Abstract
This article reviews work from the author dating back to 1978 and focuses on the structural basis of cytochrome P450 (P450) function using available contemporary techniques. Early studies used mechanism-based inactivators that bound to the protein moiety of hepatic P450s to try to localize the active site. Subsequent studies used cDNA cloning, heterologous expression, site-directed mutagenesis, and homology modeling based on multiple bacterial P450 X-ray crystal structures to predict the active sites of CYP2B enzymes with considerable accuracy. Breakthroughs in engineering and expression of mammalian P450s enabled us to determine our first X-ray crystal structure of ligand-free rabbit CYP2B4. To date, we have solved 11 CYP2B4 and three human CYP2B6 structures, which represent four significantly different conformations. The plasticity of CYP2B4 has been confirmed by deuterium exchange mass spectrometry and is substantiated by molecular dynamics simulations. In addition to major movement of secondary structure elements, more subtle reorientation of active site side chains, especially Phe206, Phe297, and Glu301, contributes to the ability of CYP2B enzymes to bind various ligands. Isothermal titration calorimetry has proven to be a useful tool for studying the thermodynamics of ligand binding to CYP2B4 and CYP2B6, and NMR has enabled study of ligand binding orientation in solution as an adjunct to X-ray crystallography. A major challenge remains to harness the power of the various approaches to facilitate prediction of CYP2B specificity and inhibition.
Introduction
It is a great honor to have been chosen as the recipient of the 2010 Bernard B. Brodie Award in Drug Metabolism. I thank the American Society for Pharmacology and Experimental Therapeutics for their recognition of the research done in my laboratory. I am indebted to the many graduate students, postdoctoral fellows, research faculty, visiting scientists, staff, and undergraduates who worked with me over the past 30 years. Finally, I am grateful to the many collaborators around the world who made research possible that could never have been done by one research group alone. Being a member of the “P450 superfamily” of scientists has been a tremendous experience and opportunity that has enriched both my scientific and personal life. I am especially fortunate to have been able to train a number of scientists who have become faculty members or leaders in the industry and are carrying the P450 tradition forward.
My Ph.D. studies at Uppsala University in Sweden under the direction of Dr. David Eaker introduced me to structure-function analysis of proteins. During my subsequent M.S. studies at the Karolinska Institute under the guidance of my good friend Dr. Magnus Ingelman-Sundberg, I was introduced to drug metabolism. At that time, P450 was just a colored contaminant in the preparations of rabbit liver epoxide hydrolase that I was trying to purify (Halpert et al., 1979). It was not until I arrived in the laboratory of Dr. Robert A. Neal1 at the Center for Environmental Toxicology at Vanderbilt University in 1978 that P450 became the focus of my research. I will always be grateful to David, Magnus, and Bob for instilling in me a love for science, emphasizing the importance of tackling important problems with scientific rigor, and recognizing just how much guidance to provide to bring out the best in me. In working with my own trainees, I have always tried to combine the best of what I learned from my own mentors.
Bob Neal's idea in 1978 was to use mechanism-based inactivators (MBIs) to label the active sites of P450s. At that time, most of these compounds labeled or destroyed the heme moiety of the enzyme, providing key insights into mechanisms of catalysis (Ortiz de Montellano, 1995) but no information about active site residues. My friend Ted Kamataki, while a visiting scientist with Bob, had performed some pioneering studies with parathion (Kamataki and Neal, 1976), suggesting that it might be one of few agents capable of labeling the protein moiety of P450s. Working with highly purified fractions of what we now call CYP2B1, I found that although parathion did indeed modify P450 protein, the mechanism of inactivation was exceedingly complex and unlikely to help elucidate the nature of the active site (Halpert et al., 1980). Dr. Chris Chengelis, another postdoctoral fellow in the laboratory, showed me an article suggesting that the antibiotic chloramphenicol might be more suitable than parathion for labeling CYP2B1 protein. That fortuitous interaction with Chris led to a series of studies involving enzyme purification, labeling, site-directed mutagenesis, homology modeling, X-ray crystallography, and solution biophysics that have kept my research going for the past 30 years. This work and recently culminated in a collaborative project with Dr. Paul Hollenberg on X-ray crystal structures of CYP2B4 with a metabolite of tert-butylphenylacetylene bound to Thr302. Thus, the work on P450 has truly been a journey from MBIs to X-ray crystal structures and back.
Mechanism-Based Inactivation of Hepatic Cytochromes P450 by Chloramphenicol and Analogs.
A key initial finding was that CYP2B1 could be inactivated by chloramphenicol with no loss of P450 detectable as the reduced CO-complex or of heme detectable as pyridine hemochromagen. The inactivation was accompanied by the covalent binding of 1.5 nmol of metabolite per mol of P450 (Halpert and Neal, 1980). In a subsequent study that Bob graciously let me publish as the sole author, I showed that there were two classes of adducts of chloramphenicol and CYP2B1, including N-ε-chloramphenicol oxamyl lysine. To our knowledge, this was the first clear identification of an adduct of a reactive intermediate of a xenobiotic and a P450 (Halpert, 1981). In later studies performed while I was a research faculty member in Jan-Åke Gustafsson's department in Huddinge, Sweden, we demonstrated the formation of this adduct after in vivo administration of chloramphenicol to phenobarbital-treated rats (Halpert et al., 1983). This finding set the stage for my first publication as an independent faculty member in the College of Pharmacy at the University of Arizona, in which CYP2B1 isolated from chloramphenicol and phenobarbital-treated rats was found to be deficient in the ability to accept electrons from NADPH-cytochrome P450 reductase (Halpert et al., 1985b). Accordingly, chloramphenicol was not suitable for identifying amino acid residues involved in ligand binding.
Concomitant with the mechanistic studies, we began to investigate the isoform selectivity of chloramphenicol in rats, in what became a very fruitful series of collaborative studies with Dr. Larry Kaminsky (Halpert et al., 1985a). Of the eight rat liver P450 enzymes known at the time, in vivo administration of chloramphenicol was found to inactivate three. Chloramphenicol proved to be a selective inactivator of the major phenobarbital-inducible dog liver P450 (CYP2B11) (Ciaccio et al., 1987; Duignan et al., 1987), suggesting that suitable modifications of the chloramphenicol structure might lead to a selective inactivator of CYP2B1 in the rat. This result was accomplished through the synthesis of N-(2-p-nitrophenethyl)chlorofluoroacetamide (Halpert et al., 1990). Most interestingly, we subsequently identified a strain variant of CYP2B1 found in Wistar-Munich (WM) rats that was refractory to inactivation by this compound due to an inability to catalyze the oxidative dehalogenation reaction required for reactive metabolite formation. The structural basis for the lack of inactivation of WM CYP2B1 was found to result from a single G478A substitution (Kedzie et al., 1991), which represented the first demonstration that a single amino acid change could render a P450 refractory to MBI. Much later, and in collaboration with Dr. Paul Hollenberg, we showed that this same substitution also rendered WM CYP2B1 refractory to N-benzyl-1-aminobenzotriazole (BBT). Molecular modeling suggested that the Ala478 resulted in steric hindrance when BBT was bound in the orientation required to inactivate CYP2B1 (Kent et al., 1997).
Initial Studies of the Structural Basis of CYP2B Substrate Specificity.
In the early 1990's, our focus broadened to encompass the structural basis of substrate specificity of CYP2B enzymes from rat, rabbit, and dog using hybrid enzymes and site-directed mutagenesis (He et al., 1992, 1994, 1996; Luo et al., 1994; Szklarz et al., 1996). These studies capitalized on the substrate recognition site (SRS) model created by Gotoh (1992). A key element of our approach was the analysis of results with CYP2B homology models based on three bacterial P450 structures (Szklarz et al., 1995, 1996). Despite the low sequence identity between the bacterial P450 and CYP2B enzymes, our approach allowed us to identify most of the active site residues involved in substrate oxidation (Szklarz and Halpert, 1997; Domanski and Halpert, 2001) long before our first X-ray crystal structure of a CYP2B-ligand complex (Scott et al., 2004). Subsequently, homology models of CYP2B1, CYP2B4, and CYP2B5 were used to rationalize selective inhibition by phenylimidazoles (Spatzenegger et al., 2001), and a combined pharmacophore and protein model of CYP2B6 was constructed to analyze substrate oxidation by this human enzyme (Wang and Halpert, 2002).
CYP2B4 Open Ligand-Free Structure.
