Abstract
We investigated the cell-death mechanisms induced in esophageal cancer cells in response to the chemotherapeutic drugs, 5-fluorouracil (5-FU) and cisplatin. Chemosensitive cell lines exhibited apoptosis whereas chemoresistant populations exhibited autophagy and a morphology resembling type II programmed cell death (PCD). Cell populations that respond with autophagy are more resistant and will recover following withdrawal of the chemotherapeutic agents. Specific inhibition of early autophagy induction with siRNA targeted to Beclin 1 and ATG7 significantly enhanced the effect of 5-FU and reduced the recovery of drug-treated cells. Pharmacological inhibitors of autophagy were evaluated for their ability to improve chemotherapeutic effect. The PtdIns 3-kinase inhibitor 3-methyladenine did not enhance the cytotoxicity of 5-FU. Disruption of lysosomal activity with bafilomycin A1 or chloroquine caused extensive vesicular accumulation but did not improve chemotherapeutic effect. These observations suggest that an autophagic response to chemotherapy is a survival mechanism that promotes chemoresistance and recovery and that selective inhibition of autophagy regulators has the potential to improve chemotherapeutic regimes. Currently available indirect inhibitors of autophagy are, however, ineffective at modulating chemosensitivity in these esophageal cancer cell lines.
Key words: esophageal cancer, chemoresistance, apoptosis, autophagy, 3-MA, bafilomycin A1, chloroquine
Introduction
Cancers of the esophagogastric region are highly malignant tumors with five-year survival rates of less than 16%.1 Research has shown that 88% of patients, selected for curative resection for esophagogastric cancer, already have disseminated tumor cells,2 that can remain dormant for variable periods, before emerging as aggressive, drug-resistant metastases.3 Improved systemic therapeutic options are therefore required to effectively eliminate primary and recurrent esophageal cancer.
Chemotherapeutic regimes must effectively induce PCD in cancer cells to overcome drug resistance and recurrence. This is a major limitation, as deregulation of cell death programs often plays a role in the development of the cancer in the first place.4 Previously, apoptosis (type I PCD) was regarded as the central mediator of PCD in response to chemotherapeutic agents and is characterized by loss of nuclear membrane, fragmentation of chromatin and cell shrinkage. However, other death programs exist in eukaryotic cells.5,6 Type II PCD is characterized by the formation of vesicles in the cytoplasm, loss of the cytoplasmic material and pyknosis of nuclear material within an intact nuclear membrane.7 Evidence suggests that this morphology is a consequence of excessive autophagy. Several studies have reported autophagic cell death in cultured mammalian cells8–12 and autophagic PCD has been demonstrated during development of Drosophila and Dictyostelium discoideum.13,14
Autophagy is a highly conserved survival response to growth limiting conditions, in which cellular components are sequestered, degraded and released for recycling.15 It is genetically regulated by a family of autophagy-related (ATG) genes, which have homologues in humans (reviewed in ref. 16). The role of autophagy in cancer remains controversial. Constitutive autophagy may be a necessary homeostatic process which removes damaged organelles and recycles macromolecules, thus protecting against cancer.17 However, when a cancer is established autophagy may take on new roles; it may help cancer cells survive in response to growth-limiting conditions such as nutrient depletion, hypoxia, absence of growth factors and the presence of cytotoxic drugs.18–22 The induction of excessive autophagy and type II PCD may also be the major cell death mechanism that takes over when apoptosis is unavailable.23 Autophagic cell death has been reported to be induced by a number of chemotherapeutic agents in mammalian cells.10,24–26 Recent studies suggest that death due to autophagy may be as unobtrusive as apoptosis and may also induce clearance signals that facilitate the removal of the dying cell.27
We investigated cell death programs initiated in esophageal cancer cells in response to the chemotherapeutic agents 5-fluorouracil (5-FU) and cisplatin. Cells that do not respond to these agents with apoptosis undergo autophagy and cell populations can recover when cytotoxic drugs are withdrawn. The ability to recover may explain recurrent disease and may be a major limiting factor in current treatment regimes. Selective inhibition of proteins involved in the formation of autophagosomes can reduce the recovery of cancer cells following cytotoxic drug treatment indicating the importance of autophagy for this recovery. Indirect inhibitors of autophagy are ineffective in improving cytotoxicity and indicate the need for the development of more selective agents.
Results
Cell death induced by 5-fluorouracil and cisplatin in esophageal cancer cells.
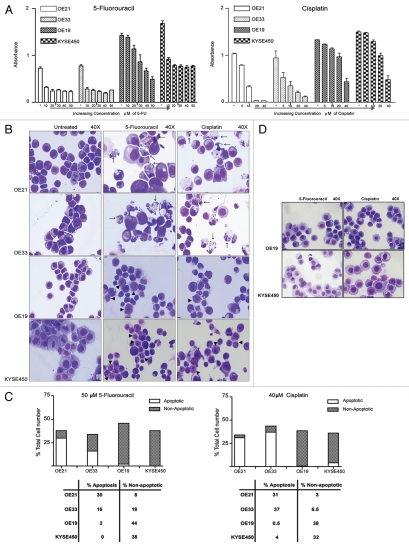
We evaluated a panel of esophageal cancer cell lines, two of squamous (OE21 and KYSE450) and two of adenocarcinoma (OE19 and OE33) backgrounds, for their sensitivity to the chemotherapeutic drugs 5-FU and cisplatin. The OE21 and OE33 cell lines are significantly more sensitive to a range of concentrations of cytotoxic drugs. Cisplatin (10 µM) induced significant effects on MTT reduction in both OE21 and OE33 cells, whereas the OE19 and KYSE450 cell lines were relatively unaffected. The KYSE450 cells were marginally more resistant to 5-FU treatment compared to OE21 and OE33 cell lines, and the OE19 cells were completely resistant to treatment with 10 µM 5-FU and only moderately affected at 20 µM (Fig. 1A).
Figure 1.
Effect of 5-fluorouracil (5-FU) and cisplatin on viability and cell death morphology in esophageal cancer cells. OE21 and KYSE 450 cells were seeded at 1 × 104 cells per cm2, OE33 and OE19 at 2 × 104 cells per cm2 (OE21 and KYSE 450 cell lines have a faster growth rate, therefore were seeded at a lower density) and were treated with a range of 5-FU (10–60 µM) or cisplatin (5–40 µM) concentrations for 48 h. (A) MTT assay was used to determine the sensitivity of each cell line to 5-FU and cisplatin. (B) Morphological features of all four esophageal cell lines (OE21, OE33, OE19 and KYSE 450) treated with 5-FU (50 µM) or cisplatin (40 µM), for 48 h. Morphological features of apoptosis are shown with arrows. Non-apoptotic cell death (shown with arrowheads), is evident in OE19 and KYSE 450 cells and is characterized by pyknosis of the nuclear material and the development of cytoplasmic vesicles (magnification 40×). (C) The extent of apoptotic (clear bars) and non-apoptotic (hatched bars) cell death in each cell line, in response to both 5-FU (50 µM) and cisplatin (40 µM) for 48 h was determined by counting three fields of view per slide, with an average of ∼100 cells per field. (D) Morphological features of OE19 and KYSE 450 cells treated with 5-FU (50 µM) and cisplatin (40 µM) for 24 h (magnification 40×). Note early formation of cytoplasmic vesicles.
The more drug sensitive esophageal cancer cell lines (OE21 and OE33) induced a predominantly apoptotic cell death morphology (type I PCD), in response to both 5-FU and cisplatin (arrows, Fig. 1B), with low levels of non-apoptotic cell death morphology. In contrast, the more drug resistant OE19 and KYSE450 cell lines displayed predominantly non-apoptotic morphology (arrowheads, Fig. 1B). OE21 cells displayed 31% apoptotic and 3% non-apoptotic cell death, whereas OE19 cells showed only 0.5% apoptosis and 38% non-apoptotic cell death, in response to cisplatin (40 µM) (Fig. 1C). This non-apoptotic morphology was characterized by the absence of a discernable plasma membrane, poorly stained and reduced cytoplasmic material, that was often highly vesicular, and a clearly pyknotic but intact nucleus. These features resemble those described for type II PCD. At earlier time points, or with lower drug concentrations, plasma membranes were discernable, but the cytoplasm was vesicular (Fig. 1D). This was not regarded as cell death; it may be the prerequisite of the type II PCD morphology that follows.
Apoptotic cell death morphology is associated with activation of caspase-3 and mitochondrial depolarization.
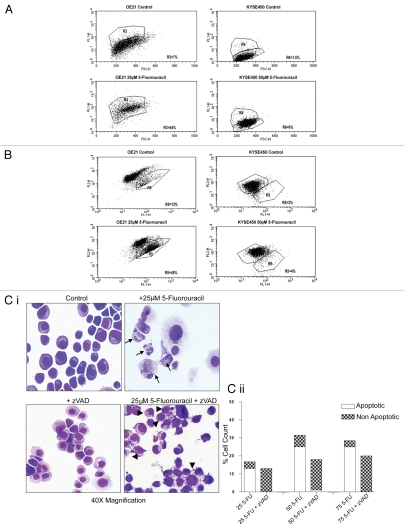
We examined typical markers of apoptotic cell death in all cell lines. Activation of caspase-3 and mitochondrial depolarization were assessed following treatment with both 5-FU and cisplatin. Cells were treated with 25 µM (OE21) and 50 µM (KYSE450) 5-FU for 48 h, and analyzed by flow cytometry. Figure 2 shows flow cytometric analysis representative of three independent experiments. Both drug sensitive (OE21 and OE33) cell lines displayed active caspase-3 (>37% and >35% active caspase-3 for OE21 and OE33 cells, respectively) and mitochondrial depolarization (>33% and >39% for OE21 and OE33 cells, respectively) in response to 5-FU (Fig. 2A and B left panels; data not shown for OE33 cells). In contrast, the more drug-resistant (OE19 and KYSE450) cell lines did not show caspase-3 activity (<4% and <2% active caspase-3 for KYSE450 and OE19 cells, respectively) or mitochondrial membrane depolarization (<1% and <2% mitochondrial depolarization for KYSE450 and OE19 cells, respectively) (Fig. 2A and B right panels). Results for cisplatin were similar (data not shown).
