Abstract
A burgeoning list of small RNAs with a variety of regulatory functions has been identified in both prokaryotic and eukaryotic cells. However, it remains difficult to identify small RNAs by sequence inspection. We used the high conservation of small RNAs among closely related bacterial species, as well as analysis of transcripts detected by high-density oligonucleotide probe arrays, to predict the presence of novel small RNA genes in the intergenic regions of the Escherichia coli genome. The existence of 23 distinct new RNA species was confirmed by Northern analysis. Of these, six are predicted to encode short ORFs, whereas 17 are likely to be novel functional small RNAs. We discovered that many of these small RNAs interact with the RNA-binding protein Hfq, pointing to a global role of the Hfq protein in facilitating small RNA function. The approaches used here should allow identification of small RNAs in other organisms.
Keywords: Hfq, rpoS, antisense regulation
In the last few years, the importance of regulatory small RNAs (sRNAs) as mediators of a number of cellular processes in bacteria has begun to be recognized. Although instances of naturally occurring antisense RNAs have been known for many years, the participation of sRNAs in protein tagging for degradation, modulation of RNA polymerase activity, and stimulation of translation are relatively recent discoveries (for review, see Wassarman et al. 1999; Wassarman and Storz 2000). These findings have raised questions about how extensively sRNAs are used, what other cellular activities might be regulated by sRNAs, and what other mechanisms of action exist for sRNAs. In addition, prokaryotic sRNAs appear to target different cellular functions than their eukaryotic counterparts that primarily act during RNA biogenesis. It is unclear whether this apparent difference between prokaryotic and eukaryotic sRNAs is accurate or stems from the incompleteness of current knowledge. Implicit in these questions is the question of how many sRNAs exist in a given organism and whether the current known sRNAs are truly representative of sRNA function in general.
To date, most known bacterial sRNAs have been identified fortuitously by the direct detection of highly abundant sRNAs (4.5S RNA, tmRNA, 6S RNA, RNaseP RNA, and Spot42 RNA), by the observation of an sRNA during studies on proteins (OxyS RNA, Crp Tic RNA, CsrB RNA, and GcvB RNA), or by the discovery of activities associated with overexpression of genomic fragments (MicF RNA, DicF RNA, DsrA RNA, and RprA RNA) (Okamoto and Freundlich 1986; Bhasin 1989; Urbanowski et al. 2000; Wassarman and Storz 2000; Majdalani et al. 2001; for review, see Wassarman et al. 1999). None of the Escherichia coli sRNAs were found as a result of mutational screens. This observation may reflect the small target size of genes encoding sRNAs compared to protein genes, or may be a consequence of the regulatory rather than essential nature of many sRNA functions. The complete genome sequence of an organism provides a rapid inventory of most encoded proteins, tRNAs, and rRNAs, but it has not led to the immediate recognition of other genes that are not translated. In particular, new bacterial sRNA genes have been overlooked because there are no identifiable classes of sRNAs that can be found based solely on sequence determinants.
We and others have previously suggested several approaches to look for new sRNAs including computer searching of complete genomes based on parameters common to sRNAs, probing of genomic microarrays, and isolating sRNAs based on an association with general RNA-binding proteins (Eddy 1999; Wassarman et al. 1999). Using a combination of these approaches, we have identified 17 novel sRNAs; in addition, we have found six small transcripts that contain short conserved open reading frames (ORFs).
Results
Identification of candidate sRNA genes by homology
As a starting point for detecting novel sRNAs in E. coli, we considered a number of common properties of the previously identified sRNAs that might serve as a guide to identify genes encoding new sRNAs. We are defining sRNAs as relatively short RNAs that do not function by encoding a complete ORF. Of the 13 small RNAs known when this work began, we were struck by the high conservation of these genes between closely related organisms. In most cases, the conservation between E. coli and Salmonella was >85%, whereas that of the typical gene encoding an ORF was frequently <70% (data not shown). Conservation tests on random noncoding regions of the genome suggested that extended conservation in intergenic regions was unusual enough to be used as an initial parameter to screen for new sRNA genes. We therefore tested this approach to look for novel sRNAs in the E. coli genome.
All known sRNAs are encoded within intergenic (Ig) regions (defined as regions between ORFs). A file (R. Overbeek, pers. comm.) containing all Ig sequences from the E. coli genome (Blattner et al. 1997) was used as a starting point for our homology search. We arbitrarily chose the 1.0- to 2.5-Mb region of the 4.6-Mb E. coli genome to test and refine our approach and developed the following steps for searching the full E. coli genome.
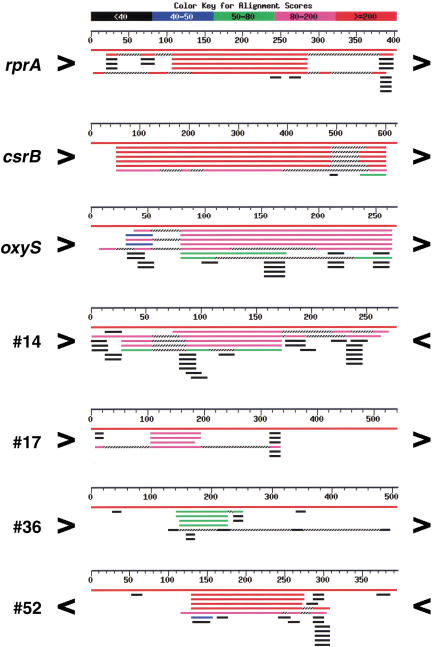
All Ig regions of 180 nucleotides or larger were compared to the NCBI Unfinished Microbial Genomes database using the BLAST program (Altschul et al. 1990). These 1097 Ig regions were rated based on the degree of conservation and length of the conserved region when compared to the closely related Salmonella and Klebsiella pneumonia species. The highest rating was given to Ig regions with a high degree of conservation (raw BLAST score of >80) over at least 80 nt (see Materials and Methods for explanation of ratings). Note that most promoters do not meet these length and conservation requirements. Figure 1 shows a set of BLAST searches for three known sRNAs (RprA RNA, CsrB RNA, and OxyS RNA), three Ig regions with high conservation (#14, #17, and #52), and one Ig region with intermediate conservation (#36). Some Ig regions had a large number of matches, often to several chromosomal regions of the same organism. These Ig regions were noted, and many were found to contain tRNAs, rRNAs, REP, or other repeated sequences. The 40 highly conserved Ig regions containing tRNAs and/or rRNAs were eliminated from our search because these regions were complicated in their patterns of conservation.
Figure 1.
BLAST alignments of representative Ig regions. The indicated Ig regions were used in a BLAST search of the NCBI Unfinished Microbial Genomes database. Each panel shows the summary figure provided by the BLAST program for matches to Salmonella enteritidis, Salmonella paratyphi A, Salmonella typhi, Salmonella typhimurium LT2, and Klebsiella pneumoniae; three contain known sRNA genes (rprA, csrB, and oxyS), and four contain sRNA candidates (#14, #17, #52, and #36; see Table 1). For each panel, the center numbered line represents the length of the full Ig region; the orientation of flanking genes is given by > (clockwise) or < (counterclockwise). The top red line in each panel is the match to Escherichia coli (full identity throughout the Ig). The other red or magenta lines resulted from the closest matches, and the other lines indicate additional less homologous matches. Location of the conserved region with respect to the borders of the Ig region also was a criterion used for the selection of our candidates; conservation 3′ to an ORF or far from the 5′ start of an ORF was considered more likely to encode an sRNA. Note that the conservation within the Ig region encoding oxyS might be interpreted as a leader sequence based on location relative to the start of the flanking gene (oxyR). However, because the conservation extends for 185 nt, candidate regions in our search in which the conservation was near the start of an ORF but was longer than 150 nt were considered further.
Next the orientation and identity of the ORFs bordering the Ig regions were determined using the Colibri database, an annotated listing of all E. coli genes and their coordinates. Inconsistencies between the Colibri database and our original file led to the reclassification of some Ig regions as shorter than 180 nt, and these were not analyzed further. Of the remaining 1006 Ig regions, 13 contained known small RNAs, 295 were in the highest conservation group, 88 showed intermediate conservation, and 610 showed no conservation.
