Abstract
It has been known for many decades that autophagy, a conserved lysosomal degradation pathway, is highly active during differentiation and development. However, until the discovery of the autophagy-related (ATG) genes in the 1990s, the functional significance of this activity was unknown. Initially, genetic knockout studies of ATG genes in lower eukaryotes revealed an essential role for the autophagy pathway in differentiation and development. In recent years, the analyses of systemic and tissue-specific knockout models of ATG genes in mice has led to an explosion of knowledge about the functions of autophagy in mammalian development and differentiation. Here we review the main advances in our understanding of these functions.
Autophagy is a process by which cytoplasmic components including macromolecules (for example proteins, glycogens, lipids and nucleotides) and organelles (for example mitochondria, peroxisomes and endoplasmic reticulum) are degraded by the lysosome (Fig. 1a)1,2. There are at least three different types of autophagy; macroautophagy (delivery of cytosolic contents to the lysosome by autophagosomes), microautophagy (inward invagination of the lysosomal membrane) and chaperone-mediated autophagy (direct translocation across the lysosomal membrane). Among these, macroautophagy (hereafter referred to as autophagy) has been the most extensively studied. Yeast genetic studies identified a set of autophagy-related (ATG) genes that are required for macroautophagy and its related processes. These genes are highly conserved among eukaryotes, allowing analyses of the roles of autophagy using reverse genetic techniques (Tables 1–3). Such analyses have revealed various physiological roles and pathological effects of autophagy, including adaptive responses to starvation, quality control of intracellular proteins and organelles, anti-aging, suppression of tumour formation, elimination of intracellular microbes, and antigen presentation2–9.
Figure 1.
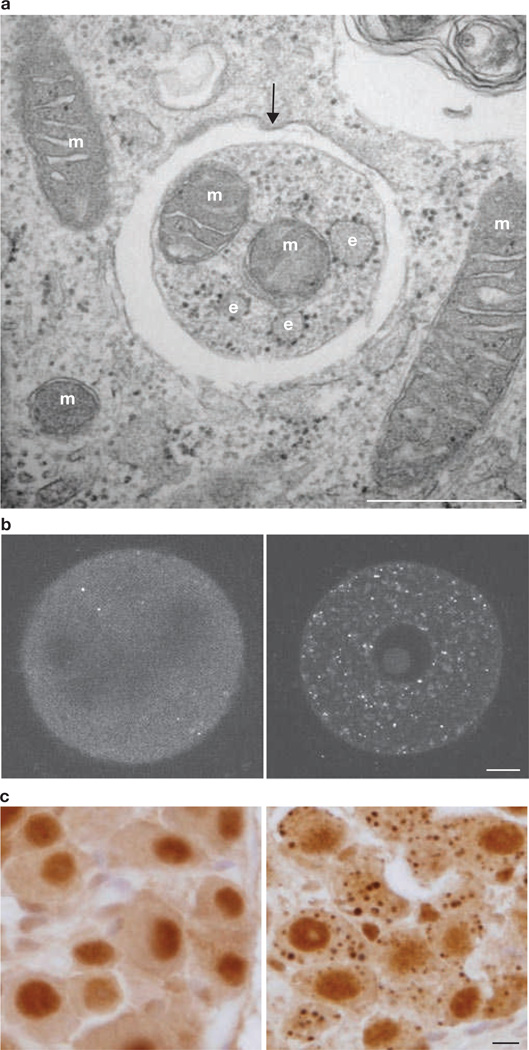
Representative images of the induction of autophagy and the outcome of its suppression. (a) Electron microscopic analysis of mouse embryonic fibroblasts during nutrient starvation. Mitochondria (m) and fragments of endoplasmic reticulum (e) are observed inside an autophagosome (arrow). Mitochondria may be randomly sequestered under conditions of starvation, but can be selectively degraded during erythroid differentiation and Parkin-mediated mitophagy. Scale bar, 500 nm. (b) Induction of autophagy after fertilization. Unfertilized (left) and fertilized (right) oocytes from female mice expressing GFP–LC3 to monitor autophagosome formation were observed by confocal microscopy. Small dots indicate GFP–LC3-positive autophagosomes. Scale bar, 10 µm. (c) Accumulation of ubiquitin-positive inclusion bodies in neurons of Atg5-deficient mice. Dorsal root ganglion neurons from wild-type (left) and systemic Atg5-deficient (right) neonates were stained with a monoclonal anti-ubiquitin antibody (1B3, purchased from MBL). Scale bar, 10 µm.
Table 1.
The roles of autophagy in development and differentiation of model organisms
| Organism | Role | Reference |
|---|---|---|
| Saccharomyces cerevisiae | Spore formation | 93 |
| Schizosaccharomyces pombe | Spore formation | 94, 95 |
| Dictyostelium discoideum | Fruiting body formation | 96, 97 |
| Sordaria macrospora | Essential for growth; fruiting body formation | 98 |
| Podospora anserina | Development of aerial hyphae; differentiation of female reproductive organs | 99 |
| Leishmania major | Differentiation into metacyclic promastigote | 100 |
| Trypanosoma cruzi | Differentiation into metacyclic trypomastigotes | 101 |
| Caenorhabditis elegans | Larval development; dauer formation; degradation of germline P granules in somatic cells | 10, 102, 103 |
| Drosophila melanogaster | Larval development; degradation of larval tissues (for example midgut and salivary gland)*; synaptic development | 104–107 |
| Arabidopsis thaliana | Dispensable | 108–110 |
| Mus musculus | See Tables 2 and 3 | |
The development-related roles of autophagy are listed. Autophagy may have other roles in each organism. For example, autophagy is important for starvation adaptation in all of these organisms.
Metamorphosis seems almost normal in the Drosophila atg7 mutant, although the pupal period is prolonged111
Table 3.
Phenotypes of tissue-specific knockout mice of ATG-related genes
| Mouse model | Target tissue/cell | Major phenotype | References |
|---|---|---|---|
| Atg5 F/F;; Nestin-Cre, Atg7 F/F; Nestin-Cre | Neural cell | Neurodegeneration; accumulation of ubiquitin and p62 | 70, 71, 80 |
| FIP200 F/F; Nestin-Cre | Neural cell | Neurodegeneration; accumulation of ubiquitin, p62 and mitochondria (more severe than Atg5/7 F/F; Nestin-Cre) | 72 |
| Atg5 F/F; Pcp2-Cre, Atg7 F/F;; Pcp2-Cre | Purkinje cell | Axonal degeneration; accumulation of p62 | 115, 116 |
| Atg5 F/F; Mx1-Cre, Atg7 F/F;; Mx1-Cre, Atg7 F/F;; Alb-Cre | Hepatocyte | Hepatic failure, hepatomegaly; accumulation of ubiquitin, p62 and mitochondria | 19, 67, 70, 80 |
| Atg5 F/F; MLC2v-Cre | Cardiomyocyte | Minimal abnormal phenotype (sensitive to pressure overload-cardiac failure) | 73 |
| Atg5 F/F; MerCreMer | Cardiomyocyte | Cardiac hypertrophy and dysfunction; accumulation of ubiquitin, p62 and mitochondria | 73 |
| Atg5 F/F; HSA-Cre | Skeletal muscle | Atrophy of fast muscle fibres; accumulation of ubiquitin and p62 | 74 |
| Atg5 F/F; Mlc1-Cre | Skeletal muscle | Muscle atrophy and weakness; accumulation of ubiquitin, p62 and mitochondria | 75 |
| Atg7 F/F; Ap2-Cre | Adipocyte | Decreased white adipose tissue mass, resistant to obesity; accumulation of p62 and mitochondria | 60, 62 |
| Atg5 F/F; EL-Cre | Pancreatic acinar cell | No abnormal phenotype (resistant to acute pancreatitis) | 117 |
| Atg7 F/F; Rip-Cre | Pancreatic β-cell | Impaired β-cell function, reduced β-cell mass; accumulation of ubiquitin, p62 and mitochondria | 76, 77 |
| Atg5 F/F; Zp3-Cre | Oocyte | Embryonic lethal at 4–8-cell stage (if fertilized with Atg5−sperm) | 11 |
| Atg5 F/F; Lck-Cre, Atg7 F/F; Lck-Cre | T cell | Decreased T-cell numbers; accumulation of mitochondria | 55, 56 |
| Atg5 F/F; CD19-Cre | B cell | Reduced B-1a B-cell numbers | 59 |
| Atg7 F/F; Vav-Cre | Haematopoietic cell | Severe anaemia, lymphopenia (T and B cells); accumulation of mitochondria | 53 |
| Atg5 F/F; CD11c-Cre | Dendritic cell | (Succumbed to viral infection due to defects in antigen presentation) | 118 |
| Atg5 F/F; Podocin-Cre | Podocyte | Late-onset glomerulosclerosis; accumulation of ubiquitin, p62 and mitochondria | 78 |
Phenotypes of tissue-specific knockout mice of ATG-related genes are listed. F, floxed allele; MLC2v, myosin light chain 2v; Mlc1, myosin light chain 1; HAS, human skeletal actin; EL, elastase I; Rip, rat insulin promoter. ‘Accumulation of ubiquitin’ refers to the accumulation of either ubiquitylated proteins or ubiquitin aggregates. ‘Accumulation of p62’ refers to the accumulation of either p62 protein or p62-positive aggregates.