Figure 2.
Analysis of caspase activity and mitochondrial membrane potential. (A) Representative flow cytometric analysis of active caspase-3 in control and 5-fluorouracil (5-FU)-treated OE21 (25 µM) and KYSE 450 (50 µM) cell lines. The percentages shown indicate the proportion of cells with active caspase-3, detected as an increase in the number of FITC (FL-1)-labeled cells. Forty four percent of OE21 cells displayed active caspase-3 following drug treatment, whereas the more drug resistant, KYSE450 cells did not show activation of caspase-3 (at a range of concentrations tested). (B) Examination of mitochondrial membrane integrity with the JC-1 probe in control and treated OE21 and KYSE 450 cell lines. Percentages denote the proportion of cells with depolarized mitochondria after a 48-h incubation with 5-FU, with similar results observed with cisplatin (data not shown). (C) (i) Morphological features of OE21 cells, treated with 5-FU alone (upper right) or pretreated with zVAD-fmk (25 µM) for 2 h prior to 5-FU treatment (lower right). Apoptosis (arrows) was completely inhibited by zVAD-fmk (lower right) resulting in a switch to a non-apoptotic morphology (arrowheads) (magnification 40×). (ii) The extent of apoptotic (clear bars) and non-apoptotic (hatched bars) cell death for each concentration of 5-FU (25 to 75 µM) in the absence or presence of zVAD-fmk (25 µM), was determined by counting three fields of view per slide, with an average of ∼100 cells per field.
In addition, we assessed whether the apoptotic morphology, induced by 5-FU in the OE21 and OE33 cell lines, was dependent upon the activation of caspases. Cells were pre-incubated with the broad spectrum caspase inhibitor benzyloxycarbonyl-Val-Ala-Asp-(O-methyl)-CH2F (zVAD-fmk) at 25 µM, 2 h prior to the addition of 5-FU. Forty-eight hours later, morphological analysis showed that the induction of apoptosis by 5-FU was completely inhibited by zVAD (lower, right-hand panel) and resulted in a switch to a non-apoptotic morphology [Fig. 2C(i)]. Quantification of cell morphology is shown in Figure 2C(ii). The inhibitory effect on the apoptotic morphology was maintained even at high concentrations of 5-FU (75 µM). zVAD does not, however, improve viability in longer term clonogenic assays (data not shown) and can, in fact, show cytotoxicity in extended incubations, by advancing either necrotic or type II PCD.28–31 Collectively, these data indicate that drug sensitivity in these esophageal cancer cell lines is associated with the induction of an apoptotic program.
Comparison of morphological features of drug-treated OE21 and OE19 esophageal cancer cells by electron microscopy.
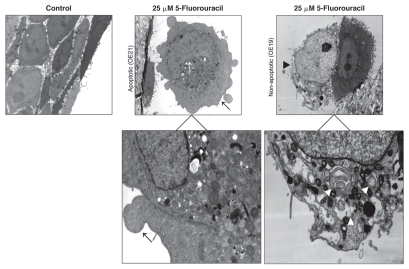
Ultrastructural features of OE21 cells, following 48-h treatment with 5-FU (25 µM), revealed morphological changes consistent with classical apoptotic cell death including marginalization of the nucleus, with an intact but blebbing plasma membrane (upper middle Fig. 3). In contrast, OE19 treated cells retained an intact nuclear membrane containing a distinct nucleolus, with areas of more electron-dense heterochromatin (upper right Fig. 3). In addition, numerous cytoplasmic vacuoles were evident, many of which appeared to surround cytoplasmic material and components, such as the mitochondria, resembling nascent autophagosomes (upper and lower right panels Fig. 3).
Figure 3.
Analysis of esophageal cells by electron microscopy. Representative electron microscopy images of vehicle control OE21 cells (upper left), 5-FU-treated (25 µM, 48 h) OE21 (upper middle) and OE19 (upper right) cells. Apoptotic features include marginalization of the nucleus (upper middle, white arrow), an intact cytoplasmic membrane, with surface blebbing (upper middle, black arrow), clear also at a higher magnification (lower left). Non-apoptotic, type II PCD features include apparent disintegration of the plasma membrane (upper right, black arrowhead), an intact nuclear membrane (lower right, white-outlined arrowhead), and numerous cytoplasmic vacuoles (lower right, white arrowheads). For each treatment or control group, transmission electron microscope images were randomly chosen, from a field of at least 100 cells.
Cytosolic redistribution of GFP-LC3 and formation of acidic vesicular organelles (AVOs) in drug-treated cells.
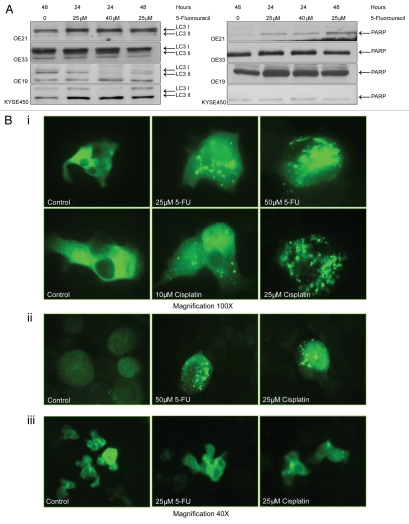
We examined expression and processing of GFP-LC3 by western blot analysis. There was no induction of a lower autophagosome-associated LC3-II band in treated OE21 or OE33 cells (Fig. 4A). In contrast, a significant increase in LC3-II levels was evident in the more drug-resistant OE19 and KYSE450 cells, following treatment with 5-FU at 24 and 48 h (Fig. 4A). Redistribution of GFP-LC3 from a diffuse cytosolic to a punctate autophagosome-associated pattern was observed in OE19 [Fig. 4B(i)] and KYSE450 [Fig. 4B(ii)] cells following treatment with 5-FU and cisplatin. Diffuse cytoplasmic localization of GFP-LC3 was observed in OE21 and OE33 cell lines, in response to both chemotherapeutic drugs [Fig. 4B(iii)].
Figure 4.
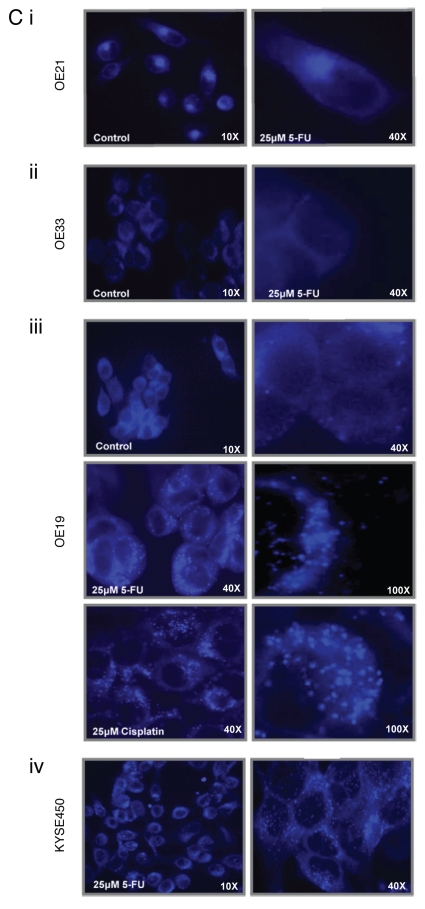
Evidence for autophagosome accumulation in drug-treated cells. (A) Cells expressing a GFP-LC3 plasmid, were cultured with 5-fluorouracil (5-FU) (25 or 40 µM), for 24 and 48 h, and analyzed by western blot with anti-LC3 antibody. Soluble LC3-I is detected at 45 kDa, while autophagosome-specific LC3-II migrates at 43 kDa. To ensure equal loading, blots were probed with PARP antibody, with corresponding blots for each cell line shown (right panel). (B) GFP-LC3 staining patterns were analyzed by fluorescence microscopy. (i) OE19 vehicle control cells (upper and lower left panels) display diffuse GFP-LC3 distributed throughout the cytoplasm. 5-FU-(25 and 50 µM; upper middle and right panels) or cisplatin (10 and 25 µM; lower middle and right panels)-treated cells, showed bright punctate patterns of GFP-LC3 fluorescence (magnification 100×). (ii) KYSE 450 vehicle control cells (left panel) displayed diffuse GFP-LC3 distribution, with both 5-FU (50 µM; middle panel) and cisplatin (25 µM; right panel), inducing a bright punctate patterns in GFP-LC3 fluorescence. (iii) OE21 control cells (left panel) displayed diffuse GFP-LC3 distributed throughout the cytoplasm. 5-FU- (25 µM; middle panel) or cisplatin (25 µM; right panel)-treated cells respectively, showed a similar diffuse GFP-LC3 distribution pattern (magnification 40×). (C) MDC-labeled vesicles in esophageal cells. Representative images of (i) OE21 control (upper left) and 5-FU (25 µM)-treated cells (upper right panel), (ii) OE33 control (left) and 5-FU (25 µM)-treated cells (right). (iii) OE19 vehicle control, 5-FU-(25 µM) and cisplatin (25 µM)-treated cells (all left panels), (iv) 5-FU (25 µM)-treated KYSE 450 cells with a higher magnification in the right panel. Images are representative of at least five independent experiments and magnification is shown in each image.
