developmental patterns but also in their cellular morphologies and profiles of gene expression. We observed that animals carrying mutations in the ubo gene (van Eeden et al. 1996) show a dramatic reduction in slow muscle fibers at 24 h as assessed with antibodies that recognize slow MyHC. Unlike wild-type sibling embryos, ubo embryos have very few and scattered slow fibers on the surface of the myotome (Fig. 3A–D). Moreover, the myofibrils in these occasional fibers are highly disorganized compared with those of their wild-type counterparts (Fig. 3E,F). Much of the residual slow MyHC expression in these embryos, however, is confined to a dispersed pattern of immunoreactivity deeper in the myotome in fibers resembling fast muscles (Fig. 3G,H). Thus, it seems that the majority of the slow myoblasts in ubo embryos are unable to migrate out to the surface and differentiate properly. Examination of the pattern of slow MyHC at earlier stages revealed that its expression is already variably reduced from the outset, suggesting that ubo function is required at the initial stages of slow myoblast induction by Hh signals from the axial mesoderm and neural tube (Fig. 3I–K). The capacity to receive such signals is not, however, compromised by ubo, as evidenced by the normal expression of myoD and ptc1 (van Eeden et al. 1996; Lewis et al. 1999; data not shown). Although the effects on slow MyHC expression are variable, we observed a dramatic and consistent effect on prox1 expression. In the myotomes of ubo embryos, no prox1 expression (either transcript or protein) is detectable at these developmental stages (Fig. 3K,L; data not shown). In contrast, the expression of Eng is not affected so obviously, consistent with the fact that Eng expression is not a general feature of slow fiber differentiation. There are, however, two important differences from wild-type embryos: First, the distinction between high-level expression in the MPs versus low levels in the adjoining fast muscles is no longer apparent (Fig. 4A,B). Second, almost all of Eng expression is observed in multinucleate syncytial fibers with rounded nuclei as opposed to wild-type embryos, where it is seen in single flattened nuclei of the MP cells and in the rounded nuclei of the surrounding multinucleate fast myotubes (Fig. 4A,B). To assess the significance of these changes in Eng expression in ubo embryos, we performed double labeling with anti-Eng and anti-slow MyHC antibodies. In wild-type embryos, slow MyHC and Eng are found to be coexpressed exclusively in the MPs, because MPs belong to the slow fiber class (Fig. 1C). The surrounding Eng-expressing multinucleate fast fibers are never observed to coexpress slow MyHC and Eng (Fig. 1C). In ubo embryos, however, we observed coexpression of residual slow MyHC and Eng in multinucleate fibers (Fig. 4C). Significantly, most of the superficially located slow MyHC-expressing fibers in ubo embryos also show bi- and trinucleate morphology (Fig. 4D). Thus, absence of wild-type ubo activity does not prevent induction of Hh target genes that are not slow muscle–specific—myoD, ptc1, and eng—but does preclude cells from executing the slow-twitch program, for example, proper initiation and maintenance of slow MyHC expression and activation of prox1 and high level expression of Eng in the MPs.
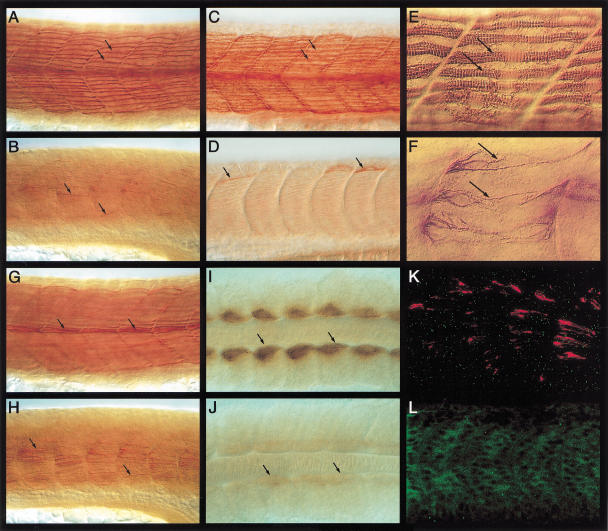
Figure 3.
Mutation in the ubo gene disrupts slow muscle pattern. (A) Surface view of the myotomal segments of a wild-type embryo at 24 h postfertilization (hpf), showing the slow fibers stained with mAb F59 (arrows). (B) Similar view of an ubo embryo, showing a drastic reduction of surface slow fibers and mAb F59 labeling. A few isolated fibers are indicated (arrows). (C, D) Similar images of a wild-type (C) and an ubo embryo (D) stained with mAb S58, showing the pattern of slow fibers. Note that mAb F59 and mAb S58 stainings appear very similar. (E) High-resolution image of wild-type surface slow fibers in A, showing the organized band of slow MyHC filaments (arrows). (F) A similar image of the surface slow fibers of an ubo embryo in B, showing the disorganized pattern of myofibrils (arrows). (G) Medial view of the wild-type embryo in A, showing the well-defined bundles of muscle pioneers (MPs; arrows). (H) Similar view of the ubo embryo in B, showing the scattered distribution of mAb F59 immunoreactivity throughout the medial region of the myotome (arrows). (I) A 10 to 12 somite wild-type embryo, showing the early pattern of slow MyHC expression in the adaxial cells (arrows). (J) A similarly staged ubo embryo, showing the dramatically low levels of slow MyHC expression (arrows). (K) Lateral view of an ubo embryo at 18 somite stage double-labeled for mAb F59 (red) and Prox1 (green), showing complete absence of Prox1 expression from the slow myoblasts and a strong reduction in slow MyHC expression. Compare with a similar-stage wild-type embryo shown in Fig. 2D. (L) Myotomal segments of an ubo embryo at 24 hpf, showing complete absence of Prox1 expression. Compare with wild-type expression shown in Figure 1A. Preparations shown in panels I and J represent dorsal views with anterior to the left. In this and all subsequent figures, comparisons of mutant or manipulated myotomes with wild-type controls are shown at identical magnifications.