Beyond these links to physiology, there is also increasing evidence that autophagy has conserved roles in differentiation and development. As a dynamic and highly inducible catabolic process that responds to environmental and hormonal cues, the autophagy pathway can drive the rapid cellular changes necessary for proper differentiation and/or development. Indeed, autophagy-defective organisms, including fungi, protozoa, worms and insects, show various abnormalities in differentiation and development (Table 1). These abnormalities may result from a deficiency in general autophagy, but they may also result from a failure to degrade specific components through selective autophagy, as shown for the degradation of germline P granules in somatic cells during Caenorhabditis elegans embryogenesis10.
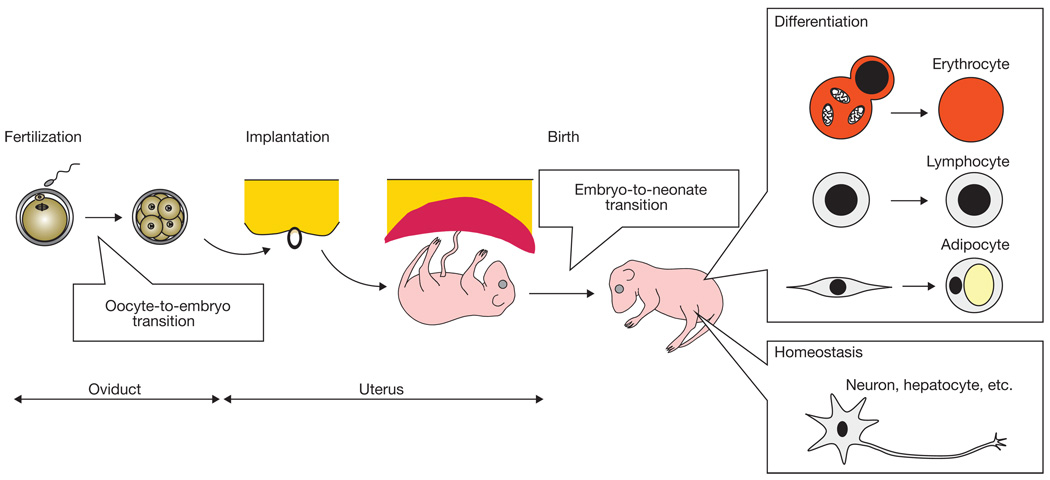
In mammals, autophagy is important for preimplantation development, survival during neonatal starvation, and cell differentiation during erythropoiesis, lymphopoeisis and adipogenesis (Fig. 2 and Tables 2 and 3). In addition to this ‘remodelling’ function, autophagy is also crucial for intracellular ‘refreshment’, and this homeostatic role is particularly important for the health of terminally differentiated cells such as neurons. In this review we discuss current knowledge about the role of autophagy in development and differentiation in mammals.
Figure 2.
The role of autophagy in development and differentiation in mammals. Autophagy has critical roles in fertilized oocytes and neonates through protein catabolism and the resulting production of the necessary amino acids. Autophagy is also important in cell remodelling (for example in mitochondrial elimination) during the differentiation of erythrocytes, lymphocytes and adipocytes. Finally, the role of autophagy in housekeeping is important, particularly in terminally differentiated cells such as neurons and hepatocytes, in which continuous renewal of cytoplasmic contents is essential.
Table 2.
Phenotypes of systemic knockout mice of ATG-related genes
| Genes | Phenotype | References |
|---|---|---|
| Atg3−/−, Atg5−/−, Atg7−/−, Atg9−/−, Atg16L1−/− | Neonatal lethal with reduced amino acid levels, suckling defect (Atg9 has an additional role in innate immune responses induced by double-stranded DNA) | 17–21 |
| beclin 1−/− | Early embryonic lethal (E7.5 or earlier) with defects in proamniotic canal closure (heterozygous mice show increased susceptibility to spontaneous tumours) | 28, 29 |
| FIP200−/− | Embryonic lethal (E13.5–E16.5) due to defective heart and liver development | 32 |
| Ambra1gt/gt | Embryonic lethal (~E14) with defects in neural tube development, and hyperproliferation of neural tissues | 30 |
| ULK1−/− | Increased reticulocyte number with delayed mitochondrial clearance | 51 |
| Atg4C−/− | Viable, fertile, increased susceptibility to carcinogen-induced fibrosarcoma | 112 |
| LC3B−/− | Normal phenotype | 113 |
| GABARAP−/− | Normal phenotype | 114 |
The phenotypes of conventional systemic knockout mice of ATG-related genes are listed. gt, gene-trapped allele.
Autophagy in mammalian embryonic development
Oocyte-to-embryo transition
The earliest autophagic event in mammalian development is observed in fertilized oocytes11 (Figs 1b and 2) and the ATG gene, Atg5, is essential during the early phase of pre-implantation development (Table 3)11. The oocyte, one of the most highly differentiated cells, suddenly changes to a highly undifferentiated state after fertilization. This ‘reprogramming’ occurs in both the nucleus and the cytoplasm. Maternal mRNA and proteins are rapidly degraded after the two-cell stage in the embryos, and new mRNA and proteins encoded by the zygotic genome are synthesized12,13, leading to marked changes in the protein species synthesized after the four-cell to eight-cell stages14. Moreover, the degradation of maternal proteins and RNAs may be necessary for the activation of the zygotic genome15. Autophagy occurs at only low levels in unfertilized oocytes but is massively induced within 4 h after fertilization (Figs 1b and 2)11. This induction of autophagy is completely dependent on fertilization and is not due to starvation after ovulation, because autophagy is not induced in the ovulated oocyte unless it has been fertilized. This type of autophagy may be triggered by calcium oscillation, because parthenogenetic activation also induces autophagy in oocytes11. Interestingly, autophagy is transiently suppressed from the late one-cell to middle two-cell stages, and then reactivated. As suppression of autophagy in mitotic phase is also observed in cultured mammalian cells16, perhaps there is a common mechanism to avoid the degradation of important nuclear factors during cell division.
Conventional Atg5−/− mice survive early embryogenesis (see below), but this is due to the presence of maternally inherited Atg5 protein in Atg5−/−oocytes. Elimination of the maternal Atg5 protein with the use of oocyte-specific Atg5-knockout mice results in embryonic lethality at the four-cell to eight-cell stages11 (Table 3). The precise role of autophagy during this process is not fully understood. Because the rate of protein synthesis is reduced in autophagy-defective embryos, normal levels of autophagy may be necessary for the production of sufficient amino acids for protein synthesis11. However, autophagy may also be required for the active elimination of unnecessary proteins and organelles that accumulate within oocytes or to facilitate ‘remodelling’ by degrading maternal suppressors of the zygotic gene program. Given that autophagy is an intracellular recycling system, these different possibilities are not mutually exclusive.
Embryo-to-neonate transition
The next wave of massive autophagy in mice is observed during the early neonatal period17 (Fig. 2). Autophagy is actively induced in all neonatal tissues except the brain until one or two days after birth17. Throughout mammalian embryogenesis, necessary nutrients are supplied through the placenta. At birth, this supply is terminated and neonates inevitably face severe starvation. Accordingly, conventional knockout of Atg3 (ref. 18), Atg5 (ref. 17), Atg7 (ref. 19), Atg9 (ref. 20) and Atg16L1 (ref. 21) causes neonatal lethality (within one day) despite almost normal appearance at birth (Table 2). Amino-acid levels are decreased in the plasma and tissues of these Atg-knockout neonates17–19, suggesting that autophagy is necessary to maintain the amino-acid pool during the early neonatal period. It remains undetermined precisely how the resultant amino acids are used in neonates. They may be used for energy production in a cell-autonomous manner to meet high energy demands in certain neonatal tissues; in support of this possibility, autophagy is highly activated in the heart and diaphragm after birth, and Atg5-deficient hearts show activation of the low-energy-sensing kinase AMPK (AMP-activated protein kinase)17.