The location of the conservation relative to the orientation of the flanking ORFs was an important consideration in choosing candidates for further analysis. In many cases (132/295 Ig regions), the conserved region was just upstream of the start of an ORF, consistent with conservation of regulatory regions, including untranslated leaders. Cases where the conserved region was more than 50 nt from an ORF start or extended over more than 150 nt in length (RprA RNA, CsrB RNA, OxyS RNA, #17, and #52 in Fig. 1), or where the bordering ORFs ended rather than started at the Ig region (#14 in Fig. 1), were considered better candidates for novel sRNAs.
Published information on promoters and other known regulatory sites within conserved regions of promising candidates was tabulated and used to eliminate many candidates in which the conservation could be attributed to previously identified promoter or 5′ untranslated leaders. Finally, the remaining candidate regions were examined for sequence elements such as potential promoters, terminators, and inverted repeat regions. We considered evidence for possible stem-loops, in particular those with characteristics of rho-independent terminators, as especially indicative of possible sRNA genes (Table 1).
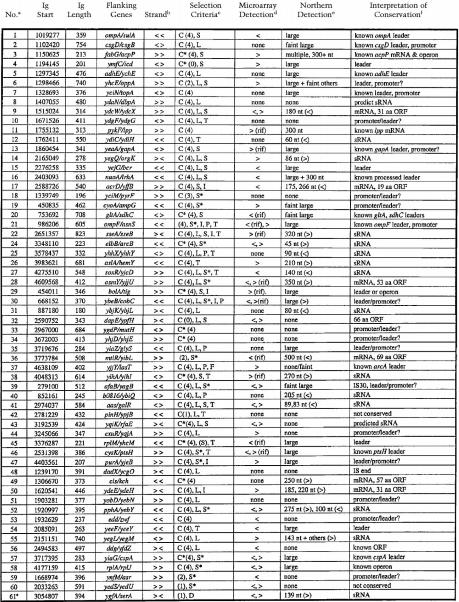
Table 1.
sRNA Candidates
Candidate numbers. #23 was not analyzed; the region of conservation corresponds to a published leader sequence. Candidate #61 was added because it is homologous to candidate #43 and the duplicated regions within #55 (see text and Table 2).
Orientation of flanking genes. > and < denote genes present on the clockwise (W) or counterclockwise (C) strand of the E. coli chromosome, respectively.
Criteria used for selection of candidates: C, conservation; C*, long conservation; (#), conservation score. Ig regions were assigned scores on the basis of BLAST searches (see Materials and Methods). #4 and #32 were rerated from 4 (conserved) to 0 on reanalysis of the endpoints of the flanking ORF (#4) and information on an ORF within the Ig region (#32). L, Location of conservation either far from 5′ end of flanking gene or near 3′ end of gene; S, signal detected in microarray experiments, S*, microarray signal on opposite strand to flanking genes; I, inverted repeat; P, predicted promoter; T, predicted terminator; D, duplicated gene.
Detection on high-density oligonucleotide probe arrays. ><, orientation of signal as in b. Rif, signals present after 20 min treatment with rifampicin.
Northern analysis of RNA extracted from MG1655 cells grown in three conditions (LB medium, exponential phase; minimal medium, exponential phase; LB medium, stationary phase). Strand specific probes were used for sRNA and mRNAs encoding novel ORFs (orientation noted < or > as in b); double stranded DNA probes were used for the rest. For #43, bands were originally detected with a double stranded probe, but appear to be from homologs (see text). Large, >400 nt.
Interpretation of high conservation was based on microarray and Northern analyses as well as literature. mRNA, small RNA transcripts predicted to encode new polypeptides (see text). “known leaders”, literature references supported the existence of leaders corresponding to conservation. For #37, conservation is consistent with the leader of the arcA gene (Compan and Touati 1994). The ORF noted for #56 is described in Seoane and Levy (1995) and Bouvier et al. (1992); see Genbank entry BAA16347.1. The IS sequence fragment in the conserved region of #48 is homologous to that described by McVeigh et al. (2000). “leaders”, A large band on Northern analysis, coupled with conservation near the 5′ end of an ORF. “promoter/leader?”, Absence of RNA signal, coupled with conservation near the 5′ end of a gene. “leader/promoter?”, RNA signal from microarray or Northern analyses suggested a leader, while the conservation is far from the expected position of a leader. “leader or operon”, (for #29) microarray analysis suggested a continuous transcript throughout Ig. “predicted sRNAs”, (for #8 and #43) Igs contain the hallmarks expected for an sRNA, but RNA transcripts were not detected. Igs encoding sRNAs also may include leaders; this is not included in the conclusion column.
Using these criteria, together with microarray expression data (see below), a set of 59 candidates was selected (Table 1). Candidates 1–18 were chosen in the first round of screening of the 1.0- to 2.5-Mb region; some of these candidates would not have met the higher criteria applied to the rest of the genome.
Selecting candidate genes by whole genome expression analysis
In an independent series of experiments, high-density oligonucleotide probe arrays were used to detect transcripts that might correspond to sRNAs from Ig regions. Total RNA isolated from MG1655 cells grown to late exponential phase in LB medium was labeled for probes or used to generate cDNA probes (see Materials and Methods). From a single RNA isolation each labeling approach was carried out in duplicate and individually hybridized to high-density oligonucleotide microarrays. The high-density oligonucleotide probe arrays used are appropriate for this analysis because they have probes specific for both the clockwise (Watson) and counterclockwise (Crick) strands of each Ig region as well as for the sense strand of each ORF. The resulting data from the four experiments were analyzed to examine global expression within Ig regions, as well as neighboring ORFs.
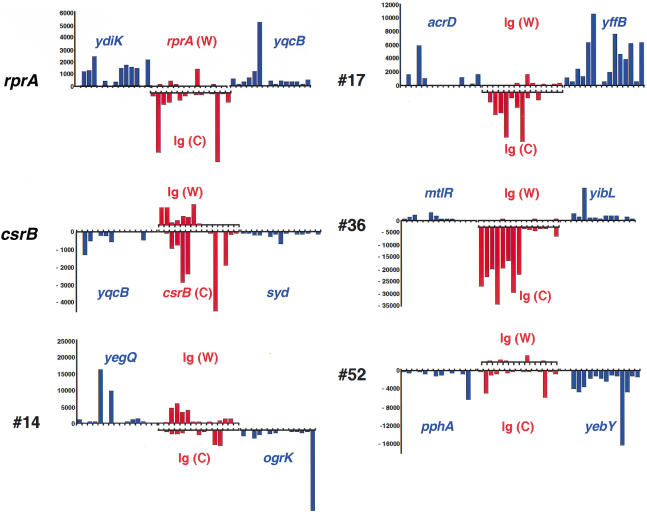
Our criteria for analyzing the microarray data evolved during the course of this analysis. Stringent criteria (longer transcripts in the Ig region, higher expression levels) identified many of the previously known sRNAs but did not uncover many strong candidates for new small RNAs. More relaxed criteria (shorter transcripts, lower expression levels) gave a very large number of candidates and therefore were not by themselves useful as the initial basis for identifying candidates. However, these data were very useful as an additional criterion for selection of candidate regions based on the conservation approach. Detection of a transcript by microarray on the strand opposite to that of surrounding ORFs was considered a strong indicator of an sRNA (S* in Table 1). Microarray data contributed to the selection of 34 of 59 candidates (Table 1). Examples of the different types of expression observed in microarray experiments are shown in Figure 2. Signal corresponding to CsrB RNA clearly is detected on the Crick (C) strand. #17 and #36 have a transcript in the Ig region on the opposing strand (C) to that for the flanking genes (Watson; W). However, the expression patterns were not as obvious in many cases, either because expression levels were low or because the pattern of expression could be interpreted in a number of ways. For instance, very little expression was detected for RprA RNA encoded on the W strand, and there is unexplained signal detected from the opposite strand of the rprA and csrB Ig regions. #14 and #52 also had some expression on each strand (Fig. 2). #14 proved to express a small RNA from the Watson strand, whereas #52 expresses sRNAs from each strand (see below and Table 2).
Figure 2.
