Abstract
Analysis of mutants defective in meiotic chromosome pairing has uncovered a role for Caenorhabditis elegans chk-2 in initial establishment of pairing between homologous chromosomes during early meiotic prophase. chk-2 is also required for the major spatial reorganization of nuclei that normally accompanies the onset of pairing, suggesting a mechanistic coupling of these two events. Despite failures in pairing, nuclear reorganization, and crossover recombination, chk-2 mutants undergo many other aspects of meiotic chromosome morphogenesis and complete gametogenesis. Although chk-2 encodes a C. elegans ortholog of the Cds1/Chk2 checkpoint protein kinases, germ-line nuclei in chk-2 mutants are competent to arrest proliferation in response to replication inhibition and to trigger DNA damage checkpoint responses to ionizing radiation. However, chk-2 mutants are defective in triggering the pachytene DNA damage checkpoint in response to an intermediate block in the meiotic recombination pathway, suggesting that chk-2 is required either for initiation of meiotic recombination or for monitoring a specific subset of DNA damage lesions. We propose that chk-2 functions during premeiotic S phase to enable chromosomes to become competent for subsequent meiotic prophase events and/or to coordinate replication with entry into prophase.
Keywords: Meiosis, chromosome pairing, checkpoint, C. elegans, Chk2, chk-2
A pivotal defining event during gamete formation in sexually reproducing organisms is the reduction in chromosome ploidy that occurs when homologous chromosomes segregate away from each other at the meiosis I division. Precise chromosome partitioning during meiosis I relies on a previously established connection between homologs that ensures their proper orientation toward opposite poles of the meiotic spindle. For most organisms, this crucial association is established and stabilized through events of the preceding meiotic prophase (Roeder 1997; Moore and Orr-Weaver 1998; Zickler and Kleckner 1999). At the onset of prophase, newly replicated chromosomes must identify and form initial associations with the appropriate partner chromosome. By the pachytene stage, the pairing process has culminated in an intimate alignment of homologous chromosomes along their entire lengths, with a highly ordered proteinaceous scaffold, the synaptonemal complex, positioned at the interface of paired homologs. Crossover recombination events between the DNA molecules of homologous chromosomes are completed within this context. Upon disassembly of the synaptonemal complex and loss of lengthwise alignment, these crossovers act in conjunction with sister chromatid cohesion to maintain temporary linkages (chiasmata) between homologs until the metaphase/anaphase transition of meiosis I.
Whereas genetic and biochemical studies have identified many of the proteins and mechanisms underlying the process of meiotic recombination (Roeder 1997; Paques and Haber 1999), much less is known about the molecular events that bring about pairing and alignment between homologous chromosomes in the first place. Much of what we do know about the homolog pairing process comes from decades of cytological observations of early meiotic prophase in a multitude of different organisms (for reviews, see Wilson 1925; Scherthan 1997; Zickler and Kleckner 1998). These studies have revealed that a dramatic spatial reorganization of the nucleus generally accompanies the process of homolog alignment. Reorganization usually includes a transient phase in which nuclei achieve a highly polarized organization, involving a tight clustering of telomeres at a limited sector of the nuclear periphery and/or a congregation of chromosomes toward one side of the nucleus. The widespread occurrence of nuclear polarization and its temporal correlation with homolog pairing suggest that it may serve to facilitate the pairing process. However, evidence from several systems indicates that telomere clustering per se is probably not strictly required for recognition and initial alignment between homologs but more likely contributes to efficient and timely completion of the pairing process (Zickler and Kleckner 1998; Trelles-Sticken et al. 2000). How the movements that reorganize chromosomes are accomplished and what their functional relationship is to homolog recognition and alignment remain unclear.
Several features of the nematode Caenorhabditis elegans make it a particularly useful experimental system for investigating mechanisms underlying meiotic prophase chromosome dynamics. C. elegans is essentially a “meiosis machine”: The germ line accounts for more than half of the cell nuclei in the adult organism, providing an abundant and easily accessible source of meiotic cells (Schedl 1997). Premeiotic nuclei and nuclei at all stages of meiotic prophase are present simultaneously in a clear temporal/spatial gradient along the distal–proximal axis of the gonad, so that each germ line represents a continuum of progression into and through meiotic prophase. This progression encompasses numerous nuclei that are actively establishing homolog alignments as well as an abundance of nuclei in which full, stable pairing and synapsis have been achieved (Albertson et al. 1997). Further, chromosome organization can be investigated in detail in the context of well-preserved three-dimensional nuclear architecture and chromosome morphology using tools such as fluorescence in situ hybridization (FISH) to monitor interactions between homologous chromosomes (Dernburg et al. 1998). Moreover, this analysis can be carried out using whole mount germ-line preparations in which the temporal/spatial context of the meiotic time course has been preserved.
The ability to perform robust cytological analyses in three-dimensionally preserved tissue is complemented by the powerful genetics of C. elegans, which has facilitated identification of components of the meiotic machinery through genetic screens for mutants defective in meiotic chromosome segregation (Villeneuve 1994; Kelly et al. 2000). For nearly all of these meiotic mutants, the segregation defects are a secondary consequence of primary defects in key meiotic prophase events. Cytological analysis of chromosome organization throughout meiotic prophase in these mutants has enabled us to identify genes required for the initial establishment of pairing as well as genes required for the subsequent maintenance of paired associations (A.J. MacQueen and A.M. Villeneuve, unpubl.). Our analysis of one of the genes defined by pairing-defective mutants has led us to discover an unanticipated role for C. elegans CHK-2, a member of the Cds1/Chk2 family of checkpoint protein kinases (for review, see Rhind and Russell 2000), in promoting normal chromosome dynamics during early meiotic prophase. We find that chk-2 is required not only for initial establishment of pairing between homologous chromosomes but also for the major spatial reorganization of chromosomes that normally accompanies the onset of pairing, providing the first molecular link between these two landmark events in the meiotic program.
Results
Identification of chk-2 mutants in a screen for defects in meiotic chromosome pairing
Ongoing genetic screens for C. elegans mutants defective in meiotic chromosome segregation (Villeneuve 1994; Kelly et al. 2000) have yielded a collection of mutants that lack chiasmata at diakinesis, the last stage of meiotic prophase (Fig. 1). Such mutants are identified in these screens because chiasmata are required to direct orderly segregation of homologs at the meiosis I division. An absence of chiasmata between homologs can result from a defect in the recombination machinery, or alternatively, it could result from a primary defect in the establishment or maintenance of homolog pairing. Because stable pairing and synapsis in C. elegans are not dependent on initiation of meiotic recombination, mutants defective in core components of the meiotic recombination machinery exhibit achiasmate chromosomes at diakinesis but appear cytologically normal during earlier stages of prophase (Dernburg et al. 1998; Zalevsky et al. 1999; Kelly et al. 2000; Chin and Villeneuve 2001). Thus, to identify components of the machinery responsible for meiotic homolog pairing, we screened mutants that contain achiasmate chromosomes at diakinesis for defects in the morphology and organization of early prophase chromosomes. To date, this analysis has identified four loci required for either the establishment or maintenance of homolog pairing.
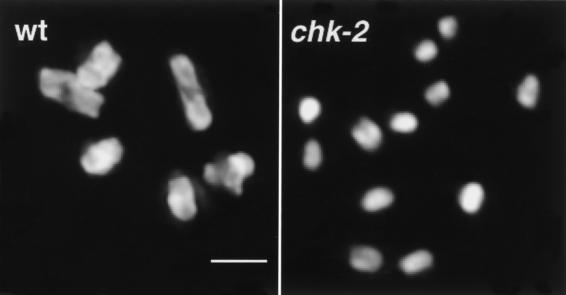
Figure 1.
Absence of chiasmata in chk-2 mutants. Each panel shows DAPI-stained chromosomes in a single oocyte nucleus at diakinesis, the last stage of meiotic prophase. (Left) The wild-type nucleus (wt) contains six bivalents, each corresponding to a pair of homologous chromosomes linked by a chiasma. (Right) The chk-2 oocyte nucleus contains 12 univalent chromosomes, indicating an absence of chiasmata. Bar, 2 μm.
Two mutations, me18 and me64, define a gene required for the initial establishment of meiotic chromosome pairing (see below). me18 was mapped to the extreme right end of chromosome V, ∼0.1 map units to the left of the unc-51 locus. We used RNA-mediated interference (RNAi) to disrupt the function of several candidate genes located within the 100-kb region immediately to the left of unc-51 and found that double-stranded RNA (dsRNA) corresponding to Y60A3A.12 elicited a robust but transient phenocopy of the me18 and me64 mutant phenotypes (see Materials and Methods). Y60A3A.12 encodes a C. elegans ortholog of Cds1/Chk2 serine/threonine kinases (Rhind and Russell 2000) and was recently designated C.e. chk-2 (GI: 10566451; Higashitani et al. 2000). Sequencing revealed a missense mutation in the me18 allele that results in an alanine to valine substitution at residue 21 of the CHK-2 protein, and a nonsense mutation in the me64 allele that terminates translation after residue 305 in the middle of the conserved protein kinase domain (Fig. 2). me64 is likely a severe loss-of-function or null allele, since this truncation is expected to render CHK-2 nonfunctional as a kinase. The me18 allele is also likely to severely reduce or eliminate chk-2 function, since me18 and me64 confer similar phenotypes both in homozygous worms and in worms heterozygous for either allele and a deficiency of the region. Phenotypic analyses reported here are for me64, but nearly all assays were also performed for me18, giving indistinguishable results.
