Abstract
DivIVA is a coiled-coil, tropomyosin-like protein of Gram-positive bacteria. Previous work showed that this protein is targeted to division sites and retained at the cell poles after division. In vegetative cells, DivIVA sequesters the MinCD division inhibitor to the cell poles, thereby helping to direct cell division to the correct midcell site. We now show that DivIVA has a second, quite separate role in sporulating cells of Bacillus subtilis. It again acts at the cell pole but in this case interacts with the chromosome segregation machinery to help position the oriC region of the chromosome at the cell pole, in preparation for polar division. We isolated mutations in divIVA that separate the protein's role in sporulation from its vegetative function in cell division. DivIVA therefore appears to be a bifunctional protein with distinct roles in division-site selection and chromosome segregation.
Keywords: Bacillus subtilis, chromosome segregation, divIVA gene, sporulation, minicell
A fundamental problem in cell division is the nature of the mechanism(s) that positions the division machinery at the correct place in the cell. In the rod-shaped, Gram-positive bacterium Bacillus subtilis, two alternative division sites can be used. In actively growing cells, division occurs by a typical binary fission process precisely at the midpoint of the rod, to give identical daughter cells. During sporulation, however (which occurs in starvation conditions), division is highly asymmetrical, occurring close to one of the cell poles (Stragier and Losick 1996; . Some progress has been made in understanding how the division site is positioned at midcell in vegetative cells, through elucidation of the MinCD/DivIVA system (Errington Jacobs and Shapiro 1999; . The MinC protein encodes an inhibitor of assembly of the division apparatus, which acts at the first step in the assembly process, the formation of a circumferential ring of the tubulin-like protein, FtsZ (Errington de Boer et al. 1990; Marston et al. 1998; Hu et al. 1999). MinC binds to, and requires for its activity, the MinD protein (de Boer et al. 1992; Huang et al. 1996; Marston and Errington 1999). MinD, in turn, recruits MinC to the cell poles, where it prevents improper assembly of the division machinery, which would otherwise produce a small anucleate minicell (Teather et al. 1974; de Boer et al. 1989; Marston and Errington 1999). Targeting of the MinCD complex to the cell poles requires DivIVA protein (Marston et al. 1998), which is a tropomyosin-like, coiled-coil protein that is targeted to nascent division sites and retained at the cell poles after division (Edwards and Errington 1997; Edwards et al. 2000). According to this model, DivIVA is a topological marker that determines the spatial activity zone of the MinCD inhibitor. MinC and MinD homologs are found in a wide range of prokaryotes and are particularly well characterized in E. coli. Curiously, in that organism, DivIVA is not present, and a quite different protein, MinE, controls the topological specificity of MinCD (de Boer et al. 1989) by a remarkable mechanism in which MinCD oscillates from pole to pole, with a cycle time of 20 seconds or so (Hu and Lutkenhaus 1999; Raskin and de Boer 1999a,b). No significant movement of DivIVA or MinD is evident in B. subtilis (M. El Karoui, unpubl.). In both E. coli and B. subtilis, deletion of the topological specificity factor (minE or divIVA) causes a cell division defect through unrestricted division inhibition by MinCD (de Boer et al. 1989; Edwards and Errington 1997). This lethal phenotype can be completely suppressed by mutation of minC or minD, suggesting that the only function for minE or divIVA lies in control of the division inhibitor. However, previous work suggested that DivIVA might have an additional function in sporulation (Cha and Stewart 1997). We have now characterized the minCD-independent sporulation phenotype of divIVA mutants in detail and show that this gene has a completely unexpected role in chromosome segregation.
Very little is known about chromosome segregation in bacterial cells. A number of genes have been identified in which mutations can perturb segregation, including mukB (Niki et al. 1991), smc (Britton et al. 1998; Moriya et al. 1998), and spo0J (Ireton et al. 1994). However, in most cases, it is not clear that the gene product is specifically involved in segregation and not in other processes such as chromosome replication, decatenation, dimer resolution, or chromosome condensation (Wake and Errington 1995).
The asymmetry of cell division during sporulation imposes unusual restraints on chromosome segregation, since, to be inherited by the small prespore cell, one chromosome needs to be moved to the extreme pole of the cell. We now know that segregation into the prespore occurs by an unusual two-step mechanism. First, the polar septum closes around the chromosome, trapping only a small part of it in the prespore compartment (Wu et al. 1995). A DNA translocator protein, SpoIIIE, then transports the remainder of the chromosome through a pore in the septum to complete the segregation process (Wu and Errington 1994; Wu and Errington 1997; Bath et al. 2000). Studies of spoIIIE mutant cells, in which DNA transfer into the prespore is blocked, revealed that the chromosome has a specific orientation at the onset of sporulation, with the oriC region always localized close to the cell pole (Wu and Errington 1994; Wu and Errington 1998). Results presented here show that DivIVA protein plays an important role in localizing or retaining the oriC region of the chromosome near the pole of the sporulating cell. The presence of DivIVA-like proteins in other Gram-positive bacteria, some of which do not have a Min system (e.g., Staphylococcus and Streptococcus) (Massidda et al. 1998), suggests that DivIVA may also participate in chromosome segregation in a range of important Gram-positive bacteria.
Results
A Min-independent role for divIVA during sporulation
Preliminary observation of cells from agar plate cultures suggested that minD divIVA cells (strain 1920) sporulated much more poorly than an isogenic minD single mutant (1901) or the wild type (SG38), in accordance with a previous report (Cha and Stewart 1997). To quantify this effect, the three strains were grown in liquid medium and induced to sporulate under more defined conditions (see Materials and Methods). Each culture was scored for formation of phase bright spores (by light microscopy) and for lysozyme-resistant colony forming units (cfu). Only a slight reduction in sporulation efficiency was seen in the minD strain compared with the wild type (86% by phase contrast and 67% by lysozyme), also as reported previously (Levin et al. 1992; Lee and Price 1993; Cha and Stewart 1997; Barák et al. 1998). However, the sporulation efficiency of the minD divIVA strain was consistently 15–20-fold lower than that of the other strains (3.4% and 4.3% of wild type), confirming that the divIVA mutation significantly impairs sporulation (at least in the absence of minD). Insertion of a wild-type copy of divIVA at the amyE locus restored sporulation efficiency in the double mutant background to that of the minD single mutant (data not shown).
min mutations produce an irregular cell length distribution, possibly because division at the cell pole to produce a minicell occurs at the expense of medial division (Teather et al. 1974; Levin et al. 1998). Since this effect could influence the measured sporulation frequency, the length distributions of the minD and minD divIVA mutants growing vegetatively and at different times after induction of sporulation were measured. No significant differences between the two strains were detected (data not shown).
DivIVA is maintained at the cell poles during sporulation but does not target to the asymmetric septum
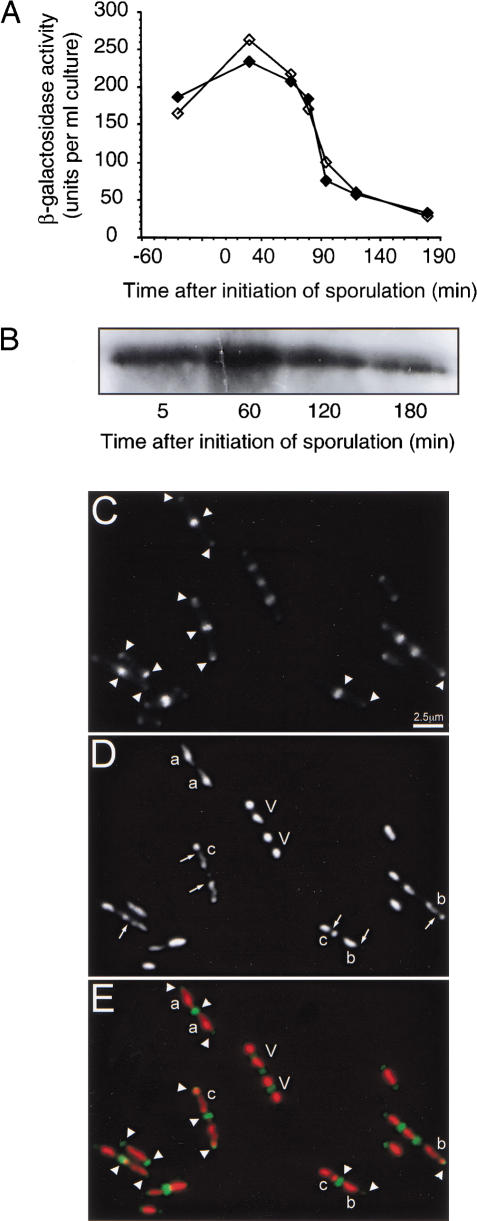
A specific requirement for DivIVA during sporulation might be reflected in an increase in expression of its gene or in the quantity of the protein. Unexpectedly, we observed the reverse, and the levels of β-galactosidase expressed from a divIVA-lacZ transcriptional fusion decreased soon after the onset of sporulation, indicated by the fall in levels of the unstable β-galactosidase enzyme (Fig. 1A). We also analyzed DivIVA protein levels by Western blotting of extracts from sporulating cells of the wild-type strain SG38. As shown in Figure 1B, the concentration of DivIVA fell slowly during sporulation, in accordance with the reduction in transcription.
