Abstract
V(D)J recombination generates a remarkably diverse repertoire of antigen receptors through the rearrangement of germline DNA. Terminal deoxynucleotidyl transferase (TdT), a polymerase that adds random nucleotides (N regions) to recombination junctions, is a key enzyme contributing to this diversity. The current model is that TdT adds N regions during V(D)J recombination by random collision with the DNA ends, without a dependence on other cellular factors. We previously demonstrated, however, that V(D)J junctions from Ku80-deficient mice unexpectedly lack N regions, although the mechanism responsible for this effect remains undefined in the mouse system. One possibility is that junctions are formed in these mice during a stage in development when TdT is not expressed. Alternatively, Ku80 may be required for the expression, nuclear localization or enzymatic activity of TdT. Here we show that V(D)J junctions isolated from Ku80-deficient fibroblasts are devoid of N regions, as were junctions in Ku80-deficient mice. In these cells TdT protein is abundant at the time of recombination, localizes properly to the nucleus and is enzymatically active. Based on these data, we propose that TdT does not add to recombination junctions through random collision but is actively recruited to the V(D)J recombinase complex by Ku80.
INTRODUCTION
V(D)J recombination generates an astonishingly diverse repertoire of antigen receptors by rearranging the germline DNA segments that encode the variable regions of immunoglobulin and T cell receptor molecules. Recombination is initiated by the lymphocyte-specific proteins RAG-1 and RAG-2, which recognize recombination signal sequences (RSS) located adjacent to the V, D and J coding segments (1,2). RAG-1 and RAG-2 cleave precisely between an RSS and its corresponding coding segment, generating two broken DNA ends: a blunt, 5′-phosphorylated signal end and a covalently sealed (hairpin) coding end (3–6). Following cleavage these ends remain associated with the RAG proteins in a post-cleavage complex (7,8) and are ultimately joined to form two characteristic junctions: a coding joint and a signal joint. It is not known whether the post-cleavage complex serves as a scaffold for assembly of the end processing and joining machinery or whether it must be disassembled to allow access of these factors.
Whereas the molecular details of site-specific DNA cleavage by the RAG proteins are becoming clear (reviewed in 9), the complex mechanisms responsible for processing and joining the broken ends remain unknown. End processing events create junctional diversity and are thus critical for production of a highly diverse repertoire of antigen receptor specificities. Coding ends frequently suffer both loss of nucleotides and addition of extra nucleotides. Two distinct mechanisms operate to add extra nucleotides. P (palindromic) nucleotides (10,11) are derived from asymmetric opening of the hairpin coding ends, producing short, self-complementary single-stranded extensions that can be incorporated into coding joints (3,12). N (non-templated) nucleotides are added by the lymphocyte-specific enzyme terminal deoxynucleotidyl transferase (TdT), which adds nucleotides randomly to 3′-ends (reviewed in 13). This is the only mechanism known to add nucleotides to signal joints (14). N nucleotides can be quite abundant at signal joints and are present at >70% of junctions at certain TCR and Ig loci (15,16). The mechanism by which TdT adds N nucleotides to V(D)J recombination intermediates, which may be held by the RAG proteins in a post-cleavage complex, remains unknown. Because TdT efficiently adds nucleotides to free DNA ends in vitro without the need for other protein cofactors it has been supposed that TdT encounters recombination intermediates by a simple process of random collision.
Genetic analyses have shown that several proteins important for repair of DNA double-strand breaks (DSB) in many cell types are employed by lymphocytes for joining V(D)J recombination intermediates, including DNA-dependent protein kinase (DNA-PK), XRCC4 protein and DNA ligase IV (reviewed in 2). DNA-PK is composed of a catalytic subunit, DNA-PKcs, and two Ku subunits, Ku70 and Ku80, that are important for targeting DNA-PK to sites of DNA damage such as DSB. Ku binds to altered DNA structures such as DSB and nicks (17–21) and can translocate internally along the DNA helix (18,22).
Previous in vivo analyses have shown that both Ku70 and Ku80 are important for coding and signal joint formation (23–30) and hairpin coding ends accumulate in Ku80-deficient cells (28). The biochemical functions of Ku in processing V(D)J recombination intermediates, however, remain unknown, and proper Ku-dependent coding and signal joint formation has not been reconstituted in a cell-free system. We have proposed that one role of Ku may be to promote disassembly or remodeling of the post-cleavage complex, helping to recruit the end processing and joining machinery (31). Consistent with this idea is the recent finding that the XRCC4–DNA ligase IV complex, which by itself has a low affinity for DNA ends, is recruited by Ku (32).
Our previous analysis of V(D)J recombination junctions from Ku80-deficient mice revealed an unexpected role for Ku80 in N nucleotide addition. Both coding and signal joints isolated from these mice are virtually devoid of N regions (33), with a defect comparable to that seen in TdT–/– mice (34,35). The addition of N regions by TdT thus seems completely dependent upon the presence of Ku80. We hypothesized that Ku80 may stimulate N region addition by recruiting TdT to the broken DNA ends (33). This proposal has received support from several recent studies. First, in vitro experiments have shown that Ku can recruit other end joining factors, such as the XRCC4–DNA ligase IV complex, to DNA (32). Ku also interacts with and stimulates the exonuclease activity of the Werner protein (36). Second, Ku and TdT can be co-immunoprecipitated from cell extracts (37), suggesting that these two proteins interact. The physiological significance of this interaction, however, remains unclear.
While the observations discussed above support the hypothesis that Ku recruits TdT to the recombination complex, we considered several alternative explanations that were difficult to test in the mouse system. In particular, we could not exclude the possibility that V(D)J junctions recovered from Ku80-deficient mice may have been formed during embryonic development, prior to the normal post-natal onset of TdT expression (38–40). We also considered the possibility that nuclear localization of TdT is defective in the absence of Ku80: several nuclear defects have been described in Ku80-deficient cells, including alterations in the attachment of chromatin to the nuclear matrix (41) and an abnormal separation between the layers of the nuclear envelope (42). Finally, Ku might be required for stable expression of TdT protein or for enzymatic activity in mammalian cells.
Here we report further investigations into the mechanism of the dependence of N region addition on Ku80. To explore the requirements for N nucleotide addition in more detail we tested the ability of Ku80-deficient fibroblasts to add N nucleotides to junctions formed by V(D)J recombination of plasmid substrates. Our results indicate that TdT protein is abundant and localized properly to the nucleus in Ku-deficient fibroblasts and is enzymatically active in extracts from these cells. Nevertheless, junctions formed by V(D)J recombination in these cells are devoid of N regions. These data suggest that Ku80 is required for proper recruitment of TdT to post-cleavage complexes containing recombination intermediates.
MATERIALS AND METHODS
Cell culture
The cell lines used in this study (xrs-6, CHOK1 4364 and RMP41) have been described previously (43–45). Cells were maintained in Dulbecco’s modified Eagle’s medium enriched with 10% fetal bovine serum and 0.1 mM MEM non-essential amino acids as described previously (46). Flasks were incubated at 37°C in a humidified chamber containing a 5% CO2 atmosphere.