Figure 2.
Changes in CHK-2 protein caused by chk-2 mutations. The positions of the conserved forkhead-associated (FHA) and serine/threonine protein kinase domains are indicated by a gray and black box, respectively. Changes to the predicted protein caused by the me18 and me64 mutations are shown above asterisks indicating the locations of these changes. (aa) amino acids.
chk-2 is required for the establishment of homolog pairing
chk-2 mutant worms display several properties typical of C. elegans mutants that lack chiasmata (Dernburg et al. 1998; Zalevsky et al. 1999; Kelly et al. 2000; Chin and Villeneuve 2001). Homozygous mutant hermaphrodites are morphologically normal and produce a large number of embryos, but an absence of chiasmata between homologs in late prophase meiocytes (Fig. 1) leads to chromosome missegregation during meiosis I and consequently to the production of aneuploid gametes and zygotes. Thus, most embryos die (95.7%, n = 787), and rare survivors are those embryos that have received a euploid (or nearly euploid) chromosome complement. A high frequency of males (45%, n = 678, compared with 0.2% for the wild type [Hodgkin et al. 1979]) is found among the surviving self progeny of chk-2 hermaphrodites; this high incidence of male (Him) phenotype (Hodgkin et al. 1979) is an additional diagnostic of mutants defective in chromosome segregation in C. elegans, in which sex is determined by the number of X chromosomes present in an otherwise diploid animal (hermaphrodites are XX, whereas males are XO). Similar evidence for chromosome missegregation and lack of chiasmata was seen in two independent RNAi studies that simultaneously targeted both C.e. chk-2 (Y60A3A.12) and a closely related gene (T08D2.7, 95% identical in coding sequence); however these preliminary analyses did not investigate the underlying basis for the observed meiotic defects (Higashitani et al. 2000; Oishi et al. 2001).
We used DAPI staining and high resolution imaging of three-dimensionally preserved germ lines to evaluate nuclei at earlier stages of meiotic prophase. In chk-2 mutants, we observed a striking disorganization of chromosomes within nuclei occupying the region of the germ line that would normally contain pachytene-stage nuclei (Fig. 3a). Wild-type pachytene-stage nuclei exhibit a distinctive organization in which parallel DAPI-stained tracks correspond to synapsed homologous chromosomes that are intimately paired along their entire lengths. In contrast, nuclei within the pachytene region of chk-2 mutant germ lines display disorganized DAPI-stained tracks that are not arranged in parallel pairs, consistent with a failure in alignment and synapsis between homologs.
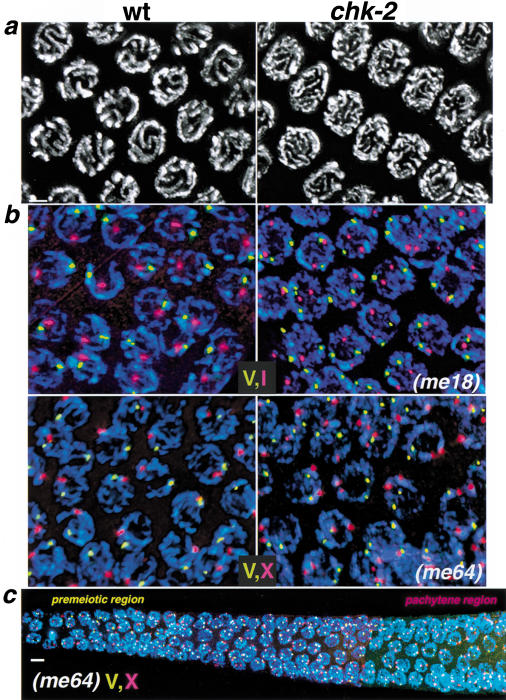
Figure 3.
Failure to establish homolog pairing and alignment in chk-2 mutants. (a) DAPI-stained nuclei from the pachytene region of wild-type (wt) and chk-2 mutant germ lines. In wild-type pachytene nuclei, parallel pairs of DAPI-stained tracks correspond to pairs of homologous chromosomes intimately aligned along their entire lengths. In chk-2 mutants, nuclei from this region of the germ line exhibit disorganized DAPI-stained tracks that are not aligned in parallel pairs. The images are projections approximately halfway through three-dimensional data stacks of whole nuclei to allow visual resolution of individual chromosomal stretches. (b,c) Two-color FISH analysis, indicating failure of homolog pairing in chk-2 mutants. (b) Nuclei shown are from the pachytene regions of the germ lines; images are projections through three-dimensional data stacks encompassing whole nuclei. DAPI signal is in blue; FISH signals corresponding to different chromosomal regions are shown in red and yellow: (I) Y13H5; (X) Y51E2; (V) 5srDNA locus. In control nuclei, either a single hybridization signal or a closely spaced doublet is observed for both probes, indicating close juxtaposition of homologous sequences. In chk-2 mutants, two separate hybridization signals are seen for each probe. (c) A composite of images taken at three overlapping positions along the distal–proximal axis of a chk-2 mutant germ line, from the premeiotic region to a region that corresponds to late pachytene stage in wild-type germ lines. For many (but not all) of the nuclei shown, the projection encompasses the full nuclear volume. Unpaired FISH signals for chromosome V and X probes are seen throughout the germ line. Bars, (a,b) 2 μm, (c) 4 μm.
Further evidence for impaired alignment and synapsis between homologous chromosomes came from examination of HIM-3 protein localization in chk-2 mutant germ lines (Fig. 4). HIM-3, a meiosis-specific chromosomal protein, normally exhibits a discrete localization along the length of chromosome axes during meiotic prophase (Zetka et al. 1999). During the pachytene stage when chromosomes are fully aligned and synapsed, the HIM-3–containing axes from each partner chromosome are in close apposition, centered at the interface between the two homologs (Zetka et al. 1999; Fig. 4a,c). In chk-2 mutant germ lines, HIM-3 assembles onto chromosomes with apparently normal timing (see below), but pachytene-region nuclei show an excess of HIM-3–labeled tracks, many of which appear thinner than those found in wild-type pachytene nuclei; taken together with the DAPI and FISH data (below), these are indicative of unsynapsed chromosome axes.
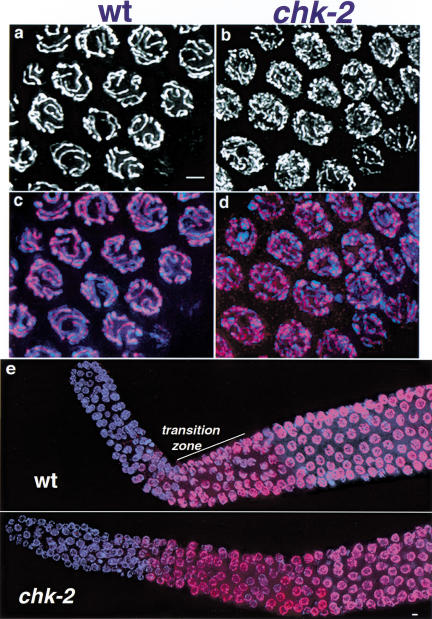
Figure 4.
Localization of HIM-3 on meiotic chromosomes. HIM-3, a meiosis-specific component of chromosome axes, was visualized by immunofluorescence. Anti-HIM-3 is shown in white (a,b) or red (c–e); DAPI is shown in blue (c–e). (a–d) High-resolution images of pachytene region nuclei; images represent projections approximately halfway through three-dimensional data stacks of whole nuclei. In wild-type (wt) pachytene nuclei (a,c), HIM-3 localizes at the interface between aligned chromosome pairs. Nuclei from the same region of the germ line in chk-2 mutants (b,d) show extensive, continuous HIM-3 localization along chromosomes, but these HIM-3 lines are more numerous and often appear thinner than those in wild-type pachytene nuclei, reflecting the fact that chromosomes are not aligned lengthwise with a homologous partner (see Fig. 3). (e) Low magnification images of distal tip through mid-pachytene region of control and chk-2 mutant germ lines; images shown are projections approximately halfway through three-dimensional data stacks of the entire germ line, encompassing whole nuclei. In wild-type germ lines, HIM-3 begins to localize to nuclei and chromosomes in the transition zone region (marked by the presence of several nuclei with crescent-shaped chromatin configurations), which corresponds to the beginning of meiotic prophase (Zetka et al. 1999). In the chk-2 mutant germ line, HIM-3 begins to localize onto chromosomes in nuclei at the same position, relative to the distal tip, as in the wild-type germ line. Bars, 2 μm.