However, it is unknown whether the decrease in amino-acid levels is the sole cause of premature death in these Atg-knockout neonates, because the mice show additional abnormalities. First, although autophagic activity is not enhanced in neurons by starvation during the neonatal period, the absence of basal autophagy in neurons may contribute to a suckling defect in Atg-knockout neonatal mice, which may exacerbate their malnutritional state17,19. However, the suckling failure alone cannot explain the early death because Atg5−/− and Atg7−/− neonates die earlier than wild-type neonates even under non-suckling conditions. Second, autophagy may also be important for the degradation of macromolecules other than proteins. For example, glycogen delivery to the lysosome results in the production of glucose and energy22,23 in newborn liver and muscle. Third, Atg5−/−neonates have less adipose tissue mass as a result of defects in adipogenesis (see below)24, which might prevent sufficient energy production. Fourth, the clearance of apoptotic corpses is defective in Atg5−/−late embryos, which might also contribute to developmental abnormalities25. Finally, as environmental conditions change markedly at birth, autophagy-mediated degradation may also contribute to cellular remodelling by generating a new set of proteins and organelles (somewhat akin to the process of metamorphosis in insects).
Other processes during embryogenesis
Although complete autophagy-defective embryos derived from oocyte-specific Atg5-deficient mice die before implantation11, conventional Atg3−/−, Atg5−/−, Atg7−/−, Atg9−/− and Atg16L1−/− embryos survive the entire embryonic period and are born at Mendelian frequency17–21. These data suggest that Atg3, Atg5, Atg7, Atg9 and Atg16L1 are not essential for embryogenesis (except during the early stages, when maternally inherited proteins allow survival in conventional knockout mice). Nonetheless, a more subtle role for these ATG genes in embryonic development cannot be ruled out. Although high levels of autophagy have not been reported during later stages of embryogenesis in mice expressing the autophagy reporter green fluorescent protein (GFP)–LC3, except in certain tissues such as the developing thymus26, certain phenotypic abnormalities of Atg5−/− mice (such as ubiquitin inclusions in embryonic neurons or decreased adipose tissue mass in neonates) indicate that wild-type levels of autophagy may be important for completely normal embryogenesis. Further studies are therefore required for the careful dissection of the role of ATG gene deficiency in more subtle aspects of embryonic development.
In contrast with the embryonic survival observed in conventional Atg3−/−, Atg5−/−, Atg7−/−, Atg9−/−, and Atg16L1−/− mice, the conventional knockout of several other ATG genes, including beclin 1, Ambra1 and FIP200, produces somewhat different phenotypes (Table 2). Beclin 1, a component of class III phosphatidylinositol-3-OH kinase (PI(3)K) complexes27, is the mammalian homologue of yeast Atg6/Vps30. Beclin 1−/− mice show early embryonic lethality28,29, and abnormally small embryos are detected at embryonic day 7.5 (E7.5)29; these embryos show massive cell death and failure to close the proamniotic canal. Beclin 1−/− embryonic stem cells are viable, although beclin 1−/− mice are early embryonic lethal, suggesting that Beclin 1 is dispensable in vitro but critically important for development in vivo. Ambra1 is a Beclin 1-interacting protein that positively regulates autophagy and is strongly expressed in developing neural tissues. Ambra1-deficient mice produced by gene-trap mutagenesis (Ambra1gt/gt) are embryonic lethal at days E10–E14 and show defective neural tube development and hyperproliferation of neural tissues30. FIP200 (focal adhesion kinase (FAK) family interacting protein of Mr 200K, also known as RB1CC1) is a ULK1 (an Atg1 homologue)-interacting protein and has a molecular function similar to that of yeast Atg17, although it shows no homology with any yeast Atg proteins31. FIP200−/− mice are also embryonic lethal between E13.5 and E16.5 as a result of defective heart and liver development32.
It is not completely understood why the phenotypes of different ATG-gene knockout mouse models vary. Because Beclin 1-including PI(3) K complexes have several distinct functions, and FIP200 has multiple interacting partners such as FAK, Pyk2, TSC1 (tuberous sclerosis complex 1) and p53, some of the abnormalities found in beclin 1−/−, Ambra1gt/gtand FIP200−/−mice may be related to other factors besides autophagy deficiency. Alternatively, the disparate phenotypes in different ATG-gene knockout mouse models may depend on the step in autophagy at which each factor functions. The Beclin 1-containing PI(3)K and the ULK1–FIP200 complexes function earlier in autophagy at the autophagosome nucleation step2,33, while Atg3, Atg5, Atg7 and Atg16L function later in autophagy in autophagosome elongation. Thus, upstream factors may show more severe phenotypes, or alternatively, as recently reported, downstream factors may be dispensable for a particular type of macro autophagy34. However, one exception to this general pattern is Atg9, which acts early (according to hierarchical analyses in yeast) but leads to a less severe phenotype when deficient35. There could also be different degrees of functional redundancy between different Atg proteins or different levels of compensatory mechanisms for the knockout of different Atg proteins. Further studies are needed to determine whether the autophagy pathway itself, or merely certain components of this pathway with autophagy-independent functions, are essential for distinct stages of embryonic development after the transition from oocyte to embryo.
Autophagy in cell differentiation
Erythrocyte differentiation and mitochondrial clearance
A long-standing question in developmental biology has been how the erythroblast loses its organelles, and to what extent the autophagy pathway functions in this process. Erythroblasts possess nuclei and intracellular organelles, which are lacking in mature erythrocytes and are replaced by haemoglobin molecules (Fig. 2). During erythroid differentiation, the nuclei are released from the cell, but the mechanism(s) by which other organelles are eliminated are poorly understood. The 15-lipoxygenase enzyme, which is highly expressed in reticulocytes, has been suggested to participate in this degradation36–38. Electron microscopy studies have also indicated a potential role for autophagy in the degradation of erythroid organelles39–43. However, organelle elimination in erythroid cells of Atg5−/− neonates seems normal44; thus, the role of autophagy in this process has been uncertain.
Several recent studies suggest that mitochondrial clearance in reticulocytes is partly dependent on autophagy. This concept first emerged from studies with cells and mice deficient in Nix (also known as BNIP3L), a Bcl-2 homology 3 (BH3)-only protein that is present on the mitochondrial outer membrane. Nix is required for the selective removal of mitochondria but not that of ribosomes or nuclei; in Nix−/− reticulocytes, autophagosomes are generated but there is a deficiency in the loss of the mitochondrial membrane potential and consequent mitochondrial engulfment by autophagosomes45–47. Nix−/− mice have lower numbers of mature erythrocytes (that is, anaemia) and a compensatory expansion of erythrocyte precursors (that is, reticulocytosis)46,48.
A direct requirement for ATG genes in mitochondrial elimination during erythroid cell maturation has also been demonstrated. There are five Atg1-related proteins in mammals, including ULK1, ULK2, ULK3, ULK4 and STK36. Because ULK3, ULK4 and STK36 lack the carboxy-terminal domain that is required for binding to Atg13 and FIP200, ULK1 and ULK2 are the primary candidate mammalian Atg1 orthologues49,50. During erythroid differentiation, the expression of ULK1, but not that of ULK2, is induced, suggesting that ULK1 may have a major role in this process51. Indeed, ULK1−/− mice show an increase in reticulocyte number. Interestingly, in addition to impaired clearance of mitochondria, the erythroid cells in ULK1−/− mice also have impaired clearance of RNA-bound ribosomes. Similarly, lethally irradiated wild-type recipients of Atg7−/− fetal liver transplants have decreased red blood cell counts and delayed mitochondrial clearance in erythroid cells52. Furthermore, haematopoietic cell-specific Atg7-knockout mice (Atg7flox/flox;Vav-Cre mice) are severely anaemic and die at 8–14 weeks of age (Table 3)53. The Atg7−/−erythrocytes accumulate selectively damaged mitochondria; this is associated with premature cell death. In contrast with ULK1−/−mice51 (but similarly to Nix−/− mice46), erythroid cells from Atg7−/−mice do not seem to accumulate any organelles besides mitochondria53.
