Abstract
casanova (cas) mutant zebrafish embryos lack endoderm and develop cardia bifida. In a substractive screen for Nodal-responsive genes, we isolated an HMG box-containing gene, 10J3, which is expressed in the endoderm. The cas phenotype is rescued by overexpression of 10J3and can be mimicked by 10J3-directed morpholinos. Furthermore, we identified a mutation within 10J3 coding sequence that cosegregates with the cas phenotype, clearly demonstrating that cas is encoded by 10J3. Epistasis experiments are consistent with an instructive role for cas in endoderm formation downstream of Nodal signals and upstream of sox17. In the absence of cas activity, endoderm progenitors differentiate into mesodermal derivatives. Thus, cas is an HMG box-containing gene involved in the fate decision between endoderm and mesoderm that acts downstream of Nodal signals.
Keywords: casanova, endoderm, nodal, cloning, zebrafish, sox
The endoderm germ layer generates the structures of the digestive and respiratory tracts. In addition, endoderm is crucial in the organization and/or induction of neighboring tissues, such as the head and the heart (Grapin-Botton and Melton 2000). In the zebrafish, endoderm derives from cells positioned at the blastoderm margin of the late blastula (Warga and Nusslein-Volhard 1999). Although endoderm and mesoderm progenitors partially overlap, most mesoderm progenitors come from positions relatively far away from the very margin at this stage.
The molecular pathway leading to endoderm formation is only partially understood. Specification of endoderm requires Nodal signaling (Kimelman and Griffin 2000). Zebrafish mutants lacking the Nodal-related factors Squint (Sqt) and Cyclops (Cyc) fail to form endoderm (Feldman et al. 1998; Sampath et al. 1998). Similarly, endoderm does not form in embryos defective in both maternal and zygotic components of one-eyed pinhead (MZoep), which encodes an EGF-CFC protein required for cells to respond to Nodal signals (Schier et al. 1997; Strähle et al. 1997; Zhang et al. 1998; Alexander and Stainier 1999; Gritsman et al. 1999). In zebrafish, Nodals induce endoderm presumably via activation of the type I TGFβ receptor TARAM-A (Tar; Renucci et al. 1996; Peyrieras et al. 1998), the mix-like homeobox transcription factor MIXER (bonnie and clyde, bon; Kikuchi et al. 2000), and the zinc-finger transcription factor GATA5 (faust; Reiter et al. 1999, 2001). Both transcription factors require a third gene, casanova(cas), to efficiently induce the endoderm-specific sox17 gene (Alexander and Stainier 1999) and to allow marginal cells to achieve the proper endodermal program. At gastrula stages, cas mutant embryos express sox17 neither in endoderm precursors nor in the forerunner cells, a small group of noninvoluting mesendodermal cells at the dorsal margin (Melby et al. 1996). At later stages, cas mutants lack a gut tube and develop a heart condition known as cardia bifida. cas activity is required cell-autonomously for endoderm development and endodermal expression of foxA2 (Alexander and Stainier 1999; Alexander et al. 1999). Thus, cas acts within endoderm precursors, downstream of the Nodal signals Cyc and Sqt and the transcription regulators MIXER and GATA5 but upstream of the transcription factors FoxA2 (Axial/Hnf3β) and SOX17.
To understand further the events controlling endoderm formation, it is essential to define, in molecular terms, the nature of the cas gene and its precise epistatic relationship with the other components of the Nodal pathway. Here, we report the molecular identification of cas and show that it encodes an HMG domain-containing SOX factor. Furthermore, we show that, in the absence of cas activity, cells normally fated to endoderm develop into mesodermal structures.
Results and Discussion
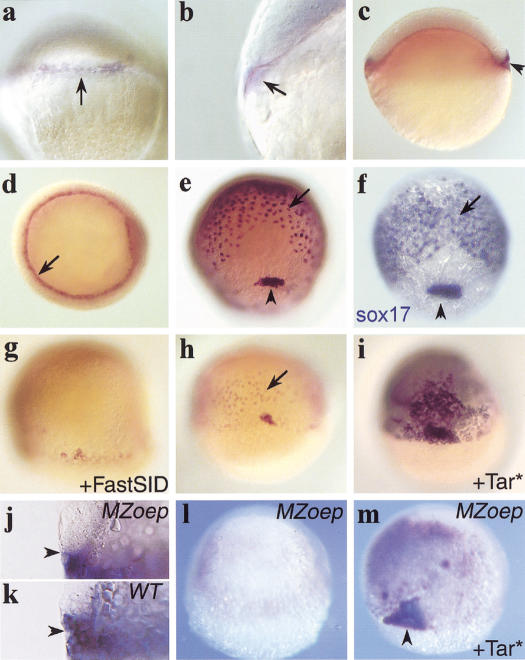
To identify new components that act downstream of Nodal/Tar in the zebrafish embryo, a subtractive screen was carried out for Tar-responsive genes (see Materials and Methods). We identified a gene, 10J3, the expression of which initiates in late blastulae (30% epiboly) at the future dorsal margin of the blastoderm cap (Fig. 1a). Not only the marginal blastomeres but also the underlying yolk syncitial layer (YSL) express 10J3 (Fig. 1b). Expression spreads around the entire margin at subsequent stages (Fig. 1c). At the onset of gastrulation, both the forerunner cells and the endoderm precursor cells, which involute all around the margin and form a population of scattered, flat cells immediately overlying the yolk cell (Warga and Nusslein-Volhard 1999), are positive for the 10J3 transcript. This pattern of expression is maintained until the end of gastrulation (Fig. 1d,e). At subsequent early somitogenesis stages, the lining of Kupfer's vesicle, a tail bud structure derived from the forerunner population, continues to express the 10J3 gene (data not shown). This pattern of expression is highly reminiscent of that of zebrafish sox17 (Fig.1f; Alexander and Stainier 1999) and suggests that the 10J3 gene could play a regulatory role in endoderm development.