Direct assessment of meiotic chromosome pairing using FISH revealed that chk-2 mutants are profoundly defective in establishing homologous pairing between any of the six pairs of C. elegans chromosomes (Fig. 3; Table 1). In wild-type animals, homologous chromosomes are unpaired in premeiotic germ-line nuclei (Dernburg et al. 1998), and thus two FISH signals per nucleus are observed for each probe (Table 1). After entry into meiotic prophase and establishment of chromosome pairing, a single hybridization signal (or two closely juxtaposed signals) is observed in each wild-type nucleus (Fig. 3b; Table 1). In contrast, chk-2 mutant germ-line nuclei contain unpaired FISH signals both before and after they have entered meiotic prophase (Fig. 3b,c; Table 1). Seven probes which together target each of the six chromosome pairs were examined, and each probe showed an absence of pairing in the chk-2 mutant.
Table 1.
Quantitative analysis of homolog pairing
| Genotype
|
Probe
|
% of nuclei with paired FISH signals (no. of nuclei scored)
|
||||
|---|---|---|---|---|---|---|
| premeiotic
|
meiotic prophase
|
|||||
| zone 1
|
zone 2
|
zone 3
|
zone 4
|
zone 5
|
||
| +/chk-2 | Y51E2 (X) | 5 (93) | 41 (104) | 89 (106) | 99 (85) | 99 (80) |
| +/chk-2 | 5S (V) | 4 (102) | 44 (91) | 95 (98) | 100 (80) | 100 (89) |
| chk-2 | Y51E2 (X) | 5 (110) | 3 (131) | 0 (109) | 1 (95) | 2 (85) |
| chk-2 | 5S (V) | 3 (118) | 3 (128) | 1 (107) | 1 (111) | 1 (102) |
Germ lines were subdivided into five equal-sized zones along the distal–proximal axis. Zone 1 contains only premeiotic nuclei; zone 2 contains a mixture of premeiotic and leptotene/zygotene stages; zone 3 may contain some leptotene/zygotene nuclei, but most are at the pachytene stage; zones 4 and 5 contain mid-late pachytene nuclei; staging is based on wild-type germlines. Pairing of FISH signals was assessed as described in Materials and Methods; for each probe, nuclei from three germ lines were scored for each genotype.
Based on the absence of pairing and chiasmata, we anticipated that measurement of meiotic recombination frequency in chk-2 mutants would reveal a severe defect in crossing over. Our expectation was borne out through examination of meiotic crossover frequency between markers near opposite ends of the X chromosome (Table 2).Although the genetic distance separating these markers in wild-type meioses is ∼42 centimorgans (cM), in the chk-2 mutant no crossovers were detected over this large interval.
Table 2.
Absence of crossover recombination in chk-2 mutants
| Genotype
|
Recombinant progeny
|
Total progeny
|
Map distance (cM)
|
|---|---|---|---|
| +/(chk-2 or +); dpy-3 unc-3/++ | 1092 | 3283 hermaphrodites | 42 |
| chk-2/chk-2; dpy-3 unc-3/++ | 0 | 349 hermaphrodites | <0.2 |
| 297 males |
chk-2 is required for spatial reorganization of the meiotic prophase nucleus
The onset of homolog pairing during wild-type meiosis is accompanied by a major spatial reorganization of chromosomes within nuclei in the transition zone region of the germ line, corresponding to the leptotene/zygotene stages of meiotic prophase (Dernburg et al. 1998). In premeiotic nuclei, DAPI-stained chromatin is widely dispersed throughout the volume between the nuclear periphery and a prominent, centrally located nucleolus (Fig. 5a–f). Upon entry into meiotic prophase, nuclei remain round but become highly polarized (Fig. 5a–f). The chromatin becomes asymmetrically localized and concentrated toward one side of the nucleus, while the nucleolus adopts an off-center position opposite that of the clustered chromosomes. This polarization imparts a distinctive crescent-shaped appearance to the DAPI-stained chromatin in transition zone nuclei that is readily evident even in low magnification images (Fig. 5a–c). As nuclei progress into the pachytene stage, they lose this polarization: newly aligned and synapsed chromosomes become dispersed about the nuclear periphery, once again surrounding the nucleolus.
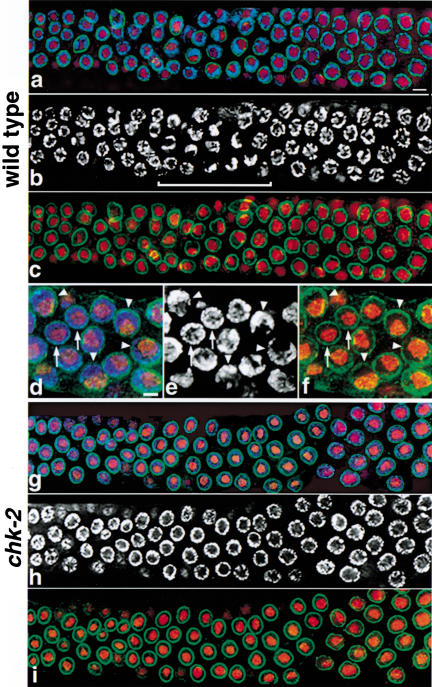
Figure 5.
Failure in spatial reorganization of early prophase nuclei in chk-2 mutants. Germ-line nuclei were triple-labeled with DAPI (blue in a,d,g, white in b,e,h) to label chromosomes, mAbD77 (red) to label nucleoli, and anti-Ce-lamin (green) to indicate nuclear outline. (a–c) Composite image of three overlapping regions along the distal–proximal axis of a wild-type germ line, extending from the premeiotic region (left) through the pachytene stage. In premeiotic nuclei, chromatin is dispersed about the periphery of most nuclei, imparting a round appearance to the DAPI signals. The transition zone, indicated by a bracket in b, contains nuclei that have recently entered meiotic prophase; the DAPI signals in many nuclei in this region have a distinct crescent-shaped appearance, reflecting a highly polarized nuclear organization (see below). DAPI signals again appear more round in pachytene nuclei (right), in which chromosomes are once more dispersed about the nuclear periphery and surround the nucleolus. For the DAPI and lamin signals, images are projections through three-dimensional data stacks encompassing 0.7- to 1.2-μm-thick sections centered slightly above or below the equatorial planes of most nuclei; for nucleolar signals, projections encompass the whole nuclei. (d–f) Detail of nuclear organization in the wild-type transition zone. These images are projections of three-dimensional data stacks that encompass whole nuclei for the DAPI and nucleolar signals; for the lamin signals, 0.7-μm-thick sections centered approximately at the nuclear equator are shown to indicate the outline of the nucleus without obstructing the other signals. Arrows point to nuclei with premeiotic morphology, in which chromatin completely surrounds a prominent, central nucleolus. Arrowheads point to nuclei with a highly polarized nuclear organization: the chromatin is concentrated toward one side of the nucleus and no longer surrounds the nucleolus, which has adopted a reciprocal position adjacent to the nuclear periphery on the opposite side of the nucleus (note that DAPI-dark regions in b, e, and h correspond to the nucleolus). (g–i) Composite image of three overlapping regions along the distal–proximal axis of a chk-2 mutant germ line. Images are projections through three-dimensional data stacks encompassing a 0.7-μm-thick equatorial section of most nuclei. In all nuclei in both premeiotic (roughly the leftmost third of the image) and meiotic regions of the germ line, the chromatin completely surrounds the nucleolus; no nuclei with polarized organization are observed. Bars, (a) 4 μm, (d) 2 μm.
chk-2 mutant germ lines do not contain any nuclei with the polarized spatial organization that normally distinguishes nuclei in the transition zone. As nuclei enter and progress through meiotic prophase in chk-2 mutants, chromosomes do not become clustered to one side of the nucleus, as is evident from the absence of crescent-shaped chromatin configurations (Fig. 5g–i). Instead, a widely dispersed, peripheral distribution of chromosomes persists until the end of what normally would be the pachytene stage. Thus, C. elegans chk-2 is required not only for homolog pairing but also for the specialized spatial reorganization of nuclear components that normally accompanies the pairing process.
chk-2 mutants initiate and complete other aspects of meiosis and gametogenesis
Despite failures in early meiotic nuclear reorganization and homolog pairing, chk-2 germ-line nuclei do initiate and complete other aspects of the meiotic program. The previously mentioned meiotic chromosomal protein HIM-3 assembles onto chromosomes in the region of chk-2 germ lines where early meiotic nuclei are normally located (Fig. 4e) and remains associated with them throughout meiotic prophase. As in wild-type germ lines, nuclei within the meiotic region of chk-2 germ lines are, on average, larger than nuclei within the premeiotic region, and mitotic figures are absent from the meiotic region. Moreover, chromosomes in chk-2 mutant germ lines complete the meiotic condensation program, giving rise to diakinesis chromosomes that lack chiasmata but appear normally condensed (Fig. 1). Finally, the survival of embryos from chk-2 hermaphrodites is no worse than that seen for other previously identified meiotic segregation mutants that lack chiasmata, indicating that chk-2 mutants produce functional (albeit aneuploid) gametes.
chk-2 mutant germ-line nuclei retain a functional DNA damage checkpoint and replication block arrest
chk-2 orthologs in other systems have been shown to participate in several distinct signaling pathways that safeguard genome integrity (for reviews, see Dasika et al. 1999; Murakami and Nurse 2000; Rhind and Russell 2000). These include DNA damage and replication block checkpoints that prevent cell cycle progression in response to genotoxic stresses. Therefore, we tested whether germ-line nuclei in chk-2 mutants are competent to trigger previously described DNA damage checkpoint responses or to arrest proliferation in response to a replication block.