Figure 1.
Fate of DivIVA protein during sporulation. (A) Expression of divIVA-lacZ in growing cells of B. subtilis 1756. Note that this strain also carries a repressible allele of divIVA Pspac–divIVA) and that it made no difference whether the strain was grown in the presence (filled symbols) or absence (empty symbols) of IPTG. (B) Western blot analysis of DivIVA protein in sporulating cells of the wild-type strain SG38. Protein samples were corrected for cell number, then separated by 12% SDS-PAGE. The gel was blotted and probed with polyclonal anti-DivIVA antiserum, and visualized by enhanced chemiluminescence. (C–E) Localization of DivIVA-GFP in sporulating cells of B. subtilis. Strain 1803 was induced to sporulate, and at t80 a sample of cells was examined by fluorescence microscopy. Panels C–E show fluorescence images of a typical field of cells. C, GFP channel; D, DNA (DAPI); E, overlay of the two images with GFP false colored green and DNA red. Arrowheads show DivIVA-GFP targeted to midcell division sites and mature cell poles of vegetative cells (V) and various types of sporulating cells. Cells at the axial filament stage (a), with a newly-formed asymmetric septum (b) or with fully segregated prespore and mother cell nucleoids (c) are shown. Arrows show the positions of asymmetric septa, which have no significant GFP signal.
To determine the fate of the DivIVA protein remaining in sporulating cells, we examined a strain harboring a divIVA-gfp fusion (1803) by fluorescence microscopy. Although this fusion does not support proper control of Min function, it targets to vegetative cell division sites, as does the wild-type protein (data not shown). In addition, we used DAPI staining to reveal the nucleoid morphology of the cells, as this is a convenient morphological marker for spore development (e.g., Hauser and Errington 1995). As shown in Figure 1 (C-D), strong accumulations of DivIVA were detected only at the extreme poles of sporulating cells (arrowheads in Fig. 1C). This was true for cells that still had a vegetative appearance (V), or that had entered the early stages of sporulation, as judged by DNA morphology (cell types a, b, and c in Fig. 1C–D). In similar experiments we used immunofluorescence microscopy (IFM) and costained cells for both native DivIVA protein and for β-galactosidase expressed from an early sporulation-specific promoter (spoIIA-lacZ; Errington and Mandelstam 1986). This allowed us to confirm that the cells were sporulating efficiently and that they had been permeabilized. Of 100 such cells examined by IFM from samples taken at three different time points after the induction of sporulation, only one cell showed DivIVA targeted near the asymmetric septum (data not shown). It seems that during sporulation, DivIVA remains at the poles to which it was recruited during earlier vegetative divisions, but that it does not target in significant amounts to the new asymmetric division septa. This is the first example of a protein that is recruited specifically to vegetative division septa; all of the other division proteins that have been tested appear to be recruited to both kinds of septa (Levin and Losick 1996; Harry and Wake 1997; Daniel et al. 2000; Sievers and Errington 2000; Feucht et al. 2001).
divIVA mutation impairs expression of ςFand ςE-dependent genes
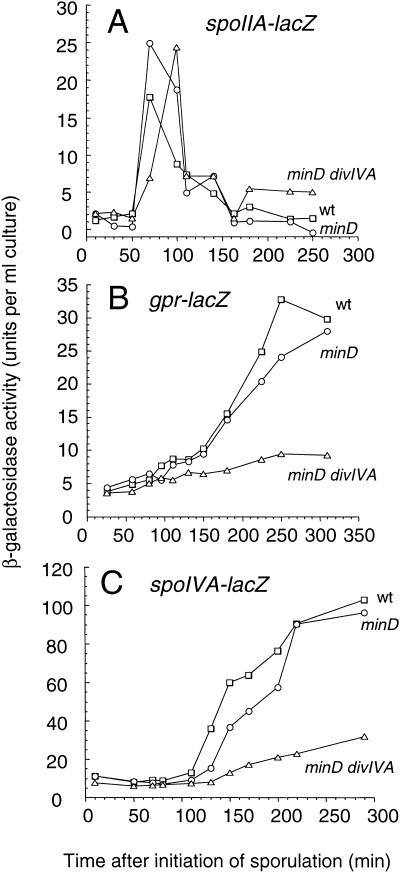
To determine when the sporulation defect was first manifested, we measured the expression of various reporter genes that are normally turned on at different times during sporulation. With a spoIIAA-lacZ fusion (fusion to the early sporulation promoter of the operon encoding ςF; Errington and Mandelstam 1986) no significant difference was seen among the wild-type, minD, and minD divIVA strains (Fig. 2A). Expression began about 1h after resuspension and peaked 30 to 60 min later, as expected. In contrast, with the gpr-lacZ fusion (controlled by active ςF early in prespore development; Sussman and Setlow 1991), a clear reduction of expression was seen in the minD divIVA mutant, compared to the other strains. Thus, in the wild-type and minD strains, β-galactosidase began to be detected after about 90 min and expression continued to climb rapidly until at least 270 min, whereas with the minD divIVA double mutant, only low levels of activity (close to the background levels of a strain with no lacZ fusion) were seen (Fig. 2B). Similar results were obtained with a different ςF-dependent fusion (spoIIQ-lacZ; Londoño-Vallejo et al. 1997; data not shown). As expected of cells deficient in ςF activity, expression of a ςE-dependent fusion, spoIVA-lacZ (Stevens and Errington 1990), was also substantially reduced in the minD divIVA double mutant, compared with minD and wild-type strains (Fig. 2C). Therefore, the divIVA mutation seemed to block sporulation at a point after the synthesis of ςF but before its activation.
Figure 2.
Effect of divIVA disruption on the expression of sporulation-specific genes. Strains bearing lacZ fusions to the sporulation genes spoIIAA (A), gpr (B), or spoIVA (C) were induced to sporulate, and β-galactosidase activities were measured. For each lacZ fusion, strains that were otherwise wild-type (□), or carried mutations in minD (○), or minD and divIVA (▵), were compared. The strains used were as follows: (A) 1806, 1807, and 1808; (B) 1810, 1813, and 1811; (C) 713, 1856, and 1857.
minD divIVA cells complete asymmetric septation
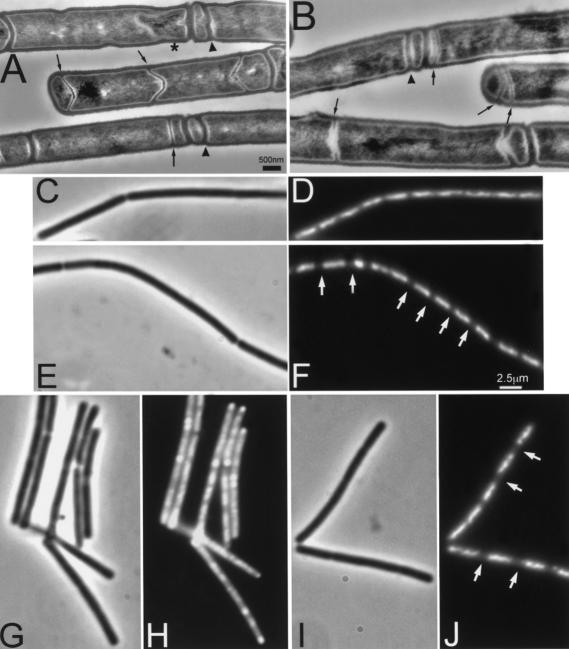
It is now well established that ςF activation occurs immediately after formation of the asymmetric septum and that activation is dependent on septum formation (Beall and Lutkenhaus 1989, 1991, 1992; Levin and Losick 1994; Daniel et al. 1998; Feucht et al. 1999). Since the min/divIVA system was known to be involved in the control of septation in vegetative cells, it seemed likely that the minD divIVA double mutant was affected in asymmetric septation. To investigate this possibility, we used transmission electron microscopy to examine samples of wild-type and appropriate mutant strains. Surprisingly, examination of sections of cells collected 70, 105 and 240 min after induction of sporulation did not reveal an obvious difference between the minD and minD divIVA strains, at least prior to prespore engulfment (Fig. 3A,B; data not shown).
