Abstract
Autophagy-related (Atg) pathways deliver cytosol and organelles to the vacuole in double-membrane vesicles called autophagosomes, which are formed at the phagophore assembly site (PAS), where most of the core Atg proteins assemble. Atg28 is a component of the core autophagic machinery partially required for all Atg pathways in Pichia pastoris. This coiled-coil protein interacts with Atg17 and is essential for micropexophagy. However, the role of Atg28 in micropexophagy was unknown. We used the yeast two-hybrid system to search for Atg28 interaction partners from P. pastoris and identified a new Atg protein, named Atg35. The atg35Δ mutant was not affected in macropexophagy, cytoplasm-to-vacuole targeting or general autophagy. However, both Atg28 and Atg35 were required for micropexophagy and for the formation of the micropexophagic apparatus (MIPA). This requirement correlated with a stronger expression of both proteins on methanol and glucose. Atg28 mediated the interaction of Atg35 with Atg17. Trafficking of overexpressed Atg17 from the peripheral ER to the nuclear envelope was required to organize a peri-nuclear structure (PNS), the site of Atg35 colocalization during micropexophagy. In summary, Atg35 is a new Atg protein that relocates to the PNS and specifically regulates MIPA formation during micropexophagy.
Key words: Atg protein, peroxisome, micropexophagy, MIPA, nucleus
Introduction
Autophagy is a conserved process, which plays a central role in the adaptation of eukaryotic cells to changing environmental conditions. Defects in this process are associated with cancer, neurodegeneration, microbial infection and aging.1 Autophagy can be selective or nonselective. Nutrient starvation induces nonselective or general autophagy, wherein portions of the cytosol, including organelles, are engulfed by double-membrane vesicles, called autophagosomes. These vesicles originate from the phagophore assembly site (PAS) and fuse with the vacuole (yeast lysosome) to release their contents (autophagic bodies) into the vacuole lumen for degradation by hydrolases.2,3
Pexophagy, the selective autophagic degradation of peroxisomes, is triggered under conditions when these organelles are no longer needed for metabolism. The methylotrophic yeast, Pichia pastoris, when grown on methanol, has clusters of large peroxisomes needed for methanol utilization. In P. pastoris, pexophagy is easily achieved by the transfer of methanol-grown cells to ethanol or glucose medium. Under ethanol adaptation, individual peroxisomes are surrounded by a phagophore originating from the PAS that eventually forms pexophagosomes in a process known as macropexophagy. When methanol-grown cells of P. pastoris are shifted to glucose medium, micropexophagy occurs. Here, clusters of peroxisomes are surrounded by extensions of the vacuolar membrane, the vacuolar sequestering membranes (VSM), and by a separate structure known as the micropexophagic apparatus (MIPA), a small cup-shaped double-membrane structure formed from the PAS on the surface of a peroxisome cluster that is engulfed by the VSM. Once the fusion of the VSM and MIPA is complete, the cluster of peroxisomes is trapped inside the vacuole for degradation.4
Another type of selective autophagy is the constitutive cytoplasm-to-vacuole targeting (Cvt) pathway that operates under vegetative conditions in Saccharomyces cerevisiae and P. pastoris. It allows the selective transport of at least two vacuolar resident hydrolases, aminopeptidase I (Ape1) and α-mannosidase (Ams1).5,6
Of the 34 known autophagy-related (Atg) proteins, 17 are components of the core autophagic machinery, required for all Atg pathways, and 17 are pathway- or species-specific adaptations. An important question is, how does the core autophagic machinery interact with pexophagy- and Cvt-specific components to ensure selective cargo delivery to the vacuole? The pexophagy-specific additions to the P. pastoris core autophagic machinery include the peroxisome receptor Atg30, its adaptors Atg11 and Atg17, the phosphatidylinositol 3-phosphate binding protein, Atg24 and the sterol glucosyltransferase Atg26.7–11 The dual interaction of Atg30 with functionally active Atg11 and Atg17 organizes the pexophagy-specific PAS in P. pastoris.8,12 Atg17 interacts and colocalizes with Atg28, the core autophagic protein partially required for all Atg pathways, providing the first link between the pexophagy-specific and general components of the PAS in P. pastoris.12,13 P. pastoris Atg28 might be related to two S. cerevisiae proteins, Atg29 and Atg31, both of which interact with Atg17, and together with Atg1 and Atg13, organize the autophagy-specific PAS in S. cerevisiae.14–18 However, knowledge of other interacting partners and the role of Atg28 was lacking.
We performed a yeast two-hybrid (YTH) library screen with Atg28 and found a novel protein, Atg35, required specifically for micropexophagy. Both deletion and overexpression of the ATG35 gene specifically inhibited MIPA formation, but not pexophagosome formation or macropexophagy. This paper provides the first genetic evidence for a difference in the molecular mechanism of MIPA and pexophagosome formation. Atg35 is also the first nuclear Atg protein known to regulate the Atg pathway in yeasts. Our data on interaction and localization of Atg35 suggest that it is a new and important component of the autophagic machinery under micropexophagy conditions.
Results
Identification of Atg35 as an Atg28 interaction partner.
Of the 34 Atg proteins playing roles in Atg pathways, many act together and form a network of interacting proteins. We sought to identify the interaction partners of P. pastoris Atg28, a component of the core autophagic machinery partially required for all Atg pathways and particularly for micropexophagy.12,13
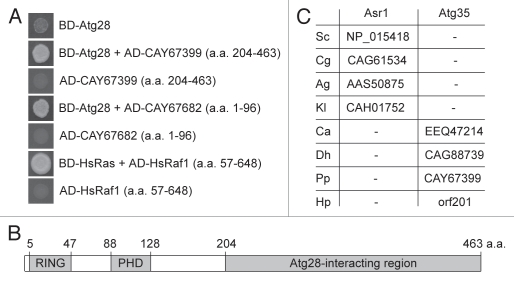
We screened a P. pastoris YTH genomic DNA library (see Materials and Methods) in S. cerevisiae cells with PpAtg28 as the bait. As a positive control, we used the interaction between human Ras and truncated Raf1 (amino acids (a.a.) 57–648) proteins.19 Sequence analysis of two positive clones revealed the C-terminal part of the hypothetical protein, CAY67399 (a.a. 204–463), and the N terminus of Rdi1, CAY67682 (a.a. 1–96) (Fig. 1A). Here we describe the functional role of CAY67399, which was named Atg35.
Figure 1.
Identification of Atg35. (A) Atg28 interacted with the C-terminal part of Atg35 (CAY67399) and the N terminus of Rdi1 (CAY67682) in the YTH library screen. The S. cerevisiae cells with indicated constructs were grown in YTH medium. Human Ras and truncated Raf1, which are known to interact with each other, were used as a positive control for interaction. (B) Structure of the P. pastoris Atg35. Atg35 has two conserved domains, the RING-finger and the PHD-finger. The Atg28-interacting region of Atg35 is also shown. (C) Atg35 co-evolved with Atg28 in yeasts. Saccharomycetales have either (i) Atg29, Atg31 and Asr1 or (ii) Atg28 and Atg35. The accession numbers of Asr1 and Atg35 are presented. Abbreviations of organisms: Sc, S. cerevisiae; Cg, C. glabrata; Ag, A. gossypii; Kl, K. lactis; Ca, C. albicans; Dh, D. hansenii; Pp, P. pastoris and Hp, H. polymorpha. For HpAtg35 (orf201) see reference 50.