Expression profile across high-density oligonucleotide arrays for representative Ig regions. Probe intensities are shown for the indicated Ig regions (red) and the flanking ORFs (blue), calculated from the perfect match minus the mismatch intensities. All negative differences were set to zero. The data shown are for one experiment using cDNA probes, but similar results were seen in the duplicate experiment and with directly labeled RNA probes. The Ig regions and each flanking gene generally contain 15 interrogating probes. Upward bars correspond to genes transcribed on the Watson (W, clockwise) strand, and downward bars correspond to genes transcribed on the Crick (C, counterclockwise) strand. The C strand signal for the CsrB Ig region corresponds well with the known location of the csrB gene. Similarly for the RprA Ig region, the W strand signal corresponds with the location of the rprA gene, but only one probe is positive. The W strand signal for #14 and the C strand signal for #17 overlap well with the conserved regions shown in the BLAST analysis in Figure 1. #36 was chosen for further analysis because of the strong C strand signal; both flanking ORFs are on the W strand. For #52, low levels of expression were seen on both strands; the very low level for probes in the middle of the Ig on the C strand overlapped best with the conserved region found by the BLAST searches (Fig. 1).
Table 2.
Novel sRNAs and Predicted Small ORFsa
| No.
|
Gene
|
Minute
|
RNA Sizeb,c,d
|
Strande
|
Expressionf
|
Hfq Bindingg
|
Effect
on rpoS-lacZh
|
Other
Informationl
|
|
|---|---|---|---|---|---|---|---|---|---|
| S
|
M
|
||||||||
| 12 | rydB | 38 | 60b | <<< | M >> S > E | NT | 0.4 | 1.0 | |
| 14 | ryeE | 47 | 86b | >>< | E, S > M | + (E) | 0.25 | 1.2 | bordered by cryptic prophage |
| 22 | ryfA | 57 | 320c | >>< | E, M | NT | NT | NT | PAIR3 (Rudd 1999) |
| 24 | ryhA | 72 | 45b | <>< | S >> M > E | + (S) | 1.0 | 1.9 | 105, 120 nt, present S >> M > E 105 nt binds Hfq (+, S) |
| 25 | ryhB | 77 | 90b | <<> | M >> S | + (M) | 1.2 | 0.4 | multicopy plasmid restricts growth on succinate |
| 26 | ryiA | 86 | 210b | <>< | E > M, S | + (E) | 0.9 | 1.5 | 155 nt, present M > E, S |
| 27 | ryjA | 92 | 140b | ><> | S >> M | − (S) | NT | NT | |
| 31 | rybB | 19 | 80b | ><< | S >> M | + (S) | 1.0 | 2.3 | |
| 38 | ryiB | 87 | 270b | <>> | M > S >> E | − (M) | 1.0 | 1.6 | CsrC (Romeo, pers. comm.) |
| 40 | rybA | 18 | 205b | ><> | S > M > E | − (S) | 1.2 | 1.5 | ladder up from 255, 300 nt, present S > M > E |
| 41-I | rygA | 64 | 89b | <<> | S >> M, E | + (S) | 1.3i | 1.7i | PAIR2 (Rudd 1999) |
| 41-II | rygB | 64 | 83b | <<> | S, E > M | + (S) | 1.3i | 1.7i | PAIR2 (Rudd 1999) |
| 52-I | ryeA | 41 | 275b | <>< | M > E > S | −/+ (M) | 1.1i | 1.0i | 148, 152, 180 nt (+ others), present M, S |
| 52-II | ryeB | 41 | 100b | <<< | S >> M | + (S) | 1.1i | 1.0i | 70 nt, present S >> M |
| 55-I | ryeC | 46 | 143c | <>> | S > M > E | NT | 1.2 | 1.6 | QUAD1a (Rudd 1999) |
| 107c | M > E, S | ||||||||
| 55-II | ryeD | 46 | 137c | <>> | M > E > S | NT | NT | NT | QUAD1b (Rudd 1999) |
| 102c | M > E | ||||||||
| 61 | rygC | 65 | 139c | >>< | S >> M > E | NT | NT | NT | QUAD1c (Rudd 1999) |
| 107c | S, M > E | ||||||||
| 8 | rydA | 30 | 139d | > (>) > | none | NT | NT | NT | Expression not detected; predicted sRNA |
| 43 | rygD | 69 | 143d | > (<) < | none | NT | NT | NT | QUAD1d (Rudd 1999) Expression not detected |
| 9 | yncL | 32 | 180b | ><> | S > M > E | +/− (S) | NT | NT | 31 aa ORF |
| 17 | ypfM | 55 | 266b | ><> | E >> M | −/+ (E) | 2.0 | 1.5 | 19 aa ORF 175 nt, present E, M |
| 28 | ytjA | 99 | 305b | >>> | S > M | NT | NT | NT | 53 aa ORF |
| 36 | yibT | 81 | 500b | ><> | S >> E, M | NT | 1.3 | 1.0 | 69 aa ORF |
| 49 | yciY | 28 | 250b | <>< | E, M | NT | NT | NT | 57 aa ORF |
| 50 | yneM | 35 | 185b | >>< | S | NT | NT | NT | 31 aa ORF |
| 220b | M > E | ||||||||
Table is divided into three sections: detected sRNAs, predicted sRNAs, and detected RNAs predicted to encode small ORFs.
b,c,dRNA sizes estimated from Northern analyses using bsingle stranded RNA probes or coligonucleotide probes, or dfrom predictions resulting from sequence analysis (see text).
> < denotes orientation of sRNA and flanking genes as in Table 1.
Relative expression in three growth conditions: E, LB medium, exponential phase; M, minimal medium, exponential phase; and S, LB medium, stationary phase.
RNA coimmunoprecipitation with Hfq as detected by Northern analysis: +, strong binding (>30% of RNA bound); +/−, weak binding (5–10%); −/+, minimal binding (<5%), and −, no detectable binding. E, M, S refer to cell growth conditions examined as in f. NT, Not tested.
Expression of rpoS–lacZ fusion in the presence of multicopy plasmids carrying intergenic regions. Activity was measured in stationary phase in LB medium (S) or minimal medium (M) and normalized to the activity of the vector control in the same experiment. In parallel experiments, cells carrying the vector alone gave 1.3–2 (S) and 0.7–2.6 (M) units, cells carrying the pRS-DsrA plasmid gave a 4.9-fold increase (S) and 12-fold increase (M); cells carrying the pRS-RprA plasmid gave 3.1-fold (S) and 3.3-fold (M) increases. Results in table are average of at least three independent assays. Values in bold were considered significantly different from the control. NT, Not tested.
Numbers 41 and 52 each express two sRNAs so it is not possible to assign a phenotype to a given small RNA. Thus far there is no evidence for a strong phenotype for either candidate.
Included is information about additional RNA bands detected in Northern analysis as well as ORF predictions.
Given that a number of the known sRNAs are relatively stable, we tested whether selection for stable RNAs might allow the microarray data to be more useful for de novo identification of sRNA candidates. The transcription inhibitor rifampicin was added to cells for 20 min prior to harvesting the RNA with the intention of enriching for stable RNAs. Many of the known sRNAs can be detected after the rifampicin treatment. Of the 59 candidates in Table 1, 12 retained a hybridization signal (marked rif in Table 1), and 4 of these proved to correspond to small transcripts (see below). Other rif-resistant transcripts detected in Ig regions appeared to be highly expressed leaders.
Small RNA transcripts detected by Northern hybridization
The final test for the presence of an sRNA gene was the direct detection of a small RNA transcript. The candidates in Table 1 were analyzed by Northern hybridization using RNA extracted from MG1655 cells harvested from three growth conditions (exponential phase in LB medium, exponential phase in M63-glucose medium, or stationary phase in LB medium). The microarray analysis discussed above used RNA isolated from cells grown to late exponential phase in LB medium, which is intermediate between the two LB growth conditions used for the Northern analysis. Initially, Northern analysis was carried out using double-stranded DNA probes containing the full Ig region for most candidates. In three cases (#8, #22, and #55) PCR amplification of the Ig region to generate a probe was not successful and therefore oligonucleotide probes were used for Northern analysis. Seventeen candidates gave distinct bands consistent with small RNAs, and one additional candidate gave a somewhat larger RNA, but the location of conservation was not consistent with a leader sequence for a flanking ORF (#36). In some of these cases, two or more RNA species were detected with a single Ig probe (Table 2; see also Fig. 3). One candidate (#43) gave a signal with the double-stranded DNA probe, but contains regions duplicated elsewhere in E. coli that probably account for this signal (see below). Of the remaining 41 candidates, 17 gave no detectable transcript. These Ig regions could encode sRNAs expressed only under very specific growth conditions. For instance, #8 has all the sequence hallmarks of an sRNA gene (a well-conserved region preceded by a possible promoter and ending with a terminator), but has not been detected. Alternatively, the observed conservation could be caused by nontranscribed regulatory regions. Fairly large RNAs were detected for another 24 candidates. Given the size of these transcripts together with data on the orientation of flanking genes and the location of conserved regions, it is likely these are leader sequences within mRNAs (Table 1).