Gartner et al. (2000) described a conserved DNA damage checkpoint pathway in the C. elegans germ line that is dependent on mrt-2, the C. elegans ortholog of checkpoint genes S. pombe rad1+ and S. cerevisiae RAD17. This checkpoint pathway is required to trigger germ cell apoptosis at the end of the pachytene stage in response to DNA damage induced by ionizing radiation (IR) and is also required for IR-induced transient arrest in proliferation of premeiotic germ-line nuclei. We tested whether this DNA-damage checkpoint pathway is functional in chk-2 mutant germ lines.
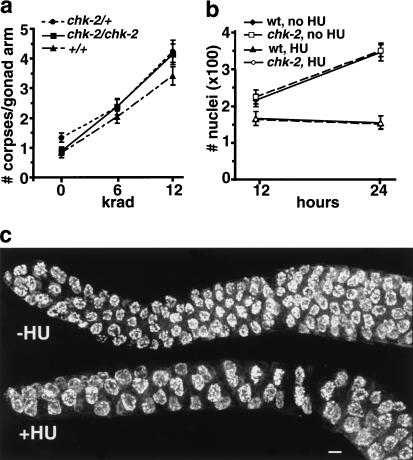
We found that exposure of chk-2 mutant germ lines to IR resulted in a dose-dependent increase in germ cell apoptosis that was indistinguishable from that seen in wild-type controls (Fig. 6a). Further, 12 krad of γ irradiation caused an arrest in the proliferation of nuclei within the premeiotic region of both wild-type and chk-2 mutant germ lines, as evidenced by a lower density of nuclei and an absence of normal mitotic figures within the premeiotic region at 12 h post-irradiation (data not shown). These results indicate that C.e. chk-2 mutants retain function of a conserved DNA damage checkpoint.
Figure 6.
Functional pachytene DNA damage checkpoint and hydroxyurea (HU)-induced proliferation arrest in chk-2 mutant germ lines. (a) chk-2 and control germ lines were tested for the ability to trigger checkpoint-induced apoptosis of late pachytene meiocytes in response to ionizing radiation. The number of germ cell corpses was scored at 22–24 h after exposure of late-stage L4 hermaphrodites to the indicated doses of γ irradiation. The Y-axis indicates the mean number of germ-line corpses scored per gonad arm; 33–47 gonad arms were scored for each data point. Error bars indicate standard error of the mean. (b) Quantitation of numbers of germ cell nuclei in wild-type and chk-2 mutant worms chronically exposed to HU and in untreated (age-matched) controls. HU treatment began at the late L4 larval stage, and germ-line nuclei in optically bisected gonad arms (see Materials and Methods) were counted 12 or 24 h later. Each data point indicates the average value from 4–10 germ lines; error bars indicate standard deviations. (c) Morphological changes in chk-2 premeiotic nuclei after HU exposure; wild-type germ lines exhibited an identical response. Images shown are projections encompassing whole DAPI-stained nuclei, from an untreated germ line (top), and from a germ line after a 12-h exposure to HU (bottom); in the HU-treated germ line, nuclei are substantially enlarged and reduced in number, and DAPI signals appear more diffuse. Bar, 4 μm.
We also assessed the response of germ-line nuclei to hydroxyurea (HU), a drug that causes DNA replication arrest through inhibition of ribonucleotide reductase and consequent depletion of dNTP pools. After 12 h of exposure to 25 mM HU, an arrest in proliferation of premeiotic nuclei was readily detected in both wild-type and chk-2 mutant worms. Germ lines exposed to HU lacked normal condensed mitotic figures and contained a reduced density of premeiotic nuclei, which appeared abnormally large and displayed more diffuse DAPI signals than did nuclei in untreated germ lines (Fig. 6c). Depletion of ribonucleotide reductase activity by RNAi (in wild-type worms) elicited a similar response. Further, HU-induced arrest in proliferation of premeiotic nuclei prevented normal expansion in the number of germ-line nuclei over time: after 24 h of exposure to HU, distal germ lines in both wild-type and chk-2 worms contained approximately half the number of nuclei present in distal germ lines of untreated animals (Fig. 6b). Most affected nuclei in both wild-type and chk-2 worms apparently have the capacity to recover from HU-induced arrest: whereas worms exposed to HU for 10 h exhibited the arrest response, worms treated in the same way but then removed from HU for 1 day contained many normal-sized nuclei and some mitotic figures in the distal germ line (data not shown). HU exposure did not elicit any change in the cytological appearance of meiotic prophase nuclei.
chk-2 mutants are defective in triggering the pachytene checkpoint in response to RAD-51 depletion
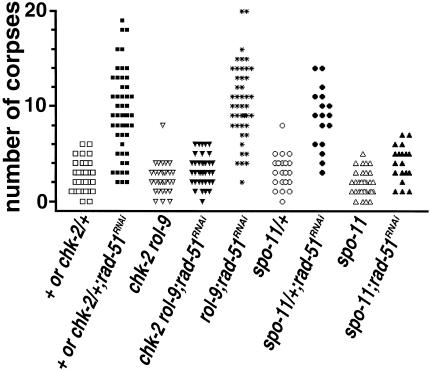
Gartner et al. (2000) showed that the conserved DNA damage checkpoint that induces apoptosis in late pachytene meiocytes in response to IR exposure can also be triggered by using RNAi to deplete germ lines of RAD-51, a conserved component of the meiotic recombination/repair machinery. Checkpoint activation in this context presumably results from accumulation of unrepaired meiotic recombination intermediates, as the checkpoint is not triggered when RAD-51 is depleted in spo-11 mutant worms in which pairing and synapsis are normal but meiotic recombination is not initiated (Dernburg et al. 1998; Gartner et al. 2000).
Whereas chk-2 mutants are competent to trigger the pachytene DNA damage checkpoint in response to IR treatment, they are defective in triggering the checkpoint in response to RAD-51 depletion (Fig. 7). rad-51 RNAi in control worms elicited a marked increase in germ cell apoptosis, confirming the previous report. In contrast, rad-51 RNAi failed to elicit a similar increase in germ cell apoptosis in chk-2 mutants. The levels of germ cell apoptosis observed after rad-51 RNAi in chk-2 worms were comparable to those observed after rad-51 RNAi in spo-11 worms. These results suggest either that chk-2 (like spo-11) is required to initiate meiotic recombination or, alternatively, that chk-2 is necessary for germ cells to recognize or respond to the presence of unrepaired meiotic recombination intermediates.
Figure 7.
chk-2 mutants are defective in triggering the pachytene checkpoint in response to rad-51 RNAi. Scatterplot depicting number of apoptotic germ cell corpses detected in individual gonad arms of control ([chk-2 or +]/+, rol-9, spo-11/+), chk-2, or spo-11 worms with or without rad-51 RNAi treatment. For these experiments, all worms were homozygous for the ced-1(e1735) mutation, which inhibits removal of corpses (Ellis et al. 1991).
Discussion
A link between premeiotic S phase and meiotic prophase?
Members of the conserved Cds1/Chk2 family of checkpoint protein kinases function in a variety of signaling contexts that monitor and safeguard genomic integrity (for reviews, see Dasika et al. 1999; Murakami and Nurse 2000; Rhind and Russell 2000). These include checkpoints that arrest the cell cycle at G1/S, G2/M or metaphase/anaphase boundaries in response to DNA damage or replication blocks, and an “Intra-S” checkpoint that slows DNA replication in response to damage incurred during S phase. Cds1/Chk2 proteins act at a critical juncture in these checkpoint pathways, linking upstream signal transduction cascades with specific cell-cycle targets. There is considerable plasticity in the way these conserved proteins have been deployed: organisms vary regarding which checkpoint-triggering stimuli activate Cds1/Chk2, and within a given organism, Cds1/Chk2 phosphorylates different molecular targets when activated in different cell-cycle contexts. Here, we add another quite distinct role to the repertoire of this versatile protein kinase, in promoting nuclear reorganization and establishment of homolog pairing at the onset of meiotic prophase.
