Introduction
Members of the myosin superfamily perform a wide variety of transport, assembly, anchoring, and signaling functions in cells (1–29). They share conserved motor domains, usually located at the N-terminus, that bind actin filaments, hydrolyze ATP, and convert the chemical energy into mechanical work. Myosin motors have highly variable tail domains that presumably are related to their localizations and functions in the cell through specific binding to cargos and membrane proteins (10–12,15,30–34). Processive myosin motors are usually dimers whose tail domains are linked in a coiled-coil (CC). Between the motor domain and the tail domain is the so-called myosin neck or lever-arm domain, which is generally believed to be involved in motor regulation and/or to act as a lever that rotates or tilts to amplify the angstrom-level conformational changes in the motor domain to the nanometer-sized power-stroke motions. The neck domain, or light chain domain (LCD), usually binds calmodulin or calmodulin-like light chains (35) (both termed CaM hereafter) at successive consensus motifs (IQ domains (1)) in a relatively long α-helical segment of the heavy chain. The length of the lever has often been assumed to be determined by the number of IQ motifs and CaMs bound in each myosin isoform. However, this assumption has been challenged by many types of evidence that other portions of the heavy chain exhibit mechanical stiffness and also contribute to the power stroke. If the lever arm is instead considered as the portion of a myosin motor that corresponds to the mechanical segment that tilts during the working stroke, it can be defined as a combination of three regions: 1), the converter (the rotating subdomain at the C-terminus of the motor domain); 2), the CaM-binding LCD region; and 3), any segment of the heavy chain between LCD and the CC, if that segment is stiff (36).
Unconventional myosins have been categorized into more than 30 distinct classes (5,14,16,37). Their structure, function, and regulation have been described and reviewed extensively (1–10,12–17,20–29,33,37–50). In the best studied of these myosins, the neck regions of the motor have properties consistent with their function as lever arms: they tilt back and forth between two main orientations during stepping (51–56), the step size and velocity of motility depend on the length of the neck (40,47,57–60), and in constructs attached to artificial neck regions, the direction of motion depends on the orientation of the artificial attachment (61). In this mini-review, we consider the specialized lever-arm structures of three myosins (myosins V, VI, and X) and the impact these structures have on the stepping characteristics and functions of these myosins. We focus here on the role of the neck regions as mechanical lever arms, although they are also involved in regulating motility (45,62) and (possibly) sensing tension (42,49,63–65). For myosins VI and X, there are open research questions regarding the length and composition of the lever arms and the consequent implications for their paths, angular motions, and processivity.
Puzzles about the Discrepancy between the Lever-Arm Length and Step Size of Myosin Motors
Myosin motors bind actin, and many of them translocate along actin filaments processively (i.e., they take several steps per diffusional encounter). An actin filament has a polar structure with barbed (plus) and pointed (minus) ends. The right-handed, double-helical structure of actin filaments with a pitch of ∼72 nm or 26–28 actin subunits, and half-pitch of ∼36 nm or 13 subunits, allows the myosin motors to take different paths—straight, wiggly, or spiral—depending on the distributions of their step sizes.
Different myosin classes, including myosins V, VI, and X, have different numbers of IQ domains and CaMs bound. Myosin V, which is by far the best-known processive cargo transporter and most-studied nonmuscle myosin motor, has six IQ motifs and six CaMs bound per heavy chain (Fig. 1 a). Myosin VI, which moves in the opposite direction (toward the minus end of actin) with respect to other myosin motors, has two CaMs per heavy chain—one bound to an IQ motif and one bound to a unique sequence termed Insert 2 (Fig. 1 b). Myosin X, which is specialized for transport on actin filaments and bundles, has three IQ-bound CaMs per heavy chain (Fig. 1 c). Although it is established that tail domains play a major role in regulating myosin motility (50,66,67), many single-molecule studies using truncated myosin constructs have shown that myosin motility is also strongly dependent on the structures of their particular lever arms (36,40,44,57–60,68,69). Therefore, differential regulation of myosin functions in the cell may be a cooperative process involving both the tail and lever-arm domains.
Figure 1.
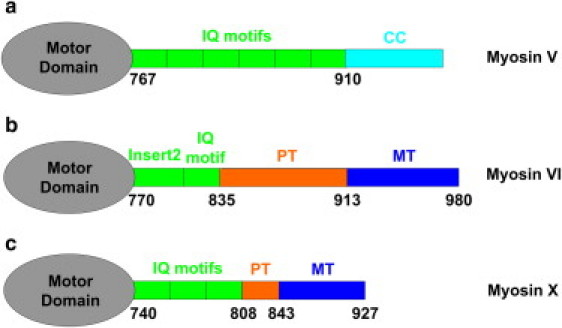
Schematic diagram of myosins V, VI, and X without their cargo-binding domains. (a) Chicken myosin V has six IQ motifs that bind CaMs or CaM-like light chains (35) and serve as lever arms. The coiled-coil (CC) domains dimerize two myosin V monomers to facilitate the hand-over-hand stepping mechanism. (b) Myosin VI has a special segment (Insert 2) that also binds CaM and turns the lever arm in the opposite direction from other myosin motors. Its proximal tail (PT, orange) is a triple α-helix bundle (TAHB), and its medial tail (MT, blue) is a stable single α-helix (SAH), both of which have been suggested to be involved in extension of the lever-arm and heavy-chain dimerization. (c) Myosin X has three IQ motifs. Its PT (orange) and MT (blue) domains have been suggested to form SAHs and act as part of the lever arm.
Several single-molecule techniques, such as optical traps (68,70–76), and fluorescence methods with amusing acronyms, such as FIONA (56,77–81), SHREC (82), and SHRImP (83), have been developed to characterize the motility of processive myosin motors as they walk along their actin tracks, revealing the center-of-mass step size as well as the kinetic rates of actomyosin interaction (see Table 1). It must be noted that due to different experimental conditions, such as the myosin construction (particularly the dimerization sequence), method of actin attachment, temperature, ATP concentration, salt concentration, and buffer pH, some values of motility parameters are highly variable among studies. Therefore, one should use caution when comparing them across studies. A systematic comparison of the processive myosin motors is needed. It has been shown that myosins V, VI, and X all step processively hand-over-hand along actin filaments (51,54–56,77–80,82,83). They are capable of taking similar step sizes (∼34–36 nm; Table 1) even though the IQ-bound CaM-containing sections of their lever arms have different lengths (Fig. 1). Therefore, it is particularly intriguing to consider how they achieve their processive motility and step sizes in spite of this rather large variability. As shown in Fig. 2a, for instance, myosin V has an average 36 nm step size, which suggests a nearly 90° angle between the two lever arms (lever-arm length = converter + 6 IQ = 26 nm) without a further heavy chain component. This geometry is consistent with the direct visualization of myosin V on actin filaments by electron microscopy (60,84) and scanning probe microscopy (85). It also implies that the two heads of myosin V can span across the 36 nm helical half-pitch of actin filaments, and the molecule can follow relatively straight paths along the filament (Fig. 2 a). Myosin V has also been shown to intermittently exhibit lever-arm tilts and actin subunit shifts without taking full steps, and sudden azimuthal (sideways) steps (53,85).
Table 1.