Figure 3.
Effect of minD and minD divIVA mutations on asymmetric septum formation and chromosome segregation during sporulation. Cultures of strains 1901 (minD; A,C,D,G,H) and 1920 (minD divIVA; B,E,F,I,J) were induced to sporulate by the resuspension method. (A,B) Samples were taken after 105 min for sectioning and transmission electron microscopy. Examples of minicell septa and thin (sporulation-type) septa are indicated by arrowheads and arrows, respectively. The star indicates a prespore being engulfed by the mother cell. (C–J) In a separate experiment, samples of sporulating cells were fixed at 90 (C–F) and 180 (G–J) min after initiation of sporulation. Samples were viewed by phase contrast (C,E,G,I) and fluorescence (DAPI) (D,F,H,J) microscopy. Arrowheads point to some condensed prespore nucleoids in the minD mutant cells, and arrows point to the larger gaps between nucleoids in the double mutant cells. Scale bar for A, 500 nm.
min mutant strains have been shown to form thin sporulation-specific septa at midcell in addition to the asymmetric wild-type position, at a low frequency (Barák et al. 1998). These septa and the minicells in the minD and minD divIVA strains (indicated by arrows and arrowheads, respectively, in Fig. 3A,B) complicated the quantitation of their morphological phenotypes. Therefore, the cells of the 105-min samples were divided into groups based on the number, width, and position of their septa. Cells with at least one thin septum (examples arrowed in Fig. 3A,B) were termed “sporulating.” All other cells, including those lacking any septum or having only a thick septum, were termed “vegetative.” Both mutant strains had proportions of vegetative and sporulating cells similar to those of the wild-type strain (Table 1). Furthermore, the minD divIVA strain had numbers of vegetative and sporulating cells in each of the subcategories equivalent to those of the minD strain. Thus, the divIVA mutation did not significantly change the frequency of formation of the sporulation septum (at least in the minD background).
Table 1.
Effect of mutations in minD and divIVA on septum formation during sporulation
| Strain
|
Genotype
|
% Cells with Phenotypea
|
|||||
|---|---|---|---|---|---|---|---|
| vegetative
|
sporulating
|
||||||
| no septum
|
medial septum
|
polar septum
|
asymmetric septum
|
medial septum
|
other
|
||
| SG38 | + | 13 | 21 | ND | 59 | ND | 7 |
| 1901 | minD | 8 | 7 | 13 | 39 | 4 | 29 |
| 1920 | midD divIVA | 14 | 10 | 13 | 35 | 9 | 19 |
A total of 180 cells (medial sections) of each strain were counted in samples taken 110 min after the initiation of sporulation. The category labeled “Other” comprises cells of various configurations containing two or more septa of any thickness. ND, not detected.
In the above measurements we did not distinguish between sporulating cells with a straight septum and those in which development was proceeding to the stage of engulfment. In fact, as expected for a strain deficient in ςF and ςE activities (Illing and Errington 1991), later morphological development was arrested in the minD divIVA mutant and almost no cells with engulfed prespores were seen, unlike the minD mutant (example indicated by a star in Fig. 3A).
We also examined the patterns of septation during sporulation of these strains by IFM using antibodies against the FtsZ division protein. Again, no significant difference was seen between the minD and minD divIVA strains (data not shown).
Disruption of divIVA blocks an early step in prespore chromosome segregation
It was surprising to find that in the minD divIVA strain the gene encoding ςF was being transcribed and the prespore septum formed, yet the expression of ςF-dependent genes was greatly reduced. Close examination of the electron micrographs suggested a possible explanation. It seemed that the prespore-like compartments of the strain with the divIVA mutation (1920) hardly ever showed signs of DNA. A failure of DNA segregation into the prespore could explain the lack of ςF-dependent gene expression. To test this possibility, we used fluorescence microscopy to examine the DNA distributions in the mutant strains. As shown in Figure 3J, even late in sporulation the nucleoids of the minD divIVA cells were arrested at or before the axial filament stage and had not developed further to give the characteristically condensed prespore and diffuse mother cell DNA staining patterns present in the minD (Fig. 3H) and wild-type strains. Close examination of the DNA earlier in sporulation (Fig. 3C-F) suggested that the nucleoids of minD divIVA cells were separated by larger gaps (arrows in Fig. 3F) than those of the minD strain. The appearance of the sporulating cells of the double mutant was reminiscent of that of spoIIIE mutants, although in the latter, the prespore compartments are clearer because they all contain a small amount of DNA (about 30% of the chromosome; Wu and Errington 1994). Interestingly, despite the clear presence of minicell-like compartments in the minD single mutant (Table 1), there was little evidence of corresponding anucleate compartments in the DNA-stained cells (Fig. 3). It therefore appears that DNA can be trapped in minicell compartments during sporulation, unlike the normal situation in vegetative cells (Sharpe and Errington 1995).
To examine the divIVA effect on chromosome segregation in more detail, we introduced a spoIIE-gfp fusion into the minD and minD divIVA strains. The SpoIIE protein is synthesized specifically in sporulating cells and is targeted to the sporulation septum (Arigoni et al. 1995; Wu et al. 1998), providing a cytological marker against which to score the position of the nucleoid. These experiments confirmed that the gaps seen between nucleoids in sporulating cells of the divIVA minD double mutant were frequently adjacent to polar septa in sporulating cells (data not shown; see below). Taken together, these results demonstrate that the primary sporulation defect arising from the divIVA mutation is due to a failure of DNA segregation into the prespore compartment. Irrespective of whether or not ςF is activated in the prespore-like compartment, the reporter gene is not localized in that compartment, so expression does not occur.
Isolation of divIVA mutations that affect sporulation but not division site selection
If DivIVA protein is bifunctional, it should be possible to isolate mutations that separate its role in sporulation from its well characterized role in division site selection. In work to be published elsewhere, we randomly mutagenized a plasmid carrying the divIVA gene (M. Dyson, H.B. Thomaides, and J. Errington, unpubl.), using the E. coli mutator strain XL1-red (Stratagene). The mutagenized plasmid library was transformed into strain 3011 with selection for plasmid integration on the basis of kanamycin resistance. Transformants should arise by a single-crossover mechanism integrating the plasmid into the host chromosome. The host strain has a deletion of divIVA, but the lethality of this is suppressed by deletion of the minD gene. MinD function can be provided by inducing a gfp–minD fusion gene with xylose. Transformants were screened for nonlethality in the presence of xylose (thus retaining the ability to topologically regulate the MinCD division inhibitor) and for reduced sporulation on the basis of colony appearance and β-galactosidase activity (expressed from a ςF-dependent spoIIQ-lacZ reporter gene; Londoño-Vallejo et al. 1997). Of 2225 transformants screened, five mutants were obtained that appeared to have a substantially reduced sporulation efficiency but more or less normal cell length. The mutants were screened by Western blot analysis, and all but one of them turned out to have reduced protein levels. DNA sequencing revealed that one of the mutants had a substitution in the −10 region of the divIVA promoter: TATAAT was converted to TG TAAT (strain 3015). Two other mutations produced substitutions in the C-terminal region of DivIVA: I136T (strain 3018) and L140F (strain 3016). The protein levels of the fourth mutant (strain 3014) were affected specifically during sporulation: its mutation would produce a substitution, V25I, in the N-terminal region of DivIVA. The isolation of at least the first three of these mutants suggested that DivIVA is required at higher levels for the sporulation function than for the vegetative function.