A commonality among previously described roles for Cds1/Chk2 family members is the job of monitoring the status of chromosomal DNA, particularly during S phase. In the Intra-S checkpoint, Cds1/Chk2 appears to regulate the replication process itself (Paulovich and Hartwell 1995; Lindsay et al. 1998), likely in part by modulating late origin utilization (Santocanale and Diffley 1998). Premeiotic S phase in C. elegans immediately precedes the spatial reorganization of the nucleus that marks entry into meiotic prophase (A. Dernburg and A.J. MacQueen, unpubl.). In light of roles for Cds1/Chk2 proteins in monitoring and/or modulating S phase in other contexts, the fact that the meiotic defects in C. elegans chk-2 mutants become apparent immediately after S phase suggests that C.e. chk-2 likely functions during premeiotic S phase to ensure the success of subsequent events during early meiotic prophase.
How might chk-2 function to couple premeiotic S phase with early prophase events? One possibility is that chk-2 could monitor the status of DNA replication to coordinate completion of replication with subsequent meiotic prophase entry. In this context, chk-2 might act as a negative regulator that prevents entry into the meiotic prophase program until replication is complete. Alternatively, chk-2 might act in a positive capacity to activate a subset of the meiotic prophase program upon sensing completion of replication.
A second possibility is that chk-2 might function during S phase to enable chromosomes to become competent for subsequent meiotic prophase events. For example, chk-2 might directly promote a restructuring of chromatin that would normally occur in conjunction with premeiotic replication. Evidence that a Cds1/Chk2 protein has the capacity to directly regulate chromatin assembly in another context was recently reported (Emili et al. 2001; Hu et al. 2001). Alternatively, chk-2 might regulate the length of premeiotic S phase. It has been found in many organisms that the duration of premeiotic S phase is considerably longer than is its mitotic S-phase counterpart, suggesting that a lengthened S phase might be functionally important for preparing chromosomes to participate in meiotic prophase events (Holm 1977; Zickler and Kleckner 1998). Further, there is evidence that Rad53, the budding yeast Cds1/Chk2 ortholog, modulates replication timing in irradiated vegetative cells by regulating late origin firing (Santocanale and Diffley 1998). Thus, C. elegans chk-2 might function to lengthen the duration of premeiotic S phase to allow the completion of chromatin modification processes that occur in conjunction with replication and are essential for meiotic prophase chromosome and nuclear dynamics.
Independent evidence for functional coupling between S phase and subsequent meiotic prophase events has emerged recently from several investigations (Borde et al. 2000; Cha et al. 2000; Merino et al. 2000; Davis et al. 2001; Smith et al. 2001). For example, Borde et al. (2000) found that in S. cerevisiae, the time of initiation of recombination events in a given chromosomal domain is directly correlated with the time of replication through that domain. Further, analysis of S. cerevisiae spo11 mutants revealed a correlation between the duration of premeiotic S phase and the ability of meiotic chromosomes to pair efficiently during early prophase; this led Cha et al. (2000) to propose that meiotic S-phase progression may be directly coupled to morphogenesis of chromosomal features required for interhomolog pairing interactions. Our analysis of C. elegans chk-2 mutants identifies CHK-2 as a candidate regulatory protein through which coupling of premeiotic replication and meiotic prophase events could occur.
Relationship between nuclear reorganization and homologous chromosome pairing
An extensive spatial reorganization of chromosomes within the nucleus is a widely conserved feature of early meiotic prophase that has been recognized for over a century (Wilson 1925; Scherthan 1997; Zickler and Kleckner 1998). Although details of this reorganization phase vary considerably among organisms that undergo synaptic meiosis, a universal feature is a transient highly polarized organization of the nucleus. In many organisms the most obvious evidence of polarization is a prominent clustering of telomeres to a limited sector of the nuclear periphery, often adjacent to a microtubule-organizing center (e.g., Moens 1969; Scherthan et al. 1996; Bass et al. 1997; Trelles-Sticken et al. 1999). Polarization can also be manifested in the form of a marked clustering of chromosomes toward one side of the nucleus and is often accompanied by displacement of the nucleolus to an off-center position (e.g., Holm 1977; Bass et al. 1997; this work). Although such nuclear reorganization events occur concurrently with meiotic homolog pairing, their importance in the process of pairing remains unclear. Telomere clustering per se does not seem to be a prerequisite for homolog recognition and alignment, since extensive alignment of homologs appears to occur prior to telomere clustering in Sordaria macrospora (Zickler and Kleckner 1998, 1999), and pairing is substantially delayed but not lost in the ndj1 mutant of S. cerevisiae, which lacks telomere clustering but retains other aspects of nuclear reorganization (Trelles-Sticken et al. 2000). These observations suggest that telomere clustering is not required for the establishment of homolog pairing but may instead facilitate timely completion and/or stabilization of intimate pairing.
The most striking defects in C.e. chk-2 mutants are the failure to establish pairing between homologous chromosomes and the absence of a polarized organization of chromosomes at the onset of meiotic prophase. We have also observed this coordinate loss of homolog pairing and nuclear spatial reorganization in worms carrying a mutation in another gene, hal-2 (homolog alignment; A.J. MacQueen and A.M. Villeneuve, unpubl.), further supporting a mechanistic coupling of these two prominent prophase events.
Our analysis has identified chk-2 as a molecular link between pairing and reorganization, and as such it provides an entry point for investigating the nature of this coupling. It may be the case that chk-2 is required for both events because it plays a role in chromosome movement and/or clustering per se, events that may in turn be prerequisites for pairing establishment. Alternatively, chromosome clustering and homolog pairing may be otherwise independent events that are coordinately regulated by chk-2, perhaps through a shared requirement for a chk-2-dependent modification of meiotic chromatin structure. Identification of targets regulated by chk-2 at this crucial cellular transition may help clarify the elusive relationship between spatial reorganization of the nucleus and homolog pairing.
Relationship of chk-2 to the pachytene checkpoint
Whereas chk-2 mutants are competent to trigger apoptosis of late pachytene meiocytes in response to IR-induced DNA damage, they are defective in activating this pachytene checkpoint in response to depletion of RAD-51, a protein required for completion of meiotic recombination. This result could indicate that chk-2 mutant germ cells are defective in initiation of meiotic recombination, because recombination intermediates that trigger the checkpoint would not accumulate if recombination were never initiated. A failure to initiate meiotic recombination in chk-2 mutants would suggest that successful pairing between homologous chromosomes may be a prerequisite for recombination initiation in C. elegans or that the initiation machinery is regulated by chk-2.
Alternatively, chk-2 mutants may be successful in initiating recombination but defective in activating the pachytene checkpoint in response to accumulated recombination intermediates. In this scenario, chk-2 would be required for checkpoint activation only in response to a subset of possible checkpoint-triggering lesions; some lesions induced by IR would trigger apoptosis in a chk-2-independent fashion. A possible role for chk-2 in the pachytene checkpoint raises the question of whether chk-2 might have two (or more) temporally separable functions, an early role in promoting nuclear reorganization and homolog pairing and a later role in monitoring recombination. Of course, a requirement for chk-2 in the pachytene checkpoint might be a secondary consequence of a failure to establish an appropriate chromosomal context required for such monitoring to occur.
Our analysis of chk-2 mutant germ lines did not identify a requirement for C. elegans chk-2 in either IR-induced DNA damage checkpoint responses or HU-induced proliferation arrest in germ-line nuclei. This result indicates either that chk-2 does not participate in these responses to genotoxic stress or that redundant functions are available. Redundancy in Cds1/Chk2 pathways has been described in other systems; for example, in S. pombe, Chk1 can compensate for loss of Cds1 in a replication block checkpoint (Lindsay et al. 1998). Moreover, a recent gene duplication (subsequent to divergence of nematodes from other metazoa) generated a second predicted C.e. chk-2 gene, T08D2.7, encoding a protein that is 91% identical to the chk-2 gene product described in this paper.
Checkpoint protein kinases and meiotic prophase chromosome dynamics
This is the first demonstration that a Cds1/Chk2 protein kinase plays a central role in promoting spatial reorganization of chromosomes during early meiotic prophase. Although it remains to be seen whether Cds1/Chk2 proteins will act in a similar capacity during meiosis in other organisms, it is intriguing to note that transcripts from a Drosophila Cds1/Chk2 homolog, Dmnk, are highly enriched in female germ cells and are present during early meiotic prophase (Oishi et al. 1998).
In mammalian cells, Cds1/Chk2 has been shown to be a substrate of the Atm protein kinase in the context of a DNA damage checkpoint (Matsuoka et al. 1998, 2000; Chaturvedi et al. 1999). The fact that Atm localizes to chromosomes during mammalian meiotic prophase (Keegan et al. 1996) suggests the possibility that Chk2 might function in mammalian meiosis as well. However, the meiotic prophase phenotype observed in Atm−/− mice is at least superficially quite distinct from the phenotype we see in C. elegans chk-2 mutants: whereas C.e. chk-2 mutant germ cells apparently fail to initiate spatial reorganization of chromosomes, Atm−/− mouse spermatocytes clearly initiate nuclear reorganization (Pandita et al. 1999; Scherthan et al. 2000). In fact, a bouquet-like configuration of chromosomes with tightly clustered telomeres, which is extremely transient during wild-type meiosis, was observed in an abnormally high fraction of meiocytes in Atm−/− mice.