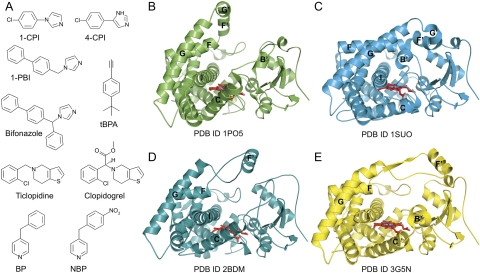
The first X-ray crystal structures of rabbit CYP2C5 were huge breakthroughs in the study of mammalian P450s (Cosme and Johnson, 2000; Williams et al., 2000; Wester et al., 2003a,b). To obtain sufficient amounts of high-quality CYP2C5 for crystallization trials, both N-terminal and internal modifications and mutations were necessary. However, it was not clear whether these modifications were necessary or sufficient to crystallize other microsomal P450s. Fortunately, expression of similarly truncated and N-terminally modified CYP2B1, CYP2B4, and CYP2B11 without further internal modifications was found to yield large amounts of catalytically active CYP2Bs with increased solubility, which were suitable for structural and functional analysis (Scott et al., 2001). These findings enabled the first crystallization of rabbit CYP2B4 (Table 1) (Scott et al., 2003). Although analyses of ligand-bound and ligand-free structures of CYP2C5 (Wester et al., 2003a,b) indicated that P450s are flexible enzymes, the wide-open structure of CYP2B4 (Fig. 1B) showed a drastic rearrangement of what were later termed “plastic regions” (Zhao et al., 2006). This reorganization of secondary structural elements creates a large substrate access cleft through movement of the F-G cassette region, which comprises helices F, F′, G′, and G on one side of the enzyme and the B′/C loop and C helix on the other (Scott et al., 2003). In contrast, regions on the proximal side of the enzyme (helices E, J, K, and L) remain relatively unchanged compared with other P450 structures. In this structure, the open conformation is stabilized through formation of a homodimer where residues of helices F′ and G′ partially fill the active site of a symmetry-related molecule, and the imidazole moiety of His226 coordinates the sixth ligand position of the heme iron of the opposite chain. Subsequently, all CYP2B4 structures have been determined with an H226Y mutant to prevent this dimerization.
TABLE 1.
Crystal structures of mammalian P450 2B enzymes
| Enzyme | Ligand | Protein Data Bank Data Code | Reference |
|---|---|---|---|
| CYP2B4 | 4-CPI | 1SUO | Scott et al., 2004 |
| CYP2B4 | 1-CPI | 2Q6N | Zhao et al., 2007 |
| CYP2B4 | 1-PBI | 3G5N | Gay et al., 2009 |
| CYP2B4 | Bifonazole | 2BDM | Zhao et al., 2006 |
| CYP2B4 | Clopidogrel | 3ME6 | Gay et al., 2010b |
| CYP2B4 | Ticlopidine | 3KW4 | Gay et al., 2010b |
| CYP2B4 | tBPA, closed | 3R1A | Gay et al., 2011 |
| CYP2B4 | tBPA, open | 3R1B | Gay et al., 2011 |
| CYP2B4 | 3MVR | Wilderman et al., 2010 | |
| CYP2B4 | 1PO5 | Scott et al., 2003 | |
| CYP2B6 | 4-CPI | 3IBD | Gay et al., 2010c |
| CYP2B6 | BP | 3QOA | M. B. Shah, J. Pascual, A. G. Roberts, Q. Zhang, C. D. Stout, and J. R. Halpert, manuscript in preparation |
| CYP2B6 | NBP | 3QUB | M. B. Shah, J. Pascual, A. G. Roberts, Q. Zhang, C. D. Stout, and J. R. Halpert, manuscript in preparation |
Fig. 1.
Structural plasticity of CYP2B enzymes observed by X-ray crystallography. A, the structural diversity of CYP2B ligands and substrates is indicated by the variability of size, shape, and chemical functionality of compounds bound in CYP2B4 and CYP2B6 structures. In binding these ligands, CYP2B enzymes have been observed to adopt four markedly different conformations. B, in the absence of ligand, CYP2B4 is able to adopt an open conformation, with a U-shaped active site cleft. C, the most populated CYP2B form is the closed conformation, which corresponds to CYP2B4 complexes of 4-CPI, 1-CPI, ticlopidine, and clopidogrel, the closed tBPA-modified and closed ligand-free CYP2B4 structures, and the CYP2B6 complexes of 4-CPI, BP, and NBP. D, CYP2B4 has also been observed in a second open conformation in complex with bifonazole that is distinct from the open ligand-free structure. The open tBPA-modified CYP2B4 structure also resembles this conformation. E, the 1-PBI complex of CYP2B4 adopts a conformation that is intermediate to the closed and bifonazole-bound structures. Flexible helices of note have been labeled in B to E.
Analysis of the open structure of CYP2B4 showed that active site residues on the I helix (Ser294, Phe297, Ala298, and Thr302) and those near β1–4 (Leu362, Ile363, and Val367) were in close proximity to the heme. However, numerous other active site residues identified by site-directed mutagenesis and comparison to the CYP2C5 structures (Ile101, Ile114, Phe115, Phe206, Ile209, Val477, and Gly478) were located far from the heme and each other. In this CYP2B4 protein conformation, it would be impossible for a substrate bound in the active site to make contact with all of these residues, implying that some structural rearrangement would be necessary upon substrate binding for these residues to move into proximity of the ligand. Crystallographic studies of a CYP2B4-ligand complex were expected to reveal the conformational change associated with the closure of the active site.
Structural Studies of CYP2B4 and Imidazoles.
To map the structural response of CYP2B4 to the presence of ligands, crystallographic studies of a series of chemically uniform imidazole-based inhibitors were undertaken. Correlating structural information to solution behavior of the enzyme and using these data to predict new ligands was also a major goal of this effort. The approaches used in these studies span a range of techniques, all designed to complement one another to provide a clearer picture of the relationship between enzyme structure and function.
4-(4-Chlorophenyl)imidazole.
The 4-(4-chlorophenyl)imidazole (4-CPI) (Fig. 1A) complex of CYP2B4 adopts a closed conformation (Table 1) (Scott et al., 2004) as predicted. Portions of the enzyme that formed the open cleft in the ligand-free structure have moved toward to each other to close the active site around the small inhibitor (Fig. 1C). The B′ helix and N terminus of the I helix move toward each other, which places the B′ helix near the G helix. Helices F, F′, G′, G and their associated loops, which form what is known as the F-G cassette, move to interact with the A helix and β1 sheet at the N terminus of the enzyme, helix B′ and the B′/C loop, the N terminus of the I helix, and the β4 sheet, thus capping the active site. The F-G cassette forms the lid of the active site, and interactions between residues of helices B′ and G appear to play a key role in closing the active site.
4-CPI was found to make contact with 11 CYP2B4 residues (Ile101, Val104, Ile114, Phe115, Phe297, Ala298, Glu301, Thr302, Ile363, Val367, and Val477) that are all within 5 Å of the bound inhibitor. This list is in agreement with those identified previously as active site residues by site-directed mutagenesis studies of CYP2B1 (Domanski et al., 2001; Domanski and Halpert, 2001; Honma et al., 2005). The CYP2B4 active site, which packs tightly around the relatively small 4-CPI (178.6 Da) inhibitor, would probably have to expand to some degree to accommodate CYP2B substrates, which are generally larger molecules.
Bifonazole.
To obtain a structure of CYP2B4 in an intermediate conformation between the ligand-free and 4-CPI bound structures, the enzyme was crystallized with the larger, branched, imidazole inhibitor bifonazole (310.4 Da) (Fig. 1A) (Zhao et al., 2006). With a wide-open active site (Fig. 1D), the resulting structure (Table 1) is actually expanded even more than the ligand-free structure. Ten residues (Ser128, Met132, Val292, Leu295, Phe296, Ala298, Gly299, Glu301, Thr302, and Ile363) were found within 5 Å of the inhibitor. Only four of them (Ala298, Glu301, Thr302, and Ile363) also make contact with 4-CPI in that structure. These changes are caused by concerted movement of several secondary structural elements, most notably the B′/C loop and C helix, F-G cassette, and N terminus of the I helix. Rather than forming the typical B′/C loop structure, which contains helix B′, residues 100 to 115 move upward toward the N terminus of the enzyme in a fully extended loop with no helix. The C helix is also pulled upward, where it moves closer to the N terminus of the I helix, which bent ∼15° to expand the ligand binding pocket. The F helix is approximately 1.5 turns shorter in the structure and the F/F′ loop is much longer. The length of this loop allows the F′ helix to move upward and away from the active site. Helix G is also pushed away from the protein core, resulting in no contact with the ligand. In addition, to protect the hydrophobic F′ helix, CYP2B4-bifonazole forms a dimer in the crystal, where the F-G regions of one molecule insert into the active site of a symmetry-related molecule and vice versa. This is thought to occur to protect hydrophobic regions exposed during crystallization. However, these portions of the protein are likely buried in the membrane in vivo.