Atg35 consists of 463 a.a. and has two putative domains: a RING-finger (a.a. 5–47) and a PHD-finger (a.a. 88–128) (Fig. 1B). The closest orthologs of Atg35 are hypothetical proteins from Pichia stipitis (PICST_60919), Pichia guilliermondii (PGUG_03993) and Clavispora lusitaniae (CLUG_01611). Recently, we reported that yeasts from the order Saccharomycetales have either one large Atg28 protein or two smaller proteins, Atg29 and Atg31.12 Here, we analyzed the genomes of the same eight yeasts and found that all contain a protein with the N-terminal RING and PHD fingers, either a longer Atg35 or a shorter Asr1 (see Fig. S1 in Sup. Material). Asr1 and Atg35 proteins also share a C-terminal nuclear localization signal.20 However, Asr1 proteins lack the internal fragment of approximately 260 a.a. that has two conserved motifs (see Fig. S2 in Sup. Material). Importantly, Candida albicans, Debaryomyces hansenii, P. pastoris and Hansenula polymorpha have both Atg28 and Atg35, but S. cerevisiae, Candida glabrata, Ashbya gossypii and Kluyveromyces lactis have Atg29, Atg31 and Asr1 (Fig. 1C). Therefore, Atg35 might have co-evolved with Atg28 in yeasts, and these two proteins might be functionally related as explained later.
Atg35 is a micropexophagy-specific protein required for efficient MIPA formation.
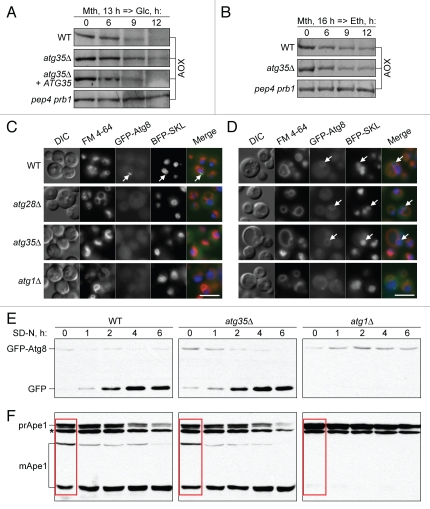
We examined the P. pastoris atg35Δ mutant for pexophagy by western blotting for peroxisomal alcohol oxidase (AOX) (Fig. 2A and B). Upon adaptation of methanol-grown cells to ethanol (induces macropexophagy), there was no difference in degradation of AOX between the wild-type (WT) and atg35Δ cells. However, upon glucose adaptation (induces micropexophagy), the levels of AOX in the atg35Δ mutant decreased more slowly and to a lesser extent than in the WT strain. The ATG35 gene complemented the micropexophagy defect in the atg35Δ mutant. Both micro- and macropexophagy were completely blocked in the pep4 prb1 mutant deficient in vacuolar proteases A and B (Fig. 2A and B). The results of our biochemical experiments suggested that Atg35 is partially required for micropexophagy, but not for macropexophagy.
Figure 2.
Atg35 is a micropexophagy-specific protein required for efficient MIPA formation. (A and B) Atg35 is required for micro- but not macropexophagy. Cells were grown overnight in SM and transferred to (A) SD or (B) SE. At the indicated time-points culture samples were collected and processed for immunoblotting for AOX. “atg35Δ + ATG35”, the atg35Δ mutant complemented with the ATG35 gene (SVN2). (C and D) Atg28 and Atg35 are specifically required for efficient MIPA formation. The WT (STN78), atg28Δ (STN87), atg35Δ (STN179) and atg1Δ (STN71) cells expressing GFP-Atg8 and BFP-SKL were grown overnight in SM with FM 4-64 and transferred to (C) SD or (D) SE for 1 h. VSM were labeled with FM 4-64, MIPA and pexophagosomes with GFP-Atg8 and peroxisomes with BFP-SKL. Arrows point to (C) MIPA and (D) pexophagosomes. Bar, 5 µm. (E and F) Atg35 is not required for the general autophagy and Cvt pathways. (E) GFP-Atg8 processing and (F) prApe1 maturation assays. The WT (STN70), atg35Δ (SVN3) and atg1Δ (STN66) cells expressing GFP-Atg8 were grown overnight in SD and transferred to SD-N. At the indicated time-points, culture samples were collected and processed for immunoblotting for (E) GFP and (F) Ape1. *, nonspecific band; red box, maturation of prApe1 under vegetative conditions.
Next, we studied the roles of Atg28 and Atg35 in micropexophagy. During micropexophagy, the VSM and the MIPA need to form.4 We compared the VSM and MIPA formation in the WT and mutants by fluorescence microscopy using: (i) Atg8 N-terminally tagged with green fluorescent protein (GFP-Atg8) to label the MIPA, (ii) blue fluorescent protein with the C-terminal tripeptide, Ser-Lys-Leu (BFP-SKL), to label peroxisomes and (iii) the dye N-(3-triethylammoniumpropyl)-4-(p-diethylaminophenylhexatrienyl) pyridinium dibromide (FM 4–64) to label the vacuole membrane and VSM. The VSM formation was normal in all tested strains. While the MIPA was found in the WT (17.6 ± 0.3% of cells), it was 1.5-fold less frequent in atg35Δ (11.5 ± 0.7% of cells), p < 0.01. Interestingly, the atg28Δ mutant was completely blocked in MIPA formation (0.8 ± 0.3% of cells with a MIPA), as was the negative control, atg1Δ (0.6 ± 0.3% of cells) (Fig. 2C). In contrast to the defects in MIPA formation, both atg28Δ and atg35Δ mutants exhibited normal formation of pexophagosomes under macropexophagy conditions (Fig. 2D). This was completely blocked in the atg1Δ strain. Our pexophagy experiments suggested that Atg28 and Atg35 are specifically required for MIPA formation during micropexophagy.
We also studied the role of Atg35 in the general autophagy and Cvt pathways. Cells of the WT, atg35Δ and atg1Δ strains expressing GFP-Atg8 were transferred from nitrogen-rich to nitrogen-starvation conditions to induce general autophagy. The kinetics of GFP-Atg8 processing is a sensitive measure of autophagy rate.21 The release of more proteolytically stable GFP after vacuolar degradation of Atg8 was examined by western blot. In contrast to atg1Δ, the atg35Δ mutant exhibited normal processing of GFP-Atg8 (Fig. 2E). These results were confirmed by the prApe1 maturation assay. The vacuolar processing of the precursor of Ape1 (prApe1) to the mature (mApe1) forms occurs by the Cvt and general autophagy pathways under vegetative and starvation conditions, respectively.5 In contrast to atg1Δ, the atg35Δ mutant had normal maturation of prApe1 under starvation conditions (Fig. 2F). The results of two biochemical assays indicate that Atg35 is not required for general autophagy.
The lack of Atg35 had no effect on the Cvt pathway because the maturation of prApe1 was normal in the atg35Δ mutant under vegetative conditions, but was blocked in the atg1Δ mutant (Fig. 2F compares the WT, atg35Δ and atg1Δ at the time-point “0 hour”). Altogether, our results indicate that Atg35 is not required for the macropexophagy, Cvt or general autophagy pathways. Instead, it plays a specific role in micropexophagy at the MIPA formation stage.
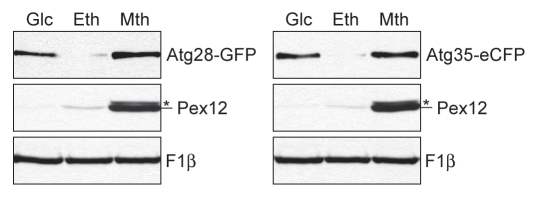
Atg28 and Atg35 are co-expressed.