There have been no reports analyzing possible roles of S. cerevisiae Rad53 during meiotic prophase. However, a gene duplication that occurred in a fungal ancestor subsequent to divergence of the animal and fungal lineages gave rise to Mek1, a meiosis-specific protein kinase that is closely related to Cds1/Chk2 proteins (Rockmill and Roeder 1991). Like Rad53 and mammalian Cds1/Chk2, activation of Mek1 kinase activity is dependent on an Atm homolog, Mec1 (Bailis and Roeder 2000). The phenotypes resulting from loss of S.c. Mek1 and C.e. CHK-2 are quite distinct, however, indicating that the two proteins likely play very different roles during early meiotic prophase. Whereas C.e. chk-2 mutants are profoundly deficient for homolog pairing, S.c. mek1 mutants are defective for synaptonemal complex morphogenesis and sister chromatid cohesion but exhibit almost wild-type levels of homolog pairing (Nag et al. 1995; Bailis and Roeder 1998). On the other hand, both Mek1 and CHK-2 are required for pachytene checkpoint responses to intermediate blocks in the recombination pathway (Xu et al. 1997; Bailis and Roeder 2000), suggesting the possibility that they may also have some functions in common.
Conclusions
Here, we have identified an unanticipated role for a member of the conserved Cds1/Chk2 protein kinase family, in promoting both spatial reorganization of nuclei and initial establishment of homologous chromosome pairing at the onset of meiotic prophase. While it has been assumed for decades that these events must be mechanistically coupled, our analysis of C. elegans chk-2 mutants now establishes a clear genetic and molecular basis for this linkage. Analyzing other mutants with the same set of meiotic prophase defects (e.g., hal-2) and investigating CHK-2 germ-line targets should prove to be fruitful routes toward uncovering the mechanisms by which early meiotic prophase nuclei reorganize their chromosomes and ultimately achieve a precise alignment of homologous chromosomes.
Materials and methods
Genetics
The following mutations and chromosome rearrangements (strain background Bristol N2) were used: LGI: ced-1(e1735); LGIV: spo-11(ok79), nT1[unc-?(n754) let-?(m435)](IV,V); LGV: chk-2(me18), chk-2(me64), unc-51(e369), rol-9(sc148), ozDf1, ozDf2; LGX: dpy-3(e27), unc-3(e151) (Riddle et al. 1997; Dernburg et al. 1998; Clifford et al. 2000; this work). The me18 mutation was generated as in Villeneuve (1994); me64 was generated as in Kelly et al. (2000). chk-2 strains were maintained as chk-2 rol-9/unc-51 or chk-2 rol-9/unc-51 rol-9. Unless otherwise noted, wild-type controls were chk-2/+.
me18 was mapped to the far right end of chromosome V using the method of Williams et al. (1992) and other standard crosses. me18 was further localized to a position ∼0.1 cM left of unc-51 as follows: 532 Rol progeny from hermaphrodites of genotype me18 rol-9/unc-51 were plated individually to identify Rol non-Him recombinants; 4/5 Rol non-Him recombinants carried the unc-51 mutation, and 1/5 did not.
Recombination frequency between dpy-3 and unc-3 was measured as in Kelly et al. (2000).
DAPI staining, FISH, and imaging
Gonad dissection, fixation for 4‘,6-diamidino-2-phenylindole (DAPI) staining, fluorescence in situ hybridization (FISH) and imaging using the DeltaVision deconvolution microscopy system were conducted as in Zalevsky et al. (1999). Data were collected as a series of optical sections in increments of 0.2 or 0.25 μm. Probe for the 5s rDNA locus (chromosome V, central right position) was generated as in Dernburg et al. (1998). All other probes were generated from yeast artificial chromosome (YAC) clones as in Zalevsky et al. (1999). The following YACs were used (chromosomal locations in parenthesis): Y51E2 (X, extreme left), Y13H5 (I, left), Y48E9 (I, right), Y13E11 (IV, central right), Y13H2 (III, right), and Y6D1 (II, central).
For quantitative analysis of pairing, germ lines were subdivided into five equal-sized zones (36 × 36 μm) along the distal–proximal axis, with zone 1 beginning approximately three nuclear diameters from the distal tip. A z series of images was collected for each zone; for nuclei that were completely contained within the data stack, distances between peak intensities of FISH signals were measured using the IVE software package (Chen et al. 1996). FISH signals were considered paired if the distance between their peak intensities was ≤0.7 μm; this distance corresponds to the maximum observed distance (∼0.6–0.7 μm) between the outer margins of the paired masses of electron-dense chromatin flanking the synaptonemal complex in electron micrographs of pachytene nuclei (Goldstein and Slaton 1982; Dernburg et al. 1998). In >95% of the cases in which FISH signals were scored as paired, the two signals were either visibly touching or almost completely overlapping. The frequencies with which homologous FISH signals appeared paired in premeiotic nuclei are comparable to the frequencies of associations between heterologous FISH signals in experiments in which two different probes were used simultaneously (data not shown), suggesting that these may reflect accidental or nonspecific associations. Pairing of FISH signals in chk-2 mutant germ lines never rose above the background premeiotic frequency.
Immunofluorescence
The following primary antibodies were used: rabbit polyclonal anti-HIM3 (Zetka et al. 1999); mouse monoclonal antibody against S. cerevisiae Nop1p (mAbD77) (Aris and Blobel 1988); and rabbit polyclonal anti-Ce-lamin (Liu et al. 2000). Cy3 or FITC anti-rabbit or anti-mouse secondary antibodies were obtained from Jackson Immunochemicals. Gonads were dissected from young adult worms (21 h post L4 stage) as for FISH and fixed as in Seydoux and Dunn (1997). Fixation was followed by several washes in PBT (1× PBS, 0.1% Tween-20; Sigma), and a 30-min incubation in 1% bovine serum albumin diluted in PBT. A hand-cut paraffin square was used to cover the tissue with 50 μL of antibody solution, applied at a 1:200 dilution. Incubation was conducted in a humid chamber for 1 h at room temperature (RT) followed by 15–18 h at 4°C. Slides were rinsed once in PBT, then incubated for 2–3 h (RT) with fluorophore-conjugated secondary antibody at a dilution of 1:200. For double-labeling experiments, two primary antibodies were applied simultaneously.
RNA interference
The template for generating double-stranded RNA (dsRNA) for Y60A3A.12 RNA interference (RNAi) was created by synthesizing first-strand cDNA using the primer 5′-TAATACGACT CACTATAGAGCTGCTGCTGAAATCG-3′ and subsequently using this primer in conjunction with 5′-TAATACGACTCAC TATAGGACCCTCGTAAATCAGG-3′ to amplify ∼1 kb of Y60A3A.12 ORF, flanked by T7 promoter sequences. rad-51 dsRNA was generated using cDNA clone yk241d12 containing the T3 and T7 promoters. dsRNA was prepared using a Promega in vitro transcription kit and purified using RNeasy kit (QIAGEN). dsRNA was delivered by injection (∼100 μg/mL) into germ lines or intestines of young adults, as in Fire et al. (1998). The chk-2 mutant phenocopy elicited by Y60A3A.12 RNAi was strongest in the germ lines of injected worms; 2 d after injection, these worms exhibited achiasmate diakinesis chromosomes, absence of nuclear polarization in the transition zone, and failure in homolog pairing (assayed by FISH). F1 progeny of injected animals initially exhibited achiasmate chromosomes in oocytes and a partial pairing defect, but in older F1 progeny the chromosome organization of prophase nuclei appeared normal, indicating recovery from RNAi.
Checkpoint-induced apoptosis
Germ cell corpses were identified using Nomarski optics as in Gartner et al. (2000). For experiments in Figure 6a, late L4 larvae were exposed to indicated doses of γ irradiation from a 137Cs source, and corpses were scored 22–24 h later. Experiments assessing germ cell apoptosis after rad-51 RNAi were conducted using strains carrying ced-1(e1735) to maximize the number of scorable corpses (consistent results were obtained in experiments performed in the absence of a ced-1 mutation). Young adults of genotype rol-9, chk-2(me64) rol-9/++, or spo-11/nT1 were injected with rad-51 dsRNA. F1 progeny arising from eggs laid 12–36 h post-injection were plated individually as L4 larvae, held for 18.5–19.5 h at 20°C, and analyzed for germ cell apoptosis. In general, only one gonad arm was scorable in any individual worm. For chk-2 experiments, chk-2/chk-2 worms were distinguished from control (chk-2/+ and +/+) siblings by using the closely linked cis marker rol-9. For spo-11 experiments, spo-11/spo-11 worms were distinguished from heterozygous siblings by lack of the dominant Unc marker on the nT1 balancer chromosome. Data from rad-51 RNAi experiments were considered only in cases in which all F1s from the same cohort produced >95% dead eggs (a diagnostic of successful depletion of rad-51 activity in +/+, chk-2/+, or spo-11/+ worms).