Thus, the autophagy pathway seems to function selectively in mitochondrial clearance during erythroid differentiation46,51–53. One proposed mechanism for the selective degradation of mitochondria (called mitophagy) is through Nix-mediated mitochondrial recognition. Nix has a typical LC3-interacting WXXL-like motif, WVEL, at the amino terminus, through which Nix interacts with LC3 family proteins such as γ-aminobutyric acid receptor-associated protein (GABARAP) and LC3A on autophagosomal membranes47,54. Thus, Nix could serve as a cargo receptor for mitophagy. However, Nix may have additional autophagy-independent roles in mitochondrial clearance. The treatment of Nix−/− reticulocytes with an uncoupling reagent restores mitochondrial engulfment by autophagosomes46, indicating that effects on mitochondrial membrane potential, rather than a specific function as a cargo receptor for mitophagy, may, at least in part, underlie Nix-mediated effects in erythroid maturation.
Future studies are required to delineate more precisely the role of mitophagy and its molecular mechanisms in erythroid differentiation. How other organelles, such as ribosomes and ER, are eliminated during erythroid differentiation is also an interesting question. The normal clearance of ribosomes in Nix−/− cells46 and of ribosomes and ER in Atg7−/−cells53 suggest the presence of unknown degradation system(s), one of which may be the recently reported Atg5/Atg7-independent macroautophagy34. Mitophagy, and/or alternative forms of macroautophagy, may also have a more general role in the differentiation of other cell lineages (see below).
Lymphocyte differentiation
Haematopoietic-specific, B-lymphocyte-specific and T-lymphocyte-specific deletion of ATG genes has revealed specific roles of autophagy in lymphocyte differentiation. In addition to severe anaemia, the haematopoietic cell-specific Atg7-knockout mice (Atg7flox/flox;Vav-Cre mice) also have a significant decrease in T and B lymphocyte counts (with no alterations in numbers of cells of the myeloid lineage)53. Both CD4 and CD8 T cells from these mice have a greater number of mitochondria, which translates into increased mitochondrial mass, higher levels of superoxide, and increased susceptibility to apoptosis after in vitro culture. The Atg7−/− fetal liver cell transplant recipient mice discussed above are also lymphopenic52. Furthermore, T-cell-specific Atg5-knockout and Atg7-knockout mice (Atg5flox/flox;Lck-Cre and Atg7flox/flox;Lck-Cre) have decreased numbers of peripheral T cells, increased T-cell accumulation of mitochondria, and enhanced apoptosis in mature T cells (Table 3)55,56. Atg5−/− chimaeric mice also show a similar decrease in T and B lymphocyte numbers and defects in stimulation-induced proliferation57. During the normal development of thymocytes to circulating mature T cells, mitochondrial content is decreased, but this developmental change is suppressed in autophagy-defective T cells55. Thus, the clearance of mitochondria may represent a developmental process that contributes to the survival of mature T cells. It is not yet fully understood why erythroid and lymphocyte, but not myeloid, survival depends partly on mitophagy.
Another recently identified role of autophagy in T cells is the elimination of autoreactive T cells in the thymus58. High levels of autophagy in thymic epithelial cells26, the only non-haematopoietic cell type that constitutively expresses major histocompatibility complex class II (MHC II) molecules, were initially described in GFP–LC3 reporter mice. Subsequently, autophagy was shown to be essential for the delivery of certain endogenously synthesized antigens to MHC II loading compartments7,8. Genetic disruption of Atg5 in thymic epithelial cells results in altered selection of certain MHC-II-restricted T-cell specificities and in autoimmunity58. Thus, this autophagy-dependent antigen presentation pathway participates in self-tolerance.
As noted above, ATG gene deletion in mice also decreases B-cell counts. Like Atg7flox/flox;Vav-Cre mice53 and Atg5−/− chimaeric mice57, B cell-specific Atg5-knockout mice (Atg5flox/flox;CD19-Cre) have decreased numbers of B cells59. In addition to its role in B-cell development in the bone marrow59, Atg5 is required for the survival of certain types of B cell in the periphery. The mechanism underlying the B-cell defects is unknown; unlike in T cells, mitochondrial content is not developmentally regulated in B cells55.
Adipocyte differentiation and energy metabolism
In a similar manner to erythrocyte maturation, adipogenesis also involves marked intracellular remodelling. Adipocytes are differentiated from preadipocytes, which emerge from multipotent mesenchymal precursors. During adipogenesis, the fibroblast-like cells differentiate into round cells containing single large lipid droplets (Fig. 2). Autophagy is actively induced during this differentiation process in vitro24, and both in vitro and in vivo studies have confirmed that autophagy contributes to cellular remodelling during adipogenesis.
Primary mouse embryonic fibroblasts (MEFs) can be differentiated into adipocytes in the presence of adipogenic factors, but this process is significantly delayed or suppressed in primary Atg5−/− MEFs24. The knockdown of Atg7 or Atg5 in 3T3-L1 preadipocytes also suppresses triglyceride accumulation and decreases protein levels of mediators and markers of adipocyte differentation60. Moreover, treatment with short interfering RNA against LC3 impairs lipid droplet formation in various mammalian cell lines such as HeLa, PC12 and HepG2 cells61.
Newborn Atg5−/− mice possess fewer subcutaneous adipocytes than wild-type neonates24, and the importance of autophagy in adipogenesis in vivo has been further confirmed in adipocyte-specific Atg7-knockout mice (Atg7flox/flox;Ap2-Cre mouse)60,62. These mutant mice have decreased white adipose tissue mass and have smaller adipocytes containing multi-locular lipid droplets, increased numbers of mitochondria and increased cytoplasmic volume. These adipocyte features are characteristic of brown adipose tissue, and several brown fat-associated enzymes such as UCP-1 and PGC-1α are expressed in the white adipose tissue of these mutant mice60 (the increase in UCP-1 was not observed in ref. 62).
Interestingly, the adipocyte-specific Atg7-knockout mice are leaner than wild-type mice, even though there is no difference in their food intake. Furthermore, they are resistant to obesity induced by a high-fat diet and show higher insulin sensitivity60,62. This may be due in part to increased β-oxidation of fatty acids and lower free fatty acid levels in their plasma. Basal physical activity is also higher in the adipocyte-specific Atg7-knockout mice62. Thus, autophagy is important not only for adipocyte differentiation but also for local and whole-body lipid metabolism.
An important question is whether the anti-obesity and insulin sensitization effects of Atg7 disruption are primarily the result of alterations in the ratios of brown adipose-like tissue versus white adipose tissue. Although the amount of brown adipose tissue declines after birth, recent evidence indicates that significant amounts are found in adult humans63–65. There is a growing consensus that the ratio of white to brown adipose tissue may influence the development of obesity, which is characterized by an increased mass of white adipose tissue66. Given the defect in adipocyte differentiation in adipocyte-specific Atg7-knockout animals, it is difficult to predict how the disruption of autophagy would affect body weight and insulin sensitivity in the fully developed animal. Studies of inducible adipocyte-specific ATG-gene knockout mice will be required to address this question.
The involvement of autophagy in hepatocyte lipid droplet accumulation seems more complex than its role in adipocyte lipid metabolism. During starvation, free fatty acids are remobilized from adipose tissue to the liver and accumulate in lipid droplets. One paper reports that there is less formation of lipid droplets in hepatocyte-specific Atg7-knockout mice (Atg7flox/flox;Alb-Cre mice)67. However, it is also known that suppression of autophagy in hepatocytes leads to an increase in hepatic triglyceride storage in lipid droplets in Atg7flox/flox;Mx1-Cre and Atg7flox/flox;Alb-Cre mouse lines19,68. This may be due to impaired autophagic degradation of lipid droplets, because small lipid droplets can be selectively engulfed by autophagosomes in cultured hepatocytes68. It is not clear why different phenotypes are observed in different investigations of liver-specific Atg7-knockout mice. Perhaps Atg7 is required for the rapid formation of lipid droplets that occurs in the liver during the acute response to starvation, whereas Atg7 may inhibit the chronic accumulation of lipid droplets during normal feeding states. Furthermore, the mechanistic basis for differential effects of autophagy on lipid metabolism in the liver versus the adipocyte remains to be determined.