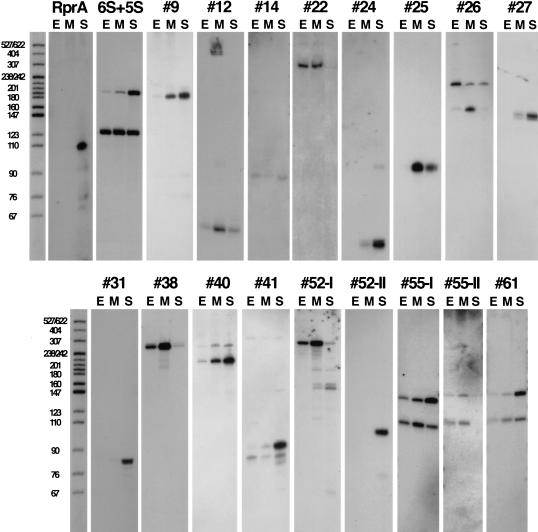
Figure 3.
Detection of novel sRNAs by Northern hybridization. Northern hybridization using strand-specific probes for each candidate was done on RNA extracted from MG1655 cells grown under three different growth conditions: (E) exponential growth in LB medium, (M) exponential growth in M63-glucose medium, and (S) stationary phase in LB medium. Five micrograms of total RNA was loaded in each lane. Exposure times were optimized for each panel for visualization here, therefore the signal intensity shown does not indicate relative abundance between sRNAs. Oligonucleotide probes were used for #12, #22, #55-I, #55-I, and #61; RNA probes were used for all other panels. DNA molecular weight markers (5′-end-labeled MspI-digested pBR322 DNA) were run with each set of samples for direct estimation of RNA transcript length. One lane of DNA molecular weight markers is shown for comparison, but these are approximate sizes because there was slight variation in the running of gels.
For candidates expressing RNAs not expected to be 5′ untranslated leaders, Northern analysis was carried out with strand-specific probes to determine gene orientation (Fig. 3). For many of the candidates, we used sequence elements (see below) as well as expression information from the microarray experiments to predict which strand was most likely expressed; both strands were tested when predictions were unclear. The results from the strand-specific probes generally agreed with predictions and were used to estimate the RNA size (Table 2). Interestingly, in one case there is an sRNA expressed from both the W and C strands within the Ig (#52; Fig. 3). For #12, although no sRNA had been detected using a double-stranded DNA probe, the presence of a potential terminator and promoter remained suggestive of the presence of an sRNA gene. Therefore, oligonucleotide probes also were used in Northern analysis of this candidate, and a small RNA transcript was detected (Fig. 3; Table 1).
Examination of expression profiles of the RNAs under different growth conditions gave an indication of specificity of expression. Some candidates were detected under all three growth conditions; others were preferentially expressed under one growth condition (Fig. 3; Table 2). For instance, #25 was present primarily during growth in minimal medium, consistent with the absence of detection in the whole genome expression experiment, which analyzed RNA isolated from cells grown in rich medium.
Sequence predictions of sRNA genes and ORFs
For the candidates expressing small RNA transcripts, the conserved sequence blocks (contigs) from K. pneumoniae, the highest conserved Salmonella species, and in a few cases Yersinia pestis, were selected from the NCBI Unfinished Microbial Genome database and aligned with the E. coli Ig region using GCG Gap (Devereux et al. 1984). Multiple alignments were assembled by hand, and the conserved regions were examined for likely promoters and terminators and other conserved structures (data not shown). Information from the alignments, together with results from strand-specific Northern and microarray expression analyses, allowed assignments of gene orientation, putative regulatory regions, and RNA length from the predicted starting and ending positions. Where a terminator sequence was very apparent (13 of 19 candidates), transcription was assumed to end at the terminator, and the observed size of the transcript was used to help identify possible promoters. The identification of promoters and terminators was less definite when there was only one species with conservation to E. coli.
As the alignments were assembled, the pattern of conservation in some cases was reminiscent of patterns expected from ORFs, with higher sequence variation in positions consistent with the third nucleotide of codons. GCG Map (Devereux et al. 1984) was used to predict translation in all frames for all of the candidate small RNAs. In six cases, the conservation and translation potential suggested the presence of a short ORF (data not shown). In these cases, a ribosome-binding site and the potential ORF were well conserved, with the most variation in the third position of codons, but other elements of the predicted RNA were less well conserved. For example, #17 expresses an RNA of ∼266 nt, containing a predicted ORF of only 19 amino acids. Within the predicted Shine-Delgarno sequence and ORF, only 9/80 positions showed variation for either Klebsiella or Salmonella, but the overall RNA is <60% conserved. We predict that for #17, as well as five others (Table 2), the detected RNA transcript is functioning as an mRNA, encoding a short, conserved ORF. An evaluation of both the new predicted ORFs and the untranslated sRNAs with GLIMMER, a program designed to predict ORFs within genomes, gave complete agreement with our designations (Delcher et al. 1999).
We have assigned gene names to all candidates that we have confirmed are expressed as RNAs (see Table 2). The genes we predict to encode ORFs were given names according to accepted practice for ORFs of unknown function (Rudd 1998). The genes that express sRNAs without evidence of conserved ORFs were named with a similar nomenclature: ryx, with ry denoting RNA of unknown function and x indicating the 10 min interval on the E. coli genetic map.
We noted one instance of overlap in sequence between our new sRNAs. The conserved region within #43 is highly homologous to a duplicated region within #55, as well as to a fourth region of the chromosome within a more poorly conserved Ig (#61 in Table 1). This repeated region was previously denoted the QUAD repeat and suggested to encode sRNAs (Rudd 1999). Each of the QUAD repeats contains a short stretch homologous to boxC, a repeat element of unknown function present in 50 copies or more within the genome of E. coli (Bachellier et al. 1996). Rudd also has detected transcripts from the QUAD regions (G. Tolun, Z. Li, and K. Rudd, pers. comm.). To determine which of the four QUAD genes was being expressed, we designed oligonucleotide probes unique for each of the four repeats. These oligonucleotide probes demonstrated expression for three of the four QUAD genes (#55-I, #55-II, and #61); furthermore, each gave two RNA bands (Fig. 3; Table 2). No signal was detected for the fourth repeat (#43). The #41 Ig region encodes another pair of repeats, PAIR2 (Rudd 1999), and we observed two RNA species, suggesting that each of the repeats may be transcriptionally active. Finally, another repeat region noted by Rudd, PAIR3, is encoded by the #22 Ig region.
Many sRNAs bind Hfq and modulate rpoS expression
Hfq is a small, highly abundant RNA-binding protein first identified for its role in replication of the RNA phage Qβ (Franze de Fernandez et al. 1968; for review, see Blumenthal and Carmichael 1979). Recently, Hfq has been shown to be involved in a number of RNA transactions in the cell, including translational regulation (rpoS), mRNA polyadenylation, and mRNA stability (ompA, mutS, and miaA) (Muffler et al. 1996; Tsui et al. 1997; Vytvytska et al. 1998; Hajndsorf and Regnier 2000; Vytvytska et al. 2000). Three of the known E. coli sRNAs regulate rpoS expression: DsrA RNA and RprA RNA positively regulate rpoS translation, whereas OxyS RNA represses its translation. In all three cases the Hfq protein is required for regulation (Zhang et al. 1998; Majdalani et al. 2001; Sledjeski et al. 2001), and binding studies have revealed a direct interaction between Hfq and the OxyS and DsrA RNAs (Zhang et al. 1998; Sledjeski et al. 2001).
