Abstract
Transcription factors (TFs) are regulatory proteins that bind DNA in promoter regions of the genome and either promote or repress gene expression. Here, we predict analytically that enhanced homooligonucleotide sequence correlations, such as poly(dA:dT) and poly(dC:dG) tracts, statistically enhance nonspecific TF-DNA binding affinity. This prediction is generic and qualitatively independent of microscopic parameters of the model. We show that nonspecific TF binding affinity is universally controlled by the strength and symmetry of DNA sequence correlations. We perform correlation analysis of the yeast genome and show that DNA regions highly occupied by TFs exhibit stronger homooligonucleotide sequence correlations, and thus a higher propensity for nonspecific binding, than do poorly occupied regions. We suggest that this effect plays the role of an effective localization potential that enhances quasi-one-dimensional diffusion of TFs in the vicinity of DNA, speeding up the stochastic search process for specific TF binding sites. The effect is also predicted to impose an upper bound on the size of TF-DNA binding motifs.
Introduction
Transcription factors (TFs) are proteins that regulate gene expression in both prokaryotic (e.g., bacteria) and eukaryotic (e.g., yeast or human) cells. TFs bind regulatory promoter regions of DNA in the genome. It is commonly accepted that each TF binds specifically a relatively small set of DNA sequences called TF binding motifs or TF binding sites (TFBSs). A TF binds its specific binding motifs with a higher affinity than other genomic sequences of the same length (1,2). A typical length of TF binding motif varies between 6 and 20 nucleotides. Recent high-throughput measurements of TF binding preferences on a genome-wide scale have challenged the classical picture of TF specificity (3,4). These experiments measured binding preferences of >100 TFs to tens of thousands of DNA sequences and demonstrated a high level of multispecificity in TF binding (3,4). It has been also pointed out that weak-affinity TF binding motifs are essential for gene-expression regulation (5).
A key question is how TFs find their specific binding sites in a background of nonspecific sites in a cell genome. This question was first addressed theoretically in seminal works of Berg, Winter, and von Hippel (6,7). The central idea of this approach, as expressed in recent reviews (8–10), is that the search process is a combination of three-dimensional and one-dimensional diffusion. It has been shown in different theoretical models that one-dimensional diffusion (termed sliding or hopping in different models) facilitates the search process under certain conditions (11–17). Despite the success of these phenomenological models, a complete understanding of the search process phenomena is still lacking (8). In particular, one of the key open questions is what makes a TF switch from three-dimensional diffusion to one-dimensional sliding in specific genomic locations (8). Invariably, an assumption is made about the existence of some nonspecific binding sites that bring TFs to the vicinity of DNA for one-dimensional sliding. This assumption is a key component of all theoretical models, yet the molecular origin of this effect is not understood (8,10). Recent single-molecule experimental studies undoubtedly show that different DNA-binding proteins spend the majority of their time nonspecifically bound and diffusing along DNA (18–22). The question is, what biophysical mechanism provides such nonspecific attraction toward genomic DNA and regulates the strength of this attraction at a given genomic location?
Here, we predict that DNA sequence correlations statistically regulate nonspecific TF-DNA binding preferences. Depending on the symmetry and lengthscale of sequence correlations, the nonspecific binding affinity can be either enhanced or reduced. In particular, we show that homooligonucleotide sequence correlations, where nucleotides of the same type are clustered together generically, reduce the nonspecific TF-DNA binding free energy, thus enhancing the binding affinity (Fig. 1). Sequence correlations in which nucleotides of different types alternate have the opposite effect, increasing the nonspecific TF-DNA binding free energy (Fig. 1). Correlation analysis of the yeast-genome regulatory sequences suggests that the predicted design principle is exploited at the genome-wide level to increase the strength of nonspecific binding at these regulatory genomic locations.
Figure 1.