Motility measurements of myosins V, VI, and X
| Myosin | Step size (nm) | Run length (μm) | Vmax (nm/s) | Pn | 2Δα (degree) | Helical path handedness | Helical pitch (μm) |
|---|---|---|---|---|---|---|---|
| V | 73.8∗ ± 5.3 (77) | 0.85 (56) (1 ATP, 25 KCl) | 453 (51) (25 KCl) | 18–35 | 0.5 ± 29.1 (54) | Left (104) | 1.7 ± 0.6 (104) |
| 37.5 ± 6.2 (79) | 0.66 (93) (2 ATP, 25 KCl) | 330 (93) (25 KCl) | |||||
| 35.0 ± 8.9 (111) (2 ATP) | 2.2 (113) (in vivo) | 710 (113) (in vivo) | |||||
| 37.0 ± 10.3 (111) (0.01 ATP) | 1.3 (113) (in vitro) | 500 (113) (in vitro) | |||||
| 40.2 ± 6.4 (112) | 1.3 (114) (1 ATP, 25 KCl) | 380 (114) (25 KCl) | |||||
| 37.0 ± 2.0 (113) (in vivo) | 0.8 (114) (1 ATP, 100 KCl) | 550 (114) (100 KCl) | |||||
| VI | 63.3∗ ± 16.7 (78) eGFP on N-term | 0.78 (65) (0.1 ATP, 25 KCl) | 7–31 | −0.5 ± 54.7 (54) | Right (54) | 1.3 ± 0.5 (54, 104) | |
| 70∗ ± 23 (80) | 0.44 (65) (2 ATP, 25 KCl) | ||||||
| 55.2∗ ± 17.2 (31) (wild-type) | 0.28 (80) (0.04 ATP, 25 KCl) | 288 (54) (25 KCl) | |||||
| 30 ± 12 (115) | 0.55 (98) (2 ATP, 25 KCl) | 291 (115) (25 KCl) | |||||
| 38 ± 16 (116) (wild -type) | 1.10 (31) (1 ATP, 50 KCl) | 550 (116) (25 KCl) | |||||
| 0.23 (115) (2 ATP, 25 KCl) | |||||||
| 0.24 (116) (5 ATP, 25 KCl) | |||||||
| X | 68∗ ± 8.4 (56) | 1.09 (56) (0.5 ATP, 25 KCl) | 235 (56) (25 KCl) | 10–32 | 3.4 ± 30.0 (56) | Left (56, 105) | 1.0 ± 0.5 (56) |
| 16.4 ± 1.8 (94) on single actin | 0.82 (56) (2 ATP, 25 KCl) | 330 (93) (25 KCl) on single actin | |||||
| 17.5 ± 1.9 (94) on bundled actin | 0.17 (93) (2 ATP, 25 KCl) on single actin | 340 (93) (25 KCl) on bundled actin | |||||
| 0.63 (93) (2 ATP, 25 KCl) on bundled actin | 578 (117) (in vivo) |
Pn is the processivity number (run length/step size). Note that some values (e.g., run length and Vmax) are highly variable among studies, probably due to different experimental conditions such as myosin constructs, actin attachment, temperature, [ATP], [salt], and pH. The values given here are examples. ATP and KCl values are in mM units. Step sizes marked with asterisks are from myosin molecules labeled near or on the motor domain, and therefore should be halved to estimate the step size of the center of mass.
Figure 2.
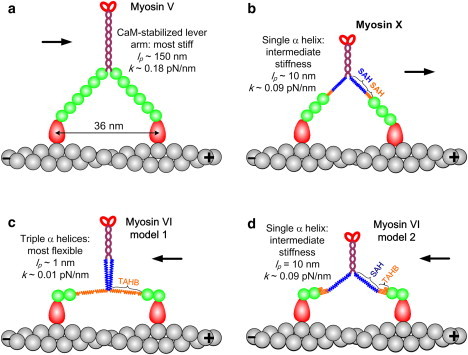
Models of myosin V, VI, and X lever arms. (a) Myosin V has six CaMs bound to IQ motifs that form its lever arm. (b) Myosin X is thought to use its PT and MT domains as stiff SAHs to extend its short LCD and form a longer lever arm. (c and d) The myosin VI lever arm may contain (c) an unfolded TAHB in the PT domain (88) or (d) mainly a stable SAH in the MT domain (87).
Myosin VI and myosin X have much shorter LCDs than myosin V (Fig. 2), although both have been shown to step hand-over-hand along actin filaments like myosin V (56,77,78,80,82,83). For myosin VI, a recent study with high-temporal-resolution tracking of fluorescent markers supplemented the simple model of hand-over-hand stepping with a mixture of hand-over-hand and inchworm-like motions that depend on the load and ADP concentration (75). When they take hand-over-hand motions, and both heads are bound on the actin filament, the LCDs of both myosins VI and X are too short to span their average step sizes. In other words, myosins VI and X should have additional lever-arm components to achieve the measured hand-over-hand step sizes. Sequence analysis and biophysical characterization led to the prediction that the proximal tail (PT) and medial tail (MT) domains of myosin X (∼120 residues; Fig. 1 c) do not dimerize into a CC, but form stable single α-helices (SAHs) (86) (Fig. 2 b). The sequence is particularly rich in glutamic acid (E), arginine (R), and lysine (K) residues that form stabilizing salt bridges outside of the α-helix (86,87). This ERK region was shown experimentally to serve as a mechanical lever with a persistence length of ∼10 nm (36). The persistence length is a measure of the bending stiffness of a rod or linear polymer chain (43).
The conformation of the myosin VI lever arm is still under debate (87–89). Mukherjea et al. (88) suggested that the PT domain of myosin VI (∼80 residues; orange band in Fig. 1 b) contains a triple α-helical bundle (TAHB) that unfolds to serve as a portion of the lever arm when triggered by myosin VI dimerization (Fig. 2 c). In contrast, Spink et al. (87) argued that the TAHB remains folded as a portion of the lever arm, and that the main contributor of extended length is the MT domain (∼70 residues; blue band in Figs. 1 b and 2, c and d), which forms a stable SAH like the one in myosin X. These two postulated lever-arm structures are expected to have different stepping characteristics, as explained below.
Step Sizes Indicate Different Lever-Arm Stiffness Values for the Three Myosin Motors
The stiffness against bending of a lever arm is given by (43)
| (1) |
where E is the Young's modulus, I is the moment of inertia (a measure of the radial distribution of material), L is the lever-arm length, lp is the persistence length, kB is Boltzmann's constant, and T is the temperature. The persistence lengths are very different for the LCD and other portions of the lever arm. The following persistence lengths have been estimated or measured: lp = 150 nm for a group of successive IQ motifs stabilized by CaMs (69), lp = 1 nm for unfolded triple α-helices linked by flexible coils (90), and lp = 10 nm for an ERK-stabilized SAH (87). In some cases, there are kinks in a SAH domain, which would lower its persistence length. In a two-head, actin-bound state, the intramolecular strain of a myosin motor is a function of the head-to-head distance and azimuthal shifts on the double-helical actin filament. Although other regions of the myosin head, such as the converter domain (91) and other pliant sections (41,54–56), likely contribute to the flexibility, the amount of lever-arm bending provides a lower limit on the range of actin subunits that are energetically favorable for two-headed binding.
Vilfan (69) and Lan and Sun (90) used an elastic lever-arm model and a worm-like chain model to quantify the mechanical energetic cost of spanning various distances for myosin V and VI, as shown in Fig. 3 a (90). The α-helix stabilized by six CaMs of the myosin V lever arm, modeled as an elastic rod, has a much sharper variation of free energy with interhead distance compared with myosin VI, which contains a much floppier lever-arm extension (Fig. 3 a). The modeled free-energy changes of myosin V and VI with distance are consistent with their measured step-size distributions (Fig. 3 b) considering that the distribution of distances should be a narrower exponential function of the free energy. Thus, a flexible lever arm or other pliant regions allows a myosin motor head to choose multiple actin sites to bind (i.e., a more variable step size) without dramatically increasing the intramolecular strain and free energy.
Figure 3.
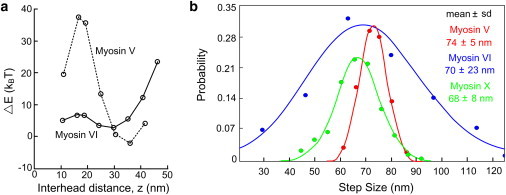
Step-size distributions are in accordance with the lever-arm flexibilities of myosin motors. (a) The elastic energy difference (ΔE) of myosins V and VI as a function of head-head distance in the two-head bound state on an actin filament (modified from Lan and Sun (90)). (b) Step-size distributions of myosins V (adapted from Yildiz et al. (77)), VI (adapted from Fig. 4 of Ökten et al. (80)), and X (from Sun et al. (56)). Step sizes are listed as mean ± SD. Note that the step sizes plotted correspond to the motion of the labeled CaM nearest the motor domain, so the distances are twice that of the motor's center of mass.