Figure 1.
Expression of 10J3, a Nodal-regulated gene in endoderm and forerunner cells and regulation by Nodal signaling. Expression of 10J3 (a–e) and sox17 (f) during early embryogenesis. (a,e,f) Dorsal view, animal pole up; (b) lateral view; (c) lateral view; dorsal right; (d) animal pole view, optical section at germ ring level; (f) posterior view, dorsal up. Expression starts at 30% epiboly in a sector of marginal blastomeres (a, arrow) and in the underlying YSL (b, arrow). (c) Until the beginning of gastrulation, both expression domains expand around the margin. (Arrowhead) Start of forerunner cell expression. (d) At the shield stage, expression is detected in hypoblastic endoderm precursor cells (arrows) and in the forerunner cells (not visible). (e) Expression in the endodermal precursors (arrows) and forerunner cells (arrowhead) continues during gastrulation. During the whole gastrulation period, 10J3 and sox17 expression domains within the blastoderm are very similar. (e,f) 70%–80% epiboly. (g–i) 10J3 expression is regulated by the nodal pathway. Embryos are at 60% epiboly. Shown is a dorsal view with the animal pole up. (g) In embryos injected with FastSID mRNA (400 pg), blastodermal 10J3 expression is strongly reduced. (h) Uninjected control embryo. (Arrow) Endodermal precursors. (i) In embryos, injection of Tar* receptor mRNA (1.2 pg), up-regulates and expands cas expression. (j–m) Expression of 10J3 in MZoep embryos. (j,k) Optical section through the marginal region at 30% epiboly. (l,m) Dorsal view at 70% epiboly. (j) 10J3 is expressed only in the YSL in MZoep embryos. (Arrowhead) The limit between the blastoderm and the YSL. (k) In wild-type embryos, expression in marginal blastomeres is observed. (l) At 70% epiboly, cas expression is lost in MZoep embryos, but can be restored ectopically by overexpression of Tar* (m).
The pattern of expression of the 10J3 gene is consistent with a role downstream of Nodal signals. To verify its dependence with regard to Nodal signals, we expressed the dominant-negative inhibitor of Nodal signaling, FAST-1SID in zebrafish embryos (Muller et al. 2000). As a consequence, 10J3 was significantly down-regulated in injected embryos (Fig. 1g) as compared with controls (Fig. 1h). Also, the 10J3 gene was not expressed during gastrulation in the blastoderm of MZoep or sqt/cyc mutants, which do not form endoderm or forerunner cells (Fig. 1l and data not shown). Conversely, to test the responsiveness of 10J3 to Nodal/Tar, we expressed the constitutively active variant of the receptor Tar* in zebrafish embryos (Renucci et al. 1996; Peyrieras et al. 1998). This led to ectopic up-regulation of the 10J3 gene in wild-type embryos (Fig. 1h,i) and strongly induced 10J3 expression in MZoep embryos in a large population of deep flat endoderm-like cells and in a small group of superficial cells highly reminiscent of the forerunner cluster (Fig. 1m; Peyrieras et al. 1998). Expression in the YSL, however, was not affected by lack or activation of Nodal signaling (Fig. 1j,k and data not shown). Thus, the expression of the 10J3 gene requires Nodal signals for expression in the endoderm precursors and forerunner cells but not in the YSL.
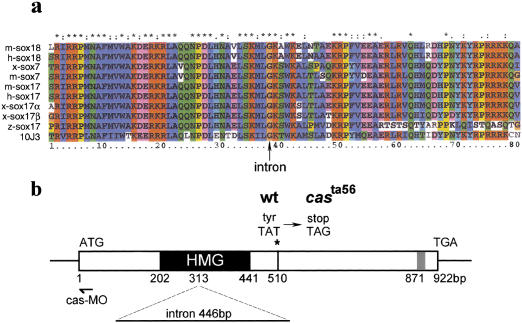
Sequencing of the 10J3 cDNA revealed that it encodes a 307-amino-acid protein belonging to the HMG-domain–containing family of SOX transcription factors (Fig. 2a). Phylogenetic analysis with different tree-building methods shows that it forms a novel member of the F subfamily of sox genes, which includes sox17, sox7, and sox18 (Fig. 2a; Bowles et al. 2000). The 10J3 gene also shares the exon–intron structure with other members of this subfamily (Wegner 1999; Bowles et al. 2000); a single intron, located at the glycine residue of the consensus sequence K(M/I)LGK in members of the F subfamily (Fig. 2a) separates the HMG domain-encoding exons (Fig. 2b; Bowles et al. 2000). Regions outside the HMG box are not strongly conserved with the exception of a short stretch located near the carboxyl terminus with the consensus sequence EF(D/E)QY. This short domain conserved between SOX7, SOX17, SOX18, and CAS resides within the roughly mapped limits of the activation domain of mouse Sox17 (Kanai et al. 1996).
