Abstract
Autophagy is a tightly regulated pathway involving the lysosomal degradation of cytoplasmic organelles or cytosolic components. This pathway can be stimulated by multiple forms of cellular stress, including nutrient or growth factor deprivation, hypoxia, reactive oxygen species, DNA damage, protein aggregates, damaged organelles or intracellular pathogens. Both specific, stimulus-dependent and more general stimulus-independent signaling pathways are activated to coordinate different phases of autophagy. Autophagy can be integrated with other cellular stress responses through parallel stimulation of autophagy and other stress responses by specific stress stimuli, through dual regulation of autophagy and other stress responses by multi-functional stress signaling molecules, and/or through mutual control of autophagy and other stress responses. Thus, autophagy is a cell biological process that is a central component of the integrated stress response.
Introduction
Eukaryotic cells must adapt continuously to fluctuations in external conditions, including physical parameters, such as temperature and ultraviolet light; chemical cues such as ion concentrations, pH, oxygen tension, redox potentials and metabolite concentrations; extracellular signals such as contact-dependent signals, hormones, cytokines and neurotransmitters; and microbial pathogens. Beyond a certain threshold, such fluctuations are considered ‘stresses’, meaning that the cell's response to this stress determines whether it can function properly and survive.
During the response to sublethal stress, cells undergo rapid changes to adapt their metabolism and protect themselves against potential damage. This is orchestrated through a multifaceted cellular program, which involves the concerted action of diverse stress response pathways. One of the key pathways that mediates stress-induced metabolic adaptation and damage control is macroautophagy, referred to simply as “autophagy”. During autophagy, cells form double-membraned vesicles, autophagosomes, that sequester organelles, proteins, or portions of the cytoplasm, for delivery to the lysosome (He and Klionsky, 2009). The sequestered contents are degraded in the lysosome, allowing cells to eliminate damaged or harmful components through catabolism and recycling to maintain nutrient and energy homeostasis. Autophagy constitutes a major protective mechanism that allows cells to survive in response to multiple stressors and that helps defend organisms against degenerative, inflammatory, infectious, and neoplastic diseases (Levine and Kroemer, 2008; Mizushima et al., 2008).
Besides autophagy, the cellular response to stress involves numerous other pathways including those that regulate nutrient uptake, intermediary metabolism, cell cycle and growth control, cell fate and lineage decisions, and cellular survival/death programs. Therefore, it is not surprising that there is a close integration between signals that regulate these cellular processes and those that regulate autophagy. In this review, we will summarize recent advances in understanding how different cellular stress signals and stress stimuli regulate autophagy.
The Core Pathway of Mammalian Autophagy
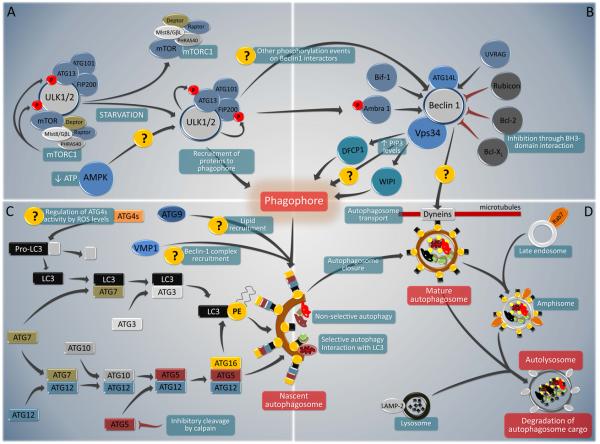
The core pathway of mammalian autophagy (Fig. 1) begins with the formation of an isolation membrane (also called a phagophore) and involves at least five molecular components, including: (1) the Atg1/unc-51-like kinase (ULK) complex; (2) the Beclin 1/class III phosphatidylinositol 3-kinase (PI3K) complex; (3) two transmembrane proteins, Atg9 and vacuole membrane protein 1 (VMP1); (4) two ubiquitin-like protein (Atg12 and Atg8/LC3) conjugation systems; and (5) proteins that mediate fusion between autophagosomes and lysosomes (Yang and Klionsky, 2010). Some of these core autophagy pathway components are directly controlled by cellular stress signals (Fig. 1).
FIGURE 1. Overview of the major components of the core pathway of mammalian autophagy.
Several key molecular components participate in the initiation, execution and completion of autophagy. Autophagy inducers such as starvation modulate the inhibitory interaction of TORC1 with the ULK1/2 complex. Through phosphorylation of Ambra1, and maybe through other putative interactions, ULK1/2 complex (A) also regulates the activity of Beclin 1/ class III phosphatidylinositol 3-kinase (PI3K) complex (B). Beclin 1 interacts with several enhancing (blue) or inhibitory (grey) factors that modulate its binding to Vps34, the catalytic unit of the PI3K, whose lipid kinase activity is essential for autophagy. In addition to these two complexes, autophagosome formation requires the participation of two ubiquitin-like protein (Atg12 and Atg8/LC3) conjugation systems and two transmembrane proteins (Atg9 and VMP-1) (C). Whereas the roles of Atg9 and VMP-1 are currently not completely understood, both conjugation systems are essential for the biogenesis of the isolation membrane, also called ‘phagophore’. In addition, the Atg8/LC3 system is required for autophagosome transport and maturation, as well as for the selection of autophagic cargo. Fully mature autophagosomes can fuse with Rab7-positive late endosomes to form amphisomes. Finally, autophagosomes or amphisomes fuse their external membranes with those from acidic lysosomes to acquire hydrolytic activity, degrade their cargo and recycle essential biomolecules to the cytoplasm (D).
Origin of the isolation membrane
The isolation membrane (also called ‘phagophore’) can be generated from multiple sources that include the ER (Axe et al., 2008; Hayashi-Nishino et al., 2009; Yla-Anttila et al., 2009), the outer mitochondrial membrane (Hailey et al., 2010), and the plasma membrane (Ravikumar et al., 2010). PI3P is required for the formation of ‘omegasomes’, the Ω-shaped structures that bud from the ER during the initial steps of vesicle nucleation/autophagosome formation (Axe et al., 2008). ATG16L1 directly interacts with clathrin, which connects the endocytic pathway to autophagy (Ravikumar et al., 2010). While it appears that general autophagy inducers such as starvation stimulate lipid recruitment from all known sources, it is not known whether specific stimuli (such as ER stress, mitochondrial damage or signals emanating from the plasma membrane) cause autophagosomes to be preferentially formed from specific membrane sources.
The Atg1/ULK Complex
The mammalian orthologs of yeast Atg1, ULK1 and ULK2, play a key role in autophagy induction, acting downstream of the mammalian target of rapamycin (mTOR) complex 1 (mTORC1, a polyprotein complex that contains mTOR, Raptor, mLST8/GßL, Deptor and PRAS40 (Efeyan and Sabatini, 2010)). In normal (nutrient-replete) conditions, mTORC1 possesses kinase activity and interacts with a complex that contains ULK1, Atg13, FIP200 and Atg101 (Fig. 1A). Upon mTORC1 inhibition, for example by starvation (see below), mTORC1 dissociates from the ULK complex, leading to dephosphorylation of specific residues within ULK1 (or ULK2) and Atg13 (which are normally phosphorylated by mTORC1), the catalytic activation of ULK1 (or ULK2) and the ULK1 (or ULK2)-mediated phosphorylation of other residues in Atg13 and FIP200 (Fig. 1A). Thus, mTORC1 inhibition is probably coupled to ULK1 (or ULK2) activation through a process that involves dissociation of a large protein complex and (de)phosphorylation events (Mizushima, 2010). An intriguing, but not yet tested, possibility, is that the ULK1/ULK2 complex might also be positively regulated in low energy conditions by its recently described interactions with several subunits of the energy-sensing kinase, AMP-activated protein kinase (AMPK) (Behrends et al., 2010). It is not entirely known how ULK1/2 activates downstream components of the autophagic machinery. However, ULK1 can phosphorylate Ambra1 (Di Bartolomeo et al., 2010), a component of the Beclin 1/Class III PI3K complex (He and Levine, 2010) (Fig. 1A).
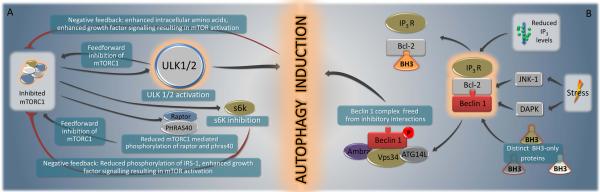
There are multiple circuits of positive and negative feedback in mTORC1-mediated autophagy regulation (Neufeld, 2010) (Fig. 2A). Feed-forward mechanisms involve Deptor, PRAS40 and ULK1/2, which are each inhibited by mTORC1-mediated phosphorylation and, in turn, inhibit mTOR activity. This may lead to amplification of initially modest changes in TOR activity. Negative feedback control is achieved via the products of autophagy including amino acids as well as via inhibition of S6 kinase, a protein that has mTORCI-dependent activity and which may be necessary for sustained autophagy (Neufeld, 2010). Thus, mTORC1 is part of a rheostat that is either switched off (to inhibit autophagy at the level of ULK1/2) or on (to induce autophagy by ULK1/2 activation, as a result of a positive amplification loop). When on, these effects are self-limited due to the presence of negative feedback loops (Fig. 2A).
FIGURE 2. Overview of selected signal transduction pathways that regulate autophagy components that function in vesicle nucleation/phagophore formation.
Selected signals that converge on ULK1/2 (A) and the Beclin 1 complex (B) are depicted. Note the multiple positive and negative feedback loops depicted in A. For details see text.
Beclin 1 and its Interactome
The “Beclin 1 core complex” involves Beclin 1, Vps15, Vps34 and likely, Ambra1 (He and Levine, 2010). This multiprotein complex must be formed for the allosteric activation of the class III PI3K Vps34 to generate phosphatidylinositol-3-phosphate (PI3P) which recruits effectors such as the double FYVE domain-containing protein 1 (DFCP1) (Axe et al., 2008) and WD-repeat protein interacting with phosphoinositides (WIPI) family proteins (Polson et al., 2010) to mediate the initial stages of vesicle nucleation/autophagosome formation. Numerous proteins that interact with Beclin 1 induce or inhibit autophagy (Fig. 1B). Atg14 (also called Atg14L or Barkor, for Beclin 1-associated autophagy-related key regulator) is essential for PI3K activity and autophagy induction. UVRAG (UV radiation resistance-associated gene) competes with Atg14 for binding to Beclin 1 and may promote PI3K activity in a cell type-specific fashion, and through interactions with class C Vps/HOPS complexes, promotes autophagosome fusion with the late endosome/lysosome. Bif-1/endophilin B1 interacts with Beclin 1 via UVRAG to function as a positive regulator of the PI3K complex, and has an N-BAR domain that may promote membrane curvature (Fig. 1B). Rubicon (RUN domain protein as Beclin 1 interacting and cysteine-rich containing) negatively regulates autophagy (as well as endocytic trafficking) through its interaction with Beclin 1/PI3K complexes (Fig. 1B).