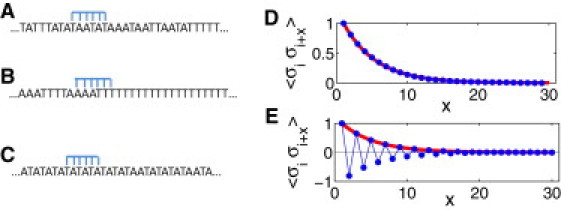
Schematic representation of the model for TF binding to DNA, and examples of DNA sequence correlation functions. (A) Random sequence. (B) Enhanced homooligonucleotide (i.e., ferromagneticlike) correlations lead to statistically enhanced nonspecific TF-DNA binding affinity. (C) Enhanced antiferromagneticlike correlations (alternating nucleotides of different types) lead to reduced nonspecific TF-DNA binding affinity. All examples of sequences (A–C) represent simulation snapshots. (D and E) Examples of the correlation function computed for sequences with enhanced ferromagneticlike correlations (D) and those with enhanced antiferromagneticlike correlations (E), where bold lines represent the exponential decay of the correlation functions.
This article is organized as follows. First, we present a simple, analytically solvable model that describes TF-DNA binding. This model uses two-nucleotide alphabet DNA sequences. We develop a stochastic procedure allowing us to design DNA sequences with a controlled symmetry and strength of sequence correlations. We analyze the free energy of nonspecific TF-DNA binding within the framework of this model, and give an intuitive explanation for the origin of the predicted effect. Second, we generalize the model to four-letter alphabet DNA sequences and show, as well, that all key conclusions hold qualitatively true in this case. Third, we compute the free energy of nonspecific TF-DNA binding for yeast genomic sequences and show that sequences highly occupied by TFs in vivo possess a statistically higher propensity for nonspecific binding to TFs compared with sequences depleted in TFs. We conclude by proposing experiments that will allow direct testing of the predicted effect.
Theory and Results
Free energy of nonspecific TF-DNA binding in model sequences
In this work, we use a simple variant of the Berg-von Hippel model to describe TF-DNA binding (1). For the analytical analysis, we apply the model to artificial DNA sequences containing two types of nucleotide rather than four. However, we show that all key conclusions hold qualitatively true for four-nucleotide alphabet sequences, as well.
The energy of a TF bound to DNA at a specific location i (see Fig. 1) can be expressed as
| (1) |
where i and j represent individual basepairs, M is the effective length of the TF (i.e., the number of contacts between TF and DNA), describes two possible nucleotide types at each position j, and K is the interaction strength. We therefore assume that the energy contributions of individual basepairs to the total binding energy, , are additive. We also assume that the energy of each contact is exclusively defined by the basepair type. The sequence of a DNA molecule of length L is uniquely defined by the set of L numbers, σj, where .
We note that Eq. 1 provides a minimal model for TF-DNA binding. It captures the recognition specificity of TF in the simplest possible way, by assigning different contact energies, and , with two possible nucleotide types. In reality, a TF recognizes DNA motifs forming a complex, cooperative network of hydrogen and electrostatic bonds (1,2). Yet we suggest that the design principle for enhanced nonspecific TF-DNA binding predicted using such a simplified model is likely to be quite general and robust with respect to microscopic details of TF-DNA interactions.
The free energy of binding of an individual TF to DNA is given by , with the partition function
| (2) |
where kB is the Boltzmann constant, T is the absolute temperature, and we imply periodic boundary conditions. We ask the question, what are the statistical properties of F as a function of the symmetry and strength of DNA sequence correlations?
To answer this question, we first design a DNA sequence using a stochastic design procedure. This procedure allows nucleotides within the DNA sequence to anneal, with each configuration being accepted with the Boltzmann probability
| (3) |
where Td is the design temperature controlling the strength of correlations (this is different from the thermodynamic temperature, T), Ed is the design intra-DNA energy. For simplicity, we take into account only the nearest-neighbor interactions in the design energy:
| (4) |
where J is the design intrasequence interaction strength, and Zd is the corresponding Ising model partition function (23),
| (5) |
where .