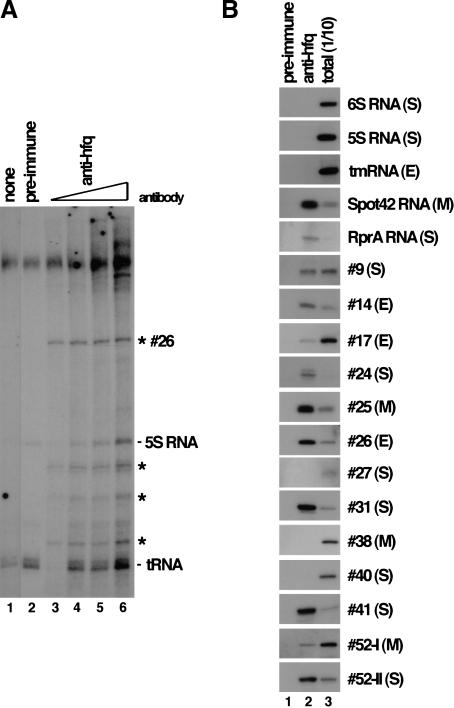
Given the interaction of the Hfq protein with at least three of the known sRNAs, we asked how many of the newly discovered sRNAs are bound by this protein. Hfq-specific antisera was used to immunoprecipitate Hfq-associated RNAs from extracts of cells grown under the conditions used for the Northern analysis. Total immunoprecipitated RNA was examined using two methods. First, RNA was 3′-end labeled and selected RNAs were visualized directly on polyacrylamide gels. Under each growth condition, several RNA species coimmunoprecipitated with Hfq-specific sera but not with preimmune sera, which suggests that many sRNAs interact with Hfq (Fig. 4A; data not shown). Second, selected RNAs were examined using Northern hybridization to determine whether other known sRNAs and any of our newly discovered sRNAs interact with Hfq. For each sRNA, Hfq binding was examined under growth conditions where the sRNA was most abundant (Fig. 4B; Table 2). sRNAs present in samples using the Hfq antisera but not preimmune sera were concluded to interact with Hfq. Comparison of levels of a selected sRNA relative to the total amount of that sRNA in the extract revealed that many of the sRNAs bound Hfq quite efficiently (>30% bound) (#14, #24, #25, #26, #31, #41, #52-II, Spot42 RNA, and RprA RNA), but other sRNAs bound Hfq less efficiently (<10% bound) (#9, #17, and #52-I), or not at all (#27, #38, #40, 6S RNA, 5S RNA, and tmRNA) (Fig. 4; Table 2). The physiological significance of the weaker interactions remains to be tested.
Figure 4.
Coimmunoprecipitation of sRNAs with the Hfq protein. (A) Immunoprecipitations using extract from MG1655 cells grown in LB medium in exponential growth (OD600 = 0.4) were done using no antibody (lane 1); 5 μL of preimmune serum (lane 2); or 0.5, 1, 5, or 10 μL of hfq antisera (lanes 3–6). Selected RNAs were fractionated on a 10% polyacrylamide urea gel after 3′-end labeling. Asterisks mark RNA bands present in the anti-hfq precipitated samples but not in the preimmune control samples and therefore represent Hfq-interacting RNAs. (B) Immunoprecipitations were done using extract from MG1655 cells grown under three different growth conditions: (E) exponential growth in LB medium, (M) exponential growth in M63–glucose medium, and (S) stationary phase in LB medium. Immunoprecipitations were carried out with 5 μL of preimmune sera (lane 1) or 5 μL of Hfq antisera (lane 2) and compared to total RNA from 1/10 extract equivalent used in the immunoprecipitations (lane 3). RNAs were fractionated on 10% polyacrylamide urea gels and analyzed by Northern hybridization using RNA probes to previously known sRNAs or our novel RNAs as indicated.
As mentioned above, at least three of the known sRNAs that interact with Hfq also regulate translation of rpoS, the stationary phase ς factor. In light of the fact that many of the new sRNAs also interact with Hfq, we examined whether these new sRNAs affect rpoS expression. Plasmids carrying the Ig regions encoding either control sRNAs (pRS-DsrA and pRS-RprA) or many of our novel sRNAs were introduced into an MG1655 Δlac derivative carrying an rpoS–lacZ translational fusion. We then compared expression of the rpoS–lacZ fusion in these cells to cells carrying the control vector by measuring β-galactosidase activity at stationary phase in LB or M63–glucose medium (Table 2). As expected, overproduction of either DsrA RNA or RprA RNA increased rpoS–lacZ expression significantly (Table 2 legend). A number of plasmids (pRS-#24, pRS-#31) led to increased rpoS–lacZ expression, whereas others (pRS-#12, pRS-#14, and pRS-#25) led to decreased expression. These results suggest that the corresponding sRNAs may directly regulate rpoS expression or indirectly affect rpoS expression by altering Hfq activity, possibly by competition. Intriguingly, there is not a complete correlation between Hfq binding and altered rpoS–lacZ expression in these studies.
As a start in defining possible functions for the sRNAs, we screened strains carrying the multicopy plasmids for effects on growth in LB medium at various temperatures as well as growth in minimal medium containing a number of different carbon sources. pRS-#25 renders cells unable to grow on succinate in agreement with predictions for #25 RNA interaction with sdh mRNA (discussed below). We were unable to isolate plasmids carrying the #27 Ig region without mutations, suggesting that overproduction of this small RNA may interfere with growth. No other growth phenotypes were observed. A caveat for the interpretation of results with the multicopy plasmids is that they contain the full intergenic region; therefore, we cannot rule out effects of sequences outside the sRNA genes but within the intergenic regions.
Discussion
In summary, a multifaceted search strategy to predict sRNA genes was validated by our discovery of 17 novel sRNAs. Northern analysis determined that 44 of 60 candidate regions express RNA transcripts, some of them expressing more than one RNA species. Of these transcripts, 24 were concluded to be 5′ untranslated leaders for mRNAs of flanking genes, and another 6 are predicted to encode new, short ORFs (Tables 1 and 2). The 17 transcripts believed to be novel, functional sRNAs range from 45 nt to 320 nt in length and vary significantly in expression levels and expression profiles under different growth conditions. More than half of the new sRNAs were found to interact with the RNA-binding protein Hfq, suggesting that Hfq binding may be a defining characteristic of a family of prokaryotic sRNAs.
Evaluation of selection criteria
Three general approaches for predicting sRNA genes were evaluated in this work. In the primary approach, Ig regions were scored for degree and length of conservation between closely related bacterial species followed by examination of sequence features. This approach proved to be very productive in identifying Ig regions encoding novel sRNAs in E. coli; >30% of the candidates selected primarily on the basis of their conservation proved to encode novel small transcripts. The availability of nearly completed genome sequences for Salmonella and Klebsiella made this approach possible. Any organism for which the genome sequences of closely related species are known can be analyzed in this way. Comparative genomics of this sort have been used before to search for regulatory sites (for review, see Gelfand 1999), but have not been employed previously to find sRNAs.
Although we found the conservation-based approach to be the most productive in identifying sRNA genes, we note a number of limitations to its use. A high level of conservation is not sufficient to indicate the presence of an sRNA gene. Many of the most highly conserved regions, not unexpectedly, were consistent with regulatory and leader sequences for flanking genes. We also did not analyze any Ig regions where conservation was attributable to sources other than an sRNA. For example, potential sRNAs processed from mRNAs, or any sRNAs encoded by the antisense strand of ORFs or leaders, may have been missed in our approach. We made the assumption that Ig regions must be ≥180 nt to encode an sRNA of ≥60nt, a 50–60-nt promoter and regulatory region to control expression of the sRNA, as well as regulatory regions for flanking genes. Any sRNA genes in smaller Ig regions would have been overlooked. We also excluded the highly conserved tRNA and rRNA operons from our consideration because of their complexity. It is certainly possible that sRNA genes may be associated with these other RNA genes. In fact, sRNA genes have been predicted to be encoded in at least one tRNA operon (R. Carter, I. Dubchak, and S. Holbrook, pers. comm.). In addition, conservation need not be a property of all sRNAs. We expect sRNAs that play a role in modulating cellular metabolism to be well conserved, as is the case for the previously identified sRNAs. Nevertheless, sRNAs may be encoded within or act upon regions for which there is no homology between E. coli, Klebsiella, and Salmonella (e.g., in cryptic prophages and pathogenicity islands), and they would be missed by this approach. Only 1 of 24 Ig regions within the e14, CP4-54, or CP4-6 prophages showed conservation. A few of these Ig regions showed evidence of transcription by microarray analysis, and RNAs have been implicated in immunity regulation in phage P4 (Ghisotti et al. 1992), which is related to the prophages CP4-54 and CP4-6. Despite the limitations listed above, however, we believe the use of conservation provides a relatively quick identification of the majority of sRNAs.