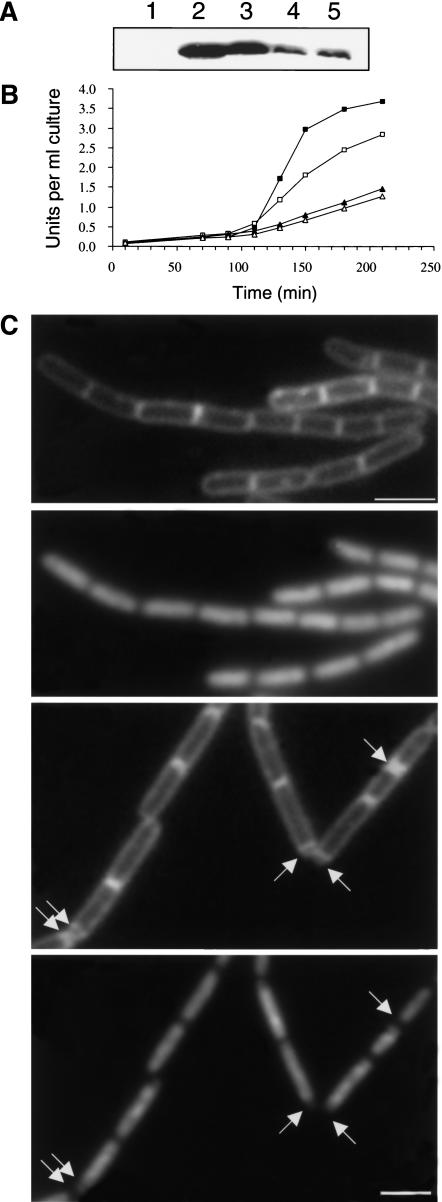
One mutant, strain 3013, produced normal levels of DivIVA in sporulating cells (Fig. 4A, cf. lane 3 with the promoter mutant and L140F mutant in lanes 4 and 5). Its mutation produced a conservative substitution, N99D, in the middle of the protein sequence. This mutant had a severe deficiency in Stage II gene expression during sporulation; a reduction equivalent to that of the divIVA null mutant (Fig. 4B). We then measured the frequency of sporulation, relative to that of the isogenic wild-type strain (3012), by phase contrast microscopy of sporulating cultures at t7. By this measurement, the 3013 mutant was, if anything, affected more than was the null mutant 3011 (2.4% vs. 4.8% of wild-type sporulation frequency).
Figure 4.
Effects of the new divIVA mutations on protein stability, sporulation, and control of MinCD function. (A) Western blot analysis of protein accumulation in sporulating cells of the divIVA null mutant 3011 (lane 1), the wild-type control 3012 (lane 2), 3013 (N99D; lane 3), 3015 (promoter mutation; lane 4), and 3016 (L140F; lane 5), taken at t210. (B) Alkaline phosphatase synthesis during sporulation in strains 3011 (divIVA::cat, ▴), 3012 (divIVA+, ▪), 3013 (N99D, ▵), and 3014 (V25I, □). (C–F) Cell division and chromosome segregation in the 3017 mutant. Strain 3017, bearing the mutant divIVA allele conferring the N99D substitution, was induced to sporulate, and samples taken before resuspension (i.e., in CH growth medium; C,D) and 85 min after initiation of sporulation (E,F) were stained and visualized for the cell membrane (with FM5–95; C,E) and for DNA (with DAPI; D,F). Arrows in panels E and F point to the positions of some prespore compartments visible with the membrane stain, some of which are clearly deficient in DNA.
Preliminary experiments suggested that the 3013 mutant was not significantly affected in control of the Min system on the basis of its vegetative cell-length frequency distribution, but also that this mutant showed the expected defect in prespore chromosome segregation (data not shown). To examine this in more detail and to avoid the complications of having minD disruption and inducible gfp-minD constructions in the cell, we crossed the mutant divIVA allele into an otherwise wild-type background (strain 1271). The overall reduction in sporulation of the new completely min+ strain (3017) was not dissimilar from that of its parent strain, 3013 (4.5% of the lysozyme resistance of a wild-type control strain, SG38). Figure 4C,D shows that vegetative cells of the new strain had a wild-type cell-length distribution, with no significant minicells (<0.4%; 250 cells counted). When induced to sporulate, it was apparent that the block in development was again in progression beyond the septation stage. As shown in Figure 4E, prespore septa were frequently formed, but as expected, the small prespore-like compartments were frequently devoid of DNA (arrows in Fig. 4F). These images show live cells stained for both membrane and DNA, so that the two features can be compared directly. Although the DNA staining in these living cells is rather less clear than in fixed cells (Fig. 3, cf. H and J), results very similar to those of Figure 3 were obtained with fixed cells (not shown).
The isolation of various types of Spo− Min+ mutants, particularly the N99D mutant, lends strong support to the notion that divIVA has two distinct functions; in control of division site selection and in chromosome segregation during sporulation.
Discussion
A second distinct function for DivIVA in chromosome segregation during sporulation
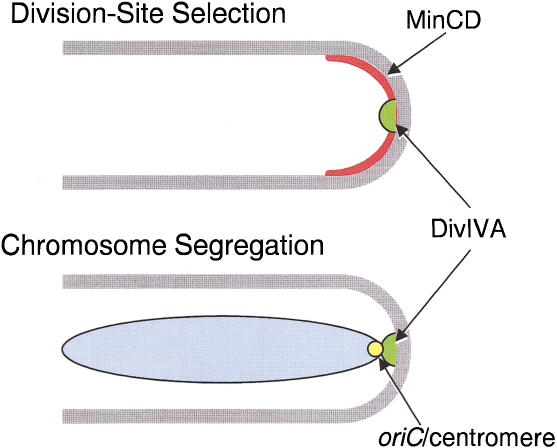
Previous work established a clear role for DivIVA protein in division site selection in B. subtilis (Edwards and Errington 1997; Marston et al. 1998). Cha and Stewart (1997) reported that cells with a point mutation in divIVA (divIVA1) had a deficiency in sporulation over and above that of minD mutants (which have a weak sporulation defect, probably due to their abnormal cell length distribution; Levin et al. 1992; Lee and Price 1993; Cha and Stewart 1997; Barák et al. 1998). We have now characterized the phenotype of a divIVA null mutant in detail and have shown that it has a very specific effect on chromosome segregation into the prespore. The defect seems to be exerted at the point when, normally, the oriC region of the chromosome becomes localized in the vicinity of the cell pole (see below). Figure 5 illustrates how DivIVA protein targeted to the cell poles could interact with MinCD to prevent polar division in vegetative cells, and with a putative centromere-like sequence in the oriC region of the chromosome, facilitate chromosome segregation in sporulating cells. This latter function probably does not come into play in vegetative cells, because depletion of DivIVA does not result in a significant chromosome segregation defect in vegetative growth (H.B. Thomaides, unpubl.). The mechanisms responsible for accurate chromosome segregation in vegetative cells are still not understood, though several candidate genes have been identified, particularly spoIIIE, spo0J, and smc (Ireton et al. 1994; Sharpe and Errington 1995; Britton et al. 1998; Moriya et al. 1998). Whatever the underlying mechanisms may be, they probably operate in such a way that the nucleoids do not normally come into close juxtaposition with the cell poles, and therefore do not require the DivIVA system, unlike sporulating cells.
Figure 5.
Dual role of DivIVA protein in division site selection and chromosome segregation during sporulation.
Implications for the function of DivIVA-like proteins in other Gram-positive bacteria
The clear presence of divIVA-like genes in other Gram-positive bacteria, including cocci, which do not appear to have min genes, could now be explained by its second quite separate role in chromosome segregation. In a preliminary report, Massidda et al. (1998) indicated that disruption of the divIVA homolog of Streptococus pneumoniae gives a phenotype in which septation is reduced and some anucleate cells are formed. In light of our findings, this would be consistent with the primary defect in these mutants being in chromosome segregation. Chromosome segregation in sporulating cells of B. subtilis seems to be a specialized process, differing from that of vegetative cells because one chromosome needs to segregate to the extreme pole of the cell. This raises the interesting possibility that Gram-positive cocci are descended from primitive spore formers that have lost the ability to sporulate.
We showed previously that DivIVA protein can be targeted to the cell division site in a broad range of organisms, including the Gram-negative bacterium E. coli, and even a eukaryote, the fission yeast Schizosaccharomyces pombe (Edwards and Errington 1997; Edwards et al. 2000). It may be that DivIVA and similar proteins act as general markers for the cell pole, possibly used for a diversity of functions in different organisms.