Hydroxyurea studies
Standard worm culture plates, pre-seeded with bacteria lawns, were overlayed with 250 μL of M9 solution containing 92 mg/mL hydroxyurea (HU) (Sigma) and held overnight at RT; the final concentration of HU in these plates was ∼25 mM. L4 larvae were picked to fresh HU plates and were maintained on the same plates during the entire time course. Morphological examination and quantitation of numbers of germ-line nuclei were performed on fixed DAPI-stained germ lines. For quantitation, germ lines were optically bisected along the longitudinal axis, and nuclei in the distal arm (from distal tip to the bend) of the half-gonad on the side nearest the coverslip were counted. The bisection boundary was defined by the position of the nucleus-deficient central rachis; boundary nuclei were included in the counts.
Acknowledgments
A.J.M. is grateful to A. Dernburg for training in cytological methods and for reagents, and to Y. Greunbaum, M. Paddy, and M. Zetka for antibodies. We thank T. Schedl, The Caenorhabditis Genetics Center, the Sanger Center, and the National Institute of Genetics (Japan) for strains and clones. We thank A. Dernburg, G. Stanfield, and M. Costa for key suggestions during the course of this work, and H. Scherthan, E. Martinez-Perez, and members of the Villeneuve laboratory for helpful discussions and critique of the manuscript. This work was supported by NIH grant GM53804 to A.M.V.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL villen@cmgm.stanford.edu; FAX (650) 725-7739.
Article and publication are at http://www.genesdev.org/cgi/doi/10.1101/gad.902601.
References
- Albertson DG, Rose AM, Villeneuve AM. Chromsome organization, mitosis, and meiosis. In: Riddle DL, Blumenthal T, Meyer BJ, Priess JR, editors. C. elegans II. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1997. pp. 47–78. [PubMed] [Google Scholar]
- Aris JP, Blobel G. Identification and characterization of a yeast nucleolar protein that is similar to a rat liver nucleolar protein. J Cell Biol. 1988;107:17–31. doi: 10.1083/jcb.107.1.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailis JM, Roeder GS. Synaptonemal complex morphogenesis and sister-chromatid cohesion require Mek1-dependent phosphorylation of a meiotic chromosomal protein. Genes & Dev. 1998;12:3551–3563. doi: 10.1101/gad.12.22.3551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailis JM, Roeder GS. Pachytene exit controlled by reversal of Mek1-dependent phosphorylation. Cell. 2000;101:211–221. doi: 10.1016/S0092-8674(00)80831-4. [DOI] [PubMed] [Google Scholar]
- Bass HW, Marshall WF, Sedat JW, Agard DA, Cande WZ. Telomeres cluster de novo before the initiation of synapsis: A three-dimensional spatial analysis of telomere positions before and during meiotic prophase. J Cell Biol. 1997;137:5–18. doi: 10.1083/jcb.137.1.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borde V, Goldman AS, Lichten M. Direct coupling between meiotic DNA replication and recombination initiation. Science. 2000;290:806–809. doi: 10.1126/science.290.5492.806. [DOI] [PubMed] [Google Scholar]
- Cha RS, Weiner BM, Keeney S, Dekker J, Kleckner N. Progression of meiotic DNA replication is modulated by interchromosomal interaction proteins, negatively by Spo11p and positively by Rec8p. Genes & Dev. 2000;14:493–503. [PMC free article] [PubMed] [Google Scholar]
- Chaturvedi P, Eng WK, Zhu Y, Mattern MR, Mishra R, Hurle MR, Zhang X, Annan RS, Lu Q, Faucette, et al. Mammalian Chk2 is a downstream effector of the ATM-dependent DNA damage checkpoint pathway. Oncogene. 1999;18:4047–4054. doi: 10.1038/sj.onc.1202925. [DOI] [PubMed] [Google Scholar]
- Chen H, Hughes DD, Chan TA, Sedat JW, Agard DA. IVE (Image Visualization Environment): A software platform for all three-dimensional microscopy applications. J Struct Biol. 1996;116:56–60. doi: 10.1006/jsbi.1996.0010. [DOI] [PubMed] [Google Scholar]
- Chin GM, Villeneuve AM. C. elegans mre-11 is required for meiotic recombination and DNA repair but is dispensable for the meiotic G2 DNA damage checkpoint. Genes & Dev. 2001;15:522–534. doi: 10.1101/gad.864101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clifford R, Lee MH, Nayak S, Ohmachi M, Giorgini F, Schedl T. FOG-2, a novel F-box containing protein, associates with the GLD-1 RNA binding protein and directs male sex determination in the C. elegans hermaphrodite germline. Development. 2000;127:5265–5276. doi: 10.1242/dev.127.24.5265. [DOI] [PubMed] [Google Scholar]
- Dasika GK, Lin SC, Zhao S, Sung P, Tomkinson A, Lee EY. DNA damage-induced cell cycle checkpoints and DNA strand break repair in development and tumorigenesis. Oncogene. 1999;18:7883–7899. doi: 10.1038/sj.onc.1203283. [DOI] [PubMed] [Google Scholar]
- Davis L, Barbera M, McDonnell A, McIntyre K, Sternglanz R, Jin Q, Loidl J, Engebrecht J. The Saccharomyces cerevisiae MUM2 gene interacts with the DNA replication machinery and is required for meiotic levels of double strand breaks. Genetics. 2001;157:1179–1189. doi: 10.1093/genetics/157.3.1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dernburg AF, McDonald K, Moulder G, Barstead R, Dresser M, Villeneuve AM. Meiotic recombination in C. elegans initiates by a conserved mechanism and is dispensable for homologous chromosome synapsis. Cell. 1998;94:387–398. doi: 10.1016/s0092-8674(00)81481-6. [DOI] [PubMed] [Google Scholar]
- Ellis RE, Jacobson DM, Horvitz HR. Genes required for the engulfment of cell corpses during programmed cell death in Caenorhabditis elegans. Genetics. 1991;129:79–94. doi: 10.1093/genetics/129.1.79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emili A, Schieltz DM, Yates JR, Hartwell LH. Dynamic interaction of DNA damage checkpoint protein Rad53 with chromatin assembly factor Asf1. Mol Cell. 2001;7:13–20. doi: 10.1016/s1097-2765(01)00150-2. [DOI] [PubMed] [Google Scholar]
- Fire A, Xu S, Montgomery MK, Kostas SA, Driver SE, Mello CC. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature. 1998;391:806–811. doi: 10.1038/35888. [DOI] [PubMed] [Google Scholar]
- Gartner A, Milstein S, Ahmed S, Hodgkin J, Hengartner MO. A conserved checkpoint pathway mediates DNA damage–induced apoptosis and cell cycle arrest in C. elegans. Mol Cell. 2000;5:435–443. doi: 10.1016/s1097-2765(00)80438-4. [DOI] [PubMed] [Google Scholar]
- Goldstein P, Slaton DE. The synaptonemal complexes of Caenorhabditis elegans: Comparison of wild-type and mutant strains and pachytene karyotype analysis of wild-type. Chromosoma. 1982;84:585–597. doi: 10.1007/BF00292857. [DOI] [PubMed] [Google Scholar]
- Higashitani A, Aoki H, Mori A, Sasagawa Y, Takanami T, Takahashi H. Caenorhabditis elegans Chk2-like gene is essential for meiosis but dispensable for DNA repair. FEBS Lett. 2000;485:35–39. doi: 10.1016/s0014-5793(00)02178-5. [DOI] [PubMed] [Google Scholar]
- Hodgkin J, Horvitz HR, Brenner S. Nondisjunction mutants of the nematode Caenorhabditis elegans. Genetics. 1979;91:67–94. doi: 10.1093/genetics/91.1.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holm P. The premeiotic DNA replication of euchromatin and heterochromatin in Lilium longiflorum (Thunb.) Carlsberg Res Commun. 1977;42:249–281. [Google Scholar]
- Hu F, Alcasabas AA, Elledge SJ. Asf1 links Rad53 to control of chromatin assembly. Genes & Dev. 2001;15:1061–1066. doi: 10.1101/gad.873201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keegan KS, Holtzman DA, Plug AW, Christenson ER, Brainerd EE, Flaggs G, Bentley NJ, Taylor EM, Meyn MS, Moss SB, et al. The Atr and Atm protein kinases associate with different sites along meiotically pairing chromosomes. Genes & Dev. 1996;10:2423–2437. doi: 10.1101/gad.10.19.2423. [DOI] [PubMed] [Google Scholar]