Figure 2.
10J3 encodes an HMG box transcription factor. (a) Comparison of the HMG box regions of several vertebrate Sox transcription factors. 10J3 and zebrafish (z) sox17 encode members of the F subfamily. Similar types of residues are color coded. (*) Residues conserved completely in the alignment (see Materials and Methods). (b) Schematic representation of the 10J3 gene structure. The position of a conserved intron in the HMG box, the casta56 mutation and the morpholino oligonucleotide hybridization region (cas-MO) are indicated. GenBank accession no.: 10J3/cas, AY027650; mouse (m)-sox18, AAG48578, human (h)-sox18, AB033888; Xenopus (x)-sox7, D83649; mouse (m)-sox7, AB023419; putative human (h)-sox17, AK025905; mouse (m)-sox17, D49474; Xenopus (x)-sox17α, AJ001730; x-sox17β, AJ001742; and zebrafish (z)-sox17, AF168614.
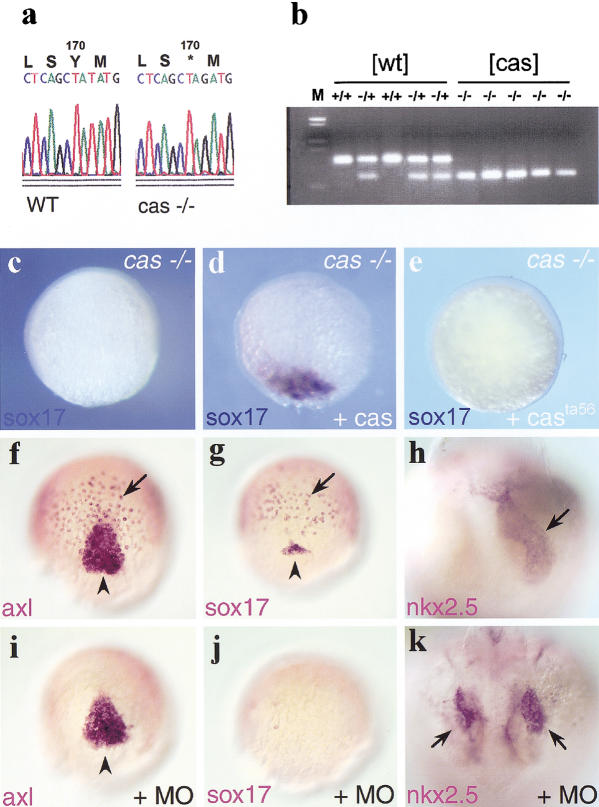
cas mutants lack endoderm and forerunner cells (Alexander et al. 1999). Expression of 10J3 within these territories prompted us to test whether this gene could be identical to cas. Genomic fragments were amplified from wild-type and cas embryos with 10J3-specific primers and sequenced. cas mutants were found to harbor a point mutation downstream of the 10J3 HMG-box, leading to premature termination of the 10J3-encoded protein (Figs. 2b and 3a). This point mutation cosegregates with the cas mutant phenotype (Fig. 3b), demonstrating that 10J3 and cas are tightly linked. Expression of the wild-type but not the mutant 10J3 protein rescued the expression of the endodermal marker sox17 in cas embryos (Fig. 3c–e). Furthermore, injection of antisense morpholino oligonucleotides specifically directed against 10J3 (cas-MO, Fig. 2b) phenocopied the cas mutant phenotype: (1) injected embryos lacked cells expressing foxA2 (axial/HNF3β; Strähle et al. 1996; Fig. 3f,i) whereas foxA2 expression was unaffected in the midline of the body axis; (2) sox17 expression (Alexander and Stainier 1999) was completely abolished by injection of cas-MO (Fig. 3g,j); and (3) injected embryos developed cardia bifida at later stages (Fig. 3h,k). In contrast, injection of control morpholinos of distinct sequence did not induce these phenotypes (data not shown). Taken together, these knockdown experiments confirmed that the 10J3 HMG box gene is identical to cas. The phenotype of the casta56allele is likely due to the deletion of the carboxyl terminus of the 10J3 protein, a region that was shown to contain a transcriptional activation domain, including the EF(D/E)QY conserved sequence, and other protein interaction domains in related HMG proteins (van de Wetering et al. 1993; Hosking et al. 1995; Kanai et al. 1996; Sudbeck et al. 1996; Zorn et al. 1999). Below, we will refer to 10J3 as the cas gene.
Figure 3.