The ferromagneticlike case, , produces sequences with homooligonucleotide stretches. The correlation length, , is the characteristic lengthscale of the correlations decay, (23). The antiferromagneticlike case, , produces sequences with a different symmetry of alternating nucleotides (Fig. 1). We define the average free energy of TF binding to DNA as the annealed average,
| (6) |
where the averaging is performed with probability (Eq. 3) and . The quenched averaging, , is analyzed numerically below, and it gives qualitatively similar results (Fig. 2). The averaging in Eq. 6 gives
| (7) |
where Zd is given by Eq. 5, and
| (8) |
We argue that the DNA correlations symmetry affects statistically the interaction free energy. It is natural therefore to analyze the free-energy difference between designed sequences and their randomized analogs, which lack symmetry:
| (9) |
where is the free energy computed for entirely random sequences (i.e., for sequences designed using a very high value of Td or, equivalently, ). The first key property of is that it is invariant with respect to the sign of the TF-DNA binding-affinity constant, K. Second, it is always satisfied that if (ferromagneticlike correlations within designed DNA sequences (see Eq. 4)), and if (antiferromagneticlike correlations). Fig. 2 shows the behavior of at different magnitudes of the design strength. The central observation here is that the behavior of critically depends on the symmetry and the lengthscale of DNA sequence correlations. The presence of homooligonucleotide stretches along DNA sequences statistically increases the propensity of such sequences for nonspecific binding to TFs. The DNA stretches with nucleotides of different types alternating produce the opposite effect: such sequences will have a reduced propensity for nonspecific binding. We note that the quenched average, , computed numerically, is in good agreement with the annealed average (Fig. 2)
Figure 2.
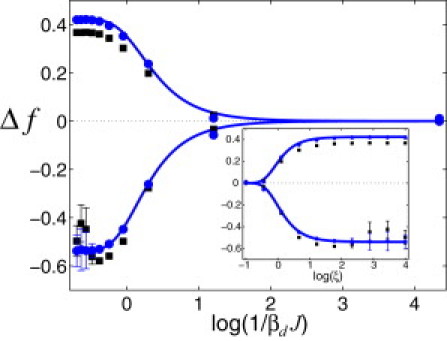
TF-DNA binding free-energy difference normalized per basepair, , computed using Eq. 7 as a function of the reduced design temperature, (solid curves). The upper and lower branches of the graph correspond to (antiferromagneticlike DNA sequence correlations) and (ferromagneticlike correlations), respectively. The results of MC simulations of the system are in excellent agreement with the analytical results (solid circles). We used the parameters , , and . In Monte Carlo simulations, we used 7.5 × 106 MC moves to design each DNA sequence at each value of Td. To generate each point in the plot, we used a set of 100 sequences. To compute error bars, we divided each set of 100 sequences randomly into 10 subsets, and then calculated the SD of the subset averages for . The error bars correspond to 1 SD. The numerically computed quenched average, , is also shown (solid squares). In the computations, we used the same parameters and definitions as specified above. (Inset) Same data for as in the main figure, but plotted as a function of ξ.
The reduction in TF-DNA binding free energy in the presence of homooligonucleotide sequence correlations can be understood intuitively in the following way. Homooligonucleotide sequence correlations generically enhance fluctuations of the TF-DNA binding energy, . This effect has to do with the symmetry: a TF sliding along correlated DNA sequences where nucleotides of the same type have the tendency to cluster, will experience homogeneous DNA islands, such as poly(dA:dT) and poly(dC:dG) tracts. Statistically, this leads to the dominant contribution of either very strong or very weak energies to the TF-DNA binding energy spectrum. This symmetry effect leads therefore to the widening of the TF-DNA binding energy spectrum, . Such widening generically leads to the reduction of the TF-DNA binding free energy, due to the fact that the dominant contribution to the partition function, Z, comes from the low-energy tail of (24). Alternatively, a DNA sequence with enhanced antiferromagneticlike correlations (i.e., with alternating nucleotides of different types) will have the opposite effect: a TF sliding along such a sequence will experience very heterogeneous binding sites. This leads to the narrowing down of and consequently to the increase of the nonspecific TF-DNA binding free energy.
We note that the predicted effect is not restricted to TFs; it is operational for any other kind of DNA-binding protein.
Extension of the model to four-letter-alphabet DNA sequences
We show in this section that four-letter alphabet DNA sequences demonstrate statistical binding properties qualitatively similar to those of the two-letter alphabet sequences analyzed above. This will allow us to extend all our insights gained from the analytical model directly to genomic DNA sequences. We argue that the same underlying physical mechanism controls the nonspecific binding propensity in both cases.
Contrary to the two-letter alphabet DNA sequences, where within our modeling framework a TF is fully described by the single parameter K, in the four-letter alphabet DNA case, a TF is characterized by four energy parameters, KA, KT, KC, and KG. Although those energy constants are generally unknown, their order of magnitude can be roughly estimated as , and in addition, we allow the TF-DNA contact energies to fluctuate. We therefore draw these energies from the Gaussian probability distributions, , with zero mean and standard deviations (SD), σα, where ; and we average the free energy over many TF realizations.