The role of DivIVA in chromosome segregation during sporulation
Previous work on chromosome segregation into the prespore of sporulating cells of B. subtilis suggested that it works in a two-step manner. First, a particular region of the chromosome, centered approximately on oriC, migrates up to the cell pole. Asymmetric cell division then occurs, separating the chromosome into two domains, the larger of which (oriC distal) is located in the large compartment. SpoIIIE protein then acts as a DNA pump that transports the misplaced DNA through the division septum into the small polar prespore compartment (Wu et al. 1995; Bath et al. 2000). The specific orientation of the chromosome in sporulating cells, with the oriC region positioned close to the cell pole, suggested the existence of a centromere-like function in this part of the chromosome (Wu and Errington 1994, 1998). We recently mapped the centromere-like sequence to a region located about 200 kbp away from oriC (L.J. Wu and J. Errington, unpubl.). The active sequences behave as a relatively diffuse region, rather than as a specific site, suggesting that they comprise multiple DNA binding sites that act in a semi-cooperative manner. The results presented here strongly suggest that DivIVA comprises part of the mechanism that is used to anchor the centromere in the vicinity of the cell pole. Since DivIVA protein is recruited to the cell poles (Edwards and Errington 1997) and retained at the poles, even after the onset of sporulation (Fig. 1C-E), it is perfectly positioned to be able to act as a polar receptor or anchor for the oriC region of the chromosome.
Examination of the sequence of DivIVA does not reveal any known motifs suggestive of an ability to bind DNA directly. Its most prominent feature is a large, central coiled-coil domain (Edwards et al. 2000), which is likely to be involved in protein-protein interactions. It therefore seems more likely that DivIVA interacts with one or more intermediate proteins that act as a bridge to the DNA. It should be possible to identify these proteins on the basis of binding to DivIVA protein, or by analysis of suppressors of the divIVA3013 mutant.
It is still not clear whether DivIVA is required to actually bring the centromere region to the pole, or whether it acts secondarily to anchor the chromosome at the pole after some other system, as yet unknown, has delivered it. Detailed phenotypic analysis of the effects of the divIVA3013 mutation, which will avoid the complications arising from the abnormal length distributions of min divIVA double mutants, should help to resolve this issue.
Materials and methods
Strains, plasmids, and bacteriophages
These are mainly listed in Table 2. Plasmid pSG1850 was constructed by cloning a segment of DNA containing about 900 bp upstream of divIVA through to the end of the divIVA coding sequence into plasmid pBEST502. The DNA segment was amplified with primers H32 (5′-ATAGGGGATCCGCAGCAGC CAAGC-3′) and H33 (5′-GATAATCGGATCCTTTATTC CTTTT-3′), each of which introduced a BamHI site, and these sites were used for the cloning. Bacteriophage φ105J19 was described previously (Errington and Mandelstam 1986).
Table 2.
Bacterial strains and plasmids
| Strain or plasmid
|
Relevant genotypea
|
Source, construction or referenceb
|
|---|---|---|
| B. subtilis strains | ||
| SG38 | spo+ | Errington and Mandelstam (1986) |
| 168 | spo+ | C. Anagnostopoulos |
| 700 | Ω(spoIIAB′-′lacZ cat) | Partridge et al. (1991) |
| 713 | spoIVA-lacZ cat | Stevens et al. (1992) |
| 1271 | Ω(yoaVW::spoIIQ-lacZ tet) | L.-J. Wu, unpub. |
| 1305 | spoIIE::pSG1902 (spoIIE-gfp cat) | Wu et al. (1998) |
| 1311 | minD::ermC spoIIE::pSG1902 (spoIIE-gfp cat) | pSG1902 → 1901 (Cm) |
| 1312 | minD::ermC divIVA::tet spoIIE::pSG1902 (spoIIE-gfp cat) | pSG1902 → 1920 (Cm) |
| 1756 | divIVA::pSG1043 (PdivIVA-lacZ ermC Pspac− divIVA) | Edwards and Errington (1997) |
| 1803 | divIVA::pSG1612 (PdivIVA-gfp divIVA+ cat) | pSG1612 → 168 (Cm) |
| 1806 | (φ105J19) spoIIAA-lacZ cat | φ105J19 → SG38 (Cm) |
| 1807 | minD::ermC (φ105J19) spoIIAA-lacZ cat | φ105J19 → 1901 (Cm) |
| 1808 | minD::ermC divIVA::tet (φ105J19) spoIIAA-lacZ cat | φ105J19 → 1920 (Cm) |
| 1809 | Ω(spoIIAB′-′lacZ cat) | 700 → 168 (Cm) |
| 1810 | gpr::pPS1326 (gpr-lacZ cat) | pPS1326 → SG38 (Cm) |
| 1813 | minD::ermC gpr::pPS1326 (gpr-lacZ cat) | pPS1326 → 1901 (Cm) |
| 1811 | minD::ermC divIVA::tet gpr::pPS1326 (gpr-lacZ cat) | pPS1326 → 1920 (Cm) |
| 1855 | spoIVA-lacZ cat | 713 → SG38 (Cm) |
| 1856 | minD::ermC spoIVA-lacZ cat | 713 → 1901 (Cm) |
| 1857 | minD::ermC divIVA::tet spoIVA-lacZ cat | 713 → 1920 (Cm) |
| 1865 | ΔdivIVA::cat minD::ermC Ω(amyE::Pxyl-gfp-minD spc) | M. Dyson and H.B.T., unpubl. |
| 1901 | minD::ermC | Edwards and Errington (1997) |
| 1920 | minD::ermC divIVA::tet | Marston and Errington (1999) |
| 3011 | ΔdivIVA::cat minD::ermC Ω(amyE::Pxyl-gfp-minD spc) Ω(yoaVW::spoIIQ-lacZ tet) | 1271 → 1865 (Tc) |
| 3012 | As 3011, chr::pSG1850 (divIVA+ kan) | pSG1850 → 1920 (Km) |
| 3013 | As 3011, chr::pSG1850 (divIVA13 kan) (N99D) | pSG1850* → 1920 (Km) |
| 3014 | As 3011, chr::pSG1850 (divIVA14 kan) (V25I) | pSG1850* → 1920 (Km) |
| 3015 | As 3011, chr::pSG1850 (divIVA15 kan) (promoter mutation) | pSG1850* → 1920 (Km) |
| 3016 | As 3011, chr::pSG1850 (divIVA16 kan) (L140F) | pSG1850* → 1920 (Km) |
| 3017 | ΔdivIVA::catΩ(yoaVW::spoIIQ-lacZ tet) chr::pSG1850 (divIVA13 kan) (N99D) | 3013 → 1271 (Km and Cm) |
| 3018 | As 3011, chr::pSG1850 (divIVA18 kan) (I136T) | pSG1850* → 1920 (Km) |
| Plasmids | ||
| pBEST502 | kan. Integration plasmid | |
| pPS1326 | gpr-lacZ cat. Integration plasmid | Sun et al. (1991) |
| pSG1612 | PdivIVA-gfp divIVA+ cat. Integration plasmid | Edwards et al. (2000) |
| pSG1850 | ylmH+ divIVA+ kan. Integration plasmid | See Material and Methods |
| pSG1902 | spoIIE-gfp cat. Integration plasmid | Wu et al. (1998) |
All strains carry the trpC2 marker.
For DNA transformations, the source of the DNA is shown, followed by an arrow, then the recipient strain, with the antibiotic selection in parentheses. Cm, chloramphenicol; Km, kanamycin; Tc, tetracycline.
General methods
B. subtilis strains were grown in transformation medium (MD; Kunst and Rapoport 1995), Oxoid antibiotic medium no. 3 (PAB), casein hydrolysate (CH; Nicholson and Setlow 1990; Partridge and Errington 1993), S medium supplemented with 1% CH (S; Sharpe et al. 1998) or sporulation medium (SM; Partridge and Errington 1993). Concentrations for antibiotic selections were as follows: kanamycin (5 μg/mL), spectinomycin (50 μg/mL), chloramphenicol (5 μg/mL), tetracycline (10 μg/mL). B. subtilis strains were transformed using the methods described by Kunst and Rapoport (1995), except that 20 min after the addition of 0.5–2.0 μg of DNA, the competent cultures were supplemented with casamino acid solution to a final concentration of 0.66%. In some experiments, the method described by Jenkinson (1983) was used. Cultures were induced to sporulate by the resuspension method of Sterlini and Mandelstam (1969), as specified by Partridge and Errington (1993). Time zero was defined as the point immediately after resuspension in sporulation medium.
E. coli strains were grown in 2× TY medium or on nutrient agar plates supplemented with ampicillin (100 μg/mL).
Cloning methods were based on those described by Sambrook et al. (1989). Amplification of DNA fragments for cloning was carried out using Pfu DNA polymerase (Stratagene) according to the manufacturer's instructions. Oligonucleotide primers were either made on an Applied Biosystems (ABI) DNA synthesizer, Model 380B by the service provided by the Chemical Pathology Unit, Oxford, or were purchased from Sigma-Genosys. DNA templates were sequenced on an Applied Biosystems 377A Sequencer by a departmental service.
