Abstract
P16INK4A (also known as P16 and MTS1), a protein consisting exclusively of four ankyrin repeats, is recognized as a tumor suppressor mainly due to the prevalence of genetic inactivation of the p16INK4A (or CDKN2A) gene in virtually all types of human cancers. However, it has also been shown that elevated expression (up-regulation) of P16 is involved in cellular senescence, aging, and cancer progression, indicating that the regulation of P16 is critical for its function. Here, we discuss the regulatory mechanisms of P16 function at the DNA level, the transcription level, and the posttranscriptional level, as well as their implications in the structure-function relationship of P16 and in human cancers.
Keywords: P16, tumor suppressor, down-regulation, over-expression, cancer
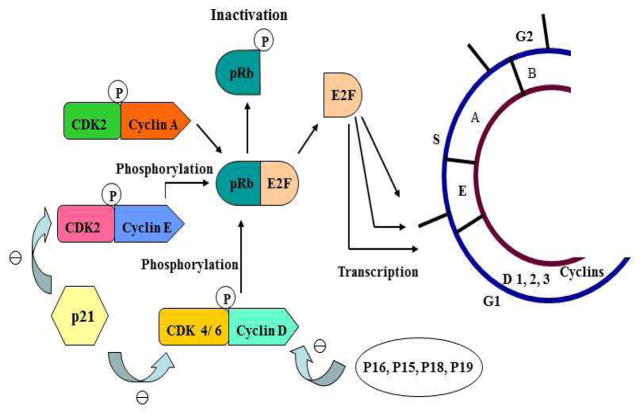
P16, also designated MTS1 and P16INK4A, is one of the most extensively studied proteins in the past decades due to its critical roles in cell cycle progression, cellular senescence, and the development of human cancers (1–5) At the G1-to-S transition, P16 specifically inhibits cyclin-dependent kinases 4 and 6 (CDK4, CDK6)-mediated phosphorylation of pRb, the retinoblastoma susceptible gene product, thus sequestering E2F transcription factors as incompetent pRb/E2F complexes and consequently blocking cell cycle progression (5) (Figure 1). Furthermore, it has also been demonstrated that elevated P16 expression induced by oncogenes, DNA damage response, or aging can trigger and accelerate cellular senescence (1–4). While genetic inactivation of the p16 gene (cyclin-dependent kinase inhibitor 2a, CDKN2A) by deletion, methylation, and point mutation has been found in nearly 50% of all human cancers (6–9), the over-expression of P16 at both mRNA and protein levels is also associated with poor prognosis for cancers including neuroblastoma, cervical, ovarian, breast, prostate tumors, and oral cancers (10). Together, these findings demonstrate that both the transcriptional level and translational status of P16 are critical for its overall ability to mediate cellular activities. However, while numerous studies have focused on defining the genetic/epigenetic status of p16 in different cancers in order to investigate the molecular mechanism(s) of carcinogenesis (6, 11), the critical regulation of P16 itself has been understudied until recently. Here we review recent findings in P16 regulation and discuss their significance in understanding the roles of P16 in cancer from a biochemical perspective.
Figure 1. The P16-CDK4/6-pRb Pathway.
Arrows and minus signs represent positive and negative regulatory effects, respectively. P, phosphorylated. This figure was modified from Reference 56.
Basic biochemical features of P16
In 1993, a truncated version of the p16 cDNA gene was first identified in a yeast two-hybrid screen for proteins that interact with human CDK4 (12, 13). The cDNA encoded a polypeptide of 148 amino acid residues with an estimated molecular mass of ~16 kD that negatively modulated the kinase activity of CDK4 (Figure 2A). Consequently, it was designated as P16INK4A (for inhibitor of CDK4) or CDKN2 (for CDK inhibitor 2). One year later, the MTS1 (Multiple Tumor Suppressor 1) locus on human chromosome 9p was described following the discovery that the p16 gene was frequently inactivated by homozygous deletion or mutation in melanomas as well as a broad spectrum of additional human cancer types (13). The full-length p16 cDNA gene was reported soon thereafter, which encodes a protein with eight additional amino acid residues at the N-terminus in comparison with the originally reported cDNA sequence (14, 15) (Figure 2A). Further studies have established that P16 is a pivotal regulator of cell cycle progression with diverse physiological functions and unique structural and biophysical properties.
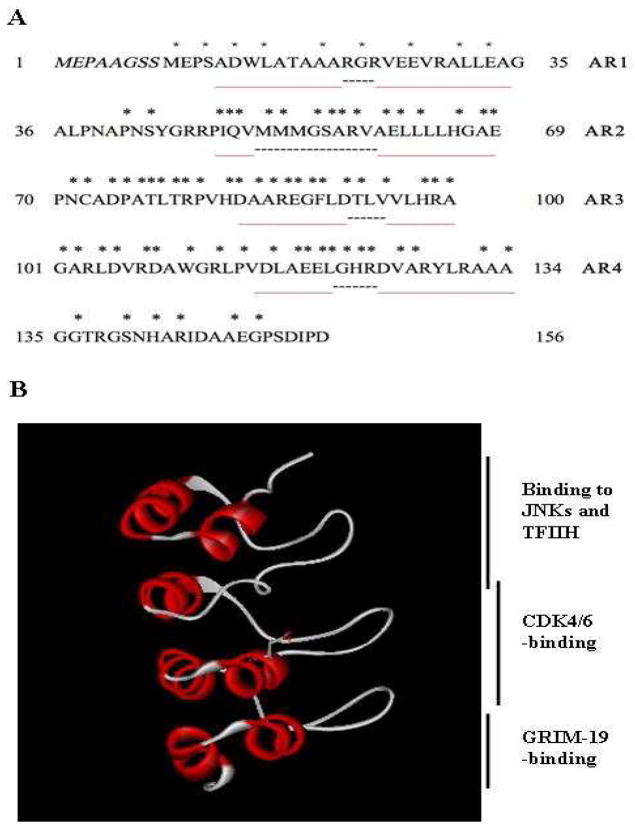
Figure 2. Primary, secondary, and tertiary structures of P16.
A, Sequence and secondary structure of P16. Italic residues at the N-terminal represent missing residues in the cDNA gene first reported. Red and dashed lines represent helical and loop regions, respectively. Residues with * marks are those with mutations in human cancers. AR, ankyrin repeat. B, Tertiary structure and domains of P16. The solution structure of P16 (PDB code: 2A5E) is presented here (30), in which D84, the residue critical for CDK4 inhibition, is highlighted. JNK, c-Jun N-terminal kinases.
Function
P16 primarily functions in cell cycle control as a negative regulator of the prominent pRb/E2F pathway (16). In G0 and early G1 phases, pRb is hypophosphorylated and forms complexes with members of the E2F family of transcription factors. These complexes sequester E2Fs and prevent their access to the promoters of proliferation-associated genes, such as cyclin B1 (CCNB), dihydrofolate reductase (DHFR), jun B proto-oncogene (JUNB), and thymidine kinase 1 (TK1) (17, 18) (Figure 1). Once committed to cell proliferation, pRb is progressively hyperphosphorylated by CDK4 and CDK6 in late G1, resulting in the entry into S phase. Binding of P16 directly down-regulates the kinase activities of CDK4 and CDK6, keeping pRb in a hypophosphorylated status. Furthermore, P16 can disrupt complexes of CDK4/6 and non-P16 CDK inhibitors such as P27 (CDKN1B), thus leading to the release of these non-P16 inhibitors, suppression of CDK2 activity, and increases in the expression of hypophosphorylated pRb (5). Consequently, these P16-dependent events culminate in cell arrest at the G1/S boundary. Interestingly, it has been reported recently that the suppressive impact of P16 on E2F-mediated gene expression can be enhanced by the physical association between P16 and GRIM-19 (Gene associated Retinoid-IFN-induced Mortality-19), a pro-apoptotic protein functioning in the IFN-β/RA-induced cell death pathway (19). A more detailed account of these findings will be addressed subsequently in this review.
In addition to the pRb/E2F pathway, P16 also contributes to cell cycle progression through alternate and independent regulatory pathways (20–22). First, phosphorylation of the carboxyl-terminal domain (CTD) of the large subunit of RNA polymerase II by the CDK7 subunit of general transcription factor TFIIH is an essential regulatory event in transcription (20, 21). P16 is able to interact with TFIIH in the preinitiation complex, inhibit phosphorylation of the CTD, and contribute to the capacity of this pathway to induce cell cycle arrest. Second, it has been reported that P16 contacts the glycine-rich loop of c-jun N-terminal kinases 1 and 3 (JNK1 and JNK3) and suppresses their kinase activities (22). The loss of JNK activity negatively regulates UV-induced c-Jun phosphorylation in melanoma cells, and consequently interferes with cell transformation promoted by the H-Ras-JNK-c-Jun-AP1 signaling axis (22).
Additionally, accumulating evidences have shown that P16 is involved in cellular senescence and aging through molecular mechanisms yet to be explicitly elucidated (4, 23, 24). The expression of P16 increases remarkably with aging in a large number of rodent and human tissues in both healthy and disease states (4). This elevated level of P16 expression has been shown to induce cellular senescence and aging in various progenitor cells and premalignant tumor cells (3), including neural progenitor cells (25), pancreas islet progenitor cells (26), and haematopoietic stem cells (27). These findings suggest that an aging-associated increase in P16 expression can contribute to the decline in replicative potential for certain self-renewing compartments, a characteristic of aging (4). Moreover, a number of recent studies have demonstrated that P16 could be involved in the cellular response to genotoxic agents (4, 12). P16-compromised or P16-deficient cells demonstrated sensitivity to ultraviolet (UV) light-induced apoptosis, suggesting that the absence of functional P16 allows propagation of proapoptotic signaling (28). Conversely, following genotoxic-induced DNA damage, elevated expression of P16 in tumor cells resulted in cell cycle arrest and inhibition of apoptotic events such as cytochrome c release, mitochondrial membrane depolarization, and activation of the caspase cascade. These findings demonstrate that P16 is able to mitigate mitochondria sensitivity to proapoptotic signals in DNA damaged cancer cells (29).
Structure
P16 is exclusively composed of four ankyrin repeat (AR) motifs (30) (Figure 2B). Ankyrin repeats are relatively-conserved motifs of about 31~ 34 residues (30, 31). They are abundantly present in proteins of plants, prokaryotes, viruses, yeast, invertebrates, and vertebrates, and are involved in numerous physiological processes through mediating protein/protein interactions (31). Like in other AR proteins, each AR motif in P16 exhibits a helix-turn-helix (HTH) conformation except that the first helix in the second AR is only composed of four residues (30). Neighboring ARs are linked by loops of varying length in such a way that the orientations of these loops are perpendicular to the helical axes. In comparison with the helical regions, the loops show less defined structure except some very short, “nascent” β sheets, thus they are fairly flexible in conformation. In solution, the four AR motifs of P16 are stacked together in a linear fashion to form a helix bundle with a concave surface in which clusters of charged groups are present for target binding (30). Interestingly, the solution structure of P16 is virtually unchanged upon binding to CDK6 (a close homologue of CDK4) (31).
As revealed in the crystal structures of P16/CDK6 (32), P19INK4D/CDK6 (33), and P18INK4C/CDK6/viral cyclin D complexes (34) (P18INK4C and P19INK4D, abbreviated as P18 and P19 hereafter, are close homologues of P16 to be discussed later), binding of CDK6 to the concave surface of P16 (or P18, P19) exposes the catalytic cleft of CDK6 to P16 so that an electrostatic interaction is formed between D84 of P16 and R31 of CDK6 (R24 in CDK4). Since R31 is located at the active site of CDK6 and its positively charged side chain could stabilize the transition state of CDK6 (32), the aforementioned D84 (P16)/R31 (CDK6) interaction could destabilize the transition state thus diminish the kinase activity. This finding is consistent with independent cellular studies showing that cancer-related mutations at either R24 of CDK4 (R24C) or D84 of P16 (D84N) abolished the inhibition in vivo, leading to uncontrollable cell proliferation (30, 35–38). Moreover, P16 could inhibit the activity of CDK4/6 by impairing the binding of its activator cyclin D, as it has been shown that P16 binding to CDK6 substantially shrinks the binding surface for cyclin D, even though the P16-binding and cyclin D-binding surfaces in CDK4/6 are opposite to each other (32, 34).
In addition to the aforementioned D84 (P16) and R24 (CDK4), binding of P16 to CDK4 involves a great number of residues that are discontinuously dispersed in both proteins (30, 31). Most of CDK4-interacting residues in P16, including D84, are located in the second and third ARs, and the loop linking these two ARs (Figure 2B) (32), which is consistent with the finding that a peptide derived from part of the second and third ARs of P16 encompassing residues 83–102 remained potent in inhibiting CDK4 in vitro and in vivo (39). Residues in the first and fourth ARs (including the flexible N- and C-termini) contribute little to P16/CDK4 association (32). They may facilitate CDK4 binding simply through stabilizing the global structure of P16 as evidenced by biochemical studies showing that point mutations and removal of the N- and C-termini caused decrease in the stability and solubility of P16 (30, 40). Notably, residues in the first and fourth ARs (including the flexible N- and C-termini) have been found to play important roles in binding to non-CDK proteins (19–22) as well as in posttranslational modification of P16 (41, 42). First, protein fragmentation experiments have demonstrated that residues 1–60 and residues in the fourth AR are responsible for binding to GRIM-19 (19) and JNKs (22), respectively. Secondly, the N- and C-termini of P16 harbor four phosphorylation sites, Ser7, Ser8, ser140, and Ser152, and phosphorylation at these four sites brings about different perturbations in the structure, function, and stability of P16 (41, 42). Thirdly, previous studies have also indicated that the first AR of P16 may be involved in inhibiting CDK7-CTD kinase, TFIIH (19, 21). Taken together, all four ARs are required for the structural integrity of P16, and these structurally similar motifs play distinct biological roles.
Conformational Stability
Due to the modular and repetitive nature of AR proteins (not including proteins composed of both ARs and non-AR motifs), their structures are mainly maintained by interactions between residues in the same AR motif or in the neighboring AR motifs. It has been well established that at least four ARs are required to pack together to form a stable and functional protein (31, 43). Evidently, P16 is just at the margin. As demonstrated in previous guanidinium hydrocholoride (GdmHCl)-induced unfolding studies (40, 43), the free energy of denaturation in water (ΔGdwater) of P16, the parameter widely used to represent the conformational stability of a protein, is only 1.94 ± 0.10 kcal*mol−1, substantially lower than the common range of 5~15 kcal*mol−1 (31). Once impeding the determination of its structure, the unstable nature of P16 could be advantageous in its role as a tumor suppressor. Studies in our group and other laboratories have shown that P16 missense mutants are prevalent in human cancers and many of these mutants may have lost their inhibitory functions due to impaired folding or stability (30, 43). From this perspective, the low conformational stability is a major cause that P16 mutations result in cancer.
Regulation at the DNA level
The locus encompassing the p16 gene, namely the INK4b/ARF/INK4a locus, is situated on human chromosome 9p21 (14). Due to its high incidence of genetic deletion found in a variety of human malignancies including melanoma, pancreatic adenocarcinoma, bladder carcinoma, and leukemia (1, 4, 13), this locus was believed to harbor promising tumor suppressor candidates long before any of these tumor suppressors was identified. It is now known that this locus instructs five established or candidate tumor suppressors, P16 (13), P15INK4B (P15) (14), P14ARF (44), P16γ (45), and P12 (46), yet the physiological functions of some of them remain to be further elucidated (6). As discussed below, the complexity of this locus and its susceptibility to genetic alterations have a bearing on P16 functions.