Anti-apoptotic family members (such as Bcl-2, Bcl-XL and Mcl-1) are also important negative regulators of autophagy through an inhibitory interaction of their BH3-binding groove with the BH3 domain of Beclin 1 (Maiuri et al., 2007; Pattingre et al., 2005) (Fig. 2B). There are several distinct mechanisms through which autophagy-inducing signals can disrupt this inhibitory interaction, including competition by pro-apoptotic BH3-only proteins (such as BNIP3, Bad, Bik, Noxa, Puma and BimEL) (Maiuri et al., 2007), phosphorylation of the BH3 domain of Beclin 1 by death-associated protein kinase (DAPK) (Zalckvar et al., 2009) or phosphorylation of Bcl-2 by c-Jun N-teminal kinase-1 (JNK1) (Pattingre et al., 2009; Wei et al., 2008). Accordingly, BH3-only proteins (such as BAD in starvation or BNIP3 in hypoxia) (Bellot et al., 2009; Maiuri et al., 2007) as well as Beclin 1/Bcl-2 dissociating kinases (such as JNK1 in starvation conditions) (Wei et al., 2008) are required for autophagy induction in response to specific forms of stress.
The inositol-1,4,5 trisphosphate (IP3) receptor, which is an IP3-activated calcium channel at the endoplasmic reticulum (ER), interacts with Beclin 1 indirectly, via Bcl-2 (Fig. 2B). Upon cellular reduction of IP3 levels (or antagonist binding) and starvation, this interaction is disrupted (Vicencio et al., 2009). This mechanism may explain how agents that cause a reduction of IP3 levels cause autophagy in an mTOR-independent fashion (Sarkar et al., 2007). The toll-like receptor (TLR) adaptor molecules MyD88 and TRIF also may interact with Beclin 1, thereby reducing the binding of Beclin 1 to Bcl-2 and promoting autophagy (Shi and Kehrl, 2008). During TLR4-induced autophagy, tumor necrosis factor receptor (TNFR)-associated factor 6, TRAF6, interacts with Beclin 1 and mediates K63-linked ubiquitination of Beclin 1, which enhances Class III PI3K activity (Shi and Kehrl, 2010). The protein PINK1, which has been studied as a serine-threonine kinase that interacts with Parkin to stimulate mitophagy, also interacts with Beclin 1 (Michiorri et al., 2010). Although it is not clear how all these proteins affect the overall composition and function of the multiprotein Beclin 1-containing complex, an attractive model is that multiple proteins directly or indirectly interact with Beclin 1 to relay extracellular and intracellular stress signals to the autophagic machinery.
Atg9 and VMP1
Atg9 may provide lipids to the phagophore membrane by cycling between distinct subcellular compartments (Fig. 1C). The cycling of Atg9 requires Atg1/ULK1 and the kinase activity of Vps34. Another possibility is that Atg9 cycling involves the UVRAG/Bif-1-containing Beclin 1 complex since Bif-1 transiently associates with Atg9 after starvation (Orsi et al., 2010). VMP1, which interacts with Beclin 1, may function as a transmembrane protein that recruits Beclin 1 (and other components of the Beclin 1 complex) to the phagophore (Fig. 1C). It also interacts with TP53INP2 (tumor protein 53-induced nuclear protein 2), which like VMP1 is essential for autophagy and the translocation of Beclin 1 and LC3 to autophagosomes (Yang and Klionsky, 2010).
Conjugation Systems
Two ubiquitin-like conjugation systems are part of the vesicle elongation process (Fig. 1C). The first pathway involves the covalent conjugation of Atg12 to Atg5, with the help of the E1-like enzyme Atg7 and the E2-like enzyme Atg10. This conjugate is organized into a complex by associating with Atg16 in a non-covalent fashion to form the multimeric Atg12-Atg5-Atg16 complex, which functions as the E3 ligase of LC3 (Yang and Klionsky, 2010). The abundance of Atg5 may be regulated by calcium-dependent activation of the cysteine protease, calpain, which cleaves and inactivates Atg5, at least in cultured cells. Thus, the reduction of cytosolic Ca2+ (or calpain inactivation) may prevent Atg5 cleavage (Fig. 1C), resulting in increased cellular levels of full-length Atg5 and the pro-autophagic Atg12-Atg5 conjugate (Xia et al., 2010).
The second pathway involves the conjugation of phosphatidylethanolamine (PE) to a glycine (Gly) residue of yeast Atg8/mammalian LC3 by the sequential action of the protease Atg4, the E1-like enzyme Atg7, and the E2-like enzyme Atg3 (Fig. 1B). This lipid conjugation leads to the conversion of the soluble form of LC3 (named LC3-I) to the autophagic vesicle-associated form, LC3-II (Yang and Klionsky, 2010). The lipidated form of LC3 is stably associated with the autophagosome membrane, and its biochemical and microscopic detection is widely used to measure cellular autophagy (Mizushima et al., 2010) (Fig. 1C). In mammals, four Atg4 (Atg4A-B) and at least six orthologues of Atg8 exist, among which LC3B (hereafter referred to simply as LC3), GABARAP and GATE16 have been most studied (Weidberg et al., 2010).
Several stress signals regulate autophagy at the level of the Atg8/LC3 conjugation system. For example, the death-associated protein kinase, DAPK, positively regulates autophagy by associating with the LC3-interacting cytoskeleton molecule MAP1B (Harrison et al., 2008) (in addition to phosphorylating Beclin 1, discussed above (Zalckvar et al. 2009)). The cellular FLICE-like inhibitor protein, c-FLIP, negatively regulates autophagy by preventing Atg3 from binding and activating LC3 (Lee et al., 2009). Moreover, reactive oxygen species may regulate the active cysteine site of Atg4 (Fig. 1C) (Scherz-Shouval R et al. 2007). The enrichment of proteins with lipid kinase, WD40, and GTPase regulatory domains in the mammalian Atg8-interaction network (Behrends et al., 2010) may provide additional clues as to how stress signals interface with autophagy control at the level of Atg8/LC3 regulation.
Several proteins that possess an LC3-interacting region (LIR) and interact with LC3 (and its paralogs) serve as adaptors to target defined structures such as ubiquitinated proteins or mitochondria to the autophagic machinery (Fig. 1D). The best-characterized examples are p62 (also known as sequestosome1, SQSTM1) and NBR1 (Neighbor of BRCA1), which both recognize ubiquitinated proteins (Kirkin et al., 2009) as well as BNIP3L (also known as NIX), which binds to mitochondrial membranes (Novak et al., 2010). The regulation of these autophagy adaptor proteins is not yet well understood, but will likely be key to understanding how specific stress stimuli trigger selective autophagy.
Autophagy, Stress Stimuli, and Stress Signals
Autophagy is induced by a variety of stress stimuli, including nutrient and energy stress, ER stress, pathogen-associated molecular patterns (PAMPs) and danger-associated molecular patterns (DAMPs), hypoxia, redox stress, and mitochondrial damage. The stimulation of autophagy by these stimuli involves diverse signals that have overlapping functions in autophagy and the control of other cellular stress responses.
Autophagy Induced by Nutrient Stress
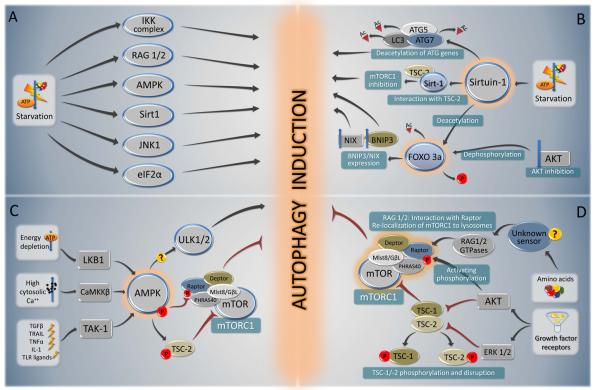
Nutrient depletion is the most potent known physiological inducer of autophagy. In the majority of cultured mammalian cells, nutrient depletion induces autophagy within minutes, with maximal levels observed when cells are cultured in the simultaneous absence of nutrients (such as amino acids and glucose) and growth factors (such as those contained in serum) (Boya et al., 2005). In mice, following starvation for 24-48 hours, most cells in most tissues display increased numbers of autophagosomes (Mizushima, 2010). Several critical molecules regulate starvation-induced autophagy (Fig. 3A); of these, mTOR and AMPK have been best characterized, and recent studies also suggest a crucial role for sirtuins.
FIGURE 3. Overview of the major signal transduction pathways that regulate autophagy in response to starvation.
A summary of starvation-induced pro-autophagic signaling (A) is followed by a schematic overview of the signaling cascades involving sirtuin-1 and Foxo 3a (B), AMPK (C) and mTORC1 (D).
Sirtuins and protein (de)acetylation
Sirtuins are a family of NAD-dependent deacetylases that sense environmental stress (Haigis and Sinclair, 2010). The induction of autophagy by starvation (but not by mTORC1 inhibition or ER stress) requires Sirt1 (Lee et al., 2008; Morselli et al., 2010). Accordingly, Sirt1−/− mice display a phenotype consistent with impaired autophagy, including increased levels of the autophagy substrate p62, accumulation of damaged organelles, disruption of energy homeostasis, and early perinatal lethality (Lee et al., 2008). The transfection of cells with sirtuin 1 (Sirt1) with intact deacetylase activity is sufficient to stimulate autophagy in cultured mammalian cells (Lee et al., 2008). In this context, it is intriguing that p300 acetyltransferase knockdown (Lee and Finkel, 2009), as well as histone acetylase inhibition by spermidine, a natural polyamine, potently induce autophagy (Eisenberg et al., 2009), suggesting that protein acetylation may play a general role in autophagy regulation.