The binding energy of TF at a given site i is expressed by
| (10) |
where is a four-component vector of the type at each DNA position j, with the position of 1 specifying one of four possible identities (A, T, C, or G) of the basepair at position j, with δαβ being the Kronecker delta. The sequence design procedure is analogous to the one introduced above, Eq. 4, with the 4 × 4 symmetric matrix of the design potentials entering the sum, . The results for the average TF-DNA binding free energy in the ensemble of different TFs are shown in Fig. 3. The key conclusion here is that, provided that in the design procedure nucleotides of the same type attract, the lower the design temperature (and thus the longer the correlation length of homooligonucleotide stretches), Td, the lower the TF-DNA binding free energy.
Figure 3.
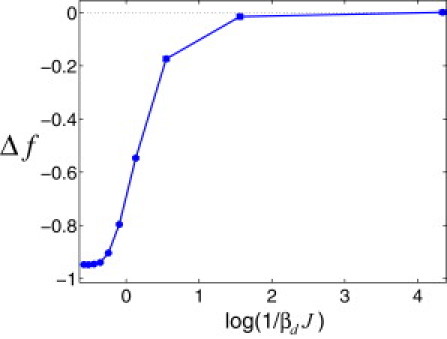
Average TF-DNA binding free energy, , numerically computed at different values of the design temperature, where , with the partition function for an entirely random DNA sequence. We designed 200 sequences with length at each Td. We performed 5 × 106 MC steps to design each sequence, attempting in each step to exchange two basepairs chosen at random. The overall nucleotide composition for each sequence was uniform and fixed. The design potential was (attraction) for identical nearest-neighbor basepairs and (repulsion) for different nearest-neighbor basepairs, with . The contact energies, , were drawn from a Gaussian distribution, , with zero mean, , and standard deviation for each nucleotide type α. We computed as an average over 250 TFs and 200 sequences at each Td and used . The error bars are calculated as specified in Fig. 2, and they are smaller than the marker size.
Free energy of nonspecific TF-DNA binding in yeast genome
We ask further the key question, is the predicted design principle for nonspecific TF-DNA binding operating in a living cell? To answer this question, we computed TF-DNA binding free energies using yeast-genome DNA. Our working hypothesis here is that if the predicted effect is operational, genomic regions that need to be highly accessible to TFs should possess a higher propensity for nonspecific TF-DNA binding than regions that need not be highly accessible to TFs. To test this hypothesis, we compiled two datasets of genomic DNA. First, we collected ∼1600 high-confidence yeast DNA regulatory promoter sequences (for organelle organization and biogenesis genes), each sequence 100 nucleotides long. We describe this dataset as upstream. These upstream sequences are experimentally known to be highly accessible to TFs. The second dataset involves a comparable number of weakly accessible genomic sequences. For this purpose, we chose the first 100 nucleotide stretches of the mRNA coding regions of those organelle organization and biogenesis genes. We describe the second dataset as downstream. The datasets were compiled by Lee et al. (25).
It turns out that upstream sequences demonstrate statistically stronger homooligonucleotide correlations in A and T compared to downstream sequences, and the difference in correlations of C and G is not significant between the datasets. The normalized correlation function, , computed for the sets of upstream and downstream sequences, is shown in Fig. 4 (blue and red, respectively). This function is defined as , where , and is obtained analogously, using the set of randomly permuted sequences averaged with respect to different random realizations. shows qualitatively similar behavior (data not shown).
Figure 4.
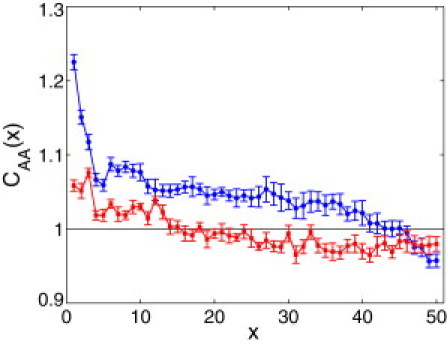
Normalized correlation function, (see the text for the definition), computed for upstream (blue circles) and downstream (red squares) sequence sets. Each set consists of 1663 sequences, with each sequence 100 nucleotides long.