The INK4b/ARF/INK4a locus
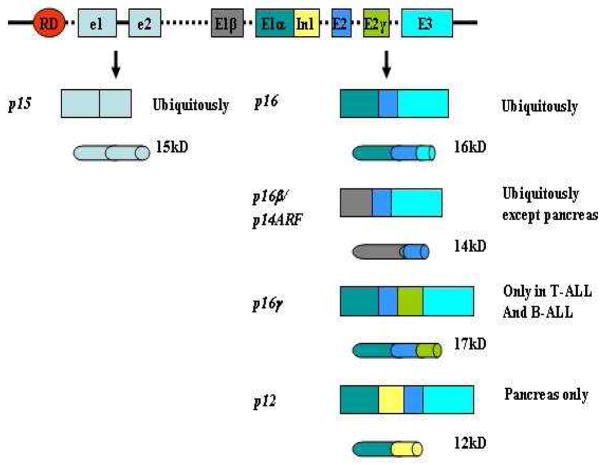
Figure 3 represents the unique architecture of the INK4b/ARF/INK4a locus (45). First, two tumor suppressor genes, p16 and p15INK4B (also known as MTS2; thereafter p15) are located in tandem in the adjacent DNA of about 35 kb such that the open reading frame of p15 is physically distinct from that of p16, and the two exons of p15 are totally different from exon 1a, exon 2, and exon 3 of p16 (1,4). However, whereas the expression of p15 is upon the induction of transformation growth factor β (TGF-β) (14), P15 and P16 are almost indistinguishable in structure and biochemical properties: like P16, P15 consists of four ARs and acts as a specific inhibitor of CDK4/6 that regulates progression through the G1 phase of the cell cycle (4, 47). Hence, it is assumed that the p16 and p15 genes arose from a duplication event in evolution. Secondly, exon 1β, an additional exon located between the p15 gene and exon 1a of p16, is spliced onto exons 2 and 3 of p16 to generate the tumor suppressor p14ARF gene (also known as p16β; p19ARF in mice) (48) (Figure 3). Interestingly, exon 1β bears no sequence resemblance to exon 1a of p16 and is transcribed from its own promoter. Moreover, the coding frame of p14ARF is offset by a single base pair relative to p16 so that exons 2 and 3 are translated in an alternating reading frame (ARF) to that for P16 (45). Accordingly, P14ARF shares no structural homology with P16 and exhibits distinct biological functions. P14ARF specifically binds to HDM2 (MDM2 in mice), a protein with the E3 ubiquitin ligase activity, and promotes degradation of the latter, thus blocking its ability to mask the transcriptional activating function of P53, which consequently suppresses oncogenic transformation in a P53-dependent manner (49). Thirdly, both p16 and p12 genes are splicing variants of p16 with potentials in tumor suppression. The p16 gene is identical to p16 except that its coding frame contains an in-frame insertion of 196 bp between exon 2 and exon 3, which leads to an additional stretch of 14 residues at the C-terminus of P16 (45). To date, no significant difference has been found between P16γ and P16 in inhibiting CDK4/6-mediated phosphorylation and repressing the E2F response. Since the expression of p16γ has been detected in the majority of p16-expressing primary T-ALL and B-ALL patient samples as well as other p16-expressing tumor specimens (45), questions remain with regard to the necessity of the co-existence of P16γ and P16 in cells. The p12 gene is a pancreas-only transcriptional variant of p16. In this transcript, an additional 274 bp on intron 1, contiguous with the 3′ end of exon 1α, is included in the normal exon 1α sequences followed by exons 2 and 3 (46). An in-frame stop codon in the intron 1-derived sequence results in a polypeptide of 116 residues with an identical N-terminus but a distinct C-terminus in comparison with P16. It has been shown that P12 fails to interact with CDK4 but exhibits a pRb-independent growth suppressing activity in cells (46). Nonetheless, the molecular mechanism underlying such growth suppressing activity is unknown.
Figure 3. Schematic structure of the INK4b/ARF/INK4a locus.
Rectangles represent DNA and mRNA, and cylinders represent proteins. RD: the regulatory sequence of the INK4/ARF locus; e1, e2: exons 1 and 2 of p15INK4B; E1β, E1α, E2, E2γ, E3: exons 1β, 1α, 2, 2γ, and 3 of p16INK4A; In1: intron 1 of p16INK4A; kD, kilo Daltons. Sizes of coding regions and proteins are not in proportion strictly. This figure was modified from Reference 45.
Since human cancers frequently harbor homozygous deletions of the whole INK4b/ARF/INK4a locus (4, 5, 13), the co-existence of the aforementioned genes in this locus once brought about considerable disputes on which of these genes, especially p16, p15, and p14ARF, was the “authentic” tumor suppressor representing the principal tumor-suppressing activity originated from chromosome 9p21. A number of studies using knockout mice have shown that mice specifically deficient for each of p16, p15, and p14ARF are more prone to spontaneous cancers than wild-type mice, but appear less tumor prone than animals deficient for both p16 and p14ARF (50, 51). Together with the fact that over-expression of each of p16, p15, and p14ARF leads to cell cycle arrest at G1/G0, these observations strongly support that P16, P15, and P14ARF potently suppress tumorigenesis individually and synergistically. While the identity of P16γ or P12 as a tumor suppressor remains to be established, it is safe to state that P16, P15, and P14ARF together constitute one of the primary anti-tumor defenses in human through strict regulation of both pRb and P53 pathways.
Susceptibility to genetic alterations
The aforementioned complexity of the INK4b/ARF/INK4a locus makes this locus exceedingly vulnerable to genetic alterations since a single genetic event, such as homozygous deletions, could simultaneously influence multiple tumor suppressors (4). For example, in addition to the aforementioned homozygous deletion of the INK4b/ARF/INK4a locus that abrogates the expression of all p16, p15, and p14ARF, some cancer-related point mutations or small deletions in exon 2 have been found to impair both p16 and p14ARF (52). It has also been reported that in some tumor specimens, multiple genetic events, such methylation of the p15 promoter and point mutations of p16, occurred concurrently and these events influenced p16, p15, and p14ARF differently (2, 8). Yet it is fair to note that a substantial portion of genetic alterations found in human cancers target only one of p16, p15, and p14ARF (53–55). Presumably, co-inactivation of two or all of p16, p15, and p14ARF may be more oncogenic in certain tissues than loss of one alone. Moreover, whereas P15, P16, and P14ARF are all potent tumor suppressors, most mutation events in the INK4b/ARF/INK4a locus, especially those point mutations and intragenic alterations, impair p16 separately or together with p14ARF (53, 54). Hence, we focus on genetic alterations of p16 and their effects on P16 functions in this review.
The p16 gene is virtually the most frequently mutated gene, only secondary to p53, in human cancers (5). The estimated frequencies of p16 inactivation in different types of human tumors are as following (2, 6, 56): breast cancer, 20%; non-small cell lung carcinoma (NSCLC), 65%; colorectal cancer, 30%; bladder cancer, 60%; squamous cell carcinoma of the head and neck (SCCHN), 50–70%; melanoma, 60%; leukemia, 60%; esophagus cancer, 70%; multiple myeloma, 60%; pancreatic carcinoma, 85% or higher. Inactivation of p16 involves four types of genetic alterations, namely, homozygous deletion, promoter hypermethylation, loss of heterozygosity (LOH), and point mutation (6, 54, 56). While homozygous deletion and promoter hypermethylation usually constitute the majority of p16 alterations (6), there arguably exists a preference for a specific type of p16 alterations in certain tumor types (57). For example, 48% of pancreatic carcinoma specimens harbored homozygous deletions of p16 (58), whereas about 30% of observed p16 alterations in SCCHN specimens were point mutations (56, 59). In primary gastric carcinoma, aberrant methylation is the major type of p16 alterations (34%) but deletions or mutations of p16 are rare (0–2%), indicative of a tendency for p16 to be inactivated through promoter hypermethylation (60). Similarly, hypermethylation of the p16 promoter has been found in about 73% of hepatocellular carcinoma (HCC) specimens (61). Interestingly, in comparison with such high incidence of p16 methylation in HCC, only 29.4% of liver cirrhosis specimens and 23.5% of chronic liver hepatitis specimens harbored p16 methylation, suggesting that p16 methylation occurs more frequently at the late stage of the development of liver cancers (61). On the contrary, promoter hypermethylation has been found to be a major mechanism to inactivate p16 in esophageal adenocarcinoma with the incidence of 61%, and about 85% of these methylation events have been observed in corresponding Barrett’s esophagus (BE) specimens (62, 63). Since Barrett’s metaplasia is well recognized as the precursor to esophageal adenocarcinoma, these results indicate that p16 methylation occurs at the early developmental stage of esophageal adenocarcinoma. Taken together, the incidence and the mechanisms of p16 inactivation vary with tumor types and tumor developmental stages.
Furthermore, the nature of a genetic alteration on p16 determines its mutagenic effect on P16 functions. While homozygous deletions and aberrant methylation-mediated silencing usually lead to complete loss of P16 function in cells, point mutations, especially missense mutations and in-frame small deletions, may only partially impair the structure and function of P16, thus their contributions to tumorigenesis need to be evaluated in caution (6, 30, 43, 56). To date, cancer-related missense mutations have been found in at least 76 residues of P16 (2, 6, 56). As shown in Figure 2A, these residues are dispersed into the whole molecule but the majority of them are located within the second, third, and fourth ARs. Residues with cancer-related missense mutations can be divided into four groups based on their mutagenic effect on the CDK4-inhibitory ability, structural stability and integrity (43, 56): (i) Residues directly involved in the association with CDK4/6, such as E26, D74, D84, and D92. These residues are located on the concave face for CDK4 binding (Figure 2B). Missense substitutions at these residues lead to unchanged structures, comparable conformational stability, but significantly-decreased CDK4-inhibitory activities. For example, D84H, a mutant frequently found in cancers, is stable and well structured but does not exhibit any detectable CDK4-inhibitory activity. (ii) The second group includes most of residues with cancer-related missense mutations, such as W15, E69, N71, F90, W110, and L121. These residues are not directly involved in CDK4 binding. Instead, they contribute to P16/CDK4 association through stabilizing the global structure of P16 or facilitating the important P16/CDK4 contacts in their neighborhood. Mutations in these residues bring about moderate decrease in CDK4-inhibitory activity and conformational stability but little structural perturbation. (iii) Residues important in forming the core structure of P16, such as L63, L78, P81, A100, G101, and P114. Mutations in these residues significantly perturb the global structure of P16, thus eliminate its CDK4-inhibitory activity. (iv) The last group includes residues whose missense substitutions do not cause any detectable changes in the CDK4-inhibitory activity, stability, or structure. Most of residues in this group are located within the first and fourth ARs and the N, C-termini. Interestingly, residues in these regions are involved in binding to targets other than CDK4/6, such as TFIIH, JNKs and GRIM-19 (19–22), or in phosphorylation (41). Hence, mutations at these residues could impair P16 functions other than modulating CDK4/6 (20). This notion is supported by the following findings. First, some tumor-derived mutants in the fourth AR and C-terminus of P16, such as S140C, H142R, and A147G, retained its CDK4-binding ability but failed to contact GRIM-19 (19). Second, N-terminal P16 mutations, such as R24P, are impaired in their inhibition of TFIIH CTD phosphorylation by CDK7, whereas mutations located in the central region of P16 have no effect on this particular interaction (64).
Regulation at the transcription level
The unique nature of the INK4b/ARF/INK4a locus also brings about complexity in the regulation of p16 transcription. On one hand, p16, p15, and p14ARF have distinct independent promoters, and the corresponding proteins function in different pathways, suggesting that these genes are independently activated or repressed under different circumstances (1, 4, 65). On the other hand, the obvious advantage of grouping genes within a single chromosomal domain is that they can be regulated en bloc by the same chromatin-remodeling event(s) (4), thus favoring the notion that the INK4b/ARF/INK4a locus is coordinately regulated. Recent studies have provided evidence in support of both regulatory mechanisms.
Independent regulation of p16
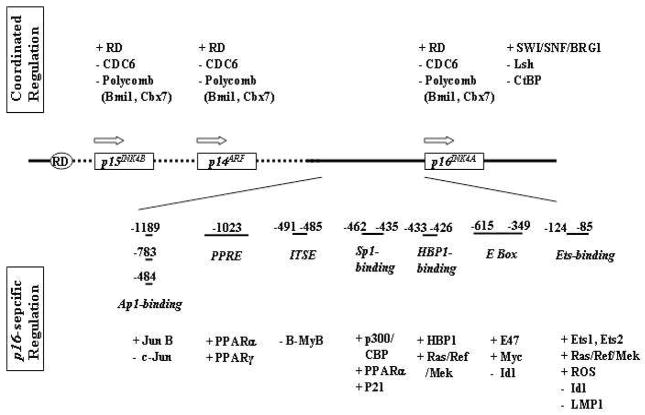
Given the importance of P16 in tumor suppression, senescence, and aging, transcriptional regulation of p16 has been an area of intense study in the past decade. While some molecular mechanisms remain to be further explored, it is clear that transcription of p16 is subject to multiple levels of control, and most of these regulations are related to diverse regulatory elements present in the p16 promoter (Figure 4).
Figure 4. The structure of the p16INK4A promoter and regulators of p16INK4A transcription.
Empty arrows represent the transcription directions, and plus and minus marks indicate positive and negative effects on the transcription of p16INK4A, respectively. RD: the regulatory sequence of the INK4b/ARF/INK4a locus; ITSE, the INK4a transcription silencing element; PPRE, the peroxisome proliferator response element. Sizes of the DNA elements are not in proportion strictly.
Ets-binding site-mediated regulation
Ets1 and Ets2 transcription factors are known to be downstream targets of Ras-Raf-Mek signaling and can be activated by MAPK-mediated phosphorylation (66). There exists a conserved Ets-binding site in the p16 promoter ranging from −124 to −85. Upon binding, Ets1 and Ets2 are able to activate the p16 promoter and induce elevated expression of p16 in human fibroblasts. Interestingly, such Ets2-mediated transactivation is neutralized by physical association of Ets2 with Id1, a helix-loop-helix (HLH) protein, whereas ectopic expression of oncogenic Ras increases Ets1/Ets2 binding to the p16 promoter in human diploid fibroblasts (66, 67). Since Ets2 is the predominant Ets protein in the transactivation of p16, it appears that Id1 functions to counterbalance the activation of the p16 promoter mediated by Ras-Raf-Mek signaling (66). In young fibroblasts, p16 is expressed at low levels due to a balance between Ets2 and Id1. However, such a steady state can be overridden by introduction of oncogenic Ras, which promotes aberrant phosphorylation of Ets2, thus activating the p16 promoter and transcription. During senescence, the Ras-Raf-Mek signaling is attenuated and the Ets2 level is low; hence, the increased expression of Ets1 and concomitant down-regulation of Id1 result in up-regulation of p16. From this perspective, the balance between Ets1/2 and Id1 seems to act as a sensor that detects aberrant growth signals (mitogenic stress/oncogenic stress). Indeed, it has been demonstrated that the contribution of reactive oxygen species (ROS) to senescence is partially attributed to its transactivation of p16 expression through the Ras-Raf-Erk-Ets signaling pathway (68).
Alternatively, LMP1, the latent membrane protein encoded by the Epstein-Barr virus, represses the p16 promoter through promoting the nuclear export of Ets2, which consequently inhibits Ets2-mediated transactivation (69).
E box-mediated regulation
Besides its effect on Ets activity, Id1 may also influence the transactivation activity of E47. Like other class I basic helix-loop-helix (bHLH) proteins (also known as E proteins) (67), E47 contains an HLH domain, which primarily mediates homo- and heterodimerization with other HLH proteins to regulate gene expression, and a basic region for binding to the consensus DNA sequence CANNTG, namely, the E-box element. There are two E-box elements in the p16 promoter, located at positions −349 (CAGGTG) and −615 (CAGGTG) (67). Upon homodimerization or heterodimerization with other E proteins, E47 binds to these two E-boxes and activate the transcription of p16 in senescent cells. Id1 contains a highly conserved HLH domain but lacks the basic DNA binding domain. As such, Id1 can only form a heterodimer with E47, and binds to E-box elements as a dominant negative regulator thus inhibiting E47-mediated activation of the p16 promoter. Similarly, TAL1, a tissue-specific class II bHLH transcription factor, is able to functionally repress the p16 promoter through heterodimerizing with E47 (70).