Homeostatic role in terminally differentiated cells
Autophagy may be dispensable for many types of rapidly proliferating cells, but its housekeeping role may become essential in differentiated or senescent cells (Fig. 2)69. Tissue-specific knockout of ATG genes causes the accumulation of ubiquitin-positive inclusion bodies/aggregates in neurons (Fig. 1c)70–72, hepatocytes19, cardiac muscle73, skeletal muscle74,75, pancreatic β-cells76,77and podocytes (Table 3)78. These inclusions are also positive for the autophagy substrate p62 (ref. 79). These aggregates are observed as early as E15.5 in neurons70. In addition to these inclusion bodies, soluble and cytosolic ubiquitylated proteins and p62 accumulate in these cells. The observations that ablation of autophagy leads to the degeneration/dysfunction of each tissue (Table 3) indicate that autophagy has a critical homeostatic role in post-mitotic differentiated cells. The accumulation of p62 may be the primary cause of the cellular toxicity, at least in the liver (but not in the brain), because hepatomegaly and hepatic dysfunction in the liver-specific Atg7-knockout mice are restored by simultaneous knockout of p62 (ref. 80). The effect of p62 overexpression is, at least in part, a result of aberrant hyperactivation of the transcription factor Nrf2, which occurs because p62 binds to and inhibits Keap1, an E3-ligase for Nrf2 (refs 81–84).
In non-dividing cells, quality control, not only of proteins but also of organelles, is important. As discussed above, mitochondrial quantity and quality can be controlled by mitophagy. Indeed, the accumulation of mitochondria, most of which are deformed or dysfunctional, is observed in autophagy-defective cells in most tissue-specific ATG-gene knockout models (Table 3)19,53,55,60,62,72,73,75–78. Recent studies have considered the mechanisms of autophagy-mediated quality control of mitochondria in the context of Parkinson’s disease. Parkin is a ubiquitin ligase whose mutation causes familial Parkinson’s disease85. On mitochondrial damage and depolarization, this cytosolic soluble protein translocates to mitochondria and subsequently induces mitophagy86. The recruitment of Parkin to mitochondria requires PINK1, another protein kinase associated with Parkinson’s disease87–92. However, it is not exactly clear how the PINK1–Parkin pathway promotes mitophagy. Because the E3-ligase activity of Parkin is a prerequisite for mitophagy, the ubiquitylation of some mitochondrial proteins is likely to be essential. So far, VDAC1 (voltage-dependent anion channel 1)88 and mitofusin (a mitochondrial pro-fusion factor)91 have been proposed as Parkin substrates. Ubiquitylation of these proteins could recruit the autophagy adaptor p62 (ref. 88), and/or affect mitochondrial fission or fusion, to mediate mitophagy91. The failure of any step in this pathway would lead to the accumulation of dysfunctional mitochondria and the excessive production of reactive oxygen species (ROS), resulting in the injury of ROS-sensitive cells such as the dopaminergic neurons.
Conclusion
Through the analysis of systemic and tissue-specific ATG-gene knockout mice, great advances have been made in our understanding of the role(s) of autophagy in mammalian development and differentiation. The autophagy pathway seems essential for two critical stages of early development: the pre-implantation period after oocyte fertilization, and the early postnatal period after disruption of the placental food supply. Autophagy could also have a role in other stages of embryogenesis, because the deletion of some ATG genes leads to lethality during mid-embryonic development, and the ATG-gene knockout mice that survive the postnatal period display some developmental abnormalities. The autophagy pathway is also involved in the differentiation of two cell types that require marked architectural remodelling — erythroid cells, presumably through mitochondrial clearance, and adipocytes — probably through a more complex mechanism. The autophagy pathway is also involved in the differentiation of lymphocytes, although this is not a cell type that undergoes as marked a remodelling. Moreover, autophagy’s homeostatic role in protein and organelle quality control may prevent the degeneration of post-mitotic cells throughout embryonic development and postnatal life. The failure of autophagy to perform this specific function in adulthood may underlie the pathogenesis of certain neurodegenerative diseases, such as familial Parkinson’s disease. Future studies are likely to reveal a more universal role for autophagy in numerous other developmental and differentiation events that require cellular self-digestion, cellular housekeeping and/or cellular recycling to meet high nutritional and energy demands.
ACKNOWLEDGEMENTS
We thank Chieko Kishi and Satoshi Tsukamoto for preparation of the images in Fig. 1a and Fig. 1b, respectively. This work was supported by Grants-in-Aid for Scientific Research from the Ministry of Education, Culture, Sports, Science and Technology, Japan (to N.M.), the Toray Science Foundation (to N.M.), the Takeda Science Foundation (to N.M.) and National Institutes of Health grants RO1 CA85254, RO1 CA109618 and U54 AI057156 (to B.L.).
Footnotes
COMPETING FINANCIAL INTERESTS
The authors declare no competing financial interests.
Reprints and permissions information is available online at http://npg.nature.com/reprintsandpermissions/
Contributor Information
Noboru Mizushima, Email: nmizu.phy2@tmd.ac.jp, Department of Physiology and Cell Biology, Tokyo Medical and Dental University, Tokyo 113-8519, Japan..
Beth Levine, Howard Hughes Medical Institute, University of Texas Southwestern Medical Center, Dallas, TX 75390-9113, USA, the Department of Internal Medicine, University of Texas Southwestern Medical Center, Dallas, TX 75390-9113, USA and the Department of Microbiology, University of Texas Southwestern Medical Center, Dallas, TX 75390-9113, USA..
References
- 1.Mizushima N. Autophagy: process and function. Genes Dev. 2007;21:2861–2873. doi: 10.1101/gad.1599207. [DOI] [PubMed] [Google Scholar]
- 2.Levine B, Kroemer G. Autophagy in the pathogenesis of disease. Cell. 2008;132:27–42. doi: 10.1016/j.cell.2007.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rubinsztein DC. The roles of intracellular protein-degradation pathways in neurodegeneration. Nature. 2006;443:780–786. doi: 10.1038/nature05291. [DOI] [PubMed] [Google Scholar]
- 4.Mizushima N, Klionsky DJ. Protein turnover via autophagy: implications for metabolism. Annu. Rev. Nutr. 2007;27:19–40. doi: 10.1146/annurev.nutr.27.061406.093749. [DOI] [PubMed] [Google Scholar]
- 5.Mizushima N, Levine B, Cuervo AM, Klionsky DJ. Autophagy fights disease through cellular self-digestion. Nature. 2008;451:1069–1075. doi: 10.1038/nature06639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cecconi F, Levine B. The role of autophagy in mammalian development: cell makeover rather than cell death. Dev. Cell. 2008;15:344–357. doi: 10.1016/j.devcel.2008.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Virgin HW, Levine B. Autophagy genes in immunity. Nat. Immunol. 2009;10:461–470. doi: 10.1038/ni.1726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Deretic V, Levine B. Autophagy, immunity, and microbial adaptations. Cell Host Microbe. 2009;5:527–549. doi: 10.1016/j.chom.2009.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.White E, Karp C, Strohecker AM, Guo Y, Mathew R. Role of autophagy in suppression of inflammation and cancer. Curr. Opin. Cell Biol. 2010;22:212–217. doi: 10.1016/j.ceb.2009.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhang Y, et al. SEPA-1 mediates the specific recognition and degradation of P granule components by autophagy in C. elegans. Cell. 2009;136:308–321. doi: 10.1016/j.cell.2008.12.022. [DOI] [PubMed] [Google Scholar]