The expression of ATG28 and ATG35 genes was compared under peroxisome proliferation and degradation conditions. The WT strains expressing Atg28-GFP and Atg35-eCFP under their own promoters were grown overnight in glucose, ethanol and methanol media. Independent of carbon source, cells of both strains had the same amount of F1β (loading control, Fig. 3). The levels of Pex12 (carbon source control) increased in the order glucose, ethanol, methanol, as expected. However, we observed an unusual expression of Atg28 and Atg35: very low levels of both proteins in ethanol and high levels of both proteins in glucose and methanol (Fig. 3). The co-expression of Atg28 and Atg35 on methanol and glucose is consistent with their interaction (Fig. 1A and shown later) and specific requirement for micropexophagy.
Figure 3.
Atg28 and Atg35 are co-expressed in methanol and glucose media. The WT strains expressing Atg28-GFP (STN120) and Atg35-eCFP (STN227) under their own promoters were grown for 16 h in glucose (SD), ethanol (SE) and methanol (SM) media. The same number of cells from each culture was TCA precipitated and immunoblotted for F1β, Pex12 and GFP/eCFP. *, nonspecific band.
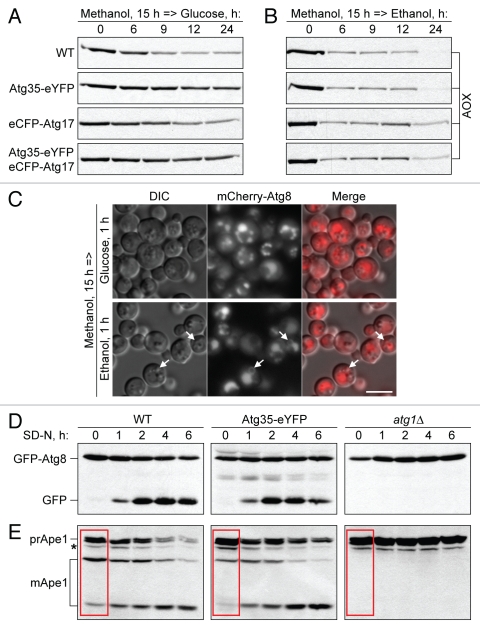
Overexpression of Atg35 specifically inhibits MIPA formation.
To further examine the role of Atg35 in autophagy-related pathways, we overexpressed Atg35-eYFP and eCFP-Atg17 (as a control) in the WT strain and followed pexophagy by western blotting for AOX (Fig. 4A and B). Overexpression of eCFP-Atg17 slightly delayed both micro- and macropexophagy. However, overexpression of Atg35-eYFP inhibited micropexophagy, but had no effect on macropexophagy. The strain that overexpressed both Atg35-eYFP and eCFP-Atg17 was blocked in micropexophagy (due to Atg35-eYFP) and slightly affected in macropexophagy (due to eCFP-Atg17) (Fig. 4A and B). These results suggest that an excess of Atg35, but not of Atg17, specifically inhibits micropexophagy. We also verified that overexpression of Atg35, but not its tag, inhibits micropexophagy. The strain expressing Atg35-eCFP under its own promoter had normal micropexophagy rates in comparison with WT, whereas the strain that overexpressed untagged Atg35 was blocked in micropexophagy, like the strain that overexpressed Atg35-eYFP (see Fig. S3 in Sup. Material).
Figure 4.
Overexpression of Atg35 inhibits MIPA formation. (A and B) Overexpression of Atg35, but not of Atg17, specifically inhibits micropexophagy. The cells from WT (PPY12 h) and WT overexpressing Atg35-eYFP (STN274), eCFP-Atg17 (STN350), or both Atg35-eYFP and eCFP-Atg17 (STN322), were grown overnight in SM and transferred to (A) SD or (B) SE. At the indicated time-points, culture samples were collected and processed for immunoblotting for AOX. (C) Overexpression of Atg35 specifically affects MIPA formation. The WT strain overexpressing Atg35-eYFP and expressing the endogenous levels of mCherry-Atg8 (STN338) was grown overnight in SM and transferred to SD or SE for 1 h. VSM, MIPA and pexophagosomes were labeled with mCherry-Atg8. Arrows point to pexophagosomes. Bar, 5 µm. (D and E) Overexpression of Atg35 does not affect general autophagy, but delays the Cvt pathway under vegetative, but not starvation, conditions. (D) GFP-Atg8 processing and (E) prApe1 maturation assays. The WT (STN70), WT overexpressing Atg35-eYFP (SJCF1376) and atg1Δ (STN66) cells, all expressing the endogenous levels of GFP-Atg8, were grown overnight in SD and transferred to SD-N. At the indicated time-points, culture samples were collected and processed for immunoblotting for (D) GFP and (E) Ape1. *, nonspecific band; red box, maturation of prApe1 under vegetative conditions.
Then, we studied the stage of micropexophagy affected by overexpression of Atg35-eYFP. Both the VSM and MIPA were visualized using mCherry-Atg8. VSM formation was not affected by overexpression of Atg35-eYFP, but MIPA formation was blocked (Fig. 4C). In contrast, pexophagosomes were normally formed in the strain that overexpressed Atg35-eYFP under macropexophagy conditions (Fig. 4C and see arrows). We also labeled the MIPA with GFP-Atg8 in the strain that overexpressed untagged Atg35 and measured the frequency of MIPA formation. Overexpression of Atg35 decreased the percentage of cells with the MIPA three-fold from 8.2 ± 1.7% (STN70) to 2.7 ± 1.1% (SJCF1473), p < 0.01. The negative control, atg1Δ (STN66), was completely blocked with only 0.6 ± 0.5% cells containing a MIPA. Combined, our results suggest that the overexpression of Atg35 specifically affects MIPA formation and interferes with micropexophagy.
We also monitored the effect of Atg35-eYFP overexpression on the general autophagy and Cvt pathways. Cells of the WT, WT overexpressing Atg35-eYFP and atg1Δ, all expressing the endogenous levels of GFP-Atg8, were transferred from nitrogen-rich to nitrogen-starvation conditions. Overexpression of Atg35-eYFP did not affect the vacuolar processing of GFP-Atg8 upon nitrogen starvation (Fig. 4D). Similar results were obtained in the prApe1 maturation assay. The strain that overexpressed Atg35-eYFP had normal vacuolar maturation of prApe1 under starvation conditions (Fig. 4E). The results of two biochemical assays indicated that the overexpression of Atg35 does not affect general autophagy. However, it did affect the Cvt pathway, as maturation of prApe1 was less efficient in the strain that overexpressed Atg35-eYFP under vegetative conditions (Fig. 4E and compare the three strains at the time-point “0 hour”). Therefore, the overexpression of Atg35 does not affect macropexophagy or general autophagy. It specifically affects the Cvt pathway under vegetative, but not starvation, conditions, while also impairing micropexophagy at the MIPA formation stage.
Atg17 recruits Atg35 to a novel perinuclear structure (PNS) during micropexophagy.