Additionally, Myc, an E-box-binding transcription factor, has been reported to bind to the promoter and the first intron of p16 and up-regulate its expression in human cells (1).
Sp1-binding site-mediated regulation
The GC-rich region within the p16 promoter contains at least five putative GC boxes (−474 to −447, −462 to −435, −380 to −355, −76 to −49, and −26 to −1), which represent the putative binding target sites for Sp transcription factors (including Sp1, Sp3, Sp4) (71–73). It has been well documented that there is a positive transcription regulatory element (position −466 to −451) in the p16 promoter, which harbors a GC box (−462 to −435) for Sp1 binding (71). Such binding is enhanced during cellular senescence mainly due to an increase in Sp1 binding affinity (not Sp1 protein amount). Moreover, ectopic expression of Sp1 induces the transcription of p16 in human fibroblasts (72). These results suggest that Sp1 positively influences p16 transcription upon binding to the corresponding GC box in the p16 promoter. Furthermore, it has been shown that P300/CBP, a transcriptional co-activator with histone acetyltransferase activity, cooperates with Sp1 to stimulate both p16 promoter activity and p16 mRNA expression. While ectopic expression of P300 is able to induce cell cycle arrest through up-regulating p16 expression, the participation of P300 in p16 transcription is Sp1-dependent. As revealed in chromatin immunoprecipitation (CHIP) assays and protein/protein interaction assays, P300 physically interacts with the N-terminal domain of Sp1 in vivo through its Q domain, and such association recruits P300 to the p16 promoter so that the histone acetyltransferase domain of P300 is able to contribute to p16 transcriptional activation through inducing hyperacetylation of histone H4 at the p16 gene (73).
HBP1-binding site-mediated regulation
It has been reported that the p16 promoter harbors a binding site at position −426 to −433 for the HMG box-containing protein 1 (HBP1) transcription factor, a downstream effector in the Ras-Raf-Mek signaling pathway (74). The sequence-specific binding of HBP1 to the p16 promoter positively regulates the expression of p16, and triggers premature senescence in primary cells. HBP1 knockdown delays Ras-induced premature senescence in WI38 cells in early passages, and also facilitates Ras-induced cell transformation through transcriptionally up-regulating TERT and Myc but down-regulating p16 (74). These findings indicate that HBP1-mdeiated regulation of p16 may be part of the premature-senescence-executing machinery upon imbalances of Ras and other signals.
ITSE-mediated regulation
The p16 promoter also harbors a negative regulatory element, the INK4a transcription silence element (ITSE), ranging from position −491 to −485 (75). The activity of the p16 promoter increased significantly in young 2BS cells when ITSE was deleted. Intriguingly, ITSE contains a binding site for Myb-related protein B (B-MYB), a transcription factor involved in the regulation of cell survival, proliferation, and differentiation (76). In human embryonic lung fibroblast cells, B-MYB down-regulates p16 expression, whereas knocking down of B-MYB has an opposite effect. Since B-MYB levels in the inner cell mass/embryonic stem cells (ICM/ESCs) are 100 times greater than those in normal proliferating cells, its repression of p16 could be important in pluripotent stem cells.
Ap1 site-mediated regulation
Mammalian AP1 proteins are homodimers and heterodimers composed of basic region-leucine zippers (bZIP) proteins including Jun proteins (c-Jun, JunB, JunD), Fos proteins, Jun dimerization partners (JDP1 and JDP2), and the closely related activating transcription factors (ATF2, LRF1/ATF3 and B-ATF). Three AP1-like sites are present in the mouse p16 promoter, including TGACTGA at −1189, TGACTTCA at −783, and TGACACA at −484 (1, 77). It has been shown that ectopic expression of JunB induces high levels of P16, leading to premature senescence in primary mouse fibroblast and reduced proliferation in 3T3 cells (77). Such induction of p16 expression is attributed to transactivation upon binding of JunB to the Ap1 sites in the p16 promoter. JunB also down-regulates the expression of cyclin D1. As such, over-expression of JunB in 3T3 cells completely abolishes the kinase activity of CDK4, resulting in reduced pRb hyperphosphorylation and extended G1 phase. Conversely, c-Jun acts as an antagonist of JunB: it up-regulates cyclin D1 but down-regulates P16, thus promoting cell proliferation. Interestingly, as described earlier, P16 is able to bind to JNKs and inhibit c-Jun phosphorylation and AP-1 activity (22). These findings indicate a putative feedback loop between P16 and c-Jun, even though the underlying molecular basis and potential clinical significance are not clear.
PPRE-mediated regulation
A peroxisome proliferator response element (PPRE) has been identified in the p16 promoter at position −1023 (78). In vascular smooth muscle cells (SMC), peroxisome proliferator-activated receptor alpha (PPARα) negatively regulates cell cycle progression at the G1/S transition through inducing p16 expression. The underlying mechanism is that PPARα specifically binds to the canonical PPRE region and interacts with Sp1 in the proximal Sp1-binding sites of the p16 promoter, thus enhancing p16 mRNA expression. Similarly, PPARγ is able to activate the p16 promoter upon binding to the PPRE region in human diploid fibroblast cells (2BS and WI38), whereas phosphorylation of PPARγ represses its transactivation function (78).
Regulation mediated by unspecified elements
SNF5 is a component of the chromatin remodeling complex SWI/SNF. The SWI/SNF complex disrupts histone-DNA interactions to regulate access of binding domains to transcription machinery (79). Re-expression of hSNF5 in malignant rhabdoid tumor cells (MRT) leads to G1 arrest with induction of p16 expression and transcriptional repression of cyclins A, D1, and E (80). Since the BAF60a subunit of the mammalian SWI/SNF complex physically interacts with JunB, the SWI/SNF complex might be directly recruited to the p16 promoter so that hSNF5 is able to activate p16 transcription. In addition, as demonstrated in genetic studies in Drosophila, SWI/SNF belongs to the trithorax group of activators, which counteracts the Polycomb group of silencers (PcG) to maintain patterns of developmental gene expression (81, 82). It appears that SWI/SNF and PcG proteins act antagonistically, yet PcG proteins regulate the entire INK4a/ARF/INK4b locus as discussed in the following sections. Interestingly, a recent study has identified BRG1, the catalytic component of the SWI/SNF complex, as a novel binding partner of P16 (83, 84). While BRG1 is not required for P16-induced cell cycle inhibition, P16/BRG1 interaction negatively modulates the chromatin remodeling activity of BRG1. Taken together, these findings indicate a putative feedback loop between P16 and the SWI/SNF complex.
Most recently, it has been demonstrated that lymphoid specific helicase (Lsh), a member of the SNF2/helicase family, is involved in p16 regulation (85). In human diploid fibroblasts, Lsh over-expression delays cell senescence by silencing p16 and such transcriptional repression is attributed to Lsh-related deacetylation of histone H3 at the p16 promoter. Lsh also physically interacts with the p16 promoter as well as histone deacetylases 1 and 2 (HDAC1 and HDAC2) in vivo, which consequently recruits a co-repressor complex containing HDAC1 and HDAC2 to the p16 promoter and represses endogenous p16 expression. Nevertheless, the factor(s) guiding the sequence-specific binding of Lsh to the p16 promoter awaits elucidation.
Coordinated regulation of p16, p14ARF, and p15
In spite of the aforementioned fact that some stimuli selectively regulate p16 but not p14ARF nor p15, these three genes are lowly expressed in normal tissues as well as a considerable number of tumor specimens (with the intact INK4b/ARF/INK4a locus), and in most of cases, such low expression is concomitant (3, 4, 6). Moreover, although they have different 5′ regulatory domains, both p16 and p14ARF mRNAs are of extraordinary stability, which is presumably attributed to their shared 3′ sequences (86). These common regulatory features led to a notion that there might be mechanisms controlling p16, p14ARF, and p15 simultaneously. This notion has been strongly supported by the following observations showing that p16, p14ARF, and p15 are concomitantly down-regulated upon expression of PcG proteins, such as Bmi1, Cbx7, Ring1b or Phc2 (7, 87). Of note, PcG proteins are a group of transcriptional repressors that function to generate and recognize histone modifications, thus transcriptionally silencing chromatin, especially genomic domains with clusters of genes (1, 11, 88). First, in knockouts of Bmi1, Bmi1-deficient mouse embryonic fibroblasts (MEFs) undergo premature senescence and accelerated accumulation of p16, p19ARF, and p15. In contrast, ectopic expression of Bmi1 increases the lifespan of both mouse and human fibroblasts, and the proliferative defects of Bmi1-deficient cells can be partially rescued if Bmi1-deficient mice are crossed into an INK4a/ARF-null background. Similar observations have been reported with knockouts of Cbx2, Mel18, PPhc2, and Ring1b. Secondly, Bmi1-mediated repression of the INK4b/ARF/INK4a locus is dependent on the continued presence of the multi-protein complex containing EZH2, a histone methytransferase, and Polycomb-Repressive Complex 2 (PRC 2) (11). While it is frequently up-regulated in cancer, EZH2 is down-regulated in stressed and senescing cells, which coincides with reduction in associated trimethylation of lysine 27 of histone 3, displacement of Bmi1, and the transcriptional activation of the INK4b/ARF/INK4a locus. Thirdly, it has been shown that Cbx7, a PcG homologue, interacts with Ring1 and is localized to nuclear Polycomb bodies (89, 90). Cbx7 extends the lifespan of a wide range of normal human cells and immortalizes mouse fibroblasts by transcriptionally repressing the INK4b/ARF/INK4a locus in a Bmi1-independent manner (91).
The recent identification of a transcriptional regulatory element in the INK4b/ARF/INK4a locus has provided insights into the mechanisms underlying the PcG-involving coordinated regulation of the INK4b/ARF/INK4a locus (7, 88). As shown in Figure 3, a putative replication origin exists at 1.5 kb upstream of the ATG start codon of the p15 gene, and the location of this replication origin coincides with a DNA element of about 350 bp that are conserved among mammalian INK4b/ARF/INK4a loci. Remarkably, this regulatory domain, RDINK4/ARF (hereafter, RD), is also a relevant transcriptional regulatory sequence that enhances the concomitant expression of p15, p16, and p14ARF. Moreover, CDC6, an essential DNA replication regulator frequently over-expressed in human oral, brain, and lung cancers as well as a subset of mantle cell lymphomas, binds to RD and recruits HDAC 1 and HDAC2 to the INK4b/ARF/INK4a locus, resulting in heterochromatinization and transcriptional silencing. Furthermore, recent studies have demonstrated that CDK6 physically interacts with Bmi1 in young MEF cells, thus recruiting PRC 1 and PRC 2 to the INK4b/ARF/INK4a locus and resulting in transcriptional silencing of this locus as well as its replication during late S phase (92). Upon senescence, Jmjd3, a histone demethylase, is over-expressed and the MLL1 protein, a histone methyltransferase, is recruited to the INK4b/ARF/INK4a locus, provoking the dissociation of PcG proteins, its transcriptional activation and replication at early S phase. Therefore, the replication, transcription and epigenetics of the INK4b/ARF/INK4a locus are integrated through modulating the RD element.
Interestingly, another transcriptional co-repressor, COOH-terminal binding protein (CtBP), is able to access the p16 promoter and enhance the PcG-based epigenetic histone mark, thus favoring p16 silencing via DNA methylation (93). Whereas CtBP-mediated repression of p16 can be reduced by stimuli, such as increased ROS, CtBP has little influence on the expression of p14ARF, indicating that even though CtBP functions through the coordinated PcG-mediated regulation mechanism, its influences on p16 and p14ARF expression are different.
Additional mechanisms transcriptionally modulating p16
The expression of p16 can also be modulated by mechanisms other than the aforementioned independent and coordinated mechanisms, such as phase-specific expression, differential splicing, and the modulation of transcript stability. First, the expression of p16 oscillates throughout the cell cycle, reaching a peak during S phase when P16 is available to inactivate those no longer-needed CDK4/6-cyclin D complexes (56). Second, partial deletions, mutations, and promoter hypermethylation have been implicated in the generation of splicing variants of p16, with most appearing to be tissue specific. For examples, a number of aberrant p16 species that lose parts of exons 1 and 2 or contain insertions of intron 2 have been identified in gastric cancer, most of which lead to dysfunctional P16 (94). In melanoma, the wild-type exon 2 donor splicing site has been found to be “removed” by an intron 2 mutation, thus generating two alternative transcripts that read into intron 2 (95, 96). Of note, these variants exist only in tumor specimens, and their sequence alterations occur at regions different from those generating p16γ and p12. Third, an instability determinant within the 3′-untranslated region (UTR) of the p16 mRNA has been identified in human diploid fibroblasts (86). This 3′-UTR site exhibits a stem-loop structure and is a specific target of AUF1, an RNA binding protein (RBP) implicated in promoting mRNA decay. siRNA-induced reductions in AUF1 increase the stability of p16 mRNA, thus leading to elevated expression of p16 as well as cellular senescence. Fourth, HuR, the ubiquitously expressed member of the Hu RNA-binding protein family, is involved in AUF1-mediated decay of p16 mRNA through recruiting the RNA-induced silencing complex (RISC) (97). Similarly, other RBPs, such as heterogeneous ribonucleoprotein particles A1 and 2 (hnRNP A1 and hnRNP2), are able to negatively regulate the stability of p16 mRNA. Last but not least, recent studies have shown that p16 is subject to miRNA-mediated regulation, yet the significance of this regulatory interaction remains to be elucidated (98). In senescing human diploid fibroblasts and cervical carcinoma cells, elevated p16 expression was associated with down-regulation of miR-24-2, a miRNA that was predicted to interact with the p16 mRNA coding and 3′ UTR regions. Interestingly, ectopic miR24-2 over-expression led to remarkable reduction in P16 protein but not p16 mRNA, suggesting that miR24-2 negatively modulates the initiation and elongation of P16 translation. This finding agrees with extensive evidences demonstrating that mammalian miRNAs suppress protein biosynthesis more commonly than promote target mRNA degradation (98).
Taken together, a fairly large number of cis and trans factors positively or negatively regulate p16 expression through diverse mechanisms. Since some of these mechanisms (pathways) are correlated with each other in cells, p16 regulation cannot be explained by a single isolated pathway. For example, while Myc binds to E-boxes of the p16 promoter and up-regulates p16 transcription, it has also been demonstrated that Myc can activate Bmi1, a potent repressor of the entire INK4b/ARF/INK4a locus (1). Evidently, the interplay between Myc and Bmi1 increases the complexity in evaluating the influence of Myc on p16 expression. Furthermore, considering that some potential regulators of p16 expression, such as melanocyte-inducing transcription factor MITF, pRb, CDK4/6, E2Fs, are not addressed here in detail simply due to their yet-to-be-defined mechanisms, the cellular machinery controlling p16 expression should be even more complicated than that presented above. It is also worthwhile to note that physiological significance of these miscellaneous factors in regulating p16 gene should not be underestimated in comparison with those regulators mentioned earlier. An interesting example is pRb. It has been known for a long time that p16 expression is negatively associated with pRb status in human tumor cell lines (99, 100), which arguably raises the possibility that p16 transcription might be modulated by E2Fs since inactivation of pRb is expected to de-repress or activate E2F-responsive genes (101). Such inverse correlation is further supported by recent findings that elevated expression of p16 is strongly associated with human papillomavirus (HPV) infection in human cervical and oral tumors (102, 103). The best-characterized activity of E7 from HPV type 16, the most frequently detected type in cervical and oral cancers, is its ability to bind to and induce ubiquitination-mediated degradation of pRb, which results in constitutive activation of several transcription factors, including members in the E2F family (104, 105). Hence, HPV infection brings about concomitant pRb inactivation, E2F activation, and up-regulation of p16 transcription. Presumably, if there is a cellular pathway in which pRb transcriptionally regulates p16, this pathway would contribute in part to HPV-induced transactivation of p16 as well as cancer development.