- Kelly KO, Dernburg AF, Stanfield GM, Villeneuve AM. Caenorhabditis elegans msh-5 is required for both normal and radiation-induced meiotic crossing over but not for completion of meiosis. Genetics. 2000;156:617–630. doi: 10.1093/genetics/156.2.617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindsay HD, Griffiths DJ, Edwards RJ, Christensen PU, Murray JM, Osman F, Walworth N, Carr AM. S-phase-specific activation of Cds1 kinase defines a subpathway of the checkpoint response in Schizosaccharomyces pombe. Genes & Dev. 1998;12:382–395. doi: 10.1101/gad.12.3.382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Ben-Shahar TR, Riemer D, Treinin M, Spann P, Weber K, Fire A, Gruenbaum Y. Essential roles for Caenorhabditis elegans lamin gene in nuclear organization, cell cycle progression, and spatial organization of nuclear pore complexes. Mol Biol Cell. 2000;11:3937–3947. doi: 10.1091/mbc.11.11.3937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuoka S, Huang M, Elledge SJ. Linkage of ATM to cell cycle regulation by the Chk2 protein kinase. Science. 1998;282:1893–1897. doi: 10.1126/science.282.5395.1893. [DOI] [PubMed] [Google Scholar]
- Matsuoka S, Rotman G, Ogawa A, Shiloh Y, Tamai K, Elledge SJ. Ataxia telangiectasia-mutated phosphorylates Chk2 in vivo and in vitro. Proc Natl Acad Sci. 2000;97:10389–10394. doi: 10.1073/pnas.190030497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merino ST, Cummings WJ, Acharya SN, Zolan ME. Replication-dependent early meiotic requirement for Spo11 and Rad50. Proc Natl Acad Sci. 2000;97:10477–10482. doi: 10.1073/pnas.190346097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moens PB. The fine structure of meiotic chromosome polarization and pairing in Locusta migratoria spermatocytes. Chromosoma. 1969;28:1–25. doi: 10.1007/BF00325986. [DOI] [PubMed] [Google Scholar]
- Moore DP, Orr-Weaver TL. Chromosome segregation during meiosis: Building an unambivalent bivalent. Curr Top Dev Biol. 1998;37:263–299. doi: 10.1016/s0070-2153(08)60177-5. [DOI] [PubMed] [Google Scholar]
- Murakami H, Nurse P. DNA replication and damage checkpoints and meiotic cell cycle controls in the fission and budding yeasts. Biochem J. 2000;349:1–12. doi: 10.1042/0264-6021:3490001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nag DK, Scherthan H, Rockmill B, Bhargava J, Roeder GS. Heteroduplex DNA formation and homolog pairing in yeast meiotic mutants. Genetics. 1995;141:75–86. doi: 10.1093/genetics/141.1.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oishi I, Sugiyama S, Otani H, Yamamura H, Nishida Y, Minami Y. A novel Drosophila nuclear protein serine/threonine kinase expressed in the germline during its establishment. Mech Dev. 1998;71:49–63. doi: 10.1016/s0925-4773(97)00200-1. [DOI] [PubMed] [Google Scholar]
- Oishi I, Iwai K, Kagohashi Y, Fujimoto H, Kariya KI, Kataoka T, Sawa H, Okano H, Otani H, Yamamura H, et al. Critical role of Caenorhabditis elegans homologs of Cds1 (Chk2)-related kinases in meiotic recombination. Mol Cell Biol. 2001;21:1329–1335. doi: 10.1128/MCB.21.4.1329-1335.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandita TK, Westphal CH, Anger M, Sawant SG, Geard CR, Pandita RK, Scherthan H. Atm inactivation results in aberrant telomere clustering during meiotic prophase. Mol Cell Biol. 1999;19:5096–5105. doi: 10.1128/mcb.19.7.5096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paques F, Haber JE. Multiple pathways of recombination induced by double-strand breaks in Saccharomyces cerevisiae. Microbiol Mol Biol Rev. 1999;63:349–404. doi: 10.1128/mmbr.63.2.349-404.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paulovich AG, Hartwell LH. A checkpoint regulates the rate of progression through S phase in S. cerevisiae in response to DNA damage. Cell. 1995;82:841–847. doi: 10.1016/0092-8674(95)90481-6. [DOI] [PubMed] [Google Scholar]
- Rhind N, Russell P. Chk1 and Cds1: Linchpins of the DNA damage and replication checkpoint pathways. J Cell Sci. 2000;113:3889–3896. doi: 10.1242/jcs.113.22.3889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riddle DL, Blumenthal T, Meyer BJ, Priess JR. C. elegans II. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1997. [PubMed] [Google Scholar]
- Rockmill B, Roeder GS. A meiosis-specific protein kinase homolog required for chromosome synapsis and recombination. Genes & Dev. 1991;5:2392–2404. doi: 10.1101/gad.5.12b.2392. [DOI] [PubMed] [Google Scholar]
- Roeder GS. Meiotic chromosomes: It takes two to tango. Genes & Dev. 1997;11:2600–2621. doi: 10.1101/gad.11.20.2600. [DOI] [PubMed] [Google Scholar]
- Santocanale C, Diffley JF. A Mec1- and Rad53-dependent checkpoint controls late-firing origins of DNA replication. Nature. 1998;395:615–618. doi: 10.1038/27001. [DOI] [PubMed] [Google Scholar]
- Schedl T. Developmental genetics of the germ line. In: Riddle DL, Blumenthal T, Meyer BJ, Priess JR, editors. C. elegans II. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1997. pp. 241–269. [PubMed] [Google Scholar]
- Scherthan H. Chromosome behaviour in earliest meiotic prophase. In: Gill JSP H, Puertas M, editors. Chromosomes today. London: Chapman and Hall; 1997. pp. 217–248. [Google Scholar]
- Scherthan H, Weich S, Schwegler H, Heyting C, Harle M, Cremer T. Centromere and telomere movements during early meiotic prophase of mouse and man are associated with the onset of chromosome pairing. J Cell Biol. 1996;134:1109–1125. doi: 10.1083/jcb.134.5.1109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scherthan H, Jerratsch M, Dhar S, Wang YA, Goff SP, Pandita TK. Meiotic telomere distribution and sertoli cell nuclear architecture are altered in atm- and atm-p53-deficient mice. Mol Cell Biol. 2000;20:7773–7783. doi: 10.1128/mcb.20.20.7773-7783.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seydoux G, Dunn MA. Transcriptionally repressed germ cells lack a subpopulation of phosphorylated RNA polymerase II in early embryos of Caenorhabditis elegans and Drosophila melanogaster. Development. 1997;124:2191–2201. doi: 10.1242/dev.124.11.2191. [DOI] [PubMed] [Google Scholar]
- Smith KN, Penkner A, Ohta K, Klein F, Nicolas A. B-type cyclins CLB5 and CLB6 control the initiation of recombination and synaptonemal complex formation in yeast meiosis. Curr Biol. 2001;11:88–97. doi: 10.1016/s0960-9822(01)00026-4. [DOI] [PubMed] [Google Scholar]
- Trelles-Sticken E, Loidl J, Scherthan H. Bouquet formation in budding yeast: Initiation of recombination is not required for meiotic telomere clustering. J Cell Sci. 1999;112:651–658. doi: 10.1242/jcs.112.5.651. [DOI] [PubMed] [Google Scholar]
- Trelles-Sticken E, Dresser ME, Scherthan H. Meiotic telomere protein Ndj1p is required for meiosis-specific telomere distribution, bouquet formation and efficient homologue pairing. J Cell Biol. 2000;151:95–106. doi: 10.1083/jcb.151.1.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villeneuve AM. A cis-acting locus that promotes crossing over between X chromosomes in Caenorhabditis elegans. Genetics. 1994;136:887–902. doi: 10.1093/genetics/136.3.887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams BD, Schrank B, Huynh C, Shownkeen R, Waterston RH. A genetic mapping system in Caenorhabditis elegans based on polymorphic sequence-tagged sites. Genetics. 1992;131:609–624. doi: 10.1093/genetics/131.3.609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson EB. The cell in development and heredity. New York: MacMillan; 1925. [Google Scholar]
- Xu L, Weiner BM, Kleckner N. Meiotic cells monitor the status of the interhomolog recombination complex. Genes & Dev. 1997;11:106–118. doi: 10.1101/gad.11.1.106. [DOI] [PubMed] [Google Scholar]
- Zalevsky J, MacQueen AJ, Duffy JB, Kemphues KJ, Villeneuve AM. Crossing over during Caenorhabditis elegans meiosis requires a conserved MutS-based pathway that is partially dispensable in budding yeast. Genetics. 1999;153:1271–1283. doi: 10.1093/genetics/153.3.1271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zetka MC, Kawasaki I, Strome S, Muller F. Synapsis and chiasma formation in Caenorhabditis elegans require HIM-3, a meiotic chromosome core component that functions in chromosome segregation. Genes & Dev. 1999;13:2258–2270. doi: 10.1101/gad.13.17.2258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zickler D, Kleckner N. The leptotene-zygotene transition of meiosis. Annu Rev Genet. 1998;32:619–697. doi: 10.1146/annurev.genet.32.1.619. [DOI] [PubMed] [Google Scholar]
- Zickler D, Kleckner N. Meiotic chromosomes: Integrating structure and function. Annu Rev Genet. 1999;33:603–754. doi: 10.1146/annurev.genet.33.1.603. [DOI] [PubMed] [Google Scholar]