Among the three motors, myosin V, with the most-rigid lever arms, shows the tightest step-size distribution, indicating a regular manner of stepping, mainly spanning 11, 13, and 15 actin subunits, consistent with electron microscopy studies (60,84). The lever arm of myosin X, including the postulated stable α-helical PT domain (86), should have about half the bending stiffness of myosin V (Fig. 2 b). This is consistent with the slightly wider step-size distribution of myosin X (Fig. 3 b). A recent experiment (92) demonstrated the importance of the SAH domain for myosin X processivity. The motor, lever, and tail domains were swapped between myosin V and myosinX. A chimera with a myosin V tail and myosin X lever arm, but without the SAH, was not motile with either type of motor domain.
Myosin VI shows much more variability of step sizes than myosin V and X (Fig. 3 b and Table 1), suggesting that it has even lower stiffness than a SAH. Rather, the much broader step-size distribution of myosin VI supports the argument that the lever-arm portion between the LCD and the CC is an unfolded TAHB with flexible linkers in between its segments, and with only ∼1/20 of myosin V's bending stiffness (Fig. 2 c). A similar conclusion follows from angular measurements on the myosin lever arms, as discussed below. Although the experimental conditions vary among the entries in Table 1, measurements of the step size are generally much more consistent than those of run length or Vmax. Therefore, we accept the consensus that myosin VI takes more-variable steps than the other two motors.
Given the dramatic difference in the lever-arm structure and flexibility, it is surprising to see that the three myosin motors have processivity values (i.e., successive steps per diffusional encounter, which are often quantified as run length/step size) that are in the same general range of 10–30 (Table 1). One possible explanation for this observation is that mechanochemical coordination between the two heads, a key to most models of processive stepping, is more impaired by off-axis strain. It has been shown that myosin V constructs with truncated lever arms move much less processively than those with full-length lever arms because the truncated motor takes shorter steps (57,59,76), necessitating binding on the side of the actin filament and possibly causing large off-axis intramolecular strain.
Myosin X has been found to localize primarily in filopodia, which are protrusive cellular structures that contain bundled actin filaments. Several recent studies (93,94) reported that myosin X preferentially selects bundled actins for motility and shows poor processivity on single filaments. This selectivity was attributed to the short, 3-CaM lever arm of myosin X, and a straddle mechanism was proposed whereby the two heads of a myosin X molecule necessarily track along two adjacent actin filaments. The improved processivity on filament bundles was attributed to the reduced energetic cost of binding to two favorably oriented actins on adjacent filaments relative to sites on a single filament, given the short lever arms. However, in cells, myosin X was found to move on both actin bundles (93) and filaments that are not tightly bundled (18,19,95). Another recent study, using similar myosin X constructs in vitro, showed robust processivity on both individual and bundled actin filaments (56). Localization measurements with nanometer resolution showed that the step size of myosin X on single actin filaments is 34 nm (Fig. 3 b), contrasting with the results of Nagy et al. (93) and supporting the hypothesis that the SAH is a part of the lever arm (86).
The myosin X constructs used by Sun et al. (56) and Nagy et al. (93) were very similar except for the dimerization sequences (GCN4 leucine zipper (93) and Myo5 CC (56)) and truncation points (residue 920 (93) and residue 939 (56); see Fig. 1 c). Residue 920 is in the MT (blue band in Fig. 1 c), which may be part of the lever arm (86). It is possible that truncation at residue 920 causes an unstable domain structure that diminishes gating within myosin X, or that a leucine zipper decreases the flexibility of the neck, inhibiting the search for appropriate actin binding and thus reducing the observed processivity. Myosin X moves more slowly on bundled actin filaments, and shows smaller and more-variable step sizes than it exhibits on single actin filaments (56), which suggests that myosin X switches onto adjacent actin filaments somewhat randomly, in contrast to the more predictable mandatory straddle mechanism suggested by Nagy et al. (93). Although the myosin X used by Sun et al. (56), artificially dimerized with Myo5 CC (M10HMM-M5CC), localizes to the tips of filopodia (18), the native tail of myosin X seems to be necessary for it to take part in forming stable and long filopodia (M. Ikebe, University of Massachusetts Medical School, personal communication, 2011).
Lever-Arm Tilt Angles also Vary with Stiffness in the Three Myosins
The double-helical nature of actin filaments and the variability of step sizes imply that the myosin lever arms explore various orientations along the actin filament (the axial angle) and around it (the azimuthal angle). Given the different effective lever-arm stiffness values, the three myosin motors should demonstrate different azimuthal variations when their heads are bound to two azimuthally variable actin subunits. A few single-molecule techniques, including polarized total internal reflection fluorescence (polTIRF) (51,54,56,96) and defocused orientation and position imaging (DOPI) (52,55), have been used to investigate these lever-arm rotational motions.
In rotational measurements on myosin motors, a bifunctional fluorophore is usually coupled to CaM through two linkers to limit rotation of the fluorescent probe relative to the protein. By modulating calcium concentration, a labeled CaM subunit is exchanged for an endogenous CaM so that angular changes of the probe report the rotational motions of the lever arm. The probe orientation can be defined by the axial angle, βP, and the azimuthal angle, αP, relative to the actin filament track (Fig. 4 d). The axial angle (βP) distributions for CaM probes on myosins V, VI, and X (Fig. 4) all show two distinct peaks, showing that the labeled lever arms populate two states (leading and trailing positions) during motility, as expected for the hand-over-hand model. In all three cases, the βP angles are measured from the axis of actin in the direction of travel, so the peak at higher angle is assigned to the leading head and at lower angle to the trailing head. One aspect of the βP distribution for myosin VI, however, shows a distinct difference. The βP angle of the leading lever arm (<βP.Lead> = 117°; Fig. 4 b) is significantly more variable than that of the trailing arm. This observation is consistent with the higher variance of position for the leading head of myosin VI compared with that of the trailing head (78) that follows from the high flexibility of myosin VI. Given that the bifunctional probe has a fixed angle relative to the myosin lever arm it is on, a coordinate transformation can be used to calculate the lever-arm angle from the measured probe angle. Myosin VI lever-arm angles deduced from the probe angles in Fig. 4 b indicate that the lever arm generally rotates ∼180° (from 0° to 180°) (54,55,74), which can be accommodated with either of the myosin VI models shown in Fig. 2, c and d.
Figure 4.
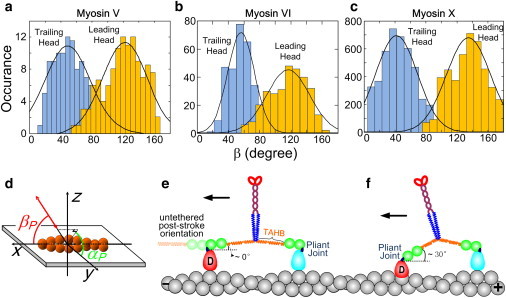
Axial angle βP distributions of (a) myosin V (54), (b) myosin VI (54), and (c) myosin X (56) during processive motility. (d) Definitions of probe axial angle, βP, and azimuthal angle, αP, relative to the actin. (e and f) Myosin VI exhibits variability of stepping distance, leading-head actin position, and leading-lever-arm angle, suggesting a pliant region near the lever-head junction (dark blue ovals). βP.Lead = 122 ± 29°,117 ± 27°, and 135 ± 28° for myosin V (a), VI (b), and X (c), respectively. βP.Trail = 49 ± 29°, 56 ± 17°, and 42 ± 27° for myosin V (a), VI (b), and X (c), respectively (mean ± SD). Note that for myosin VI, the axial angle distribution of the leading head is wider than that of the trailing head. Also note that the SD of the myosin VI trailing-head distribution (at 56°) is significantly narrower than those of myosin V and X, perhaps because of its unique single exchangeable CaM per head.