Regulation at the posttranslational level
Phosphorylation
It has been reported that senescence in human prostatic epithelial cells (HPEC) does not only induce elevated expression of P16 protein but promotes phosphorylation of P16 (106). Interestingly, senescence-related forms of phosphorylated P16 exhibit increased binding affinity with CDK4/6 in comparison with unphosphorylated P16, indicating that phosphorylation of specific sites on P16 in senescent HEPC facilitates the binding of P16 to target CDKs and contribute thereby to G1 arrest in senescence. Additional studies also demonstrate that P16 is phosphorylated at four specific serine sites, Ser7, Ser8, Ser140, and Ser152, in human fibroblast cells (41). These four Ser residues do not directly contact CDK4/6 as revealed in the crystal structure of the P16/CDK6 complex (32), however, mutations involving these residues have been found in familial and sporadic melanomas, indicative of the importance of P16 phosphorylation in cancer. Remarkably, in WI38 cells, only Ser152 is phosphorylated in CDK4/6-bound P16 (41), suggesting that the physiological effects of phosphorylation at these four residues may be different from each other. This notion is further supported by a recent study, in which phosphomimetic Ser→Glu substitutions were used to evaluate the biochemical and biophysical effects upon phosphorylation (42). The results show that the phosphomimetic substitution at Ser8 of P16 eliminates the majority of its CDK4-inhibitory activity but does not perturb the core structure or the conformational stability. Contrarily, Ser→Glu substitutions at Ser 7, Ser140, and Ser152 do not bring about ant detectable changes in the core structure or the CDK4-inhibitory activity except that substitutions at Ser140 and Ser152 moderately destabilize P16 in heat-induced unfolding. In addition, it has been shown that P16 specifically binds to IKKβ, the primary kinase to phsosphorylate I Bα, in human fibroblast cells (42). Such binding leads to phosphorylation of P16 at Ser8, which consequently abolishes its CDK4-inhibitory activity as described above. These findings strongly support that phosphorylation of P16 represents an important mechanism of P16 regulation, whereas the corresponding kinases and physiological effects upon phosphorylation need to be further investigated. Thus, any pathways that influence phosphorylation of P16 might have an impact on P16 function. For example, the activation of IKKβ as a result of inflammatory cytokine signaling, infectious agents, and DNA damage, may potentially up-regulate CDK4-mediated phosphorylation of pRb through phosphorylating and inactivating P16. Moreover, protein phosphorylation is closely related to the level of intracellular oxidative stress (23, 106). Oxidative stress may induce P16 phosphorylation, which may enable tumor cells to enter cell division arrest and premature senescence, thus keeping them from progressing into malignant ones.
Degradation
P16 has a relatively short half life ranging from 30 minutes to 3.5 hours in a variety of cancer cell lines (107), yet the molecular mechanisms underlying such rapid turnover are unknown. Since it has been shown that P16 is degraded in a proteasome-dependent manner in vivo (108, 109), one might assume that like P53, P16 is led to ubiquitination-mediated proteasomal degradation upon phosphorylation. However, whilst conjugation of ubiquitin to an internal lysine is the initial event in ubiquitination-mediated proteasomal degradation of most of proteins, P16 is lysine-free (109). Moreover, endogenous P16 is completely acetylated at its N-terminus thus making P16 not suitable for non-lysine polyubiquitination at the N-terminal residue (108). In addition, the degradation of P16 is independent of ubiquitination and it only requires the 20S catalytic core, not the entire 26S proteasome, suggesting that both polyubiquitination and the 19S proteasome do not contribute to P16 turnover (108). Of note, the 26S proteasome consists of the 20S and 19S subunits. The 19S proteasome functions to bind polyubiquitinated polypeptides and drive them to the 20S catalytic core in an ATP-dependent process. Intriguingly, it has been demonstrated recently (108) that REG, an ATP- and ubiquitin-independent proteasome activator that interacts with the 20S catalytic core and enhances the latter’s activity, is physically associated with P16 in vivo, and loss of such association in REGγ-deficient cells stabilizes P16. These findings indicate the potential involvement of the REGγ pathway in P16 degradation.
Coordination with other proteins
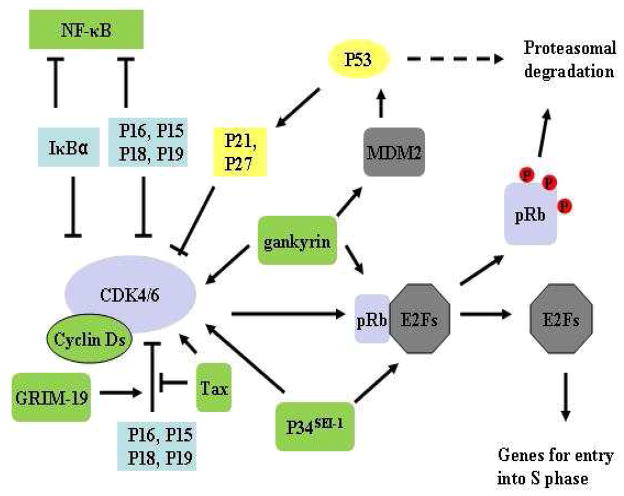
As described earlier, P16 functions through protein/protein interactions with diverse target proteins (16, 31). Any proteins that are able to influence the interactions between P16 and its targets could potentially contribute to the regulation of P16. Indeed, a number of proteins have been found to positively/negatively modulate the P16/CDK4 association as well as subsequent CDK4-mediated phosphorylation of pRb, most of which are relevant to human cancer. Some of these proteins are discussed below (Figure 5).
Figure 5. Coordination of P16 and other proteins in modulating CDK4/6-mediated phosphorylation of pRb.
Regulators to be investigated in our proposed studies are in green shadow. Arrows and bars represent positive and negative regulation, respectively. Dotted lines, genomic DNA; P, phosphorylated.
Cyclin Ds
In mammalian cells, CDK4 or 6 itself only exhibits minimal pRb-phosphorylating activity and becomes fully functional only after being charged with cyclin Ds, including D1, D2, and D3 (5, 16). The expression of cyclin Ds is controlled by the influence of the extracellular growth factors; when these mitogens are removed, the accumulation of cyclin Ds is prevented and cyclin Ds are rapidly degraded; thus, cell cycle progression is halted at the restriction point in G1 phase. Over-expression of cyclin Ds, especially cyclin D1, has been frequently found in many human cancers, such as esophageal carcinomas, non-small cell lung cancers, and breast cancers (110). Elevated expression of cyclin Ds in cells leads to increased CDK4-mediated phosphorylation of pRb thus promoting cell cycle progression. Since CDK4-P16 and CDK4-cyclin D binary complexes, not the P16/CDK4/cylin D ternary complex, are present in most of cells, over-expression of cyclin Ds could decrease the CDK4-P16 complex (30, 32).
Other INK4 proteins
Besides the aforementioned P15 and P16, the INK4 family contains another two proteins, P18 and P19, both of which are exclusively composed of five ARs (111). While all INK4 proteins have similar structures and are indistinguishable in CDK4 binding and inhibition (30, 40, 43, 112), there are notable differences among these proteins. First, the genes encoding these four proteins are located within distinct chromosomal regions and are regulated differently in transcription. The p18 and p19 genes are located within chromosomes 1p32 and 19p13.2, respectively (113), and their transcription is regulated periodically during the cell cycle: the expression levels of both genes are very low during the G1 phase, but increase rapidly at the G1/S transition with a maximum at the S phase (47, 114). In contrast, the expression level of p16 reaches its peak at the late G1 phase (56); the expression of p15 is upon induction by TGF-β (14). Secondly, INK4 proteins may play different roles in differentiation and senescence promotion. It has been reported that both p18 and p19 were widely expressed in different tissues during mouse embryogenesis while expression of p15 and p16 was not detected (115). Previous studies also showed that the expression of p16 and p18 increased as cells approached senescence (116). Therefore, these INK4 proteins are not regarded as functionally redundant in general. However, emerging evidences indicate that there exits certain redundancy among INK4 proteins in tumor suppression. It has been reported that loss of p18 in mice can induce elevated expression of p16 in certain tissues and deletion of both p18 and p16 brings synergistic effects in the development of pituitary tumors (117, 118). Similarly, acute suppression of p16 in primary human astrocytes results in enhanced proliferation and E2F-mediated induction of p18 expression, indicative of a potential compensatory mechanism in cells (117). Additionally, P15 is able to substitute P16 in tumor suppression in p16-deficient MEF cells (119). Under stress, loss of p16 leads to a significant increase in P15 in MEF cells, however, such increases occurs only at the protein level, indicating that P15 is stabilized in the absence of p16. Consistently, the expression of p16 promotes the proteasomal degradation of P15 (119). Hence, P15 functionally compensates for the loss of P16 through a mechanism different from the one underlying P18 compensation. Together, these findings strongly support that a hierarchy of tumor suppressive roles for INK4 proteins exists, wherein P15 and P18 likely serve as back-ups in the absence of P16.
KIP inhibitors
KIP proteins (CDK inhibitor proteins), including P21, P27, and P57, are universal inhibitors competent in inhibiting most of the CDK-cyclin complexes as well as some kinases unrelated to CDKs (5, 6). While CDK2 is their primary target, they also bind and inhibit CDK4 and CDK6 in cells. Binding of KIP proteins to CDK4/6 arguably leads to the displacement of P16 from CDK4/6, thus enabling P16 function in pathways other than CDK4/6-mediated phosphorylation of pRb or directly driving P16 into degradation. The distribution of KIPs between CDK4/6 and other CDKs also provides a mechanism in which P53 can indirectly influence CDK-mediated phosphorylation of pRb through modulating P21 (120). Additionally, it has been reported that P21 is able to activate the promoter of p16 in HeLa cells, and such activation involves Sp1 and the corresponding GC-boxes in the promoter (73) (Figure 4).
GRIM-19
As described earlier, GRIM-19 is able to suppress STAT3-dependent transcription and oncogenic transformation in HeLa cells upon IFN-β/RA-induction (19). Interestingly, ectopic expression of GRIM-19 is able to suppress the expression of genes controlled by E2F1. Furthermore, such suppression is achieved via physically associating with P16 and boosting the latter’s inhibition of CDK4/6-mediated phosphorylation of pRb. In cells, ectopic expression of GRIM-19 leads to the formation of a ternary complex containing CDK4, P16, and GRIM-19, and the presence of GRIM-19 enhances the binding of P16 to CDK4 (19). In contrast, over-expression of cyclin D1 led to loss of the CDK4-P16-GRIM-19 ternary complex and prevalence of the cyclin D1/CDK4 binary complex, suggesting that binding of cyclin D1 disrupts CDK4 interactions with P16/GRIM-19.
NF-κB
NF-κB is a transcription factor controlling vital genes required for immune response and inflammation, cell growth and differentiation, cell adhesion, and apoptosis (121). Emerging evidences have demonstrated a crosstalk between the INK4-CDK4-pRb and IKK-NF-κB pathways (122, 123). Whereas P16 is able to bind to and suppress the transactivational activity of NF-κB, IκBα, a specific inhibitor of NF-κB, competes with P16 for CDK4 binding and inhibits CDK4-mediated phosphorylation of pRb as potently as P16 does (122). Moreover, IKKβ, the primary kinase for IκBα phosphorylation, is capable of phosphorylating P16 at Ser8 (42). Such crosstalk leads to a potential correlation between two major molecular events in human cancer, namely, down-regulation of p16 and activation (over-expression) of NF-κB. While inhibition of CDK4 by IκBα could serve as a safety back-up in the absence of P16, over-expressed NF-κB may compete with CDK4 for binding P16, thus promoting cell cycle progression through the pRb pathway (42).
Gankyrin
Gankyrin is a newly defined regulatory subunit associated with the 26S proteasome (124, 125). Over-expression of gankyrin has been found to be associated with many human cancers, including HCCs (126), esophagus SCCs (ESCCs) (127), colorectal (128), pancreatic (129) and lung cancers (130). Gankyrin functions as a dual-negative regulator of the two most prominent tumor suppressor pathways: (i) INK4-CDK4-pRb, and (ii) ARF-MDM2-p53 pathways. On one hand, gankyrin binds to the ubiquitin ligase MDM2 and promotes P53 ubiquitination and subsequent proteasomal degradation (131). On the other hand, gankyrin physically interacts with pRb and facilitates the latter’s phosphorylation and degradation (126). In parallel, gankyrin competes with P16 for binding CDK4, but gankyrin binding to CDK4 does not inhibit the CDK4 activity, thus leading to cell cycle progression (132). Furthermore, gankyrin is a key regulator of oncogenic Ras-mediated activation of Akt through inhibiting the downstream RhoK/ROCK/PTEN pathway in mouse and human cells, thus playing an essential role in Ras-induced tumorigenesis (130).
SEI-1/TRIP-Br1
The SEI-1 gene is a candidate oncogene located within human chromosome 19q13.1, a region frequently amplified in human breast, esophagus, ovarian, lung, and pancreatic cancers (133). The SEI-1 gene product P34SEI-1 (also named SERTAD1 and TRIP-Br1) functions in multiple physiological processes (134). First, through up-regulating the SEI-1/SET/NM23H1 pathway, ectopic expression of SEI-1 markedly increases the frequencies of chromosomal alteration and micronuclei formation, thus inducing chromosomal instability (135). Secondly, P34SEI-1 is able to inhibit apoptosis through stabilizing the X-linked inhibitor of apoptosis protein (136). Thirdly, P34SEI-1 specifically binds to CDK4 (but not CDK6) in vitro and in vivo, and this binding appears to antagonize the function of P16, thus rendering CDK4-mediated phosphorylation of pRb resistant to the inhibitory effect of P16 during late G1 phase (137). P34SEI-1 is also able to contact DP-1 and stimulate E2F1/DP-1 transcriptional activity (134). Since both inactivation of p16 and over-expression of SEI-1 are prevalent in the aforementioned human tumor types (138), these two events are synergistic in regard to their influences on the CDK4-pRb-E2F pathway.
Tax
Encoded by human T lymphotropic virus 1 (HTLV-1) genome DNA exon 2, Tax is a transcription activator crucial for both HTLV-1 viral gene expression and transcription regulation in HTLV-1 infected cells (139, 140). Tax is believed to be responsible for adult T-cell leukemia and other HTLV-1 related diseases, such as tropical spastic paraparesis. The expression of Tax protein in HTLV-1 infected cells is correlated with an increase in CDK4 activity (139, 141). The underlying mechanisms involve the following two interactions. First, Tax is able to form a binary complex with P16 in vitro and in vivo, thus counteracting the CDK4/6-inhibitory activity of P16, resulting in cell cycle progression (141). Secondly, Tax protein directly binds to CDK4, and stimulates the latter’s activity in phosphorylating pRb (141). In addition, Tax binds to cyclin D3 and induces a novel hyperphosphorylated cyclin D3 protein and the concomitant increase in CDK4 kinase activity (140).