An alternative genomic sequence-based strategy for identifying sRNAs would be to search for orphan promoter and terminator elements as well as other potential RNA structural elements. Potential promoter elements were generally too abundant to be useful predictors without other information on their expected location and orientation. We found sequences predicted to be rho-independent terminators a more useful indicator of sRNAs; such sequences were clearly present for 13/17 of the sRNAs and 3/6 of the new mRNAs. In a number of cases, it appears that the sRNAs share a terminator with a convergent gene for an ORF. In other cases, either no terminator was detected or it appeared to be in a neighboring ORF. A search using promoter and terminator sequences as the requirements for identifying sRNAs might therefore have found two-thirds of the sRNAs described here. Phage integration target sequences also could be scanned for nearby sRNA genes. Many phage att sites overlap tRNAs (for review, see Campbell 1992), and ssrA, encoding the tmRNA, has a 3′ structure like a tRNA and overlaps the att site of a cryptic prophage (Kirby et al. 1994). In this work, we found that the 3′ end and terminator of #14 overlaps the previously mapped phage P2 att site (Barreiro and Haggard-Ljungquist 1992). #14 sRNA does not obviously resemble a tRNA, suggesting that the overlap between phage att sites and RNA genes extends beyond tRNAs and related molecules and may be common to additional sRNAs.
Our second approach, high-density oligonucleotide probe array expression analysis, proved to be more useful in confirming the presence of sRNA genes first found by the conservation approach than in identifying new sRNA genes de novo. Further consideration of the location of microarray signal compared to flanking genes as well as analysis of microarray signals after a variety of growth conditions should expand the ability to detect sRNAs in this manner. Under a single growth condition, signal consistent with the RNA identified by Northern analysis was detected for 5/15 of the Ig regions proven to encode new sRNAs and for 4/6 of the new mRNAs. Thus, a similar analysis of microarray data in nonconserved genomic regions might help in the identification of sRNAs missed by the conservation-based approaches. We predict that sRNAs from any organism expressed at reasonably high levels under normal growth conditions will be detected by microarrays that interrogate the entire genome, inclusive of noncoding regions.
One clear limitation in detecting sRNAs with microarray or Northern analyses is the fact that some sRNAs may be expressed only under limited growth conditions or at extremely low levels. We chose three growth conditions to scan our samples. Although most of the previously known sRNAs were seen under these conditions, OxyS RNA, which is induced by oxidative stress, was not detectable. For a few of our candidates in which no RNA was detected, it is possible that an sRNA is encoded but is not expressed sufficiently to be detected under any of our growth conditions. Another possible limitation of hybridization-based approaches is that highly structured sRNAs may be refractory to probe generation. sRNA transcripts may not remain quantitatively represented after the fragmentation used in the direct labeling approach here. cDNA labeling also may underrepresent sRNAs because they are a small target for the oligonucleotide primers, and secondary structure can interfere with efficiency of extension.
As our third approach, sRNAs were selected on the basis of their ability to bind to the general RNA-binding protein Hfq. Northern analysis revealed that many of our novel sRNAs interact with Hfq. In preliminary microarray analysis of Hfq-selected RNAs to look for additional unknown sRNAs, DsrA RNA, DicF RNA, Spot42 RNA, #14, #24, #25, #31, #41, and #52-II were detected among those RNAs with the largest difference in levels between Hfq-specific sera and preimmune sera (data not shown). This preliminary experiment suggests that microarray analysis of selected RNAs will be very valuable on a genome-wide basis. Interestingly, a large number of genes with leaders and a number of RNAs for operons were found to coimmunoprecipitate with Hfq (including the known Hfq target nlpD-rpoS mRNA) (Brown and Elliott 1996). It seems likely that the subset of sRNAs binding a common protein will represent a subset in terms of function; the sRNAs of known function associated with Hfq in our experiments appear to be those involved in regulating mRNA translation and stability. Other sRNAs have been shown to interact with specific prokaryotic RNA-binding proteins, for example, tmRNA with SmpB (Karzai et al. 1999), and the possibility of other sRNAs interacting with these proteins or other general sRNA-binding proteins should be tested. This approach is adaptable to all organisms, and, in fact, binding to Sm and Fibrillarin proteins has been the basis for identification of several sRNAs in eukaryotic cells (Montzka and Steitz 1988; Tyc and Steitz 1989).
All the criteria we used to identify sRNAs also will detect short genes encoding new small peptides, and we have found six conserved short ORFs. Although our approach was intended to develop methods to identify nontranslated genes within the genome, short ORFs also are missing from annotated genome sequences. The combination of a requirement for conservation and/or transcription with sequence predictions for ORFs should add significantly to our ability to recognize short ORFs. Small polypeptides have been shown to have a variety of interesting cellular roles. It is tempting to speculate that some of the short ORFs we have found may be involved in signaling pathways, akin to those of B. subtilis peptides that enter the medium and carry out cell–cell signaling (for review, see Lazazzera 2000).
Characteristics and possible functions of new sRNAs
The current work serves as a blueprint for the initial prediction, detection, and characterization of a large group of novel sRNAs. Although we do not have definitive information on function yet, some characteristics that may provide clues regarding the cellular roles of these new sRNAs are noted. Several known sRNAs that bind the Hfq protein act via base pairing to target mRNAs. The finding that a number of our new sRNAs bind Hfq may suggest a similar mechanism of action for this subset of sRNAs. We searched the E. coli genome for possible complementary target sequences and examined phenotypes associated with multicopy plasmids containing new sRNA genes. Intriguingly, #25, an sRNA preferentially expressed in minimal medium, has extended complementarity to a sequence near the start of sdhD, the second gene of the succinate dehydrogenase operon (data not shown). When the #25 Ig region is present on a multicopy plasmid, it interferes with growth on succinate minimal medium (Table 2), consistent with #25 sRNA acting as an antisense RNA for sdhD. Complementarity to potential target mRNAs was found for a number of other novel sRNAs, but the validity of these possible interactions remains to be confirmed by experimentation.
As outlined in the evaluation of each of our approaches, we do not expect our searches to have been exhaustive. sRNAs also have been detected by others using a variety of approaches. The sRNA encoded by #38 was independently identified as a regulatory RNA (CsrC RNA; T. Romeo, pers. comm.), and others have found additional sRNAs using variations of the approaches used here (Argaman et al. 2001). Nevertheless, we think it unlikely that there are many more than 50 sRNAs encoded by the E. coli chromosome and by closely related bacteria. We expect such sRNAs to be present and playing important regulatory roles in all organisms. Using the approaches described here, it is feasible to search all sequenced organisms for these important regulatory molecules. We anticipate that study of the expanded list of sRNAs in E. coli will allow a more complete understanding of the range of roles played by regulatory sRNAs.
Materials and methods
Computer searches
Ig regions are defined here as sequences between two neighboring ORFs. We compared Ig regions of ≥180 nt against the NCBI Unfinished Microbial Genomes database (http://www.ncbi.nlm.nih.gov/Microb_blast/unfinishedgenome.html) using the BLAST program (Altschul et al. 1990). Salmonella enteritidis sequence data were from the University of Illinois, Department of Microbiology (http://www.salmonella.org). Salmonella typhi and Yersinia pestis sequence data were from the Sanger Centre (http://www.sanger.ac.uk/Projects/S_typhi/ and http://www.sanger.ac.uk/Projects/Y_pestis/, respectively). Salmonella typhimurium, Salmonella paratyphi, and Klebsiella pneumoniae sequences were from the Washington University Genome Sequencing Center (Genome Sequencing Center, pers. comm.).
Each Ig region was rated based on the best match to Salmonella or K. pneumoniae species. Ig regions containing previously identified sRNAs were rated 5 (each of them met the criteria to be rated 4). Ig regions were rated 4 if the raw BLAST score was >200 (red in Fig. 1) or 80–200 (magenta in Fig. 1) extending for more than 80 nt; 3 if the raw BLAST score was 80–200 (magenta) extending for 60–80 nt; 2 if the raw BLAST score was 50–80 (green) extending for more than 65 nt; and 1 if the raw BLAST score was <50 (blue, black, or none) or <65 nt. The location of the longest conserved section(s) within each Ig and the number of matches to the NCBI Unfinished Microbial database were recorded. Note that the computer searches were done from May 2000 to December 2000; more sequences are expected to match as the database continues to expand. The identity and orientation of genes flanking each Ig region were determined from the Colibri database (http://genolist.pasteur.fr/Colibri). Ig regions that the Colibri database predicted to be <180 nt in length and Ig regions containing tRNA and/or rRNAs were rated 0 and removed from further consideration. An Excel document containing the full set of data from this analysis is available at http://dir2.nichd.nih.gov/nichd/cbmb/segr/segrPublications.html.