Sirt1 can deacetylate Atg5, Atg7 and LC3 (Lee et al., 2008) (Fig. 3A), while p300 can acetylate Atg5, Atg7, LC3 and Atg12 (Lee and Finkel, 2009). Furthermore, Sirt1 deacetylates the transcription factor forkhead box O3a, FOXO3a, yet another hub of autophagy regulation, leading to enhanced expression of pro-autophagic Bnip3 (Kume et al., 2010). Akt inhibition resulting from growth factor depletion also causes FOXO3 activation, although via a different mechanism. Following its dephosphorylation, FOXO3 translocates into the nucleus (Fig. 3A) and upregulates multiple autophagy-related genes such as ULK2, Beclin 1, VPS34, BNIP3 and BNIP3L, ATG12, ATG4B, LC3, and GABARAPL1 (Mammucari et al., 2007). Another sirtuin, Sirt2, dissociates from FOXO1 upon serum starvation, thus causing the acetylation of FOXO1, favoring its interaction with Atg7 in the cytoplasm and stimulating autophagy (Zhao et al., 2010). Thus, protein (de)acetylation reactions influenced by sirtuins and other enzymes may control autophagy at multiple levels.
AMPK in starvation, energy depletion, and beyond
AMPK acts as a central node that integrates several stress stimuli with autophagy initiation (Fig. 3C). AMPK monitors the energy status of the cell by sensing its AMP:ATP ratio. Several upstream kinases, including liver kinase B1 (LKB1, which is activated by energy depletion), calcium/calmodulin kinase kinase-ß (CaMKKß, which is activated by cytosolic Ca2+), and TGFß-activated kinase-1 (TAK-1, which is also involved in IKK activation), can activate AMPK by phosphorylating a threonine residue on its catalytic α-subunit (Ruderman et al., 2010). In addition, Sirt1 and AMPK engage in a coordinated positive amplification loop (the “Sirt1/AMPK cycle”) (Ruderman et al., 2010), that acts to initiate autophagy in nutrient deprivation conditions. Sirt1-mediated deacetylation of LKB1 increases its serine-threonine kinase activity and stimulates its translocation from the nucleus to the cytoplasm where it activates AMPK. AMPK can also indirectly stimulate Sirt1 activation by the reduction of nicotinamide (NAM) and an increase in NAD+ that may be catalyzed by NAM phosphoribosyltransferase upregulation, thus completing the feed forward circuitry (Lan et al., 2008; Ruderman et al., 2010).
The best-studied mechanism by which AMPK induces autophagy is through mTORC1 inhibition (see below), via phosphorylation of the tuberous sclerosis complex 2 (TSC2) and the regulatory associated protein of mTOR, Raptor (Fig. 3D). The recent identification of an interaction between AMPK and ULK1 (Behrends et al., 2010) raises the speculative possibility that AMPK may also act more directly on core components of the autophagy machinery to initiate autophagy.
mTORC1 inhibition in starvation-induced autophagy
Besides inhibition by AMPK, mTORC1 is also inhibited upon withdrawal of growth factors such as insulin or insulin-like growth factor (Fig. 3C,D). In response to growth factors, Akt becomes catalytically active (due to the sequential stimulation of growth factor-activated class I PI3K and PI3K-dependent protein kinase 1, PDK1, which phosphorylates Akt). Moreover, growth factors activate Ras, which stimulates a cascade involving Raf-1, MEK1/2 and ERK1/2. Both Akt and ERK1/2 can phosphorylate one of two subunits of the tuberous sclerosis complex 1/2 (TSC1/TSC2), and Akt can phosphorylate Raptor, thus causing the activation of mTOR (Fig. 3D) (Neufeld, 2010). Moreover, amino acids activate mTORC1 independently of the Akt-TSC1/TSC2 axis, through the Rag family of GTPases, which directly interact with Raptor and recruit mTORC1 to the lysosomal surface (Efeyan and Sabatini, 2010) (Fig. 3D). Thus, mTOR is inhibited through multiple mechanisms during starvation conditions (Sengupta et al., 2010).
Additional kinases involved in starvation-induced autophagy
Several kinases besides AMPK function in starvation-induced upregulation of autophagy. This includes JNK1, which both phosphorylates Bcl-2, reducing its affinity for the BH3 domain of Beclin 1 (Wei et al., 2008), and phosphorylates Sirt1, promoting its enzymatic activity (Nasrin et al., 2009) (Fig. 3A). Moreover, as noted below, the phosphorylation of eIF2α (Kouroku et al., 2007; Talloczy et al., 2002) and the activation of IKK (Criollo et al., 2010) are essential for starvation-induced autophagy. Other kinases such as p38α mitogen-activated protein kinase (p38α MAPK, also known as MAPK14) potently inhibit starvation-induced autophagy. p38α MAPK acts on p38IP, which is required for starvation-induced mAtg9 trafficking and autophagosome formation (Webber and Tooze, 2010). Thus, the coordinated activation (and inhibition) of several kinases is essential for the induction of autophagy by nutrient depletion. The precise details of such coordination, however, remain poorly understood.
ER Stress and Autophagy
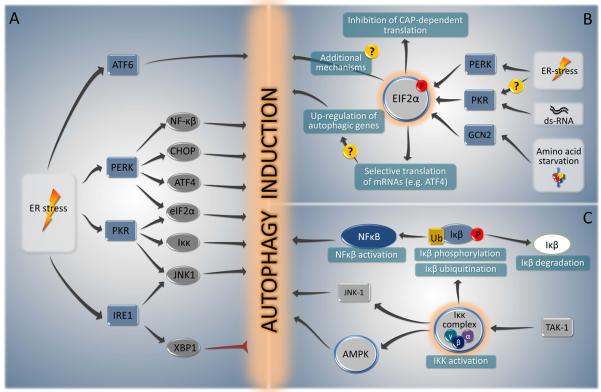
The ER is not only involved in protein synthesis and maturation (including correct folding), but may also constitute a major source/scaffold of the autophagic isolation membrane (Hayashi-Nishino et al., 2009; Yla-Anttila et al., 2009). The unfolded protein response (UPR), the major ER stress pathway (Buchberger et al., 2010), is a potent stimulus of autophagy. The three canonical branches of the UPR are mediated by three ER membrane-associated proteins, PERK (PKR-like eIF2α kinase, also known as EIF2AK3), ATF6 (activating transcription factor-6), and IRE1 (inositol requiring enzyme 1) (Fig. 4A), all of which are normally bound to and inactivated by the chaperone BiP/GRP78. The occupancy of BiP/GRP78 by misfolded proteins releases PERK, IRE1 and ATF6 from their inhibition. Among these, PERK and ATF6 acts as autophagy inducers, while IRE1 acts a negative regulator of autophagy.
FIGURE 4. Summary of the major signal transduction pathways that connect autophagy to ER stress (A), eIF2α phosphorylation (B) and IKK activation (C).
For details see text.
PERK and eIF2α phosphorylation
PERK mediates the transcriptional activation of the proteins LC3 and Atg5 in hypoxic responses through the action of the transcription factors ATF4 and CHOP, respectively (Rouschop et al., 2010). PERK may also reduce the translation of IkBα (Deng et al., 2004), thereby activating NF-κB, which also could contribute to autophagy (Fig. 4B). PERK phosphorylates the eukaryotic initiation factor 2α (eIF2α) on residue serine 51, which then initiates a cascade of events that decreases the overload of misfolded proteins, thereby alleviating ER stress (Harding et al., 2003). eIF2α phosphorylation initiates both a general inhibition of protein translation, as well as the selective translation of some stress-responsive transcripts including that of the ATF4 transcription factor and certain autophagy genes (Hotamisligil, 2010).
Cells that carry a non-phosphorylatable mutant of eIF2α (S51A) fail to induce autophagy in response to starvation, suggesting that eIF2α phosphorylation on serine 51 (and by extension eIF2α kinases) plays a major role in autophagy regulation (Kouroku et al., 2007; Talloczy et al., 2002). This phosphorylation event integrates various types of environmental and endogenous stress signals beyond ER stress, such as amino acid deprivation, exposure to double-stranded viral RNA, osmotic stress, UV light exposure, heme deficiency, hypoxia and oxidative stress (Harding et al., 2003). These divergent signals activate four different eIF2α kinases including PERK (which is activated by ER stress, radiation or hypoxia), general control non-derepressible-2 (GCN2, which is activated by uncharged tRNAs in amino acid-starved cells), heme-regulated inhibitor (HRI, which is activated by heme deficiency in erythroid precursor cells), and PKR (which is activated by double-stranded RNA and in some contexts, ER stress (Nakamura et al., 2010)) (Fig. 4B). Yeast GCN2 and mammalian PKR and PERK have been shown to be required for autophagy induced by starvation, viral infection, and ER stress, respectively (He and Klionsky, 2009). These findings illustrate the importance of eIF2α kinases in autophagy control, not only in the UPR but also in other conditions of stress.
How eIF2α phosphorylation contributes to autophagy initiation is not known. One highly speculative possibility is that eIF2α phosphorylation might somehow affect the ER in a manner that promotes the physical formation of the isolation membrane. Another possibility is that eIF2α phosphorylation stimulates autophagy via its effects on the transactivation of autophagy genes. eIF2α phosphorylation stimulates the selective translation of the ATF4 transcription factor (although general translation is shut off), which stimulates LC3 expression (which is necessary for sustained autophagy) (Milani et al., 2009) (Fig. 4B). Moreover, there are direct interactions between eIF2α subunits and core autophagy proteins, although it is not yet known whether these interactions are biologically significant (Behrends et al., 2010).
IRE1
IRE1 is a serine/threonine kinase (which activates JNK1) and also an endoribonuclease, which catalyzes the splicing of the mRNA encoding form of the transcription factor XBP1, which then transactivates UPR genes involved in ERAD, protein folding and protein quality control (Hetz and Glimcher, 2009; Hotamisligil, 2010). Perhaps unexpectedly, the inhibition of IRE1 and its downstream effector XBP1 enhances autophagy induction. Mice lacking XBP1 in neurons exhibit increased levels of baseline autophagy, which leads to increased turnover of an autophagic substrate, mutant superoxide dismutase-1 (SOD1), and protection against mutant SOD1-induced amyotrophic lateral sclerosis (Hetz et al., 2009). Since the predominant outcome of ER stress is the induction (rather than inhibition) of autophagy, it is possible that IRE1/XBP1 dependent signals have the function to dampen excessive autophagy triggered via the PERK/eIF2α pathway (Fig. 4A).