- 11.Tsukamoto S, et al. Autophagy is essential for preimplantation development of mouse embryos. Science. 2008;321:117–120. doi: 10.1126/science.1154822. [DOI] [PubMed] [Google Scholar]
- 12.Merz EA, Brinster RL, Brunner S, Chen HY. Protein degradation during preimplantation development of the mouse. J. Reprod. Fertil. 1981;61:415–418. doi: 10.1530/jrf.0.0610415. [DOI] [PubMed] [Google Scholar]
- 13.Stitzel ML, Seydoux G. Regulation of the oocyte-to-zygote transition. Science. 2007;316:407–408. doi: 10.1126/science.1138236. [DOI] [PubMed] [Google Scholar]
- 14.van Blerkom J, Brockway GO. Qualitative patterns of protein synthesis in the pre-implantation mouse embryo. I. Normal pregnancy. Dev. Biol. 1975;44:148–157. doi: 10.1016/0012-1606(75)90382-6. [DOI] [PubMed] [Google Scholar]
- 15.Schier AF. The maternal-zygotic transition: death and birth of RNAs. Science. 2007;316:406–407. doi: 10.1126/science.1140693. [DOI] [PubMed] [Google Scholar]
- 16.Eskelinen E-L, et al. Inhibition of autophagy in mitotic animal cells. Traffic. 2002;3:878–893. doi: 10.1034/j.1600-0854.2002.31204.x. [DOI] [PubMed] [Google Scholar]
- 17.Kuma A, et al. The role of autophagy during the early neonatal starvation period. Nature. 2004;432:1032–1036. doi: 10.1038/nature03029. [DOI] [PubMed] [Google Scholar]
- 18.Sou YS, et al. The Atg8 conjugation system is indispensable for proper development of autophagic isolation membranes in mice. Mol. Biol. Cell. 2008;19:4762–4775. doi: 10.1091/mbc.E08-03-0309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Komatsu M, et al. Impairment of starvation-induced and constitutive autophagy in Atg7-deficient mice. J. Cell Biol. 2005;169:425–434. doi: 10.1083/jcb.200412022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Saitoh T, et al. Atg9a controls dsDNA-driven dynamic translocation of STING and the innate immune response. Proc. Natl Acad. Sci. USA. 2009;106:20842–20846. doi: 10.1073/pnas.0911267106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Saitoh T, et al. Loss of the autophagy protein Atg16L1 enhances endotoxin-induced IL-1β production. Nature. 2008;456:264–268. doi: 10.1038/nature07383. [DOI] [PubMed] [Google Scholar]
- 22.Kotoulas OB, Kalamidas SA, Kondomerkos DJ. Glycogen autophagy in glucose homeostasis. Pathol. Res. Pract. 2006;202:631–638. doi: 10.1016/j.prp.2006.04.001. [DOI] [PubMed] [Google Scholar]
- 23.Schiaffino S, Mammucari C, Sandri M. The role of autophagy in neonatal tissues: just a response to amino acid starvation? Autophagy. 2008;4:727–730. doi: 10.4161/auto.6143. [DOI] [PubMed] [Google Scholar]
- 24.Baerga R, Zhang Y, Chen PH, Goldman S, Jin S. Targeted deletion of autophagy-related 5 (atg5) impairs adipogenesis in a cellular model and in mice. Autophagy. 2009;5:1118–1130. doi: 10.4161/auto.5.8.9991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Qu X, et al. Autophagy gene-dependent clearance of apoptotic cells during embryonic development. Cell. 2007;128:931–946. doi: 10.1016/j.cell.2006.12.044. [DOI] [PubMed] [Google Scholar]
- 26.Mizushima N, Yamamoto A, Matsui M, Yoshimori T, Ohsumi Y. In vivo analysis of autophagy in response to nutrient starvation using transgenic mice expressing a fluorescent autophagosome marker. Mol. Biol. Cell. 2004;15:1101–1111. doi: 10.1091/mbc.E03-09-0704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.He C, Levine B. The Beclin 1 interactome. Curr. Opin. Cell Biol. 2010;22:140–149. doi: 10.1016/j.ceb.2010.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Qu X, et al. Promotion of tumorigenesis by heterozygous disruption of the beclin 1 autophagy gene. J. Clin. Invest. 2003;112:1809–1820. doi: 10.1172/JCI20039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yue Z, Jin S, Yang C, Levine AJ, Heintz N. Beclin 1, an autophagy gene essential for early embryonic development, is a haploinsufficient tumor suppressor. Proc. Natl Acad. Sci. USA. 2003;100:15077–15082. doi: 10.1073/pnas.2436255100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fimia GM, et al. Ambra1 regulates autophagy and development of the nervous system. Nature. 2007;447:1121–1125. doi: 10.1038/nature05925. [DOI] [PubMed] [Google Scholar]
- 31.Hara T, et al. FIP200, a ULK-interacting protein, is required for autophagosome formation in mammalian cells. J. Cell Biol. 2008;181:497–510. doi: 10.1083/jcb.200712064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gan B, et al. Role of FIP200 in cardiac and liver development and its regulation of TNFα and TSC–mTOR signaling pathways. J. Cell Biol. 2006;175:121–133. doi: 10.1083/jcb.200604129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Longatti A, Tooze SA. Vesicular trafficking and autophagosome formation. Cell Death Differ. 2009;16:956–965. doi: 10.1038/cdd.2009.39. [DOI] [PubMed] [Google Scholar]
- 34.Nishida Y, et al. Discovery of Atg5/Atg7-independent alternative macroautophagy. Nature. 2009;461:654–658. doi: 10.1038/nature08455. [DOI] [PubMed] [Google Scholar]
- 35.Suzuki K, Kubota Y, Sekito T, Ohsumi Y. Hierarchy of Atg proteins in pre-autophagosomal structure organization. Genes Cells. 2007;12:209–218. doi: 10.1111/j.1365-2443.2007.01050.x. [DOI] [PubMed] [Google Scholar]
- 36.van Leyen K, Duvoisin RM, Engelhardt H, Wiedmann M. A function for lipoxygenase in programmed organelle degradation. Nature. 1998;395:392–395. doi: 10.1038/26500. [DOI] [PubMed] [Google Scholar]
- 37.Grullich C, Duvoisin RM, Wiedmann M, van Leyen K. Inhibition of 15-lipoxygenase leads to delayed organelle degradation in the reticulocyte. FEBS Lett. 2001;489:51–54. doi: 10.1016/s0014-5793(01)02080-4. [DOI] [PubMed] [Google Scholar]
- 38.Vijayvergiya C, et al. High-level expression of rabbit 15-lipoxygenase induces collapse of the mitochondrial pH gradient in cell culture. Biochemistry. 2004;43:15296–15302. doi: 10.1021/bi048745v. [DOI] [PubMed] [Google Scholar]
- 39.Tooze J, Davies HG. Cytolysosomes in amphibian erythrocytes. J. Cell Biol. 1965;24:146–150. doi: 10.1083/jcb.24.1.146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kent G, Minick OT, Volini FI, Orfei E. Autophagic vacuoles in human red cells. Am. J. Pathol. 1966;48:831–857. [PMC free article] [PubMed] [Google Scholar]
- 41.Gronowicz G, Swift H, Steck TL. Maturation of the reticulocyte in vitro. J. Cell Sci. 1984;71:177–197. doi: 10.1242/jcs.71.1.177. [DOI] [PubMed] [Google Scholar]
- 42.Heynen MJ, Tricot G, Verwilghen RL. Autophagy of mitochondria in rat bone marrow erythroid cells. Relation to nuclear extrusion. Cell Tissue Res. 1985;239:235–239. doi: 10.1007/BF00214924. [DOI] [PubMed] [Google Scholar]
- 43.Takano-Ohmuro H, Mukaida M, Kominami E, Morioka K. Autophagy in embryonic erythroid cells: its role in maturation. Eur. J. Cell Biol. 2000;79:759–764. doi: 10.1078/0171-9335-00096. [DOI] [PubMed] [Google Scholar]
- 44.Matsui M, Yamamoto A, Kuma A, Ohsumi Y, Mizushima N. Organelle degradation during the lens and erythroid differentiation is independent of autophagy. Biochem. Biophys. Res. Commun. 2006;339:485–489. doi: 10.1016/j.bbrc.2005.11.044. [DOI] [PubMed] [Google Scholar]
- 45.Schweers RL, et al. NIX is required for programmed mitochondrial clearance during reticulocyte maturation. Proc. Natl Acad. Sci. USA. 2007;104:19500–19505. doi: 10.1073/pnas.0708818104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sandoval H, et al. Essential role for Nix in autophagic maturation of erythroid cells. Nature. 2008;454:232–235. doi: 10.1038/nature07006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Novak I, et al. Nix is a selective autophagy receptor for mitochondrial clearance. EMBO Rep. 2010;11:45–51. doi: 10.1038/embor.2009.256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Diwan A, et al. Unrestrained erythroblast development in Nix−/− mice reveals a mechanism for apoptotic modulation of erythropoiesis. Proc. Natl Acad. Sci. USA. 2007;104:6794–6799. doi: 10.1073/pnas.0610666104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chan EYW, Tooze SA. Evolution of Atg1 function and regulation. Autophagy. 2009;5:758–765. doi: 10.4161/auto.8709. [DOI] [PubMed] [Google Scholar]