We now compare the TF-DNA binding free energies for those two datasets. To get rid of the compositional bias, for a given TF interacting with a given DNA sequence, we always compare the difference, , between the actual free energy, F, and the free energy computed for the randomized sequence (preserving the nucleotide composition of each sequence), averaged over several random realizations, : . We therefore compute numerically the probability distribution, , for these two datasets of sequences interacting with a model set of TFs. The TF-DNA binding contact energies, , are drawn from the Gaussian distributions, , as described above. We stress that the only external parameters entering the model are the SDs, σα, of . In our calculations, we set for all α. The computed values for upstream and downstream DNA sequences are shown in Fig. 5 A. We also show the cumulative probability at different values of the selectivity cutoff (Fig. 5 B). The central conclusion here is that due to the presence of enhanced homooligonucleotide (i.e., ferromagneticlike) sequence correlations, nonspecific TF-DNA binding is statistically enhanced. At the maximal selectivity cutoff, where per basepair, the probability of TF binding with the free energy below is >30% higher to upstream DNA regions than to downstream regions (Fig. 5 B). This effect leads to a shift of the thermodynamic equilibrium toward enhanced occupancy of TFs binding upstream regions rather than downstream regions. The average strength of the effect on TF occupancy can be estimated from the difference of the average TF-DNA binding free energies, per basepair, between upstream and downstream DNA regions (difference between the peak positions in Fig. 5 A). For a TF forming M contacts within the TF-DNA binding site, this difference will produce shift in the relative binding occupancy, where nup and ndown are the numbers of bound TFs in the upstream and downstream regions, respectively. For a typical TF forming contacts with 10 DNA basepairs, this leads to . We emphasize that the latter estimate provides only a lower-bound limit for the strength of the predicted correlational effect. We suggest, therefore, that the predicted mechanism for enhanced nonspecific TF-DNA binding is operational in promoter regions of a significant fraction of yeast genes.
Figure 5.
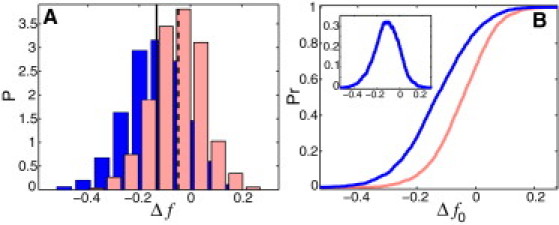
(A) Computed for 1663 upstream (blue) and downstream (red) yeast genomic sequences, where , and . For each given TF, is computed as an average over 50 randomized sequence replicas (randomization preserves the nucleotide composition of each sequence). For each sequence, we computed for 250 TFs and then took the average of these 250 values, . We used . The TF-DNA contact energies, , are drawn from a Gaussian probability distribution, , with zero mean and standard deviation , where α represents four possible nucleotides. Vertical lines show the mean of . (B) The cumulative probability, , computed using from A. (Inset) Difference between upstream and downstream .
Finally, we note that our findings suggest the existence of an upper bound for the TF-DNA binding motif size, imposed by the maximal possible strength of nonspecific binding. It is predicted (11) that if the free energy of TF-DNA nonspecific binding falls below , this significantly slows down the sliding diffusion of TFs along DNA. Our estimates therefore suggest that such slowing down is likely when the binding motif approaches the size of 20 basepairs.
Discussion and Conclusion
Here, we predict a generic biophysical mechanism that statistically regulates the strength of nonspecific TF-DNA binding in a genome. We showed analytically and numerically, using both artificially designed and genomic DNA sequences, that homooligonucleotide correlations statistically enhance nonspecific TF-DNA binding affinity. We described the symmetry of such correlations as ferromagneticlike. Alternatively, DNA sequences possessing enhanced correlations of alternating nucleotides of different types (referred to here as antiferromagneticlike) have a reduced propensity for nonspecific binding to TFs.
Our model description of TF-DNA binding is highly simplified. Yet we suggest that the design principle for enhanced nonspecific TF-DNA binding predicted in this work is likely to be quite general, it is operational in genomic locations highly occupied by TFs, and it is likely to be the rule rather than the exception. The robustness of our conclusions with respect to the details of the model stems from the fact that the predicted effect arises exclusively due to DNA sequence symmetry and its strength (which is determined by the lengthscale of the correlations decay). Computational analysis of the TF-DNA binding free energy in ∼1600 yeast genomic DNA regions highly occupied by TFs shows that those regions possess much higher propensity for nonspecific binding compared with regions depleted in TFs. In our analysis, we used a simple procedure to get rid of the DNA compositional bias, allowing us to fairly compare the relative free energies of nonspecific binding in different genomic locations.