After careful analysis of the CYP2B4 ligand-free structure as well as the 4-CPI and bifonazole complexes, it was found that approximately half of the protein structure is conserved regardless of the presence or absence of ligand or the size of any bound ligand. The regions correspond to the N-terminal proline-rich region, the C-terminal half of helix A, helix B, the C-terminal half of helix I, helices J through L, and β-sheets 1, 2, and 3. These regions form the protein core, including most of the secondary structural elements involved in heme binding. Many secondary structural elements on the proximal side of the protein are also conserved, consistent with their role in binding of electron transfer partners NADPH-cytochrome P450 reductase and cytochrome b5. However, residues in five regions were subject to structural variability and defined as plastic regions (PR). PR1 (residues 39–57) comprises a loop preceding helix A as well as the N-terminal half of helix A. PR2 (residues 101–140) includes helix B′, the B′/C loop, helix C, and the C/D loop. PR3 (residues 177–188) comprises the C-terminal half of helix E and the following loop. The largest plastic region, PR4 (residues 203–298), includes the C-terminal half of helix F through the N-terminal half of helix I. The smallest plastic region, PR5 (residues 474–480), forms the β4 hairpin in the CYP2B4-4-CPI structure. These plastic regions of CYP2B4 undergo correlated conformational changes in response to ligand binding (Fig. 1), enabling the enzyme to dramatically reshape its active site without perturbing the overall P450 fold.
1-(4-Chlorophenyl)imidazole.
Based on results from our laboratory and others, it was becoming apparent that microsomal P450s exhibit remarkable conformational diversity and plasticity (Johnson and Stout, 2005; Poulos, 2005), which results in significant differences in the structure and placement of several secondary structural elements. These differences were apparent even in highly related P450s of the same subfamily, but the largest degree of structural plasticity had been observed in the CYP2B4 structures from our laboratory. These results suggest that microsomal P450s adopt an “induced fit” mechanism for ligand binding, where plastic regions are able to reorganize to accommodate the wide variety of P450 substrates without perturbing the overall P450 fold (Fig. 1). However, it was unknown what changes to the structure and active site of CYP2B4 would occur upon binding a molecule similar to one already observed in a crystal structure.
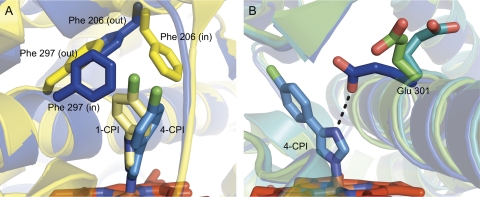
During the search for new crystallization candidates, 1-(4-chlorophenyl)imdazole (1-CPI) (Fig. 1A) displayed significant differences in binding affinity and thermodynamics, compared with its isomer 4-CPI. To determine the structural basis for these differences, the crystal structure of a CYP2B4-1-CPI complex was determined (Table 1) (Zhao et al., 2007). Overall, the tertiary structure of the complex was very similar to that of 4-CPI-bound CYP2B4 (Fig. 1C). However, repositioning of the side chains of Phe206, Phe297, and Glu301 led to a significantly different active site landscape. Phe206 and Phe297 exchanged positions, with Phe206 becoming a ligand-contact residue (Fig. 2A), while Glu301, rather than hydrogen bonding to the ligand, flips away from the active site (Fig. 2B) and interacts with His172. This causes an increase in active site volume from 200 Å3 in the 4-CPI complex to 280 Å3 in the 1-CPI complex. These findings highlight the ability of CYP2B4 to change the shape of its active site through the movement of a relatively small number of amino acid side chains.
Fig. 2.
Movement of active site residues in CYP2B-ligand complexes. To accommodate their various substrates and inhibitors, the closed forms of CYP2B enzymes show a reorientation of Phe206, Phe297, and Glu301. A, an overlay of the 1-CPI (yellow) and 4-CPI (blue) CYP2B4 complexes highlights the movement of Phe206 and Phe297. In the CYP2B4 complexes of 1-CPI and clopidogrel, the closed tBPA-modified CYP2B4 structure, and the CYP2B6 complexes of NBP and BP, Phe206 enters the active site, while Phe297 rotates out. In the 4-CPI complexes of CYP2B6 and CYP2B4, and the closed ligand-free and CYP2B4-ticlopidine structures, these two residues swap positions so that Phe206 is out and Phe297 swings in. B, an overlay of the CYP2B4 complexes of 4-CPI (blue), ticlopidine (green), and clopidogrel (teal) shows how Glu301 enters the active site to hydrogen-bond with 4-CPI and moves out of the active site to varying degrees in other CYP2B structures.
1-Biphenyl-4-methyl-1H-imidazole.
After observing the marked differences in tertiary structure in the CYP2B4 complexes of 1-CPI and 4-CPI compared with the bifonazole complex, it was unclear how the enzyme made the transition for such large structural reorganizations. To probe this mechanism, CYP2B4 was crystallized in the presence of another imidazole-based inhibitor of intermediate size (Table 1). With two phenyl rings, 1-biphenyl-4-methyl-1H-imidazole (1-PBI) (Fig. 1A) is larger than 1-CPI and 4-CPI but smaller than bifonazole, which is branched and contains three phenyl rings. Like CYP2B4-bifonazole, this structure also forms a crystallographic dimer, where the F′ helix of one monomer is buried in the active site of its partner. As previously observed, the protein core of CYP2B4-1-PBI does not change compared with other structures, whereas movement of plastic regions allows the active site to accommodate the inhibitor (Fig. 1E) (Gay et al., 2009). In this case, the B′/C loop is shifted away from the active site and contains two small helices. This conformation is different from those observed in the CPI and bifonazole complexes of CYP2B4 as well as the ligand-free open structure. The active site lid is formed by the F-G cassette and adopts a conformation intermediate to those seen in the CPI and bifonazole structures. With its biphenyl rings pointing toward β-sheet 1, the ligand interacts with six residues (Ala298, Thr302, Ile363, Val367, Pro368, and Val477) in addition to coordinating to the heme iron. This represents a hybrid of the residues involved in bifonazole binding and those involved in 4-CPI binding, with three being common in each structure of CYP2B4-ligand complexes (Ala298, Thr302, and Ile363).
Analysis of the overall CYP2B4-1-PBI complex and comparison with the other imidazole-bound structures of CY2B4 indicated that a cluster of 10 phenylalanine residues at the intersection of the F, G, and I helices could be the key to allowing the F-G cassette to adopt new conformations during ligand binding. These are Phe184, Phe188, Phe195, Phe202, Phe203, Phe206, Phe244, Phe264, Phe296, and Phe297. A sequence alignment of seven other family 2 P450s (data not shown) showed that 8 of these 10 residues are either completely or partially conserved. Before determining the 1-PBI structure, it was not entirely clear how CYP2B4 accomplished the dramatic transition from the tightly packed 1-CPI and 4-CPI structures to the wide-open bifonazole conformation. Monitoring the position of these residues in the CYP2B4 inhibitor complexes reveals significant variation as the F-G cassette pivots across the I helix, which allows the enzyme to maintain a well packed interior despite large structural changes. Hence, the intermediate 1-PBI-bound conformation appears to provide a low-energy pathway between the open and closed forms of CYP2B4.
Closed Ligand-Free Conformation.
Advances in the crystallization of microsomal P450s in our laboratory led to the ability to crystallize monomeric CYP2B4 in the absence of ligand (Table 1). This new ligand-free structure (Fig. 1C) (Wilderman et al., 2010) differed significantly from the previously reported open ligand-free structure (Fig. 1B) (Scott et al., 2003), showing that CYP2B4 is able to adopt a closed conformation in the absence of ligand that closely resembles the 4-CPI and 1-CPI complexes. Whereas the backbone maintains an RMSD of <0.8 Å compared with the other CYP2B4 closed structures, the active site cavity is intermediate to the smaller volume of the 4-CPI active site and the larger volume of the 1-CPI active site (Gay et al., 2010a; Wilderman et al., 2010). This change in volume is mostly due to the movement of Glu301 out of the active site (Fig. 2B), which also occurs in the 1-CPI complex. However, Phe206 and Phe297 retain the orientations observed in the 4-CPI structure (Fig. 2A).
Isothermal Titration Calorimetry.
The four X-ray structures of CYP2B4-imidazole complexes reveal a highly plastic enzyme that is able to reorganize flexible regions in response to the size of the inhibitor bound to the heme. To compare these structural transitions with the thermodynamics of ligand binding, our laboratory pioneered the use of ITC in studying P450-ligand interactions and coupled these results with spectroscopic studies. ITC is able to provide a complete report of the thermodynamics of protein-ligand interactions, yielding data on binding stoichiometry, affinity, change in enthalpy (ΔH), and change in entropy (ΔS). In these studies, six imidazole inhibitors of varying size and chemistry were used: 4-CPI, 1-CPI, 1-benzylimidazole, 4-phenylimidazole 1-phenylimidazole, and 1-(2-(benzyloxy)ethyl)imidazole (Muralidhara et al., 2006). Titrations of CYP2B4 with these imidazoles yielded 1:1 binding stoichiometry, with KD values ranging from 0.3 to 4.8 μM and following the same order as IC50 values. Binding of all the compounds was enthalpically driven, as indicated by negative ΔH values. The largest difference in TΔS was observed between 4-CPI (1.75 kcal/mol) and 1-(2-(benzyloxy)ethyl)imidazole (−1.21.kcal/mol) with a 2-fold difference in heat capacity changes. The larger favorable ΔH of binding for bulkier imidazoles presumably reflects strong hydrophobic interaction, but it was often compensated for by negative TΔS values. Overall, the results demonstrate the potential of ITC for delineating the thermodynamic details of ligand binding to P450s. This solution approach should provide a powerful tool for analyzing P450 binding properties of potential drug candidates and minimizing drug-drug interactions.