Monodansylcadaverine (MDC) dye was employed to assess levels of mature autophagic vesicle formation in all esophageal cancer cell lines following drug treatment. Hydrophobic interactions with lipids enhance the fluorescence of MDC, and autophagosomes contain a high level of unhydrolyzed membrane lipids.32 OE21 and OE33 cells did not show a punctate staining pattern in response to 5-FU (25 µM) (or cisplatin; data not shown), but displayed diffuse MDC staining, similar to vehicle control treated cells. Labeling of a fluorescent region immediately adjacent to the nucleus is typical for the recycling endosome and trans-Golgi network [Fig. 4C(i and ii)]. In contrast, the more drug-resistant, autophagic, OE19 and KYSE450 cell lines demonstrated extensive bright blue punctate staining in response to both chemotherapeutic drugs, consistent with accumulation of MDC in acidic vesicles [Fig. 4C(iii and iv)].
These results collectively (EM, GFP-LC3 redistribution and MDC) suggest that the cytoplasmic vesicles that develop following incubation with either 5-FU or cisplatin, in the OE19 and KYSE450 cells originate from autophagosomes.
The induction of autophagy in esophageal cancer cell lines is associated with the ability of the OE19 and KYSE450 cell lines to recover, following the removal of drugs.
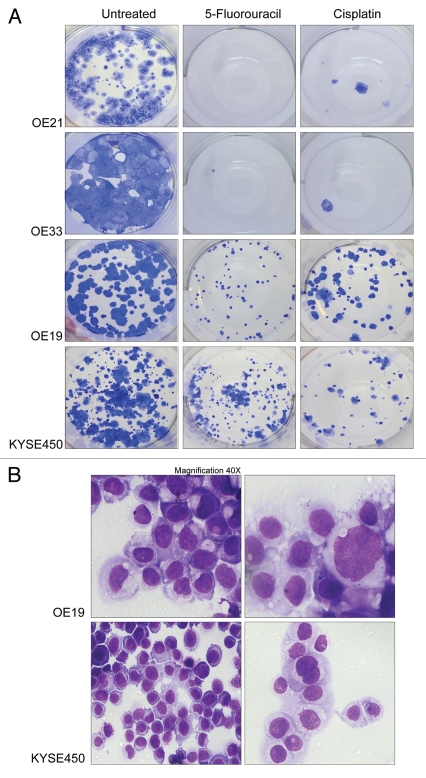
As the autophagic process is associated with survival and recovery from adverse conditions we examined whether drug-treated populations had the ability to recover, following withdrawal of drug. In order to assess this, we examined the regrowth of colonies from equal numbers of treated cells from each cell line. Cells were treated with 40 µM 5-FU or 20 µM cisplatin for 48 h. All floating cells were then discarded and adherent cells counted. One thousand viable cells (those that excluded PI) from each treatment were plated in triplicate into six-well plates (without drug) and were incubated for up to two weeks. The colonies that developed are shown in Figure 5A. Colonies were extremely rare in OE21 and OE33 cells (upper panels) treated with 5-FU or cisplatin (from concentrations as low as 5 µM). Colonies that recover from drug treatments were in excess of 80 per well in KYSE450 and OE19 (lower panels), clearly demonstrating resistance and recovery from cytotoxic drug treatment. The apoptosis-inducing cell line, OE21, was 10-fold more sensitive to 5-FU than KYSE450 cells (apoptosis resistant), as determined by clonogenic assay (Sup. Fig. 1). OE19 and KYSE450 cells displayed morphological features of autophagy when recovering (Fig. 5B). While type II cell PCD is present within the population (Fig. 1B and C), sufficient autophagic, yet viable cells were capable of recovery when the cytotoxic insult was removed.
Figure 5.
Recovery of esophageal cancer cells following withdrawal of drug. The ability of esophageal cell lines to recover following drug withdrawal was assessed with a colony formation assay (clonogenic assay). (A) Esophageal cell lines were treated with 5-fluorouracil (40 µM; center panel) or cisplatin (20 µM; right panel) for 48 h. Viable, adherent cells were counted and re-seeded (1,000 cell per well) into a well of a six-well plate (in triplicate), in the absence of drug. Ten to fourteen days later, colonies were fixed and stained. Each well shown is a representative image of at least 15 similar wells (five independent experiments). (B) Depicts the morphological features of OE19 and KYSE 450 cells, 48 h after drug (5-FU) withdrawal, indicating that autophagy was present in those cells used for the clonogenic assay. Magnification 40× with an enlarged view shown in right panels.
Role of autophagy in resistance and recovery.
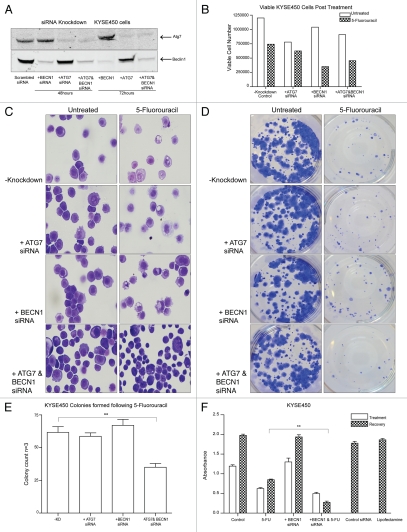
To assess whether autophagy contributed to the resistance and recovery of the KYSE450/‘type II’ cell line, we depleted two key regulators of autophagy, Beclin 1 and ATG7 with short interfering RNA (siRNA). Beclin 1 and ATG7 are required at vesicle nucleation and elongation steps, respectively (reviewed in ref. 33 and 34). These genes were knocked down separately and together in the KYSE450 cell line. Cells were seeded at equal density in six-well plates and transfected with siRNA. The levels of knockdown achieved for each gene, were greater than 85% at 48 and 72 h (Fig. 6A). Twenty-four hours after addition of siRNA, cells were treated with a high concentration of 5-FU (50 µM) for 48 h. Nonadherent cells were discarded and adherent cells were removed and counted. The numbers of viable cells following each treatment is shown in Figure 6B. These cell counts indicated that the knockdown (KD) of beclin 1 alone or together with ATG7 reduced the viable cell count at 48 h post treatment. ATG7 KD alone had only marginal effects on cell number in treated cells.
Figure 6.
Effects of beclin 1 and/or ATG7 siRNA knockdown, on recovery and morphology of drug treated cells. (A) Western blot analysis of Atg7 and Beclin 1 levels in KYSE 450 cells. The protein levels of both Atg7 (upper blot, Lanes 3 and 6) and Beclin 1 (BECN1) (lower blot, lanes 2 and 5) in single- and double-siRNA knockdown cells (upper blot and lower blots, lanes 4 and 7) were reduced by >85% at both 48 and 72 h (lane 1: scrambled siRNA control). (B) Propidium iodide (PI)-excluding, viable cells were counted to determine the initial effect of 5-FU on cell number, in single- and double-knockdown cells, compared to control untransfected cells (-KD). Cells were treated 24 h post-transfection, for 48 h with 50 µM 5-FU and data presented are representative of cell counts from three independent experiments. (C) Morphological features of these cells were examined 48 h post-treatment (50 µM 5-FU) to assess the levels of autophagic vesicles in single ATG7 and beclin 1 knockdown cells (second and third, right panels) and in double-knockdown treated cells (lower right panel) compared to control 5-FU-treated cells (upper right panel) (magnification 40×). (D) A colony formation assay was used to assess recovery, following 48 h drug treatment (5-FU 50 µM) in control (scrambled siRNA) and ATG7 and/or beclin 1 siRNA transfected cells. 2,000 cells per well (in triplicate) were re-seeded into a well of a six-well plate, allowed to adhere and grow for 12 d. Wells presented are representative of at least three similar wells. (E) Colonies were fixed, stained and counted to determine the effect of 5-FU on single- and double-knockdown transfected cells. Data are presented as mean colony count ± SEM of three independent experiments. Asterisks indicate a significant difference in the number of colonies formed in double-knockdown 5-FU treated cells, when compared to control 5-FU treated cells (**p < 0.005). (F) MTT assay was used to assess the effect of beclin 1 knockdown (BECN1), on viability following 48 h incubation with 5-FU (50 µM) and also on recovery (96 h post drug treatment) of KYSE450 cells. The recovery of 5-FU-treated cells, in which beclin 1 is silenced, is statistically different from 5-FU treated cells (**p < 0.005, *p < 0.05) (paired t-test).
Cells from each 48-h 5-FU treatment without and with siRNA were morphologically examined to assess whether vesicle formation was reduced by the siRNA treatment (Fig. 6C). Vesicles were still evident in the single beclin 1 and ATG7 KD cells suggesting that these genes are not necessarily required for vesicle formation (other proteins may compensate). In the double-knockdown cells, fewer vesicles could be detected, as the type II PCD morphology was more advanced. This would be consistent with a role for these proteins in survival and rescue from type II PCD.