Strains and plasmids
Strains were grown at 37°C in Luria-Bertani (LB) medium or M63 minimal medium supplemented with 0.2% glucose and 0.002% vitamin B1 (Silhavy et al. 1984) except for phenotype testing of strains carrying multicopy plasmids as described below. Ampicillin (50 μg/mL) was added where appropriate. E. coli MG1655 was the parent for all strains used in this study. MG1655 Δlac (DJ480, obtained from D. Jin, NCI), was lysogenized with a λ phage carrying an rpoS–lacZ translational fusion (Sledjeski et al. 1996) to create strain SG30013.
To generate clones containing the Ig region of each candidate (pCR-#N, where N refers to candidate number; see Table 1), Ig regions were amplified by PCR from a MG1655 colony and cloned into the pCRII vector using the TOPO TA cloning kit (Invitrogen). Oligonucleotides were designed so the entire conserved region and in most cases the full Ig region was included. In a few cases, repeated sequences or other irregularities required a reduction in the Ig regions cloned. See http://dir2.nichd.nih.gov/nichd/cbmb/segr/segrPublications.html for a list of all oligonucleotides used in this paper. Ig regions encoding sRNAs also were cloned into multicopy expression vectors (pRS-#N) in which each Ig region is flanked by several vector-encoded transcription terminators. To generate pRS-#N plasmids, pCR-#N plasmids were digested with BamHI and XhoI, and the Ig-containing fragments were cloned into the BamHI and SalI sites of pRS1553 (Pepe et al. 1997), replacing the lacZ-α peptide. To construct pBS-spot42, the Spot42-containing fragment was amplified by PCR from K12 genomic DNA, digested with EcoRI and BamHI, and cloned into corresponding sites in pBluescript II SK+ (Stratagene). All DNA manipulations were carried out using standard procedures. All clones were confirmed by sequencing.
RNA analysis
RNA for Northern analysis was isolated directly from ∼3 × 109 cells in exponential growth (OD600 = 0.2–0.4) or stationary phase (overnight growth) as described previously (Wassarman and Storz 2000). Then 5-μg RNA samples were fractionated on 10% polyacrylamide urea gels and transferred to Hybond N membrane as described previously (Wassarman and Storz 2000). For Northern analysis of candidate regions, double-stranded DNA probes were generated by PCR from a colony of MG1655 cells or from the pCR-#N plasmids with oligonucleotides used for cloning the pCR-#N plasmids. PCR amplification was done with 52°C annealing for 30 cycles in 1× PCR buffer (1 mM each dATP, dGTP, and dTTP; 2.5 μM dCTP; 100 μCi [α32P]dCTP; 10 ng plasmid; 1 U taq polymerase) (Perkin Elmer). Probes were purified over G-50 microspin columns (Amersham Pharmacia Biotech) prior to use. Northern membranes were prehybridized in a 1:1 mixture of Hybrisol I and Hybrisol II (Intergen) at 40°C. DNA probes with 500 μg sonicated salmon sperm DNA were heated for 5 min to 95°C and added to prehybridization solution; membranes were hybridized overnight at 40°C. Membranes were washed by rinsing twice with 4× SSC/0.1% SDS at room temperature followed by three washes with 2× SSC/0.1% SDS at 40°C. Northern blot analysis using RNA probes was done as described previously (Wassarman and Steitz 1992). RNA probes were generated by in vitro transcription according to manufacturer protocols (Roche Molecular Biochemicals) from pCR-#N plasmids linearized with EcoRV or HindIII using SP6 RNA polymerase or T7 RNA polymerase, respectively; pBS-6S (pGS0112; Wassarman and Storz 2000) or pBS-spot42 were linearized with EcoRI using T3 RNA polymerase; pGEM-5S (pG5019; Altuvia et al. 1997) or pGEM-10Sa (Altuvia et al. 1997) were linearized with EcoRI using SP6 RNA polymerase. Oligonucleotide probes were labeled by polynucleotide kinase according to manufacturer protocols (New England Biolabs) using [γ32P]ATP (>5000 Ci/mmole; Amersham Pharmacia Biotech). For oligonucleotide probes, Northern membranes were prehybridized in Ultrahyb (Ambion) at 40°C followed by addition of labeled oligonucleotide probe and hybridization overnight at 40°C. Membranes were washed twice with 2× SSC/0.1% SDS at room temperature followed by two washes with 0.1× SSC/0.1% SDS for 15 min each at 40°C.
Immunoprecipitation
Immunoprecipitations were carried out using extracts from cells in exponential growth (OD600 = 0.2–0.4) or stationary phase (overnight growth) as described previously (Wassarman and Storz 2000), using rabbit antisera against the Hfq protein (A. Zhang and G. Storz, unpubl.) or preimmune serum. After immunoprecipitation, RNA was isolated from Protein A Sepharose-antibody pellets by extraction with phenol:chloroform:isoamyl alcohol (50:50:1), followed by ethanol precipitation. RNA was examined on gels directly after 3′-end labeling or analyzed by Northern hybridization after fractionation on 10% polyacrylamide urea gels as described previously (Wassarman and Storz 2000).
rpoS–lacZ expression
Effects on rpoS–lacZ expression by multicopy plasmids containing the novel sRNAs were determined from a single colony of SG30013 transformed with pRS-#N, grown for 18 h in 5 mL of LB–ampicillin medium or M63–ampicillin medium supplemented with 0.2% glucose at 37°C. β-Galactosidase activity in the culture was assayed as described previously (Zhou and Gottesman 1998). The numbers provided in Table 2 were calculated as the ratio between pRS-#N and the pRS1553 vector control.
Phenotype testing
To test carbon source utilization or temperature sensitivity associated with the multicopy plasmids containing the novel sRNAs, a single colony of MG1655 transformed with a given pRS-#N was grown for 6 h in 5 mL of LB–ampicillin medium at 37°C. Then 10 μL of serial dilutions (10−2, 10−4, and 10−6) was spotted on M63–ampicillin plates containing 0.2% of the carbon source being tested (glucose, arabinose, lactose, glycerol, ribose, or succinate) and grown at 37°C; or on LB plates incubated at room temperature or 42°C. Plates were analyzed after both 1 d and 2 d. Failure to grow in Table 2 indicates an efficiency of plating of <10−3.
Microarray analysis
RNA for microarray analysis was isolated using the MasterPure RNA purification kit according to the manufacturer protocols (Epicentre) from MG1655 cells grown to OD600 = 0.8 in LB medium at 37°C. DNA was removed from RNA samples by digestion with DNase I for 30 min at 37°C. Probes for microarray analysis were generated by one of two methods: direct labeling of enriched mRNA or generation of labeled cDNA.
To generate direct labeled RNA probes, mRNA enrichment and labeling was done as described in the Affymetrix expression handbook (Affymetrix). Oligonucleotide primers complementary to 16S and 23S rRNA were annealed to total RNA followed by reverse transcription to synthesize cDNA strands complementary to 16S and 23S rRNA species. 16S and 23S were degraded with RNase H followed by DNase I treatment to remove cDNA and oligonucleotides. Enriched RNA was fragmented for 30 min at 95°C in 1× T4 polynucleotide kinase buffer (New England Biolabs), followed by labeling with γ-S-ATP and T4 polynucleotide kinase and ethanol precipitation. The biotin label was introduced by resuspending RNA in 96 μL of 30 mM MOPS (pH 7.5), 4 μL of a 50 mM Iodoacetylbiotin solution, and incubating at 37°C for 1 h. RNA was purified using the RNA/DNA Mini Kit according to manufacturer protocols (QIAGEN).