Transfections
Cells were transfected using FuGene 6 Transfection Reagent according to the manufacturer’s protocol (Boehringer Mannheim). Each subconfluent T-25 flask (∼106 cells) was co-transfected with 1.5 µg recombination substrate, 1.8 µg truncated (core) RAG-1 (pMS127) (47), 2.1 µg truncated (core) RAG-2 (pMS216E) (48) and 0.2–4.5 µg SV40TdTs (kindly provided by Dr Susan Gilfillan). After 48 h, DNA was recovered using the method of Hirt (49). For complementation experiments, xrs-6 cells were also transfected with 1.5 µg hamster Ku80 cDNA in pcDNA3 (a kind gift of Dr Penny Jeggo).
Protein sample preparation and western blotting
Protein from cell lysates. Cells were lysed in Laemmli sample buffer (2% SDS, 10% glycerol, 60 mM Tris–HCl pH 6.8, 0.1 M DTT, 0.001% bromophenol blue). A volume equivalent to one-fiftieth of a confluent flask was analyzed for each sample; equal loading of lanes was verified by Coomassie staining.
Protein from Hirt preparations. Cells were lysed in 0.6% SDS, 10 mM Tris pH 8.0, 1 mM EDTA, followed by precipitation of proteins and genomic DNA by the addition of one-quarter vol 5 M NaCl, 10 mM Tris pH 8.0, 1 mM EDTA (49). The pellet was resuspended by douncing in Laemmli sample buffer. The equivalent of one-fiftieth of a transfection was loaded.
Western blotting. Samples were electrophoresed through 8% SDS–polyacrylamide gels, transferred to PVDF, blocked in 5% non-fat milk TBS-T (50 mM Tris–HCl pH 7.4, 150 mM NaCl, 1% Tween-20). Blots were probed for myc-tagged RAG proteins with anti-human c-myc antibody (Pharmingen) diluted 1:1000 in TBS-T or with monoclonal anti-TdT clone 8-1E4 antibody (Sigma) diluted 1:400. Antibody was detected using enhanced chemifluorescence (ECF) (Amersham) and a Storm 860 imaging system (Molecular Dynamics).
Isolation of junctions
For hybrid joints, DNA was amplified by PCR using DR55 and ML68 primers as described previously (50). Amplified products were cloned using a TA Cloning Kit (Invitrogen) and sequenced using a Thermosequenase radiolabeled terminator cycle sequencing kit (Amersham).
For signal and coding joints we took advantage of the design of the recombination substrates, which allow selection for recombination events. The pJH289 and pJH290 substrates contain a chloramphenicol acetyltransferase (CAT) gene that is separated from its promoter by a prokaryotic transcription terminator (51). Formation of a signal joint (pJH289) or a coding joint (pJH290) removes the terminator, allowing expression of the CAT gene and chloramphenicol resistance when the recombined plasmid is introduced into bacterial cells. A β-lactamase gene elsewhere on the plasmid confers resistance to ampicillin whether or not recombination occurs. Thus, the rare plasmids containing signal or coding joints can be detected by transforming bacteria with DNA from transfections and plating on selective medium containing both ampicillin (100 µg/ml) and chloramphenicol (11 µg/ml), while unrecombined plasmids allow growth only on ampicillin-containing medium. Colonies were screened by colony PCR using the DR99 and DR100 primers (50). Sequencing was performed as described above.
Immunofluorescence
TdT-transfected and mock-transfected RMP41 and xrs-6 cells were removed from flasks 48 h after transfection by treatment with PBS + 3 mM EDTA. The cells were washed once and resuspended in PBS supplemented with 12% fetal calf serum. Cytocentrifuge smears were dried and desiccated until fixed and permeabilized for 15 min at 4°C in absolute methanol. The smears were dried and stained with a 1/40 dilution of rabbit anti-TdT antibody according to the manufacturer’s instructions (Supertechs, MD). The anti-TdT was developed with anti-rabbit IgG (Southern Biotechnology Associates, Birmingham, AL) coupled to Alexa 488 (Molecular Probes, Eugene, OR). Smears were mounted in Fluormount G (Southern Biotechnology Associates) and viewed with a Leica DMRB fluorescence microscope equipped with appropriate filter cubes (Chromatechnology, Battleboro, VT). Images were acquired with a C5810 digital color camera (Hamamatsu Photonic System, Bridgewater, NJ). Images were processed using Adobe PhotoShop and IPLab Spectrum software (Signal Analytics Software, Vienna, VA).
TdT assays
Approximately 107 cells were co-transfected with 22 µg TdT expression vector (SV40TdTs) and 6 µg each truncated RAG-1 and RAG-2 expression vector (pMS127 and pMS216E). After 48 h cells were collected and resuspended in 25 mM HEPES pH 7.5, 260 mM KCl, 0.1 M NaCl, 20% glycerol, 0.1% NP-40, 1 mM DTT, 0.5 mM PMSF and 1 µg/ml each chymotrypsin, leupeptin and peptstatin. Cells were extracted by gentle rotation at 4°C for 1 h, followed by centrifugation at 25 500 g for 25 min. The supernatant was dialyzed against 0.1 M Tris–acetate buffer pH 7.2, 10% glycerol and stored at –80°C. Protein concentration was determined with a Micro BCA kit (Pierce) using BSA as the standard. TdT was detected by western blotting, using monoclonal anti-TdT antibody (Sigma) and ECF (Amersham). Cell extracts were adjusted to equal concentrations of TdT per µg total protein by dilution with untransfected extract from the appropriate cell line.
A 20mer primer (ML68) (50) was 5′-end-labeled using T4 polynucleotide kinase and [γ-32P]ATP. Nine pmoles of primer were incubated at 37°C with 30 µg TdT/sample in a buffer containing 0.2 M potassium cacodylate pH 7.2, 4 mM MgSO4, 0.1 mM DTT, 100 µg/ml BSA, 10 µM ZnSO4, 2 mM dNTPs (52). Reactions were stopped at the indicated time points by addition of a one-half vol 95% formamide, 20 mM EDTA. Samples were denatured and loaded on a 7.4% acrylamide denaturing gel.
RESULTS
TdT expression coincides with recombination in transfected Ku80-deficient cells
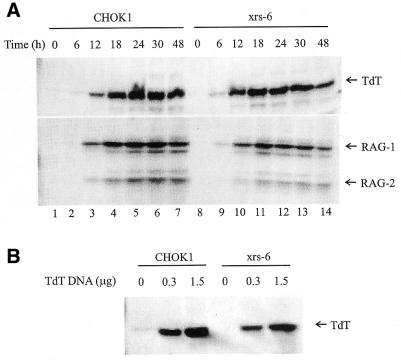
We examined TdT protein expression in the Ku80-deficient Chinese hamster ovary cell line xrs-6 and in the parental wild-type cell line CHOK1 4364 (hereafter referred to as CHOK1). In CHOK1 cells TdT protein is first detectable 12 h after transfection and is maximal at 24–30 h (Fig. 1A). In xrs-6 cells the time course of expression is slightly earlier, with TdT detectable at 6 h and maximal at 18–30 h (Fig. 1A). Thus, TdT protein is stable in Ku80-deficient fibroblasts; these data, coupled with the previous observation that TdT mRNA levels are undiminished in Ku80-deficient mice (33), show that Ku80 is not required for expression of TdT.