We estimated that in yeast, the predicted effect leads to a free energy reduction per DNA basepair in contact with a TF of at least (on average) in DNA regions with enhanced propensity for nonspecific binding. This leads to at least a threefold concentration enrichment in TFs (on average) of such highly promiscuous DNA regions in yeast. We suggest, therefore, that in addition to all known signals, genomic DNA might also encode its intrinsic propensity for nonspecific binding to TFs. The predicted effect plays the role of an effective, nonspecific localization potential, enhancing the level of one-dimensional diffusion of TFs along genomic DNA at the genome-wide level and thus speeding up the search process for specific TF binding sites (6–11). We stress that all our conclusions are obtained assuming a quasiequilibrium nature of TF-DNA binding. It would be important to investigate the dynamic aspects of the predicted phenomena.
It is important to note that too high a level of nonspecific TF-DNA binding impairs the overall search efficiency (11,12). This suggests that the strength of the predicted effect in vivo might be subject to both positive and negative regulation. It has been pointed out in a seminal work of Iyer and Struhl (26) that activity of poly(dA:dT) tracts increases with their length. We suggest that this observation is a direct consequence of the effect of enhanced nonspecific TF-DNA binding by poly(dA:dT), predicted here. Another key observation of Iyer and Struhl (26), that poly(dC:dG) functions in a similar manner to poly(dA:dT), further strengthens our prediction.
Extensive correlation analysis of different organismal genomes and direct, large-scale measurements of TF-DNA binding preferences using DNA sequences with controlled strength and symmetry of correlations, should provide an ultimate test of the phenomenon predicted here. Protein-DNA binding arrays (4) and high-throughput microfluidics technology (3) allow a direct experimental test of our predictions in vitro. A key experiment would measure the TF-DNA binding affinity in different sets of DNA, each set containing DNA sequences with a specific TF-DNA binding motif embedded in a background of nonspecific sequences with varying symmetry and strength of correlations between DNA sets. We expect that DNA sequences with enhanced homooligonucleotide correlations in background sequences will generically possess a higher binding affinity to different TFs compared with background sequences either lacking such correlations or having correlations with alternating nucleotides of different types.
Acknowledgments
We thank Noa Musa-Lempel for help in compiling yeast genomic sequences. D.B.L. acknowledges financial support from Israel Science Foundation grant 1014/09.
References
- 1.Berg O.G., von Hippel P.H. Selection of DNA binding sites by regulatory proteins. Statistical-mechanical theory and application to operators and promoters. J. Mol. Biol. 1987;193:723–750. doi: 10.1016/0022-2836(87)90354-8. [DOI] [PubMed] [Google Scholar]
- 2.Stormo G.D., Fields D.S. Specificity, free energy and information content in protein-DNA interactions. Trends Biochem. Sci. 1998;23:109–113. doi: 10.1016/s0968-0004(98)01187-6. [DOI] [PubMed] [Google Scholar]
- 3.Fordyce P.M., Gerber D., Quake S.R. De novo identification and biophysical characterization of transcription-factor binding sites with microfluidic affinity analysis. Nat. Biotechnol. 2010;28:970–975. doi: 10.1038/nbt.1675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Badis G., Berger M.F., Bulyk M.L. Diversity and complexity in DNA recognition by transcription factors. Science. 2009;324:1720–1723. doi: 10.1126/science.1162327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Segal E., Raveh-Sadka T., Gaul U. Predicting expression patterns from regulatory sequence in Drosophila segmentation. Nature. 2008;451:535–540. doi: 10.1038/nature06496. [DOI] [PubMed] [Google Scholar]
- 6.Berg O.G., Winter R.B., von Hippel P.H. Diffusion-driven mechanisms of protein translocation on nucleic acids. 1. Models and theory. Biochemistry. 1981;20:6929–6948. doi: 10.1021/bi00527a028. [DOI] [PubMed] [Google Scholar]