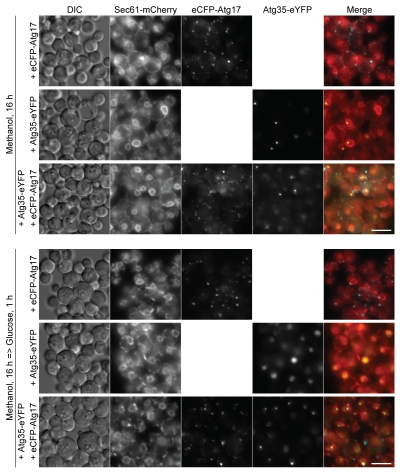
Atg35 has a putative nuclear localization signal near the C terminus (see Fig. S1 in Sup. Material). We tried to address the localization of Atg35-eCFP expressed under its own promoter, but the expression of Atg35-eCFP was below the level of detection by fluorescence microscopy even in methanol medium, wherein it is induced maximally (Fig. 3 and data not shown). We therefore examined the localization of overexpressed Atg35-eYFP relative to the peripheral ER/nuclear envelope marker, Sec61-mCherry,22 and the PAS marker, overexpressed eCFP-Atg17.7 In methanol-grown cells, eCFP-Atg17 localized to several dot-like structures scattered along the peripheral ER. In some cells, one of the eCFP-Atg17 dots could also be found at the nuclear envelope (Fig. 5 and top panel). After the transfer of cells to glucose medium, eCFP-Atg17 accumulated on two to three dots/cell, as previously described in reference 7 and 23. Interestingly, one to two of these dots localized at the nuclear envelope and the rest on the peripheral ER or in the cytosol (Fig. 5 and third panel from bottom). Co-overexpression of Atg35-eYFP did not affect the trafficking of eCFP-Atg17: the majority of the protein still moved from several dots on the peripheral ER to one to two dots on the nuclear envelope under micropexophagy conditions (Fig. 5).
Figure 5.
Atg17 recruits Atg35 at the PNS during micropexophagy. The WT strains overexpressing eCFP-Atg17 (STN373), Atg35-eYFP (STN343) or both eCFP-Atg17 and Atg35-eYFP (STN346), and expressing the endogenous levels of Sec61-mCherry were grown overnight in SM (three top panels) and transferred to SD (three bottom panels) for 1 h. The peripheral ER and nuclear envelope were labeled with Sec61-mCherry and the PAS with eCFP-Atg17. Bar, 5 µm.
At the same time, Atg35-eYFP localized to the nucleus and a single dot-like structure on the nuclear envelope, the PNS, in methanol medium. The transfer of cells to glucose medium resulted in homogenous distribution of Atg35-eYFP in the nucleus (Fig. 5 and second panel from bottom). However, co-overexpression of eCFP-Atg17 stimulated relocalization of Atg35-eYFP at the PNS during micropexophagy. Moreover, Atg35-eYFP at the PNS necessarily colocalized with one of the eCFP-Atg17 dots in glucose medium (Fig. 5 and bottom panel). We would like to stress that, in methanol medium, Atg35-eYFP and eCFP-Atg17 seldom colocalized at the PNS. Accumulation of eCFP-Atg17 on the nuclear envelope under micropexophagy conditions was required to organize the PNS and recruit Atg35-eYFP to this structure.
The latter conclusion was confirmed with Atg35-eYFP expressed from the moderate YPT1 promoter. First, we showed that moderate expression of Atg35-eYFP did not inhibit micropexophagy in the WT strain (see Fig. S4A in Sup. Material). Then, we found that moderately expressed Atg35-eYFP was diffuse in the nucleus and did not form the PNS, independent of eCFP-Atg17 overexpression, in methanol medium. However, after the transfer of cells to glucose medium, eCFP-Atg17 stimulated localization of Atg35-eYFP at the PNS (see Fig. S4B in Sup. Material). Therefore, localization of overexpressed Atg35-eYFP at the PNS in methanol medium (Fig. 5) was most probably an artifact of protein overexpression, whereas localization of Atg35-eYFP at the PNS in glucose medium (Figs. 5 and S4B) depends on eCFP-Atg17 and might be relevant for micropexophagy.
Atg28 mediates the interaction between Atg17 and Atg35.
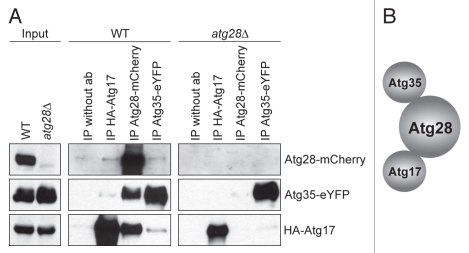
Previously, we found that Atg28 interacts with Atg17,12 and the C-terminal part of Atg35 (Fig. 1A) in the YTH system. To further study the interaction between these three proteins we overexpressed Atg28-mCherry, Atg35-eYFP and HA-Atg17 in the WT strain. Cells were transferred from methanol to glucose medium to induce micropexophagy, and protein-protein interactions were studied by co-immunoprecipitation. Although with each of the proteins, we could co-immunoprecipitate the other two, the highest amounts of Atg35-eYFP and HA-Atg17 were pulled down with Atg28-mCherry (Fig. 6A and middle panel). These results suggested that the Atg17-Atg35 interaction might be indirect and mediated by Atg28. To check this possibility, we introduced Atg35-eYFP and HA-Atg17 in the atg28Δ mutant. Without Atg28, Atg35-eYFP and HA-Atg17 failed to co-immunoprecipitate with each other (Fig. 6A and right panel). Taken together, our YTH and co-immunoprecipitation results support a model where Atg28 plays a central role by mediating the interaction between Atg17 and Atg35 (Fig. 6B).
Figure 6.
Atg28 mediates the interaction of Atg35 with Atg17. (A) Co-immunoprecipitation of Atg17, Atg28 and Atg35. The WT strain overexpressing Atg28-mCherry, Atg35-eYFP and HA-Atg17 (STN401) and the atg28Δ mutant overexpressing Atg35-eYFP and HA-Atg17 (STN387) were grown for 16 h in SM and transferred for 1 h to SD. Immunoprecipitation was performed as described in Materials and Methods. (B) Schematic view of Atg17, Atg28 and Atg35 interactions.
Discussion
Pexophagy can proceed by two different modes, micropexophagy and macropexophagy. There are many morphological differences between the two, including VSM and MIPA formation during micropexophagy, in contrast to pexophagosome formation during macropexophagy.4 However, the genetic dissection of the two pathways is at its infancy and this analysis is complicated by the fact that most of the known Atg proteins are required for both modes of pexophagy at the stage of pexophagy-specific PAS formation or its elongation into the MIPA or pexophagosome. Until now only two proteins were shown to be required for micro-, but not macropexophagy, the α-subunit of phosphofructokinase, Pfk1 and the vacuolar membrane protein, Vac8. However, Pfk1 might be implicated in glucose signaling events and Vac8 may be involved in VSM formation and homotypic VSM fusion.24–26 Here, we present the first evidence that MIPA and pexophagosome formation are genetically distinct. We show that the core autophagic protein, Atg28, together with its novel interaction partner Atg35, represent the micropexophagy-specific branch of the autophagic machinery required specifically for MIPA, but not for pexophagosome formation.
Atg28 interacts with Atg17 and is at least partially required for all Atg pathways with a stronger requirement for micro-, than for macropexophagy.12,13 Here we show that Atg28 is essential for MIPA, but dispensable for pexophagosome, formation (Fig. 2C and D). Therefore, it is not surprising that a newly identified interaction partner of Atg28, Atg35, is also micropexophagy-specific and required for MIPA formation. However, in contrast to the partial necessity of Atg28, Atg35 is not required for the macropexophagy, general autophagy and Cvt pathways (Fig. 2). This can explain why the phenotype of the atg35Δ mutant in micropexophagy is less severe than that of atg28Δ. Indeed, in addition to its specific role in MIPA formation together with Atg35, Atg28 might be also involved at the earlier and more general step of PAS organization together with Atg17. Under micropexophagy conditions, Atg28 could be involved in both functions and may therefore become indispensable.