CAS is encoded by the 10J3 gene. (a) Sequencing of casta56 and sibling genomic DNA reveals a T-to-G mutation at position 510 that leads to an amber stop codon instead of a tyrosine. (b) Segregation analysis with a CAPS marker reveals cosegregation of the mutation in the homozygous state with cas mutant embryos. Cosegregation was observed in all 160 cas embryos tested. Representative restriction patterns of five siblings with a wild-type phenotype and five siblings with a cas phenotype are shown. (c–e) 10J3 mRNA rescues sox17 expression in cas mutants. Embryos are at 60% epiboly. Shown is a dorsal view with the animal pole up. (e) Uninjected cas embryo. (d) cas embryo injected with 100 pg of wild-type 10J3 mRNA. (e) cas embryo injected with 200 pg of mutant cas mRNA. (f–k) Injection of 1 pmole of a morpholino antisense oligonucleotide against 10J3 (cas-MO) results in loss of endoderm and forerunner cells and cardia bifida. (f,g,i,j) Dorsal view, 60% epiboly. (h,k) Dorsal view of heart region, anterior down, about 1.5 d of development. (f,i) Expression of foxA2/axial in cas-MO injected embryos (i) and uninjected wild-type embryos (f). The endodermal expression domain of foxA2/axial (arrow) is missing in the cas-MO injected embryos, whereas the domain representing the nascent axis remains (arrowhead). (g,j) Expression of sox17 in cas-MO injected embryos (j) and uninjected wild-type embryos (g). Expression in the endoderm (arrow) and forerunner cells (arrowhead) is strongly reduced or absent in cas-MO injected embryos. (h,k) cas-MO injected embryos exhibit cardia bifida. nkx2.5 staining reveals two separated heart tubes (arrows) in the cas-MO injected embryos (k), whereas the wild-type control shows a single heart tube (h, arrow).
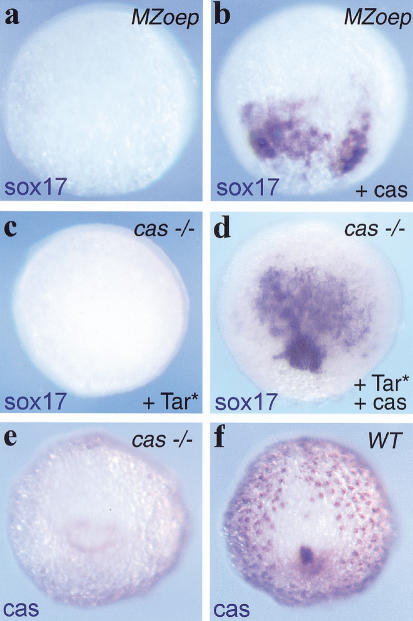
Although previous epistatic analyses have shown that cas is a component of the Nodal pathway, essential for endoderm formation, the way it acts within this pathway is not clear. In particular, we wished to understand whether cas acts in a permissive or an instructive manner downstream of Nodal signals. MZoep embryos, which lack both maternal and zygotic oep, are deficient in Nodal signaling as cells cannot respond to Nodal signals in the absence of Oep (Gritsman et al. 1999). As a consequence, these embryos fail to activate endodermal marker genes (Strähle et al. 1997; Schier and Talbot 1998; Alexander and Stainier 1999). To test whether cas can by-pass the requirement of Nodal signaling for endoderm formation, we injected cas mRNA into MZoep embryos. casmRNA led to a strong ectopic activation of sox17 (Fig. 4a,b) and gata5 (data not shown) in MZoep embryos. Some sox17-positive cells adopted a deep position, suggesting they had involuted (data not shown). Later endoderm differentiation markers were not rescued in these experiments, suggesting that cas activity or the activity of other genes might be required during postgastrulation stages to allow proper endoderm differentiation in Nodal-deficient mutants. In contrast to the induction of early endodermal markers, cas mRNA did not induce the mesendodermal marker goosecoid (data not shown), showing that the effect of cas is restricted to the induction of endoderm. Furthermore, cas plays a central downstream role in the transduction of Nodal signals, as Tar*-activated cells are unable to activate the downstream endodermal gene sox17 in cas embryos (Fig. 4c; Alexander and Stainier 1999). As expected, expression of cas within Tar*-activated cells restored the capacity of these cells to express sox17 (Fig. 4d). Altogether, these results are in agreement with previous epistasis experiments (Alexander and Stainier 1999; Reiter et al. 2001) but also demonstrate that cas acts in an instructive manner with regard to endoderm formation as it can induce endodermal markers in the absence of Nodal signals.
Figure 4.
cas acts in an instructive manner downstream of nodal signals during gastrulation. (a–d) CAS acts in an instructive manner within the Nodal pathway during endoderm induction. Sox17 expression at 60% epiboly, dorsal view. (a,b) Injection of MZoep mutant embryos with 100 pg of cas mRNA restores sox17 expression (b), which is missing in the uninjected control MZoep mutant embryo (a). (c,d) Coinjection of 1.2 pg of Tar* and 40 pg of cas mRNA restores the endoderm inducing activities of TAR* in the cas mutant background (d). No sox17 expression can be detected in control cas mutant embryos injected with 1.2 pg of Tar* RNA only. (e,f) cas regulates its own expression from the beginning of gastrulation onward. Embryos are at the bud stage. Shown is a posterior view with the dorsal side up. (e) Blastodermal expression is lacking in cas mutants. (f) Sibling control. The genotype of each embryo is indicated in the upper right corner of each panel.