Regulation of P16 and human cancer
Carcinogenesis is a multi-step process through which normal cells are sequentially transformed via the activation of proto-oncogenes and inactivation of tumor suppressor genes into their malignant derivatives. While genetic inactivation of p16 has been one of the most prominent genetic changes identified in human cancers to date, some frequent molecular events, such as activation (over-expression) of cyclin D1 (CCND1), gankyrin (PSMD10), SEI-1 (SERTAD1), CDC6, and NF-κB, also lead to the deregulation of P16 function through diverse mechanisms. Consequently, the physiological up-regulation or down-regulation of P16 in human cancers is subject to the coordination and summation of well-known genetic alterations of p16 and activation of aforementioned oncogenes as well as changes in related tumor suppressors. On one hand, as described earlier, functional redundancy allows P15 or P18 (or IκBα) to successfully substitute for P16 in tumor suppression in the presence of genetic inactivation of p16 (deletion and silencing) (117, 119), whereas partial loss of P16 function due to missense p16 mutations can be compensated by elevated expression as observed in some tumor specimens (10, 142). On the other hand, even in the presence of intact p16, other molecular events, such as over-expression of CDC6, cyclin D1, gankyrin, and SEI-1 have the potential to “functionally inactivate” P16, thus promoting cancer progression. Hence, the genetic status of p16 does not automatically reflect the status of P16 function in human cancers, and the contributions of P16 function to human cancer may vary with individual cases. For example, deletion or silencing of p16 could be the primary cause for aberrant cell proliferation in the presence of intact and normally expressed p15 and p18, whereas synergistic effect is likely observed on aberrant cell proliferation in the absence of p16, p15, and/or p18 (117, 119). In contrast, loss of p16 may not contribute significantly to carcinogenesis in the presence of increased expressed p15 and p18.
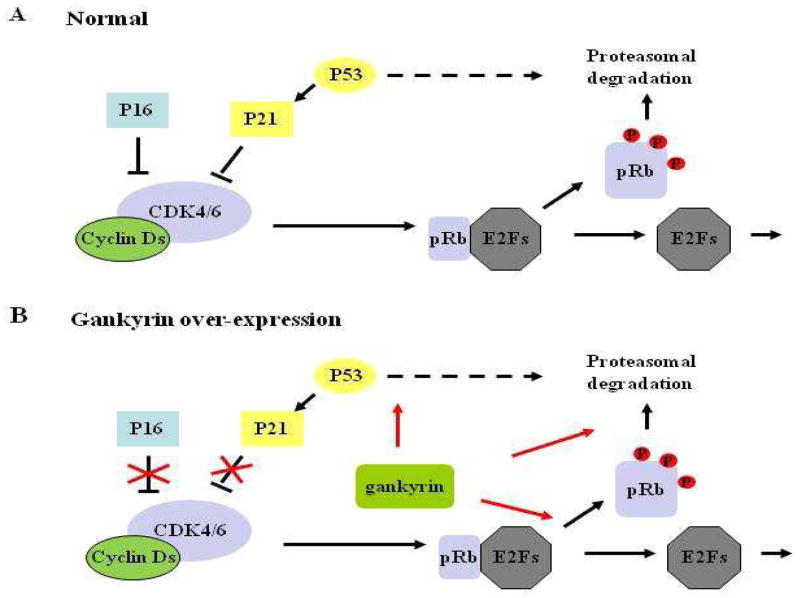
The coordination between genetic inactivation of p16 and oncogene-mediated deregulation of P16 may also vary with the developmental stages of human cancers (125). Even though genetic inactivation of p16 has been established as a landmark during epithelial cancer progression, it remains to be elucidated whether this event occurs at the earliest, “initiating” stages of carcinogenesis. Emerging evidences show that deregulation of P16 through activation of the aforementioned oncogenes occurs during the earliest stages of cancer development, possibly in advance to genetic alterations of p16 (143–145). For example, in a chemically-induced rodent hepatocarcinogenesis model, hypermethylation of the p16 gene appears at later HCC stages, whereas gankyrin is over-expressed soon much after the initial carcinogen exposure (liver fibrosis), preceding the loss of pRb (cirrhosis) and hepatocellular adenoma formation (HCA) (143). Moreover, it has been reported that the frequency of gankyrin over-expression in human HCC decreases as Edmondson-Steiner grades increase, indicating that aberrant gankyrin over-expression is an early initiating event in HCC development (144, 145). Recent studies in our laboratory have also demonstrated that gankyrin is over-expressed in all tested human OSCC specimens and about 50% of premalignant oral lesions (146). Hence, aberrant gankyrin expression, independent of genetic alterations of p16, may represent a valuable biomarker for early cancer detection and intervention.
Furthermore, oncogene-mediated deregulation of P16 function may have more profound effects than genetic inactivation of p16. On one hand, oncoproteins such as gankyrin and P34SEI-1 have diverse physiological functions, and their aberrations may generate multifactorial consequences capable of modulating cell growth and carcinogenic progression independently (125, 137). For example, aberrant gankyrin expression may bring about changes in the cell cycle via CDK4 interference, ubiquitination-mediated protein degradation of pRb and P53 via the proteasome complex, and p53-mediated apoptotic suppression (125) (Figure 6). On the other hand, perturbations in RD/CDC6 interaction, such as genetic alterations of RD and elevated expression of CDC6, may have a global effect on all three tumor suppressors (P16, P15, and P14ARF) encoded by the INK4b/ARF/INK4a locus, thus providing an inclusive effect greater than the individual genetic inactivation events of the p16 or p14ARF genes (88). Moreover, in most of human cancers, genetic alterations occur at the p16 gene while the p15 gene remains intact (6). However, perturbations in RD/CDC6 interaction are able to abolish the aforementioned functional redundancy between P15 and P16 in tumor suppression.
Figure 6. Multifactorial effects of aberrant gankyrin over-expression on cell growth and carcinogenic progression.
In both A and B, arrows and crosses represent positive and negative regulatory effects, respectively. In B, red crosses indicate that over-expressed gankyrin preludes the inhibition of P16 and P21 on CDK4/6, while red arrows represent the enhancing effects on phosphorylation of pRb, ubiquitination of P53, and proteasome-mediated degradation of both pRb and P53. P, phosphorylated. This figure was modified from Reference 125.
While down-regulation of P16 mainly contributes to cancer progression through promoting aberrant cell proliferation, the involvement of up-regulation (over-expression) of P16 in human cancers is poorly documented. Once regarded as a characteristic of stress-induced cellular senescence and aging (3, 23), over-expression of P16 (including both wild-type and mutant P16) has been reported to be associated with poor prognosis for cancers including neuroblastoma, cervical, ovarian, breast, and prostate tumors as well as OSCCs (10). Specifically, the incidence of over-expression of P16 is up to 55% in human OSCCs. Apparently, even though over-expression of some partially-impaired P16 mutants may represent an unidentified feedback mechanism to render P16 functions in some tumor specimens, the involvement of over-expression of P16 in cancers deviates from its conventional role as a tumor suppressor. It is very likely that over-expression of P16 is induced by stress or oncogenic environmental risk factors through an undefined feedback loop, but its inhibition of cell proliferation is bypassed or counteracted by other molecular events, such as aberrant SEI-1 expression (138) or the expression of viral Tax protein in the host cells upon HLTV-1 infection (141), so that cell transformation and aberrant cell proliferation occur in the presence of elevated P16. Like in stress-induced senescence and aging, over-expression of p16 in human cancers could be mainly ascribed to the transactivation of the p16 gene through Ras-Raf-Mek signaling as well as other mechanisms (3, 23). However, some alterations in RD/CDC6 interaction, such as amplification of RD or down-regulation of CDC6, may underlie up-regulation of p16 in human cancers (88) as evidenced by our recent finding that p16 mRNA over-expression in HeLa cells is correlated with the amplification of RD (unpublished data).
It is also important to note that the involvement of P16 in the age-associated decline in function of certain tissue-specific stem cells (25–27, 147) is of significance in emerging anti-cancer stem cell therapeutics. Cancer stem cells (CSCs) are a small, resilient subset (less than 1 in 10,000) of cells found within many tumors or hematological cancers that possess characteristics associated with normal stem cells, especially the capacity to self-renew and differentiate, leading to tumor initiation and driving its growth, recurrence and metastasis (147–155). CSCs are pluripotent, which allows these cells to adapt and to resist to standard chemotherapy, radiotherapy, and even current molecular targeted therapies (147). Hence, for cancer therapy, it is best to eliminate CSCs, as the dormant CSCs may re-enter proliferative phase once the proliferation-inhibiting drugs are cleared (156, 157). Recent studies have shown that repression of p16 (as well as p14ARF) by aforementioned Bmi1 (Figure 4) and other regulators is a key requirement for self-renewal of stem cells, and this Bmi1-P16 signaling pathway appears to be active in CSCs (147, 158–160). Presumably, up-regulating P16 or down-regulating Bmi1 in CSCs negatively modulates the proliferation and self-renewal of CSCs, thus reducing cancer incidence. From this perspective, P16 represents a potential target for cancer eradication by the elimination of CSCs.
In conclusion, the regulation of P16 function is multifactorial. It integrates mechanisms that target the DNA, RNA, and protein levels through independent and overlapping pathways, some of which remain to be further explored. While the general role of P16 in tumor suppression is well established, the specific contributions of p16 deregulation to the development of a particular tumor depend on the nature of the p16 deficiency and the coordination of other mediating molecular events occurring in the same tumor microenvironment. This complex orchestration of direct and indirect mechanism of growth control derived from alterations in P16 function may best be addressed by a “personalized” assessment of the intricate roles of P16 in cancer progression. As the approach of personalized diagnosis or personalized medicine is becoming a trend for cancer treatment, it is a challenging mission for biochemists to facilitate this process by uncovering more of the molecular details of the “regulatory web” of P16.
Acknowledgments
The authors would like to thank Dr. Thomas J. Knobloch in Division of Environmental Health Sciences, College of Public Health for critical reading of this manuscript. The authors also apologize to many colleagues whose work could not be cited here due to space constraints.
Abbreviations
- AR
ankyrin repeat
- BE
Barrett’s esophagus carcinoma
- CDK
cyclin-depedent kinase
- CDKN2A
CDK inhibitor 2a, also P16INK4A (P16)
- CDKN1B
CDK inhibitor 1b, also P27
- CSC
cancer stem cell
- ESCC
esophagus squamous cell carcinoma
- GdmHCl
guanidinium hydrochloride
- GRIM-19
gene associated retinoid-IFN-induced mortality-19
- HCA
hepatocellular adenoma formation
- HCC
hepatocellular carcinoma
- HDAC
histone deacetylase
- HLH
helix-loop-helix
- HPV
human papillomavirus
- HTH
helix-turn-helix
- HTLV-1
human T lymphotropic virus 1
- IFN
interferon
- INK4
inhibitors of cyclin-dependent kinases 4 and 6, including P16, P15INK4B (P15), P18INK4C (P18) and P19INK4D (P19)
- JUNK
c-jun N-terminal kinase
- KIP
universal CDK inhibitors, including P21, P27, and P57
- MEF
mouse embryonic fibroblasts
- MRT
malignant rhaboid tumor cell
- MTS 1and 2
multiple tumor suppressors 1 and 2, also referred to P16 and P15, respectively
- NSCLC
non-small cell lung carcinoma
- p14ARF
the alternate reading frame of the p16 gene, also p16β
- OSCC
oral squamous cell carcinoma
- PcG
the Polycomb group of silencers
- pRb
the retinoblastoma susceptible gene product
- PRC
the Polycomb-repressive complex
- RD
the enhancer element of the INK4b/ARF/INk4a locus
- RISC
the RNA-induced silencing complex
- ROS
reactive oxygen species
- SCCHN
squamous cell carcinoma of the head and neck
- SMC
smooth muscle cell
- UTR
untranslated region
Footnotes
The work on P16 in the author’s laboratory was supported by NIH RO1 CA69472.