Measurement of sporulation frequency by phase contrast microscopy
Samples of unfixed sporulating cultures taken at t7 were examined by phase contrast microscopy. The number of phase bright spores in a total of at least 800 cells was counted and expressed as a percentage of the value obtained for the equivalent wild-type culture. Control strains SG38 and 3012 produced, respectively, 50% and 42% spores.
Measurement of lysozyme-resistant spores
A 100 μL sample from a culture 7 h after resuspension was diluted 1/50 in sporulation medium (Sterlini and Mandelstam 1969), treated with lysozyme (250 μg/mL) for 15 min at 37°C, and plated on nutrient agar, as described by Jenkinson et al. (1980). Wild-type strain SG38 produced 7.0 × 107 lysozyme-resistant cfu/mL.
Enzyme assays
β-galactosidase was assayed using a method based on the hydrolysis of o-nitrophenol-β-D-galactopyranoside (ONPG) or 4-methylumbelliferyl-β-D-galactopyranoside (MUG) as described by Daniel et al. (1996) and Errington and Mandelstam (1986), respectively.
Alkaline phosphatase was assayed as described by Errington and Mandelstam (1983).
Western blot analysis
The cell pellets from 500 μL culture samples were lysed in volumes adjusted to compensate for differences in culture cell density. Hybridization to Hybond-C nitrocellulose membranes (Amersham) was carried out essentially as described by Wu and Errington (1994) using an enhanced chemiluminescence detection system (Amersham). Polyclonal anti-DivIVA serum was used at the concentration described previously (1/5000; Marston et al. 1998).
Transmission electron microscopy
Samples of sporulating culture (15 mL) were collected 110 min after induction of sporulation and centrifuged at 6000 rpm. The cell pellet was resuspended in 1.5 mL of 4% gluteraldehyde and left without shaking at 4°C for 4 h. The cells were then washed once in 1× Kellenberger solution (Kellenberger et al. 1958) and then resuspended in 200 μL of fresh solution. A small drop of this suspension was placed on a coverslip (1 index thickness) treated with 0.01% poly-L-lysine (Sigma), to create a single cell monolayer. Excess 4% gluteraldehyde was added over the cells on the coverslip, which was then left at 25°C for 1 h, before being carefully washed several times with 1× Kellenberger solution. The samples were stained and embedded on epon/resin stubs by a modification of the method described by Illing and Errington (1991). Briefly, the cells were placed in osmium tetroxide for 1 h in the dark at 25°C, then washed twice in AnalaR water (BDH). A 0.5% solution of the aqueous magnesium salt of uranyl acetate was added onto the coverslip, which was then left in the dark at 25°C overnight. The cells were then washed twice in water and subjected to a dehydration series in ethanol, followed by a fixation series in epon/resin. The cells were air dried and a thin layer (1–1.5 mL) of epon/resin was added to them before curing at 65°C overnight. Stubs were then added and again the samples were cured at 65°C overnight. Finally, the coverslip was removed from the stub and the embedded cells were serially sectioned at 100 nm intervals. Sections were stained with lead citrate (Reynolds 1963) and viewed on carbon grids on a Zeiss 912 Omega electron microscope (accelerating voltage 80 kV). Images were collected on a CCD camera (1024 × 1024 chip; depth 14 bits). The lengths of cellular compartments and the widths of septa for 180 cells were analyzed. Septa with a width from 28 to 86 nm (averaging 57±11 nm) were defined as thick (vegetative-type), and those ranging from 6 to 24 nm (averaging 14 ± 3 nm) were defined as thin (sporulation-type). Digital images and length measurements were obtained using Esivision AnaLysis 2.1 software. The quantitative data were then analyzed with Microsoft Excel 5.
Light microscopy
For phase contrast microscopy, samples of live cells in culture medium were examined on poly-lysine-treated microscope slides. For other live-cell applications, cell samples were placed on a thin film of agarose as described previously (Glaser et al. 1997). For examination of septa and DNA, cells were fixed with ethanol or gluteraldehyde as described (Hauser and Errington 1995; Glaser et al. 1997). Fluorescent staining of the membranes of live cells was done with FM 5–95 (Molecular Probes). Typically, 8 μL of FM 5–95 (200 μg/mL in H2O) was added to a 4 mL culture 30 min before sampling for microscopy. At the time of sampling, 6 μL of culture was added to a further 1.2 μL FM 5–95 (2 μg/mL), and, if necessary, 2 μL DAPI solution (1 μg/mL), and 5 μL of this mixture was placed on an agarose-coated slide.
For immunofluorescence microscopy, cells were fixed, permeabilized and labeled with antibody as described previously (Pogliano et al. 1995; Lewis et al. 1996; Reznekov et al. 1996), except that gluteraldehyde was used at 0.00005%. Staining for DivIVA was done as described previously (Marston et al. 1998). Mouse anti-β-galactosidase antibody (Sigma) and anti-mouse IgG Cy3 conjugate (Sigma) were used at 1/1200 and 1/250 dilutions, respectively.
Digital images were collected and processed essentially as described by Lewis and Errington (1997), except that for some experiments Metamorph v 3.6 was used for image processing. To ensure accurate overlay of images obtained with different fluorescence settings, the images were acquired automatically using a Biopoint filter wheel (Digital Pixel). Final images were assembled using Adobe Photoshop V. 3.05 or Microsoft Powerpoint V. 4.0, at 300 pixels per inch resolution.
The cell length analysis was carried out using Metamorph, or Object-Image 1.57b-fat5 software (Vischer et al. 1994), standardized against a digital image of a 10 μm graticule slide.
Acknowledgments
This work was supported by grants from the Biotechnology and Biological Sciences Research Council. J.E. is the recipient of a BBSRC Senior Research Fellowship. M.E.K. was the recipient of a long-term EMBO Fellowship (reference ALTF 209–1999). We thank Jeremy Sanderson for assistance with the electron microscopy and Mark Dyson for providing mutagenized pSG1850.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL erring@molbiol.ox.ac.uk; FAX 44-1865-275556.
Article and publication are at http://www.genesdev.org/cgi/doi/10.1101/gad.197501.