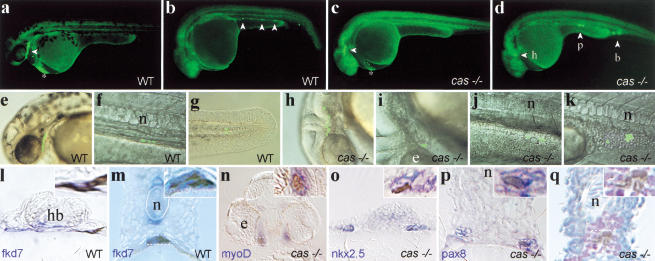
To understand how the absence of cas activity affects the behavior and fate of marginal cells that express cas and that are normally fated to endoderm, we first assessed the expression of cas mRNA in cas mutants. cas mutant embryos express cas mRNA in a pattern similar to wild-type embryos at the onset of gastrulation (data not shown). At the end of gastrulation (bud stage), cas mRNA was no longer detectable in the blastoderm of mutant embryos (Fig. 4e,f). However expression was still evident in the YSL at the bud stage. Thus, cas expression is initiated normally in cas mutants. Lack of cas blastodermal expression at bud stages could be due to specific loss of the cas-expressing cell population in the mutant during gastrulation; however, increased cell death was not apparent. Alternatively, cas mutant cells may take up another cell fate. To investigate the fate acquired by endodermal progenitors in cas embryos, fate mapping experiments were carried out by injection of the photoactivatable, fluorescent tracer Fluorescein-Dextran (FD) and uncaging of the dye at the 40% epiboly stage in a small group (1 to 5) of blastomeres located immediately at the blastoderm margin (Bally-Cuif et al. 2000). Consistent with published results (Warga and Nusslein-Volhard 1999), we found that most labeled blastomeres in wild-type or cas embryos involuted at the onset of gastrulation and migrated within the inner hypoblast germ layer as expected from marginal cells. Around 30 h, clones of labeled cells from wild-type embryos contributed predominantly to endodermal derivatives (45% endodermal cells, 43 % mesendodermal cells, 12% mesodermal cells, n = 361 cells in 53 wild-type embryos), such as the pharyngeal endoderm (Fig. 5a,e,l), the gut (Fig. 5b,f,m) as well as head (hatching gland; Fig. 5a, *) and tail mesendodermal derivatives (Fig. 5g), but infrequently to mesodermal structures. In contrast, labeled clones developed mostly into mesodermal derivatives in cas mutant embryos (0% endodermal cells, 38% mesendodermal cells, 62% mesodermal cells, n = 269 cells in 31 cas mutant embryos), such as head mesenchymal cells (Fig. 5c,h,n), the heart primordia (Fig. 5d,i,o), the pronephros (Fig. 5d,j,p), blood (Fig. 5d,k,q), and somites (data not shown). cas mutant cells also contributed to the hatching gland, which is present in cas embryos. We conclude that, in the absence of a functional cas gene, the marginalmost endodermal progenitors are respecified to a mesodermal/mesendodermal fate and that the function of cas in this process is to ensure the proper allocation of endodermal progenitors to endoderm-derived tissues.
Figure 5.
Marginalmost blastomeres are transfated to mesoderm in cas embryos. One (b,d) or five (a,c) blastomeres of the most marginal row were labeled at 40% epiboly by uncaging the photoactivatable FD. At a later stage (30–36 h), embryos were photographed under low magnification epifluorescence (a–d, whole-mount side views, anterior to the left) or high magnification (e–k) to visualize cells harboring uncaged FD. Higher magnification views were superimposed on Nomarski pictures (e–k). (Open arrowheads) Fluorescent cells or groups of cells. (a,b,e–g) Fate acquired by marginalmost precursors in wild-type embryos. Progenitors contributed cells to the pharyngeal endoderm in the head (a,e) and the gut in the trunk (b,f), in addition to the hatching gland (a, *) and tail mesendoderm (g; see Bally-Cuif et al. 2000) as well as occasional mesoderm cells (blood in b). (c,d,h–k) Principal fates acquired by marginalmost precursors in cas embryos. Progenitors contributed mostly to head mesenchymal cells (c,h), myocardial cells (d,i), trunk pronephric duct cells (d,j), and blood (d,k) as well as hatching gland cells (c, *). (l–q) Then, embryos were fixed, stained with antifluorescein antibodies to reveal uncaged FD (brown/black) and by in situ hybridization with tissue specific markers (blue/purple) and analyzed by cross-section. A close-up view is provided in the upper right corner of each section and corresponds to the region included in the dotted rectangle. Wild-type cells are located in the pharyngeal endoderm (l) and the gut (m) and expressfkd7 (l,m). fkd7 is also expressed in the hypochord and the ventral brain. cas cells are located in mesoderm-derived tissues including head muscles (n), the heart primordia (o), the pronephric duct (p), and blood (q). These tissues are revealed by the markers indicated in the lower left corner, except for in q, in which blood is revealed by counterstaining with hematoxylin and eosin. (b) Blood; (e) eye; (h) heart; (hb) hindbrain; (n) notochord; (p) pronephros.