Interestingly, Atg35 could have co-evolved with Atg28 in yeasts, as the species with Atg29 and Atg31 have a much shorter RING- and PHD-finger containing protein, Asr1 (see Figs. S1 and S2 in Sup. Material). Atg35 and Atg28 are also co-expressed in media that induce peroxisomes and their degradation by micropexophagy (Fig. 3). Finally, Atg28 mediates the interaction of Atg35 with Atg17 (Fig. 6), a scaffold protein that together with Atg30 and Atg11 organizes the pexophagy-specific PAS.8,12 In summary, our data indicate that Atg28 is a branching point of the core autophagic machinery, which engages the micropexophagy-specific Atg35 for efficient MIPA formation.
Like the S. cerevisiae Asr1,20,27 the P. pastoris Atg35 is a very low-abundance nuclear protein. Under peroxisome proliferation conditions, Atg35 localizes to the nucleus. At this stage, Atg17 is mainly localized to several dot-like structures on the peripheral ER. We note that since Atg17 does not have any discernible transmembrane domains, it must be peripherally associated with the ER membranes. The induction of micropexophagy stimulates relocalization of Atg17 from multiple spots on the peripheral ER to two to three dots, one to two of which are necessarily localized on the nuclear envelope. We do not understand how this relocation occurs, but under micropexophagy conditions, Atg35 always colocalizes with Atg17 at a single dot-like PNS. Trafficking of Atg17 plays an important role in the localization of Atg35 to the PNS, as without Atg17 overexpression, Atg35 is diffuse in the nucleus (Figs. 5 and S4B). Several previous studies used Atg17 as a marker of the pexophagy-specific PAS in P. pastoris and also found two Atg17-containing dots.7,23 Similar to our finding, only one of the Atg17 dots colocalized with one of the Atg24 perivacuolar spots, probably the PAS.7 To determine whether the identified PNS and PAS are the same or different Atg17-containing structures, further studies will be required.
The autophagy-specific PAS in S. cerevisiae is organized by two Atg17 complexes, Atg1-Atg13-Atg17 and Atg17-Atg29-Atg31.14,15,18 Interestingly, when Atg17 is artificially targeted to the plasma membrane, the PAS is organized at that location.28 Here, we show that Atg17 dots are mostly localized to the nuclear envelope under micropexophagy conditions (Fig. 5). Therefore, independent of whether the PNS and PAS are the same or different structures organized by Atg17, the nuclear envelope might still play an important role in MIPA formation. Recently, it was shown that vesicular trafficking from and to the ER mediated by Sar1 is essential for MIPA formation.23 Our Atg17 and Atg35 colocalization studies are in line with these observations and point to the possibility that the nuclear envelope is the specific region of the ER involved in this process. However, clarification of this possibility must await further studies.
Importantly, overexpression of Atg35 inhibits micropexophagy, but not macropexophagy or general autophagy (Fig. 4). One possibility is that the excess of Atg35 at the PNS interferes with retrograde transport of ER proteins involved in MIPA formation. Specific inhibition of MIPA formation was recently described upon overexpression of the dominant-negative mutant of Sar1, Sar1H79G.23 This mutant, Sar1H79G, has defective GTPase activity and, therefore, inhibits the recycling of proteins back to the ER.29,30 However, the lack of sufficient Atg17 leads to diffuse localization of overexpressed Atg35 in the nucleus (Fig. 5 and second panel from bottom) and Atg35 still inhibits micropexophagy (Fig. 4A). This result suggests a second possibility, that Atg35 might negatively regulate micropexophagy at the level of transcription. Recently, it was shown that the S. cerevisiae Asr1 can directly bind and inactivate the RNA polymerase II complex.27 In addition, protein synthesis is required for the progression of micro- but not macropexophagy.31,32 Therefore, we hypothesize that aside from its positive role in MIPA formation in glucose medium, Atg35 might negatively regulate the expression of genes critical for the progression of micropexophagy in methanol medium. This point will be addressed in future studies.
Although Atg35 is the first known nuclear protein required for the Atg pathway in yeasts, nucleo-cytoplasmic shuttling of Atg proteins is increasingly recognized as a regulatory mechanism of autophagy in mammals.33 Nuclear export of both Beclin 1 (corresponds to Atg6/Vps30 in yeasts) and TP53INP2/DOR (does not have a yeast homolog) was shown to be required for autophagy in mammalian cells.34–36 Therefore, nuclear localization of these proteins might play a role in preventing autophagy in the cytoplasm before its activation by starvation. Further studies of Atg35 and its mammalian homolog rA9 that, like Asr1, directly binds the C-terminal domain of the largest subunit of RNA polymerase II27,37 will shed light on the role of the nucleus in the regulation of autophagy.
Materials and Methods
Strains and plasmids.
For routine cloning and propagation of plasmid DNA the Escherichia coli DH5α strain was used. The S. cerevisiae strain SKY48 (MATα trp1 his3 ura3 6lexAop-LEU2 3cIop-LYS2) (Invitrogen, C833-00) was used for the YTH. The P. pastoris strains and plasmids used in this study are listed in Table 1.
Table 1.
P. pastoris strains and plasmids used in this study
| Name | Description | Genotype and plasmid | Source |
| GS115 | WT | his4 | 38 |
| GS200 | WT | arg4 his4 | 39 |
| PPS2 | atg28Δ | GS200 atg28Δ::ScARG4 | 13 |
| PPY12h | WT | arg4 his4 | 40 |
| R12 | atg1Δ | GS115 atg1-1::ZeocinR | 41 |
| SJCF1376 | WT | STN274 his4::pJCF419 (PATG8-GFP-ATG8, HIS4, HygromycinR) | This study |
| SJCF1472 | WT | PPY12h his4::pJCF549 (PGAPDH-ATG35, HIS4) | This study |
| SJCF1473 | WT | SJCF1472 HIS4::pJCF419 (PATG8-GFP-ATG8, HygromycinR) | This study |
| SJCF1474 | WT | PPY12h his4::pJCF552 (PYPT1-ATG35-eYFP, HIS4) | This study |
| SJCF1475 | WT | STN350 his4::pJCF552 (PYPT1-ATG35-eYFP, HIS4) | This study |
| SMD542 | WT | GS200 PAOX-β-GAL, ARG4 | 13 |
| SMD1163 | pep4 prb1 | pep4 prb1 his4 | 32 |
| STN66 | atg1Δ | R12 his4::pJCF208 (PATG8-GFP-ATG8, HIS4) | 12 |
| STN70 | WT | GS115 his4::pJCF208 (PATG8-GFP-ATG8, HIS4) | 12 |
| STN71 | atg1Δ | STN66 PAOX1::pRDM054 (PAOX1-BFP-SKL, HygromycinR) | This study |
| STN77 | atg28Δ | PPS2 his4::pJCF208 (PATG8-GFP-ATG8, HIS4) | 12 |
| STN78 | WT | STN70 PAOX1::pRDM054 (PAOX1-BFP-SKL, HygromycinR) | This study |
| STN87 | atg28Δ | STN77 PAOX1::pRDM054 (PAOX1-BFP-SKL, HygromycinR) | This study |
| STN120 | WT | PPY12h ATG28::pTYN5 (ATG28-GFP, HIS4) | This study |
| STN179 | atg35Δ | SVN3 PAOX1::pRDM054 (PAOX1-BFP-SKL, HygromycinR) | This study |
| STN227 | WT | PPY12h ATG35::pTYN14 (ATG35-eCFP, G418R) | This study |
| STN274 | WT | PPY12h his4::pTYN17 (PGAPDH-ATG35-eYFP, HIS4) | This study |
| STN315 | atg28Δ | PPS2 his4::pTYN17 (PGAPDH-ATG35-eYFP, HIS4) | This study |
| STN322 | WT | STN274 arg4::pJCF170 (PGAPDH-eCFP-ATG17, ARG4) | This study |
| STN338 | WT | STN274 arg4::pJCF477 (PATG8-mCHERRY-ATG8, ARG4) | This study |
| STN343 | WT | STN274 SEC61::pKSN256 (SEC61-mCHERRY, HygromycinR) | This study |
| STN346 | WT | STN322 SEC61::pKSN256 (SEC61-mCHERRY, HygromycinR) | This study |
| STN350 | WT | PPY12h arg4::pJCF170 (PGAPDH-eCFP-ATG17, ARG4) | This study |
| STN373 | WT | STN350 SEC61::pKSN256 (SEC61-mCHERRY, HygromycinR) | This study |
| STN385 | WT | STN274 his4::pTYN26 (PGAPDH-HA-ATG17, HIS4, HygromycinR) | This study |
| STN387 | atg28Δ | STN315 his4::pTYN26 (PGAPDH-HA-ATG17, HIS4, HygromycinR) | This study |
| STN401 | WT | STN385 arg4::pJCF500 (PGAPDH-ATG28-mCHERRY, ARG4) | This study |
| SVN1 | atg35Δ | GS200 atg35Δ::ScARG4 | This study |
| SVN2 | atg35Δ | SVN1 his4::pTYN10 (PATG35-ATG35, HIS4) | This study |
| SVN3 | atg35Δ | SVN1 his4::pJCF208 (PATG8-GFP-ATG8, HIS4) | This study |
| Y11430 | WT | Prototroph | Collection strain |
Media and growth conditions.