Immune Signals
Infection (or exposure of cells to microbial products) constitutes a specialized form of cellular stress that, in many cases, results in autophagy induction (Sumpter and Levine, 2010). Autophagy induction during infection is regulated by cytokines such as IFNγ (and downstream immunity-related GTPases) as well as pathogen recognition receptors (PRRs) that recognize conserved components of pathogens or products of their replication (PAMPs). The PRRs include families of Toll-like receptors (TLRs) that recognize PAMPS to activate pro-inflammatory cytokines and type I interferon production via NF-κB-, MAPK- and interferon-regulatory pathways; NOD-like receptors (NLRs) that primarily activate signaling via NF-κB and MAPK and components of the inflammasome; RIG-I like receptors (RLRs) that recognize cytoplasmic viral RNA and dA:dT rich dsDNA; C-type lectins, and the double-stranded RNA binding protein kinase PKR (Sumpter and Levine, 2010). Since these PRRs recognize not only PAMPS, but also DAMPs (which include products of necrotic cells, hypoxia, abnormal intracellular ion gradients, reactive oxygen species, and accumulation of misfolded proteins), this family of “immune signals” that regulates autophagy may represent a more generalized system that cells use to elicit autophagy in response to different forms of stress.
TAK1, IKK and NF-κB
The transcription factor, NF-κB, and some of its upstream regulators, function to integrate diverse stress signals including immune signals with the autophagy pathway. NF-κB is activated when the inhibitor of NF-κB (IκB) is phosphorylated by the IκB kinase (IKK), a kinase that is composed of one regulatory subunit (IKKγ, also known as NEMO) and two catalytic subunits (IKKα, IKKβ). The IKK complex is activated in response to multiple different stressors such as reactive oxygen species, DNA damage, and ligation of death receptors and PRRs. Frequently, this IKK activation is mediated by another upstream kinase, TAK1 (Fig. 4C). (Baud and Karin, 2009). In murine and human cells, knockout and/or knockdown of Tak1 or any of the Ikk subunits (but not that of the Nf-κb subunit p65) prevents the induction of autophagy in response to diverse stimuli including starvation, rapamycin, p53 inhibition, or ER stress (Criollo et al., 2010; Herrero-Martin et al., 2009). Conversely, expression of constitutively active IKK subunits stimulate autophagy through an NF-κB-independent pathway that relies on the activation of AMPK and JNK1 (Criollo et al., 2010). However, the NF-κB family member p65/RelA can upregulate beclin 1 expression and autophagy during T cell activation (Copetti et al., 2009) and NF-κB-incompetent cells are deficient in heat shock-induced autophagy (Nivon et al., 2009). Thus, there may be both IKK-dependent, NF-κB-independent as well as NF-κB-dependent mechanisms for autophagy induction. In addition, the p105 subunit of NF-κB can interact with certain autophagy proteins, including Beclin 1 (Behrends et al., 2010), but the significance of these interactions has yet to be explored.
Aside from the regulation of autophagy by NF-κB, autophagy may, in turn, regulate NF-κB signaling. For example, NF-κB signaling may be negatively regulated by targeting IKK and the NF-κB inducing kinase, NIK, for autophagic degradation (Qing et al., 2007). Furthermore, when autophagy is inhibited, p62 accumulates and activates the NF-κB signaling pathway (Mathew et al., 2009), suggesting that normal levels of autophagy may provide a “break” on NF-κB signaling.
PKR and the “meta-inflammasome”
An interesting recent study uncovers a potential relationship between an autophagy-regulatory eIF2α kinase, PKR, previously believed to respond primarily to virus infection (Talloczy et al., 2002; Talloczy et al., 2006), and the coordinate regulation of JNK1 and insulin signaling with ER stress and fatty acid exposure. PKR was found to respond to ER stress and fatty acid exposure, and be required for the JNK1 phosphorylation, serine phosphorylation of the insulin receptor substrate 1 (IRS1) and consequent insulin resistance that occurs in response to these stimuli (Nakamura et al., 2010). Based on these findings and a previous report that PKR activates the IKKβ-NF-κB pathway (Bonnet et al., 2000), Hotamisligil et al. (Hotamisligil, 2010) proposed the existence of a ‘meta-inflammasome’ composed of PKR, eIF2α, JNK, IRS, IKK and other components that link ER stress to more global stress responses, including inflammatory signaling and metabolic dysfunction. In the postulated “meta-inflammasome”, PKR phosphorylates eIF2α, (which induces autophagy), activates the ‘inflammatory kinases’ IKKß and JNK1 (which both induce autophagy), and phosphorylates (and inhibits) IRS1 (which also would induce autophagy) (Nakamura et al., 2010). Thus, this hypothetical molecular platform might explain the functional overlap and crosstalk between the JNK1, IKK and eIF2α pathways in the induction of autophagy.
It is, however, unclear whether this autophagy-promoting activity, rather than other downstream effects of the signaling molecules, would explain the pro-inflammatory and adverse metabolic effects of the “metainflammasome”, since genetic deletions of autophagy genes generally activates inflammatory signaling (Sumpter and Levine, 2010; Virgin and Levine, 2009) and a recent study found that hepatic suppression of the Atg7 autophagy gene results in increased ER stress and insulin resistance (Yang et al., 2010). Perhaps, the activation of autophagy by the metainflammasome serves as a negative feedback mechanism to limit ER stress; this hypothesis would reconcile the speculated role of the meta-inflammasome in autophagy activation with the cytoprotective roles of autophagy.
Hypoxia and Anoxia
Hypoxia and anoxia (with oxygen concentrations <3% and <0.1%, respectively) both cause autophagy through a variety of different mechanisms. Hypoxia-induced autophagy depends on hypoxia-inducible factor, HIF, while anoxia-induced autophagy is HIF-independent (Majmundar et al., 2010; Mazure and Pouyssegur, 2010). HIF is a heterodimer of a constitutive ß subunit and an oxygen-regulated α subunit that only becomes stabilized (and hence expressed) when oxygen concentration declines below a threshold of ~5%. Upon moderate hypoxia (1-3% oxygen), HIF activates the transcription of BNIP3 and BNIP3L (NIX), two BH3-only proteins that can disrupt the inhibitory interaction between Beclin 1 and Bcl-2 (Bellot et al., 2009) (Fig. 5A). Moreover, BNIP3L, which often is present at the outer surface of mitochondria, possesses a WXXL motif that binds to LC3 and its homolog GABARAP (Novak et al., 2010), thereby targeting mitochondria for autophagic destruction. The transcription of BNIP3 is also upregulated by the transcription factor FOXO3, provided that it is deacetylated by Sirt1 (Kume et al., 2010).
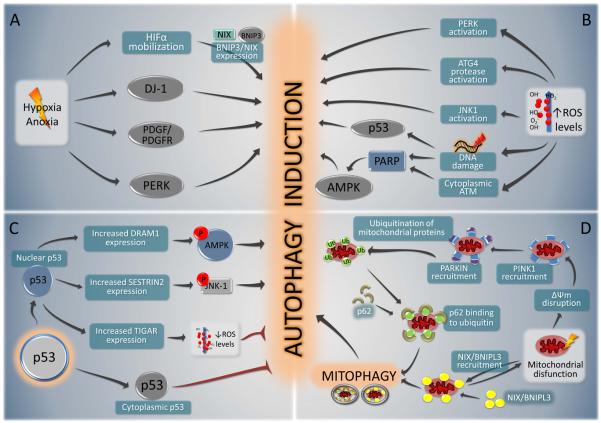
FIGURE 5. Overview of selected stress pathways that induce autophagy.
The mechanisms involved in autophagy induction by hypoxia or anoxia (A); increased oxidative damage (reactive oxygen species, ROS) (B); perturbation of the p53 system (C) or mitochondrial dysfunction D) are represented schematically.
Additional pathways have been implicated in hypoxia-induced autophagy, especially in cases of severe hypoxia or anoxia (Fig. 5A). These include the protein DJ-1 (also called CAP1/RS/PARK7, which regulates autophagy by an unknown mechanism), the autocrine stimulation of a PDGFR-dependent pathway, the stimulation of AMPK through metabolic stress, and the UPR of the ER (Mazure and Pouyssegur, 2010). Maximal induction of autophagy by hypoxia also requires PERK-mediated phosphorylation of eIF2α (Rouschop et al., 2010), further underscoring the importance of eIF2α phosphorylation as a general autophagy regulator. Hypoxia increases the transcription of the essential autophagy genes LC3 and Atg5 through the transcription factors ATF4 and CHOP, respectively, which are both regulated by PERK (Rouschop et al., 2010).
Redox Stress and p53
Oxidative stress induces autophagy through multiple mechanisms (Fig. 5B). Exogenous hydrogen peroxide reportedly can activate PERK (and thereby stimulate eIF2α phosphorylation) (Liu et al., 2008), directly oxidize and activate Atg4 proteases (and thereby accelerate the production of proteolytically mature LC3) (Scherz-Shouval et al., 2007), and inhibit mTOR either directly (Liu et al., 2008) or indirectly, via activation of PARP1 (as a result of DNA damage) and/or via activation of a cytoplasmic pool of ATM and the subsequent activation of LKB1 and AMPK1(Alexander et al., 2010; Huang et al., 2009). The cellular response to an increase in ROS often involves the activation of mitogen-activated protein kinases (MAPKs), including JNK1, which can activate autophagy (Wong et al., 2010). Finally, DNA damage also stimulates the expression of pro-autophagic p53-induced target genes.
The tumor suppressor protein p53, which can be activated by different types of stress including redox stresss, has a dual effect on autophagy (Fig. 5C), acting as a positive regulator of autophagy via its transcriptional activity and as a negative regulator of autophagy via its cytoplasmic functions (Green and Kroemer, 2009). As a transcription factor, p53 transactivates several autophagy inducers including DRAM1 (which may operate through JNK1 activation) and Sestrin2 (which binds to the ternary complex TSC1/TSC2-AMPK, inducing phosphorylation and activation of TSC2 by AMPK). One of the few p53-induced genes that inhibits autophagy is TIGAR, a fructose-2,6-bisphosphatase that stimulates the pentose phosphate shunt and helps to lower intracellular ROS (Bensaad et al., 2009). When present in the cytoplasm, p53 acts as a tonic inhibitor of autophagy. In several contexts of autophagy induction, p53 must be depleted from the cytoplasm for optimal autophagy induction to ensue. This may involve the activation of the p53-specific ubiquitin ligase, HDM2, which targets p53 to proteasome-mediated destruction (Green and Kroemer, 2009). However, it is not yet fully clear how cytoplasmic p53 inhibits autophagy. The only essential autophagy protein that is known to interact with p53 is FIP200, which is a multifunctional protein that is present in the ULK1 complex, yet also binds to Pyk2, FAK, TSC2, ASK1 and TRAF2 and plays a role in the control of cell adhesion, migration, proliferation and cell death (Gan and Guan, 2008). Thus, an interesting question is whether p53 somehow negatively regulates the ULK1 complex through its interaction with FIP200.