Figure 1.
Expression of TdT. (A) Cells were transfected with expression vectors encoding TdT (1.5 µg), RAG-1 (1.8 µg) and RAG-2 (2.1 µg). Cells were harvested at the time points indicated. Total cell lysate was analyzed on duplicate gels which were transferred to membranes and probed with monoclonal anti-TdT (upper) or anti-myc antibodies (which detect the myc tag on the expressed RAG proteins) (lower). (B) Cells were transfected with different amounts of TdT expression vector, as indicated, and collected at 48 h. Protein was isolated from the Hirt preparation.
RAG-1 and RAG-2 proteins are first detectable at 12 h in CHOK1 cells and 6–12 h in xrs-6 cells (Fig. 1A). Consistent with the time course of RAG protein expression, V(D)J recombination is first detectable at 12 h following transfection (D.B.Roth, unpublished observations). It is important to note that levels of TdT are comparable in xrs-6 and CHOK1 cells during the period of maximal RAG expression (Fig. 1A, compare lanes 4–6 with lanes 11–13). Whereas TdT may not be expressed until after recombination occurs in Ku-deficient mice (if, for example, the junctions analyzed were formed in fetal life), in the fibroblast system we can be confident that TdT is present while recombination is occurring. Furthermore, in this system it is possible to measure the efficiency of N nucleotide addition at various levels of TdT expression, because protein levels can be varied by altering the amount of expression vector added (Fig. 1B).
Junctions from Ku80-deficient fibroblasts lack N regions
Having established that TdT protein expression is not significantly diminished in Ku80-deficient cells, we next assessed TdT-dependent N region addition. We transfected TdT into two different wild-type Chinese hamster ovary cell lines, CHOK1 and RMP41, as well as Ku80-deficient xrs-6 cells. As discussed above, formation of the standard V(D)J recombination products, coding and signal joints, is severely impaired in cells lacking Ku80. In an attempt to isolate these extremely rare junctions we utilized the design of the plasmid substrates pJH289 and pJH290, which allows products of V(D)J recombination to be scored in bacteria (see Materials and Methods). Unfortunately, we could not detect even a single coding joint in >216 000 bacterial colonies recovered from several transfections of xrs-6 cells with the pJH290 substrate (corresponding to a recombination frequency of <0.0005%). This is consistent with previous reports indicating that coding joints could not be isolated from this cell line (24,27).
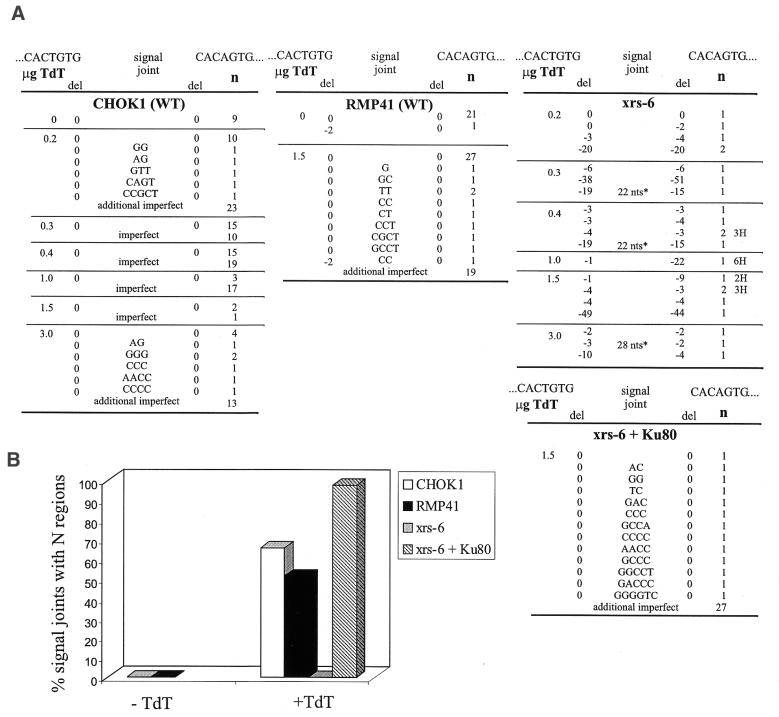
We were able to recover rare signal joints from xrs-6 cells, in agreement with earlier reports (24,27). We expected that N regions might be frequent in signal joints formed in wild-type fibroblasts, because N regions occur at frequencies of 14–70% in signal joints of adult mice (depending on the loci) and in pre-B cells and cell lines expressing TdT (14–16,33). For analysis of signal joints we made use of the observation that a precise signal joint generates an ApaL1 restriction site. Because signal joints from wild-type cells rarely contain deletions (1,14,53), we presumed that ApaL1 resistance signified the presence of N nucleotides, an assumption that was validated by nucleotide sequence analysis of selected junctions. In two different wild-type cells lacking TdT (CHOK1 and RMP41) only one out of 31 joints was ApaL1 resistant; nucleotide sequence analysis of this junction revealed a 2 nt deletion (Fig. 2A).
Figure 2.
Analysis of signal joints. (A) Nucleotide sequence analysis of signal joints. Letters in the center indicate extra nucleotides. The amount of TdT transfected is shown. The number of nucleotides deleted from each end is shown under del. The number of junctions with the indicated sequences is shown under n. The data for xrs-6 cells were collected from two independent transfections at each TdT concentration. Imperfect and additional imperfect describe junctions that were ApaL1-resistant but were not sequenced. 2H, 3H and 6H indicate 2, 3 or 6 nt of homology, respectively, at the junction. (B) Summary of frequency of N regions. Because signal joints from wild-type cells very rarely contain deletions (see text), ApaL1-resistant junctions were plotted as junctions containing N regions. All signal joints recovered from xrs-6 cells were sequenced. *The sequences of the long inserts are as follows: 22 nt, ACAGCCAGACAGTGGAGTACTA; 28 nt, ACAGTGTACAGTGTACAGTGGAGTACTA. The underlined sequence is from plasmid pJH289. The sequence in italics is a triplet of the sequence ACAGTGT, which occurs in the RAG-1 and RAG-2 expression vectors, pMS127 and pMS216E.