References
- Arigoni F, Pogliano K, Webb CD, Stragier P, Losick R. Localization of protein implicated in establishment of cell type to sites of asymmetric division. Science. 1995;270:637–640. doi: 10.1126/science.270.5236.637. [DOI] [PubMed] [Google Scholar]
- Barák I, Prepiak P, Schmeisser F. MinCD proteins control the septation process during sporulation of Bacillus subtilis. J Bacteriol. 1998;180:5327–5333. doi: 10.1128/jb.180.20.5327-5333.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bath J, Wu L, Errington J, Wang JC. Role of Bacillus subtilis SpoIIIE in DNA transport across the mother cell-prespore division septum. Science. 2000;290:995–997. doi: 10.1126/science.290.5493.995. [DOI] [PubMed] [Google Scholar]
- Beall B, Lutkenhaus J. FtsZ in Bacillus subtilis is required for vegetative septation and for asymmetric septation during sporulation. Genes & Dev. 1991;5:447–455. doi: 10.1101/gad.5.3.447. [DOI] [PubMed] [Google Scholar]
- Beall B, Lutkenhaus J. Impaired cell division and sporulation of a Bacillus subtilis strain with the ftsA gene deleted. J Bacteriol. 1992;174:2398–2403. doi: 10.1128/jb.174.7.2398-2403.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beall B, Lutkenhaus J. Nucleotide sequence and insertional inactivation of a Bacillus subtilis gene that affects cell division, sporulation, and temperature sensitivity. J Bacteriol. 1989;171:6821–6834. doi: 10.1128/jb.171.12.6821-6834.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Britton RA, Lin DC-H, Grossman AD. Characterization of a prokaryotic SMC protein involved in chromosome partitioning. Genes & Dev. 1998;12:1254–1259. doi: 10.1101/gad.12.9.1254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cha J-H, Stewart GC. The divIVA minicell locus of Bacillus subtilis. J Bacteriol. 1997;179:1671–1683. doi: 10.1128/jb.179.5.1671-1683.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniel RA, Harry EJ, Errington J. Role of penicillin-binding protein PBP 2B in assembly and functioning of the division machinery of Bacillus subtilis. Mol Microbiol. 2000;35:299–311. doi: 10.1046/j.1365-2958.2000.01724.x. [DOI] [PubMed] [Google Scholar]
- Daniel RA, Harry EJ, Katis VL, Wake RG, Errington J. Characterization of the essential cell division gene ftsL (yllD) of Bacillus subtilis and its role in the assembly of the division apparatus. Mol Microbiol. 1998;29:593–604. doi: 10.1046/j.1365-2958.1998.00954.x. [DOI] [PubMed] [Google Scholar]
- Daniel RA, Williams AM, Errington J. A complex four-gene operon containing essential cell division gene pbpB in Bacillus subtilis. J Bacteriol. 1996;178:2343–2350. doi: 10.1128/jb.178.8.2343-2350.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Boer PAJ, Crossley RE, Rothfield LI. Central role for the Escherichia coli minC gene product in two different cell division-inhibition systems. Proc Natl Acad Sci. 1990;87:1129–1133. doi: 10.1073/pnas.87.3.1129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Boer PAJ, Crossley RE, Rothfield LI. A division inhibitor and a topological specificity factor coded for by the minicell locus determine proper placement of the division septum in E. coli. Cell. 1989;56:641–649. doi: 10.1016/0092-8674(89)90586-2. [DOI] [PubMed] [Google Scholar]
- de Boer PAJ, Crossley RE, Rothfield LI. Roles of MinC and MinD in the site-specific septation block mediated by the MinCDE system of Escherichia coli. J Bacteriol. 1992;174:63–70. doi: 10.1128/jb.174.1.63-70.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards DH, Errington J. The Bacillus subtilis DivIVA protein targets to the division septum and controls the site specificity of cell division. Mol Microbiol. 1997;24:905–915. doi: 10.1046/j.1365-2958.1997.3811764.x. [DOI] [PubMed] [Google Scholar]
- Edwards DH, Thomaides HB, Errington J. Promiscuous targeting of Bacillus subtilis cell division protein DivIVA to division sites in Escherichia coli and fission yeast. EMBO J. 2000;19:2719–2727. doi: 10.1093/emboj/19.11.2719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Errington J, Daniel RA. Cell division during growth and sporulation. In: Sonenshein L, Losick R, Hoch JA, editors. Bacillus subtilis and its closest relatives: From genes to cells. Washington, D.C.: ASM Press; 2002. [Google Scholar]
- Errington J, Mandelstam J. Variety of sporulation phenotypes resulting from mutations in a single regulatory locus, spoIIA, in Bacillus subtilis. J Gen Microbiol. 1983;129:2091–2101. doi: 10.1099/00221287-129-7-2091. [DOI] [PubMed] [Google Scholar]
- ————— Use of a lacZ gene fusion to determine the dependence pattern of sporulation operon spoIIA in spo mutants of Bacillus subtilis. J Gen Microbiol. 1986;132:2967–2976. doi: 10.1099/00221287-132-11-2967. [DOI] [PubMed] [Google Scholar]
- Feucht A, Daniel RA, Errington J. Characterization of a morphological checkpoint coupling cell-specific transcription to septation in Bacillus subtilis. Mol Microbiol. 1999;33:1015–1026. doi: 10.1046/j.1365-2958.1999.01543.x. [DOI] [PubMed] [Google Scholar]
- Feucht A, Lucet I, Yudkin MD, Errington J. Cytological and biochemical characterization of the FtsA cell division protein of Bacillus subtilis. Mol Microbiol. 2001;40:115–125. doi: 10.1046/j.1365-2958.2001.02356.x. [DOI] [PubMed] [Google Scholar]
- Glaser P, Sharpe ME, Raether B, Perego M, Ohlsen K, Errington J. Dynamic, mitotic-like behavior of a bacterial protein required for accurate chromosome partitioning. Genes & Dev. 1997;11:1160–1168. doi: 10.1101/gad.11.9.1160. [DOI] [PubMed] [Google Scholar]
- Harry EJ, Wake RG. The membrane-bound cell division protein DivIB is localized to the division site in Bacillus subtilis. Mol Microbiol. 1997;25:275–283. doi: 10.1046/j.1365-2958.1997.4581822.x. [DOI] [PubMed] [Google Scholar]
- Hauser PM, Errington J. Characterization of cell cycle events during the onset of sporulation in Bacillus subtilis. J Bacteriol. 1995;177:3923–3931. doi: 10.1128/jb.177.14.3923-3931.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Z, Lutkenhaus J. Topological regulation of cell division in Escherichia coli involves rapid pole to pole oscillation of the division inhibitor MinC under the control of MinD and MinE. Mol Microbiol. 1999;34:82–90. doi: 10.1046/j.1365-2958.1999.01575.x. [DOI] [PubMed] [Google Scholar]
- Hu Z, Mukherjee A, Pichoff S, Lutkenhaus J. The MinC component of the division site selection system in Escherichia coli interacts with FtsZ to prevent polymerization. Proc Natl Acad Sci. 1999;96:14819–14824. doi: 10.1073/pnas.96.26.14819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang J, Cao C, Lutkenhaus J. Interaction between FtsZ and inhibitors of cell division. J Bacteriol. 1996;178:5080–5085. doi: 10.1128/jb.178.17.5080-5085.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Illing N, Errington J. Genetic regulation of morphogenesis in Bacillus subtilis: Roles of ςE and ςF in prespore engulfment. J Bacteriol. 1991;173:3159–3169. doi: 10.1128/jb.173.10.3159-3169.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ireton K, Gunther NWI, Grossman AD. spo0J is required for normal chromosome segregation as well as the initiation of sporulation in Bacillus subtilis. J Bacteriol. 1994;176:5320–5329. doi: 10.1128/jb.176.17.5320-5329.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobs C, Shapiro L. Bacterial cell division: A moveable feast. Proc Natl Acad Sci. 1999;96:5891–5893. doi: 10.1073/pnas.96.11.5891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenkinson HF. Altered arrangement of proteins in the spore coat of a germination mutant of Bacillus subtilis. J Gen Microbiol. 1983;129:1945–1958. doi: 10.1099/00221287-129-6-1945. [DOI] [PubMed] [Google Scholar]
- Jenkinson HF, Kay D, Mandelstam J. Temporal dissociation of late events in Bacillus subtilis sporulation from expression of genes that determine them. J Bacteriol. 1980;141:793–805. doi: 10.1128/jb.141.2.793-805.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kellenberger E, Ryter A, Sechaud J. Electron microscope study of DNA containing plasms. II. Vegetative and mature phage DNA as compared with normal bacterial nucleoids in different physiological states. J Biophysic Biochem Cytol. 1958;4:671–676. doi: 10.1083/jcb.4.6.671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunst F, Rapoport G. Salt stress is an environmental signal affecting degradative enzyme synthesis in Bacillus subtilis. J Bacteriol. 1995;177:2403–2407. doi: 10.1128/jb.177.9.2403-2407.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S, Price CW. The minCD locus of Bacillus subtilis lacks the minE determinant that provides topological specificity to cell division. Mol Microbiol. 1993;7:601–610. doi: 10.1111/j.1365-2958.1993.tb01151.x. [DOI] [PubMed] [Google Scholar]
- Levin PA, Losick R. Characterization of a cell division gene from Bacillus subtilis that is required for vegetative and sporulation septum formation. J Bacteriol. 1994;176:1451–1459. doi: 10.1128/jb.176.5.1451-1459.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levin PA, Losick R. Transcription factor Spo0A switches the localization of the cell division protein FtsZ from a medial to a bipolar pattern in Bacillus subtilis. Genes & Dev. 1996;10:478–488. doi: 10.1101/gad.10.4.478. [DOI] [PubMed] [Google Scholar]