Mitochondrial Damage
Cells must remove damaged mitochondria to prevent the accumulation of ROS. This process of mitochondrial quality control is mediated by mitophagy, the selective autophagic removal of mitochondria. Considerable advances have been made in understanding the mechanisms by which damaged mitochondria are targeted for autophagy, as well as the functional significance of mitochondrial quality control in preventing aging, neurodegenerative diseases, and other pathologies.
In response to potentially lethal stress or damage, mitochondrial membranes can be permeabilized through multiple distinct biochemical routes. Indeed, mitochondrial membrane permeabilization (MMP) constitutes one of the hallmarks of imminent apoptotic or necrotic cell death (Kroemer et al., 2007). However, if only a fraction of mitochondria are permeabilized, autophagic removal of the damaged mitochondria can rescue the cell. The autophagic recognition of depolarized mitochondria is mediated by a refined voltage sensor, involving the mitochondrial kinase, PINK1. Under normal circumstances, PINK1 is continuously recruited to the mitochondrial outer membrane, but subject to voltage-dependent proteolysis, which leads to its removal from mitochondria and to proteasome-mediated degradation (Narendra et al., 2010). Upon mitochondrial depolarization, PINK1 rapidly accumulates on the mitochondrial surface, facilitates the recruitment of the E3 ubiquitin ligase Parkin (Narendra et al., 2010) which ubiquitinates mitochondrial substrates including the outer membrane protein VDAC1, recruits the autophagy adaptor molecule, p62/SQSTM1, and thereby targets mitochondria for autophagic removal (Geisler et al., 2010) (Fig. 5D). Both PINK1 and Parkin genes were originally identified because loss-of-function mutations affecting either of them cause familial Parkinson's disease in humans. Disease-causing mutations in PINK1 and Parkin disrupt Parkin recruitment and Parkin-induced mitophagy (Geisler et al., 2010; Narendra et al., 2010). At least in some cell types (MEFs), depolarization-induced mitophagy involves BNIP3L (also known as NIX) (Novak et al., 2010) (Fig. 5D) and in mice, BNIP3L/Nix is required for the removal of mitochondria during erythroid maturation (Sandoval et al., 2008). At present, the functional relationship between NIX-dependent mitophagy and the PINK1/Parkin mitophagy pathway has not been elucidated.
Interestingly, the maturation of autophagosomes during mitophagy may be controlled differently than during starvation-induced autophagy. The ubiquitin-binding deacetylase, histone deacetylase-6 (HDAC6) promotes autophagosomal maturation by recruiting a cortactin-dependent, actin–remodelling machinery to ubiquitinated structures, where assembly of the F-actin network facilitates autophagosome-lysosome fusion. (Lee et al., 2010). However, HDAC6 deficiency leads to a failure of autophagosomal maturation only in the context of quality control autophagy (mitophagy and protein aggregate removal), but not in starvation-induced autophagy (Lee et al., 2010). This specific requirement for a ubiquitin-binding deacetylase in quality control autophagy suggests that ubiquitin modification may be a key signal underlying distinct steps in selective autophagy.
Autophagy Regulatory Pathways in the General Stress Response
In the preceding section, we reviewed specific stress stimuli and stress signals that regulate autophagy. Certain general concepts regarding the interrelationship between autophagy regulatory pathways and the general stress response are beginning to emerge.
Integration of Autophagy and other Cellular Stress Responses
The integration of autophagy and other cellular stress responses can be conceptualized in the framework of three broad concepts. First, a single type of stress stimulus elicits a variety of signals that trigger distinct cellular responses (one of which is autophagy) that cooperate for the sake of optimal cellular repair and adaptation. Second, distinct stress responses are often integrated through the ability of a single molecular event to stimulate multiple adaptive pathways (one of which is autophagy). Third, distinct cellular stress response pathways, including autophagy, mutually control other stress response pathways.
Some representative examples of the first concept (Fig. 6A) include (i) redox stress, which induces transcriptional reprogramming through HIF, NF-κB and p53 activation, elicits the UPR, and stimulates both general and selective autophagy; (ii) hypoxia, which induces adaptive responses including the transcriptional activation of angiogenic and cytoprotective cytokines in parallel with autophagy stimulation; and (iii) DNA damage, which elicits nuclear p53 translocation, cell cycle checkpoint activation, cell cycle arrest, and autophagy.
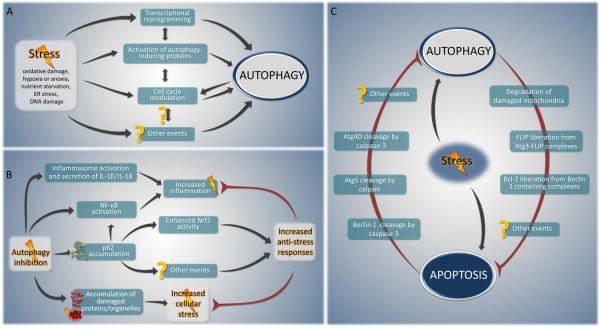
FIGURE 6. Hypothetical models of key cellular stress response networks.
A. Particular stress stimuli (e.g. oxidative damage, hypoxia or anoxia, nutrient starvation, ER stress) can elicit different responses that cooperate to achieve optimal cellular repair and adaptation. A diverse range of stressors activate interconnected cytoprotective mechanisms able to modulate autophagy at different levels, such as transcriptional reprogramming, protein modifications (phosphorylation, acetylation, etc.) or cell cycle modulation. B. Autophagy inhibition and stress. Autophagy impairment leads to the accumulation of damaged proteins and organelles, which in turn can elicit cellular stress. Moreover, disabled autophagy can increase the abundance of p62, resulting in an enhanced activity of NF-κB, which leads to enhanced inflammation. By contrast, p62 accumulation leads to the activation of Nrf2 transcription factor and in a consequent increase in the expression of stress response enzymes. C. Mutual exclusion between autophagy and apoptosis. Autophagy, as a cytoprotective pathway, eliminates potential sources of pro-apoptotic stimuli such as damaged mitochondria, thereby setting a higher threshold against apoptosis induction. By contrast, the apoptosis-associated activation of proteases such as calpain and caspase-3 may destroy autophagy-specific factors (Atg4D, Beclin 1 or Atg5), thereby suppressing autophagy.
Many of the signals discussed in preceding sections illustrate the second concept. For example, eIF2α phosphorylation participates in the UPR in stress granule formation, in general translational control, and in autophagy induction (Kimball et al., 2003; Talloczy et al., 2002). Similarly, activation of TAK1 and IKK coordinates the activation of NF-κB signaling and that of autophagy (Criollo et al., 2010; Herrero-Martin et al., 2009). Activation of LKB1-AMPK stimulates autophagy, as well as the phosphorylation and activation of p27kip1, a cyclin-dependent kinase inhibitor that leads to cell cycle arrest (Liang et al., 2007). mTOR inhibition results in autophagy induction, as well as in the inhibition of anabolic reactions including IRES-independent mRNA translation. FOXO3 activation induces the transcription of genes whose products increase both proteasomal and autophagic degradation (Mammucari et al., 2007).
The control of other stress pathways by autophagy was discussed briefly in previous sections. For example, as noted above, deficient autophagy leads to p62 accumulation, which activates NF-κB signaling (Mathew et al., 2009). In addition, p62 accumulation in the setting of deficient autophagy activates the stress responsive transcription factor Nrf2 (Komatsu et al., 2010). High levels of p62 disrupt the interaction between the Cul3/Rbx1 ubiquitin ligase complex, Keap1 (which normally targets Nrf2 for proteasomal degradation) and Nrf2, resulting in enhanced Nrf2 activity and an increase in expression of Nrf2-regulated stress response enzymes (Komatsu et al., 2010) (Fig. 6B). In addition, several different autophagy genes regulate different aspects of inflammatory signaling, although, in some instances, it is not known whether such regulation is a consequence of autophagy or alternative functions of the autophagy genes (Sumpter and Levine, 2010; Virgin and Levine, 2009). Autophagy genes negatively regulate RLR-mediated induction of type I interferon both via conjugation of Atg12-Atg5 to CARD domains of RLR signaling molecules and through elimination of dysfunctional mitochondria. Atg16L1 and Atg7 negatively regulate inflammatory signaling, including IL-1β and IL-18 secretion. Moreover, Atg9, but not Atg7, negatively regulates activation of STING, a recently discovered transmembrane protein that is required for efficient activation of type I IFN and pro-inflammatory cytokine production in response to interferon stimulatory DNA (Saitoh et al., 2009).
These examples underscore the mutual control that exists between autophagy and inflammatory signaling pathways. Another important axis of mutual control of different stress responses pathways is between autophagy and cell cycle regulation. Autophagy is blocked during G2 phase and mitosis, and a recent study indicates that this may be mediated by cyclin-dependent kinase-mediated phosphorylation of Vps34, which negatively regulates its interaction with Beclin 1 during mitosis (Furuya et al., 2010). Moreover, there is a strong correlation between mitogenic signaling and autophagy inhibition (Levine and Kroemer, 2008). This correlation is further strengthened by a recent genome-wide screen, which found that a large percentage of genes that negatively regulate autophagy are positively involved in cell growth and proliferation (Lipinski et al., 2010). We speculate that this mutually exclusive regulation of autophagy and cell growth may constitute a fundamental mechanism that cells use to appropriately adapt their metabolism to stressful environmental conditions.
Links among General Autophagy and Quality Control Mechanisms
There is a growing consensus that autophagy is stimulated either in response to a central “command” of the cell (for example in response to nutrient depletion), in which case it is non-selective and general, or as a result of ubiquitination reactions that specifically target ‘garbage’, i.e. protein aggregates or damaged mitochondria or intracellular pathogens, for autophagic degradation. Nonetheless, these two types – general versus selective – of autophagy may not be completely separable from each other. Indeed, stimulation of the general autophagic pathway, either by inhibiting mTOR or by stimulating mTOR-independent autophagy pathways, leads to reduced accumulation of mutated, aggregation-prone proteins (such as mutant α-synyclein- or huntingtin protein) and damaged mitochondria (Sarkar et al., 2007; Williams et al., 2008). Similarly, starvation or mTOR inhibition leads to increased autophagic targeting of intracellular bacteria (Gutierrez et al., 2004). One possible interpretation of these findings is that a general increase in protein and organelle turnover may be sufficient to avoid protein aggregation and organellar degeneration. Another attractive interpretation is that general autophagy is not completely non-selective and may preferentially degrade proteins and structures that are on the “verge” of aggregation or damage. This yet unproven concept predicts that an increase in general autophagy would reset the threshold of quality control so that organelles that exhibit only minor alterations (and that normally would not be removed by autophagy) are subject to autophagic turnover. This would explain why an increase in general autophagy can accelerate bacterial clearance, delay the development of multiple neurodegenerative diseases, and mediate a general anti-aging effect (Madeo et al., 2010).