Endodermal progenitors appear to be initially specified in the casmutant. Indeed, cas embryos normally express the early endodermal markers gata5 (Reiter et al. 1999, 2001), mixer (bon) (Kikuchi et al. 2000), her5 (Bally-Cuif et al. 2000), and cas itself before the onset of gastrulation, indicating that the field from which endodermal progenitors originate has been properly defined in the mutant. However, cas embryos do not express the late endoderm specification markers sox17 and foxA2 in deep, nonaxial, cells (Alexander and Stainier 1999; Alexander et al. 1999), suggesting that cells from the endodermal field are unable to acquire a proper endodermal fate in subsequent stages. cas might control the capacity of cells to involute during gastrulation and reach the blastoderm, as is observed in Nodal mutants (Feldman et al. 2000). This hypothesis is not consistent with our observation that marginal cells do involute in cas mutant embryos. On the contrary, the fact that marginal cas cells normally fated to endoderm adopt a mesodermal fate, combined with the fact that cas induces endodermal markers in Nodal-deficient mutants strongly supports the idea that the function of cas is to specify the endodermal identity of marginal cells downstream of Nodal signals.
Materials and methods
Fish stocks and embryo production
Mutant alleles used were: casz321, a mutation identified among the progeny of swirl heterozygous fish obtained from the Tuebingen stock center. This allele proved identical to the casta56 allele and was renamed casta56 (Chen et al. 1996) and oepm134(Schier et al. 1996). MZoep was generated as described previously (Gritsman et al. 1999).
In situ analysis
In situ hybridization was performed as described (Hauptmann and Gerster 1994).
Cloning of cas and mapping of the mutation
The 10J3 cDNA was identified in a subtractive screen for TARAM-A inducible genes in zebrafish gastrula. A partial cDNA (MPMGP637J0310) was identified on a macroarray of a shield stage library (RZPD library no. 637; Clark et al. 1999). Details of the screen will be published elsewhere. A gastrula-stage cDNA library (provided by T. Lepage) was screened with the 10J3 probe to obtain full-length cDNAs, which were confirmed by RACE on mRNA and analysis of genomic DNA. A stop codon resides 42 bp upstream of the first ATG, indicating that the isolated cDNA clone most likely contains the entire open reading frame.
Alignments and phylogenetic tree building were carried out by use of the CLUSTAL X program (Thompson et al. 1997).
To identify the mutation in the casta56 allele, genomic DNA from mutants was amplified by PCR and sequenced. Cosegregation of the point mutation with the cas mutant phenotype was monitored by use of a CAPS marker (codominant cleavable amplified polymorphic sequences): A reverse oligonucleotide carries a point mutation generating a BglII site in conjunction with the point mutation in casta56 but not with the wild-type cas allele. By use of a forward oligonucleotide, a 112-bp fragment is amplified irrespective of the genotype, which is not cleaved in the wild-type allele but is cleaved into 82- and 30-bp fragments in the mutant allele.
Oligonucleotide sequences were as follows: reverse (mutated) primer, 5′-GGGCCGCTGAGGGGCTTG ACCTTGAAAGAT-3′ and forward primer, 5′-ACGAAAGTGCAACAAGCGGTGCAGCAAGATG-3′.
Microinjection and cell fate determination
The cas cDNA was subcloned into pCS2 (Turner and Weintraub 1994). The casta56 mutation was introduced into this plasmid. Synthesis of capped mRNA and microinjection into zebrafish early embryos were as described (Peyrieras et al. 1998).
Morpholinos (Nasevicius and Ekker 2000) purchased from GeneTools (Corvalis, USA) were as follows: cas-MO, 5′-CAGGGAGCATCCGGTC GAGATACAT-3′ and control-MO, 5′-CCTCTTACCTCAGTTACAAT TTATA-3′.
These oligonucleotides were diluted in H2O to a concentration of 0.5 mM and mixed 2 : 1 with phenol red prior to injection.
To map the fate of cells at the blastoderm margin, embryos were injected with caged-Fluorescein Dextran (FD, 20 ng, Molecular Probes). At 40% epiboly, FD was uncaged with a microbeam laser (photonics instruments) in one cell at the blastoderm margin. Embryos were doubly stained by immunohistochemistry with antifluorescein antibody coupled to peroxidase (Feldman 2000) and by in situ hybridization with the relevant probes (Hauptmann 1994). References for the probes can be obtained at http://www.zfish.uoregon.edu/cgibin/ZFINjump?record=JUMPTOGENE.
Acknowledgments
We thank T. Lepage, D. Stainier, A. Schier, and the Tuebingen stock centre for plasmids and fish strains, L. Bally-Cuif for a critical reading of this manuscript and D. Biellmann, A. Karmin, O. Nkundwa, F. Chelgoum, and F. Bouallague for fish care. This work was supported by a Boehringer Ingelheim fellowship to T.D. and a LNCC grant to P.M. We are also grateful to AFM, ARC, and ACI.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL rosa@wotan.ens.fr; FAX 33-01-44-32-39-88.