Deuterium Exchange Mass Spectrometry.
Deuterium exchange mass spectrometry (DXMS) is a solution method used to observe protein structural dynamics, protein-protein interactions, and protein-ligand interactions (Eyles and Kaltashov, 2004; Ferguson et al., 2009). Amide protons that are more exposed to solvent are exchanged for deuterons in deuterated buffer more readily than those protons that are protected. DXMS was used to correlate the wide range of CYP2B4 conformations observed in the 4-CPI and 1-PBI complexes (Scott et al., 2004; Gay et al., 2009) and ligand-free structures (Scott et al., 2003; Wilderman et al., 2010), to solution behavior of the enzyme. These experiments indicate that, in the absence of ligand, CYP2B4 adopts a predominantly open conformation in solution (Wilderman et al., 2010). Furthermore, upon ligand binding, amide protons of plastic regions are provided varying degrees of solvent protection depending on the identity of that ligand. Peptides covering large portions of PR2 and PR4 showed significant slowing of the H-D exchange rate in the presence of 4-CPI compared with ligand-free CYP2B4, consistent with closed and open structures, respectively. In contrast, CYP2B4 with and without 1-PBI shows little difference in the H-D exchange rate across the entire protein. This finding indicates that binding of 1-PBI does not increase protection from amide proton exchange for the plastic regions of the protein in the B′/C loop and the F-G cassette, but it does not contradict the differences in conformation between the crystal structures of open ligand-free and 1-PBI-bound CYP2B4. In addition, because DXMS reports the average conformation in solution, these results also suggest that small populations of ligand-free CYP2B4 could adopt the compact conformation observed in the closed crystal structure (Wilderman et al., 2010).
Molecular Dynamics.
To explore the energetically accessible conformations of CYP2B4 in the absence of ligand, a molecular dynamics (MD) simulation was performed. The results showed that over time, the closed ligand-free structure shifts to adopt a structure very different from the starting model. A comparison of the final MD model to the closed structure indicated that they differed by an RMSD value similar to comparisons of the closed and 1-PBI-bound structures. These changes in protein structure correlated to movements of the F-G cassette and B′/C loop during the simulation (Wilderman et al., 2010). In solution, flexibility of these regions would allow CYP2B4 to bind large compounds, such as 1-PBI and bifonazole, while also easily accommodating smaller compounds such as 4-CPI.
CYP2B6.
Despite accounting for only 2 to 10% of hepatic P450 content, CYP2B6 takes part in the metabolism of 3 to 12% of all drugs (Wang and Tompkins, 2008), including artemisinin, diazepam, nicotine, and bupropion (Zanger et al., 2007). With 28 known alleles, this highly polymorphic enzyme varies in expression levels and drug metabolism across populations (Zanger et al., 2007). These differences can lead to serious consequences during treatment with drugs such as efavirenz, which has a narrow therapeutic index and is primarily cleared by CYP2B6 (Ward et al., 2003). CYP2B6 activation of the prodrug cyclophosphamide may also be compromised because of altered enzyme activity (Xie et al., 2003; de Jonge et al., 2005). Based on these reports and others, we set out to crystallize a genetic variant of CYP2B6 bound to 4-CPI, which yielded the highest resolution ligand complex of CYP2B4. The crystallized protein (Table 1) contains the naturally occurring K262R mutation as well as an engineered Y226H mutation, which improves thermal stability.
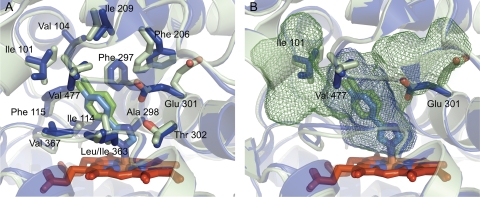
The closed, compact structure of the CYP2B6-4-CPI complex (Fig. 1C) is very similar to that of CYP2B4 (Gay et al., 2010c). An overlay of the two structures yields an RMSD of 0.65 Å, which is lower than an overlay of the 1-CPI and 4-CPI complexes of CYP2B4 (0.76 Å). This indicates that ligand identity may have a greater impact on CYP2B protein conformations than does the primary sequence. The structural similarities between CYP2B4-4-CPI and CYP2B6-4-CPI are also evident in the active site (Fig. 3A), where the only difference in side chain identity is residue 363, which is Leu in CYP2B6 and Ile in CYP2B4. With regard to 4-CPI binding, the identity of residue 363 does not affect ligand orientation. However, studies have shown that mutating this residue in CYP2B6 affects oxidation of 7-butoxycoumarin, p-nitrophenol, and 7-ethoxy-4-trifluorocoumarin (Domanski et al., 1999; Spatzenegger et al., 2003), indicating that variability at this location could be key to explaining species differences in metabolism and product profiles among CYP2B enzymes. Despite their similarities, the 4-CPI complexes of CYP2B6 and CYP2B4 differ significantly in their active site cavity volumes, which are 582 Å3 and 253 Å3, respectively (Gay et al., 2010a). Rotation of Glu301 out of the CYP2B6 active site compared with CYP2B4 allows for the active site cavity to extend upward toward the F helix (Fig. 3B). On the opposite side of the active site, small rotations of Ile101 and Val477 facilitate the inclusion of another small pocket near the A helix and β-sheet 1. In light of the observed global plasticity of CYP2B4, it is interesting to note that the size and shape of CYP2B active sites can also change dramatically due to movement of a small number of side chains. It is also noteworthy that additional, unpublished crystal structures of CYP2B6 (Table 1) in complex with 4-benzylpyridine (BP) and 4-(4-nitrobenzyl)pyridine (NBP) (Fig. 1A) also show rearrangement of active sites residues Phe206 and Phe297 to accommodate the new ligands (Fig. 2A), rather than using larger, global movements of secondary structural elements as observed in some CYP2B4 complexes. These changes are similar to those observed in the 1-CPI and 4-CPI complexes of CYP2B4.
Fig. 3.
Active site comparison of CYP2B4 (blue) and CYP2B6 (light green) closed structures. A, an overlay of the 4-CPI complexes of both enzymes shows a remarkably similar active site. The only difference in amino acid side chains is at position 363, which is Ile in CYP2B4 and Leu in CYP2B6. B, small changes in the active sites of these two enzymes result in significant differences in cavity volumes. Rotations of Ile101, Glu301, and Val477 result in active site volumes of 253 Å3 and 582 Å3 for CYP2B4 and CYP2B6, respectively.
The K262R natural variant of CYP2B6 (allele CYP2B6*4) results from a single nucleotide polymorphism. In the X-ray crystal structure, the substituted Arg262 enters into a hydrogen-bonding network with nearby residues His252, Thr255, Asp263, and Asp266. These residues all occur on helices G and H, part of PR4, which has been observed in multiple positions in CYP2B4 structures. However, despite the movement of this region, this network of hydrogen bonds remains intact. An alignment of 10 CYP2B enzymes (data not shown) indicates that Lys262 in CYP2B6 is the only residue that is not conserved among these hydrogen-bonding partners. Alleles that contain a Lys at position 262, as in wild-type CYP2B6, could have altered positioning of the G helix as it moves above the I helix. These effects could be passed on to active site residues, changing the enzyme's binding pocket. This may explain how polymorphic mutations in P450s occurring far from the active site affect ligand binding and catalysis.
CYP2B4 and Mobile Ligands.
The degree of structural flexibility observed in the series of imidazole-bound structures of CYP2B4 is the largest reported among mammalian P450s. The movement of five plastic regions has shown that CYP2B4 is able to reorganize several secondary structural elements in response to ligand binding. However, it was unknown how these structures related to conformations of enzyme complexes of pharmaceutically relevant drugs. To explore the behavior of CYP2B4 drug complexes, we chose two compounds, clopidogrel and ticlopidine (Fig. 1A), which were expected to have greater freedom of movement in the binding pocket. Both are potent thienopyridine antiplatelet prodrugs, which are also strong MBIs of human CYP2B6 (Richter et al., 2004). Both drugs require activation to their thiol metabolites, which then block ADP binding to its platelet receptor P2Y12 by an irreversible modification of a cysteine residue.
X-Ray Crystallography of Ticlopidine and Clopidogrel Complexes of CYP2B4.
Electron density maps for a CYP2B4-ticlopidine complex were equally valid for fitting the drug in two distinct orientations (Gay et al., 2010b), both of which are consistent with metabolic data gathered with other mammalian P450 enzymes (Tuong et al., 1981; Picard-Fraire, 1984; Ha-Duong et al., 2001; Dalvie and O'Connell, 2004; Ruan and Zhu, 2010). One orientation places the chlorophenyl moiety closer to the heme, while the other places the opposite end of the molecule bearing a thiophene ring closest to the heme. Because of its stereocenter, clopidogrel was easier to fit in the electron density and exhibited a single orientation, which points the chlorophenyl ring toward the heme. The Cα traces of both complexes aligned very well with each other and revealed a compact, closed structure (Fig. 1C) that resembles the conformation observed in the two previously determined CYP2B4 structures with the 4-CPI and 1-CPI (Scott et al., 2004; Zhao et al., 2007) and the closed ligand-free structure (Wilderman et al., 2010). The active site of CYP2B4 is able to accommodate these small ligands by moving only a small number of side chains.