Mutual Exclusion between Autophagy and Apoptosis
Autophagy is usually a self-limited process that protects cells from cell death by multiple mechanisms that include, but are not limited to, maintainance of bioenergetic homeostasis, recycling of misfolded and aggregate-prone proteins, and removal of uncoupled or permeabilized mitochondria. The intrinsic pathway of apoptosis is initiated by mitochondrial membrane permeabilization (MMP) (Kroemer et al., 2007). If MMP is limited to a fraction of mitochondria, this will result in the selective autophagic removal of depolarized mitochondria and the avoidance of cell death. Thus, the efficacy of autophagy may set a higher threshold for the ability of MMP to constitute an irreparable, lethal event. In addition, it is plausible that the liberation of Bcl-2 and FLIP from activated autophagy protein complexes may free up these molecules to block the intrinsic and extrinsic pathways of apoptosis, respectively (Lee et al., 2009; Pattingre et al., 2005).
Given the predominantly cytoprotective role of autophagy, it seems (teleo)logical that induction of apoptosis would be coupled to the inactivation of autophagy (Fig. 6C). There are some examples of molecular events that support this concept. Caspase-3 cleaves Beclin 1, thereby destroying its pro-autophagic activity. The C-terminal fragment of Beclin 1 that results from this cleavage acquires a new function and can amplify mitochondrion-mediated apoptosis (Wirawan et al., 2010). Caspase-3 activation also cleaves and activates Atg4D, an enzyme that catalyzes the delipidation of the LC3 paralog GABARPL1. This proteolytic activation increases Atg4D targeting to mitochondria via a putative BH3 domain and enhances its cytotoxic activity (Betin and Lane, 2009). Similarly, the proteolytic activity of calpain can destroy the pro-autophagic function of Atg5 (Xia et al., 2010) while it generates an Atg5-derived pro-apoptotic mitochondrion-permeabilizing fragment of Atg5 (Yousefi et al., 2006). All these results underscore that concept that autophagy and apoptosis are antagonistic events that tend to inhibit each other.
Concluding Remarks
In this review, we have described stimulus-dependent, as well as common regulatory and execution steps of autophagy, with a focus on the close links that exist between autophagy and different types of adaptive and repair responses to stress. These links consist of multiple and intricate mechanisms that are intertwined in complex intersecting pathways, which often function in positive and negative amplification loops, thus composing molecular switches and homeostatic devices. A more detailed molecular comprehension of these regulatory networks, as well as their biomathematical integration using systems biology approaches, may furnish testable models for more refined experimental and therapeutic manipulations of autophagy. At present, the comprehension of autophagic regulation has only just begun, and its multiple links to cell growth, proliferation, senescence and apoptosis await further exploration. We expect that additional major circuits of autophagy regulation will emerge and perhaps supersede in importance the pathways that we currently know – or believe to know.
Acknowledgments
GK is supported by the Ligue Nationale contre le Cancer (Equipes labellisée), Agence Nationale pour la Recherche (ANR), European Commission (Apo-Sys, ChemoRes), Fondation pour la Recherche Médicale (FRM), Institut National du Cancer (INCa) and Cancéropôle Ile-de-France. GM is supported by EMBO. BL is supported by the National Institutes of Health (NIH)/National Cancer Institute (NCI) and the Howard Hughes Medical Institute (HHMI).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Alexander A, Cai SL, Kim J, Nanez A, Sahin M, MacLean KH, Inoki K, Guan KL, Shen J, Person MD, et al. ATM signals to TSC2 in the cytoplasm to regulate mTORC1 in response to ROS. Proc Natl Acad Sci U S A. 2010;107:4153–4158. doi: 10.1073/pnas.0913860107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Axe EL, Walker SA, Manifava M, Chandra P, Roderick HL, Habermann A, Griffiths G, Ktistakis NT. Autophagosome formation from membrane compartments enriched in phosphatidylinositol 3-phosphate and dynamically connected to the endoplasmic reticulum. J Cell Biol. 2008;182:685–701. doi: 10.1083/jcb.200803137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baud V, Karin M. Is NF-kappaB a good target for cancer therapy? Hopes and pitfalls. Nat Rev Drug Discov. 2009;8:33–40. doi: 10.1038/nrd2781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behrends C, Sowa ME, Gygi SP, Harper JW. Network organization of the human autophagy system. Nature. 2010;466:68–76. doi: 10.1038/nature09204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellot G, Garcia-Medina R, Gounon P, Chiche J, Roux D, Pouyssegur J, Mazure NM. Hypoxia-induced autophagy is mediated through hypoxiainducible factor induction of BNIP3 and BNIP3L via their BH3 domains. Mol Cell Biol. 2009;29:2570–2581. doi: 10.1128/MCB.00166-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bensaad K, Cheung EC, Vousden KH. Modulation of intracellular ROS levels by TIGAR controls autophagy. EMBO J. 2009;28:3015–3026. doi: 10.1038/emboj.2009.242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Betin VM, Lane JD. Caspase cleavage of Atg4D stimulates GABARAPL1 processing and triggers mitochondrial targeting and apoptosis. J Cell Sci. 2009;122:2554–2566. doi: 10.1242/jcs.046250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonnet MC, Weil R, Dam E, Hovanessian AG, Meurs EF. PKR stimulates NF-kappaB irrespective of its kinase function by interacting with the IkappaB kinase complex. Mol Cell Biol. 2000;20:4532–4542. doi: 10.1128/mcb.20.13.4532-4542.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boya P, Gonzalez-Polo RA, Casares N, Perfettini JL, Dessen P, Larochette N, Metivier D, Meley D, Souquere S, Yoshimori T, et al. Inhibition of macroautophagy triggers apoptosis. Mol Cell Biol. 2005;25:1025–1040. doi: 10.1128/MCB.25.3.1025-1040.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchberger A, Bukau B, Sommer T. Protein quality control in the cytosol and the Endoplasmic Reticulum: Brothers in Arms. Mol Cell. 2010;40 doi: 10.1016/j.molcel.2010.10.001. [DOI] [PubMed] [Google Scholar]
- Copetti T, Bertoli C, Dalla E, Demarchi F, Schneider C. p65/RelA modulates BECN1 transcription and autophagy. Mol Cell Biol. 2009;29:2594–2608. doi: 10.1128/MCB.01396-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Criollo A, Senovilla L, Authier H, Maiuri MC, Morselli E, Vitale I, Kepp O, Tasdemir E, Galluzzi L, Shen S, et al. The IKK complex contributes to the induction of autophagy. EMBO J. 2010;29:619–631. doi: 10.1038/emboj.2009.364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng J, Lu PD, Zhang Y, Scheuner D, Kaufman RJ, Sonenberg N, Harding HP, Ron D. Translational repression mediates activation of nuclear factor kappa B by phosphorylated translation initiation factor 2. Mol Cell Biol. 2004;24:10161–10168. doi: 10.1128/MCB.24.23.10161-10168.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Bartolomeo S, orazzari M, Nazio F, Oliverio S, Lisi G, Antonioli M, Pagliarini V, Matteoni S, Fuoco C, Giunta L, et al. The dynamic interaction of Ambra1 with the dynein motor complex regulates mammalian autophagy. J Cell Biol. 2010 doi: 10.1083/jcb.201002100. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Efeyan A, Sabatini DM. mTOR and cancer: many loops in one pathway. Curr Opin Cell Biol. 2010;22:169–176. doi: 10.1016/j.ceb.2009.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisenberg T, Knauer H, Schauer A, Buttner S, Ruckenstuhl C, Carmona-Gutierrez D, Ring J, Schroeder S, Magnes C, Antonacci L, et al. Induction of autophagy by spermidine promotes longevity. Nat Cell Biol. 2009;11:1305–1314. doi: 10.1038/ncb1975. [DOI] [PubMed] [Google Scholar]
- Furuya T, Kim M, Lipinski M, Li J, Kim D, Lu T, Shen Y, Rameh L, Yankner B, Tsai LH, et al. Negative regulation of Vps34 by Cdk mediated phosphorylation. Mol Cell. 2010;38:500–511. doi: 10.1016/j.molcel.2010.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gan B, Guan JL. FIP200, a key signaling node to coordinately regulate various cellular processes. Cell Signal. 2008;20:787–794. doi: 10.1016/j.cellsig.2007.10.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geisler S, Holmstrom KM, Skujat D, Fiesel FC, Rothfuss OC, Kahle PJ, Springer W. PINK1/Parkin-mediated mitophagy is dependent on VDAC1 and p62/SQSTM1. Nat Cell Biol. 2010;12:119–131. doi: 10.1038/ncb2012. [DOI] [PubMed] [Google Scholar]
- Green DR, Kroemer G. Cytoplasmic functions of the tumour suppressor p53. Nature. 2009;458:1127–1130. doi: 10.1038/nature07986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutierrez MG, Master SS, Singh SB, Taylor GA, Colombo MI, Deretic V. Autophagy is a defense mechanism inhibiting BCG and Mycobacterium tuberculosis survival in infected macrophages. Cell. 2004;119:753–766. doi: 10.1016/j.cell.2004.11.038. [DOI] [PubMed] [Google Scholar]
- Haigis MC, Sinclair DA. Mammalian sirtuins: biological insights and disease relevance. Annu Rev Pathol. 2010;5:253–295. doi: 10.1146/annurev.pathol.4.110807.092250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hailey DW, Rambold AS, Satpute-Krishnan P, Mitra K, Sougrat R, Kim PK, Lippincott-Schwartz J. Mitochondria supply membranes for autophagosome biogenesis during starvation. Cell. 2010;141:656–667. doi: 10.1016/j.cell.2010.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harding HP, Zhang Y, Zeng H, Novoa I, Lu PD, Calfon M, Sadri N, Yun C, Popko B, Paules R, et al. An integrated stress response regulates amino acid metabolism and resistance to oxidative stress. Mol Cell. 2003;11:619–633. doi: 10.1016/s1097-2765(03)00105-9. [DOI] [PubMed] [Google Scholar]