Two thousand cells from each treatment were then plated to assess effects on long-term colony formation (5-FU concentration (50 µM) is slightly higher here than in Figure 5 (40 µM) to facilitate enumeration of colonies). While the siRNA would not be effective for long duration studies, this assay would indicate the viability of the cells that remained attached (and excluded PI) after 48 h drug treatment, and the effects of short term (>72 h) ATG7 and beclin 1 knockdown. Knockdown of ATG7 or beclin 1, alone or in combination did not alter colony formation in control untreated cells. Following 5-FU treatment, single knockdown of ATG7 or beclin 1 did not affect the numbers of colonies that were established, whereas the double-knockdown significantly reduced colony formation (p < 0.005) (Fig. 6D and E). Therefore, while beclin 1 KD alone reduced viable cell numbers at 48 h, those cells that remained attached were still capable of recovering and reforming colonies in the absence of drugs. Attached cells with both ATG7 and beclin 1 knockdown formed significantly fewer colonies, consistent with autophagy playing a protective role and facilitating recovery of the treated cells.
As beclin 1 knockdown alone had early effects on the 48 h viable cell counts, we evaluated its effects by MTT assay (Fig. 6F); 5-FU was withdrawn at 48 h and cultures allowed to recover for 96 h. beclin 1 KD attenuated the ability of KYSE450 cells to recover from the 5-FU (96 h) possibly due to an early enhancement of cytotoxicity (at 48 h) as suggested by the viable cell counts above (Fig. 6B).
A combination of beclin 1 and ATG7 knockdown clearly reduced the overall survival of drug-treated cultures, and was associated with more advanced features of type II PCD (Fig. 6C). Therefore, if autophagy plays a role in the type II PCD process, it must utilize alternative regulators.
Pharmacological inhibitors of autophagy: kinase inhibitors.
We assessed whether reported pharmacological inhibitors of components of autophagy pathways could modulate chemosensitivity and reduce the resistance and recovery of 5-FU-treated cells. PtdIns 3-kinase inhibitors, selective for the class III PtdIns 3-kinases, inhibit autophagy in other cells. We evaluated the effects of two inhibitors; 3-methyladenine (3-MA), (primarily a class III kinase inhibitor) and LY294002 (primarily a class I kinase inhibitor). OE21 (apoptosis inducing) and KYSE450 (apoptosis resistant) cell lines were treated with 3-MA (0.1–10.0 mM) with and without 5-fluorouracil (30 or 50 µM) for 48 h and viability was assessed using the MTT assay [Sup. Fig. 2A(i)]. Recovery data were acquired 48 h after drug withdrawal. Both cells lines (OE21 and KYSE450) recovered from treatment with 3-MA alone. Combination treatments of 3-MA (0.1–10 mM) and 5-FU did not significantly influence sensitivity or recovery of OE21 or KYSE450 cells. Recovery was only slightly diminished at a high concentration of 3-MA (10 mM) in KYSE450 cells. 3-MA induced mixed morphologies (apoptosis and type II death) in OE21 cells and induced both an accumulation of vesicles and enhancement of type II morphologies in KYSE450 cells [Sup. Fig. 2A(ii)]. It is possible that the marginally enhanced cytotoxicity in KYSE450 cells at the higher 3-MA concentration was due to reduced autophagy and promotion of type II PCD. However, in KYSE450 cells, LC3-II accumulates within 24 h in response to 3-MA alone and when combined with 5-FU [Sup. Fig. 2A(iii)]. This is not consistent with this compound acting as an inhibitor of autophagy. Similar results were obtained with LY294002, which would be expected to induce autophagy through inhibition of AKT and mTOR. LY294002 was better at reducing recovery in drug-treated cells than 3-MA, 10 µM LY294002 combined with 5-FU reduced recovery of KYSE450 cells by 50% [Sup. Fig. 2B(i)]. Vesicles were evident in treated cells [Sup. Fig. 2B(ii)] and phospho-Akt was diminished (Sup. Fig. 2C). These data are indicative of autophagy induction by both compounds.
Lysosomal antagonist's: bafilomycin A1 and chloroquine.
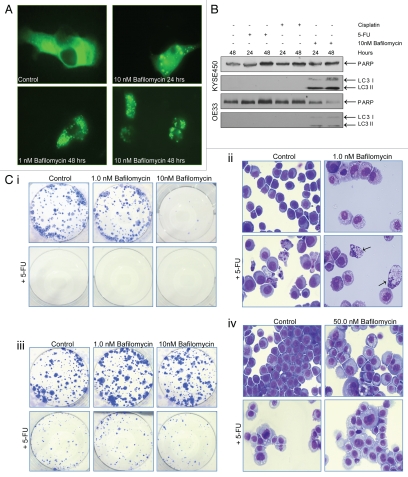
Inhibitors of lysosomal function were assessed in drug combinations. Bafilomycin A1 is a macrolide antibiotic that inhibits the vacuolar or V-type ATPase, an enzyme required for lysosomal acidification. This has been reported to inhibit the fusion between autophagosomes and lysosomes.35 To confirm this activity of bafilomycin A1, OE19 cells expressing a GFP-LC3 plasmid were cultured in the presence of 10 nM bafilomycin A1 for 24 and 48 h. Cytoplasmic distribution of GFP-LC3 was diffuse in vehicle control cells, whereas cells treated with bafilomycin A1 accumulated GFP-LC3-tagged autophagosomes, evident as bright punctate staining at 24 (10 nM) and 48 h (1 and 10 nM) (Fig. 7A). Bafilomycin A1 (10 nM) also induced an accumulation of endogenous LC3-II in KYSE450 and OE33 esophageal cells (Fig. 7B). The effects of bafilomycin A1 alone (1, 10, 50 and 100 nM) or in combination with 5-FU (40 µM) on viability were then assessed by clonogenic assay in one apoptosis-sensitive (OE21) and one apoptosis-resistant cell line (KYSE450).
Figure 7.
Effect of bafilomycin A1 (Baf) alone and in combination with 5-FU, on cell viability and morphology. (A) OE19 cells expressing a GFP-LC3 plasmid cultured in the presence of 10 nM bafilomycin A1 for 24 and 48 h. Bafilomycin A1 induced accumulation of GFP-LC3-tagged autophagosomes at 24 (10 nM, upper right) and 48 h (1 and 10 nM, lower panels). Images are representative of three individual experiments and similar data were obtained with KYSE 450 cells. (B) Bafilomycin A1 (10 nM) induced accumulation of endogenous LC3 in both KYSE 450 and OE33 esophageal cells, 24 and 48 h post treatment, as determined by western blot analysis. Lane 1: vehicle control, lanes 2 and 3:40 µM 5-FU (24 or 48 h), lanes 4 and 5:25 µM cisplatin (24 or 48 h), lanes 6 and 7: 10 nM bafilomycin A1 (24 or 48 h). Endogenous LC3 is not apparent in drug-treated cells with this antibody and standard RIPA buffer; blots in Figure 3 were GFP-LC3 transfected. (C) The ability of both esophageal cell lines to recover following drug withdrawal was assessed with a colony formation assay. OE21 (i) and KYSE 450 (iii) cells were treated with bafilomycin A1 (1 nM and 10 nM), alone (upper middle and right panels) and in combination with 5-FU (40 µM) for 48 h (lower middle and right panels). Similar effects were observed with 50 and 100 nM bafilomycin A1 (not shown). In combination treatments, bafilomycin A1 was added 2 h prior to 5-FU. Viable, adherent cells were counted and re-seeded (1,000 cells per well) into a well of a six-well plate (in triplicate), in the absence of drug. Ten to fourteen days later, colonies were fixed and stained. Each well shown is a representative image of at least 12 similar wells. The morphological features of OE21 (ii) and KYSE 450 (iv) cells, 48 h after treatment with bafilomycin A1 or 5-FU alone or in combination. Arrows highlight the presence of both an apoptotic and autophagic morphology within the same cell, identifiable in treated OE21 cells (lower right panel) [C(ii)]. KYSE 450 cells show an expanded vesicular compartment [C(iv)].
OE21 cells treated with bafilomycin A1 alone, at concentrations over 1 nM underwent significant cell death and recovering colonies were rare (<10) [Fig. 7C(i)]. As these cells are sensitive to 5-FU and underwent apoptosis, there was no benefit to adding bafilomycin A1 with 5-FU. Examination of the morphology of treated OE21 cells revealed that, on its own, bafilomycin A1 induced significant levels of vesicular accumulation in OE21 (Fig. 7C(ii), upper right). When combined with 5-FU (40 µM), OE21 cells exhibited both vesicular accumulation and apoptosis, and both morphologies could clearly be identified in the same cells (nuclear fragmentation with extensive cytoplasmic vacuolization) (lower right, arrows), indicating co-existence of apoptosis and autophagy in apoptosis-sensitive cells. KYSE450 cells were unaffected by bafilomycin A1 treatments (1 to 100 nM, data not shown for 50 or 100 nM), and combination treatments did not alter their ability to recover from treatment with 5-FU [Fig. 7C(iii)]. The morphology of KYSE450 cells treated with bafilomycin A1 alone indicated an expanded vesicular compartment. Following treatment with a combination of bafilomycin A1 and 5-FU, the morphology was the same as 5-FU alone, without any evidence of apoptosis or advancement of type II PCD (Fig. 7C(iv), lower right). Therefore, there was no benefit to combining bafilomycin A1 and 5-FU in either cell line.