In the presence of TdT, however, 29 out of 56 (52%) of the junctions isolated from RMP41 cells and 131 out of 184 (71%) of the junctions isolated from CHOK1 cells were ApaL1 resistant (Fig. 2B). Of 21 of these junctions sequenced, all contained extra nucleotides. These N regions were GC-rich (72% GC) and 1–5 nt in length (Fig. 2A), as expected for TdT-mediated additions (13,54,55). Efficient N region addition was observed at all concentrations of TdT transfected, from 0.2 to 3.0 µg (Fig. 2A), demonstrating that the concentration of TdT was not limiting in our experiments.
Analysis of signal joints from xrs-6 cells transfected with TdT revealed a strikingly different picture. From 329 400 bacterial colonies we isolated and sequenced 22 signal joints; only three contained insertions. These junctional inserts were exceptionally long (22–28 nt) and corresponded to blocks of sequence from the transfected recombination substrate and expression vectors (Fig. 2A), suggesting that they resulted from a TdT-independent mechanism such as oligonucleotide capture (56–58). Twenty-one junctions contained deletions, but only six contained short sequence homologies, in agreement with previous studies in xrs-6 cells lacking TdT (23,24). Transfection of the cDNA for hamster Ku80 into xrs-6 cells fully complemented the defect, yielding abundant N regions (12 out of 13 sequences, 92%) (Fig. 2A and B). These data show that Ku80 is required for N nucleotide addition to signal joints, even in the presence of high levels of TdT.
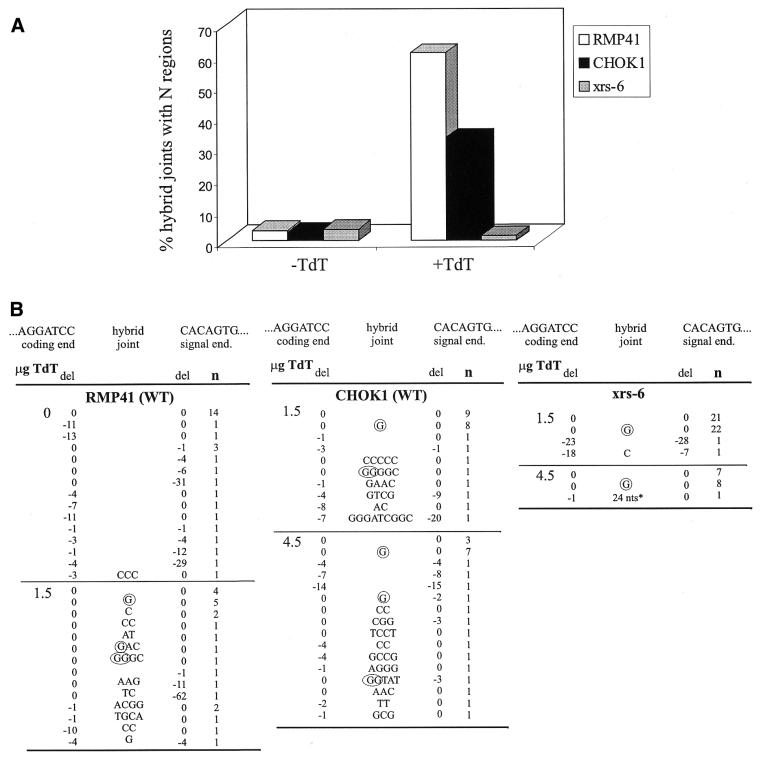
We next analyzed hybrid joints, which are non-standard products that arise by joining of a signal end to a coding end. These joints frequently contain N regions in wild-type cells (59) and are formed efficiently in Ku80-deficient cells (33,50). We thus sought to determine whether these junctions would exhibit N regions in the absence of Ku80.
Hybrid joints formed from the recombination substrate pJH299 can be readily detected using a PCR assay (50). In wild-type cells N nucleotides were present in 57 (RMP41) and 33% (CHOK1) of hybrid joints (Fig. 3A). These N regions were comparable in length (between 1 and 9 nt) and GC content (71%) (Fig. 3B) to those isolated from wild-type mice (13,54,55). In xrs-6 cells, however, only two out of 61 junctions from four independent transfections contained insertions, even when large quantities of TdT were transfected. One of these insertions was quite long (24 nt) and 20 nt corresponded to a block of 20 nt in the substrate pJH299, suggesting that this insertion was not formed by random addition of nucleotides by TdT. Thus, the frequency of N regions at hybrid joints formed in Ku80-deficient cells (one out of 61 junctions, 1.6%) is similar to the frequency of N regions in cells not transfected with TdT (2.7–3.7%) (Fig. 3A). These data demonstrate that hybrid joints containing N regions are extremely rare in Ku80-deficient cells. This may reflect a requirement for Ku in N region addition, as observed for signal joints. Another possibility, however, is that most hybrid joints isolated from these cells may be formed by a RAG-mediated joining process that does not involve free DNA ends (33,50) and which, therefore, may be incompatible with addition of nucleotides by TdT.
Figure 3.
Analysis of hybrid joints. (A) Frequency of junctions containing N regions [sequences are shown in (B)]. Data for RMP41 cells at 0 µg TdT were reported previously (69), but were generated in parallel with experiments using TdT. Data for CHOK1 and xrs-6 cells at 0 µg TdT have been reported previously (50). (B) Nucleotide sequence analysis of hybrid joints (details are as in Fig. 2A legend). Circled letters are presumptive P nucleotides. *The sequence of this insert (24 nt) is TTTCACACCGCATATGGTGCCGAC. The underlined sequence is from the recombination substrate pJH299.
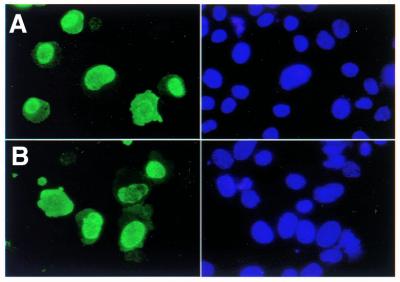
TdT is localized to the nucleus in Ku80-deficient cells
Although TdT protein is abundant in xrs-6 cells (Fig. 1), we considered the possibility that the absence of N regions might be due to failure of TdT to localize to the nucleus in the absence of Ku80. This defect could result from the aberrant nuclear membrane organization found in Ku80-deficient cells (41,42). Another possibility is that Ku80 is essential for DNA-PK-mediated phosphorylation of target proteins required for nuclear entry or retention of TdT. Alternatively, Ku- or DNA-PK-dependent gene expression may be required for TdT nuclear localization.
To examine the nuclear localization of TdT we fixed cells 48 h after transfection, stained them with anti-TdT antibody and probed with a secondary immunoflourescent antibody. Nuclei were stained with Hoechst 22358. TdT is clearly located in the nuclei of both RMP41 (Fig. 4A) and xrs-6 cells (Fig. 4B). In some cells TdT is also present in the cytoplasm, but these appeared at the same frequency in RMP41 and xrs-6 colonies. The xrs-6 cells are larger and the Hoechst staining of nuclear material was much more condensed and less heterogeneous than the staining of wild-type nuclei. This was apparent in the mock-transfected cells as well and was not a result of TdT expression (data not shown), and is consistent with the previously reported morphology of Ku80-deficient fibroblasts (41,42). These data demonstrate that the absence of N regions in xrs-6 cells does not result from a defect in nuclear localization.