- Harrison B, Kraus M, Burch L, Stevens C, Craig A, Gordon-Weeks P, Hupp TR. DAPK-1 binding to a linear peptide motif in MAP1B stimulates autophagy and membrane blebbing. J Biol Chem. 2008;283:9999–10014. doi: 10.1074/jbc.M706040200. [DOI] [PubMed] [Google Scholar]
- Hayashi-Nishino M, Fujita N, Noda T, Yamaguchi A, Yoshimori T, Yamamoto A. A subdomain of the endoplasmic reticulum forms a cradle for autophagosome formation. Nat Cell Biol. 2009;11:1433–1437. doi: 10.1038/ncb1991. [DOI] [PubMed] [Google Scholar]
- He C, Klionsky DJ. Regulation mechanisms and signaling pathways of autophagy. Annu Rev Genet. 2009;43:67–93. doi: 10.1146/annurev-genet-102808-114910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He C, Levine B. The Beclin 1 interactome. Curr Opin Cell Biol. 2010;22:140–149. doi: 10.1016/j.ceb.2010.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrero-Martin G, Hoyer-Hansen M, Garcia-Garcia C, Fumarola C, Farkas T, Lopez-Rivas A, Jaattela M. TAK1 activates AMPK-dependent cytoprotective autophagy in TRAIL-treated epithelial cells. EMBO J. 2009;28:677–685. doi: 10.1038/emboj.2009.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hetz C, Glimcher LH. Fine-tuning of the unfolded protein response: Assembling the IRE1alpha interactome. Mol Cell. 2009;35:551–561. doi: 10.1016/j.molcel.2009.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hetz C, Thielen P, Matus S, Nassif M, Court F, Kiffin R, Martinez G, Cuervo AM, Brown RH, Glimcher LH. XBP-1 deficiency in the nervous system protects against amyotrophic lateral sclerosis by increasing autophagy. Genes Dev. 2009;23:2294–2306. doi: 10.1101/gad.1830709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hotamisligil GS. Endoplasmic reticulum stress and the inflammatory basis of metabolic disease. Cell. 2010;140:900–917. doi: 10.1016/j.cell.2010.02.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Q, Wu YT, Tan HL, Ong CN, Shen HM. A novel function of poly(ADP-ribose) polymerase-1 in modulation of autophagy and necrosis under oxidative stress. Cell Death Differ. 2009;16:264–277. doi: 10.1038/cdd.2008.151. [DOI] [PubMed] [Google Scholar]
- Kimball SR, Horetsky RL, Ron D, Jefferson LS, Harding HP. Mammalian stress granules represent sites of accumulation of stalled translation initiation complexes. Am J Physiol Cell Physiol. 2003;284:C273–284. doi: 10.1152/ajpcell.00314.2002. [DOI] [PubMed] [Google Scholar]
- Kirkin V, McEwan DG, Novak I, Dikic I. A role for ubiquitin in selective autophagy. Mol Cell. 2009;34:259–269. doi: 10.1016/j.molcel.2009.04.026. [DOI] [PubMed] [Google Scholar]
- Komatsu M, Kurokawa H, Waguri S, Taguchi K, Kobayashi A, Ichimura Y, Sou YS, Ueno I, Sakamoto A, Tong KI, et al. The selective autophagy substrate p62 activates the stress responsive transcription factor Nrf2 through inactivation of Keap1. Nat Cell Biol. 2010;12:213–223. doi: 10.1038/ncb2021. [DOI] [PubMed] [Google Scholar]
- Kouroku Y, Fujita E, Tanida I, Ueno T, Isoai A, Kumagai H, Ogawa S, Kaufman RJ, Kominami E, Momoi T. ER stress (PERK/eIF2alpha phosphorylation) mediates the polyglutamine-induced LC3 conversion, an essential step for autophagy formation. Cell Death Differ. 2007;14:230–239. doi: 10.1038/sj.cdd.4401984. [DOI] [PubMed] [Google Scholar]
- Kroemer G, Galluzzi L, Brenner C. Mitochondrial membrane permeabilization in cell death. Physiol Rev. 2007;87:99–163. doi: 10.1152/physrev.00013.2006. [DOI] [PubMed] [Google Scholar]
- Kume S, Uzu T, Horiike K, Chin-Kanasaki M, Isshiki K, Araki S, Sugimoto T, Haneda M, Kashiwagi A, Koya D. Calorie restriction enhances cell adaptation to hypoxia through Sirt1-dependent mitochondrial autophagy in mouse aged kidney. J Clin Invest. 2010;120:1043–1055. doi: 10.1172/JCI41376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lan F, Cacicedo JM, Ruderman N, Ido Y. SIRT1 modulation of the acetylation status, cytosolic localization, and activity of LKB1. Possible role in AMP-activated protein kinase activation. J Biol Chem. 2008;283:27628–27635. doi: 10.1074/jbc.M805711200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee IH, Cao L, Mostoslavsky R, Lombard DB, Liu J, Bruns NE, Tsokos M, Alt FW, Finkel T. A role for the NAD-dependent deacetylase Sirt1 in the regulation of autophagy. Proc Natl Acad Sci U S A. 2008;105:3374–3379. doi: 10.1073/pnas.0712145105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee IH, Finkel T. Regulation of autophagy by the p300 acetyltransferase. J Biol Chem. 2009;284:6322–6328. doi: 10.1074/jbc.M807135200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JS, Li Q, Lee JY, Lee SH, Jeong JH, Lee HR, Chang H, Zhou FC, Gao SJ, Liang C, et al. FLIP-mediated autophagy regulation in cell death control. Nat Cell Biol. 2009;11:1355–1362. doi: 10.1038/ncb1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JY, Koga H, Kawaguchi Y, Tang W, Wong E, Gao YS, Pandey UB, Kaushik S, Tresse E, Lu J, et al. HDAC6 controls autophagosome maturation essential for ubiquitin-selective quality-control autophagy. EMBO J. 2010;29:969–980. doi: 10.1038/emboj.2009.405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine B, Kroemer G. Autophagy in the pathogenesis of disease. Cell. 2008;132:27–42. doi: 10.1016/j.cell.2007.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang J, Shao SH, Xu ZX, Hennessy B, Ding Z, Larrea M, Kondo S, Dumont DJ, Gutterman JU, Walker CL, et al. The energy sensing LKB1-AMPK pathway regulates p27(kip1) phosphorylation mediating the decision to enter autophagy or apoptosis. Nat Cell Biol. 2007;9:218–224. doi: 10.1038/ncb1537. [DOI] [PubMed] [Google Scholar]
- Lipinski MM, Hoffman G, Ng A, Zhou W, Py BF, Hsu E, Liu X, Eisenberg J, Liu J, Blenis J, et al. A Genome-Wide siRNA Screen Reveals Multiple mTORC1 Independent Signaling Pathways Regulating Autophagy under Normal Nutritional Conditions. Dev Cell. 2010 doi: 10.1016/j.devcel.2010.05.005. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L, Wise DR, Diehl JA, Simon MC. Hypoxic reactive oxygen species regulate the integrated stress response and cell survival. J Biol Chem. 2008;283:31153–31162. doi: 10.1074/jbc.M805056200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madeo F, Tavernarakis N, Kroemer G. Does autophagy induction prolong longevity? Nat Cell Biol. 2010 doi: 10.1038/ncb0910-842. in press. [DOI] [PubMed] [Google Scholar]
- Maiuri MC, Le Toumelin G, Criollo A, Rain JC, Gautier F, Juin P, Tasdemir E, Pierron G, Troulinaki K, Tavernarakis N, et al. Functional and physical interaction between Bcl-X(L) and a BH3-like domain in Beclin-1. EMBO J. 2007;26:2527–2539. doi: 10.1038/sj.emboj.7601689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majmundar AJ, Wong WJ, Simon CM. Hypoxia inducible factors and the response to hypoxic stress. Mol Cell. 2010;40 doi: 10.1016/j.molcel.2010.09.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mammucari C, Milan G, Romanello V, Masiero E, Rudolf R, Del Piccolo P, Burden SJ, Di Lisi R, Sandri C, Zhao J, et al. FoxO3 controls autophagy in skeletal muscle in vivo. Cell Metab. 2007;6:458–471. doi: 10.1016/j.cmet.2007.11.001. [DOI] [PubMed] [Google Scholar]
- Mathew R, Karp CM, Beaudoin B, Vuong N, Chen G, Chen HY, Bray K, Reddy A, Bhanot G, Gelinas C, et al. Autophagy suppresses tumorigenesis through elimination of p62. Cell. 2009;137:1062–1075. doi: 10.1016/j.cell.2009.03.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazure NM, Pouyssegur J. Hypoxia-induced autophagy: cell death or cell survival? Curr Opin Cell Biol. 2010;22:177–180. doi: 10.1016/j.ceb.2009.11.015. [DOI] [PubMed] [Google Scholar]
- Michiorri S, Gelmetti V, Giarda E, Lombardi F, Romano F, Marongiu R, Nerini-Molteni S, Sale P, Vago R, Arena G, et al. The Parkinson-associated protein PINK1 interacts with Beclin1 and promotes autophagy. Cell Death Differ. 2010 doi: 10.1038/cdd.2009.200. [DOI] [PubMed] [Google Scholar]
- Milani M, Rzymski T, Mellor HR, Pike L, Bottini A, Generali D, Harris AL. The role of ATF4 stabilization and autophagy in resistance of breast cancer cells treated with Bortezomib. Cancer Res. 2009;69:4415–4423. doi: 10.1158/0008-5472.CAN-08-2839. [DOI] [PubMed] [Google Scholar]
- Mizushima N. The role of the Atg1/ULK1 complex in autophagy regulation. Curr Opin Cell Biol. 2010;22:132–139. doi: 10.1016/j.ceb.2009.12.004. [DOI] [PubMed] [Google Scholar]
- Mizushima N, Levine B, Cuervo AM, Klionsky DJ. Autophagy fights disease through cellular self-digestion. Nature. 2008;451:1069–1075. doi: 10.1038/nature06639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizushima N, Yoshimori T, Levine B. Methods in mammalian autophagy research. Cell. 2010;140:313–326. doi: 10.1016/j.cell.2010.01.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morselli E, Maiuri MC, Markai M, Megalou E, Pasparaki A, Paliaras K, Galluzzi L, Criollo A, Malik SA, Madeo F, et al. Caloric restriction and resveratrol prolong longevity via the sirtuin-1 dependent induction of autophagy. Cell Death Disease. 2010;1:e10. doi: 10.1038/cddis.2009.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura T, Furuhashi M, Li P, Cao H, Tuncman G, Sonenberg N, Gorgun CZ, Hotamisligil GS. Double-stranded RNA-dependent protein kinase links pathogen sensing with stress and metabolic homeostasis. Cell. 2010;140:338–348. doi: 10.1016/j.cell.2010.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narendra DP, Jin SM, Tanaka A, Suen DF, Gautier CA, Shen J, Cookson MR, Youle RJ. PINK1 is selectively stabilized on impaired mitochondria to activate Parkin. PLoS Biol. 2010;8:e1000298. doi: 10.1371/journal.pbio.1000298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nasrin N, Kaushik VK, Fortier E, Wall D, Pearson KJ, de Cabo R, Bordone L. JNK1 phosphorylates SIRT1 and promotes its enzymatic activity. PLoS One. 2009;4:e8414. doi: 10.1371/journal.pone.0008414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neufeld TP. TOR-dependent control of autophagy: biting the hand that feeds. Curr Opin Cell Biol. 2010;22:157–168. doi: 10.1016/j.ceb.2009.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nivon M, Richet E, Codogno P, Arrigo AP, Kretz-Remy C. Autophagy activation by NFkappaB is essential for cell survival after heat shock. Autophagy. 2009;5:766–783. doi: 10.4161/auto.8788. [DOI] [PubMed] [Google Scholar]