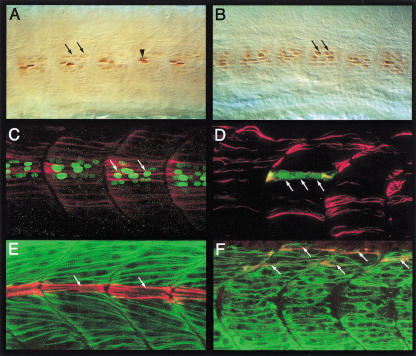
Figure 4.
Slow muscles in ubo embryos transfate into fast. (A) Engrailed (Eng) expression in the mononucleate muscle pioneers (MPs; arrow head) and surrounding syncytial fast muscles (arrows) in a wild-type embryo. Note the high levels of expression in MPs and lower levels in the fast muscles. (B) Eng expression in an ubo embryo (arrows). Note the predominant expression in multinucleate fibers and the uniform levels of expression. (C) Myotome of an ubo embryo double labeled with mAb F59 (red) and mAb 4D9 (green) showing coexpression of slow MyHC and Eng in multinucleate fibers (arrows). Compare with the wild-type expressions shown in Fig. 1C. (D) Surface view of an ubo embryo injected with α-actin GFP plasmid (GFP in green) and counterstained with mAb F59 (red) showing the syncytial nature of a surface slow fiber (arrows). Compare with the similar image of a wild-type embryo shown in Fig. 1E. (E) Medial view of a wild-type embryo, showing mAb S58 immunoreaction in the MPs (red, arrows) and fast MyHC expression in the fast muscles (green). (F) A similar view of an ubo embryo, showing the lack of a defined band of MPs with mAb S58 immunoreactivity. Instead, this pattern is replaced by dispersed colocalization of fast MyHC and mAb S58 labeling in fast muscle–like fibers (arrows).
The complete absence of the pan-slow muscle marker prox1 and the changes in the morphologies of cells expressing Eng and slow MyHC suggest that in ubo, the slow myoblasts could be arrested at a critical choice-point in the slow-twitch fiber differentiation program and consequently transfate to become multinucleate fibers resembling fast muscles. To explore this hypothesis, we examined the expression of fast MyHC in the presumptive slow myoblasts of ubo embryos. In wild-type embryos, mutually exclusive patterns of slow and fast MyHC expression are always observed (Fig. 4E). In contrast, we found that all of the residual slow MyHC expression seen in the myotome of ubo embryos colocalizes with fast MyHC in fibers resembling fast muscles, consistent with the postulated progressive transformation of slow myoblasts into fast (Fig. 4E,F).
Ubo acts cell autonomously to control the response of mesodermal cells to Hh signaling
The results presented thus far cannot exclude the possibility that the ubo phenotype reflects an abnormal behavior of some fast muscle cells. To test the transfating hypothesis further, we examined the effect of loss of ubo function in circumstances in which all the cells in the myotome are induced to form slow fibers and MPs by ectopic expression of Sonic hedgehog (Shh) or a dominant negative (dn) version of protein kinase A (PKA, an intracellular inhibitor of Hh signaling). In wild-type embryos, this response is manifested at early stages by the ectopic expression of myoD and ptc1 ubiquitously in the paraxial mesoderm and at later stages by the presence of numerous ectopic slow fibers in the myotome (Concordet et al. 1996; Currie and Ingham 1996; Hammerschmidt et al. 1996; Weinberg et al. 1996; Blagden et al. 1997; Du et al. 1997). Like normal slow muscles, these ectopic slow fibers are mononucleate and express all slow muscle-specific markers, including slow MyHC and Prox1 as well as high levels of Eng (Fig. 5A,C,E). As expected, such transformed myotomes are associated with little or no fast MyHC expression (Fig. 5G). In ubo embryos, we find that similar to wild-type siblings, there is ubiquitous induction of ptc1 and myoD throughout the paraxial mesoderm, confirming that the pathway involved in the induction of these early markers in the adaxial cells is intact and functional (data not shown). However at 24 h, we observed substantial differences compared to wild-type embryos. Although there was some ectopic expression of slow MyHC relative to control ubo embryos, the level of this misexpression was dramatically lower than that observed in wild-type sibling embryos (Fig. 5B). The extent of ectopic Eng expression was similar to that observed in wild-type embryos manipulated under identical conditions, but unlike the latter, the levels of this expression were lower and were mostly associated with multinucleate fibers (Fig. 5D). Quite strikingly, we found that even in the presence of ectopic Hh signaling, no Prox1 expression could be detected in the ubo myotome (Fig. 5F). Most significantly, however, ectopic Hh signaling failed to suppress the expression of fast MyHC in the myotomes of ubo mutant embryos (Fig. 5H), suggesting that even though these embryos respond to elevated Hh signaling with ectopic induction of myogenic precursors throughout the paraxial mesoderm (ectopic myoD expression) as well as with a limited ectopic induction of slow MyHC expression, these cells are unable to complete their differentiation as slow fibers and switch to the fast muscle developmental pathway. Because these responses of ubo embryos to extracellular (ectopic Shh) and intracellular (ectopic dnPKA) elevation of Hh signaling were indistinguishable, we conclude that ubo is required in the responding cells to interpret the Hh signal.
Figure 5.