Adjacent subunits on the long-pitch helix of actin differ from each other in azimuthal orientation by ∼28°. Thus, the variable step size and interhead distance imply a variable azimuthal angle of the motor domain bound stereospecifically to the actin. The implications of the extra variability of the leading lever-arm position and angle in myosin VI are indicated in Fig. 4, e and f. The light blue pear-shaped motor domains represent the trailing head, and the red ones show the leading head at different distances. At low ATP concentration (e.g., 150 μM for a polTIRF experiment), ATP binding is the rate-limiting step and the trailing head is mostly nucleotide-free. According to the crystal structure of apo-myosin VI (97), the trailing lever arm is nearly parallel to the actin filament. The leading head probably releases Pi rapidly (98) and contains ADP (indicated by D in Fig. 4, e and f) (99) or it may be empty (97), so its untethered (zero-stress) position is also parallel to actin facing forward (faint green balls and faint orange spring in Fig. 4 e, left). However, it is tethered to the trailing head and thus cannot adopt its untethered position. Rather, the leading lever arm must adopt different orientations when its head binds at various interhead distances. The variability of the leading-head lever-arm angle is due, on the one hand, to the highly flexible lever arms that allow binding to multiple actin sites, and on the other hand suggests a pliant region between the fluorescent probe on the 2nd CaM and the motor domain. The pliant joint is probably near the converter domain (the dark blue segments in Fig. 4, e and f). We used the term “wiggly” (54) to describe the erratic side-to-side motions of myosin VI during motility, as measured using polTIRF and implied by its variable step size (78).
Reifenberger et al. (55) recently measured lever-arm angles of myosin VI using DOPI, and their results suggest that myosin VI takes nearly constant step sizes and 180° rotations of its lever arm. These results are inconsistent with the broad step-size distribution of myosin VI previously reported in several studies (68,78,80,83) (Fig. 3 b), as well as the different widths of βP angle distributions of its leading and trailing heads (Fig. 4 b). Reifenberger et al. (55) pointed out that the 180° position of the leading-head lever arm is compatible with the pre-powerstroke x-ray crystal structure containing ADP and a phosphate analog (99). However, phosphate and (possibly) ADP are both released rapidly from the leading head upon binding to actin (98), so the crystal structure with phosphate bound is not likely to represent the leading lever arm during motility (for a further discussion of this point, see Dunn et al. (65)). Further differences between the studies by Sun et al. (54) and Reifenberger et al. (55) regarding myosin VI tilting are discussed by Sun et al. (100,101). However, in both of these studies the authors concluded that the lever arm of the leading head is mechanically uncoupled partially (54) or completely (55) from the motor domain at a pliant region at the base of the lever arm (Fig. 4, e and f).
The azimuthal angle, αP (Fig. 4 d), also indicates differences among the three myosin motors. If the motor walks straight along an actin filament during hand-over-hand stepping, the probe angle returns to its original orientation every other step. This behavior depends on the axial angle, βP, returning to the original value after two steps, but is independent of the (usually unknown) local probe angle relative to the lever arm (54,56). Therefore, the calculated difference of αP after two steps, termed 2Δα, can be used to indicate the regularity of myosin's stepping path. The 2Δα values for myosin VI have a much broader distribution than those of myosin V and X (Fig. 5). This clear difference between the motors provides strong support for the notion that myosin VI has high structural flexibility and walks azimuthally wiggly on actin filaments (54).
Figure 5.
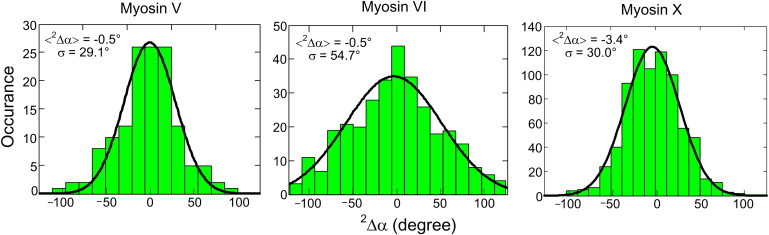
2Δα distributions of myosin V (54), myosin VI (54), and myosin X (56). The lever arm alternates between leading and trailing positions, and returns to its original state after two transitions. Because the relative orientation between the probe and labeled lever arm is fixed, the measured azimuthal change of the probe after two steps, termed 2Δα, should be same as the azimuthal change of the lever arm irrespective of the relative angle between the probe and lever arm. When 2Δα is zero or small, the molecule is walking overall straight along the axis of actin, and the SD of its 2Δα distribution denotes the regularity of the myosin stepping.
Myosin Motors Walk Spirally along Actin Filaments in Long Runs
Given the double-helical structure of actin filaments, it is not surprising to find that myosin motors can adopt a helical path when actin filaments are supported away from a hard surface. Several different types of experiments have enabled observation of these spiral paths (54,56,94,102–105) (Fig. 6). Ali et al. (102,103) attached bead duplexes to single myosin V and VI motors and watched their unconstrained motion along actin filaments suspended between pedestal beads (Fig. 6 a). They found that myosin V walks in a left-handed helical path with a pitch of ∼2.2 μm, and myosin VI molecules mostly walk straight, but a small fraction follow a right-handed helical path, also with pitch ∼2.2 μm. A gliding filament twirling assay was used to study the spiral motion of myosins II, V, and VI (54,104) (Fig. 6 b), and the results showed that all three of these motors twirl actin filaments about their axis during motility, with myosin II and V spiraling to the left, and myosin VI spiraling to the right. The helical paths are geometrical consequences of the step sizes of the motors, quantized by the actin subunit positions, and the resulting helical arrangement of the actomyosin-binding sites along the filament. Thus, the opposite handedness of myosin VI is not caused by its unusual motion toward the pointed end of actin, but rather is due to its difference in step sizes.
Figure 6.
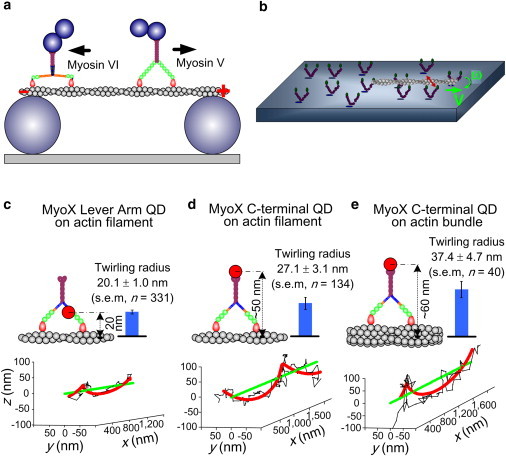
Myosin motors walk spirally around actin filaments. (a) Duplex bead assays show that myosins move along suspended actin filaments either straight or on a left-handed (myosin V (102)) or right-handed (myosin VI (103)) helical path. The duplex beads attached to the C-terminus are much bigger than depicted. (b) A gliding filament twirling assay shows spiral motion of myosins II, V, and VI (54,104). (c–e) 3D single-molecule nanometer tracking on suspended actin filaments shows the helical path of QD-labeled myosin X molecules (56). QD655 is bound to a lever-arm CaM in panel c, and to the C-terminus of myosin X in panels d and e. The molecules walk on single suspended actin filaments in panels c and d, and on a suspended fascin-actin bundle in panel e.
The values of step size listed in Table 1 are generally shorter for myosin VI and X than for myosin V. These values were all measured with actin firmly attached to a glass surface, presumably restricting the subunits available for myosin binding, and thus cannot be directly related to the handedness of motion on a freely suspended filament. The differences in the average step sizes, as well as the variance, can reflect whether a motor steps more often to the right side or the left side of the actin, leading to a long-range right- or left-handed helical path. Surprisingly, surface attachment of actin filaments does not noticeably reduce the run length compared with that on suspended actin filaments, at least for myosin X (56). The motors have flexibility in their stepping pattern so that they can walk straight on surface-attached actin, and the helical path is not obligatory.
In two other assays, myosin X was also found to follow a left-handed helical path with pitch ∼1 μm (56,105). Typical three-dimensional (3D) tracks of quantum dots (QDs) labeling myosin X at different locations on the molecule show left-handed helical paths. The radius of the helix increases when the QD is moved from the lever arm (Fig. 6 c) to the C-terminus of the construct (Fig. 6 d) and again when the molecule translocates on a larger-diameter structure, ∼10 filaments bundled together by fascin (Fig. 6 e). However, there are clear discrepancies between measured (blue bars in Fig. 6, c–e) and predicted (cartoons) helical path radii, suggesting that although myosin X has much more rigid lever arms than myosin VI, there are also some pliant regions in its structure, possibly near the junction between the lever arm and the tail (56). It should be noted, though, that a different construct, presumably more rigid, apparently follows a right-handed helical path with a much shorter pitch (94).