Chloroquine is a lysosomotropic agent that acts as a weak base in lysosomes and compromises their degradative and recycling capacity. Several studies have now reported the use of chloroquine to inhibit autophagy. We assessed whether chloroquine could enhance the effects of 5-FU in clonogenic assays as described above. There was no added cytotoxic effect when 5-FU and chloroquine were combined, in either cell line (Sup. Fig. 3).
Taken together, these data suggest that in contrast to the specific inhibition of autophagy with siRNA-mediated knockdown, indirect inhibitors, such as 3-MA and lysosomal antagonists do not increase chemosensitivity of esophageal cancer cells.
Discussion
In this study, we investigated how cell death mechanisms can influence chemotherapeutic responses in esophageal cancer cells. We present evidence that a lack of apoptosis and ability to induce autophagy can significantly limit the efficacy of chemotherapy and provide an opportunity for cancer recurrence. Furthermore, we have evaluated potential strategies to improve the effectiveness of chemotherapeutic regimes.
A key contributor to drug resistance in autophagic cancer cells is undoubtedly their failure to engage in apoptosis. In addition, autophagic survival mechanisms can further undermine cytotoxic treatment. Recently, this has also been reported to limit the efficacy of 5-FU treatment in colorectal cancer cells.36–38 In a comparative study of 5-FU and a different thymidylate synthase inhibitor [trifluorothymidine (TFT)], 5-FU was found to be less potent than TFT as it induced less apoptosis and activated autophagic survival. In addition, a non-apoptotic cell death was reported, that involved the lysosomal protease cathepsin B.38
In this study, we examined the contribution of autophagy to survival and recovery by inhibiting two key regulators, beclin 1 and ATG7 with siRNA. beclin 1 single-knockdown added to cytotoxicity at the 48 h time point, but there was no difference in the recovery of colonies after this point. This suggests that Beclin 1 can mediate early protective responses in the presence of the drug, but when equal numbers of surviving cells are then plated without drug at the 48 h time point, this single knockdown does not further affect survival (NB beclin 1 expression was still depleted at the 72 h time point). Single knockdown of ATG7 had no effect on initial treatment or recovery. A double-knockdown of both genes reduced the ability of drug-treated colonies to recover, suggesting that autophagy indeed played a role in their recovery. It is not clear why the single-knockdown of either gene did not influence recovery, whereas the double-knockdown did. If this was simply due to redundancy of both proteins in the same linear pathway then the double-knockdown should be as ineffective as the single knockdowns. It is possible there are two (or more pathways) that selectively involve these proteins. It has recently been reported, that ATG7 is not required for DNA damage-induced autophagy, but it is required for autophagy induced by starvation.39 Beclin 1-independent autophagy has also been reported in reference 40 and 41.
The reduction in colonies in the double-knockdown also indicated that knockdown of ATG7 and beclin 1 does not prevent type II PCD; this late morphology was enhanced. Therefore, if autophagy plays a role in the death process, then it must involve an alternative form of autophagy (or non-canonical autophagy) that is independent of these genes. The redundancy noted in single-knockdown experiments indicates that this is possible. Other studies have also suggested that while Beclin 1 may be involved in autophagic survival, it is not required for autophagic cell death. Non-canonical autophagy (where autophagosomes can be formed without Beclin 1 or hVps34,40) has been reported in breast cancer cells treated with reservatol11 and in neuronal cells treated with a neurotoxin.42 A recent study has also reported that Atg5−/− and Atg7−/− mice can induce autophagy in response to etoposide and clear the mitochondria from erythroid cells by ‘alternative’ macroautophagy.39 If this type of extreme mitophagy was induced in other mammalian cells it would lead to cellular demise and could be regarded as autophagy-induced cell death. Further mechanistic studies are required on autophagy pathways and the mechanisms by which various compounds selectively induce them. It is possible that some autophagy pathways may never lead to cellular demise, whereas others may compromise viability and either be part of, or accompany, a death process.
As activation of hVps34/PtdInsKC3 is important for the development of the autophagosome, we evaluated whether the class III PtdIns3K inhibitor, 3-MA, could inhibit autophagy and improve the cytotoxic effect of 5-flurouracil. A marginal enhancement of cytotoxicity was only apparent at high concentrations of 3-MA that showed significant toxicity alone. 3-MA did not inhibit autophagy in the esophageal cancer cells, in fact, it appeared to induce it (enhanced vesicle formation and LC3 accumulation). The apoptosis-sensitive cells exhibited a mixture of apoptosis and autophagy in response to both PtdIns3K inhibitors, and the autophagic group exhibited only autophagy. Other studies have also reported that 3-MA can induce autophagy43 and inhibit both class I and III PtdIns 3-kinases and possibly other survival-related enzymes (reviewed in ref. 44). In addition, class III PtdIns3K can be important for the activation of mTOR45 and thus inhibition could promote autophagy. LY294002 a more selective class I inhibitor had similar effects to 3-MA. Inhibition of PtdIns3K class I will deactivate Akt and mTOR, promoting autophagy. LY294002 has been reported to directly target the catalytic domain of mTOR, providing an alternative explanation for autophagy induction.46 Interestingly, this agent reduced the recovery of KYSE450 cells treated with 5-FU, suggesting that an autophagy inducer may be of more therapeutic value in combination regimes. LY294002 augments rapamycin-induced autophagy and cytotoxicity in malignant glioma cells.26
Inhibition of lysosomal function was evaluated as a possible strategy to reduce the survival effects of autophagy. Lysosomal antagonists did not enhance chemosensitivity in drug-treated cells. Sensitivity to bafilomycin A1 followed the same pattern as all other cytotoxic drugs tested. The apoptosis inducing cell lines were the most sensitive, and underwent both apoptotic and non-apoptotic cell death. The apoptosis-resistant cell line, KYSE450, was resistant to a 10-fold higher level of bafilomycin A1 and combination treatments with bafilomycin A1 and 5-FU did not reduce the recovery of these cells in assays of clonogenic growth. Chloroquine was less toxic to both types of cells, but clearly induced vesicular accumulation. Combination treatments did not enhance the effects of 5-FU in apoptosis-inducing cells or affect the recovery of apoptosis-resistant and/or autophagy-inducing cells. Therefore, bafilomycin A1 shows interesting activity alone in an apoptosis inducing cell line but it is not better than 5-FU at eliminating regrowth in these cells, and there is no benefit to adding either of the lysosomal inhibitors to regimens with 5-FU.
Several studies have reported activity with these agents in other cell lines, and in many instances an apoptotic response was induced.47,48 Chloroquine has been reported to induce apoptosis due to lysosomal disruption, mitochondrial membrane permeabilization and decreased protein degradation.20,49 Others have reported both apoptosis and necrosis which is concentration dependent,50 or indeed involvement of apoptosis-independent death.51–53 Activity can be influenced by p53 status, c-myc status and caspase-3 status.54–56 Choroquine has also been reported to promote tumor regression, enhance apoptosis and inhibit autophagy in a Myc-induced lymphoma model with inducible p53.20 It can also enhance the effects of Abl and Src kinase inhibitors57,58 and has been reported to enhance the effects of 5-FU in a colorectal cancer cell line.59 These indications have led to the initiation of clinical trials with chloroquine combined with other agents (reviewed in ref. 60). As other activities of these agents have been reported in reference 60 and 61, further mechanistic work is required. It is possible that their activity may be limited to certain types of cancer or specific genetic backgrounds that can influence cell death and autophagy susceptibility.
Ideally, more selective inhibitors of autophagy need to be developed. Clearly, this would also require a better understanding of the signaling involved in autophagic survival and the involvement of autophagy in type II PCD. If autophagy plays a role in type II PCD then this must be tightly and independently regulated. The genes that are involved may differ, dependent on the nature of a stimulus and type of autophagy induced. In addition, there may be defects in autophagy in cancer cells and several novel autophagy regulators have been implicated in tumor suppression.62,63 The challenge is to identify the important regulators that can be modulated effectively, to promote chemosensitivity. In this study, two genes had to be inhibited to reduce the survival effects of autophagy, suggesting some level of redundancy. The ideal drugable target would be required for survival (and not death) and would be nonredundant. In the absence of this target, other indirect strategies to interfere with autophagy may prove to have efficacy in therapeutic regimens.
This work has shown that the induction of autophagy can play a major role in the resistance and recovery of drug-treated esophageal cancer cells. We propose that unless we can find another way to induce apoptosis (or other death mechanism), selective inhibition of autophagic survival may be of major therapeutic benefit in apoptosis-resistant cancers.
Materials and Methods
Cell culture.
Established human esophageal cancer cell lines OE19, OE21 and OE33,64 were obtained from the European Collection of Cell Cultures (96071721, 96062201 and 96070808). KYSE450 cells were from DSMZ (Deutsche Sammlung von Mikroorganismen und Zellkulturen GmbH). OE19, OE21 and OE33 cell lines were maintained in RPMI 1640 medium, KYSE450 cells were maintained in 50:50 RPMI 1640:F-12 HAMS medium, all supplemented with 1% penicillin/streptomycin, 10% (v/v) fetal calf serum (Gibco, 21875-034, 15070-063, 10270) at 37°C, 5% CO2.