Figure 4.
Detection of TdT in cells. TdT was detected by rabbit anti-TdT antibodies (green fluorescence). Nuclei are stained with Hoechst 33258 (blue fluorescence). (A) Wild-type RMP41 cells; (B) xrs-6 cells. Original magnification 400×.
TdT is enzymatically active in Ku80-deficient cells
While the data described above show that TdT is abundant and correctly localized to the nucleus in Ku80-deficient cells, the formal possibility remains that Ku80 is required for enzymatic activity. For example, Ku80 could be required for a critical post-translational modification of TdT. In fact, TdT has been found to be phosphorylated in preparations from human lymphoblastic leukemias (60–62). Alternatively, an inhibitor of TdT enzymatic activity could accumulate in Ku80-deficient cells. Therefore, we directly compared the activity of TdT in crude extracts from wild-type and xrs-6 cells using standard oligonucleotide tailing assays (52,63).
A 5′-32P-labeled 20mer was incubated with dNTPs in the presence of whole cell extracts. Incubation with an extract from CHOK1 cells expressing TdT generated tailed products with reduced mobility (Fig. 5, lanes 3 and 4). Certain lengths of product appear to be over-represented, particularly a 31mer product (top arrow). It is possible that the secondary structure of this product inhibits further lengthening by TdT or confers resistance to nucleases. Several shorter tailed products are also present (lower arrows). In extracts from xrs-6 cells a similar amount of tailing was observed (Fig. 5, compare lanes 7 and 8 with lanes 3 and 4). The 31mer product is abundant and the overall distribution of products is not appreciably different in extracts from wild-type and Ku80-deficient cells. These data demonstrate that the lack of Ku80 does not appreciably affect the activity of TdT as assessed using oligonucleotide substrates.
Figure 5.
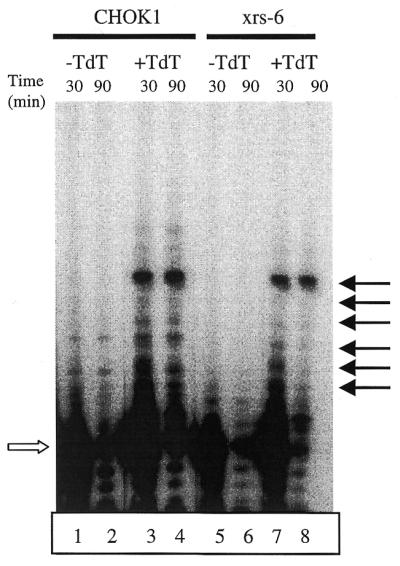
Primer extension assay for TdT activity in extracts. A 32P-labeled 20mer (open arrow) was incubated with extract from CHOK1 (lanes 1–4) or xrs-6 (lanes 5–8) cells either transfected (+TdT) or not transfected (–TdT) with TdT expression vector. Reactions were stopped at the time points indicated. Arrows at the right indicate predominant TdT-dependent extension products. All lanes are from the same gel.
DISCUSSION
We have investigated the mechanisms underlying our previous finding that N regions are absent from V(D)J recombination junctions formed in Ku80-deficient mice. Ku could potentially be required for the expression of TdT protein, its enzymatic activity or its nuclear localization. We examined the dependence of these critical steps on Ku80 by expressing TdT in Ku80-deficient fibroblast cells that undergo V(D)J recombination when transiently transfected with RAG expression vectors and appropriate recombination substrates. Our results show that despite the presence of abundant nuclear, active TdT in Ku80-deficient cells while V(D)J recombination is ongoing, recombinant junctions do not contain TdT-dependent N regions. These data suggest that Ku80 plays a novel role in either the process of N nucleotide addition or in the joining of ends to which N nucleotides have been added. Several alternative models are discussed below.
Does addition of N regions block joining?
One possibility, as we previously suggested (33), is that N nucleotides are added but that the resulting single-stranded 3′-extensions cannot be joined in the absence of Ku80. Analysis of another type of junctional insert, P nucleotides, which are derived from short single-stranded extensions generated by asymmetric opening of the hairpin coding end, argues against this model. P nucleotide insertions of 1–7 nt are found in 32% of coding and hybrid joints from Ku80-deficient cells, compared with 27% of these same joints in wild-type cells (33,50; this work), indicating that joining of ends with short single-stranded extensions is not impaired in these cells. While the polarity of the extensions that lead to P nucleotides has not been established, most opened hairpins in vivo have 3′-extensions (64,65). Thus, the frequent presence of P nucleotides suggests that ends with short 3′-extensions are capable of joining in Ku80-deficient cells.
A second possibility is that TdT adds N nucleotides aberrantly in the absence of Ku80, forming intermediates containing long 3′-extensions that are not joined efficiently. Recent in vitro studies have shown that DNA-PKcs binds TdT and negatively regulates its activity, raising the possibility that unusually long N regions might be added in the absence of Ku80 (66). However, our analysis of TdT activity in vitro argues against this model, as we did not detect any difference in the lengths of products generated by TdT from Ku80-deficient and wild-type cell extracts.
Are N regions added but then removed?
Another possibility (33) is that N regions are added to V(D)J recombination intermediates in Ku80-deficent cells but fail to appear in the final junctions because they are removed prior to joining. For example, N regions could be removed by excessive nuclease activity in the absence of Ku80. However, a large number of coding joints which lack N regions from Ku80–/– mice are full-length and contain P nucleotides, indicating that these products were not subject to degradation (33). Furthermore, the majority of hybrid joints formed in xrs-6 cells were not deleted from either end (Fig. 3B) (50). These data argue against the removal of N nucleotides by nuclease activity.
Another possible explanation for the absence of N regions is that they disappear as a consequence of the joining process. For example, joining mediated by short sequence homologies flanking the N additions would generate a junction that lacks N regions but is characterized by homology. Our analysis of junction sequences from Ku80-deficient cells is inconsistent with this model, as only six out of 22 signal joints and one out of 61 hybrid joints display homology. A related possibility is that the N nucleotides themselves could be used as homologies to direct joining. When the entire N addition is used as a homology, it would disappear in the final junction. However, one would not expect joining to always employ all N nucleotides as homologies. Thus, this model predicts that some junctions should have N regions, but none were observed. Furthermore, the frequent presence of P nucleotides at junctions from Ku80-deficient cells argues against both models, as these nucleotides should also be removed by homology-dependent joining.