- Novak I, Kirkin V, McEwan DG, Zhang J, Wild P, Rozenknop A, Rogov V, Lohr F, Popovic D, Occhipinti A, et al. Nix is a selective autophagy receptor for mitochondrial clearance. EMBO Rep. 2010;11:45–51. doi: 10.1038/embor.2009.256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orsi A, Polson HE, Tooze SA. Membrane trafficking events that partake in autophagy. Curr Opin Cell Biol. 2010;22:150–156. doi: 10.1016/j.ceb.2009.11.013. [DOI] [PubMed] [Google Scholar]
- Pattingre S, Bauvy C, Carpentier S, Levade T, Levine B, Codogno P. Role of JNK1-dependent Bcl-2 phosphorylation in ceramide-induced macroautophagy. J Biol Chem. 2009;284:2719–2728. doi: 10.1074/jbc.M805920200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pattingre S, Tassa A, Qu X, Garuti R, Liang XH, Mizushima N, Packer M, Schneider MD, Levine B. Bcl-2 antiapoptotic proteins inhibit Beclin 1-dependent autophagy. Cell. 2005;122:927–939. doi: 10.1016/j.cell.2005.07.002. [DOI] [PubMed] [Google Scholar]
- Polson HE, de Lartigue J, Rigden DJ, Reedijk M, Urbe S, Clague MJ, Tooze SA. Mammalian Atg18 (WIPI2) localizes to omegasome-anchored phagophores and positively regulates LC3 lipidation. Autophagy. 2010;6 doi: 10.4161/auto.6.4.11863. [DOI] [PubMed] [Google Scholar]
- Qing G, Yan P, Qu Z, Liu H, Xiao G. Hsp90 regulates processing of NF-kappa B2 p100 involving protection of NF-kappa B-inducing kinase (NIK) from autophagy-mediated degradation. Cell Res. 2007;17:520–530. doi: 10.1038/cr.2007.47. [DOI] [PubMed] [Google Scholar]
- Ravikumar B, Moreau K, Jahreiss L, Puri C, Rubinsztein DC. Plasma membrane contributes to the formation of pre-autophagosomal structures. Nat Cell Biol. 2010;12:747–757. doi: 10.1038/ncb2078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rouschop KM, van den Beucken T, Dubois L, Niessen H, Bussink J, Savelkouls K, Keulers T, Mujcic H, Landuyt W, Voncken JW, et al. The unfolded protein response protects human tumor cells during hypoxia through regulation of the autophagy genes MAP1LC3B and ATG5. J Clin Invest. 2010;120:127–141. doi: 10.1172/JCI40027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruderman NB, Xu XJ, Nelson L, Cacicedo JM, Saha AK, Lan F, Ido Y. AMPK and SIRT1: a long-standing partnership? Am J Physiol Endocrinol Metab. 2010;298:E751–760. doi: 10.1152/ajpendo.00745.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saitoh T, Fujita N, Hayashi T, Takahara K, Satoh T, Lee H, Matsunaga K, Kageyama S, Omori H, Noda T, et al. Atg9a controls dsDNA-driven dynamic translocation of STING and the innate immune response. Proc Natl Acad Sci U S A. 2009;106:10842–20846. doi: 10.1073/pnas.0911267106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandoval H, Thiagarajan P, Dasgupta SK, Schumacher A, Prchal JT, Chen M, Wang J. Essential role for Nix in autophagic maturation of erythroid cells. Nature. 2008;454:232–235. doi: 10.1038/nature07006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarkar S, Perlstein EO, Imarisio S, Pineau S, Cordenier A, Maglathlin RL, Webster JA, Lewis TA, O'Kane CJ, Schreiber SL, et al. Small molecules enhance autophagy and reduce toxicity in Huntington's disease models. Nat Chem Biol. 2007;3:331–338. doi: 10.1038/nchembio883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scherz-Shouval R, Shvets E, Fass E, Shorer H, Gil L, Elazar Z. Reactive oxygen species are essential for autophagy and specifically regulate the activity of Atg4. EMBO J. 2007;26:1749–1760. doi: 10.1038/sj.emboj.7601623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sengupta S, Peterson TR, Sabatini DM. Regulation of the mTor complex 1 by nutrients, growth factors and stress. Mol Cell. 2010;40 doi: 10.1016/j.molcel.2010.09.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi CS, Kehrl JH. MyD88 and Trif target Beclin 1 to trigger autophagy in macrophages. J Biol Chem. 2008;283:33175–33182. doi: 10.1074/jbc.M804478200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi CS, Kehrl JH. TRAF6 and A20 regulate lysine 63-linked ubiquitination of Beclin-1 to control TLR4-induced autophagy. Sci Signal. 2010;3:ra42. doi: 10.1126/scisignal.2000751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sumpter R, Jr., Levine B. Autophagy and innate immunity: Triggering, targeting and tuning. Semin Cell Dev Biol. 2010 doi: 10.1016/j.semcdb.2010.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talloczy Z, Jiang W, Virgin H.W.t., Leib DA, Scheuner D, Kaufman RJ, Eskelinen EL, Levine B. Regulation of starvation- and virus-induced autophagy by the eIF2alpha kinase signaling pathway. Proc Natl Acad Sci U S A. 2002;99:190–195. doi: 10.1073/pnas.012485299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talloczy Z, Virgin H.W.t., Levine B. PKR-dependent autophagic degradation of herpes simplex virus type 1. Autophagy. 2006;2:24–29. doi: 10.4161/auto.2176. [DOI] [PubMed] [Google Scholar]
- Vicencio JM, Ortiz C, Criollo A, Jones AW, Kepp O, Galluzzi L, Joza N, Vitale I, Morselli E, Tailler M, et al. The inositol 1,4,5-trisphosphate receptor regulates autophagy through its interaction with Beclin 1. Cell Death Differ. 2009;16:1006–1017. doi: 10.1038/cdd.2009.34. [DOI] [PubMed] [Google Scholar]
- Virgin HW, Levine B. Autophagy genes in immunity. Nat Immunol. 2009;10:461–470. doi: 10.1038/ni.1726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webber JL, Tooze SA. Coordinated regulation of autophagy by p38alpha MAPK through mAtg9 and p38IP. EMBO J. 2010;29:27–40. doi: 10.1038/emboj.2009.321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei Y, Pattingre S, Sinha S, Bassik M, Levine B. JNK1-mediated phosphorylation of Bcl-2 regulates starvation-induced autophagy. Mol Cell. 2008;30:678–688. doi: 10.1016/j.molcel.2008.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weidberg H, Shvets E, Shpilka T, Shimron F, Shinder V, Elazar Z. LC3 and GATE-16/GABARAP subfamilies are both essential yet act differently in autophagosome biogenesis. EMBO J. 2010;29:1792–1802. doi: 10.1038/emboj.2010.74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams A, Sarkar S, Cuddon P, Ttofi EK, Saiki S, Siddiqi FH, Jahreiss L, Fleming A, Pask D, Goldsmith P, et al. Novel targets for Huntington's disease in an mTOR-independent autophagy pathway. Nat Chem Biol. 2008;4:295–305. doi: 10.1038/nchembio.79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wirawan E, Vande Walle L, Kersse K, Cornelis S, Claerhour S, Vanoverbergher I, Roelandt R, de Rycke R, Verspuren J, Declercq W, et al. Caspase-mediated cleavage of Beclin-1 inactivates Beclin-1-induced autophagy and enhances apoptosis by promoting the release of proapoptotic factors from mitochondria. Cell Death Disease. 2010;1:e18. doi: 10.1038/cddis.2009.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong CH, Iskandar KB, Yadav SK, Hirpara JL, Loh T, Pervaiz S. Simultaneous induction of non-canonical autophagy and apoptosis in cancer cells by ROS-dependent ERK and JNK activation. PLoS One. 2010;5:e9996. doi: 10.1371/journal.pone.0009996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia HG, Zhang L, Chen G, Zhang T, Liu J, Jin M, Ma X, Ma D, Yuan J. Control of basal autophagy by calpain1 mediated cleavage of ATG5. Autophagy. 2010;6:61–66. doi: 10.4161/auto.6.1.10326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang L, Li P, Fu S, Calay ES, Hotamisligil GS. Defective hepatic autophagy in obesity promotes ER stress and causes insulin resistance. Cell Metab. 2010;11:467–478. doi: 10.1016/j.cmet.2010.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z, Klionsky DJ. Mammalian autophagy: core molecular machinery and signaling regulation. Curr Opin Cell Biol. 2010;22:124–131. doi: 10.1016/j.ceb.2009.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yla-Anttila P, Vihinen H, Jokitalo E, Eskelinen EL. 3D tomography reveals connections between the phagophore and endoplasmic reticulum. Autophagy. 2009;5:1180–1185. doi: 10.4161/auto.5.8.10274. [DOI] [PubMed] [Google Scholar]
- Yousefi S, Perozzo R, Schmid I, Ziemiecki A, Schaffner T, Scapozza L, Brunner T, Simon HU. Calpain-mediated cleavage of Atg5 switches autophagy to apoptosis. Nat Cell Biol. 2006;8:1124–1132. doi: 10.1038/ncb1482. [DOI] [PubMed] [Google Scholar]
- Zalckvar E, Berissi H, Mizrachy L, Idelchuk Y, Koren I, Eisenstein M, Sabanay H, Pinkas-Kramarski R, Kimchi A. DAP-kinase-mediated phosphorylation on the BH3 domain of beclin 1 promotes dissociation of beclin 1 from Bcl-XL and induction of autophagy. EMBO Rep. 2009;10:285–292. doi: 10.1038/embor.2008.246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y, Yang J, Liao W, Liu X, Zhang H, Wang S, Wang D, Feng J, Yu L, Zhu WG. Cytosolic FoxO1 is essential for the induction of autophagy and tumour suppressor activity. Nat Cell Biol. 2010 doi: 10.1038/ncb2069. [DOI] [PubMed] [Google Scholar]