Speculations on the Functional Relevance of Myosin Structural Flexibility
Myosin VI and X have flexible portions in their lever arms, and there may be other pliant regions near the converter domain or the lever-arm/tail junction. All three myosin motors take helical paths around actin filaments, although they can walk straight if the helical route is blocked. How are these features of the three myosin motors related to their functions in the cell?
To compare them, it is useful to consider all three motors in the same picture (Fig. 7). Myosin V has been found to be a proficient cargo transporter in lamellipodia, where cargos are switched from microtubules to actin tracks and transported toward the cell periphery. Myosin VI and X are found to transport cargos in actin-bundle rich structures, such as microvilli (9) and filopodia (12), where the filaments are much more crowded than they are in lamellipodia. For many of the myosins, structural flexibility, especially at their lever-arm/tail junctions, allows motor autoregulation by interactions between the head and tail domains (32,48,50,66,67,106–108). Both myosin VI and X exhibit proximity-triggered dimerization (31,56,109). It has been proposed that in cells, compact monomer states would facilitate diffusion of deactivated myosins VI and X to sites of dimerization and derepression (110).
Figure 7.
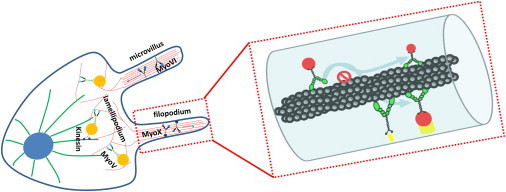
Three myosin motors have distinct cellular functions and localizations. (Left) Myosin V transports cargos from microtubule tracks to actin tracks. Myosins VI and X transport cargos in actin-bundle-rich structures such as microvilli and filopodia, which have a more crowded cellular environment than lamellipodia. (Right) Spiral paths allow myosin motors to pass around obstacles along the actin track and search for their target partners (yellow) over circumferentially broader regions.
The flexible lever arms of myosin VI and X can probably help them navigate actively through crowded environments as well. Because the actin tracks in microvilli and filopodia are bundled filaments, flexible lever arms should facilitate the motility of myosins VI and X when they need to switch onto nearby actin filaments or straddle along neighboring filaments. Myosins VI and X have also been found to act as tethers to maintain the morphology of the Golgi apparatus (11) and mitotic spindle (95), respectively. Their structural flexibility may help them adapt better as tethering struts by facilitating linkage between two separate filaments. Myosin X is involved in initiating the formation of filopodial actin bundles (18), possibly by pulling actin filaments together at the cell periphery. The semiflexible nature of the SAH domains and tail may allow myosin X to stretch, span across, and pull multiple actin filaments into bundles at the base of a filopodium. In addition, some myosin motors have been found to be force or strain sensors (42,49,63–65). The different levels of structural flexibility generated by the various types of lever-arm components may enable myosins to sense strain at different amounts of deflections or to detect force with sensitivities tuned to their specific roles.
It is also intriguing to consider that all three myosin motors can adopt helical paths around their filament tracks even though a straight path would generate more transport distance per expenditure of energy. The seemingly inefficient helical paths may be indispensible for fulfilling the functions of myosin motors. Helical paths, as well as wiggly stepping (such as for myosin VI), may help myosin motors pass around obstacles on their actin tracks. Helical paths are spatially advantageous because they provide a full 3D search range for the motors to find specific docking or unloading sites (Fig. 7, right). The target sites for myosin motors and their associated cargos have specific spatial distributions. For instance, many are membrane receptor proteins (e.g., integrins) on the ventral side of the cell, in which case a motor moving straight along the dorsal side would forfeit accurate delivery. It is thus conceivable that the spiral motion of a myosin X molecule carrying integrins, growth factor receptors, or other cargos into a filopodium would ensure the location of a substrate delivery site. It has been proposed for neural growth cones that the helical path of myosin may twist actin and thereby help to steer the growing neurone (118).
Summary
In this work, we compared myosins V, VI, and X from the perspective of the flexibility of their lever arms. Current data from various types of experiments suggest that both myosin VI and myosin X use flexible extensions to their CaM LCDs to facilitate their processive motility along actin filaments. This structural flexibility may be necessary for myosin motors to perform cellular functions such as tethering, force sensing, and navigating through crowded cellular spaces as processive transporters.
Acknowledgments
We thank Dr. E. Michael Ostap for a critical reading of the manuscript and suggestions.
This work was supported by the National Institutes of Health (grants P01-GM087253 and R01-GM086352) and the Nanoscale Science and Engineering Center, National Science Foundation (grant DMR-0425780 through the Nano/Bio Interface Center, University of Pennsylvania).
References
- 1.Cheney R.E., Mooseker M.S. Unconventional myosins. Curr. Opin. Cell Biol. 1992;4:27–35. doi: 10.1016/0955-0674(92)90055-h. [DOI] [PubMed] [Google Scholar]
- 2.Howard J. Molecular motors: structural adaptations to cellular functions. Nature. 1997;389:561–567. doi: 10.1038/39247. [DOI] [PubMed] [Google Scholar]
- 3.Sellers J.R. Myosins: a diverse superfamily. Biochim. Biophys. Acta. 2000;1496:3–22. doi: 10.1016/s0167-4889(00)00005-7. [DOI] [PubMed] [Google Scholar]
- 4.Wu X., Jung G., Hammer J.A., 3rd Functions of unconventional myosins. Curr. Opin. Cell Biol. 2000;12:42–51. doi: 10.1016/s0955-0674(99)00055-1. [DOI] [PubMed] [Google Scholar]
- 5.Berg J.S., Powell B.C., Cheney R.E. A millennial myosin census. Mol. Biol. Cell. 2001;12:780–794. doi: 10.1091/mbc.12.4.780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rêdowicz M.J. Myosins and pathology: genetics and biology. Acta Biochim. Pol. 2002;49:789–804. [PubMed] [Google Scholar]
- 7.Schliwa M., Woehlke G. Molecular motors. Nature. 2003;422:759–765. doi: 10.1038/nature01601. [DOI] [PubMed] [Google Scholar]
- 8.Brown M.E., Bridgman P.C. Myosin function in nervous and sensory systems. J. Neurobiol. 2004;58:118–130. doi: 10.1002/neu.10285. [DOI] [PubMed] [Google Scholar]
- 9.Buss F., Spudich G., Kendrick-Jones J. Myosin VI: cellular functions and motor properties. Annu. Rev. Cell Dev. Biol. 2004;20:649–676. doi: 10.1146/annurev.cellbio.20.012103.094243. [DOI] [PubMed] [Google Scholar]
- 10.Krendel M., Mooseker M.S. Myosins: tails (and heads) of functional diversity. Physiology (Bethesda) 2005;20:239–251. doi: 10.1152/physiol.00014.2005. [DOI] [PubMed] [Google Scholar]
- 11.Sahlender D.A., Roberts R.C., Buss F. Optineurin links myosin VI to the Golgi complex and is involved in Golgi organization and exocytosis. J. Cell Biol. 2005;169:285–295. doi: 10.1083/jcb.200501162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sousa A.D., Cheney R.E. Myosin-X: a molecular motor at the cell's fingertips. Trends Cell Biol. 2005;15:533–539. doi: 10.1016/j.tcb.2005.08.006. [DOI] [PubMed] [Google Scholar]
- 13.Eichler T.W., Kögel T., Gerdes H.H. The role of myosin Va in secretory granule trafficking and exocytosis. Biochem. Soc. Trans. 2006;34:671–674. doi: 10.1042/BST0340671. [DOI] [PubMed] [Google Scholar]