References
- 1.Gil J, Peters G. Regulation of the INK4b-ARF-INK4a tumor suppressor locus: all for one or one for all. Nature Reviews. 2006;7:667–677. doi: 10.1038/nrm1987. [DOI] [PubMed] [Google Scholar]
- 2.Sharpless NE. INK4a/ARF: A multifunctional tumor suppressor locus. Mut Res. 2005;576:22–38. doi: 10.1016/j.mrfmmm.2004.08.021. [DOI] [PubMed] [Google Scholar]
- 3.Collado M, Blasco MA, Serrano M. Cellular senescence in cancer and aging. Cell. 2007;130:223–233. doi: 10.1016/j.cell.2007.07.003. [DOI] [PubMed] [Google Scholar]
- 4.Kim WY, Sharpless NE. The regulation of INK4/ARF in cancer and aging. Cell. 2006;127:265–275. doi: 10.1016/j.cell.2006.10.003. [DOI] [PubMed] [Google Scholar]
- 5.Sherr CJ, Roberts JM. Living with or without cyclins and cyclin-dependent kinases. Genes Dev. 2004;18:2699–2711. doi: 10.1101/gad.1256504. [DOI] [PubMed] [Google Scholar]
- 6.Ortega S, Malumbres M, Barbacid M. Cyclin D-dependent kinases, INK4 inhibitors and cancer. Biochim Biophys Acta. 2002;1602:73–89. doi: 10.1016/s0304-419x(02)00037-9. [DOI] [PubMed] [Google Scholar]
- 7.Gonzalez S, Serrano M. A new mechanism of inactivation of the INK4/ARF locus. Cell Cycle. 2006;5:1382–1384. doi: 10.4161/cc.5.13.2901. [DOI] [PubMed] [Google Scholar]
- 8.Esteller M, Corn PG, Baylin SB, Herman JG. A gene hypermethylation profile of human cancer. Cancer Res. 2001;61:3225–3229. [PubMed] [Google Scholar]
- 9.Ruas M, Brooks S, McDonald NQ, Peters G. functional evaluation of tumor-specific variants of p16INK4a/CDKN2A: correlation with protein structure information. Oncogene. 1999;18:5423–5434. doi: 10.1038/sj.onc.1202918. [DOI] [PubMed] [Google Scholar]
- 10.Lang JC, Borchers J, Danahey D, Smith S, Stover DG, Agrawal A, Malone JP, Schuller DE, Weghorst CM, Holinga AJ, Lingam K, Patel CR, Esham B. Mutational status of overexpressed p16 in head and neck cancer: evidence for germline mutation of p16/p14ARF. Int J Oncol. 2002;21:401–408. doi: 10.3892/ijo.21.2.401. [DOI] [PubMed] [Google Scholar]
- 11.Canepa ET, Scassa ME, Ceruti JM, Marazita MC, Carcagno AL, Sirkin PF, Ogara MF. INK4 proteins, a family of mammalian CDK inhibitors with novel biological functions. IUBMB Life. 2007;59:419–426. doi: 10.1080/15216540701488358. [DOI] [PubMed] [Google Scholar]
- 12.Serrano M, Hannon GJ, Beach D. A new regulatory motif in cell-cycle control causing specific inhibition of cyclin D/CDK4. Nature. 1993;366:704–707. doi: 10.1038/366704a0. [DOI] [PubMed] [Google Scholar]
- 13.Kamb A, Gruis NA, Weaver-Feldhaus J, Liu Q, Harshman K, Tavigian SV, Stockert E, Day RS, Johnson EE, Skolnik MH. A cell cycle regulator potentially involved in genesis of many tumor types. Science. 1994;264:436–440. doi: 10.1126/science.8153634. [DOI] [PubMed] [Google Scholar]
- 14.Hannon GJ, Beach D. p15INK4B is a potential effector of TGF-beta-induced cell cycle arrest. Nature. 1994;371:257–261. doi: 10.1038/371257a0. [DOI] [PubMed] [Google Scholar]
- 15.Quelle DE, Ashmun RE, Hannon GJ, Renberger PA, Trono D, Richter KH, Walker C, Beach D, Sherr CJ, Serrano M. Cloning and characterization of murine p16INK4a and p15INK4b genes. Oncogene. 1995;11:635–645. [PubMed] [Google Scholar]
- 16.Sherr CJ, Roberts JM. CDK inhibitors: positive and negative regulators of G1-phase progression. Genes Dev. 1999;13:1501–1512. doi: 10.1101/gad.13.12.1501. [DOI] [PubMed] [Google Scholar]
- 17.Iaquinta PJ, Lees JA. Life and death decisions by the E2F transcription factors. Curr Opin Cell biol. 2007;19:649–657. doi: 10.1016/j.ceb.2007.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nevins JR. E2F: a link between the Rb tumor suppressor protein and viral oncoproteins. Science. 1992;258:424–429. doi: 10.1126/science.1411535. [DOI] [PubMed] [Google Scholar]
- 19.Sun P, Nallar SC, Raha A, Kalakonda S, Velalar CN, Reddy SP, Kalvakolanu DV. GRIM-19 and p16INK4a synergistically regulate cell cycle progression and E2F1-responsive gene expression. J Biol Chem. 2010;285:27545–27552. doi: 10.1074/jbc.M110.105767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nishiwaki E, Turner SL, Harju S, Miyazaki S, Kashiwagi M, Koh J, Serizawa H. Regulation of CDK7-carboxyl-terminal domain kinase activity by the tumor suppressor p16INK4A contributes to cell cycle regulation. Mol Cell Biol. 2000;20:7726–7734. doi: 10.1128/mcb.20.20.7726-7734.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Serizawa S. Cyclin-dependent kinase inhibitor p16INK4A inhibits phosphorylation of RNA polymerase II by general transcription factor TFIIH. J Biol Chem. 1998;273:5427–5430. doi: 10.1074/jbc.273.10.5427. [DOI] [PubMed] [Google Scholar]
- 22.Choi BY, Choi HS, Ko K, Cho YY, Zhu F, Kang BS, Ermakova SP, Ma WY, Bode AM, Dong Z. The tumor suppressor p16INK4a prevents cell transformation through inhibition of c-Jun phosphorylation and AP-1 activity. Nature Struct Cell Biol. 2005;12:699–707. doi: 10.1038/nsmb960. [DOI] [PubMed] [Google Scholar]
- 23.Yang DG, Liu L, Zheng XY. Cyclin-dependent kinase inhibitor p16INK4a and telomerase may co-modulate endothelial progenitor cells senescence. Ageing Res Reviews. 2008;7:137–146. doi: 10.1016/j.arr.2008.02.001. [DOI] [PubMed] [Google Scholar]
- 24.Krishnamurthy J, Torrice C, Ramsey MR, Kovalev GI, Al-Regaiey K, Su L, Sharpless NE. Ink4a/Arf expression is a biomarker of aging. J Clin Invest. 2004;114:1299–1307. doi: 10.1172/JCI22475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Krishnamurthy J, Ramsey MR, Ligon KL, Torrice C, Koh A, Nonner-Weir S, Sharpless NE. p16INK4a induces an age-dependent decline in islet regerative potential. Nature. 2006;443:453–457. doi: 10.1038/nature05092. [DOI] [PubMed] [Google Scholar]
- 26.Molofsky AV, Slutsky SG, Joseph NM, He S, Pardal R, Krishnamurthy J, Sharpless NE, Morrison RJ. Increasing p16INk4a expression decreases forebrain progenitors and neurogenesis during aging. Nature. 2006;443:448–452. doi: 10.1038/nature05091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Janzen V, Forkert R, Fleming HE, Saito Y, Waring MT, Dombkowski DM, Cheng T, DePinho RA, Sharpless NE, Scadden DT. Stem-cell ageing modified by the cyclin-dependent kinase inhibitor p16INK4a. Nature. 2006;443:421–426. doi: 10.1038/nature05159. [DOI] [PubMed] [Google Scholar]
- 28.Al-Mohanna MA, Manogaran PS, Al-Mukhalafi ZK, Al-hussein A, Aboussekhra A. The tumor suppressor p16(INK4a) gene is a regulator of apoptosis induced by ultraviolet light and cisplatin. Oncogene. 2004;23:201–212. doi: 10.1038/sj.onc.1206927. [DOI] [PubMed] [Google Scholar]
- 29.Le HV, Minn AJ, Massague J. Cyclin-dependent kinase inhibitors uncouple cell cycle progression from mitochondrial apoptotic functions in DNA-damaged cancer cells. J Biol Chem. 2005;280:32018–32025. doi: 10.1074/jbc.M504689200. [DOI] [PubMed] [Google Scholar]
- 30.Byeon IJ, Li J, Ericson K, Selby TL, Tevelev A, Kim HJ, O’Maille P, Tsai M-D. Tumor suppressor p16INK4A: determination of solution structure and analyses of its interaction with cyclin-dependent kinase 4. Mol Cell. 1998;1:421–431. doi: 10.1016/s1097-2765(00)80042-8. [DOI] [PubMed] [Google Scholar]
- 31.Li J, Mahajan A, Tsai M-D. Ankyrin repeat: a unique motif mediating protein-protein interactions. Biochemistry. 2006;45:15168–15178. doi: 10.1021/bi062188q. [DOI] [PubMed] [Google Scholar]
- 32.Russo AA, Tong L, Lee JO, Jeffrey PD, Pavletich NP. Structural basis for inhibition of the cyclin-dependent kinase Cdk6 by the tumour suppressor p16INK4a. Nature. 1998;395:237–243. doi: 10.1038/26155. [DOI] [PubMed] [Google Scholar]
- 33.Brotherton DH, Dhanaraj V, Wick S, Brizuela L, Domaille PJ, Volyanik E, Xu X, Parisin E, Smith BO, Archer SJ, Serrano M, Brenner SL, Blundell TL, Laue ED. Crystal structure of the complex of the cyclin D-dependent kinase Cdk6 bound to the cell-cycle inhibitor p19INK4d. Nature. 1998;395:244–250. doi: 10.1038/26164. [DOI] [PubMed] [Google Scholar]
- 34.Jeffrey PD, Tong L, Pavletich NP. Structural basis of inhibition of CDK-cyclin complexes by INK4 inhibitors. Genes Dev. 2000;14:3115–3125. doi: 10.1101/gad.851100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sotillo R, Dubus P, Martin J, de la Cueva E, Ortega S, Malumbres M, Barbacid M. Wide spectrum of tumors in knock-in mice carrying a Cdk4 protein insensitive to INK4 inhibitors. EMBO J. 2001;20:6637–6647. doi: 10.1093/emboj/20.23.6637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wolfel T, Hauer M, Schneider J, Serrano M, Wolfel C, Klehmann-Hieb E, De Plaen E, Hankeln T, Meyer zum Buschenfelde KH, Beach D. A p16INK4a-insensitive CDK4 mutant targeted by cytolytic T lymphocytes in a human melanoma. Science. 1995;269:1281–1284. doi: 10.1126/science.7652577. [DOI] [PubMed] [Google Scholar]
- 37.Rane SG, Cosenza SC, Mettus RV, Reddy EP. Germ line transmission of the Cdk4(R24C) mutation facilitates tumorigenesis and escape from cellular senescence. Mol Cell Biol. 2002;22:644–656. doi: 10.1128/MCB.22.2.644-656.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hayashi N, Sugimota Y, Tsuchiya E, Ogawa M, Nakamura Y. Somatic mutations of the MTS (multiple tumor suppressor) 1/CDK4I (cyclin-dependent kinase inhibitor) gene in human primary non-small cell lung carcinomas. Biochem Biophys Res Commun. 1994;202:1426–1430. doi: 10.1006/bbrc.1994.2090. [DOI] [PubMed] [Google Scholar]
- 39.Fåhraeus R, Paramio JM, Ball KL, Laín S, Lane DP. Inhibition of pRb phosphorylation and cell-cycle progression by a 20-residue peptide derived from p16CDKN2/INK4A. Curr Biol. 1996;6:84–91. doi: 10.1016/s0960-9822(02)00425-6. [DOI] [PubMed] [Google Scholar]
- 40.Tevelev A, Byeon IJ, Selby T, Ericson K, Kim HJ, Kraynov V, Tsai M-D. Tumor suppressor p16INK4A: structural characterization of wild-type and mutant proteins by NMR and circular dichroism. Biochemistry. 1996;35:9475–9487. doi: 10.1021/bi960211+. [DOI] [PubMed] [Google Scholar]
- 41.Gump J, Stokoe D, McCormick F. Phosphorylation of p16INK4A correlates with Cdk4 association. J Biol Chem. 2003;278:6619–6622. doi: 10.1074/jbc.C200622200. [DOI] [PubMed] [Google Scholar]
- 42.Guo Y, Yuan C, Weghorst CM, Li J. IKKβ specifically binds to P16 and phosphorylates Ser8 of P16. Biochem Biophy Res Commun. 2010;393:504–508. doi: 10.1016/j.bbrc.2010.02.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Guo Y, Mahajan A, Yuan C, Joo SH, Weghorst WM, Tsai MD, Li J. Comparisons of the conformational stability of cyclin-dependent kinase (CDK) 4-interacting ankyrin repeat (AR) proteins. Biochemistry. 2009;48:4050–4062. doi: 10.1021/bi802247p. [DOI] [PubMed] [Google Scholar]
- 44.Mao L, Merlo A, Bedi G, Shapiro GI, Edwards CD, Rollins BJ, Sidransky D. A novel p16INK4A transcript. Cancer Res. 1995;55:2995–2997. [PubMed] [Google Scholar]
- 45.Lin YC, Diccianni MB, Kim Y, Lin HH, Lee CH, Lin RJ, Joo SH, Li J, Chuang TJ, Yang AS, Kuo HH, Tsai MD, Yu AL. Human p16γ, a novel transcriptional variant of p16INK4A, coexpresses with p16INK4A in cancer cells and inhibits cell-cycle progression. Oncogene. 2007;26:7017–7027. doi: 10.1038/sj.onc.1210507. [DOI] [PubMed] [Google Scholar]
- 46.Robertson KD, Jones PA. Tissue-specific alternative splicing in the human INK4a/ARF cell cycle regulatory locus. Oncogene. 1999;18:3810–3820. doi: 10.1038/sj.onc.1202737. [DOI] [PubMed] [Google Scholar]
- 47.Reynisdottir I, Polyak K, Iavarone A, Massague J. Kip/Cip and Ink4 Cdk inhibitors cooperate to induce cell cycle arrest in response to TGF-beta. Genes Dev. 1995;9:1831–1845. doi: 10.1101/gad.9.15.1831. [DOI] [PubMed] [Google Scholar]
- 48.Quelle DE, Zindy F, Ashmun RA, Sherr CJ. Alternative reading frames of the INK4a tumor suppr essor gene encode two unrelated proteins capable of inducing cell cyle arrest. Cell. 1995;83:993–1000. doi: 10.1016/0092-8674(95)90214-7. [DOI] [PubMed] [Google Scholar]
- 49.Zhang Y, Xiong Y, Yarbrough WG. ARF promotes MDM2 degradation and stabilizes p53: ARF-INK4a locus deletion impairs both the Rb and p53 tumor suppression pathways. Cell. 1998;92:725–734. doi: 10.1016/s0092-8674(00)81401-4. [DOI] [PubMed] [Google Scholar]
- 50.Latres E, Malumbres M, Sotillo R, Martin J, Ortega S, Martin-Caballero J, Flores JM, Cordon-Cardo C, Barbacid M. Limited overlapping roles of P15(INK4b) and P18(INK4c) cell cycle inhibitors in proliferation and tumorigenesis. EMBO J. 2000;19:3496–3506. doi: 10.1093/emboj/19.13.3496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bardeesy N, Aguirre AJ, Chu GC, Cheng KH, Lopez LV, Hezel AF, Feng B, Brennan C, Weissleder R, Mahmood U, Hanahan D, Redston MS, Chin Li, Depinho RA. Both p16(Ink4a) and the p19(Arf)-p53 pathway constrains progression of pancreatic adenocarcinoma in the mouse. Proc Natl Acad Sci USA. 2006;103:5947–5952. doi: 10.1073/pnas.0601273103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kresty LA, Mallery SR, Knobloch TJ, Li J, Lloyd M, Casto BC, Weghorst CM. Frequent alterations of p16INK4a and p14ARF in oral proliferative verrucous leukoplakia. Cancer Epidemiol Biomarkers Prev. 2008;17:3179–3187. doi: 10.1158/1055-9965.EPI-08-0574. [DOI] [PubMed] [Google Scholar]
- 53.Ruas M, Peters G. The p16INK4a/CDKN2A tumor suppressor and its relatives. Biochim Biophys Acta. 1998;1378:F115–177. doi: 10.1016/s0304-419x(98)00017-1. [DOI] [PubMed] [Google Scholar]
- 54.Forbes S, Clements J, Dawson E, Bamford S, Webb T, Dogan A, Flanagan A, Teague J, Wooster R, Futreal PA, Stratton MR. Cosmic 2005. Br J Cancer. 2006;94:318–322. doi: 10.1038/sj.bjc.6602928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Greenblatt MS, Beaudet JG, Gump JR, Godin KS, Trombley L, Koh J, Bond JP. Detailed computational study of p53 and p16: using evolutionary sequence analysis and disease-associated mutations to predict the functional consequences of allelic variants. Oncogene. 2003;22:1150–1163. doi: 10.1038/sj.onc.1206101. [DOI] [PubMed] [Google Scholar]