In the clopidogrel complex, Phe206 is part of the active site, where it interacts with the drug, whereas Phe297 is pointed out of the active site (Fig. 2A). These residues switch positions to accommodate the binding of ticlopidine in that structure, which is similar to the repositioning of these side chains in the 1-CPI and 4-CPI complexes. These small differences result in active site cavity volumes of 415.4 Å3 and 373.5 Å3 for the clopidogrel and ticlopidine complexes, respectively. Analysis of the CYP2B4-4-CPI complex suggested that substrate-bound CYP2B4 would adopt an expanded conformation, which is intermediate to the closed 4-CPI structure and open ligand-free structure. However, that was not the case. A comparison of the ticlopidine and clopidogrel complexes to the 4-CPI structure reveals a similar backbone, while highlighting the ability of CYP2B4 to accommodate ligands through rearrangement of active site residues. This suggests that ligand reorientation is energetically favored over protein conformational changes for binding of these similarly sized molecules. Adjusting both protein conformation and ligand orientation in the active site gives CYP2B4 the flexibility to bind to the widest range of molecules, while also being energetically favorable.
NMR Relaxation of CYP2B4-Bound Ticlopidine.
Traditionally, protein NMR has been used to determine the structure of entire polypeptides, proteins, and enzymes. However, alternative uses of NMR have used longitudinal (T1) relaxation measurements coupled with existing crystal structures, to probe ligand interaction with P450s (Regal and Nelson, 2000; Cameron et al., 2005, 2007), where distances from ligand protons to the heme iron are generated. Using MD that takes these distance restraints into consideration, a model of the compound bound in the active site can be produced. This proved to be an excellent technique to investigate the preferred orientation of ticlopidine in the CYP2B4 active site (Gay et al., 2010b), where crystallographic studies yielded an ambiguous result. The results of these NMR experiments showed that T1 paramagnetic relaxation rates for protons on the chlorophenyl ring were 71% higher than those of the thiophene group, indicating that the drug favors an orientation that places the chlorophenyl moiety closer to the heme than the thiophene group. Compared with the tight-binding imidazole inhibitors used in previous crystallographic studies, which are anchored to the heme, ticlopidine is in “fast exchange” in solution and likely adopts multiple conformations within the enzyme active site.
Metabolism of Ticlopidine by Human CYP2B6 and Rabbit CYP2B4.
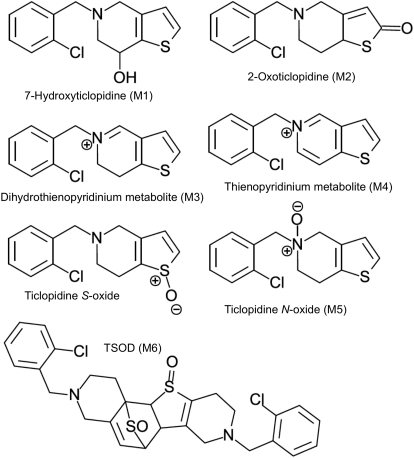
Although complementary NMR T1 relaxation and MD simulations indicated that placing the chlorophenyl group of ticlopidine closer to the heme was the preferred orientation in CYP2B4, several metabolic studies of other P450s, including CYP2B6, have suggested that oxidation would occur preferentially from the other substrate orientations (Ha-Duong et al., 2001; Dalvie and O'Connell, 2004; Richter et al., 2004; Hagihara et al., 2008). Therefore, the oxidation of ticlopidine by reconstituted CYP2B4 was studied and compared with CYP2B6, in which the thiophene portion of the molecule was expected to orient toward the heme. In vitro incubation of both enzymes with ticlopidine resulted in the same set of metabolites: 7-hydroxyticlopidine (M1), 2-oxoticlopidine (M2), 5-(2-chlorobenzyl)thieno[3,2-c]pyridin-5-ium metabolite (M3), 5-(2-chlorobenzyl)thieno[3,2-c]pyridin-5-ium metabolite (M4), ticlopidine N-oxide (M5), and ticlopidine S-oxide dimer (TSOD), a dimerization product of ticlopidine S-oxide (M6) (Fig. 4) (Talakad et al., 2011). The rates of metabolite formation deviated significantly from linearity over time, which is consistent with the known inactivation of CYP2B6 by ticlopidine. Fitting data to a first-order equation resulted in similar rate constants (kobs) for both enzymes. However, the amount of M1 and M6 formation was four to five times greater in CYP2B6 than CYP2B4, indicating a longer residence time of ticlopidine with its thiophene ring closer to heme in CYP2B6. In contrast, CYP2B4 generated larger amounts of M4 and M5 than CYP2B6, indicating an alternate orientation with the nitrogen pointed toward the heme. These results indicate that the orientation of ticlopidine in the active site of CYP2B4 predicted by X-ray crystallography and NMR, which places the chlorophenyl moiety closest to the heme, is unproductive and that the drug possibly reorients within CYP2B4 to a more metabolically productive mode.
Fig. 4.
Ticlopidine oxidation by CYP2B enzymes results in a complex mixture of products. CYP2B4 and CYP2B6 produce, to varying degrees, six major metabolites depending on the site of oxidation. These are M1, M2, a dihydrothienopyridinium metabolite (M3), a thienopyridinium metabolite (M4), M5, and M6, which is formed from the dimerization of ticlopidine S-oxide.
CYP2B4 and Mechanism-Based Inactivators.
Mechanism-based inactivation by several classes of compounds has been used to probe the active site topology or catalytic mechanism of numerous P450s (Hollenberg et al., 2008). In CYP2Bs, acetylene-based compounds have been the most widely studied (Roberts et al., 1993, 1994, 1998; Kent et al., 2002). In particular, tert-butyl acetylenes have proven especially useful as potent CYP2B inactivators, where they form adducts with the heme or active site residue Thr302 (Blobaum et al., 2005; Lin et al., 2009, 2010; Zhang et al., 2009). Despite complete modification of the threonine residue, partial enzymatic activity toward testosterone, 7-ethoxy-4-trifluorocoumarin, and benzphetamine was observed in the tBPA adduct of CYP2B4 (Zhang et al., 2009). To investigate the structural basis for these results, we used X-ray crystallography, DXMS, and molecular ligand docking to examine CYP2B4-tBPA (Gay et al., 2011) (Fig. 1A).
Two crystal structures of CYP2B4-tBPA (Table 1) were determined, where each shows a distinct protein conformation. In one form, the protein-tBPA adduct adopts a closed conformation similar to the CPI, ticlopidine, and clopidogrel complexes of CYP2B4 (Fig. 1C). In the other structure, large movements of the B′/C loop and F-G cassette result in an open conformation similar to Fig. 1D, which dimerizes in the crystal. DXMS and dynamic light scattering experiments indicated that CYP2B4-tBPA exists predominantly as the closed form in solution. However, this form is not particularly useful in explaining the partial enzymatic activity because the active site is completely filled by the inactivator molecule. To probe the heme accessibility to testosterone and 7-ethoxy-4-trifluorocoumarin, we used ligand docking in the open form of CYP2B4-tBPA in the absence of its dimer partner. These results indicated that the bound tBPA was able to rotate away from the heme, allowing substrates to bind in metabolically productive poses.
Conclusions
Accurate prediction of the metabolism of drug candidates is one of the long-term goals in the studies of P450s. This is challenging due to the conformational diversity and plasticity of drug-metabolizing P450s. X-ray crystal structures of CYP2B4 and CYP2B6 complemented by solution biophysical methods such as NMR, DXMS, and ITC have shown that CYP2B enzymes are able to accommodate their wide range of substrates and ligands in two ways, global and local plasticity. Similar to other family 2 enzymes, several ligand-bound structures of CYP2Bs have revealed a dynamic, closed active site that is able to reposition amino acid side chains, most notably Phe206, Phe297, and Glu301, to bind a large variety of chemically diverse compounds. However, more striking structural rearrangements of CYP2B4 have been observed in the presence of large imidazoles. These complexes result in markedly different conformations, which rely on the reorganization of highly mobile plastic regions, mainly the B′/C loop and F-G cassette region. The large structural differences that have been captured in CYP2B4 crystals are also corroborated by DXMS results that show the enzyme is able to adopt several distinct conformations in solution that are dependent on the presence or absence of ligand and the identity of the ligand. Furthermore, ITC and NMR applications have given insight into the variability in binding of CYP2B4 ligands. Overall, the combination of these techniques has revealed a highly dynamic, highly adaptable subfamily of P450s. Applying these results to studies of P450-ligand interactions should aid in the design of safer, more efficient drugs.