Evaluation of caspase-3 activity.
Following fixation in 4% para-formaldehyde, cells were washed in a permeabilization buffer (0.1% Triton X-100, 0.1% sodium azide, 10 mM HEPES, 4% FCS, 150 mM NaCl) and incubated with a primary rabbit polyclonal anti-active caspase-3 antibody (BD Biosciences, 557035) on ice for 1 h. This was detected with an anti-rabbit FITC conjugated secondary antibody, and samples were analyzed by FACScan at 530 nm (FL-1).
Detection of mitochondrial depolarization (ΔΓm).
Mitochondrial membrane potential was determined using the JC-1 probe (Molecular Probes, T-3168). JC1 accumulates as aggregates in normal mitochondria, which stain red (FL2; 590 nm). A loss of mitochondrial membrane potential (ΔΓm) releases the aggregated JC1 to its monomeric form, which stains the cytosol green (FL1; 530 nm). Therefore, fluorescence of JC-1 in the FL-2 channel decreases as mitochondrial membrane integrity is lost and fluorescence in the FL-1 channel increases. Cells were incubated in JC1 (7.5 µg/ml) at 37°C for 15 min, and washed prior to analysis by flow cytometry FACScan (Becton Dickinson).
Visualization of monodansylcadaverine (MDC)-labeled vacuoles.
MDC is an autofluorescent weak base that accumulates in acidic lysosomal vacuoles, showing high selectivity for autophagosomes, due to the high level of unhydrolyzed membrane lipids from engulfed organelles. Cells were incubated with 0.1 mM MDC (Sigma, 30432) in PBS at 37°C for 10 min,32 washed and immediately analyzed by fluorescence microscopy.
MTT viability assay.
Cells were seeded at 2 × 104 (OE33 and OE19) and 1 × 104 (OE21 and KYSE450) cells per cm2, treated for 48 to 96 h and incubated for an additional 60 min at 37°C in 0.5 mg/ml MTT dye (Sigma, M2128). Viable, metabolizing cells reduce MTT dye, producing a dark formazan product, with absorbance read at 562 nm, reference wavelength 620 nm. To assess recovery, at 48 h post treatment, in replicate plates (identical seeding and treatment times), all drugs were removed, culture medium replaced, and cells were cultured for a further 48 to 96 h and re-assessed by MTT.
Statistical analysis.
Values are presented as the mean absorbance ± standard error of the mean (SEM) for three or four independent experiments. We performed paired t-test (two-tailed) statistical analysis, p < 0.05 was significant. Asterisks indicate the level of significance.
Evaluation of morphology.
Morphological features of cells treated with 5-fluorouracil (5-FU) (Sigma, F6627), cisplatin (Sigma, P4394), bafilomycin A1 (Sigma, B1793), 3-methyladenine (3-MA) (Sigma, M9281), chloroquine (Sigma, C6628) and LY294002 (Calbiochem, 440202) were examined by light microscopy. Aliquots of vehicle control and drug-treated cells were cytospun onto glass slides and stained with Rapi-Diff (Braidwood Laboratories, 22007, 22008, 22009). The extent of apoptotic and non-apoptotic cell death was determined by counting the cells in at least three fields of view per slide, with an average of ∼100 cells per field. Apoptotic cell death was characterized by the presence of two or more of the following morphological features: cell shrinkage, chromatin condensation, DNA degradation and fragmentation into ‘apoptotic bodies,’ within an intact plasma membrane. Type II PCD was defined as follows: Absence of a discernable plasma membrane, with a highly vesicular cytoplasm which is often associated with extensive loss of cytoplasmic material. The nucleus remains intact, with clear pyknosis (condensed regions). Cytospin images are representative of at least three independent experiments.
Electron microscopy.
Cells were seeded on semi-porous membranes and incubated in 5-fluoruracil (5-FU) for 48 h. Cells were then fixed in a 0.165 mM phosphate buffer (pH 7.4), containing 2.0% glutaraldehyde, at room temperature (RT) for 40 min. Cells were post-fixed in osmium tetroxide (OsO4) at RT for 60 min, dehydrated in ascending grades of ethanol solutions (50%, 70%, 95%, 100% and 100% dry), prior to embedding in araldite resin (Agar Scientific , R1040). Samples were subjected to a graded infiltration process with araldite (epoxy resin) before being set and sectioned. Representative areas were chosen for ultra-thin sectioning and samples were examined by electron microscopy.
Western blotting and antibodies.
Total cellular protein extracts were prepared by scraping the cells into modified RIPA buffer (50 mM Tris HCl (pH 7.4), 150 mM NaCl, 0.25% sodium deoxycholate, 1% Igepal, 1 mM EDTA, 1× Pefabloc, 1× protease inhibitor cocktail, 1 mM Na3VO4, 1 mM NaF). Alternatively, for LC3 detection, cells were lysed on ice in NP40 lysis buffer [50 mM HEPES, pH 7.0, 150 mM NaCl, 2 mM EDTA, 0.1–1% NP-40, protease inhibitor mix (Complete™, Roche 04 693 116 001)]. All protein samples were separated on NuPAGE 4–12%, Bis-Tris gels (Invitrogen, NP0322) and electrophoretically transferred onto either nitrocellulose or PVDF membrane. All primary antibodies were incubated overnight at 4°C: anti-Beclin 1 (Cell Signaling Technologies, 3738), anti-Atg7 (Cell Signaling, 2631), anti-LC3 (Medical & Biological Laboratories, PD014), anti-phospho-Akt (Cell Signaling, 9271) and anti-PARP (Cell Signaling, 9532). Protein expression was visualized using either chemiluminescence (ECL Amersham, RPN2106), or the Odyssey IR imaging system (Li-Cor goat anti-rabbit IgG 926 32211 IRDy 800; relative protein expression levels were calculated using this system).
Vacuolar redistribution of GFP-LC3.
To visualize the formation of autophagic vesicles, the green fluorescent protein (GFP)-LC3 (pEGFP-LC3) expression vector, kindly supplied by Dr. T. Yoshimori (Osaka University, Japan) was used. Cells were transiently transfected with the Amaxa electroporation system according to the supplier's protocol. Twenty-four h post-transfection, cells were treated with 5-FU or cisplatin, fixed in 4% paraformaldehyde in PBS and transferred onto slides using a nonfluorescent fixative for analysis by fluorescence microscopy. Alternatively, western blot analysis was used to assess the expression and processing of LC3. Upon stimulation of autophagy, LC3 is upregulated and processed from soluble GFP-LC3-I (45 kDa) to the autophagosome-associated form GFP-LC3-II (43 kDa). The membrane sequestered, lipid-conjugated form of LC3-II remains with the autophagosome membrane after the vesicle has formed, and levels of both isoform are detected by western blot.65,66 Transfection efficiency was consistent for a given cell line, OE33 and KYSE450 cell lines (∼70 to 80%) compared to OE19 and OE21 cell lines (∼30%).
siRNA knockdown of ATG7 and beclin 1.
siRNA knockdown was used to inhibit mammalian beclin 1 (ortholog of ATG6) and ATG7. Cells were transfected with a pre-designed siRNA (50 nM) against beclin 1 (Dharmacon ON-TARGETplus SMARTpool Human BECN 1, NM_003766) and ATG7 (Dharmacon ON-TARGETplus SMARTpool Human ATG7, NM_006395) using the transfection reagent Lipofectamine 2000 (Invitrogen 11668-027). The transfection efficiency was greater than 85% (transfection efficiency was assessed visually using fluorescently-labeled siRNAs, Ambion AM4620) and the extent of beclin 1 and ATG7 knockdown was determined by western blot analysis of protein levels.
Colony formation assay.
We assessed the ability of cells to recover from treatments and form colonies on a monolayer surface. Following treatment, all adherent cells were trypsinized, counted and viability determined. Of those viable cells, either 1,000 or 2,000 cells (depending on the experiment) were reseeded into a well of a six-well plate (in triplicate). Cells were allowed to adhere and grow for between 10 to 14 d. To visualize colonies, media was removed, cells were fixed in 96% ethanol for 10 min and stained with Prodiff solution C (Braidwood laboratories 22009). Where possible, colonies were counted, and are presented as the mean number of colonies ±SEM from at least three independent experiments.
Acknowledgements
This work was funded by the Higher Education Authority of Ireland, Science Foundation Ireland (RFP/NSC1655) and the Cork Cancer Research Centre. We are very grateful to Dr. Simon Rajendran and Dr. Michelle Nyhan of the CCRC for excellent technical assistance and in manuscript preparation. We are also grateful to Don O'Leary Dept. of Anatomy UCC for assistance with EM.