Ubo is essential for the Hh-mediated specification of slow fibers and genetically acts downstream from shh and gli2. (A, C, E, G) Myotomes of wild-type embryos injected with shh / dnPKA mRNA, showing ectopic mAb S58 immunoreactivity (A, green), supernumerary Engrailed (Eng)-expressing muscle pioneers (MPs) (C, red), ectopic Prox1 (E, green), and loss of fast MyHC (G, green; residual expression is indicated by the arrow). The mononucleate nature of the ectopic MPs shown in C is indicated (arrows). Note the flattened, elongated nuclei of these ectopic fibres. (B,D,F, H) Myotomal segments of similarly injected and stained ubo embryos. (B) The levels of ectopic slow MyHC expression are strikingly low. (D) Ectopic Eng is induced to an extent similar to that in wild-type embryos, but the levels are comparatively low. Prox1 expression cannot be induced under such conditions in the mutant embryos (F), and they also exhibit high levels of fast MyHC expression (H). The multinucleate nature of the fibers are indicated (arrows in D and H). Note the rounded shape of the nuclei of these fibers. Embryos shown in panels E and F were coinjected with mRNA-encoding nuclear-localized β-galactosidase as a tracer (red) to control for the distribution of injected RNAs; those in panels G and H were counterstained with propidium iodide (red) to reveal the nuclei, whereas those in C and D were coinjected with the α-actin-GFP plasmid to individually label the muscle fibers (green). (I) The pattern of slow fibers in a shh (syu) mutant embryo. Note that the numbers of fibers are reduced, and there are no MP cells (compare with wild-type embryo in Fig. 3A). The pattern of slow MyHC expression in these fibers, however, appears normal and the muscles are mononucleate (arrow in inset). (J) Similar image of an syu;ubo double mutant embryo, showing the pattern of slow fibers which is similar to ubo single mutants and even more dramatically affected (compare with Fig. 3B). Note that the few slow MyHC-associated fibers seen in these situations, like in ubo mutants, are multinucleated (arrows in inset). (K) The myotomal segments of an yot;ubo double mutant embryo, showing complete absence of slow MyHC expression, a phenotype indistinguishable from yot single mutants.
To test this possibility further, we generated genetic mosaics by cell transplantation. If Ubo acts cell autonomously in the regulation of slow muscle development, then labeled wild-type cells, when transplanted into ubo embryos, should be able to differentiate into slow fibers in an otherwise ubo environment, that is, in the absence of wild-type clones in neighboring tissues like the notochord and the neural tube (which, as discussed earlier, profoundly influence slow muscle development through secreted Hh proteins). Indeed in such situations, we were able to rescue slow muscles in ubo embryos with wild-type cells: These cells were mononucleate in morphology, migrated out to the surface, expressed Prox1, and always derived from wild-type donor cells (Fig. 6A–D; van Eeden et al. 1996). This result clearly shows that in the absence of ubo, signaling from the midline is intact, as are all the possible extrinsic cues required for migration of the slow myoblasts, but that a specific defect in these cells interrupts their developmental program.
Figure 6.
Ubo function is required cell autonomously in the slow myoblasts. (A–C) Wild-type donor cells from the α-actin-GFP host transplanted into ubo hosts, when fated to form slow myoblasts, can autonomously develop into fully differentiated slow fibers in the ubo myotome. The donor cells (identifiable by GFP expression, A, green) express Prox1 (B, red, arrow) and form mononucleate fibers on the surface of the myotome (merged image in C, arrow). (D) Wild-type donor cells fated to be fast myoblasts make syncytial, Prox1-negative medial fast fibers (arrow heads) in the ubo myotome. (E–L) Donor cells from ubo embryos (labeled with rhodamine-dextran; E, blue) transplanted into wild-type hosts, when fated to form slow myoblasts (mAb F59 immunoreactivity; F, red, arrow; arrowheads indicate mAb F59 staining of host MPs and edges of the surface slow fibers), remain medial and do not express Prox1 (G, green, arrow; arrowhead indicates endogenous Prox1 in the MPs of the wild-type host). H represents the merged view of panels E–G. When similar transplantations are done into α-actin-GFP hosts (I–L; GFP expression [green] in K,L), such donor (I, arrowheads), mAb F59 immunoreactive ubo cells (J, arrows) can be traced to syncytial fibers (K,L; arrowheads indicate the nuclei; arrows, indicate F59 labeling). Such fibers are either composite, derived from fusion of mutant ubo slow myoblasts with host fast myoblasts (Fiber 1; GFP expression, rhodamine labeling and mAb F59 immunoreactivity), or formed exclusively by the fusion mutant ubo slow myoblasts with each other (Fiber 2; no GFP expression, rhodamine labeling, and mAb F59 immunoreactivity).
In a converse set of experiments, we transplanted labeled ubo cells into wild-type hosts and assayed for the developmental fates of these cells in the myotome. As we expected, if Ubo acts cell autonomously, we observed that slow myoblasts derived from ubo donors always displayed migration defects, never expressed Prox1, and, in most instances, could be traced to multinucleate syncytia that arose either from the fusion of mutant ubo slow myoblasts with each other or, more significantly, with resident fast muscle precursors (Fig. 6E–L).
Ubo acts downstream from shh and gli2 in the genetic pathway that specifies slow muscle precursors
Although the preceding observations provide substantial evidence for the cell-autonomous requirement of Ubo in slow myoblast induction, we wished to explore further whether it functions in this capacity in conjunction with the Hh signaling pathway or influences slow muscle precursor specification through a parallel mechanism. We therefore examined the slow fiber phenotypes of embryos doubly mutant for ubo and two known components of the Hh pathway in the zebrafish embryo that profoundly influence slow muscle induction—shh itself and gli2, one member of the gli gene family–encoding transcription factors that mediate the immediate response of target cells to Hh signals. Mutations in sonic you (syu; encoding Shh) impair the specification of slow myoblasts, thereby reducing the numbers of slow fibers, and completely block the induction of MP cells (van Eeden et al. 1996; Schauerte et al. 1998; Lewis et al. 1999). However, the precursors that do get specified appear to be able to differentiate into surface slow fibers and express all the slow fiber-specific genes (Fig. 5I; Lewis et al. 1999; data not shown). In contrast, mutations in you-too (yot; encoding Gli2) almost completely inhibit transduction of all Hh signaling in the paraxial mesoderm, consequently resulting in the absence of adaxial myogenesis and a concomitant lack of differentiated slow fibers (Fig. 5K; van Eeden et al. 1996; Karlstrom et al. 1999; Lewis et al. 1999). As expected from previous results on the overexpression of Shh in ubo, double mutant syu;ubo embryos showed slow muscle defects that were similar to ubo single mutants (Fig. 5J), although the numbers of slow fibers in these situations were even more reduced than in ubo mutants themselves, in keeping with the reduction in the numbers of adaxial precursors induced in these embryos in the absence of Shh activity. Thus, the combined effects of syu and ubo on slow fiber development are additive and not synergistic, consistent with ubo acting downstream from shh in the specification of the slow muscles. On similar lines, consistent with the lack of initial Hh response and concurrent absence of adaxial myogenesis in yot, double mutant yot;ubo embryos resemble yot and almost completely lack all traces of slow fiber development (Fig. 5K). These observations, considered in conjunction with results from the earlier section, allow us definitively to conclude that Ubo acts after the Gli-mediated induction of adaxial myoblasts by Hh signaling, directing these cells to the slow fiber development pathway.