- 50.Mizushima N. The role of the Atg1/ULK1 complex in autophagy regulation. Curr. Opin. Cell Biol. 2010;22:132–139. doi: 10.1016/j.ceb.2009.12.004. [DOI] [PubMed] [Google Scholar]
- 51.Kundu M, et al. Ulk1 plays a critical role in the autophagic clearance of mitochondria and ribosomes during reticulocyte maturation. Blood. 2008;112:1493–1502. doi: 10.1182/blood-2008-02-137398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhang J, et al. Mitochondrial clearance is regulated by Atg7-dependent and -independent mechanisms during reticulocyte maturation. Blood. 2009;114:157–164. doi: 10.1182/blood-2008-04-151639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Mortensen M, et al. Loss of autophagy in erythroid cells leads to defective removal of mitochondria and severe anemia in vivo. Proc. Natl Acad. Sci. USA. 2010;107:832–837. doi: 10.1073/pnas.0913170107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Schwarten M, et al. Nix directly binds to GABARAP: a possible crosstalk between apoptosis and autophagy. Autophagy. 2009;5:690–698. doi: 10.4161/auto.5.5.8494. [DOI] [PubMed] [Google Scholar]
- 55.Pua HH, Guo J, Komatsu M, He YW. Autophagy is essential for mitochondrial clearance in mature T lymphocytes. J. Immunol. 2009;182:4046–4055. doi: 10.4049/jimmunol.0801143. [DOI] [PubMed] [Google Scholar]
- 56.Stephenson LM, et al. Identification of Atg5-dependent transcriptional changes and increases in mitochondrial mass in Atg5-deficient T lymphocytes. Autophagy. 2009;5:625–635. doi: 10.4161/auto.5.5.8133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Pua HH, Dzhagalov I, Chuck M, Mizushima N, He YW. A critical role for the autophagy gene Atg5 in T cell survival and proliferation. J. Exp. Med. 2007;204:25–31. doi: 10.1084/jem.20061303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Nedjic J, Aichinger M, Emmerich J, Mizushima N, Klein L. Autophagy in thymic epithelium shapes the T-cell repertoire and is essential for tolerance. Nature. 2008;455:396–400. doi: 10.1038/nature07208. [DOI] [PubMed] [Google Scholar]
- 59.Miller BC, et al. The autophagy gene ATG5 plays an essential role in B lymphocyte development. Autophagy. 2008;4:309–314. doi: 10.4161/auto.5474. [DOI] [PubMed] [Google Scholar]
- 60.Singh R, et al. Autophagy regulates adipose mass and differentiation in mice. J. Clin. Invest. 2009;119:3329–3339. doi: 10.1172/JCI39228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Shibata M, et al. LC3, a microtubule-associated protein1A/B light chain3, is involved in cytoplasmic lipid droplet formation. Biochem. Biophys. Res. Commun. 2010;393:274–279. doi: 10.1016/j.bbrc.2010.01.121. [DOI] [PubMed] [Google Scholar]
- 62.Zhang Y, et al. Adipose-specific deletion of autophagy-related gene 7 (atg7) in mice reveals a role in adipogenesis. Proc. Natl Acad. Sci. USA. 2009;106:19860–19865. doi: 10.1073/pnas.0906048106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Cypess AM, et al. Identification and importance of brown adipose tissue in adult humans. N. Engl. J. Med. 2009;360:1509–1517. doi: 10.1056/NEJMoa0810780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.van Marken Lichtenbelt WD, et al. Cold-activated brown adipose tissue in healthy men. N. Engl. J. Med. 2009;360:1500–1508. doi: 10.1056/NEJMoa0808718. [DOI] [PubMed] [Google Scholar]
- 65.Virtanen KA, et al. Functional brown adipose tissue in healthy adults. N. Engl. J. Med. 2009;360:1518–1525. doi: 10.1056/NEJMoa0808949. [DOI] [PubMed] [Google Scholar]
- 66.Gesta S, Tseng YH, Kahn CR. Developmental origin of fat: tracking obesity to its source. Cell. 2007;131:242–256. doi: 10.1016/j.cell.2007.10.004. [DOI] [PubMed] [Google Scholar]
- 67.Shibata M, et al. The MAP1–LC3 conjugation system is involved in lipid droplet formation. Biochem. Biophys. Res. Commun. 2009;382:419–423. doi: 10.1016/j.bbrc.2009.03.039. [DOI] [PubMed] [Google Scholar]
- 68.Singh R, et al. Autophagy regulates lipid metabolism. Nature. 2009;458:1131–1135. doi: 10.1038/nature07976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kuma A, Matsui M, Mizushima N. LC3, an autophagosome marker, can be incorporated into protein aggregates independent of autophagy: caution in the interpretation of LC3 localization. Autophagy. 2007;3:323–328. doi: 10.4161/auto.4012. [DOI] [PubMed] [Google Scholar]
- 70.Hara T, et al. Suppression of basal autophagy in neural cells causes neurodegenerative disease in mice. Nature. 2006;441:885–889. doi: 10.1038/nature04724. [DOI] [PubMed] [Google Scholar]
- 71.Komatsu M. Loss of autophagy in the central nervous system causes neurodegeneration in mice. Nature. 2006;441:880–884. doi: 10.1038/nature04723. [DOI] [PubMed] [Google Scholar]
- 72.Liang CC, Wang C, Peng X, Gan B, Guan JL, et al. Neural specific deletion of FIP200 leads to cerebellar degeneration caused by increased neuronal death and axon degeneration. J. Biol. Chem. 2010;285:3499–3509. doi: 10.1074/jbc.M109.072389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Nakai A, et al. The role of autophagy in cardiomyocytes in the basal state and in response to hemodynamic stress. Nat. Med. 2007;13:619–624. doi: 10.1038/nm1574. [DOI] [PubMed] [Google Scholar]
- 74.Raben N, et al. Suppression of autophagy in skeletal muscle uncovers the accumulation of ubiquitinated proteins and their potential role in muscle damage in Pompe disease. Hum. Mol. Genet. 2008;17:3897–3908. doi: 10.1093/hmg/ddn292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Masiero E, et al. Autophagy is required to maintain muscle mass. Cell Metab. 2009;10:507–515. doi: 10.1016/j.cmet.2009.10.008. [DOI] [PubMed] [Google Scholar]
- 76.Ebato C, et al. Autophagy is important in islet homeostasis and compensatory increase of beta cell mass in response to high-fat diet. Cell Metab. 2008;8:325–332. doi: 10.1016/j.cmet.2008.08.009. [DOI] [PubMed] [Google Scholar]
- 77.Jung HS, et al. Loss of autophagy diminishes pancreatic beta cell mass and function with resultant hyperglycemia. Cell Metab. 2008;8:318–324. doi: 10.1016/j.cmet.2008.08.013. [DOI] [PubMed] [Google Scholar]
- 78.Hartleben B, et al. Autophagy influences glomerular disease susceptibility and maintains podocyte homeostasis in aging mice. J. Clin. Invest. 2010;120:1084–1096. doi: 10.1172/JCI39492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Bjørkøy G, et al. p62/SQSTM1 forms protein aggregates degraded by autophagy and has a protective effect on huntingtin-induced cell death. J. Cell Biol. 2005;171:603–614. doi: 10.1083/jcb.200507002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Komatsu M, et al. Homeostatic levels of p62 control cytoplasmic inclusion body formation in autophagy-deficient mice. Cell. 2007;131:1149–1163. doi: 10.1016/j.cell.2007.10.035. [DOI] [PubMed] [Google Scholar]
- 81.Komatsu M, et al. The selective autophagy substrate p62 activates the stress responsive transcription factor Nrf2 through inactivation of Keap1. Nat. Cell Biol. 2010;12:213–223. doi: 10.1038/ncb2021. [DOI] [PubMed] [Google Scholar]
- 82.Lau A, et al. A non-canonical mechanism of Nrf2 activation by autophagy deficiency: a direct interaction between Keap1 and p62. Mol. Cell. Biol. 2010;30:3275–3285. doi: 10.1128/MCB.00248-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Jain A, et al. p62/SQSTM1 is a target gene for transcription factor NRF2 and creates a positive feedback loop by inducing antioxidant response element-driven gene transcription. J. Biol. Chem. 2010;285:22576–22591. doi: 10.1074/jbc.M110.118976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Copple IM, et al. Physical and functional interaction of sequestosome 1 with Keap1 regulates the Keap1–Nrf2 cell defence pathway. J. Biol. Chem. 2010;285:16782–16788. doi: 10.1074/jbc.M109.096545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Kitada T, et al. Mutations in the parkin gene cause autosomal recessive juvenile parkinsonism. Nature. 1998;392:605–608. doi: 10.1038/33416. [DOI] [PubMed] [Google Scholar]