Yeast strains were grown at 30°C in a rich YPD (1% w/v yeast extract, 2% w/v peptone, 2% w/v glucose) or YPDhyg (YPD with 0.8 mg/ml of hygromycin B) medium. The YTH medium contained 0.67% w/v yeast nitrogen base without amino acids (YNB), 2% w/v galactose and 1% w/v raffinose. Peroxisome biogenesis was induced in SM (0.67% w/v YNB, 0.5% v/v methanol for liquid medium and 1% v/v methanol for plates) and degradation in SE (0.67% w/v YNB, 0.5% v/v ethanol for liquid medium and 1% v/v ethanol for plates) or SD (0.67% w/v YNB, 2% w/v glucose for liquid medium and 1% w/v glucose for plates) medium. For nitrogen starvation, SD-N (0.17% w/v YNB without ammonium sulfate, 2% w/v glucose) was used. For solid media, 2% w/v agar was added.
The YTH genomic DNA library screening.
We used the Interaction Trap based YTH system (a gift from Prof. Serebriiskii from Fox Chase Cancer Center, Philadelphia, PA USA) with two reporter genes, lacZ and LEU2.42,43 A LexA-Atg28 “bait fusion” was created and transformed into the S. cerevisiae SKY48 strain. The expression of LexA-Atg28 was verified by western blotting.
To create the P. pastoris YTH genomic DNA library (a set of “prey fusions”) the P. pastoris Y11430 genomic DNA was extracted and partially digested with Tsp509I. The resulting digestion mix was purified using the QIAquick Gel Extraction Kit (QIAGEN, 28704) to exclude genomic DNA fragments larger than 10 kb and ligated with YTH vector pJG4-5,44 which was digested with EcoRI and dephosphorylated. The constructed YTH genomic DNA library consisted of about 260,000 independent clones, 100,000 of which carried an insert of genomic DNA larger than 1 kb and covered the P. pastoris genome more than 10 times.
The S. cerevisiae strain with LexA-Atg28 was transformed with the P. pastoris YTH genomic DNA library. Screening for the “positive” clones with protein-protein interactions was performed with both lacZ and LEU2 reporters. The library plasmids from “positive” clones were isolated, sequenced and reintroduced into the S. cerevisiae strain with LexA-Atg28 to confirm the identified interactions.
Construction of the atg35Δ mutant.
The deletion cassette containing 1.5 kbp fragment upstream of the PpATG35 ORF, the ScARG4 gene and a 0.9 kbp 3′-fragment of the PpATG35 ORF together with the downstream region was created to knock out the first 0.7 kbp of the PpATG35 ORF by the gene replacement method. The ATG35 gene was deleted in the P. pastoris GS200 strain and the atg35Δ mutant was verified by PCR and Southern blotting. To complement the atg35Δ mutant, the plasmid pTYN10 containing the PpATG35 ORF with a 508 bp promoter region was constructed for integration in the HIS4 locus.
Biochemical studies of pexophagy.
The AOX plate assay was performed, as described previously in reference 45. For pexophagy assays in a liquid culture, cells were grown overnight in 3 ml of YPD. On the next day, 0.15 ml of the overnight culture was transferred into 2.85 ml of fresh YPD and pre-grown for 6 h. Then cells were washed twice with distilled water and transferred into 3 ml of SM medium with starting OD600 0.3 to induce peroxisome biogenesis for 13–16. Cells were pelleted, washed with distilled water and transferred into 3 ml of SD or SE with starting OD600 0.5. An equal volume of each culture (0.5 ml) was withdrawn at each time point and prepared for immunoblotting, as previously described in reference 46. For detection, anti-AOX antibodies were used.47 Anti-Pex12,48 anti-F1β49 and anti-GFP (Roche, 11-814-460-001) antibodies were used in Atg28 and Atg35 expression studies.
MIPA and pexophagosome formation.
Endogenously expressed GFP-Atg8 or mCherry-Atg8 was used as MIPA and pexophagosome markers. Cells expressing mCherry-Atg8 were grown in SM and cells with GFP-Atg8 were grown in SM with 2.5 µg/ml of FM 4-64 (Invitrogen, T-3166) for 15 h and transferred to SD or SE for 1 h to induce micro- or macropexophagy, respectively. The number of cells with MIPA was counted on maximum intensity projections of the deconvolved (inverse filter algorithm) Z-stacks (20 slices, 0.255 µm apart) of GFP-Atg8 images. Data are presented as average ± standard deviation of three independent experiments. Student's t-test for unpaired data sets was used to estimate statistical significance.
General autophagy and Cvt pathways.
The GFP-Atg8 processing and prApe1 maturation assays to study the rates of general autophagy and steady-state of the Cvt pathway were described previously in reference 12.
Co-immunoprecipitation studies.
Cells were grown for 16 h in SM and transferred for 1 h to SD medium before extraction. Fifty OD600 equivalents of yeast cells were washed in phosphate-buffered saline (pH 7.4) and lysed with glass beads (vortexed five times for 2 min at 4°C) in 1 ml IP lysis buffer (50 mM HEPES-KOH, pH 7.4, 0.1 M NaCl, 1% v/v Triton, 10% v/v glycerol, 5 mM NaF, 1 mM phenylmethylsulfonyl fluoride and protease inhibitor cocktail [Sigma-Aldrich, P8215]) and centrifuged (300 g, 3 min). Membrane protein solubilization was performed together with a pre-clearing step. For this purpose 50 µl of GammaBind G Sepharose (GE Healthcare, 17-0885-01) was added to the supernatant and incubated at 4°C for 4.5 h with rotation. Next, the sample was centrifuged (21,000 g, 15 min) and the supernatant fraction was incubated overnight with the selected primary antibody at 4°C in a rotating tube (1 µl anti-HA, Roche, 11-666-606-001; 10 µl anti-GFP, Roche, 11-814-460-001; 10 µl anti-DsRed, Clontech, 632496). After the overnight incubation, 100 µl of GammaBind G Sepharose was added to the supernatant fraction with the primary antibody and incubated at 4°C for 3 h with rotation. Beads were washed two times (5 ml) with the IP lysis buffer for 15 min and 1 time (5 ml) with IP lysis buffer supplemented with extra NaCl (0.5 M final concentration) for 5 min. Bound protein was eluted with 100 µl of sample buffer, boiled in a water bath for 5 min, resolved by SDS-PAGE (loading was as follows: input, 0.5 OD600 equivalent and immunopurified proteins (IP), 10 OD600 equivalents) and visualized by immunoblotting. After blotting, the membranes were probed with the same primary antibodies. The cross reactions between the denatured IgG heavy chains from the primary antibodies used for immunoprecipitation and the secondary antibodies were avoided by using the HRP-conjugated anti-IgG light chain secondary antibodies (anti-mouse, Jackson Immuno Research, 115-035-174; anti-rabbit, Jackson Immuno Research, 211-032-171).