Article and publication are at http://www.genesdev.org/cgi/doi/10.1101/gad.196901.
References
- Alexander J, Stainier DY. A molecular pathway leading to endoderm formation in zebrafish. Curr Biol. 1999;9:1147–1157. doi: 10.1016/S0960-9822(00)80016-0. [DOI] [PubMed] [Google Scholar]
- Alexander J, Rothenberg M, Henry GL, Stainier DY. casanova plays an early and essential role in zebrafish. Dev Biol. 1999;215:343–357. doi: 10.1006/dbio.1999.9441. [DOI] [PubMed] [Google Scholar]
- Bally-Cuif L, Goutel C, Wassef M, Wurst W, Rosa F. Coregulation of anterior and posterior mesendodermal development by a hairy-related transcriptional repressor. Genes & Dev. 2000;14:1664–1677. [PMC free article] [PubMed] [Google Scholar]
- Bowles J, Schepers G, Koopman P. Phylogeny of the SOX family of developmental transcription factors based on sequence and structural indicators. Dev Biol. 2000;227:239–255. doi: 10.1006/dbio.2000.9883. [DOI] [PubMed] [Google Scholar]
- Chen JN, Haffter P, Odenthal J, Vogelsang E, Brand M, van Eeden FJ, Furutani-Seiki M, Granato M, Hammerschmidt M, Heisenberg CP, et al. Mutations affecting the cardiovascular system and other internal organs in zebrafish. Development. 1996;123:293–302. doi: 10.1242/dev.123.1.293. [DOI] [PubMed] [Google Scholar]
- Clark MD, Panopoulou GD, Cahill DJ, Büssow K, Lehrach H. Construction and analysis of cDNA libraries. Methods Enzymol. 1999;303:205–233. doi: 10.1016/s0076-6879(99)03015-3. [DOI] [PubMed] [Google Scholar]
- Feldman B, Gates MA, Egan ES, Dougan ST, Rennebeck G, Sirotkin HI, Schier AF, Talbot WS. Zebrafish organizer development and germ-layer formation require nodal-related signals. Nature. 1998;395:181–185. doi: 10.1038/26013. [DOI] [PubMed] [Google Scholar]
- Feldman B, Dougan ST, Schier AF, Talbot WS. Nodal-related signals establish mesendodermal fate and trunk neural identity in zebrafish. Curr Biol. 2000;10:531–534. doi: 10.1016/s0960-9822(00)00469-3. [DOI] [PubMed] [Google Scholar]
- Grapin-Botton A, Melton DA. Endoderm development: From patterning to organogenesis. Trends Genet. 2000;16:124–130. doi: 10.1016/s0168-9525(99)01957-5. [DOI] [PubMed] [Google Scholar]
- Gritsman K, Zhang J, Cheng S, Heckscher E, Talbot WS, Schier AF. The EGF-CFC protein one-eyed pinhead is essential for nodal signaling. Cell. 1999;97:121–132. doi: 10.1016/s0092-8674(00)80720-5. [DOI] [PubMed] [Google Scholar]
- Hauptmann G, Gerster T. Two-color whole-mount in situ hybridization to vertebrate and Drosophila embryos. Trends Genet. 1994;10:266. doi: 10.1016/0168-9525(90)90008-t. [DOI] [PubMed] [Google Scholar]
- Hosking BM, Muscat GE, Koopman PA, Dowhan DH, Dunn TL. Trans-activation aned DNA-binding properties of the transcription factor, Sox-18. Nucleic Acids Res. 1995;23:2626–2628. doi: 10.1093/nar/23.14.2626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanai Y, Kanai-Azuma M, Noce T, Saido TC, Shiroishi T, Hayashi Y, Yazaki K. Identification of two Sox17 messenger RNA isoforms, with and without the high mobility group box region, and their differential expression in mouse spermatogenesis. J Cell Biol. 1996;133:667–681. doi: 10.1083/jcb.133.3.667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kikuchi Y, Trinh LA, Teiter JF, Alexander J, Yelon D, Stainier DY. The zebrafish bonnie and clyde gene encodes a Mix family homeodomain protein that regulates the generation of endodermal precursors. Genes & Dev. 2000;14:1279–1289. [PMC free article] [PubMed] [Google Scholar]
- Kimelman D, Griffin KJ. Vertebrate mesendoderm induction and patterning. Curr Opin Genet Dev. 2000;10:350–356. doi: 10.1016/s0959-437x(00)00095-2. [DOI] [PubMed] [Google Scholar]
- Melby AF, Warga RM, Kimmel CB. Specification of cell fates at the dorsal margin of the zebrafish gastrula. Development. 1996;122:2225–2237. doi: 10.1242/dev.122.7.2225. [DOI] [PubMed] [Google Scholar]
- Muller G, Albert S, Blader P, Fischer N, Hallonet M, Strahle U. Direct action of the nodal-related signal cyclops in induction of sonic hedgehog in the ventral midline of the CNS. Development. 2000;127:3889–3897. doi: 10.1242/dev.127.18.3889. [DOI] [PubMed] [Google Scholar]
- Nasevicius A, Ekker SC. Effective targeted gene ‘knockdown’ in zebrafish. Nature Genet. 2000;26:216–220. doi: 10.1038/79951. [DOI] [PubMed] [Google Scholar]