A recruitment model
The arguments presented above suggest that rather than leading to removal of N regions, the absence of Ku80 impairs the ability of TdT to add N nucleotides. It has been presumed that N nucleotides are added to free DNA ends created during V(D)J recombination. Our data show that extracts from Ku-deficient cells expressing TdT readily add nucleotides to the free 3′-ends of oligonucleotide substrates. How then can we explain the complete absence of N nucleotides at V(D)J recombination junctions formed in Ku80-deficient cells? We suggest that rather than encountering ends by random collision, TdT is specifically recruited to post-cleavage complexes containing coding and signal ends. In the absence of Ku this recruitment process may not occur, blocking the addition of N regions. Ku80 has been identified in post-cleavage complexes (7), but recruitment could also involve interactions between TdT and other components of the complex (e.g. the RAG proteins) or Ku80-dependent disassembly of the complex to allow TdT access to DNA ends.
A reviewer has pointed out that junctions generated in the absence of Ku80 may be formed by an alternative pathway, dissimilar to that used in V(D)J recombination, that excludes TdT. Although our results do not exclude this possibility, the recruitment model remains the most economical hypothesis to explain the available data. Furthermore, if the recruitment model is incorrect and TdT encounters ends by random collision, it is difficult to imagine how alternative DNA repair pathways might exclude TdT.
Precedents for our proposal that Ku recruits TdT to V(D)J recombination complexes are provided by the observation that Ku targets DNA-PKcs to DNA ends (19) and by more recent studies in yeast which suggest that yeast Ku70 may be involved in recruiting the silencing factor Sir4p to broken DNA ends to repress transcription in the vicinity of DSB (67). A role for Ku in recruiting other end joining factors is supported by recent in vitro observations that both the XRCC4–ligase IV complex and the Werner protein are recruited and stimulated by Ku (32,36). It is noteworthy that TdT purified from rat liver is found in a large multiprotein complex that includes DNA ligase and DNA polymerase activities (68). This observation further supports the intriguing possibility that TdT may be recruited in a Ku-dependent fashion to the post-cleavage complex, perhaps along with other end joining and end processing activities.
Acknowledgments
ACKNOWLEDGEMENTS
We thank S.Gilfillan for generously providing the TdT expression vector sv40TdTs and P.Jeggo for providing the Ku80/pcDNA3 expression vector. We are grateful to Vicky Brandt, Leslie Huye, Mark Landree and Jian Qiu for criticisms of the manuscript and members of the Roth laboratory for helpful discussions. Suzanne Robertson provided secretarial assistance. Monica Calicchio and Kerstin Blankenburg provided technical help. This work was supported by National Institutes of Health grants AI36420 to D.B.R and AI45794-01 to J.F.K. M.M.P. is a fellow of the Cancer Research Institute.
References
- 1.Lewis S.M. (1994) The mechanism of V(D)J joining: lessons from molecular, immunological and comparative analyses. Adv. Immunol., 56, 27–150. [DOI] [PubMed] [Google Scholar]
- 2.Bogue M. and Roth,D.B. (1996) Mechanism of V(D)J recombination. Curr. Opin. Immunol., 8, 175–180. [DOI] [PubMed] [Google Scholar]
- 3.Roth D.B., Menetski,J.P., Nakajima,P.B., Bosma,M.J. and Gellert,M. (1992) V(D)J recombination: broken DNA molecules with covalently sealed (hairpin) coding ends in scid mouse thymocytes. Cell, 70, 983–991. [DOI] [PubMed] [Google Scholar]
- 4.Roth D.B., Zhu,C. and Gellert,M. (1993) Characterization of broken DNA molecules associated with V(D)J recombination. Proc. Natl Acad. Sci. USA, 90, 10788–10792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Schlissel M., Constantinescu,A., Morrow,T., Baxter,M. and Peng,A. (1993) Double-strand signal sequence breaks in V(D)J recombination are blunt, 5′-phosphorylated, RAG-dependent, and cell cycle regulated. Genes Dev., 7, 2520–2532. [DOI] [PubMed] [Google Scholar]
- 6.McBlane J.F., van Gent,D.C., Ramsden,D.A., Romeo,C., Cuomo,C.A., Gellert,M. and Oettinger,M.A. (1995) Cleavage at a V(D)J recombination signal requires only RAG1 and RAG2 proteins and occurs in two steps. Cell, 83, 387–395. [DOI] [PubMed] [Google Scholar]
- 7.Agrawal A. and Schatz,D.G. (1997) RAG1 and RAG2 form a stable postcleavage synaptic complex with DNA containing signal ends in V(D)J recombination. Cell, 89, 43–53. [DOI] [PubMed] [Google Scholar]
- 8.Hiom K. and Gellert,M. (1997) A stable RAG1–RAG2–DNA complex that is active in V(D)J cleavage. Cell, 88, 65–72. [DOI] [PubMed] [Google Scholar]
- 9.Fugmann S.D., Lee,A.I., Shockett,P.E., Villey,I.J. and Schatz,D.G. (2000) The RAG proteins and V(D)J recombination: complexes, ends, and transposition. Annu. Rev. Immunol., 18, 495–527. [DOI] [PubMed] [Google Scholar]
- 10.Lafaille J.J., DeCloux,A., Bonneville,M., Takagaki,Y. and Tonegawa,S. (1989) Junctional sequences of T cell receptor γδ genes: implications for γδ T cell lineages and for a novel intermediate of V(D)J joining. Cell, 59, 859–870. [DOI] [PubMed] [Google Scholar]
- 11.McCormack W.T., Tjoelker,L.W., Carlson,L.M., Petryniak,B., Barth,C.F., Humphries,E.H. and Thompson,C.B. (1989) Chicken IgL gene rearrangement involves deletion of a circular episome and addition of single nonrandom nucleotides to both coding segments. Cell, 56, 785–791. [DOI] [PubMed] [Google Scholar]
- 12.Lieber M.R. (1991) Site-specific recombination in the immune system. FASEB J., 5, 2934–2944. [DOI] [PubMed] [Google Scholar]
- 13.Gilfillan S., Benoist,C. and Mathis,D. (1995) Mice lacking terminal deoxynucleotidyl transferase: adult mice with a fetal antigen receptor repertoire. Immunol. Rev., 148, 201–219. [DOI] [PubMed] [Google Scholar]