- Levin PA, Margolis PS, Setlow P, Losick R, Sun D. Identification of Bacillus subtilis genes for septum placement and shape determination. J Bacteriol. 1992;174:6717–6728. doi: 10.1128/jb.174.21.6717-6728.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levin PA, Shim JJ, Grossman AD. Effect of minCD on FtsZ ring position and polar septation in Bacillus subtilis. J Bacteriol. 1998;180:6048–6051. doi: 10.1128/jb.180.22.6048-6051.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis PJ, Errington J. Direct evidence for active segregation of oriC regions of the Bacillus subtilis chromosome and co-localization with the Spo0J partitioning protein. Mol Microbiol. 1997;25:945–954. doi: 10.1111/j.1365-2958.1997.mmi530.x. [DOI] [PubMed] [Google Scholar]
- Lewis PJ, Magnin T, Errington J. Compartmentalized distribution of the proteins controlling the prespore-specific transcription factor ςF of Bacillus subtilis. Genes Cells. 1996;1:881–894. doi: 10.1046/j.1365-2443.1996.750275.x. [DOI] [PubMed] [Google Scholar]
- Londoño-Vallejo J-A, Frehel C, Stragier P. spoIIQ, a forespore-expressed gene required for engulfment in Bacillus subtilis. Mol Microbiol. 1997;24:29–39. doi: 10.1046/j.1365-2958.1997.3181680.x. [DOI] [PubMed] [Google Scholar]
- Marston AL, Errington J. Selection of the midcell division site in Bacillus subtilis through MinD-dependent polar localization and activation of MinC. Mol Microbiol. 1999;33:84–96. doi: 10.1046/j.1365-2958.1999.01450.x. [DOI] [PubMed] [Google Scholar]
- Marston AL, Thomaides HB, Edwards DH, Sharpe M, Errington J. Polar localization of the MinD protein of Bacillus subtilis and its role in selection of the mid-cell division site. Genes & Dev. 1998;12:3419–3430. doi: 10.1101/gad.12.21.3419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Massidda O, Anderluzzi D, Friedli L, Feger G. Unconventional organization of the division and cell wall gene cluster of Streptococcus pneumoniae. Microbiol. 1998;144:3069–3078. doi: 10.1099/00221287-144-11-3069. [DOI] [PubMed] [Google Scholar]
- Moriya S, Tsujikawa E, Hassan AKM, Asai K, Kodama T, Ogasawara N. A Bacillus subtilis gene-encoding protein homologous to eukaryotic SMC motor protein is necessary for chromosome partition. Mol Microbiol. 1998;29:179–187. doi: 10.1046/j.1365-2958.1998.00919.x. [DOI] [PubMed] [Google Scholar]
- Nicholson WL, Setlow P. Sporulation, germination and outgrowth. In: Harwood CR, Cutting SM, editors. Molecular Biological Methods for Bacillus. Chichester, UK: Wiley; 1990. pp. 391–450. [Google Scholar]
- Niki H, Jaffe A, Imamura R, Ogura T, Hiraga S. The new gene mukB codes for a 177 kd protein with coiled-coil domains involved in chromosome partitioning of E. coli. EMBO J. 1991;10:183–193. doi: 10.1002/j.1460-2075.1991.tb07935.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Partridge SR, Errington J. The importance of morphological events and intercellular interactions in the regulation of prespore-specific gene expression during sporulation in Bacillus subtilis. Mol Microbiol. 1993;8:945–955. doi: 10.1111/j.1365-2958.1993.tb01639.x. [DOI] [PubMed] [Google Scholar]
- Partridge SR, Foulger D, Errington J. The role of ςF in prespore-specific transcription in Bacillus subtilis. Mol Microbiol. 1991;5:757–767. doi: 10.1111/j.1365-2958.1991.tb00746.x. [DOI] [PubMed] [Google Scholar]
- Pogliano K, Harry E, Losick R. Visualization of the subcellular location of sporulation proteins in Bacillus subtilis using immunofluorescence microscopy. Mol Microbiol. 1995;18:459–470. doi: 10.1111/j.1365-2958.1995.mmi_18030459.x. [DOI] [PubMed] [Google Scholar]
- Raskin DM, De Boer PAJ. MinDE-dependent pole-to-pole oscillation of division inhibitor MinC in Escherichia coli. J Bacteriol. 1999a;181:6419–6424. doi: 10.1128/jb.181.20.6419-6424.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ————— Rapid pole-to-pole oscillation of a protein required for directing division to the middle of Escherichia coli. Proc Natl Acad Sci. 1999b;96:4971–4976. doi: 10.1073/pnas.96.9.4971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reynolds ES. The use of lead citrate at high pH as an electron-opaque stain in electron microscopy. J Cell Biol. 1963;17:208–212. doi: 10.1083/jcb.17.1.208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reznekov O, Alper S, Losick R. Subcellular localization of proteins governing the proteolytic activation of a developmental transcription factor in Bacillus subtilis. Genes Cells. 1996;1:529–542. doi: 10.1046/j.1365-2443.1996.d01-262.x. [DOI] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T. Molecular cloning: A laboratory manual. 2nd ed. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- Sharpe ME, Errington J. Postseptational chromosome partitioning in bacteria. Proc Natl Acad Sci. 1995;92:8630–8634. doi: 10.1073/pnas.92.19.8630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharpe ME, Hauser PM, Sharpe RG, Errington J. Bacillus subtilis cell cycle as studied by fluorescence microscopy: Constancy of the cell length at initiation of DNA replication and evidence for active nucleoid partitioning. J Bacteriol. 1998;180:547–555. doi: 10.1128/jb.180.3.547-555.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sievers J, Errington J. The Bacillus subtilis cell division protein FtsL localizes to sites of septation and interacts with DivIC. Mol Microbiol. 2000;36:846–855. doi: 10.1046/j.1365-2958.2000.01895.x. [DOI] [PubMed] [Google Scholar]
- Sterlini JM, Mandelstam J. Commitment to sporulation in Bacillus subtilis and its relationship to the development of actinomycin resistance. Biochem J. 1969;113:29–37. doi: 10.1042/bj1130029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevens CM, Daniel R, Illing N, Errington J. Characterization of a sporulation gene, spoIVA, involved in spore coat morphogenesis in Bacillus subtilis. J Bacteriol. 1992;174:586–594. doi: 10.1128/jb.174.2.586-594.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevens CM, Errington J. Differential gene expression during sporulation in Bacillus subtilis: Structure and regulation of the spoIIID gene. Mol Microbiol. 1990;40:543–551. doi: 10.1111/j.1365-2958.1990.tb00622.x. [DOI] [PubMed] [Google Scholar]
- Stragier P, Losick R. Molecular genetics of sporulation in Bacillus subtilis. Annu Rev Genet. 1996;30:297–341. doi: 10.1146/annurev.genet.30.1.297. [DOI] [PubMed] [Google Scholar]
- Sun D, Fajardo-Cavazos P, Sussman MD, Tovar-Rojo F, Cabrera-Martinez R-M, Setlow P. Effect of chromosome location of Bacillus subtilis forespore genes on their spo gene dependence and transcription by EςF: Identification of features of good EςF-dependent promoters. J Bacteriol. 1991;173:7867–7874. doi: 10.1128/jb.173.24.7867-7874.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sussman MD, Setlow P. Cloning, nucleotide sequence, and regulation of the Bacillus subtilis gpr gene, which codes for the protease that initiates degradation of small, acid-soluble proteins during spore germination. J Bacteriol. 1991;173:291–300. doi: 10.1128/jb.173.1.291-300.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teather RM, Collins JF, Donachie WD. Quantal behaviour of a diffusible factor which initiates septum formation at potential division sites in Escherichia coli. J Bacteriol. 1974;118:407–413. doi: 10.1128/jb.118.2.407-413.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vischer NOE, Huls PG, Woldringh CL. Object-Image: An interactive image analysis program using structured point collection. Binary. 1994;6:160–166. [Google Scholar]
- Wake RG, Errington J. Chromosome partitioning in bacteria. Annu Rev Genet. 1995;29:41–67. doi: 10.1146/annurev.ge.29.120195.000353. [DOI] [PubMed] [Google Scholar]
- Wu LJ, Errington J. Bacillus subtilis SpoIIIE protein required for DNA segregation during asymmetric cell division. Science. 1994;264:572–575. doi: 10.1126/science.8160014. [DOI] [PubMed] [Google Scholar]
- Wu LJ, Errington J. Septal localization of the SpoIIIE chromosome partitioning protein in Bacillus subtilis. EMBO J. 1997;16:2161–2169. doi: 10.1093/emboj/16.8.2161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu LJ, Errington J. Use of asymmetric cell division and spoIIIE mutants to probe chromosome orientation and organization in Bacillus subtilis. Mol Microbiol. 1998;27:777–786. doi: 10.1046/j.1365-2958.1998.00724.x. [DOI] [PubMed] [Google Scholar]
- Wu LJ, Lewis PJ, Allmansberger R, Hauser PM, Errington J. A conjugation-like mechanism for prespore chromosome partitioning during sporulation in Bacillus subtilis. Genes & Dev. 1995;9:1316–1326. doi: 10.1101/gad.9.11.1316. [DOI] [PubMed] [Google Scholar]
- Wu LJ, Feucht A, Errington J. Prespore-specific gene expression in Bacillus subtilis is driven by sequestration of SpoIIE phosphatase to the prespore side of the asymmetric septum. Genes & Dev. 1998;12:1371–1380. doi: 10.1101/gad.12.9.1371. [DOI] [PMC free article] [PubMed] [Google Scholar]