Acknowledgments
I especially thank Dr. Sean C. Gay for expert help in preparing this manuscript and all the figures. The studies described represent the work of many talented postdoctoral fellows, visiting scientists, graduate students, and research assistants including Celia Balfour, Paul Ciaccio, Tammy Domanski, Dave Duignan, Sean Gay, Scott Grimm, You-ai He, You Qun He, Karen Kedzie, Santosh Kumar, Keiko Maekawa, B. K. Muralidhara, Art Roberts, Emily Scott, Manish Shah, Margit Spatzenegger, Jeff Stevens, Ling Sun, Grazyna Szklarz, Jyothi Talakad, Qinmi Wang, Ross Wilderman, and Yonghong Zhao. I also thank my many collaborators including Werner Braun, Chris Chin, Deepak Dalvie, Paul Hollenberg, Eric Johnson, Laurence Kaminsky, Edward Morgan, Surendra Negi, Mike Wester, Mark White, Virgil Woods, and Qinghai Zhang. Special thanks go to Dave Stout who has been the senior crystallographer on all of our structures and who has done a fantastic job of teaching and mentoring a number of postdoctoral fellows.
Biography
 James Halpert earned a B.A. in Scandinavian Languages from the University of California at Los Angeles in 1971, a PhD. in Biochemistry from Uppsala University, Sweden in 1977, and a M.S. in Toxicology from the Karolinska Institute, Stockholm, Sweden in 1978. After 15 years on the faculty at the University of Arizona, he assumed the position of Professor and Chair of the Department of Pharmacology and Toxicology at the University of Texas Medical Branch in 1998. There he also served from 2003 to 2008 as Director of the National Institute on Environmental Health Sciences Center and Interim Director of the Sealy Center for Environmental Health and Medicine. He also held the Mary Gibbs Jones Distinguished Chair in Environmental Toxicology. In March 2008, Dr. Halpert joined University of California, San Diego's Skaggs School of Pharmacy and Pharmaceutical Sciences as Professor and Associate Dean for Scientific Affairs. He was a member of the National Institutes of Health Pharmacology Study Section from 1992 to 1995 and served as Chairman from 1993 to 1995. He was Editor for Drug Metabolism and Disposition from 2000 to 2005 and is currently President of the American Society for Pharmacology and Experimental Therapeutics. Dr. Halpert's research for the past 32 years has focused on the structure and function of cytochromes P450, especially the 2B and 3A subfamilies. The work has used a wide variety of biochemical, biophysical, and structural approaches and has focused on the mechanistic and structural basis of P450 inhibition and allostery.
James Halpert earned a B.A. in Scandinavian Languages from the University of California at Los Angeles in 1971, a PhD. in Biochemistry from Uppsala University, Sweden in 1977, and a M.S. in Toxicology from the Karolinska Institute, Stockholm, Sweden in 1978. After 15 years on the faculty at the University of Arizona, he assumed the position of Professor and Chair of the Department of Pharmacology and Toxicology at the University of Texas Medical Branch in 1998. There he also served from 2003 to 2008 as Director of the National Institute on Environmental Health Sciences Center and Interim Director of the Sealy Center for Environmental Health and Medicine. He also held the Mary Gibbs Jones Distinguished Chair in Environmental Toxicology. In March 2008, Dr. Halpert joined University of California, San Diego's Skaggs School of Pharmacy and Pharmaceutical Sciences as Professor and Associate Dean for Scientific Affairs. He was a member of the National Institutes of Health Pharmacology Study Section from 1992 to 1995 and served as Chairman from 1993 to 1995. He was Editor for Drug Metabolism and Disposition from 2000 to 2005 and is currently President of the American Society for Pharmacology and Experimental Therapeutics. Dr. Halpert's research for the past 32 years has focused on the structure and function of cytochromes P450, especially the 2B and 3A subfamilies. The work has used a wide variety of biochemical, biophysical, and structural approaches and has focused on the mechanistic and structural basis of P450 inhibition and allostery.
This work was supported in part by the National Institutes of Health National Institute of Environmental Health Sciences [ES003619].
Article, publication date, and citation information can be found at http://dmd.aspetjournals.org.
doi:10.1124/dmd.111.039719.
1 I would like to express my sorrow at the recent passing of Bob Neal after a long, courageous battle with cancer. Bob was truly an outstanding scientist, leader, mentor, and gentleman.
- P450
- cytochrome P450
- MBI
- mechanism-based inactivator
- WM
- Wistar-Munich
- BBT
- N-benzyl-1-aminobenzotriazole
- SRS
- substrate recognition site
- 4-CPI
- 4-(4-chlorophenyl)imidazole
- PR
- plastic region
- 1-CPI
- 1-(4-chlorophenyl)imidazole
- 1-PBI
- 1-biphenyl-4-methyl-1H-imidazole
- RMSD
- root mean square deviation
- ITC
- isothermal titration calorimetry
- DXMS
- deuterium exchange mass spectrometry
- MD
- molecular dynamics
- BP
- 4-benzylpyridine
- NBP
- 4-(4-nitrobenzyl)pyridine
- tBPA
- tert-butylphenylacetylene
- M1
- 7-hydroxyticlopidine
- M2
- 2-oxoticlopidine
- M3
- 5-(2-chlorobenzyl)thieno[3,2-c]pyridin-5-ium metabolite
- M4
- 5-(2-chlorobenzyl)thieno[3,2-c]pyridin-5-ium metabolite
- M5
- ticlopidine N-oxide
- TSOD
- ticlopidine S-oxide dimer
- M6
- dimerization product of ticlopidine S-oxide.
Authorship Contributions
Participated in research design: Halpert.
Conducted experiments: Halpert.
Contributed new reagents or analytic tools: Halpert.
Performed data analysis: Halpert.
Wrote or contributed to the writing of the manuscript: Halpert.
References
- Blobaum AL, Harris DL, Hollenberg PF. (2005) P450 active site architecture and reversibility: inactivation of cytochromes P450 2B4 and 2B4 T302A by tert-butyl acetylenes. Biochemistry 44:3831–3844 [DOI] [PubMed] [Google Scholar]
- Cameron MD, Wen B, Allen KE, Roberts AG, Schuman JT, Campbell AP, Kunze KL, Nelson SD. (2005) Cooperative binding of midazolam with testosterone and alpha-naphthoflavone within the CYP3A4 active site: a NMR T1 paramagnetic relaxation study. Biochemistry 44:14143–14151 [DOI] [PubMed] [Google Scholar]
- Cameron MD, Wen B, Roberts AG, Atkins WM, Campbell AP, Nelson SD. (2007) Cooperative binding of acetaminophen and caffeine within the P450 3A4 active site. Chem Res Toxicol 20:1434–1441 [DOI] [PubMed] [Google Scholar]
- Ciaccio PJ, Duignan DB, Halpert JR. (1987) Selective inactivation by chloramphenicol of the major phenobarbital-inducible isozyme of dog liver cytochrome P-450. Drug Metab Dispos 15:852–856 [PubMed] [Google Scholar]
- Cosme J, Johnson EF. (2000) Engineering microsomal cytochrome P450 2C5 to be a soluble, monomeric enzyme. Mutations that alter aggregation, phospholipid dependence of catalysis, and membrane binding. J Biol Chem 275:2545–2553 [DOI] [PubMed] [Google Scholar]
- Dalvie DK, O'Connell TN. (2004) Characterization of novel dihydrothienopyridinium and thienopyridinium metabolites of ticlopidine in vitro: role of peroxidases, cytochromes P450, and monoamine oxidases. Drug Metab Dispos 32:49–57 [DOI] [PubMed] [Google Scholar]
- de Jonge ME, Huitema AD, Rodenhuis S, Beijnen JH. (2005) Clinical pharmacokinetics of cyclophosphamide. Clin Pharmacokinet 44:1135–1164 [DOI] [PubMed] [Google Scholar]
- Domanski TL, Halpert JR. (2001) Analysis of mammalian cytochrome P450 structure and function by site-directed mutagenesis. Curr Drug Metab 2:117–137 [DOI] [PubMed] [Google Scholar]
- Domanski TL, He YQ, Scott EE, Wang Q, Halpert JR. (2001) The role of cytochrome 2B1 substrate recognition site residues 115, 294, 297, 298, and 362 in the oxidation of steroids and 7-alkoxycoumarins. Arch Biochem Biophys 394:21–28 [DOI] [PubMed] [Google Scholar]