- Peyrieras N, Strahle U, Rosa F. Conversion of zebrafish blastomeres to an endodermal fate by TGF-beta-related signaling. Curr Biol. 1998;8:783–786. doi: 10.1016/s0960-9822(98)70303-3. [DOI] [PubMed] [Google Scholar]
- Reiter JF, Alexander J, Rodaway A, Yelon D, Patient R, Holder N, Stainier DY. Gata5 is required for the development of the heart and endoderm in zebrafish. Genes & Dev. 1999;13:2983–2995. doi: 10.1101/gad.13.22.2983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reiter JF, Kikuchi Y, Stainier DY. Multiple roles for Gata5 in zebrafish endoderm formation. Development. 2001;128:125–135. doi: 10.1242/dev.128.1.125. [DOI] [PubMed] [Google Scholar]
- Renucci A, Lemarchandel V, Rosa F. An activated form of type I serine/threonine kinase receptor TARAM-A reveals a specific signalling pathway involved in fish head organiser formation. Development. 1996;122:3735–3743. doi: 10.1242/dev.122.12.3735. [DOI] [PubMed] [Google Scholar]
- Sampath K, Rubinstein AL, Cheng AM, Liang JO, Fekany K, Solnica-Krezel L, Korzh V, Halpern ME, Wright CV. Induction of the zebrafish ventral brain and floorplate requires cyclops/nodal signalling. Nature. 1998;395:185–189. doi: 10.1038/26020. [DOI] [PubMed] [Google Scholar]
- Schier AF, Talbot WS. The zebrafish organizer. Curr Opin Genet Dev. 1998;8:464–471. doi: 10.1016/s0959-437x(98)80119-6. [DOI] [PubMed] [Google Scholar]
- Schier AF, Neuhauss SC, Harvey M, Malicki J, Solnica-Krezel L, Stainier DY, Zwartkruis F, Abdelilah S, Stemple DL, Rangini Z, et al. Mutations affecting the development of the embryonic zebrafish brain. Development. 1996;123:165–178. doi: 10.1242/dev.123.1.165. [DOI] [PubMed] [Google Scholar]
- Schier AF, Neuhauss SC, Helde KA, Talbot WS, Driever W. The one-eyed pinhead gene functions in mesoderm and endoderm formation in zebrafish and interacts with no tail. Development. 1997;124:327–342. doi: 10.1242/dev.124.2.327. [DOI] [PubMed] [Google Scholar]
- Strähle U, Blader P, Ingham PW. Expression of axial and sonic hedgehog in wildtype and midline defective zebrafish embryos. Int J Dev Biol. 1996;40:929–940. [PubMed] [Google Scholar]
- Strähle U, Jesuthasan S, Blader P, Garcia-Vilalba P, Hatta K, Ingham P. one-eyed pinhead is required for floor plate development in the zebrafish embryo. Genes & Function. 1997;1:131–148. doi: 10.1046/j.1365-4624.1997.00010.x. [DOI] [PubMed] [Google Scholar]
- Sudbeck P, Schmitz ML, Baeuerle PA, Scherer G. Sex reversal by loss of the C-terminal transactivation domain of human SOX9. Nature Genet. 1996;13:230–232. doi: 10.1038/ng0696-230. [DOI] [PubMed] [Google Scholar]
- Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG. The CLUSTAL X windows interface: Flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 1997;25:4876–4882. doi: 10.1093/nar/25.24.4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner DL, Weintraub H. Expression of achaete-scute homolog 3 in Xenopusembryos converts ectodermal cells to a neural fate. Genes & Dev. 1994;8:1434–1447. doi: 10.1101/gad.8.12.1434. [DOI] [PubMed] [Google Scholar]
- van de Wetering M, Oosterwegel M, van Norren K, Clevers H. Sox-4, an Sry-like HMG box protein, is a transcriptional activator in lymphocytes. EMBO J. 1993;12:3847–3854. doi: 10.1002/j.1460-2075.1993.tb06063.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warga RM, Nusslein-Volhard C. Origin and development of the zebrafish endoderm. Development. 1999;126:827–838. doi: 10.1242/dev.126.4.827. [DOI] [PubMed] [Google Scholar]
- Wegner M. From head to toes: The multiple facets of Sox proteins. Nucleic Acids Res. 1999;27:1409–1420. doi: 10.1093/nar/27.6.1409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Talbot WS, Schier AF. Positional cloning identifies zebrafish one-eyed pinhead as a permissive EGF-related ligand required during gastrulation. Cell. 1998;92:241–251. doi: 10.1016/s0092-8674(00)80918-6. [DOI] [PubMed] [Google Scholar]
- Zorn AM, Barish GD, Williams BO, Lavender P, Klymkowsky MW, Varmus HE. Regulation of Wnt signaling by Sox proteins: XSox17 alpha/beta and XSox3 physically interact with beta-catenin. Mol Cell. 1999;4:487–498. doi: 10.1016/s1097-2765(00)80200-2. [DOI] [PubMed] [Google Scholar]