Acknowledgements
A.A.S. and O.V.S. were supported by the CRDF Award UB1-2447-LV-02. V.Y.N. was supported by the JSTF Grant under the CRDF Award UB1-2447-LV-02 and by the INTAS Grant for Young Scientists 04-83-3342. S.S. was supported by an NIH grant GM069373.
Supplementary Material
References
- 1.Mizushima N, Levine B, Cuervo AM, Klionsky DJ. Autophagy fights disease through cellular self-digestion. Nature. 2008;451:1069–1075. doi: 10.1038/nature06639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Klionsky DJ. The molecular machinery of autophagy: unanswered questions. J Cell Sci. 2005;118:7–18. doi: 10.1242/jcs.01620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Suzuki K, Ohsumi Y. Molecular machinery of autophagosome formation in yeast, Saccharomyces cerevisiae. FEBS Lett. 2007;581:2156–2161. doi: 10.1016/j.febslet.2007.01.096. [DOI] [PubMed] [Google Scholar]
- 4.Manjithaya R, Nazarko TY, Farré JC, Subramani S. Molecular mechanism and physiological role of pexophagy. FEBS Lett. 2010;584:1367–1373. doi: 10.1016/j.febslet.2010.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Farré JC, Vidal J, Subramani S. A cytoplasm to vacuole targeting pathway in P. pastoris. Autophagy. 2007;3:230–234. doi: 10.4161/auto.3905. [DOI] [PubMed] [Google Scholar]
- 6.Strømhaug PE, Klionsky DJ. Cytoplasm to vacuole targeting. In: Klionsky DJ, editor. Autophagy. Georgetown: Landes Bioscience; 2003. pp. 84–106. [Google Scholar]
- 7.Ano Y, Hattori T, Oku M, Mukaiyama H, Baba M, Ohsumi Y, et al. A sorting nexin PpAtg24 regulates vacuolar membrane dynamics during pexophagy via binding to phosphatidylinositol-3-phosphate. Mol Biol Cell. 2005;16:446–457. doi: 10.1091/mbc.E04-09-0842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Farré JC, Manjithaya R, Mathewson RD, Subramani S. PpAtg30 tags peroxisomes for turnover by selective autophagy. Dev Cell. 2008;14:365–376. doi: 10.1016/j.devcel.2007.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Oku M, Warnecke D, Noda T, Müller F, Heinz E, Mukaiyama H, et al. Peroxisome degradation requires catalytically active sterol glucosyltransferase with a GRAM domain. EMBO J. 2003;22:3231–3241. doi: 10.1093/emboj/cdg331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Stasyk OV, Nazarko TY, Stasyk OG, Krasovska OS, Warnecke D, Nicaud JM, et al. Sterol glucosyltransferases have different functional roles in Pichia pastoris and Yarrowia lipolytica. Cell Biol Int. 2003;27:947–952. doi: 10.1016/j.cellbi.2003.08.004. [DOI] [PubMed] [Google Scholar]
- 11.Xie Z, Klionsky DJ. Autophagosome formation: core machinery and daptations. Nat Cell Biol. 2007;9:1102–1109. doi: 10.1038/ncb1007-1102. [DOI] [PubMed] [Google Scholar]
- 12.Nazarko TY, Farré JC, Subramani S. Peroxisome size provides insights into the function of autophagy-related proteins. Mol Biol Cell. 2009;20:3828–3839. doi: 10.1091/mbc.E09-03-0221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Stasyk OV, Stasyk OG, Mathewson RD, Farré JC, Nazarko VY, Krasovska OS, et al. Atg28, a novel coiledcoil protein involved in autophagic degradation of peroxisomes in the methylotrophic yeast Pichia pastoris. Autophagy. 2006;2:30–38. doi: 10.4161/auto.2226. [DOI] [PubMed] [Google Scholar]
- 14.Cao Y, Cheong H, Song H, Klionsky DJ. In vivo reconstitution of autophagy in Saccharomyces cerevisiae. J Cell Biol. 2008;182:703–713. doi: 10.1083/jcb.200801035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cheong H, Nair U, Geng J, Klionsky DJ. The Atg1 kinase complex is involved in the regulation of protein recruitment to initiate sequestering vesicle formation for nonspecific autophagy in Saccharomyces cerevisiae. Mol Biol Cell. 2008;19:668–681. doi: 10.1091/mbc.E07-08-0826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kabeya Y, Kawamata T, Suzuki K, Ohsumi Y. Cis1/Atg31 is required for autophagosome formation in Saccharomyces cerevisiae. Biochem Biophys Res Commun. 2007;356:405–410. doi: 10.1016/j.bbrc.2007.02.150. [DOI] [PubMed] [Google Scholar]
- 17.Kawamata T, Kamada Y, Suzuki K, Kuboshima N, Akimatsu H, Ota S, et al. Characterization of a novel autophagy-specific gene, ATG29. Biochem Biophys Res Commun. 2005;338:1884–1889. doi: 10.1016/j.bbrc.2005.10.163. [DOI] [PubMed] [Google Scholar]
- 18.Kawamata T, Kamada Y, Kabeya Y, Sekito T, Ohsumi Y. Organization of the pre-autophagosomal structure responsible for autophagosome formation. Mol Biol Cell. 2008;19:2039–2050. doi: 10.1091/mbc.E07-10-1048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Serebriiskii I, Khazak V, Golemis EA. A two-hybrid dual bait system to discriminate specificity of protein interactions. J Biol Chem. 1999;274:17080–17087. doi: 10.1074/jbc.274.24.17080. [DOI] [PubMed] [Google Scholar]
- 20.Fries T, Betz C, Sohn K, Caesar S, Schlenstedt G, Bailer SM. A novel conserved nuclear localization signal is recognized by a group of yeast importins. J Biol Chem. 2007;282:19292–19301. doi: 10.1074/jbc.M700217200. [DOI] [PubMed] [Google Scholar]
- 21.Shintani T, Klionsky DJ. Cargo proteins facilitate the formation of transport vesicles in the cytoplasm to vacuole targeting pathway. J Biol Chem. 2004;279:29889–29894. doi: 10.1074/jbc.M404399200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yan M, Rachubinski DA, Joshi S, Rachubinski RA, Subramani S. Dysferlin domain-containing proteins, Pex30p and Pex31p, localized to two compartments, control the number and size of oleate-induced peroxisomes in Pichia pastoris. Mol Biol Cell. 2008;19:885–898. doi: 10.1091/mbc.E07-10-1042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schroder LA, Ortiz MV, Dunn W., Jr The membrane dynamics of pexophagy are influenced by Sar1p in Pichia pastoris. Mol Biol Cell. 2008;19:4888–4899. doi: 10.1091/mbc.E07-09-0868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fry MR, Thomson JM, Tomasini AJ, Dunn W., Jr Early and late molecular events of glucose-induced pexophagy in Pichia pastoris require Vac8. Autophagy. 2006;2:280–288. doi: 10.4161/auto.3164. [DOI] [PubMed] [Google Scholar]
- 25.Oku M, Nishimura T, Hattori T, Ano Y, Yamashita S, Sakai Y. Role of Vac8 in formation of the vacuolar sequestering membrane during micropexophagy. Autophagy. 2006;2:272–279. doi: 10.4161/auto.3135. [DOI] [PubMed] [Google Scholar]