- 14.Foth B.J., Goedecke M.C., Soldati D. New insights into myosin evolution and classification. Proc. Natl. Acad. Sci. USA. 2006;103:3681–3686. doi: 10.1073/pnas.0506307103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.O'Connell C.B., Tyska M.J., Mooseker M.S. Myosin at work: motor adaptations for a variety of cellular functions. Biochim. Biophys. Acta. 2007;1773:615–630. doi: 10.1016/j.bbamcr.2006.06.012. [DOI] [PubMed] [Google Scholar]
- 16.Odronitz F., Kollmar M. Drawing the tree of eukaryotic life based on the analysis of 2,269 manually annotated myosins from 328 species. Genome Biol. 2007;8:R196. doi: 10.1186/gb-2007-8-9-r196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Taylor K.A. Regulation and recycling of myosin V. Curr. Opin. Cell Biol. 2007;19:67–74. doi: 10.1016/j.ceb.2006.12.014. [DOI] [PubMed] [Google Scholar]
- 18.Tokuo H., Mabuchi K., Ikebe M. The motor activity of myosin-X promotes actin fiber convergence at the cell periphery to initiate filopodia formation. J. Cell Biol. 2007;179:229–238. doi: 10.1083/jcb.200703178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Toyoshima F., Nishida E. Integrin-mediated adhesion orients the spindle parallel to the substratum in an EB1- and myosin X-dependent manner. EMBO J. 2007;26:1487–1498. doi: 10.1038/sj.emboj.7601599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Buss F., Kendrick-Jones J. How are the cellular functions of myosin VI regulated within the cell? Biochem. Biophys. Res. Commun. 2008;369:165–175. doi: 10.1016/j.bbrc.2007.11.150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ikebe M. Regulation of the function of mammalian myosin and its conformational change. Biochem. Biophys. Res. Commun. 2008;369:157–164. doi: 10.1016/j.bbrc.2008.01.057. [DOI] [PubMed] [Google Scholar]
- 22.Loubéry S., Coudrier E. Myosins in the secretory pathway: tethers or transporters? Cell. Mol. Life Sci. 2008;65:2790–2800. doi: 10.1007/s00018-008-8350-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bridgman P.C. Myosin motor proteins in the cell biology of axons and other neuronal compartments. Results Probl. Cell Differ. 2009;48:91–105. doi: 10.1007/400_2009_10. [DOI] [PubMed] [Google Scholar]
- 24.Chibalina M.V., Puri C., Buss F. Potential roles of myosin VI in cell motility. Biochem. Soc. Trans. 2009;37:966–970. doi: 10.1042/BST0370966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Vicente-Manzanares M., Ma X., Horwitz A.R. Non-muscle myosin II takes centre stage in cell adhesion and migration. Nat. Rev. Mol. Cell Biol. 2009;10:778–790. doi: 10.1038/nrm2786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Woolner S., Bement W.M. Unconventional myosins acting unconventionally. Trends Cell Biol. 2009;19:245–252. doi: 10.1016/j.tcb.2009.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nambiar R., McConnell R.E., Tyska M.J. Myosin motor function: the ins and outs of actin-based membrane protrusions. Cell. Mol. Life Sci. 2010;67:1239–1254. doi: 10.1007/s00018-009-0254-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sweeney H.L., Houdusse A. Structural and functional insights into the myosin motor mechanism. Annu. Rev. Biophys. 2010;39:539–557. doi: 10.1146/annurev.biophys.050708.133751. [DOI] [PubMed] [Google Scholar]
- 29.Kögel T., Bittins C.M., Gerdes H.H. Versatile roles for myosin Va in dense core vesicle biogenesis and function. Biochem. Soc. Trans. 2010;38:199–204. doi: 10.1042/BST0380199. [DOI] [PubMed] [Google Scholar]
- 30.Iwaki M., Tanaka H., Yanagida T. Cargo-binding makes a wild-type single-headed myosin-VI move processively. Biophys. J. 2006;90:3643–3652. doi: 10.1529/biophysj.105.075721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Park H., Ramamurthy B., Sweeney H.L. Full-length myosin VI dimerizes and moves processively along actin filaments upon monomer clustering. Mol. Cell. 2006;21:331–336. doi: 10.1016/j.molcel.2005.12.015. [DOI] [PubMed] [Google Scholar]
- 32.Li J.-F., Nebenführ A. The tail that wags the dog: the globular tail domain defines the function of myosin V/XI. Traffic. 2008;9:290–298. doi: 10.1111/j.1600-0854.2007.00687.x. [DOI] [PubMed] [Google Scholar]
- 33.Trybus K.M. Myosin V from head to tail. Cell. Mol. Life Sci. 2008;65:1378–1389. doi: 10.1007/s00018-008-7507-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Phichith D., Travaglia M., Sweeney H.L. Cargo binding induces dimerization of myosin VI. Proc. Natl. Acad. Sci. USA. 2009;106:17320–17324. doi: 10.1073/pnas.0909748106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Terrak M., Rebowski G., Dominguez R. Structure of the light chain-binding domain of myosin V. Proc. Natl. Acad. Sci. USA. 2005;102:12718–12723. doi: 10.1073/pnas.0503899102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Baboolal T.G., Sakamoto T., Peckham M. The SAH domain extends the functional length of the myosin lever. Proc. Natl. Acad. Sci. USA. 2009;106:22193–22198. doi: 10.1073/pnas.0909851106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Richards T.A., Cavalier-Smith T. Myosin domain evolution and the primary divergence of eukaryotes. Nature. 2005;436:1113–1118. doi: 10.1038/nature03949. [DOI] [PubMed] [Google Scholar]
- 38.Houdusse A., Sweeney H.L. Myosin motors: missing structures and hidden springs. Curr. Opin. Struct. Biol. 2001;11:182–194. doi: 10.1016/s0959-440x(00)00188-3. [DOI] [PubMed] [Google Scholar]
- 39.Tyska M.J., Warshaw D.M. The myosin power stroke. Cell Motil. Cytoskeleton. 2002;51:1–15. doi: 10.1002/cm.10014. [DOI] [PubMed] [Google Scholar]
- 40.Warshaw D.M. Lever arms and necks: a common mechanistic theme across the myosin superfamily. J. Muscle Res. Cell Motil. 2004;25:467–474. doi: 10.1007/s10974-004-1767-z. [DOI] [PubMed] [Google Scholar]
- 41.Houdusse A., Szent-Gyorgyi A.G., Cohen C. Three conformational states of scallop myosin S1. Proc. Natl. Acad. Sci. USA. 2000;97:11238–11243. doi: 10.1073/pnas.200376897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yanagida T., Esaki S., Tokunaga M. Single-motor mechanics and models of the myosin motor. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2000;355:441–447. doi: 10.1098/rstb.2000.0585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Howard J. Sinauer Associates; Sunderland, MA: 2001. Mechanics of motor proteins and the cytoskeleton. [Google Scholar]
- 44.Geeves M.A. Stretching the lever-arm theory. Nature. 2002;415:129–131. doi: 10.1038/415129a. [DOI] [PubMed] [Google Scholar]
- 45.Krementsov D.N., Krementsova E.B., Trybus K.M. Myosin V: regulation by calcium, calmodulin, and the tail domain. J. Cell Biol. 2004;164:877–886. doi: 10.1083/jcb.200310065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Moore J.R., Krementsova E.B., Warshaw D.M. Does the myosin V neck region act as a lever? J. Muscle Res. Cell Motil. 2004;25:29–35. doi: 10.1023/b:jure.0000021394.48560.71. [DOI] [PubMed] [Google Scholar]
- 47.Sellers J.R., Veigel C. Walking with myosin V. Curr. Opin. Cell Biol. 2006;18:68–73. doi: 10.1016/j.ceb.2005.12.014. [DOI] [PubMed] [Google Scholar]
- 48.Jung H.S., Komatsu S., Craig R. Head-head and head-tail interaction: a general mechanism for switching off myosin II activity in cells. Mol. Biol. Cell. 2008;19:3234–3242. doi: 10.1091/mbc.E08-02-0206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yanagida T., Iwaki M., Ishii Y. Single molecule measurements and molecular motors. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2008;363:2123–2134. doi: 10.1098/rstb.2008.2265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Li X.D., Jung H.S., Ikebe M. The globular tail domain puts on the brake to stop the ATPase cycle of myosin Va. Proc. Natl. Acad. Sci. USA. 2008;105:1140–1145. doi: 10.1073/pnas.0709741105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Forkey J.N., Quinlan M.E., Goldman Y.E. Three-dimensional structural dynamics of myosin V by single-molecule fluorescence polarization. Nature. 2003;422:399–404. doi: 10.1038/nature01529. [DOI] [PubMed] [Google Scholar]