- 14.Lieber M.R., Hesse,J.E., Mizuuchi,K. and Gellert,M. (1988) Lymphoid V(D)J recombination: nucleotide insertion at signal joints as well as coding joints. Proc. Natl Acad. Sci. USA, 85, 8588–8592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shimizu T. and Yamagishi,H. (1992) Biased reading frames of pre-existing DH–JH coding joints and preferential nucleotide insertions at VH–DJH signal joints of excision products of immunoglobulin heavy chain gene rearrangements. EMBO J., 11, 4869–4875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Iwasoto T. and Yamagishi,H. (1992) Novel excision products of T cell receptor γ gene rearrangements and developmental stage specificity implied by the frequency of nucleotide insertions at signal joints. Eur. J. Immunol., 22, 101–106. [DOI] [PubMed] [Google Scholar]
- 17.Mimori T. and Hardin,J.A. (1986) Mechanism of interaction between Ku protein and DNA. J. Biol. Chem., 261, 10375–10379. [PubMed] [Google Scholar]
- 18.Paillard S. and Strauss,F. (1991) Analysis of the mechanism of interaction of simian Ku protein with DNA. Nucleic Acids Res., 19, 5619–5624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gottlieb T.M. and Jackson,S.P. (1993) The DNA-dependent protein kinase: requirement for DNA ends and association with Ku antigen. Cell, 72, 131–142. [DOI] [PubMed] [Google Scholar]
- 20.Blier P., Griffith,A.J., Craft,J. and Hardin,J.A. (1993) Binding of Ku protein to DNA: measurement of affinity for ends and demonstration of binding to nicks. J. Biol. Chem., 268, 7594–7601. [PubMed] [Google Scholar]
- 21.Morozov V.E., Falzon,M., Anderson,C.W. and Kuff,E.L. (1994) DNA-dependent protein kinase is activated by nicks and larger single-stranded gaps. J. Biol. Chem., 269, 16684–16688. [PubMed] [Google Scholar]
- 22.de Vries E., van Driel,W., Bergsma,W.G., Arnberg,A.C. and van der Vliet,P.C. (1989) HeLa nuclear protein recognizing DNA termini and translocating on DNA forming a regular DNA–multimeric protein complex. J. Mol. Biol., 208, 65–78. [DOI] [PubMed] [Google Scholar]
- 23.Pergola F., Zdzienicka,M.Z. and Lieber,M.R. (1993) V(D)J recombination in mammalian mutants defective in DNA double-strand break repair. Mol. Cell. Biol., 13, 3464–3471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Taccioli G.E., Rathbun,G., Oltz,E., Stamato,T., Jeggo,P.A. and Alt,F.W. (1993) Impairment of V(D)J recombination in double-strand break repair mutants. Science, 260, 207–210. [DOI] [PubMed] [Google Scholar]
- 25.Getts R.C. and Stamato,T.D. (1994) Absence of a Ku-like DNA end binding activity in the xrs double-strand DNA repair-deficient mutant. J. Biol. Chem., 269, 15981–15984. [PubMed] [Google Scholar]
- 26.Rathmell W.K. and Chu,G. (1994) A DNA end-binding factor involved in double-strand break repair and V(D)J recombination. Mol. Cell. Biol., 14, 4741–4748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Taccioli G.E., Gottlieb,T.M., Blunt,T., Priestley,A., Demengeot,J., Mizuta,R., Lehmann,A.R., Alt,F.W., Jackson,S.P. and Jeggo,P.A. (1994) Ku80: product of the XRCC5 gene and its role in DNA repair and V(D)J recombination. Science, 265, 1442–1445. [DOI] [PubMed] [Google Scholar]
- 28.Zhu C., Bogue,M.A., Lim,D.-S., Hasty,P. and Roth,D.B. (1996) Ku86-deficient mice exhibit severe combined immunodeficiency and defective processing of V(D)J recombination intermediates. Cell, 86, 379–389. [DOI] [PubMed] [Google Scholar]
- 29.Nussenzweig A., Chen,C., da Costa Soares,V., Sanchez,M., Sokol,K., Nussenzweig,M.C. and Li,G.C. (1996) Requirement for Ku80 in growth and immunoglobulin V(D)J recombination. Nature, 382, 551–555. [DOI] [PubMed] [Google Scholar]
- 30.Gu Y., Seidl,K.J., Rathbun,G.A., Zhu,C., Manis,J.P., van der Stoep,N., Davidson,L., Cheng,H.-L., Sekiguchi,J.M., Frank,K. et al. (1997) Growth retardation and leaky SCID phenotype of Ku70-deficient mice. Immunity, 7, 653–665. [DOI] [PubMed] [Google Scholar]
- 31.Zhu C., Bogue,M.A. and Roth,D.B. (1996) Thymocyte differentiation in γ-irradiated severe-combined immunodeficient mice: characterization of intermediates and products of V(D)J recombination at the T cell receptor α locus. Eur. J. Immunol., 26, 2859–2865. [DOI] [PubMed] [Google Scholar]
- 32.Nick M.S., Snowden,C.M., McCarville,J. and Ramsden,D.A. (2000) Ku recruits the XRCC4–ligase IV complex to DNA ends. Mol. Cell. Biol., 20, 2996–3003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bogue M.A., Wang,C., Zhu,C. and Roth,D.B. (1997) V(D)J recombination in Ku86-deficient mice: distinct effects on coding, signal, and hybrid joint formation. Immunity, 7, 37–47. [DOI] [PubMed] [Google Scholar]
- 34.Gilfillan S., Dierich,A., Lemeur,M., Benoist,C. and Mathis,D. (1993) Mice lacking TdT: mature animals with an immature lymphocyte repertoire. Science, 261, 1175–1178. [DOI] [PubMed] [Google Scholar]
- 35.Komori T., Okada,A., Stewart,V. and Alt,F.W. (1993) Lack of N regions in antigen receptor variable region genes of TdT-deficient lymphocytes. Science, 261, 1171–1175. [DOI] [PubMed] [Google Scholar]
- 36.Cooper M.P., Machwe,A., Orren,D.K., Brosh,R.M., Ramsden,D. and Bohr,V.A. (2000) Ku complex interacts with and stimulates the Werner protein. Genes Dev., 14, 907–912. [PMC free article] [PubMed] [Google Scholar]
- 37.Mahajan K.N., Gangi-Peterson,L., Sorscher,D.H., Wang,J., Gathy,K.N., Mahajan,N.P., Reeves,W.H. and Mitchell,B.S. (1999) Association of terminal deoxynucleotidyl transferase with Ku. Proc. Natl Acad. Sci. USA, 96, 13926–13931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Feeney A.J. (1990) Lack of N regions in fetal and neonatal mouse immunoglobulin V-D-J junctional sequences. J. Exp. Med., 172, 1377–1390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gu H., Forster,I. and Rajewsky,K. (1990) Sequence homologies, N sequence insertion and JH gene utilization in VHDJH joining: implications for the joining mechanism and the ontogenetic timing of Ly1 B cell and B-CLL progenitor generation. EMBO J., 9, 2133–2140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bogue M., Gilfillan,S., Benoist,C. and Mathis,D. (1992) Regulation of N-region diversity in antigen receptors through thymocyte differentiation and thymus ontogeny. Proc. Natl Acad. Sci. USA, 89, 11011–11015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Schwartz J.L., Cowen,J.M., Moan,E., Sedita,B.A., Stephens,J. and Vaughan,A.T.M. (1993) Metaphase chromosome and nucleoid differences between CHO-K1 and its radiosensitive derivative xrs-5. Mutagenesis, 8, 105–108. [DOI] [PubMed] [Google Scholar]