Specific inhibition of Prox1 activity affects myofibril organization and terminal differentiation of slow fibers
The expression of the regulatory protein Prox1 has been observed in myotomal cells of other vertebrates but the significance of this expression has not been elucidated (Oliver et al. 1993; Tomarev et al. 1996). As described above, its expression in the zebrafish myotome is dependent on ubo activity and is exclusive to slow fibers, where it is initiated at the time these cells first begin their migration through the somite. The most significant defects in the development of the slow myoblasts in ubo mutants are their inability to migrate to the surface, loss of slow MyHC expression, and disrupted myofibrillar organization, all of which occur in conjunction with their failure to activate prox1. Concomitant with these defects is the acquisition of fast muscle properties, such as fusion competence and expression of fast MyHC isoforms. The complete absence of prox1 expression in the myotome of ubo mutant embryos is dramatic—it uncovers the dependence of prox1 expression on ubo function and suggests that at least some of the defects of the slow fibers could be because of the loss of Prox1 activity in these cells.
To investigate whether any of the slow muscle defects observed in ubo are caused by loss of Prox1 activity, we took advantage of the recently established technique of morpholino oligonucleotide–mediated translation inhibition (Nasevicius and Ekker 2000). Embryos injected with antisense morpholino oligonucleotides directed against prox1 mRNA were assayed for the inhibition of Prox1 expression using the anti-Prox1 antibodies (see Materials and Methods). We found that such prox1 antisense oligo–injected embryos showed a dramatic reduction or loss of Prox1 protein in the slow muscle fibers (Fig. 7A,B). We analyzed these embryos for defects in slow fiber formation by monitoring the expression patterns of markers like Eng and MyHC isoforms. The expression pattern of Eng in the MP cells, the levels of which were unaffected in such embryos, revealed that even on drastic reduction of Prox1 activity the slow fibers retain their mononucleate morphology (Fig. 7C). We also did not observe any alterations in the expression pattern of fast MyHC suggesting that Prox1 is not required for repressing fast MyHC expression in slow fibers and that the loss of Prox1 activity does not have any discernible effect on the development of the fast muscle cells (Fig. 7D). The expression of slow MyHC, however, was more revealing. In contrast to ubo mutant embryos, the peripheral migration of slow-twitch myoblasts appeared unaffected in Prox1 negative embryos (Fig. 7A,E,F). Strikingly, however, the arrangement of the slow myofibrils in these fibers appeared dramatically altered (Fig. 7A,E,F); instead of the well-organized band of slow myofibrils characteristic of wild-type slow muscle cells (Fig. 3E), the myofibrils of the slow fibers in anti-prox1 morpholino–injected embryos are disorganized and randomly oriented in the muscle cell cytoplasm (Fig. 7E,F). This phenotype closely resembles the disrupted pattern of slow MyHC expression seen in the stray surface fibers of ubo embryos (Fig. 3F). Thus activation of prox1 by Ubo in the slow myoblasts is essential for the terminal stages of fiber maturation for the proper assembly of the slow myofibrils.
Figure 7.
Suppression of Prox1 activity affects terminal differentiation of slow fibers. (A) Absence of Prox1 expression in a prox1 morpholino-injected wild-type embryo labeled with mAb F59 (red) and anti-Prox1 (green). Compare with wild-type pattern shown in Figure 1A and a control morpholino injected embryo in (B). Note that the slow fibers are able to migrate out to the surface. The shape of the myotome is slightly altered in these embryos, possibly arising as a secondary effect of the improper differentiation of the slow fibers. (C) Expression of Engrailed (Eng) in the muscle pioneers (MPs) of a prox1 morpholino-injected embryo (arrows), showing that the slow fibers retain their mononucleate morphology. (D) Fast MyHC expression (green) in a prox1 morpholino-injected embryo. There is no ectopic expression, and the pattern in the fast muscles appears normal. Compare with wild-type pattern in Figure 1D. (E,F) High-resolution DIC image of mAb F59 (E) and mAb S58 (F) stainings, showing the disrupted pattern of slow myofibrils (arrows) in the surface slow muscles of a prox1 morpholino-injected embryo. The nuclei of the fibers are indicated (arrowheads). Compare with the wild-type pattern shown in Figure 3E.