- 52.Toprak E., Enderlein J., Selvin P.R. Defocused orientation and position imaging (DOPI) of myosin V. Proc. Natl. Acad. Sci. USA. 2006;103:6495–6499. doi: 10.1073/pnas.0507134103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Syed S., Snyder G.E., Goldman Y.E. Adaptability of myosin V studied by simultaneous detection of position and orientation. EMBO J. 2006;25:1795–1803. doi: 10.1038/sj.emboj.7601060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sun Y., Schroeder H.W.I., 3rd, Goldman Y.E. Myosin VI walks “wiggly” on actin with large and variable tilting. Mol. Cell. 2007;28:954–964. doi: 10.1016/j.molcel.2007.10.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Reifenberger J.G., Toprak E., Selvin P.R. Myosin VI undergoes a 180° power stroke implying an uncoupling of the front lever arm. Proc. Natl. Acad. Sci. USA. 2009;106:18255–18260. doi: 10.1073/pnas.0900005106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sun Y., Sato O., Goldman Y.E. Single-molecule stepping and structural dynamics of myosin X. Nat. Struct. Mol. Biol. 2010;17:485–491. doi: 10.1038/nsmb.1785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Purcell T.J., Morris C., Sweeney H.L. Role of the lever arm in the processive stepping of myosin V. Proc. Natl. Acad. Sci. USA. 2002;99:14159–14164. doi: 10.1073/pnas.182539599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sakamoto T., Wang F., Sellers J.R. Neck length and processivity of myosin V. J. Biol. Chem. 2003;278:29201–29207. doi: 10.1074/jbc.M303662200. [DOI] [PubMed] [Google Scholar]
- 59.Sakamoto T., Yildez A., Sellers J.R. Step-size is determined by neck length in myosin V. Biochemistry. 2005;44:16203–16210. doi: 10.1021/bi0512086. [DOI] [PubMed] [Google Scholar]
- 60.Oke O.A., Burgess S.A., Trinick J. Influence of lever structure on myosin 5a walking. Proc. Natl. Acad. Sci. USA. 2010;107:2509–2514. doi: 10.1073/pnas.0906907107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Tsiavaliaris G., Fujita-Becker S., Manstein D.J. Molecular engineering of a backwards-moving myosin motor. Nature. 2004;427:558–561. doi: 10.1038/nature02303. [DOI] [PubMed] [Google Scholar]
- 62.Nguyen H., Higuchi H. Motility of myosin V regulated by the dissociation of single calmodulin. Nat. Struct. Mol. Biol. 2005;12:127–132. doi: 10.1038/nsmb894. [DOI] [PubMed] [Google Scholar]
- 63.Esaki S., Ishii Y., Yanagida T. Cooperative actions between myosin heads bring effective functions. Biosystems. 2007;88:293–300. doi: 10.1016/j.biosystems.2006.03.013. [DOI] [PubMed] [Google Scholar]
- 64.Laakso J.M., Lewis J.H., Ostap E.M. Myosin I can act as a molecular force sensor. Science. 2008;321:133–136. doi: 10.1126/science.1159419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Dunn A.R., Chuan P., Spudich J.A. Contribution of the myosin VI tail domain to processive stepping and intramolecular tension sensing. Proc. Natl. Acad. Sci. USA. 2010;107:7746–7750. doi: 10.1073/pnas.1002430107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Li X.D., Jung H.S., Ikebe M. The globular tail domain of myosin Va functions as an inhibitor of the myosin Va motor. J. Biol. Chem. 2006;281:21789–21798. doi: 10.1074/jbc.M602957200. [DOI] [PubMed] [Google Scholar]
- 67.Thirumurugan K., Sakamoto T., Knight P.J. The cargo-binding domain regulates structure and activity of myosin 5. Nature. 2006;442:212–215. doi: 10.1038/nature04865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Rock R.S., Ramamurthy B., Sweeney H.L. A flexible domain is essential for the large step size and processivity of myosin VI. Mol. Cell. 2005;17:603–609. doi: 10.1016/j.molcel.2005.01.015. [DOI] [PubMed] [Google Scholar]
- 69.Vilfan A. Elastic lever-arm model for myosin V. Biophys. J. 2005;88:3792–3805. doi: 10.1529/biophysj.104.046763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Veigel C., Wang F., Molloy J.E. The gated gait of the processive molecular motor, myosin V. Nat. Cell Biol. 2002;4:59–65. doi: 10.1038/ncb732. [DOI] [PubMed] [Google Scholar]
- 71.Altman D., Sweeney H.L., Spudich J.A. The mechanism of myosin VI translocation and its load-induced anchoring. Cell. 2004;116:737–749. doi: 10.1016/s0092-8674(04)00211-9. [DOI] [PubMed] [Google Scholar]
- 72.Uemura S., Higuchi H., Ishiwata S. Mechanochemical coupling of two substeps in a single myosin V motor. Nat. Struct. Mol. Biol. 2004;11:877–883. doi: 10.1038/nsmb806. [DOI] [PubMed] [Google Scholar]
- 73.Veigel C., Schmitz S., Sellers J.R. Load-dependent kinetics of myosin-V can explain its high processivity. Nat. Cell Biol. 2005;7:861–869. doi: 10.1038/ncb1287. [DOI] [PubMed] [Google Scholar]
- 74.Bryant Z., Altman D., Spudich J.A. The power stroke of myosin VI and the basis of reverse directionality. Proc. Natl. Acad. Sci. USA. 2007;104:772–777. doi: 10.1073/pnas.0610144104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Nishikawa S., Arimoto I., Yanagida T. Switch between large hand-over-hand and small inchworm-like steps in myosin VI. Cell. 2010;142:879–888. doi: 10.1016/j.cell.2010.08.033. [DOI] [PubMed] [Google Scholar]
- 76.Oguchi Y., Mikhailenko S.V., Ishiwata S. Robust processivity of myosin V under off-axis loads. Nat. Chem. Biol. 2010;6:300–305. doi: 10.1038/nchembio.322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Yildiz A., Forkey J.N., Selvin P.R. Myosin V walks hand-over-hand: single fluorophore imaging with 1.5-nm localization. Science. 2003;300:2061–2065. doi: 10.1126/science.1084398. [DOI] [PubMed] [Google Scholar]
- 78.Yildiz A., Park H., Sweeney H.L. Myosin VI steps via a hand-over-hand mechanism with its lever arm undergoing fluctuations when attached to actin. J. Biol. Chem. 2004;279:37223–37226. doi: 10.1074/jbc.C400252200. [DOI] [PubMed] [Google Scholar]
- 79.Snyder G.E., Sakamoto T., Selvin P.R. Nanometer localization of single green fluorescent proteins: evidence that myosin V walks hand-over-hand via telemark configuration. Biophys. J. 2004;87:1776–1783. doi: 10.1529/biophysj.103.036897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Ökten Z., Churchman L.S., Spudich J.A. Myosin VI walks hand-over-hand along actin. Nat. Struct. Mol. Biol. 2004;11:884–887. doi: 10.1038/nsmb815. [DOI] [PubMed] [Google Scholar]
- 81.Sakamoto T., Webb M.R., Sellers J.R. Direct observation of the mechanochemical coupling in myosin Va during processive movement. Nature. 2008;455:128–132. doi: 10.1038/nature07188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Churchman L.S., Ökten Z., Spudich J.A. Single molecule high-resolution colocalization of Cy3 and Cy5 attached to macromolecules measures intramolecular distances through time. Proc. Natl. Acad. Sci. USA. 2005;102:1419–1423. doi: 10.1073/pnas.0409487102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Balci H., Ha T., Selvin P.R. Interhead distance measurements in myosin VI via SHRImP support a simplified hand-over-hand model. Biophys. J. 2005;89:413–417. doi: 10.1529/biophysj.105.060608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Walker M.L., Burgess S.A., Knight P.J. Two-headed binding of a processive myosin to F-actin. Nature. 2000;405:804–807. doi: 10.1038/35015592. [DOI] [PubMed] [Google Scholar]