- 42.Yasui L.S., Ling-Indeck,L., Johnson-Wint,B., Fink,T.J. and Molsen,D. (1991) Changes in the nuclear structure in the radiation-sensitive CHO mutant cell xrs-5. Radiat. Res., 127, 269–277. [PubMed] [Google Scholar]
- 43.Jeggo P.A. (1985) X-ray sensitive mutants of Chinese hamster ovary cell line: radio-sensitivity of DNA synthesis. Mutat. Res., 145, 171–176. [DOI] [PubMed] [Google Scholar]
- 44.Kao F.-T., Chasin,L. and Puck,T.T. (1969) Genetics of somatic mammalian cells. X. Complementation analysis of glycine-requiring mutants. Proc. Natl Acad. Sci. USA, 64, 1284–1291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Merrihew R.V., Sargent,R.G. and Wilson,J.H. (1995) Efficient modification of the APRT gene by FLP/FRT site-specific targeting. Somat. Cell Mol. Genet., 21, 299–307. [DOI] [PubMed] [Google Scholar]
- 46.Steen S.B., Gomelsky,L., Speidel,S.L. and Roth,D.B. (1997) Initiation of V(D)J recombination in vivo: role of recombination signal sequences in formation of single and paired double-strand breaks. EMBO J., 16, 2656–2664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sadofsky M.J., Hesse,J.E., McBlane,J.F. and Gellert,M. (1993) Expression and V(D)J recombination activity of mutated RAG-1 proteins. Nucleic Acids Res., 21, 5644–5650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sadofsky M.J., Hesse,J.E. and Gellert,M. (1994) Definition of a core region of RAG-2 that is functional in V(D)J recombination. Nucleic Acids Res., 22, 1805–1809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hirt B. (1967) Selective extraction of polyoma DNA from infected mouse cell cultures. J. Mol. Biol., 26, 365–369. [DOI] [PubMed] [Google Scholar]
- 50.Han J.-O., Steen,S.B. and Roth,D.B. (1997) Ku86 is not required for protection of signal ends or for formation of nonstandard V(D)J recombination products. Mol. Cell. Biol., 17, 2226–2234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hesse J.E., Lieber,M.R., Gellert,M. and Mizuuchi,K. (1987) Extrachromosomal DNA substrates in pre-B cells undergo inversion or deletion at immunoglobulin V(D)J joining signals. Cell, 49, 775–783. [DOI] [PubMed] [Google Scholar]
- 52.Chang L.M.S. and Bollum,F.J. (1990) Multiple roles of divalent cation in the terminal deoxynucleotidyltransferase reaction. J. Biol. Chem., 265, 17436–17440. [PubMed] [Google Scholar]
- 53.Lieber M.R., Hesse,J.E., Lewis,S., Bosma,G.C., Rosenberg,N., Mizuuchi,K., Bosma,M.J. and Gellert,M. (1988) The defect in murine severe combined immune deficiency: joining of signal sequences but not coding segments in V(D)J recombination. Cell, 55, 7–16. [DOI] [PubMed] [Google Scholar]
- 54.Carroll A.M., Slack,J.K. and Chang,W.-T. (1993) Biased T-cell receptor δ element recombination in scid thymocytes. Mol. Cell. Biol., 13, 3632–3640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Iwasato T. and Yamagishi,H. (1992) Novel excision products of T cell receptor γ gene rearrangements and developmental stage specificity implied by the frequency of nucleotide insertions at signal joints. Eur. J. Immunol., 22, 101–106. [DOI] [PubMed] [Google Scholar]
- 56.Roth D.B., Chang,X.-B. and Wilson,J.H. (1989) Comparison of filler DNA at immune, nonimmune, and oncogenic rearrangements suggests multiple mechanisms of formation. Mol. Cell. Biol., 9, 3049–3057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lieber M.R. (1992) The mechanism of V(D)J recombination: a balance of diversity, specificity, and stability. Cell, 70, 873–876. [DOI] [PubMed] [Google Scholar]
- 58.Carroll A.M., Slack,J.K. and Mu,X. (1993) V(D)J recombination generates a high frequency of nonstandard TCR Dδ-associated rearrangements in thymocytes. J. Immunol ., 150, 2222–2230. [PubMed] [Google Scholar]
- 59.Lewis S.M., Hesse,J.E., Mizuuchi,K. and Gellert,M. (1988) Novel strand exchanges in V(D)J recombination. Cell, 55, 1099–1107. [DOI] [PubMed] [Google Scholar]
- 60.Elias L., Longmire,J., Wood,A. and Ratliff,R. (1982) Phosphorylation of terminal deoxynucleotidyl transferase in leukemic cells. Biochem. Biophys. Res. Commun., 106, 458–465. [DOI] [PubMed] [Google Scholar]
- 61.Chang L.M.S. and Bollum,F.J. (1982) Cyclic AMP-dependent phosphorylation of terminal deoxynucleotidyl transferase. J. Biol. Chem., 257, 9584–9592. [PubMed] [Google Scholar]
- 62.Chang L.M.S. and Bollum,F.J. (1986) Molecular biology of terminal transferase. Crit. Rev. Biochem ., 21, 27–52. [DOI] [PubMed] [Google Scholar]
- 63.Bentolila L.A., D’Andon,M.F., Nguyen,Q.T., Martinez,O., Rougeon,F. and Doyen,N. (1995) The two isoforms of mouse terminal deoxynucleotidyl transferase differ in both the ability to add N regions and subcellular localization. EMBO J., 14, 4221–4229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Schlissel M. (1998) Structure of nonhairpin coding-end DNA breaks in cells undergoing V(D)J recombination. Mol. Cell. Biol., 18, 2029–2037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Livak F. and Schatz,D.G. (1997) Identification of V(D)J recombination coding end intermediates in normal thymocytes. J. Mol. Biol., 267, 1–9. [DOI] [PubMed] [Google Scholar]
- 66.Mickelsen S., Snyder,C., Trujillo,K., Roth,D.B. and Meek,K. (1999) Modulation of terminal deoxynucleotidyl transferase activity by the DNA dependent protein kinase. J. Immunol ., 163, 834–843. [PubMed] [Google Scholar]
- 67.Tsukamoto Y., Kato,J. and Ikeda,H. (1997) Silencing factors participate in DNA repair and recombination in Saccharomyces cerevisiae. Nature, 388, 900–903. [DOI] [PubMed] [Google Scholar]
- 68.Pandey V.N., Dave,V.P., Patil,M.S., Pradhan,D.S., Amrute,S.B. and Modak,M.J. (1990) Terminal deoxynucleotidyl transferase containing megadalton complex from young rat thymus nuclei: identification and characterization. Biochemistry, 29, 4037–4041. [DOI] [PubMed] [Google Scholar]
- 69.Han J.-O., Erskine,L.A., Purugganan,M.M., Stamato,T.D. and Roth,D.B. (1998) V(D)J recombination intermediates and non-standard products in XRCC4-deficient cells. Nucleic Acids Res., 26, 3769–3775. [DOI] [PMC free article] [PubMed] [Google Scholar]