- 56.Li J, Byeon I-J, Poi MJ, Ericson K, Selby T, O’Maille P, Qin D, Tsai M-D. Structure-function relationship of the INK4 family of tumor suppressors. In: Ehrlich M, editor. DNA Alterations in Cancer: Genetic and Epigenetic Changes. Eaton Publishing; Massachusetts: 2000. pp. 71–83. [Google Scholar]
- 57.Sanchez_beato M, Sanchez-Aguilera A, Piris MA. Cell cycle deregulation in B-cell lymphomas. Blood. 2003;101:1220–1235. doi: 10.1182/blood-2002-07-2009. [DOI] [PubMed] [Google Scholar]
- 58.Schutte M, Hruban RH, Geradts J, Maynard R, Hilgers W, Rabindran SK, Moskaluk CA, Hahn SA, Schwarte-Waldhoff I, Schmiegel W, Baylin SB, Kern SE, Herman JG. Abrogation of the Rb/p16 tumor-suppressive pathway in virtually all pancreatic carcinomas. Cancer Res. 1997;57:3126–3130. [PubMed] [Google Scholar]
- 59.Poi MJ, Yen T, Li J, Song H, Lang JC, Schuller DE, Tsai M-D, Weghorst CM. Somatic INK4a-ARF Locus Mutations: A significant mechanism of gene inactivation in squamous cell carcinomas of the head and neck. Mol Carcinogenesis. 2001;30:26–36. doi: 10.1002/1098-2744(200101)30:1<26::aid-mc1010>3.0.co;2-e. [DOI] [PubMed] [Google Scholar]
- 60.Rocco JW, Didransky D. p16(MTS-1/CDKN2/INK4a) in cancer progression. Exp Cell Res. 2001;264:42–55. doi: 10.1006/excr.2000.5149. [DOI] [PubMed] [Google Scholar]
- 61.Kaneto H, Sasaki S, Yamamoto H, Itoh F, Toyota M, Suzuki H, Ozeki I, Iwata N, Ohmura T, Satoh T, Karino Y, Satoh T, Toyota J, Satoh M, Endo T, Omata M, Imai K. Detection of hypermethylation of the p16(INK4A) gene promoter in chronic hepatitis and cirrhosis associated with hepatitis B or C virus. Gut. 2001;48:372–377. doi: 10.1136/gut.48.3.372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Bian YS, Osterheld MC, Fontolliet C, Bosman FT, Benhattar J. p16 inactivation by methylation of the CDKN2A promoter occurs early during neoplstic progression in Barrett’s esophagus. Gastroenterology. 2002;122:1113–1121. doi: 10.1053/gast.2002.32370. [DOI] [PubMed] [Google Scholar]
- 63.Hardie LJ, Darnton SJ, Wallis YL, Chauhan A, Hainaut P, Wild CP, Casson AG. p16 expression in Barrett’s esophagus and esophageal adenocarcinoma: association with genetic and epigenetic alterations. Cancer Lett. 2005;217:221–230. doi: 10.1016/j.canlet.2004.06.025. [DOI] [PubMed] [Google Scholar]
- 64.Becker TM, Rizos H, Kefford RF, Mann G. Functional impairment of mealanoma-associated p16INK4a mutants in melanoma cells despite retention of cyclin-dependent kinase 4 binding. Clin Cancer Res. 2001;7:3282–3288. [PubMed] [Google Scholar]
- 65.Sharpless NE, DePinha RA. How stem cells age and why this makes us grow old. Nature Reviews. 2007;8:703–713. doi: 10.1038/nrm2241. [DOI] [PubMed] [Google Scholar]
- 66.Ohtani N, Zebedee Z, Huot TJG, Stinson JA, Sugimoto M, Ohashi Y, Sharrocks AD, Peters G, Hara E. Opposing effects of Ets and Id proteins on p16INk4a expression during cellular senescence. Nature. 2001;409:1067–1070. doi: 10.1038/35059131. [DOI] [PubMed] [Google Scholar]
- 67.Zheng W, Wang H, Xue L, Zhang Z, Tong X. Regulation of cellular senescence and p16INK4a expression by Id1 and E47 proteins in human diploid fibroblast. J Biol Chem. 2004;279:31524–31532. doi: 10.1074/jbc.M400365200. [DOI] [PubMed] [Google Scholar]
- 68.Schmid G, Kramer MP, Maurer M, Wandl S, Wesierska-Gadek J. Cellular and organismal ageing: role of the p53 tumor suppressor protein in the induction of transient and terminal senescence. J Cell Biochem. 2007;101:1355–1369. doi: 10.1002/jcb.21383. [DOI] [PubMed] [Google Scholar]
- 69.Yang X, He Z, Xin B, Cao L. LMP1 of Epstein-Barr virus suppresses cellular senescence associated with the inhibition of p16INK4a expression. Oncogene. 2000;19:2002–2013. doi: 10.1038/sj.onc.1203515. [DOI] [PubMed] [Google Scholar]
- 70.Hansson A, Manetopoulos C, Jonsson J, Axelson H. The basic helix-loop-helix transcription factor TAL1/SCL inhibits the expression of the p16INK4A and pTα genes. Biochem Biophys Res Commun. 2003;312:1073–1081. doi: 10.1016/j.bbrc.2003.11.030. [DOI] [PubMed] [Google Scholar]
- 71.Wu J, Xue L, Weng M, Sun Y, Zhang Z, Wang W, Tong T. Sp1 is essential for p16INK4a expression in human diploid fibroblasts during senescence. PLoS One. 2007;2:e164. doi: 10.1371/journal.pone.0000164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Wang X, Pan L, Feng Y, Wang Y, Han Q, Han L, Han S, Guo J, Huang B, Lu J. p300 plays a role in p16INK4a expression and cell cycle arrest. Oncogene. 2008;27:1894–1904. doi: 10.1038/sj.onc.1210821. [DOI] [PubMed] [Google Scholar]
- 73.Xue L, Wu J, Zheng W, Wang P, Li J, Zhang Z, Tong T. Sp1 is involved in the transcriptional activation of p16INK4 by p21Waf1 in HeLa cells. FEBS Lett. 2004;564:199–204. doi: 10.1016/S0014-5793(04)00352-7. [DOI] [PubMed] [Google Scholar]
- 74.Li H, Wang W, Liu X, Paulson KE, Yee AS, Zhang X. Transcriptional factor HBP1 targets P16INK4A, upregulating its expression and consequently is incolved in Ras-induced premature senescence. Oncogene. 2010;29:5083–5094. doi: 10.1038/onc.2010.252. [DOI] [PubMed] [Google Scholar]
- 75.Wang W, Wu J, Zhang Z, Tong T. Characterization of regulatory elements on the promoter region of p16INK4a that contributes to overexpression of p16 in senescent fibroblasts. J Biol Chem. 2001;276:48655–48661. doi: 10.1074/jbc.M108278200. [DOI] [PubMed] [Google Scholar]
- 76.Huang Y, Wu J, Li R, Wang P, Han L, Zhang Z, Tong T. B-MYB delays cell aging by repressing p16 (INK4α) transcription. Cell Mol Life Sci. 2011;68:893–901. doi: 10.1007/s00018-010-0501-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Shaulian E, Karin M. AP-1 in cell proliferation and survival. Oncogene. 2001;20:2390–2400. doi: 10.1038/sj.onc.1204383. [DOI] [PubMed] [Google Scholar]
- 78.Gan Q, Huang J, Zhou R, Niu J, Zhu X, Wang J, Zhang Z, Tong T. PPARγ accelerates cellular senescence by inducing p16INK4a expression in human diploid fibrobroblasts. J Cell Science. 2008;121:2235–2245. doi: 10.1242/jcs.026633. [DOI] [PubMed] [Google Scholar]
- 79.Betz BL, Strobeck MW, Reisman DN, Knudsen ES, Weissman BE. Re-expression of hSNF5/INI1/BAF47 in pediatric tumor cells leads to G1 arrest associated wuth indcution of p16ink4a and activation of RB. Oncogene. 2002;21:5193–5203. doi: 10.1038/sj.onc.1205706. [DOI] [PubMed] [Google Scholar]
- 80.Oruetxebarria I, Venturini F, Kekarainen T, Houweling A, Zuijderduijn LMP, Mohhd-Sarip A, Vries RGJ, Hoeben RC, Verrijzer CP. p16INK4a is required for hSNF5 chromatin remodeler-induced cellular senescence in malignant rhaboid tumor cells. J Biol Chem. 2004;279:3807–3816. doi: 10.1074/jbc.M309333200. [DOI] [PubMed] [Google Scholar]
- 81.Orlando V, Jane EP, Chinwalla V, Harte PJ, Paro R. Binding of trithorax and Polycomb proteins to the bithorax complex: dynamic changes during early Drosophila embryogenesis. EMBO J. 1998;17:5141–5150. doi: 10.1093/emboj/17.17.5141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Francis NJ, Saurin AJ, Shao Z, Kingston RE. Reconstitution of a functional core polycomb repressive complex. Mol Cell. 2001;8:545–556. doi: 10.1016/s1097-2765(01)00316-1. [DOI] [PubMed] [Google Scholar]
- 83.Medina PP, Romero OA, Kohno T, Montuenga LM, Pio R, Yokota J, Sanchez-Cespedes M. Frequent BRG1/SMARCA4-inacrivating mutations in human lung cancer cell lines. Human Mut. 2008;29:617–622. doi: 10.1002/humu.20730. [DOI] [PubMed] [Google Scholar]
- 84.Becker TM, Haferkamp S, Dijkstra MK, Scurr LL, Frausto M, Diefenbach E, Scolyer RA, Reisman DN, Mann GJ, Kefford RF, Rizos H. The chromatin remodeling factor BRG1 is a novel binding partner of the tumor suppressor p16INK4a. Mol Cancer. 2009;8:4–15. doi: 10.1186/1476-4598-8-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Zhou R, Han L, Li G, Tong T. Senescence delay and repression of p16INK4a by Lsh via recruitment of histone deacetylases in human diploid fibroblasts. Nucleic Acid Res. 2009;37:5183–5196. doi: 10.1093/nar/gkp533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Guo GE, Ma LW, Jiang B, Yi J, Tong TJ, Wang WG. Hydrogen peroxide induces p16INK4a through an AUF1-dependent manner. J Cell Biochem. 2010;109:1000–1005. doi: 10.1002/jcb.22474. [DOI] [PubMed] [Google Scholar]
- 87.Jacobs JL, Kieboom K, Marino S, DePinho RA, van Lohuizen M. The oncogene and Polycomb-group gene bmi-1 regulates cell proliferation and senescence through the ink4a locus. Nature. 1999;397:164–168. doi: 10.1038/16476. [DOI] [PubMed] [Google Scholar]
- 88.Gonzalez S, Klatt P, Delgado S, Conde E, Lopez-Rios F, Sanchez-Cespedes M, Mendez J, Antequera F, Serrano M. Oncogenic activity of Cdc6 through repression of the INK4/ARF locus. Nature. 2006;440:702–706. doi: 10.1038/nature04585. [DOI] [PubMed] [Google Scholar]
- 89.Bezsonova I, Walker JR, Bacik JP, Duan S, Dhe-Paganon S, Arrowsmith CH. Ring1B contains a ubiquitin-like docking module for interaction with Cbx proteins. Biochemistry. 2009;48:10542–10548. doi: 10.1021/bi901131u. [DOI] [PubMed] [Google Scholar]
- 90.Maertens GN, El Messaoudi-Aubert S, Racek T, Stock JK, Nicholls J, Rodriguez-Niedenfuhr M, Gil J, Peters G. Several distinct polycomb complexes regulate and co-localize on the INK4a tumor suppressor locus. PLoS One. 2009;4:e6380. doi: 10.1371/journal.pone.0006380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Gil J, Bernard D, Martinez D, Beach D. Polycomb CBX7 has a unifying role in cellular lifespan. Nature Cell Biol. 2004;6:67–72. doi: 10.1038/ncb1077. [DOI] [PubMed] [Google Scholar]
- 92.Agherbi H, Gaussmann-Wenger A, Verthuy C, Chasson L, Serrano M, Djabali M. Polycomb mediated epigenetic silencing and replication timing at the INK4a/ARF locus during senescence. PLoS One. 2009;4:e5622. doi: 10.1371/journal.pone.0005622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Mroz EA, Baird AH, Michaud WA, Rocco JW. COOH-terminal binding protein regulates expression of the p16INK4A tumor suppressor and senescence in primary human cells. Cancer Res. 2008;68:6049–6053. doi: 10.1158/0008-5472.CAN-08-1279. [DOI] [PubMed] [Google Scholar]
- 94.Chen YJ, Chang JG, Shih LS, Chen PH, Endo M, Whang-Peng J, Chen JM. Frequent detection of aberrant RNA transcripts of the CDKN2 gene in human gastric adenocarcinoma. Int J Cancer. 1997;71:350–354. doi: 10.1002/(sici)1097-0215(19970502)71:3<350::aid-ijc7>3.0.co;2-x. [DOI] [PubMed] [Google Scholar]
- 95.Harland M, Taylor CF, Bass S, Churchman M, Randerson-Moor JA, Holland EA, Mann GJ, Bishop DT, Newton Bishop JA. Intronic sequence variants of the CDKN2A gene in melanoma pedigrees. Genes Chromosomes Cancer. 2005;43:128–136. doi: 10.1002/gcc.20177. [DOI] [PubMed] [Google Scholar]
- 96.Loo JC, Liu L, Hao A, Gao L, Agatep R, Shennan M, Summers A, Goldstein AM, Tucker MA, Deters C, Fusaro R, Blazer K, Weitzel J, Lassam N, Lynch H, Hogg D. Germline splicing mutations of CDKN2A predispose to melanoma. Oncogene. 2003;22:6387–6394. doi: 10.1038/sj.onc.1206736. [DOI] [PubMed] [Google Scholar]
- 97.Chang N, Yi J, Guo G, Liu X, Shang Y, Tong T, Cui Q, Zhan M, Gorospe M, Wang W. HuR uses AUF1 as a cofactor to promote p16INK4 mRNA decay. Mol Cell Biol. 2010;30:3875–3886. doi: 10.1128/MCB.00169-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Lal A, Kim HH, Abdelmohsen K, Kuwano Y, Pillmann P, Jr, Srikantan S, Subrahmanyam R, Martindale JL, Yang X, Ahmed F, Navarro F, Dykxhoorn D, Lieberman J, Gorospe M. p16INK4a translation supresse by miR-24. PLoS One. 2008;3:e1864. doi: 10.1371/journal.pone.0001864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Muirhead DM, Hoffman HT, Robinson RA. Correlation of clinicopathological features with immunohistochemical expression of cell cycle regulatory proteins p16 and retinoblstoma: distinct association with keratinisation and differentiation in oral cavity squamous cell carcinoma. J Clin Pathol. 2006;59:711–715. doi: 10.1136/jcp.2005.030502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Parry D, Bates S, Mann DJ, Peters G. Lack of cyclin D-Cdk complexes in Rb-negative cells correlates with high levels of p16INK4/MTS1 tumor suppressor gene product. EMBO J. 1995;14:503–511. doi: 10.1002/j.1460-2075.1995.tb07026.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.DeGregori J, Leone G, Miron A, Jakoi L, Nevins JR. Distinct roles for E2F proteins in cell growth control and apoptosis. Proc Natl Acad Sci USA. 1997;94:7245–7250. doi: 10.1073/pnas.94.14.7245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Syrjanen S. Human papillomavirus (HPV) in head and neck cancer. J Clin Virol. 2005;32(Suppl 1):S59–66. doi: 10.1016/j.jcv.2004.11.017. [DOI] [PubMed] [Google Scholar]
- 103.Cuschieri K, Wentzensen N. human papillomavirus mRNA and p16 detection as biomarkers for the improved diagnosis of cervical neoplasia. Cacner Epidemiol Biomarkers Prev. 2008;17:2536–2545. doi: 10.1158/1055-9965.EPI-08-0306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Buitrago-Perez A, Garaulet G, Vazquez-Carballo A, Paramio JM, Garcia-Escudero R. Molecular signature of HPV-induced carcinogenesis: pRb, p53 and gene expression profiling. Curr Genomics. 2009;10:26–34. doi: 10.2174/138920209787581235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Narisawa-Saito M, Kiyono T. Basic mechanisms of high-risk human papillomavirus-induced carcinogenesis: roles of E6 and E7 proteins. Cancer Sci. 2007;98:1505–1511. doi: 10.1111/j.1349-7006.2007.00546.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Sandhu C, Peehl DM, Slingerland J. p16INK4A mediates cyclin dependent kinase 4 and 6 inhibition in senescent prostatic epithelial cells. Cancer Res. 2000;60:1616–1622. [PubMed] [Google Scholar]
- 107.Gombart AF, Yang R, Campbell MJ, Berman JD, Koeffler HP. Inhibition of growth of human leukemia cell lines by retrovirally expressed wild-type p16INK4A. Leukemia. 2007;11:1673–1680. doi: 10.1038/sj.leu.2400840. [DOI] [PubMed] [Google Scholar]