- 86.Narendra D, Tanaka A, Suen DF, Youle RJ. Parkin is recruited selectively to impaired mitochondria and promotes their autophagy. J. Cell Biol. 2008;183:795–803. doi: 10.1083/jcb.200809125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Vives-Bauza C, et al. PINK1-dependent recruitment of Parkin to mitochondria in mitophagy. Proc. Natl Acad. Sci. USA. 2010;107:378–383. doi: 10.1073/pnas.0911187107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Geisler S, et al. PINK1/Parkin-mediated mitophagy is dependent on VDAC1 and p62/SQSTM1. Nat. Cell Biol. 2010;12:119–131. doi: 10.1038/ncb2012. [DOI] [PubMed] [Google Scholar]
- 89.Narendra DP, et al. PINK1 is selectively stabilized on impaired mitochondria to activate Parkin. PLoS Biol. 2010;8 doi: 10.1371/journal.pbio.1000298. e1000298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Kawajiri S, et al. PINK1 is recruited to mitochondria with parkin and associates with LC3 in mitophagy. FEBS Lett. 2010;584:1073–1079. doi: 10.1016/j.febslet.2010.02.016. [DOI] [PubMed] [Google Scholar]
- 91.Ziviani E, Tao RN, Whitworth AJ. Drosophila parkin requires PINK1 for mitochondrial translocation and ubiquitinates mitofusin. Proc. Natl Acad. Sci. USA. 2010;107:5018–5023. doi: 10.1073/pnas.0913485107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Matsuda N, et al. PINK1 stabilized by mitochondrial depolarization recruits Parkin to damaged mitochondria and activates latent Parkin for mitophagy. J. Cell Biol. 2010;189:211–221. doi: 10.1083/jcb.200910140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Tsukada M, Ohsumi Y. Isolation and characterization of autophagy-defective mutants of Saccharomyces cerevisiae. FEBS Lett. 1993;333:169–174. doi: 10.1016/0014-5793(93)80398-e. [DOI] [PubMed] [Google Scholar]
- 94.Kohda TA, et al. Fission yeast autophagy induced by nitrogen starvation generates a nitrogen source that drives adaptation processes. Genes Cells. 2007;12:155–170. doi: 10.1111/j.1365-2443.2007.01041.x. [DOI] [PubMed] [Google Scholar]
- 95.Mukaiyama H, et al. Autophagy-deficient Schizosaccharomyces pombe mutants undergo partial sporulation during nitrogen starvation. Microbiology. 2009;155:3816–3826. doi: 10.1099/mic.0.034389-0. [DOI] [PubMed] [Google Scholar]
- 96.Otto GP, Wu MY, Kazgan N, Anderson OR, Kessin RH. Macroautophagy is required for multicellular development of the social amoeba Dictyostelium discoideum. J. Biol. Chem. 2003;278:17636–17645. doi: 10.1074/jbc.M212467200. [DOI] [PubMed] [Google Scholar]
- 97.Otto GP, Wu MY, Kazgan N, Anderson OR, Kessin RH. Dictyostelium macroautophagy mutants vary in the severity of their developmental defects. J. Biol. Chem. 2004;279:15621–15629. doi: 10.1074/jbc.M311139200. [DOI] [PubMed] [Google Scholar]
- 98.Nolting N, Bernhards Y, Poggeler S. SmATG7 is required for viability in the homothallic ascomycete Sordaria macrospora. Fungal Genet. Biol. 2009;46:531–542. doi: 10.1016/j.fgb.2009.03.008. [DOI] [PubMed] [Google Scholar]
- 99.Pinan-Lucarre B, Paoletti M, Dementhon K, Coulary-Salin B, Clave C. Autophagy is induced during cell death by incompatibility and is essential for differentiation in the filamentous fungus Podospora anserina. Mol. Microbiol. 2003;47:321–333. doi: 10.1046/j.1365-2958.2003.03208.x. [DOI] [PubMed] [Google Scholar]
- 100.Besteiro S, Williams RA, Morrison LS, Coombs GH, Mottram JC. Endosome sorting and autophagy are essential for differentiation and virulence of Leishmania major. J. Biol. Chem. 2006;281:11384–11396. doi: 10.1074/jbc.M512307200. [DOI] [PubMed] [Google Scholar]
- 101.Alvarez VE, et al. Autophagy is involved in nutritional stress response and differentiation in Trypanosoma cruzi. J. Biol. Chem. 2008;283:3454–3464. doi: 10.1074/jbc.M708474200. [DOI] [PubMed] [Google Scholar]
- 102.Melendez A, et al. Autophagy genes are essential for dauer development and life-span extension in C. elegans. Science. 2003;301:1387–1391. doi: 10.1126/science.1087782. [DOI] [PubMed] [Google Scholar]
- 103.Takacs-Vellai K, et al. Inactivation of the autophagy gene bec-1 triggers apoptotic cell death in C. elegans. Curr. Biol. 2005;15:1513–1517. doi: 10.1016/j.cub.2005.07.035. [DOI] [PubMed] [Google Scholar]
- 104.Juhasz G, Csikos G, Sinka R, Erdelyi M, Sass M. The Drosophila homolog of Aut1 is essential for autophagy and development. FEBS Lett. 2003;543:154–158. doi: 10.1016/s0014-5793(03)00431-9. [DOI] [PubMed] [Google Scholar]
- 105.Berry DL, Baehrecke EH. Growth arrest and autophagy are required for salivary gland cell degradation in Drosophila. Cell. 2007;131:1137–1148. doi: 10.1016/j.cell.2007.10.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Denton D, et al. Autophagy, not apoptosis, is essential for midgut cell death in Drosophila. Curr. Biol. 2009;19:1741–1746. doi: 10.1016/j.cub.2009.08.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Shen W, Ganetzky B. Autophagy promotes synapse development in Drosophila. J. Cell Biol. 2009;187:71–79. doi: 10.1083/jcb.200907109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Hanaoka H, et al. Leaf senescence and starvation-induced chlorosis are accelerated by the disruption of an Arabidopsis autophagy gene. Plant Physiol. 2002;129:1181–1193. doi: 10.1104/pp.011024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Doelling JH, Walker JM, Friedman EM, Thompson AR, Veirstra RD. The APG8/12-activating enzyme APG7 is required for proper nutrient recycling and senescence in Arabidopsis thaliana. J. Biol. Chem. 2002;277:33105–33114. doi: 10.1074/jbc.M204630200. [DOI] [PubMed] [Google Scholar]
- 110.Yoshimoto K, et al. Processing of ATG8s, ubiquitin-like proteins, and their deconjugation by ATG4s are essential for plant autophagy. Plant Cell. 2004;16:2967–2983. doi: 10.1105/tpc.104.025395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Juhasz G, Erdi B, Sass M, Neufeld TP. Atg7-dependent autophagy promotes neuronal health, stress tolerance, and longevity but is dispensable for metamorphosis in Drosophila. Genes Dev. 2007;21:3061–3066. doi: 10.1101/gad.1600707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Mariño G, et al. Tissue-specific autophagy alterations and increased tumorigenesis in mice deficient in ATG4C/autophagin-3. J. Biol. Chem. 2007;282:18573–18583. doi: 10.1074/jbc.M701194200. [DOI] [PubMed] [Google Scholar]
- 113.Cann GM, et al. Developmental expression of LC3α and β: absence of fibronectin or autophagy phenotype in LC3β knockout mice. Dev. Dyn. 2007;237:187–195. doi: 10.1002/dvdy.21392. [DOI] [PubMed] [Google Scholar]
- 114.O’Sullivan GA, Kneussel M, Elazar Z, Betz H. GABARAP is not essential for GABA receptor targeting to the synapse. Eur. J. Neurosci. 2005;22:2644–2648. doi: 10.1111/j.1460-9568.2005.04448.x. [DOI] [PubMed] [Google Scholar]
- 115.Komatsu M, et al. Essential role for autophagy protein Atg7 in the maintenance of axonal homeostasis and the prevention of axonal degeneration. Proc. Natl Acad. Sci. USA. 2007;104:14489–14494. doi: 10.1073/pnas.0701311104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Nishiyama J, Miura E, Mizushima N, Watanabe M, Yuzaki M. Aberrant membranes and double-membrane structures accumulate in the axons of Atg5-null Purkinje cells before neuronal death. Autophagy. 2007;3:591–596. doi: 10.4161/auto.4964. [DOI] [PubMed] [Google Scholar]
- 117.Hashimoto D, et al. Involvement of autophagy in trypsinogen activation within the pancreatic acinar cells. J. Cell Biol. 2008;181:1065–1072. doi: 10.1083/jcb.200712156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Lee HK, et al. In vivo requirement for Atg5 in antigen presentation by dendritic cells. Immunity. 2010;32:227–239. doi: 10.1016/j.immuni.2009.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]