- Domanski TL, Schultz KM, Roussel F, Stevens JC, Halpert JR. (1999) Structure-function analysis of human cytochrome P-450 2B6 using a novel substrate, site-directed mutagenesis, and molecular modeling. J Pharmacol Exp Ther 290:1141–1147 [PubMed] [Google Scholar]
- Duignan DB, Sipes IG, Leonard TB, Halpert JR. (1987) Purification and characterization of the dog hepatic cytochrome P450 isozyme responsible for the metabolism of 2,2′,4,4′,5,5′-hexachlorobiphenyl. Arch Biochem Biophys 255:290–303 [DOI] [PubMed] [Google Scholar]
- Eyles SJ, Kaltashov IA. (2004) Methods to study protein dynamics and folding by mass spectrometry. Methods 34:88–99 [DOI] [PubMed] [Google Scholar]
- Ferguson PL, Kuprowski MC, Boys BL, Wilson DJ, Pan JX, Konermann L. (2009) Protein folding and protein-ligand interactions monitored by electrospray mass spectrometry. Curr Anal Chem 5:186–204 [Google Scholar]
- Gay SC, Roberts AG, Halpert JR. (2010a) Structural features of cytochromes P450 and ligands that affect drug metabolism as revealed by x-ray crystallography and NMR. Future Med Chem 2:1451–1468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gay SC, Roberts AG, Maekawa K, Talakad JC, Hong WX, Zhang Q, Stout CD, Halpert JR. (2010b) Structures of cytochrome P450 2B4 complexed with the antiplatelet drugs ticlopidine and clopidogrel. Biochemistry 49:8709–8720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gay SC, Shah MB, Talakad JC, Maekawa K, Roberts AG, Wilderman PR, Sun L, Yang JY, Huelga SC, Hong WX, et al. (2010c) Crystal structure of a cytochrome P450 2B6 genetic variant in complex with the inhibitor 4-(4-chlorophenyl)imidazole at 2.0-Å resolution. Mol Pharmacol 77:529–538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gay SC, Sun L, Maekawa K, Halpert JR, Stout CD. (2009) Crystal structures of cytochrome P450 2B4 in complex with the inhibitor 1-biphenyl-4-methyl-1H-imidazole: ligand induced structural response through α-helical repositioning. Biochemistry 48:4762–4771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gay SC, Zhang H, Wilderman PR, Roberts AG, Liu T, Li S, Lin HL, Zhang Q, Woods VL, Stout CD, et al. (2011) Structural analysis of mammalian cytochrome P450 2B4 covalently bound to the mechanism-based inactivator tert-butylphenylacetylene: insight into partial enzymatic activity. Biochemistry doi:10.1021/bi100914z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gotoh O. (1992) Substrate recognition sites in cytochrome P450 family 2 (CYP2) proteins inferred from comparative analyses of amino acid and coding nucleotide sequences. J Biol Chem 267:83–90 [PubMed] [Google Scholar]
- Ha-Duong NT, Dijols S, Macherey AC, Goldstein JA, Dansette PM, Mansuy D. (2001) Ticlopidine as a selective mechanism-based inhibitor of human cytochrome P450 2C19. Biochemistry 40:12112–12122 [DOI] [PubMed] [Google Scholar]
- Hagihara K, Nishiya Y, Kurihara A, Kazui M, Farid NA, Ikeda T. (2008) Comparison of human cytochrome P450 inhibition by the thienopyridines prasugrel, clopidogrel, and ticlopidine. Drug Metab Pharmacokinet 23:412–420 [DOI] [PubMed] [Google Scholar]
- Halpert J. (1981) Covalent modification of lysine during the suicide inactivation of rat liver cytochrome P-450 by chloramphenicol. Biochem Pharmacol 30:875–881 [DOI] [PubMed] [Google Scholar]
- Halpert J, Glaumann H, Ingelman-Sundberg M. (1979) Isolation and incorporation of rabbit liver epoxide hydrase into phospholipid vesicles. J Biol Chem 254:7434–7441 [PubMed] [Google Scholar]
- Halpert J, Hammond D, Neal RA. (1980) Inactivation of purified rat liver cytochrome P-450 during the metabolism of parathion (diethyl p-nitrophenyl phosphorothionate). J Biol Chem 255:1080–1089 [PubMed] [Google Scholar]
- Halpert J, Jaw JY, Balfour C, Kaminsky LS. (1990) Selective inactivation by chlorofluoroacetamides of the major phenobarbital-inducible form(s) of rat liver cytochrome P-450. Drug Metab Dispos 18:168–174 [PubMed] [Google Scholar]
- Halpert J, Näslund B, Betnér I. (1983) Suicide inactivation of rat liver cytochrome P-450 by chloramphenicol in vivo and in vitro. Mol Pharmacol 23:445–452 [PubMed] [Google Scholar]
- Halpert J, Neal RA. (1980) Inactivation of purified rat liver cytochrome P-450 by chloramphenicol. Mol Pharmacol 17:427–431 [PubMed] [Google Scholar]
- Halpert J, Balfour C, Miller NE, Morgan ET, Dunbar D, Kaminsky LS. (1985a) Isozyme selectivity of the inhibition of rat liver cytochromes P-450 by chloramphenicol in vivo. Mol Pharmacol 28:290–296 [PubMed] [Google Scholar]
- Halpert JR, Miller NE, Gorsky LD. (1985b) On the mechanism of the inactivation of the major phenobarbital-inducible isozyme of rat liver cytochrome P-450 by chloramphenicol. J Biol Chem 260:8397–8403 [PubMed] [Google Scholar]
- He Y, Luo Z, Klekotka PA, Burnett VL, Halpert JR. (1994) Structural determinants of cytochrome P450 2B1 specificity: evidence for five substrate recognition sites. Biochemistry 33:4419–4424 [DOI] [PubMed] [Google Scholar]
- He YA, Balfour CA, Kedzie KM, Halpert JR. (1992) Role of residue 478 as a determinant of the substrate specificity of cytochrome P450 2B1. Biochemistry 31:9220–9226 [DOI] [PubMed] [Google Scholar]
- He YQ, Szklarz GD, Halpert JR. (1996) Interconversion of the androstenedione hydroxylase specificities of cytochromes P450 2B4 and 2B5 upon simultaneous site-directed mutagenesis of four key substrate recognition residues. Arch Biochem Biophys 335:152–160 [DOI] [PubMed] [Google Scholar]
- Hollenberg PF, Kent UM, Bumpus NN. (2008) Mechanism-based inactivation of human cytochromes P450s: experimental characterization, reactive intermediates, and clinical implications. Chem Res Toxicol 21:189–205 [DOI] [PubMed] [Google Scholar]
- Honma W, Li W, Liu H, Scott EE, Halpert JR. (2005) Functional role of residues in the helix B' region of cytochrome P450 2B1. Arch Biochem Biophys 435:157–165 [DOI] [PubMed] [Google Scholar]
- Johnson EF, Stout CD. (2005) Structural diversity of human xenobiotic-metabolizing cytochrome P450 monooxygenases. Biochem Biophys Res Commun 338:331–336 [DOI] [PubMed] [Google Scholar]
- Kamataki T, Neal RA. (1976) Metabolism of diethyl p-nitrophenyl phosphorothionate (parathion) by a reconstituted mixed-function oxidase enzyme system: studies of the covalent binding of the sulfur atom. Mol Pharmacol 12:933–944 [PubMed] [Google Scholar]
- Kedzie KM, Balfour CA, Escobar GY, Grimm SW, He YA, Pepperl DJ, Regan JW, Stevens JC, Halpert JR. (1991) Molecular basis for a functionally unique cytochrome P450IIB1 variant. J Biol Chem 266:22515–22521 [PubMed] [Google Scholar]
- Kent UM, Hanna IH, Szklarz GD, Vaz AD, Halpert JR, Bend JR, Hollenberg PF. (1997) Significance of glycine 478 in the metabolism of N-benzyl-1-aminobenzotriazole to reactive intermediates by cytochrome P450 2B1. Biochemistry 36:11707–11716 [DOI] [PubMed] [Google Scholar]
- Kent UM, Mills DE, Rajnarayanan RV, Alworth WL, Hollenberg PF. (2002) Effect of 17-alpha-ethynylestradiol on activities of cytochrome P450 2B (P450 2B) enzymes: characterization of inactivation of P450s 2B1 and 2B6 and identification of metabolites. J Pharmacol Exp Ther 300:549–558 [DOI] [PubMed] [Google Scholar]
- Lin HL, Zhang H, Jushchyshyn M, Hollenberg PF. (2010) Covalent modification of Thr302 in cytochrome P450 2B1 by the mechanism-based inactivator 4-tert-butylphenylacetylene. J Pharmacol Exp Ther 333:663–669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin HL, Zhang H, Noon KR, Hollenberg PF. (2009) Mechanism-based inactivation of CYP2B1 and its F-helix mutant by two tert-butyl acetylenic compounds: covalent modification of prosthetic heme versus apoprotein. J Pharmacol Exp Ther 331:392–403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo Z, He YA, Halpert JR. (1994) Role of residues 363 and 206 in conversion of cytochrome P450 2B1 from a steroid 16-hydroxylase to a 15α-hydroxylase. Arch Biochem Biophys 309:52–57 [DOI] [PubMed] [Google Scholar]