- 26.Yuan W, Tuttle DL, Shi YJ, Ralph GS, Dunn W., Jr Glucose-induced microautophagy in Pichia pastoris requires the α-subunit of phosphofructokinase. J Cell Sci. 1997;110:1935–1945. doi: 10.1242/jcs.110.16.1935. [DOI] [PubMed] [Google Scholar]
- 27.Daulny A, Geng F, Muratani M, Geisinger JM, Salghetti SE, Tansey WP. Modulation of RNA polymerase II subunit composition by ubiquitylation. Proc Natl Acad Sci USA. 2008;105:19649–19654. doi: 10.1073/pnas.0809372105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Suzuki K, Kubota Y, Sekito T, Ohsumi Y. Hierarchy of Atg proteins in pre-autophagosomal structure organization. Genes Cells. 2007;12:209–218. doi: 10.1111/j.1365-2443.2007.01050.x. [DOI] [PubMed] [Google Scholar]
- 29.Altan-Bonnet N, Sougrat R, Lippincott-Schwartz J. Molecular basis for Golgi maintenance and biogenesis. Curr Opin Cell Biol. 2004;16:364–372. doi: 10.1016/j.ceb.2004.06.011. [DOI] [PubMed] [Google Scholar]
- 30.Ward TH, Polishchuk RS, Caplan S, Hirschberg K, Lippincott-Schwartz J. Maintenance of Golgi structure and function depends on the integrity of ER export. J Cell Biol. 2001;155:557–570. doi: 10.1083/jcb.200107045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sakai Y, Koller A, Rangell LK, Keller GA, Subramani S. Peroxisome degradation by microautophagy in Pichia pastoris: identification of specific steps and morphological intermediates. J Cell Biol. 1998;141:625–636. doi: 10.1083/jcb.141.3.625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tuttle DL, Dunn W., Jr Divergent modes of autophagy in the methylotrophic yeast Pichia pastoris. J Cell Sci. 1995;141:625–636. doi: 10.1242/jcs.108.1.25. [DOI] [PubMed] [Google Scholar]
- 33.Drake KR, Kang M, Kenworthy AK. Nucleocytoplasmic distribution and dynamics of the autophagosome marker EGFP-LC3. PLoS One. 2010;5:9806. doi: 10.1371/journal.pone.0009806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Liang XH, Yu J, Brown K, Levine B. Beclin 1 contains a leucine-rich nuclear export signal that is required for its autophagy and tumor suppressor function. Cancer Res. 2001;61:3443–3449. [PubMed] [Google Scholar]
- 35.Mauvezin C, Orpinell M, Francis VA, Mansilla F, Duran J, Ribas V, et al. The nuclear cofactor DOR regulates autophagy in mammalian and Drosophila cells. EMBO Rep. 2010;11:37–44. doi: 10.1038/embor.2009.242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nowak J, Archange C, Tardivel-Lacombe J, Pontarotti P, Pébusque MJ, Vaccaro MI, et al. The TP53INP2 protein is required for autophagy in mammalian cells. Mol Biol Cell. 2009;20:870–881. doi: 10.1091/mbc.E08-07-0671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yuryev A, Patturajan M, Litingtung Y, Joshi RV, Gentile C, Gebara M, et al. The C-terminal domain of the largest subunit of RNA polymerase II interacts with a novel set of serine/arginine-rich proteins. Proc Natl Acad Sci USA. 1996;93:6975–6980. doi: 10.1073/pnas.93.14.6975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cregg JM, Barringer KJ, Hessler AY, Madden KR. Pichia pastoris as a host system for transformations. Mol Cell Biol. 1985;5:3376–3385. doi: 10.1128/mcb.5.12.3376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Waterham HR, de Vries Y, Russel KA, Xie W, Veenhuis M, Cregg JM. The Pichia pastoris PER6 gene product is a peroxisomal integral membrane protein essential for peroxisome biogenesis and has sequence similarity to the Zellweger syndrome protein PAF-1. Mol Cell Biol. 1996;16:2527–2536. doi: 10.1128/mcb.16.5.2527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gould SJ, McCollum D, Spong AP, Heyman JA, Subramani S. Development of the yeast Pichia pastoris as a model organism for a genetic and molecular analysis of peroxisome assembly. Yeast. 1992;8:613–628. doi: 10.1002/yea.320080805. [DOI] [PubMed] [Google Scholar]
- 41.Strømhaug PE, Bevan A, Dunn W., Jr GSA11 encodes a unique 208 kDa protein required for pexophagy and autophagy in Pichia pastoris. J Biol Chem. 2001;276:42422–42435. doi: 10.1074/jbc.M104087200. [DOI] [PubMed] [Google Scholar]
- 42.Estojak J, Brent R, Golemis EA. Correlation of two-hybrid affinity data with in vitro measurements. Mol Cell Biol. 1995;15:5820–5829. doi: 10.1128/mcb.15.10.5820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gyuris J, Golemis EA, Chertkov H, Brent R. Cdi1, a human G1 and S phase protein phosphatase that associates with Cdk2. Cell. 1993;75:791–803. doi: 10.1016/0092-8674(93)90498-f. [DOI] [PubMed] [Google Scholar]
- 44.Toby GG, Golemis EA. Using the yeast interaction trap and other two-hybrid based approaches to study protein-protein interactions. Methods. 2001;24:201–217. doi: 10.1006/meth.2001.1182. [DOI] [PubMed] [Google Scholar]
- 45.Stasyk OV, Nazarko TY, Sibirny AA. Methods of plate pexophagy monitoring and positive selection for ATG gene cloning in yeasts. Methods Enzymol. 2008;451:229–239. doi: 10.1016/S0076-6879(08)03216-3. [DOI] [PubMed] [Google Scholar]
- 46.Baerends RJ, Faber KN, Kram AM, Kiel JAKW, van der Klei IJ, Veenhuis M. A stretch of positively charged amino acids at the N terminus of Hansenula polymorpha Pex3p is involved in incorporation of the protein into the peroxisomal membrane. J Biol Chem. 2000;275:9986–9995. doi: 10.1074/jbc.275.14.9986. [DOI] [PubMed] [Google Scholar]
- 47.McCollum D, Monosov E, Subramani S. The pas8 mutant of Pichia pastoris exhibits the peroxisomal protein import deficiencies of Zellweger syndrome cells—the PAS8 protein binds to the COOH-terminal tripeptide peroxisomal targeting signal, and is a member of the TPR protein family. J Cell Biol. 1993;121:761–774. doi: 10.1083/jcb.121.4.761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Snyder WB, Koller A, Choy AJ, Subramani S. The peroxin Pex19p interacts with multiple, integral membrane proteins at the peroxisomal membrane. J Cell Biol. 2000;149:1171–1178. doi: 10.1083/jcb.149.6.1171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Snyder WB, Faber KN, Wenzel TJ, Koller A, Lüers GH, Rangell L, et al. Pex19p interacts with Pex3p and Pex10p and is essential for peroxisome biogenesis in Pichia pastoris. Mol Biol Cell. 1999;10:1745–1761. doi: 10.1091/mbc.10.6.1745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ramezani-Rad M, Hollenberg CP, Lauber J, Wedler H, Griess E, Wagner C, et al. The Hansenula polymorpha (strain CBS4732) genome sequencing and analysis. FEMS Yeast Res. 2003;4:207–215. doi: 10.1016/S1567-1356(03)00125-9. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.