Discussion
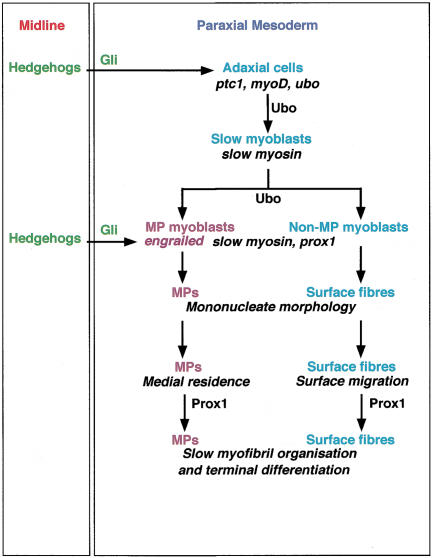
The earliest event in the development of the slow fibers in the zebrafish embryo is the precocious delineation of a distinct set of muscle progenitors from the paraxial mesoderm by Hh signaling from the midline. The primal myogenic response in these adaxial cells to Hhs is the induction of myoD expression (Weinberg et al. 1996). This appears to be directly mediated by the Gli family of transcription factors, the intracellular transducers of the Hh signal, because mutations in zebrafish Gli2 reduce or completely eliminate adaxial myoD expression, thereby inhibiting the process of myogenesis in the adaxial mesoderm (van Eeden et al. 1996; Karlstrom et al. 1999; Lewis et al. 1999). Although Hh-induced myoD expression commits the adaxial cells as muscle progenitors, this is unlikely to confer fiber-type development pathway specifications on them. Thus, myoD, which is part of a general set of information required to make muscle, is also later deployed independent of Hh signaling in a second episode of myogenesis in the embryo, as part of the fast muscle developmental program (Weinberg et al. 1996). We therefore propose that in conjunction with components of the general myogenic pathway, Hh signaling also induces the expression of the ubo gene in the adaxial cells. Ubo then acts as a selector molecule that shunts the adaxial cells toward the slow muscle development pathway (Fig. 8). Earlier preliminary studies (van Eeden et al. 1996; Lewis et al. 1999) and our present analyses show that ubo affects slow muscle development without disrupting the general myogenic response of the adaxial cells to Hhs, unlike mutations in components of the Hh pathway (van Eeden et al. 1996; Schauerte et al. 1998; Lewis et al. 1999; Barresi et al. 2000). These observations, together with epistasis analysis using mutations in known constituents of the Hh pathway, convincingly position ubo downstream from Hh signaling in the developmental program of the slow muscles. In addition, ubo does not alter Hh-mediated developmental processes elsewhere in the embryo, such as patterning of the ventral neural tube (S. Roy, C. Wolff, and P.W. Ingham, unpubl.). Thus, at least in the myotome, ubo appears to uncover a cell type–specific function for a gene acting downstream from Hh signaling and dedicated to the specification and differentiation of a distinct lineage of muscle cells. Apart from the differences in patterns of MyHC protein expression, the slow and fast fiber populations have a number of contrasting properties, including their cellular morphologies and developmental behavior as well as profiles of regulatory gene expression. The gradual loss of slow fiber attributes in ubo embryos, concomitant with the gain of fast muscle characteristics, strengthens this model of ubo function and suggests that such transdetermination of the slow myoblasts can indeed effectively occur at a relatively late point in their development. Even under conditions of ectopic Hh signaling, which is able to induce precocious myogenesis throughout the somite of ubo embryos, the ectopically specified muscle precursors, unlike in wild-type embryos, are unable to differentiate as slow fibers and mature instead as fast muscles. One limitation of our analysis is that it is restricted to a single mutant allele of ubo; it is possible that this allele represents a hypomorphic mutation, which might explain the limited induction of slow MyHC expression seen in the mutant embryos as well as under conditions of elevated Hh signaling, the progressive transfating of the induced slow myoblasts and also the ability of a few mutant slow fibers to migrate out to the surface. In this view, a null allele of ubo should completely block initiation of slow muscle development and prevent expression of slow MyHC, sparing only the Hh-mediated induction of general regulators of myogenesis like myoD in the adaxial cells.
Figure 8.
Schematic representation of the genetic pathway that regulates specification and differentiation of the slow muscle fibers. Hh proteins from the midline signal to the paraxial mesodermal cells by a Gli-dependent mechanism. This results in the induction of general markers of Hh response–like ptc1 and in the induction of myogenesis through the expression of myogenic regulatory genes in the most proximal cells, the adaxial cells. We propose that Hh signaling also induces the expression of the ubo gene in these cells, which allows them to interpret this signal and adopt the slow fiber-specific developmental program. In a separate inductive event, Hh signaling acts to induce high levels of Engrailed (Eng) expression in the muscle pioneer (MP) precursors. The requirement of the homeobox gene prox1 is restricted to the final stages for terminal differentiation of the slow fibers.
Eng and the specification of MP cells
Our analysis of the ubo mutation also unravels an added dimension of regulation in the diversification of the slow fibers—the specification of the MP cells. Although it is clear from lineage analysis that the MPs are drawn from the slow muscle precursor pool, the mechanism by which they are segregated and acquire properties that distinguish them from the surface slow fibers is obscure. Perhaps the most widely used marker to distinguish the MPs from the surface slow fibers is the expression of Eng proteins in the former; earlier studies have provided evidence that at least this aspect of MP development is directed by Hh signaling (Halpern et al. 1993; Currie and Ingham 1996; Hammerschmidt et al. 1996; van Eeden et al. 1996; Du et al. 1997; Lewis et al. 1999). We have shown that although the ubo mutation affects the developmental pattern of all the slow fibers, including the MPs, it does not interfere with the induction of Eng expression in the myotome. However, it does affect the levels of Eng expression, because in ubo embryos the high level aspect of the expression is lost and cannot be induced even under conditions of ectopic Hh signaling. Therefore, it would seem that we are looking at layers of developmental pathways superimposed on one another: a Hh-mediated ubo-driven slow fiber–specific pathway on a general myogenic program, together with a Hh-mediated ubo-dependent pathway layered on the slow muscle development program for the high-level expression of Eng in MP cells (Fig. 8). In ubo mutants, because the MPs adopt fast fiber properties, this is also reflected faithfully in the altered pattern of Eng expression observed in these embryos.