- 7.von Hippel P.H., Berg O.G. Facilitated target location in biological systems. J. Biol. Chem. 1989;264:675–678. [PubMed] [Google Scholar]
- 8.Kolomeisky A.B. Physics of protein-DNA interactions: mechanisms of facilitated target search. Phys. Chem. Chem. Phys. 2011;13:2088–2095. doi: 10.1039/c0cp01966f. [DOI] [PubMed] [Google Scholar]
- 9.Halford S.E., Marko J.F. How do site-specific DNA-binding proteins find their targets? Nucleic Acids Res. 2004;32:3040–3052. doi: 10.1093/nar/gkh624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mirny L., Slutsky M., Kosmrlj A. How a protein searches for its site on DNA: the mechanism of facilitated diffusion. J. Phys. A. 2009;42:434013. [Google Scholar]
- 11.Slutsky M., Mirny L.A. Kinetics of protein-DNA interaction: facilitated target location in sequence-dependent potential. Biophys. J. 2004;87:4021–4035. doi: 10.1529/biophysj.104.050765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Slutsky M., Kardar M., Mirny L.A. Diffusion in correlated random potentials, with applications to DNA. Phys. Rev. E. 2004;69:061903. doi: 10.1103/PhysRevE.69.061903. [DOI] [PubMed] [Google Scholar]
- 13.Mirny L.A. Nucleosome-mediated cooperativity between transcription factors. Proc. Natl. Acad. Sci. USA. 2010;107:22534–22539. doi: 10.1073/pnas.0913805107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cherstvy A.G., Kolomeisky A.B., Kornyshev A.A. Protein-DNA interactions: reaching and recognizing the targets. J. Phys. Chem. B. 2008;112:4741–4750. doi: 10.1021/jp076432e. [DOI] [PubMed] [Google Scholar]
- 15.Das R.K., Kolomeisky A.B. Facilitated search of proteins on DNA: correlations are important. Phys. Chem. Chem. Phys. 2010;12:2999–3004. doi: 10.1039/b921303a. [DOI] [PubMed] [Google Scholar]
- 16.Hu T., Grosberg A.Yu., Shklovskii B.I. How proteins search for their specific sites on DNA: the role of DNA conformation. Biophys. J. 2006;90:2731–2744. doi: 10.1529/biophysj.105.078162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sheinman M., Kafri Y. The effects of intersegmental transfers on target location by proteins. Phys. Biol. 2009;6:016003. doi: 10.1088/1478-3975/6/1/016003. [DOI] [PubMed] [Google Scholar]
- 18.Wang Y.M., Austin R.H., Cox E.C. Single molecule measurements of repressor protein 1D diffusion on DNA. Phys. Rev. Lett. 2006;97:048302. doi: 10.1103/PhysRevLett.97.048302. [DOI] [PubMed] [Google Scholar]
- 19.Blainey P.C., Luo G., Xie X.S. Nonspecifically bound proteins spin while diffusing along DNA. Nat. Struct. Mol. Biol. 2009;16:1224–1229. doi: 10.1038/nsmb.1716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Elf J., Li G.-W., Xie X.S. Probing transcription factor dynamics at the single-molecule level in a living cell. Science. 2007;316:1191–1194. doi: 10.1126/science.1141967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tafvizi A., Huang F., Leith J.S., Fersht A.R., van Oijen A.M. Tumor suppressor p53 slides on DNA with low friction and high stability. Biophys. J. 2008;95:L01–L03. doi: 10.1529/biophysj.108.134122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liu S., Abbondanzieri E.A., Zhuang X. Slide into action: dynamic shuttling of HIV reverse transcriptase on nucleic acid substrates. Science. 2008;322:1092–1097. doi: 10.1126/science.1163108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Plischke M., Bergersen B. World Scientific; Singapore: 1994. Equilibrium Statistical Physics. [Google Scholar]
- 24.Lukatsky, D. B., and M. Elkin. 2011. Energy fluctuations shape free energy of biomolecular interactions. arXiv:1101.4529v1.
- 25.Lee W., Tillo D., Nislow C. A high-resolution atlas of nucleosome occupancy in yeast. Nat. Genet. 2007;39:1235–1244. doi: 10.1038/ng2117. [DOI] [PubMed] [Google Scholar]
- 26.Iyer V., Struhl K. Poly(dA:dT), a ubiquitous promoter element that stimulates transcription via its intrinsic DNA structure. EMBO J. 1995;14:2570–2579. doi: 10.1002/j.1460-2075.1995.tb07255.x. [DOI] [PMC free article] [PubMed] [Google Scholar]