- 85.Kodera N., Yamamoto D., Ando T. Video imaging of walking myosin V by high-speed atomic force microscopy. Nature. 2010;468:72–76. doi: 10.1038/nature09450. [DOI] [PubMed] [Google Scholar]
- 86.Knight P.J., Thirumurugan K., Peckham M. The predicted coiled-coil domain of myosin 10 forms a novel elongated domain that lengthens the head. J. Biol. Chem. 2005;280:34702–34708. doi: 10.1074/jbc.M504887200. [DOI] [PubMed] [Google Scholar]
- 87.Spink B.J., Sivaramakrishnan S., Spudich J.A. Long single α-helical tail domains bridge the gap between structure and function of myosin VI. Nat. Struct. Mol. Biol. 2008;15:591–597. doi: 10.1038/nsmb.1429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Mukherjea M., Llinas P., Sweeney H.L. Myosin VI dimerization triggers an unfolding of a three-helix bundle in order to extend its reach. Mol. Cell. 2009;35:305–315. doi: 10.1016/j.molcel.2009.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Lister I., Schmitz S., Kendrick-Jones J. A monomeric myosin VI with a large working stroke. EMBO J. 2004;23:1729–1738. doi: 10.1038/sj.emboj.7600180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Lan G., Sun S.X. Flexible light-chain and helical structure of F-actin explain the movement and step size of myosin-VI. Biophys. J. 2006;91:4002–4013. doi: 10.1529/biophysj.106.089888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Köhler J., Winkler G., Kraft T. Mutation of the myosin converter domain alters cross-bridge elasticity. Proc. Natl. Acad. Sci. USA. 2002;99:3557–3562. doi: 10.1073/pnas.062415899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Nagy S., Rock R.S. Structured post-IQ domain governs selectivity of myosin X for fascin-actin bundles. J. Biol. Chem. 2010;285:26608–26617. doi: 10.1074/jbc.M110.104661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Nagy S., Ricca B.L., Rock R.S. A myosin motor that selects bundled actin for motility. Proc. Natl. Acad. Sci. USA. 2008;105:9616–9620. doi: 10.1073/pnas.0802592105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Ricca B.L., Rock R.S. The stepping pattern of myosin X is adapted for processive motility on bundled actin. Biophys. J. 2010;99:1818–1826. doi: 10.1016/j.bpj.2010.06.066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Woolner S., O'Brien L.L., Bement W.M. Myosin-10 and actin filaments are essential for mitotic spindle function. J. Cell Biol. 2008;182:77–88. doi: 10.1083/jcb.200804062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Rosenberg S.A., Quinlan M.E., Goldman Y.E. Rotational motions of macro-molecules by single-molecule fluorescence microscopy. Acc. Chem. Res. 2005;38:583–593. doi: 10.1021/ar040137k. [DOI] [PubMed] [Google Scholar]
- 97.Ménétrey J., Bahloul A., Houdusse A. The structure of the myosin VI motor reveals the mechanism of directionality reversal. Nature. 2005;435:779–785. doi: 10.1038/nature03592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Sweeney H.L., Park H., Rosenfeld S.S. How myosin VI coordinates its heads during processive movement. EMBO J. 2007;26:2682–2692. doi: 10.1038/sj.emboj.7601720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Ménétrey J., Llinas P., Houdusse A. The structural basis for the large powerstroke of myosin VI. Cell. 2007;131:300–308. doi: 10.1016/j.cell.2007.08.027. [DOI] [PubMed] [Google Scholar]
- 100.Sun Y., Schroeder H.W., 3rd, Goldman Y.E. Myosin VI lever arm rotation: fixed or variable? Proc. Natl. Acad. Sci. USA. 2010;107:E63–, author reply E64. doi: 10.1073/pnas.0914990107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Sun, Y., H. W. I. Schroeder, J. F. Beausang, K. Homma, M. Ikebe, …. 2010. Myosin VI lever arm rotation: Fixed or variable? Available from Nature Preceedings athttp://dx.doi.org/10.1038/npre.2010.4182.1 [DOI] [PMC free article] [PubMed]
- 102.Ali M.Y., Uemura S., Ishiwata S. Myosin V is a left-handed spiral motor on the right-handed actin helix. Nat. Struct. Biol. 2002;9:464–467. doi: 10.1038/nsb803. [DOI] [PubMed] [Google Scholar]
- 103.Ali M.Y., Homma K., Ikebe M. Unconstrained steps of myosin VI appear longest among known molecular motors. Biophys. J. 2004;86:3804–3810. doi: 10.1529/biophysj.103.037416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Beausang J.F., Schroeder H.W.I., 3rd, Goldman Y.E. Twirling of actin by myosins II and V observed via polarized TIRF in a modified gliding assay. Biophys. J. 2008;95:5820–5831. doi: 10.1529/biophysj.108.140319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Arsenault M.E., Sun Y., Goldman Y.E. Using electrical and optical tweezers to facilitate studies of molecular motors. Phys. Chem. Chem. Phys. 2009;11:4834–4839. doi: 10.1039/b821861g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Liu J., Taylor D.W., Taylor K.A. Three-dimensional structure of the myosin V inhibited state by cryoelectron tomography. Nature. 2006;442:208–211. doi: 10.1038/nature04719. [DOI] [PubMed] [Google Scholar]
- 107.Umekia N., Jung H.S., Watanabea S., Sakai T., Li X.-d. The tail binds to the head–neck domain, inhibiting ATPase activity of myosin VIIA. Proc. Natl. Acad. Sci. USA. 2007;106:8483–8488. doi: 10.1073/pnas.0812930106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Sellers J.R., Knight P.J. Folding and regulation in myosins II and V. J. Muscle Res. Cell Motil. 2007;28:363–370. doi: 10.1007/s10974-008-9134-0. [DOI] [PubMed] [Google Scholar]
- 109.Yu C., Feng W., Zhang M. Myosin VI undergoes cargo-mediated dimerization. Cell. 2009;138:537–548. doi: 10.1016/j.cell.2009.05.030. [DOI] [PubMed] [Google Scholar]
- 110.Sweeney H.L., Houdusse A. Myosin VI rewrites the rules for myosin motors. Cell. 2010;141:573–582. doi: 10.1016/j.cell.2010.04.028. [DOI] [PubMed] [Google Scholar]
- 111.Mehta A.D., Rock R.S., Cheney R.E. Myosin-V is a processive actin-based motor. Nature. 1999;400:590–593. doi: 10.1038/23072. [DOI] [PubMed] [Google Scholar]
- 112.Rief M., Rock R.S., Spudich J.A. Myosin-V stepping kinetics: a molecular model for processivity. Proc. Natl. Acad. Sci. USA. 2000;97:9482–9486. doi: 10.1073/pnas.97.17.9482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Pierobon P., Achouri S., Cappello G. Velocity, processivity, and individual steps of single myosin V molecules in live cells. Biophys. J. 2009;96:4268–4275. doi: 10.1016/j.bpj.2009.02.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Baker J.E., Krementsova E.B., Warshaw D.M. Myosin V processivity: multiple kinetic pathways for head-to-head coordination. Proc. Natl. Acad. Sci. USA. 2004;101:5542–5546. doi: 10.1073/pnas.0307247101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Rock R.S., Rice S.E., Sweeney H.L. Myosin VI is a processive motor with a large step size. Proc. Natl. Acad. Sci. USA. 2001;98:13655–13659. doi: 10.1073/pnas.191512398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Nishikawa S., Homma K., Ikebe M. Class VI myosin moves processively along actin filaments backward with large steps. Biochem. Biophys. Res. Commun. 2002;290:311–317. doi: 10.1006/bbrc.2001.6142. [DOI] [PubMed] [Google Scholar]
- 117.Kerber M.L., Jacobs D.T., Cheney R.E. A novel form of motility in filopodia revealed by imaging myosin-X at the single-molecule level. Curr. Biol. 2009;19:967–973. doi: 10.1016/j.cub.2009.03.067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Tamada A., Kawase S., Kamiguchi H. Autonomous right-screw rotation of growth cone filopodia drives neurite turning. J. Cell Biol. 2010;188:429–441. doi: 10.1083/jcb.200906043. [DOI] [PMC free article] [PubMed] [Google Scholar]