Prox1 regulates terminal differentiation of the slow fibers
Another remarkable aspect of the ubo phenotype is highlighted by the complete absence of prox1 expression in the embryonic slow myoblasts. Even in situations in which supernumerary slow muscle precursors are induced by ectopic activation of the Hh pathway, prox1 expression is never induced in these cells. Although this is certainly strong evidence that prox1 expression in the embryonic slow myoblasts is critically dependent on Ubo, it is unclear at this point whether this is a direct response or mediated through intermediate steps. Although recent studies in the mouse using targeted inactivation of the prox1 locus have helped to clarify the function of this gene in the development of the lens, liver, and the lymphatic system, its role in muscle cells has not been investigated (Wigle and Oliver 1999; Wigle et al. 1999; Sosa-Pineda et al. 2000). The absence of Prox1 in the embryonic slow fibers in ubo suggests that one or more features of these muscles—their fusion incompetence and consequently their mononucleate morphology, migratory behavior, expression profiles of MyHC isoforms, and terminal differentiation—could be regulated by Prox1 activity. Using antisense oligonucleotides to specifically “knock down” Prox1 activity, we have found that Prox1 functions at a rather late stage in the maturation of the slow fibers; its absence resulting in disrupted myofibrillar organization. Because such situations were not associated with any other obvious phenotypic defects in the slow fibers, it would suggest that this represents the singular role of Prox1 in slow muscle cells. This result provides evidence that the disrupted myofibrillar phenotype observed in a few slow fibers in ubo that escape early transfating and succeed in migrating to the surface is likely to arise because of the inability of these mutant fibers to activate prox1 expression. The focus of Prox1 function in the ontogeny of the slow muscle cells revealed by this analysis is in keeping with the onset of its expression at a relatively late stage in the slow myoblasts (at 18-somite stage, Fig. 2D) after the synthesis of slow MyHC is well underway in these cells. Although the slow myoblasts begin to elaborate slow MyHC molecules precociously, even while they are resident adaxially within the somite, much or all of this protein appears as randomly oriented filaments in the cytoplasm of the myoblasts (Fig. 2A–C; Devoto et al. 1996). The first indication of myofibrillar organization and assumption of fiber-like morphology is evident at the 18 somite stage when the myoblasts have begun to express Prox1 (Fig. 2D). A number of studies have alluded to the distinctive architecture of the contractile apparatus of teleost slow fibers (Waterman 1969; Felsenfeld et al. 1990; Stoiber et al. 1998). Therefore, in its capacity as a regulatory protein, Prox1 could function to modulate the expression of specific kinds of myofibril organizing components that are unique to the terminal differentiation program of the slow fibers. Interestingly, defects in terminal differentiation and improper assembly of contractile protein filaments are associated with a number of skeletal muscle and cardiac myopathies (Engel 1999; Gregorio and Antin 2000). Although many of these disorders are caused by mutations in components of the myofibrillar apparatus itself, little is known about the possible roles of dysfunctional regulatory proteins in the generation of these pathological conditions. In this light, our results on prox1 function in the muscle cells of the zebrafish may be of particular significance.
Hh signaling and fiber-type diversification in amniote embryos
Although the segregation of myoblasts into slow versus fast subtypes has been observed during embryonic myogenesis in the somites of amniotes, analysis of Hh-mediated fiber-type specification in the zebrafish embryo has provided the first mechanistic insight into the kinds of developmental signals that drive this early diversification (Currie and Ingham 1996; Blagden et al. 1997; Du et al. 1997). Hh signaling has, however, been shown to play a crucial role in regulating somitic myogenesis in amniotes (Borycki and Emerson 2000). Moreover, recent studies in the chick have suggested that Hh may have a similar effect on fiber-type diversification (Cann et al. 1999). In this regard, it is striking that, as in the zebrafish, the earliest (primary) muscle fibers that emerge in the amniote somite are mononucleate cells and express slow isoforms of muscle MyHC (Holtzer et al. 1957; Cann et al. 1999). Therefore, it is tempting to speculate that even in amniote embryos, similar genetic mechanisms, involving an ubo homolog, could link the segregation and differentiation of the primary fibers with the inductive activities of Hh proteins.
Materials and methods
Fish strains and embryos
The ubotp39, syut4, and yotty119 mutations were isolated in a large-scale mutagenesis screen at the Max-Planck Institut for Entwicklungsbiologie, Tübingen (van Eeden et al. 1996). The skeletal muscle α-actin-GFP transgenic strain was a gift from S. Higashijima.
Immunohistochemistry
Antibody labeling was done essentially as previously described (Westerfield 1995; Devoto et al. 1996; Bladgen et al. 1997; Barresi et al. 2000). The following primary antibodies were used: mAb F59 and mAb S58 (Devoto et al. 1996), mAb EB165 (anti-fast MyHC; Bladgen et al. 1997), mAb 4D9 (anti-Engrailed; Patel et al. 1989), mAb anti-βgalactosidase (Promega), and a human Prox1 antiserum (Glasgow and Tomarev 1998). Three different anti-MyHC antibodies were used to label the slow fibers—mAb F59 and mAb S58 (Devoto et al. 1996; Barresi et al. 2000), as well as BA-D5 (Bladgen et al. 1997; data not shown). The specificities of these antibodies to muscle fiber types in the zebrafish and higher vertebrates have been discussed previously (Devoto et al. 1996; Bladgen et al. 1997).
Embryo injections and cell transplantations
Embryos were injected with capped mRNA, antisense prox1/control morpholino oligonucleotides, the α-actin-GFP plasmid (Higashijima et al. 1997) or dextran-coupled rhodamine/biotin (Molecular Probes) using standard procedures. Because of the highly mosaic segregation of injected DNA constructs in zebrafish embryos, injection of the α-actin-GFP plasmid fortuitously allowed us to randomly label the muscle cells independently of their lineage. In all instances analysed (n > 50), we found that all slow GFP-positive fibers were mononucleate, whereas all nonslow GFP-positive fibers (hence, fast) were syncytial. The morpholinos were purchased from Gene Tools, LLC. The sequences of the prox1 and control morpholinos are 5′-ATGTGCTGTCATGGTCAGGCATCAC-3′ and 5′-CTTTTT TTTGGGACTTTTCTCTTTG-3′, respectively. The prox1 morpholino was used at a concentration of 1mM and consistently produced reproducible phenotypes with an injection volume around 8 nL that manifested in a drastic reduction of Prox1 expression and strong fibrillation defects in the slow muscles. The control morpholino at the same dose produced no discernible effects. For our analysis of the effect of Prox1 “knock down” on slow muscle development, the results we have presented are representative of approximately 100 injected embryos that were processed for each experiment, of which 75% to 80% showed the specific effects. For generating mosaic embryos, approximately 20 to 30 cells from rhodamine/biotin dextran–injected donor embryos at the high or dome stage were transferred to each wild-type or α-actin-GFP host embryo using a micromanipulator as described previously (Ho and Kane 1990).