- 108.Chen X, Barton LF, Chi Y, Clurman BE, Roberts JM. Ubiquitin-independent degradation of cell-cycle inhibitors by the REGγ proteasome. Mol Cell. 2007;26:843–852. doi: 10.1016/j.molcel.2007.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Ben-Saadon R, Fajerman I, Ziv T, Hellman U, Schwartz AL, Clechanover A. The tumor suppressor p16(INK4a) and the human papillomavirus oncoprotein-58 E7 are naturally occurring lysine-less proteins that are degraded by the ubiquitin system. Direct evidence for ubiquitination of the N-terminal residue. J Biol Chem. 2004;279:41414–41421. doi: 10.1074/jbc.M407201200. [DOI] [PubMed] [Google Scholar]
- 110.Bates S, Peters G. Cyclin D1 as a cellular proto-oncogene. Semin Cancer Biol. 1995;6:73–82. doi: 10.1006/scbi.1995.0010. [DOI] [PubMed] [Google Scholar]
- 111.Hirai H, Roussel MF, Kato JY, Ashmun RA, Sherr CJ. Novel INK4 proteins, p19 and p18, are specific inhibitors of the cyclin D-dependent CDK4 and CDK6. Mol Cell Biol. 1995;15:2672–2681. doi: 10.1128/mcb.15.5.2672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Li J, Poi MJ, Qin D, Selby TL, Byeon IJ, Tsai M-D. Tumor suppressor INK4: quantitative structure-function analyses of p18INK4C as an inhibitor of cyclin-dependent kinase 4. Biochemistry. 2000;39:649–657. doi: 10.1021/bi991281u. [DOI] [PubMed] [Google Scholar]
- 113.Carnero A, Hannon GJ. The INK4 family of CDK inhibitors. Curr Top Microbiol Immunol. 1998;227:43–55. doi: 10.1007/978-3-642-71941-7_3. [DOI] [PubMed] [Google Scholar]
- 114.Okuda T, Hirai H, Valentine VA, Shurtleff SA, Kidd VJ, Lahti JM, Sherr CJ, Downing JR. Molecular cloning, expression pattern, and chromosomal localization of human CDK2D/INK4d, an inhibitor of cyclin D-dependent kinases. Genomics. 1995;29:623–630. doi: 10.1006/geno.1995.9957. [DOI] [PubMed] [Google Scholar]
- 115.Zindy F, Quelle DE, Roussel MF, Sherr CJ. Expression of the p16INK4a tumor suppressor versus other INk4 family members during mouse development and aging. Oncogene. 1997;15:203–211. doi: 10.1038/sj.onc.1201178. [DOI] [PubMed] [Google Scholar]
- 116.Serrano M, Lin AWE, Beach D, Lowe SW. Oncogenic ras provokes premature cell senescence associated with accumulation of p53 and p16INK4a. Cell. 1997;88:593–602. doi: 10.1016/s0092-8674(00)81902-9. [DOI] [PubMed] [Google Scholar]
- 117.Wiedemeyer R, Brennan C, Heffernan TP, Xiao Y, Mahoney J, Protopopov A, Zheng H, Bignell G, Furnari F, Cavenee WK, Hahn WC, Ichimura K, Collins VP, Chu GC, Stratton MR, Ligon K, Futreal PA, Chin L. Feedback circiut among INK4 tumor suppressors constrains human glioblastoma development. Cancer Cell. 2008;13:355–364. doi: 10.1016/j.ccr.2008.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Ramsey MR, Krishnamurthy J, Pei XH, Torrice C, Lin W, Carrasco DR, Ligon KL, Xiong Y, Sharpless NE. Expression of p16Ink4a compensates for p18Ink4c loss in cyclin-dependent kinase 4/6-dependent tumors and tissues. Cancer Res. 2007;67:4732–4741. doi: 10.1158/0008-5472.CAN-06-3437. [DOI] [PubMed] [Google Scholar]
- 119.Krimpenfort P, Upenberg A, Song JY, van der Valk M, Nawijin M, Zevenhoven J, Berns A. p15Ink4b is a critical tumor suppressor in the absence of p16Ink4a. Nature. 2007;448:943–946. doi: 10.1038/nature06084. [DOI] [PubMed] [Google Scholar]
- 120.Massague J. G1 cell-cycle control and cancer. Nature. 2004;432:198–306. doi: 10.1038/nature03094. [DOI] [PubMed] [Google Scholar]
- 121.Jacobs MD, Harrison SC. Structure of an IκBα/NF-κB complex. Cell. 1998;95:749–758. doi: 10.1016/s0092-8674(00)81698-0. [DOI] [PubMed] [Google Scholar]
- 122.Li J, Joo SH, Tsai M-D. An NF-κB-specific inhibitor, IκBα, binds to and inhibits cyclin-dependent kinase 4. Biochemistry. 2003;42:13476–12383. doi: 10.1021/bi035390r. [DOI] [PubMed] [Google Scholar]
- 123.Wolff B, Naumann M. INK4 cell cycle inhibitors direct transcriptional inactivation of NF-κB. Oncogene. 1999;18:2663–2666. doi: 10.1038/sj.onc.1202617. [DOI] [PubMed] [Google Scholar]
- 124.Higashitsuji H, Liu Y, Mayer RJ, Fujita J. The oncoprotein gankyrin negatively regulates both p53 and RB by enhancing proteasomal degradation. Cell Cycle. 2005;4:1335–1337. doi: 10.4161/cc.4.10.2107. [DOI] [PubMed] [Google Scholar]
- 125.Li J, Guo Y. Gankyrin oncoprotein: structure, function, and involvement in cancer. Current Chemical Biology. 2010;4:13–19. doi: 10.2174/2212796811004010013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Higashitsuji H, Itoh K, Nagao T, Dawson S, Nonoguchi K, Kido T, Mayer RJ, Arii S, Fujita J. Reduced stability of retinoblastoma protein by gankyrin, an oncogenic ankyrin-repeat protein overexpressed in hepatomas. Nat Med. 2000;6:96–99. doi: 10.1038/71600. [DOI] [PubMed] [Google Scholar]
- 127.Oritz CM, Ito T, Tanaka E, Tsunoda S, Nagayama S, Sakai Y, Higashitsuji H, Fujita J, Shimada Y. Gankyrin oncoprotein overexpression as a critical factor for tumor growth in human esophageal squamous cell carcinoma and its clinical significance. Int J Cancer. 2008;122:325–332. doi: 10.1002/ijc.23106. [DOI] [PubMed] [Google Scholar]
- 128.Tang S, Yang G, Meng Y, Du R, Li X, Fan R, Zhao L, Bi Q, Jin J, Gao L, Zhang L, Li H, Fan M, Wang Y, Wu K, Liu J, Fan D. Overexpression of a novel gene gankyrin correlates with the malignant phenotype of colorectal cancer. Cancer Biol Ther. 2010;9:88–95. doi: 10.4161/cbt.9.2.10283. [DOI] [PubMed] [Google Scholar]
- 129.Meng Y, He L, Guo X, Tang S, Zhao X, Du R, Jin J, Bi Q, Li H, Nie Y, Liu J, Fan D. Gankyrin promotes the proliferation of human pancreatic cancer. Cancer Letters. 2010;297:9–17. doi: 10.1016/j.canlet.2010.04.019. [DOI] [PubMed] [Google Scholar]
- 130.Man J-H, Liang B, Gu Y-X, Zhou T, Li A-Li, Li T, Jin B-F, Bai B, Zhang H-Y, Zhang W-N, Li W-H, Gong W-L, Li H-Y, Zhang X-M. Gankyrin plays an essential role in Ras-induced tumorigenesis through regulation of the RhoA/ROCK pathway in mammalian cells. J Clin Invest. 2010;120:2829–2841. doi: 10.1172/JCI42542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Higashitsuji H, Higashitsuji H, Itoh K, Sakurai T, Nagao T, Sumitomo Y, Masuda T, Dawson S, Shimada Y, Mayer RJ, Fujita J. The oncoprotein gankyrin binds to MDM2/HDM2, enhancing ubiquitylation and degradation of p53. Cancer Cell. 2005;8:75–87. doi: 10.1016/j.ccr.2005.06.006. [DOI] [PubMed] [Google Scholar]
- 132.Li J, Tsai M-D. Novel insights into the INK4-CDK4/6-Rb pathway: counter action of gankyrin against INK4 proteins regulates the CDK4-mediated phosphorylation of Rb. Biochemistry. 2002;41:3977–3983. doi: 10.1021/bi011550s. [DOI] [PubMed] [Google Scholar]
- 133.Tang TC, Sham JS, Xie D, Fang Y, Huo KK, Wu QL, Guan XY. Identification of a candidate oncogene SEI-1 within a minimal amplified region at 19q13.1 in ovarian cancer cell liens. Cancer Res. 2002;62:7157–7161. [PubMed] [Google Scholar]
- 134.Hsu SI, Yang CM, Sim KG, Hentschel DM, O’Leary E, Bonventre JV. TRIP-Br: a novel family of PHD zinc finger- and bromodomain-interacting protein that regulate the transcriptional activity of E2F-1/DP-1. EMBO J. 2001;20:2273–2285. doi: 10.1093/emboj/20.9.2273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Li Y, Nie CJ, Hu L, Qin Y, Liu HB, Zeng TT, Chen L, Fu L, Deng W, Chen SP, Jia WH, Zhang C, Xie D, Guan XY. Characterization of a novel mechansim of genomic instability involving the SEI1/SET/NM23H1 pathway in esophageal cancers. Cancer Res. 2010;70:5695–5705. doi: 10.1158/0008-5472.CAN-10-0392. [DOI] [PubMed] [Google Scholar]
- 136.Hong SW, Kim CJ, Park WS, Shin JS, Lee SD, Ko SG, Jung SI, Park IC, An SK, Lee WK, Lee WJ, Jin KH, Lee MS. p34SEI-1 inhibits apoptosis through the stablization of the X-linked inhibitor of apoptosis protein: p34SEI-1 as a novel target for anti-breast cancer strategies. Cancer Res. 2009;69:741–746. doi: 10.1158/0008-5472.CAN-08-1189. [DOI] [PubMed] [Google Scholar]
- 137.Li J, Melvein WS, Tsai MD, Weghorst CM, Muscarealla P. The nuclear protein p34SEI-1 regulates the kinase activity of cyclin-dependent kinase 4 in a concentration-dependent manner. Biochemistry. 2004;43:4394–4399. doi: 10.1021/bi035601s. [DOI] [PubMed] [Google Scholar]
- 138.Li J, Muscarella P, Joo SH, Knobloch TJW, Melvin S, Weghorst CM, Tsai M-D. Dissection of CDK4-binding and transactivation activities of p34SEI-1 and comparisons between functions of p34SEI-1 and p16INK4A. Biochemistry. 2005;44:13246–13256. doi: 10.1021/bi0504658. [DOI] [PubMed] [Google Scholar]
- 139.Higuchi M, Fujii M. Distinct functions of HTLV-1 Tax1 from HTLV-2 Tax2 contribute key roles to viral pathogenesis. Retrovirology. 2009;6:117. doi: 10.1186/1742-4690-6-117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Matsuoka M, Jeang KT. Human T-cell leukemia virus type 1 (HTLV-1) and leukemic transformation: viral infectivity, tax, HBZ and therapy. Oncogene. 2011;30:1379–1389. doi: 10.1038/onc.2010.537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Li J, Li H, Tsai MD. Direct binding of the N-terminus of HTLV-1 Tax oncoprotein to cyclin-dependent kinase 4 is a dominant path to stimulate the kinase activity. Biochemistry. 2003;42:6821–6928. doi: 10.1021/bi034369n. [DOI] [PubMed] [Google Scholar]
- 142.Muscarella P, Bloomston M, Brewer AR, Mahajan A, Frankel WL, Ellison CE, Farrar WB, Weghorst CM, Li J. Expression of the p16INK4A/Cdkn2a gene is prevalently down-regulated in human pheochromocytoma tumor specimens. Gene Expression. 2008;14:207–216. doi: 10.3727/105221608786883825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Park TJ, Kim HS, Byun KH, Jang JJ, Lee YS, Lim IK. Sequential changes in hepatocarcinogenesis induced by diethylnitrosamine plus thioacetamide in Fisher 344 rats: Induction of gankyrin expression in liver fibrosis, pRB degradation in cirrhosis, and methylation of p16INK4a exon 1 in hepatocellular carcinoma. Mol Carcinogenesis. 2001;30:138–150. doi: 10.1002/mc.1022. [DOI] [PubMed] [Google Scholar]
- 144.Tan I, Fu XY, Liu SQ, Li HH, Hong Y, Wu MC, Wang HY. Expression of p28GANK and its correlation with RB in human hepatocellular carcinoma. Liver Int. 2005;25:667–676. doi: 10.1111/j.1478-3231.2005.01003.x. [DOI] [PubMed] [Google Scholar]
- 145.Umemura A, Itoh Y, Itoh K, Nakajima T, Higashitsuji H, Onoue H, Fukumoto M, Okanoue T, Fujita J. Association of gankyrin protein expression with early clinical stages and insulin-like growth factor-binding protein 5 expression in human hepatocellular carcinoma. Hepatology. 2008;47:493–502. doi: 10.1002/hep.22027. [DOI] [PubMed] [Google Scholar]
- 146.Li J, Knobloch TJ, Kresty LA, Zhang X, Lang JC, Schuller DE, Weghorst CM. Gankyrin, a biomarker for epithelial carcinogenesis, is overexpressed in human oral cancers. Anticancer Res. 2011 in press. [PubMed] [Google Scholar]
- 147.Grinstein E, Wernet P. Cellular signaling in normal and cancerous stem cells. Cellular Signaling. 2007;19:2428–2433. doi: 10.1016/j.cellsig.2007.06.021. [DOI] [PubMed] [Google Scholar]
- 148.Bonnet D, Dick JE. Human acute myeloid leukemia is organized as a hierarchy that originates from a primitive hematopoietic cell. Nat Med. 1997;3:730–737. doi: 10.1038/nm0797-730. [DOI] [PubMed] [Google Scholar]
- 149.Singh SK, Clarke ID, Terasaki M, Bonn VE, Hawkins C, Squire J, Dirks PB. Identification of a cancer stem cell in human brain tumors. Cancer Res. 2003;63:5821–5828. [PubMed] [Google Scholar]
- 150.Al-Hajj M, Wicha MS, Benito-Hernandez A, Morrison SJ, Clarke MF. Prospective identification of tumorigenic breast cancer cells. Proc Natl Acad Sci USA. 2003;100:3983–3988. doi: 10.1073/pnas.0530291100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.O’Brien CA, Pollett A, Gallinger S, Dick JE. A human colon cancer cell capable of initiating tumor growth in immunodeficient mice. Nature. 2007;445:106–110. doi: 10.1038/nature05372. [DOI] [PubMed] [Google Scholar]
- 152.Zhang S, Balch C, Chan MW, Lai HC, Matei D, Schilder JM, Yan PS, Huang TH, Nephew KP. Identification and characterization of ovarian cancer-initiating cells from primary human tumors. Cancer Res. 2008;68:4311–4320. doi: 10.1158/0008-5472.CAN-08-0364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Li C, Heidt DG, Dalerba P, Burant CF, Zhang L, Adsay V, Wicha M, Clarke MF, Simeone DM. Identification of pancreatic cancer stem cells. Cancer Res. 2007;67:1030–1037. doi: 10.1158/0008-5472.CAN-06-2030. [DOI] [PubMed] [Google Scholar]
- 154.Maitland NJ, Collins AT. Prostate cancer stem cells: a new target for therapy. J Clin Oncol. 2008;26:299–306. doi: 10.1200/JCO.2007.15.1472. [DOI] [PubMed] [Google Scholar]
- 155.Schatton T, Murphy GF, Frank NY, Yamaura K, Waaga-Gasser AM, Gasser M, Zhan Q, Jordan S, Duncan LM, Weishaupt C, Fuhlbrigge RC, Kupper TS, Sayegh MH, Frank MH. Identification of cells initiating human melanomas. Nature. 2008;451:345–349. doi: 10.1038/nature06489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Wang Y, Liu Y, Malek SN, Zheng P, Liu Y. Targeting HIF1α eliminates cancer stem cells in hematological malignancies. Cell Stem Cell. 2011;8:399–411. doi: 10.1016/j.stem.2011.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Schmidt P, Kopecky C, Hombach A, Zigrino P, Mauch C, Abken H. Eradication of melanomas by targeted elimination of a minor subset of tumor cells. Proc Natl Acad Sci USA. 2011;108:2474–2479. doi: 10.1073/pnas.1009069108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Park IK, Qian D, Kiel M, Becker MW, Pihalja M, Weissman IL, Morrison SJ, Clarke MF. Bmi-1 is required for maintenance of adult self-renewing haematopoietic stem cells. Nature. 2003;423:302–305. doi: 10.1038/nature01587. [DOI] [PubMed] [Google Scholar]
- 159.Molofsky AV, Pardal R, Iwashita T, Park IK, Clarke MF, Morrison SJ. Bmi-1 dependence distinguishes neural stem cell self-renewal from progenitor proliferation. Nature. 2003;425:962–967. doi: 10.1038/nature02060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Hemmati HD, Nakano I, Lazareff JA, Masterman-Smith M, Geschwind DH, Bronner-Fraser M, Kornblum HI. Cancerous stem cells can arise from pediatric brain tumors. Proc Natl Acad Sci USA. 2003;100:15178–15183. doi: 10.1073/pnas.2036535100. [DOI] [PMC free article] [PubMed] [Google Scholar]