Abbreviations
- 5-FU
5-fluorouracil
- PCD
programmed cell death
- 3-MA
3-methyladenine
- PtdIns 3-kinase
phosphatidylinositol-3-kinase
- ATG
autophagy-related
- zVAD-fmk
benzyloxycarbonyl-Val-Ala-Asp-(O-methyl)-CH2F
- GFP
green fluorescent protein
- LC3
microtubule-associated protein 1 light chain 3
- MDC
monodansylcadaverine
- PI
propidium iodide
- siRNA
small interfering RNA
- Baf
bafilomycin A1
- TFT
trifluorothymidine
- ATP
adenosine triphosphate
- AVO
acidic vesicular organelles
Supplementary Material
References
- 1.Sant M, Aareleid T, Berrino F, Lasota BM, Carli PM, Faivre J, et al. EUROCARE-3: survival of cancer patients diagnosed 1990–94—results and commentary. Ann Oncol. 2003;14:61–118. doi: 10.1093/annonc/mdg754. [DOI] [PubMed] [Google Scholar]
- 2.O'Sullivan GC, Sheehan D, Clarke A, Stuart R, Kelly J, Kiely MD, et al. Micrometastases in esophagogastric cancer: high detection rate in resected rib segments. Gastroenterology. 1999;116:543–548. doi: 10.1016/s0016-5085(99)70175-7. [DOI] [PubMed] [Google Scholar]
- 3.Ryan P, McCarthy S, Kelly J, Collins JK, Dunne C, Grogan L, et al. Prevalence of bone marrow micrometastases in esophagogastric cancer patients with and without neoadjuvant chemoradiotherapy. J Surg Res. 2004;117:121–126. doi: 10.1016/j.jss.2003.12.008. [DOI] [PubMed] [Google Scholar]
- 4.Raguz S, Yague E. Resistance to chemotherapy: new treatments and novel insights into an old problem. Br J Cancer. 2008;99:387–391. doi: 10.1038/sj.bjc.6604510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ricci MS, Zong WX. Chemotherapeutic approaches for targeting cell death pathways. Oncologist. 2006;11:342–357. doi: 10.1634/theoncologist.11-4-342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Degterev A, Yuan J. Expansion and evolution of cell death programmes. Nat Rev. 2008;9:378–390. doi: 10.1038/nrm2393. [DOI] [PubMed] [Google Scholar]
- 7.Clarke PG. Developmental cell death: morphological diversity and multiple mechanisms. Anat Embryol. 1990;181:195–213. doi: 10.1007/BF00174615. [DOI] [PubMed] [Google Scholar]
- 8.Pattingre S, Tassa A, Qu X, Garuti R, Liang XH, Mizushima N, et al. Bcl-2 antiapoptotic proteins inhibit Beclin 1-dependent autophagy. Cell. 2005;122:927–939. doi: 10.1016/j.cell.2005.07.002. [DOI] [PubMed] [Google Scholar]
- 9.Yu L, Wan F, Dutta S, Welsh S, Liu Z, Freundt E, Baehrecke EH, Lenardo M. Autophagic programmed cell death by selective catalase degradation. Proc Natl Acad Sci USA. 2006;103:4952–4957. doi: 10.1073/pnas.0511288103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Opipari AW, Jr, Tan L, Boitano AE, Sorenson DR, Aurora A, Liu JR. Resveratrol-induced autophagocytosis in ovarian cancer cells. Cancer Res. 2004;64:696–703. doi: 10.1158/0008-5472.can-03-2404. [DOI] [PubMed] [Google Scholar]
- 11.Scarlatti F, Maffei R, Beau I, Codogno P, Ghidoni R. Role of non-canonical Beclin 1-independent autophagy in cell death induced by resveratrol in human breast cancer cells. Cell Death Differ. 2008;15:1318–1329. doi: 10.1038/cdd.2008.51. [DOI] [PubMed] [Google Scholar]
- 12.Debnath J, Baehrecke EH, Kroemer G. Does autophagy contribute to cell death? Autophagy. 2005;1:66–74. doi: 10.4161/auto.1.2.1738. [DOI] [PubMed] [Google Scholar]
- 13.Berry DL, Baehrecke EH. Growth arrest and autophagy are required for salivary gland cell degradation in Drosophila. Cell. 2007;131:1137–1148. doi: 10.1016/j.cell.2007.10.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lam D, Kosta A, Luciani MF, Golstein P. The inositol 1,4,5-trisphosphate receptor is required to signal autophagic cell death. Molecular biology of the cell. 2008;19:691–700. doi: 10.1091/mbc.E07-08-0823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yorimitsu T, Klionsky DJ. Autophagy: molecular machinery for self-eating. Cell Death Differ. 2005;12:1542–1552. doi: 10.1038/sj.cdd.4401765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mizushima N. Autophagy: process and function. Genes Dev. 2007;21:2861–2873. doi: 10.1101/gad.1599207. [DOI] [PubMed] [Google Scholar]
- 17.Mizushima N, Levine B, Cuervo AM, Klionsky DJ. Autophagy fights disease through cellular self-digestion. Nature. 2008;451:1069–1075. doi: 10.1038/nature06639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jin S, White E. Tumor suppression by autophagy through the management of metabolic stress. Autophagy. 2008;4:563–566. [PMC free article] [PubMed] [Google Scholar]
- 19.Degenhardt K, Mathew R, Beaudoin B, Bray K, Anderson D, Chen G, et al. Autophagy promotes tumor cell survival and restricts necrosis, inflammation and tumorigenesis. Cancer Cell. 2006;10:51–64. doi: 10.1016/j.ccr.2006.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Amaravadi RK, Yu D, Lum JJ, Bui T, Christophorou MA, Evan GI, et al. Autophagy inhibition enhances therapy-induced apoptosis in a Myc-induced model of lymphoma. J Clin Inves. 2007;117:326–336. doi: 10.1172/JCI28833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Vousden KH, Ryan KM. p53 and metabolism. Nat Rev Cancer. 2009;9:691–700. doi: 10.1038/nrc2715. [DOI] [PubMed] [Google Scholar]
- 22.Rosenfeldt MT, Ryan KM. The role of autophagy in tumour development and cancer therapy. Expert Rev Mol Med. 2009;11:36. doi: 10.1017/S1462399409001306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Scarlatti F, Granata R, Meijer AJ, Codogno P. Does autophagy have a license to kill mammalian cells? Cell Death Differ. 2009;16:12–20. doi: 10.1038/cdd.2008.101. [DOI] [PubMed] [Google Scholar]
- 24.Kanzawa T, Kondo Y, Ito H, Kondo S, Germano I. Induction of autophagic cell death in malignant glioma cells by arsenic trioxide. Cancer Res. 2003;63:2103–2108. [PubMed] [Google Scholar]
- 25.Kanzawa T, Germano IM, Komata T, Ito H, Kondo Y, Kondo S. Role of autophagy in temozolomide-induced cytotoxicity for malignant glioma cells. Cell Death Differ. 2004;11:448–457. doi: 10.1038/sj.cdd.4401359. [DOI] [PubMed] [Google Scholar]
- 26.Takeuchi H, Kondo Y, Fujiwara K, Kanzawa T, Aoki H, Mills GB, Kondo S. Synergistic augmentation of rapamycin-induced autophagy in malignant glioma cells by phosphatidylinositol-3-kinase/protein kinase B inhibitors. Cancer Res. 2005;65:3336–3346. doi: 10.1158/0008-5472.CAN-04-3640. [DOI] [PubMed] [Google Scholar]
- 27.Krysko DV, Vandenabeele P. From regulation of dying cell engulfment to development of anti-cancer therapy. Cell Death Differ. 2008;15:29–38. doi: 10.1038/sj.cdd.4402271. [DOI] [PubMed] [Google Scholar]
- 28.Scaringi L, Cornacchione P, Ayroldi E, Corazzi L, Capodicasa E, Rossi R, Marconi P. Omeprazole induces apoptosis in jurkat cells. Int J Immunopathol Pharmacol. 2004;17:331–342. doi: 10.1177/039463200401700313. [DOI] [PubMed] [Google Scholar]
- 29.Vandenabeele P, Vanden Berghe T, Festjens N. Caspase inhibitors promote alternative cell death pathways. Sci STKE. 2006;2006:44. doi: 10.1126/stke.3582006pe44. [DOI] [PubMed] [Google Scholar]
- 30.Wu YT, Tan HL, Huang Q, Kim YS, Pan N, Ong WY, et al. Autophagy plays a protective role during zVAD-induced necrotic cell death. Autophagy. 2008;4:457–466. doi: 10.4161/auto.5662. [DOI] [PubMed] [Google Scholar]
- 31.Yu L, Alva A, Su H, Dutt P, Freundt E, Welsh S, et al. Regulation of an ATG7-beclin 1 program of autophagic cell death by caspase-8. Science. 2004;304:1500–1502. doi: 10.1126/science.1096645. [DOI] [PubMed] [Google Scholar]
- 32.Biederbick A, Kern HF, Elsasser HP. Monodansylcadaverine (MDC) is a specific in vivo marker for autophagic vacuoles. Eur J Cell Biol. 1995;66:3–14. [PubMed] [Google Scholar]
- 33.Takahashi Y, Meyerkord CL, Wang HG. Bif-1/endophilin B1: a candidate for crescent driving force in autophagy. Cell Death Differ. 2009;16:947–955. doi: 10.1038/cdd.2009.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Geng J, Klionsky DJ. The Atg8 and Atg12 ubiquitin-like conjugation systems in macroautophagy. EMBO Rep. 2008;9:859–864. doi: 10.1038/embor.2008.163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Boya P, Gonzalez-Polo RA, Casares N, Perfettini JL, Dessen P, Larochette N, et al. Inhibition of macroautophagy triggers apoptosis. Mol Cell Biol. 2005;25:1025–1040. doi: 10.1128/MCB.25.3.1025-1040.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Li J, Hou N, Faried A, Tsutsumi S, Takeuchi T, Kuwano H. Inhibition of autophagy by 3-MA enhances the effect of 5-FU-induced apoptosis in colon cancer cells. Ann Surg Oncol. 2009;16:761–771. doi: 10.1245/s10434-008-0260-0. [DOI] [PubMed] [Google Scholar]