Acknowledgments
We thank C. Nüsslein-Volhard and the Max-Planck Institut for Entwicklungsbiologie for providing the mutant strains; F. Stockdale, S. Tomarev, E. Bandman, and the Developmental Studies Hybridoma Bank for antibodies; and F. Wilson for excellent technical assistance. This work was supported by a Wellcome Trust International Travelling Research Fellowship (S.R.) and by grants from the European Union Training and Mobility of Researchers network (T.M.R.) and the Wellcome Trust (P.W.I.). The confocal microscope facility is funded by the Yorkshire Cancer Research (Y.C.R.).
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL p.w.ingham@sheffield.ac.uk; FAX 44-(0) 114-222-2788.
Article and publication are at http://www.genesdev.org/cgi/doi/10.1101/gad.195801.
References
- Arnold HH, Braun T. Genetics of muscle determination and development. Curr Top Dev Biol. 2000;48:129–164. doi: 10.1016/s0070-2153(08)60756-5. [DOI] [PubMed] [Google Scholar]
- Barresi MJ, Stickney HL, Devoto SH. The zebrafish slow-muscle–omitted gene product is required for Hedgehog signal transduction and the development of slow muscle identity. Development. 2000;127:2189–2199. doi: 10.1242/dev.127.10.2189. [DOI] [PubMed] [Google Scholar]
- Blagden CS, Currie PD, Ingham PW, Hughes SM. Notochord induction of zebrafish slow muscle mediated by Sonic hedgehog. Genes & Dev. 1997;11:2163–2175. doi: 10.1101/gad.11.17.2163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borycki AG, Emerson CP. Multiple tissue interactions and signal transduction pathways control somite myogenesis. Curr Top Dev Biol. 2000;48:165–224. doi: 10.1016/s0070-2153(08)60757-7. [DOI] [PubMed] [Google Scholar]
- Cann GM, Lee JW, Stockdale FE. Sonic hedgehog enhances somite cell viability and formation of primary slow muscle fibres in avian segmented mesoderm. Anat Embryol. 1999;200:239–252. doi: 10.1007/s004290050276. [DOI] [PubMed] [Google Scholar]
- Concordet JP, Lewis KE, Moore JW, Goodrich LV, Johnson RL, Scott MP, Ingham PW. Spatial regulation of a zebrafish patched homologue reflects the roles of Sonic hedgehog and protein kinase A in neural tube and somite patterning. Development. 1996;122:2835–2846. doi: 10.1242/dev.122.9.2835. [DOI] [PubMed] [Google Scholar]
- Currie PD, Ingham PW. Induction of a specific muscle cell type by a Hedgehog-like protein in zebrafish. Nature. 1996;382:452–455. doi: 10.1038/382452a0. [DOI] [PubMed] [Google Scholar]
- ————— The generation and interpretation of positional information within the vertebrate myotome. Mech Dev. 1998;73:3–21. doi: 10.1016/s0925-4773(98)00036-7. [DOI] [PubMed] [Google Scholar]
- Devoto SH, Melancon E, Eisen JS, Westerfield M. Identification of separate slow and fast muscle precursor cells in vivo, prior to somite formation. Development. 1996;122:3371–3380. doi: 10.1242/dev.122.11.3371. [DOI] [PubMed] [Google Scholar]
- Du SJ, Devoto SH, Westerfield M, Moon RT. Positive and negative regulation of muscle cell identity by members of the Hedgehog and TGF-β gene families. J Cell Biol. 1997;139:145–156. doi: 10.1083/jcb.139.1.145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engel AG. Myofibrillar myopathy. Ann Neurol. 1999;46:681–683. doi: 10.1002/1531-8249(199911)46:5<681::aid-ana1>3.0.co;2-b. [DOI] [PubMed] [Google Scholar]
- Felsenfeld AL, Walker C, Westerfield M, Kimmel C, Streisinger G. Mutations affecting skeletal muscle myofibril structure in the zebrafish. Development. 1990;108:443–459. doi: 10.1242/dev.108.3.443. [DOI] [PubMed] [Google Scholar]
- Frasch M. Controls in patterning and diversification of somatic muscles during Drosophila embryogenesis. Curr Opin Genet Dev. 1999;9:522–529. doi: 10.1016/s0959-437x(99)00014-3. [DOI] [PubMed] [Google Scholar]
- Glasgow E, Tomarev SI. Restricted expression of the homeobox gene prox 1 in developing zebrafish. Mech Dev. 1998;76:175–178. doi: 10.1016/s0925-4773(98)00121-x. [DOI] [PubMed] [Google Scholar]
- Gregorio CC, Antin PB. To the heart of myofibril assembly. Trends Cell Biol. 2000;10:355–362. doi: 10.1016/s0962-8924(00)01793-1. [DOI] [PubMed] [Google Scholar]
- Halpern ME, Ho RK, Walker C, Kimmel CB. Induction of muscle pioneers and floor plate is distinguished by the zebrafish no tail mutation. Cell. 1993;75:99–111. [PubMed] [Google Scholar]
- Hammerschmidt M, Bitgood MJ, McMahon AP. Protein kinase A is a common negative regulator of Hedgehog signalling in the vertebrate embryo. Genes & Dev. 1996;10:647–658. doi: 10.1101/gad.10.6.647. [DOI] [PubMed] [Google Scholar]
- Hatta K, Bremiller R, Westerfield M, Kimmel CB. Diversity of expression of Engrailed-like antigens in zebrafish. Development. 1991;112:821–832. doi: 10.1242/dev.112.3.821. [DOI] [PubMed] [Google Scholar]