- Muralidhara BK, Negi S, Chin CC, Braun W, Halpert JR. (2006) Conformational flexibility of mammalian cytochrome P450 2B4 in binding imidazole inhibitors with different ring chemistry and side chains. Solution thermodynamics and molecular modeling. J Biol Chem 281:8051–8061 [DOI] [PubMed] [Google Scholar]
- Ortiz de Montellano PR. (1995) The 1994 Bernard B. Brodie Award Lecture. Structure, mechanism, and inhibition of cytochrome P450. Drug Metab Dispos 23:1181–1187 [PubMed] [Google Scholar]
- Picard-Fraire C. (1984) Pharmacokinetic and metabolic characteristics of ticlopidine in relation to its inhibitory properties on platelet function. Agents Actions Suppl 15:68–75 [PubMed] [Google Scholar]
- Poulos TL. (2005) Structural and functional diversity in heme monooxygenases. Drug Metab Dispos 33:10–18 [DOI] [PubMed] [Google Scholar]
- Regal KA, Nelson SD. (2000) Orientation of caffeine within the active site of human cytochrome P450 1A2 based on NMR longitudinal (T1) relaxation measurements. Arch Biochem Biophys 384:47–58 [DOI] [PubMed] [Google Scholar]
- Richter T, Mürdter TE, Heinkele G, Pleiss J, Tatzel S, Schwab M, Eichelbaum M, Zanger UM. (2004) Potent mechanism-based inhibition of human CYP2B6 by clopidogrel and ticlopidine. J Pharmacol Exp Ther 308:189–197 [DOI] [PubMed] [Google Scholar]
- Roberts ES, Alworth WL, Hollenberg PF. (1998) Mechanism-based inactivation of cytochromes P450 2E1 and 2B1 by 5-phenyl-1-pentyne. Arch Biochem Biophys 354:295–302 [DOI] [PubMed] [Google Scholar]
- Roberts ES, Hopkins NE, Alworth WL, Hollenberg PF. (1993) Mechanism-based inactivation of cytochrome P450 2B1 by 2-ethynylnaphthalene: identification of an active-site peptide. Chem Res Toxicol 6:470–479 [DOI] [PubMed] [Google Scholar]
- Roberts ES, Hopkins NE, Zaluzec EJ, Gage DA, Alworth WL, Hollenberg PF. (1994) Identification of active-site peptides from 3H-labeled 2-ethynylnaphthalene-inactivated P450 2B1 and 2B4 using amino acid sequencing and mass spectrometry. Biochemistry 33:3766–3771 [DOI] [PubMed] [Google Scholar]
- Ruan Q, Zhu M. (2010) Investigation of bioactivation of ticlopidine using linear ion trap/orbitrap mass spectrometry and an improved mass defect filtering technique. Chem Res Toxicol 23:909–917 [DOI] [PubMed] [Google Scholar]
- Scott EE, He YA, Wester MR, White MA, Chin CC, Halpert JR, Johnson EF, Stout CD. (2003) An open conformation of mammalian cytochrome P450 2B4 at 1.6-Å resolution. Proc Natl Acad Sci USA 100:13196–13201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott EE, Spatzenegger M, Halpert JR. (2001) A truncation of 2B subfamily cytochromes P450 yields increased expression levels, increased solubility, and decreased aggregation while retaining function. Arch Biochem Biophys 395:57–68 [DOI] [PubMed] [Google Scholar]
- Scott EE, White MA, He YA, Johnson EF, Stout CD, Halpert JR. (2004) Structure of mammalian cytochrome P450 2B4 complexed with 4-(4-chlorophenyl)imidazole at 1.9-Å resolution: insight into the range of P450 conformations and the coordination of redox partner binding. J Biol Chem 279:27294–27301 [DOI] [PubMed] [Google Scholar]
- Spatzenegger M, Liu H, Wang Q, Debarber A, Koop DR, Halpert JR. (2003) Analysis of differential substrate selectivities of CYP2B6 and CYP2E1 by site-directed mutagenesis and molecular modeling. J Pharmacol Exp Ther 304:477–487 [DOI] [PubMed] [Google Scholar]
- Spatzenegger M, Wang Q, He YQ, Wester MR, Johnson EF, Halpert JR. (2001) Amino acid residues critical for differential inhibition of CYP2B4, CYP2B5, and CYP2B1 by phenylimidazoles. Mol Pharmacol 59:475–484 [DOI] [PubMed] [Google Scholar]
- Szklarz GD, Halpert JR. (1997) Use of homology modeling in conjunction with site-directed mutagenesis for analysis of structure-function relationships of mammalian cytochromes P450. Life Sci 61:2507–2520 [DOI] [PubMed] [Google Scholar]
- Szklarz GD, He YA, Halpert JR. (1995) Site-directed mutagenesis as a tool for molecular modeling of cytochrome P450 2B1. Biochemistry 34:14312–14322 [DOI] [PubMed] [Google Scholar]
- Szklarz GD, He YQ, Kedzie KM, Halpert JR, Burnett VL. (1996) Elucidation of amino acid residues critical for unique activities of rabbit cytochrome P450 2B5 using hybrid enzymes and reciprocal site-directed mutagenesis with rabbit cytochrome P450 2B4. Arch Biochem Biophys 327:308–318 [DOI] [PubMed] [Google Scholar]
- Talakad JC, Shah MB, Walker GS, Xiang C, Halpert JR, Dalvie D. (2011) Comparison of in vitro metabolism of ticlopidine by human cytochrome P450 2B6 and rabbit cytochrome P450 2B4. Drug Metab Dispos 39:539–550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuong A, Bouyssou A, Paret J, Cuong TG. (1981) Metabolism of ticlopidine in rats: identification and quantitative determination of some its metabolites in plasma, urine and bile. Eur J Drug Metab Pharmacokinet 6:91–98 [DOI] [PubMed] [Google Scholar]
- Wang H, Tompkins LM. (2008) CYP2B6: new insights into a historically overlooked cytochrome P450 isozyme. Curr Drug Metab 9:598–610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q, Halpert JR. (2002) Combined three-dimensional quantitative structure-activity relationship analysis of cytochrome P450 2B6 substrates and protein homology modeling. Drug Metab Dispos 30:86–95 [DOI] [PubMed] [Google Scholar]
- Ward BA, Gorski JC, Jones DR, Hall SD, Flockhart DA, Desta Z. (2003) The cytochrome P450 2B6 (CYP2B6) is the main catalyst of efavirenz primary and secondary metabolism: implication for HIV/AIDS therapy and utility of efavirenz as a substrate marker of CYP2B6 catalytic activity. J Pharmacol Exp Ther 306:287–300 [DOI] [PubMed] [Google Scholar]
- Wester MR, Johnson EF, Marques-Soares C, Dansette PM, Mansuy D, Stout CD. (2003a) Structure of a substrate complex of mammalian cytochrome P450 2C5 at 2.3-Å resolution: evidence for multiple substrate binding modes. Biochemistry 42:6370–6379 [DOI] [PubMed] [Google Scholar]
- Wester MR, Johnson EF, Marques-Soares C, Dijols S, Dansette PM, Mansuy D, Stout CD. (2003b) Structure of mammalian cytochrome P450 2C5 complexed with diclofenac at 2.1-Å resolution: evidence for an induced fit model of substrate binding. Biochemistry 42:9335–9345 [DOI] [PubMed] [Google Scholar]
- Wilderman PR, Shah MB, Liu T, Li S, Hsu S, Roberts AG, Goodlett DR, Zhang Q, Woods VLJ, Stout CD, et al. (2010) Plasticity of cytochrome P450 2B4 as investigated by hydrogen-deuterium exchange mass spectrometry and X-ray crystallography. J Biol Chem 285:38602–38611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams PA, Cosme J, Sridhar V, Johnson EF, McRee DE. (2000) Mammalian microsomal cytochrome P450 monooxygenase: structural adaptations for membrane binding and functional diversity. Mol Cell 5:121–131 [DOI] [PubMed] [Google Scholar]
- Xie HJ, Yasar U, Lundgren S, Griskevicius L, Terelius Y, Hassan M, Rane A. (2003) Role of polymorphic human CYP2B6 in cyclophosphamide bioactivation. Pharmacogenomics J 3:53–61 [DOI] [PubMed] [Google Scholar]
- Zanger UM, Klein K, Saussele T, Blievernicht J, Hofmann MH, Schwab M. (2007) Polymorphic CYP2B6: molecular mechanisms and emerging clinical significance. Pharmacogenetics 8:743–759 [DOI] [PubMed] [Google Scholar]
- Zhang H, Lin HL, Walker VJ, Hamdane D, Hollenberg PF. (2009) tert-Butylphenylacetylene is a potent mechanism-based inactivator of cytochrome P450 2B4: inhibition of cytochrome P450 catalysis by steric hindrance. Mol Pharmacol 76:1011–1018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y, Sun L, Muralidhara BK, Kumar S, White MA, Stout CD, Halpert JR. (2007) Structural and thermodynamic consequences of 1-(4-chlorophenyl)imidazole binding to cytochrome P450 2B4. Biochemistry 46:11559–11567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y, White MA, Muralidhara BK, Sun L, Halpert JR, Stout CD. (2006) Structure of microsomal cytochrome P450 2B4 complexed with the antifungal drug bifonazole: insight into P450 conformational plasticity and membrane interaction. J Biol Chem 281:5973–5981 [DOI] [PubMed] [Google Scholar]