- 37.Xiong HY, Guo XL, Bu XX, Zhang SS, Ma NN, Song JR, et al. Autophagic cell death induced by 5-FU in Bax or PUMA deficient human colon cancer cell. Cancer Lett. 2010;288:68–74. doi: 10.1016/j.canlet.2009.06.039. [DOI] [PubMed] [Google Scholar]
- 38.Bijnsdorp IV, Peters GJ, Temmink OH, Fukushima M, Kruyt FA. Differential activation of cell death and autophagy results in an increased cytotoxic potential for trifluorothymidine compared to 5-fluorouracil in colon cancer cells. Int J Cancer. 2010;126:2457–2468. doi: 10.1002/ijc.24943. [DOI] [PubMed] [Google Scholar]
- 39.Nishida Y, Arakawa S, Fujitani K, Yamaguchi H, Mizuta T, Kanaseki T, et al. Discovery of Atg5/Atg7-independent alternative macroautophagy. Nature. 2009;461:654–658. doi: 10.1038/nature08455. [DOI] [PubMed] [Google Scholar]
- 40.Scarlatti F, Maffei R, Beau I, Ghidoni R, Codogno P. Non-canonical autophagy: an exception or an underestimated form of autophagy? Autophagy. 2008;4:1083–1085. doi: 10.4161/auto.7068. [DOI] [PubMed] [Google Scholar]
- 41.Priault M, Hue E, Marhuenda F, Pilet P, Oliver L, Vallette FM. Differential dependence on Beclin 1 for the regulation of pro-survival autophagy by Bcl-2 and Bcl-xL in HCT116 colorectal cancer cells. PLoS One. 5:8755. doi: 10.1371/journal.pone.0008755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhu JH, Horbinski C, Guo F, Watkins S, Uchiyama Y, Chu CT. Regulation of autophagy by extracellular signal-regulated protein kinases during 1-methyl-4-phenylpyridinium-induced cell death. Am J Pathol. 2007;170:75–86. doi: 10.2353/ajpath.2007.060524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Petiot A, Ogier-Denis E, Blommaart EF, Meijer AJ, Codogno P. Distinct classes of phosphatidylinositol-3′-kinases are involved in signaling pathways that control macroautophagy in HT-29 cells. J Biol Chem. 2000;275:992–998. doi: 10.1074/jbc.275.2.992. [DOI] [PubMed] [Google Scholar]
- 44.Tolkovsky AM, Xue L, Fletcher GC, Borutaite V. Mitochondrial disappearance from cells: a clue to the role of autophagy in programmed cell death and disease? Biochimie. 2002;84:233–240. doi: 10.1016/s0300-9084(02)01371-8. [DOI] [PubMed] [Google Scholar]
- 45.Backer JM. The regulation and function of Class III PI3Ks: novel roles for Vps34. Biochem J. 2008;410:1–17. doi: 10.1042/BJ20071427. [DOI] [PubMed] [Google Scholar]
- 46.Brunn GJ, Williams J, Sabers C, Wiederrecht G, Lawrence JC, Jr, Abraham RT. Direct inhibition of the signaling functions of the mammalian target of rapamycin by the phosphoinositide-3-kinase inhibitors, wortmannin and LY294002. EMBO J. 1996;15:5256–5267. [PMC free article] [PubMed] [Google Scholar]
- 47.Jiang BH, Liu LZ. PI3K/PTEN signaling in tumorigenesis and angiogenesis. Biochim Biophys Acta. 2008;1784:150–158. doi: 10.1016/j.bbapap.2007.09.008. [DOI] [PubMed] [Google Scholar]
- 48.Zheng Y, Zhao YL, Deng X, Yang S, Mao Y, Li Z, et al. Chloroquine inhibits colon cancer cell growth in vitro and tumor growth in vivo via induction of apoptosis. Cancer Invest. 2009;27:286–292. doi: 10.1080/07357900802427927. [DOI] [PubMed] [Google Scholar]
- 49.Boya P, Gonzalez-Polo RA, Poncet D, Andreau K, Vieira HL, Roumier T, et al. Mitochondrial membrane permeabilization is a critical step of lysosome-initiated apoptosis induced by hydroxychloroquine. Oncogene. 2003;22:3927–3936. doi: 10.1038/sj.onc.1206622. [DOI] [PubMed] [Google Scholar]
- 50.Fan C, Wang W, Zhao B, Zhang S, Miao J. Chloroquine inhibits cell growth and induces cell death in A549 lung cancer cells. Bioorg Med Chem. 2006;14:3218–3222. doi: 10.1016/j.bmc.2005.12.035. [DOI] [PubMed] [Google Scholar]
- 51.Bissonnette RP, Echeverri F, Mahboubi A, Green DR. Apoptotic cell death induced by c-myc is inhibited by bcl-2. Nature. 1992;359:552–554. doi: 10.1038/359552a0. [DOI] [PubMed] [Google Scholar]
- 52.Maclean KH, Dorsey FC, Cleveland JL, Kastan MB. Targeting lysosomal degradation induces p53-dependent cell death and prevents cancer in mouse models of lymphomagenesis. J Clin Invest. 2008;118:79–88. doi: 10.1172/JCI33700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wei LH, Kuo ML, Chen CA, Chou CH, Cheng WF, Chang MC, et al. The anti-apoptotic role of interleukin-6 in human cervical cancer is mediated by upregulation of Mcl-1 through a PI 3-K/Akt pathway. Oncogene. 2001;20:5799–5809. doi: 10.1038/sj.onc.1204733. [DOI] [PubMed] [Google Scholar]
- 54.Loehberg CR, Thompson T, Kastan MB, Maclean KH, Edwards DG, Kittrell FS, et al. Ataxia telangiectasia-mutated and p53 are potential mediators of chloroquine-induced resistance to mammary carcinogenesis. Cancer Res. 2007;67:12026–12033. doi: 10.1158/0008-5472.CAN-07-3058. [DOI] [PubMed] [Google Scholar]
- 55.McNeil CM, Sergio CM, Anderson LR, Inman CK, Eggleton SA, Murphy NC, et al. c-Myc overexpression and endocrine resistance in breast cancer. J Steroid Biochem Mol Biol. 2006;102:147–155. doi: 10.1016/j.jsbmb.2006.09.028. [DOI] [PubMed] [Google Scholar]
- 56.Yang S, Zhou Q, Yang X. Caspase-3 status is a determinant of the differential responses to genistein between MDA-MB-231 and MCF-7 breast cancer cells. Biochim Biophys Acta. 2007;1773:903–911. doi: 10.1016/j.bbamcr.2007.03.021. [DOI] [PubMed] [Google Scholar]
- 57.Bellodi C, Lidonnici MR, Hamilton A, Helgason GV, Soliera AR, Ronchetti M, et al. Targeting autophagy potentiates tyrosine kinase inhibitor-induced cell death in Philadelphia chromosome-positive cells, including primary CML stem cells. J Clin Invest. 2009;119:1109–1123. doi: 10.1172/JCI35660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wu Z, Chang PC, Yang JC, Chu CY, Wang LY, Chen NT, et al. Autophagy Blockade Sensitizes Prostate Cancer Cells towards Src Family Kinase Inhibitors. Genes Cancer. 1:40–49. doi: 10.1177/1947601909358324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Sasaki K, Tsuno NH, Sunami E, Tsurita G, Kawai K, Okaji Y, et al. Chloroquine potentiates the anti-cancer effect of 5-fluorouracil on colon cancer cells. BMC Cancer. 2010;10:370. doi: 10.1186/1471-2407-10-370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Solomon VR, Lee H. Chloroquine and its analogs: a new promise of an old drug for effective and safe cancer therapies. Eur J Pharmacol. 2009;625:220–233. doi: 10.1016/j.ejphar.2009.06.063. [DOI] [PubMed] [Google Scholar]
- 61.Shacka JJ, Klocke BJ, Roth KA. Autophagy, bafilomycin and cell death: the “a-B-cs” of plecomacrolide-induced neuroprotection. Autophagy. 2006;2:228–230. doi: 10.4161/auto.2703. [DOI] [PubMed] [Google Scholar]
- 62.Hoyer-Hansen M, Jaattela M. Autophagy: an emerging target for cancer therapy. Autophagy. 2008;4:574–580. doi: 10.4161/auto.5921. [DOI] [PubMed] [Google Scholar]
- 63.Eisenberg-Lerner A, Kimchi A. The paradox of autophagy and its implication in cancer etiology and therapy. Apoptosis. 2009;14:376–391. doi: 10.1007/s10495-008-0307-5. [DOI] [PubMed] [Google Scholar]
- 64.Rockett JC, Larkin K, Darnton SJ, Morris AG, Matthews HR. Five newly established oesophageal carcinoma cell lines: phenotypic and immunological characterization. Br J Cancer. 1997;75:258–263. doi: 10.1038/bjc.1997.42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kabeya Y, Mizushima N, Ueno T, Yamamoto A, Kirisako T, Noda T, et al. LC3, a mammalian homologue of yeast Apg8p, is localized in autophagosome membranes after processing. EMBO J. 2000;19:5720–5728. doi: 10.1093/emboj/19.21.5720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Yoshimori T. Autophagy: a regulated bulk degradation process inside cells. Biochem Biophys Res Commun. 2004;313:453–458. doi: 10.1016/j.bbrc.2003.07.023. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.