- Higashijima S, Okamoto H, Ueno N, Hotta Y, Eguchi G. High-frequency generation of transgenic zebrafish which reliably express GFP in whole muscles or the whole body by using promoters of zebrafish origin. Dev Biol. 1997;192:289–299. doi: 10.1006/dbio.1997.8779. [DOI] [PubMed] [Google Scholar]
- Ho RK, Kane DA. Cell-autonomous action of zebrafish spt-1 mutation in specific mesodermal precursors. Nature. 1990;348:728–730. doi: 10.1038/348728a0. [DOI] [PubMed] [Google Scholar]
- Holtzer H, Marshall JM, Finck H. An analysis of myogenesis by the use of fluorescent antimyosin. J Biophys Biochem Cytol. 1957;3:705–729. doi: 10.1083/jcb.3.5.705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes SM, Salinas PC. Control of muscle fibre and motorneuron diversification. Curr Opin Neurobiol. 1999;9:54–64. doi: 10.1016/s0959-4388(99)80007-5. [DOI] [PubMed] [Google Scholar]
- Karlstrom RO, Talbot WS, Schier AF. Comparative synteny cloning of zebrafish you-too: Mutations in the Hedgehog target gli2 affect ventral forebrain patterning. Genes & Dev. 1999;13:388–393. doi: 10.1101/gad.13.4.388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimmel CB, Warga RM. Cell lineages generating axial muscle in the zebrafish embryo. Nature. 1987;327:234–237. doi: 10.1038/327234a0. [DOI] [PubMed] [Google Scholar]
- Lewis KE, Currie PD, Roy S, Schauerte H, Haffter P, Ingham PW. Control of muscle cell-type specification in the zebrafish embryo by Hedgehog signalling. Dev Biol. 1999;216:469–480. doi: 10.1006/dbio.1999.9519. [DOI] [PubMed] [Google Scholar]
- Nasevicius A, Ekker SC. Effective targeted gene “knockdown” in zebrafish. Nat Genet. 2000;26:216–220. doi: 10.1038/79951. [DOI] [PubMed] [Google Scholar]
- Oliver G, Sosa-Pineda B, Geisendorf S, Spana EP, Doe CQ, Gruss P. Prox 1, a prospero-related homeobox gene expressed during mouse development. Mech Dev. 1993;44:3–16. doi: 10.1016/0925-4773(93)90012-m. [DOI] [PubMed] [Google Scholar]
- Patel NH, Martin-Blanco E, Coleman KG, Poole SJ, Ellis MC, Kornberg TB, Goodman CS. Expression of Engrailed proteins in arthropods, annelids, and chordates. Cell. 1989;58:955–968. doi: 10.1016/0092-8674(89)90947-1. [DOI] [PubMed] [Google Scholar]
- Roy S, VijayRaghavan K. Muscle pattern diversification in Drosophila: The story of imaginal myogenesis. Bioessays. 1999;21:486–498. doi: 10.1002/(SICI)1521-1878(199906)21:6<486::AID-BIES5>3.0.CO;2-M. [DOI] [PubMed] [Google Scholar]
- Schauerte HE, van Eeden FJ, Fricke C, Odenthal J, Strahle U, Haffter P. Sonic hedgehog is not required for the induction of medial floor plate cells in the zebrafish. Development. 1998;125:2983–2893. doi: 10.1242/dev.125.15.2983. [DOI] [PubMed] [Google Scholar]
- Sosa-Pineda B, Wigle JT, Oliver G. Hepatocyte migration during liver development requires Prox1. Nat Genet. 2000;25:254–255. doi: 10.1038/76996. [DOI] [PubMed] [Google Scholar]
- Stockdale FE. Myogenic cell lineages. Dev Biol. 1992;154:284–298. doi: 10.1016/0012-1606(92)90068-r. [DOI] [PubMed] [Google Scholar]
- Stoiber W, Haslett JR, Goldschmid A, Sanger AM. Patterns of superficial fibre formation in the European pearlfish (Rutilus frisii meidingeri) provide a general template for slow muscle development in teleost fish. Anat Embryol. 1998;197:485–496. doi: 10.1007/s004290050159. [DOI] [PubMed] [Google Scholar]
- Tomarev SI, Sundin O, Banerjee-Basu S, Duncan MK, Yang JM, Piatigorsky J. Chicken homeobox gene Prox 1 related to Drosophila prospero is expressed in the developing lens and retina. Dev Dyn. 1996;206:354–367. doi: 10.1002/(SICI)1097-0177(199608)206:4<354::AID-AJA2>3.0.CO;2-H. [DOI] [PubMed] [Google Scholar]
- van Eeden FJ, Granato M, Schach U, Brand M, Furutani-Seiki M, Haffter P, Hammerschmidt M, Heisenberg CP, Jiang YJ, Kane DA, et al. Mutations affecting somite formation and patterning in the zebrafish, Danio rerio. Development. 1996;123:153–164. doi: 10.1242/dev.123.1.153. [DOI] [PubMed] [Google Scholar]
- Waterman RE. Development of the lateral musculature in the teleost, Brachydanio rerio: A fine structural study. Am J Anat. 1969;125:457–493. doi: 10.1002/aja.1001250406. [DOI] [PubMed] [Google Scholar]
- Weinberg ES, Allende ML, Kelly CS, Abdelhamid A, Murakami T, Andermann P, Doerre OG, Grunwald DJ, Riggleman B. Developmental regulation of zebrafish myoD in wild-type, no tail and spadetail embryos. Development. 1996;122:271–280. doi: 10.1242/dev.122.1.271. [DOI] [PubMed] [Google Scholar]
- Westerfield M. The Zebrafish Book. Oregon: University of Oregon Press; 1995. [Google Scholar]
- Wigle JT, Chowdhury K, Gruss P, Oliver G. Prox1 function is crucial for mouse lens-fibre elongation. Nat Genet. 1999;21:318–322. doi: 10.1038/6844. [DOI] [PubMed] [Google Scholar]
- Wigle JT, Oliver G. Prox1 function is required for the development of the murine lymphatic system. Cell. 1999;98:769–778. doi: 10.1016/s0092-8674(00)81511-1. [DOI] [PubMed] [Google Scholar]