To generate cDNA probes, 5 μg of total RNA was reverse transcribed using the Superscript II system for first strand cDNA synthesis (Life Technologies) and 500-ng random hexamers. RNA and primers were heated to 70°C and cooled to 25°C; reaction buffer was then added, followed by addition of Superscript II and incubation at 42°C. RNA was removed by RNase H and RNase A. The cDNA was purified using the Qiaquick cDNA purification kit (QIAGEN) and fragmented by incubation of up to 5 μg cDNA and 0.2 U DNase I for 10 min at 37°C in 1× one-phor-all buffer (Amersham Pharmacia Biotech). The reaction was stopped by incubation for 10 min at 99°C, and fragmentation was confirmed on a 0.7% agarose gel to verify that average length fragments were 50–100 nt. Fragmented cDNA was 3′-end-labeled with terminal transferase (Roche Molecular Biochemicals) and biotin-N6-ddATP (DuPont/NEN) in 1× TdT buffer (Roche Molecular Biochemicals) containing 2.5 mM cobalt chloride for 2 h at 37°C.
Hybridization to microarrays and staining procedures were done according to the Affymetrix expression manual (Affymetrix). The arrays were read at 570 nm with a resolution of 3 μm using a laser scanner.
The expression of genes was analyzed using the Affymetrix Microarray Suite 4.01 software program. Detection of transcripts in intergenic regions was done using the intensities of each probe designed to be a perfect match and the corresponding probe designed to be the mismatch. If the perfect match probe showed an intensity that was 200 units higher than the mismatch probe, the probe pair was called positive. Two neighboring positive probe pairs were considered evidence of a transcript. The location and length of the transcripts were estimated based on the first and last identified positive probe pair within an Ig region.
Acknowledgments
We thank R. Overbeek for the file of intergenic sequences, D. Jin for MG1655 ΔlacZ, A. Zhang for Hfq antibodies, R.M. Saxena for technical assistance, and S. Salzberg for running the GLIMMER program. We made extensive use of the NCBI Unfinished Microbial Genome database. In particular, the authors thank the Sanger Center; the Genome Sequencing Center; Washington University, St. Louis; and the University of Illinois, Department of Microbiology for communication of DNA sequence data to that database prior to publication. We thank S. Altuvia, S. Holbrook, T. Romeo, K. Rudd, and their collaborators for permission to quote unpublished results; and B. Peculis, T. Romeo, K. Rudd, R. Weisberg, and members of our laboratories for comments on the manuscript.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL storz@helix.nih.gov; FAX (301) 402-0078.
E-MAIL susang@helix.nih.gov; FAX (301) 496-3875.
Article and publication are at http://www.genesdev.org/cgi/doi/10.1101/gad.901001.
References
- Altschul SF, Gish W, Miller W, Myers EW, Lipman D. A basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Altuvia S, Weinstein-Fischer D, Zhang A, Postow L, Storz G. A small stable RNA induced by oxidative stress: Role as a pleiotropic regulator and antimutator. Cell. 1997;90:43–53. doi: 10.1016/s0092-8674(00)80312-8. [DOI] [PubMed] [Google Scholar]
- Argaman, L., Hershberg, R., Vogel, J., Bejerano, G., Wagner, E.G.H., Margalit, H., and Altuvia, S. 2001. Novel small RNA-encoding genes in the intergenic regions of Escherichia coli. Curr. Biol. (in press). [DOI] [PubMed]
- Bachellier S, Gilson E, Hofnung M, Hill CW. Repeated sequences. In: Neidhardt FC, et al., editors. Escherichia coli and Salmonella: Cellular and molecular biology. Washington, D.C.: American Society for Microbiology; 1996. pp. 2012–2040. [Google Scholar]
- Barreiro V, Haggard-Ljungquist E. Attachment sites for bacteriophage P2 on the Escherichi coli chromosome: DNA sequences, localization on the physical map, and detection of a P2-like remnant in E. coliK-12 derivatives. J Bacteriol. 1992;174:4086–4093. doi: 10.1128/jb.174.12.4086-4093.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhasin RS. “Studies on the mechanism of the autoregulation of the crp operon of E. coli K12.” Ph.D. thesis. Stony Brook, NY: State University of New York at Stony Brook; 1989. [Google Scholar]
- Blattner FR, Plunkett G, Bloch CA, Perna NT, Burland V, Riley M, Collado-Vides J, Glasner JD, Rode CK, Mayhew GF, et al. The complete genome sequence of Escherichia coliK-12. Science. 1997;277:1453–1474. doi: 10.1126/science.277.5331.1453. [DOI] [PubMed] [Google Scholar]
- Blumenthal T, Carmichael GG. RNA replication: Function and structure of Qβ-replicase. Annu Rev Biochem. 1979;48:525–548. doi: 10.1146/annurev.bi.48.070179.002521. [DOI] [PubMed] [Google Scholar]
- Bouvier J, Richaud C, Higgins W, Bogler O, Stragier P. Cloning, characterization, and expression of the dapE gene of Escherichia coli. J Bacteriol. 1992;174:5265–5271. doi: 10.1128/jb.174.16.5265-5271.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown L, Elliott T. Efficient translation of the RpoS ς factor in Salmonella typhimurium requires Host Factor I, an RNA-binding protein encoded by the hfqgene. J Bacteriol. 1996;178:3763–3770. doi: 10.1128/jb.178.13.3763-3770.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell AM. Chromosomal insertion sites for phages and plasmids. J Bacteriol. 1992;174:7495–7499. doi: 10.1128/jb.174.23.7495-7499.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Compan I, Touati D. Anaerobic activation of arcA transcription in Escherichia coli: Roles of Fnr and ArcA. Mol Microbiol. 1994;11:955–964. doi: 10.1111/j.1365-2958.1994.tb00374.x. [DOI] [PubMed] [Google Scholar]
- Delcher AL, Harmon D, Kasif S, White O, Salzberg SL. Improved microbial gene identification with GLIMMER. Nucleic Acids Res. 1999;27:4636–4641. doi: 10.1093/nar/27.23.4636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devereux J, Haeberli P, Smithies O. A comprehensive set of sequence analysis programs for the VAX. Nucleic Acids Res. 1984;12:387–395. doi: 10.1093/nar/12.1part1.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eddy SR. Noncoding RNA genes. Curr Opin Genet Dev. 1999;9:695–699. doi: 10.1016/s0959-437x(99)00022-2. [DOI] [PubMed] [Google Scholar]
- Franze de Fernandez M, Eoyang L, August J. Factor fraction required for the synthesis of bacteriophage Qβ RNA. Nature. 1968;219:588–590. doi: 10.1038/219588a0. [DOI] [PubMed] [Google Scholar]
- Gelfand MS. Recognition of regulatory sites by genome comparison. Res Microbiol. 1999;150:755–771. doi: 10.1016/s0923-2508(99)00117-5. [DOI] [PubMed] [Google Scholar]
- Ghisotti D, Chiaramonte R, Forti F, Zangrossi S, Sironi G, Deho G. Genetic analysis of the immunity region of phage-plasmid P4. Mol Microbiol. 1992;6:3405–3413. doi: 10.1111/j.1365-2958.1992.tb02208.x. [DOI] [PubMed] [Google Scholar]
- Hajndsorf E, Regnier P. Host factor Hfq of Escherichia colistimulates elongation of poly(A) tails by poly(A) polymerase I. Proc Natl Acad Sci. 2000;97:1501–1505. doi: 10.1073/pnas.040549897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karzai AW, Susskind MM, Sauer RT. SmpB, a unique RNA-binding protein essential for the peptide-tagging activity of SsrA (tmRNA) EMBO J. 1999;18:3793–3799. doi: 10.1093/emboj/18.13.3793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirby JE, Trempy JE, Gottesman S. Excision of a P4-like cryptic prophage leads to Alp protease expression in Escherichia coli. J Bacteriol. 1994;176:2068–2081. doi: 10.1128/jb.176.7.2068-2081.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazazzera BA. Quorum sensing and starvation: Signals for entry into stationary phase. Curr Opin Microbiol. 2000;3:177–182. doi: 10.1016/s1369-5274(00)00072-2. [DOI] [PubMed] [Google Scholar]
- Majdalani N, Chen S, Murrow J, St. John K, Gottesman S. Regulation of RpoS by a novel small RNA: The characterization of RprA. Mol Microbiol. 2001;39:1382–1394. doi: 10.1111/j.1365-2958.2001.02329.x. [DOI] [PubMed] [Google Scholar]
- McVeigh A, Fasano A, Scott DA, Jelacic S, Moseley SL, Robertson DC, Savarino SJ. IS1414, an Escherichia coliinsertion sequence with a heat-stable enterotoxin gene embedded in a transposase-like gene. Infect Immun. 2000;68:5710–5715. doi: 10.1128/iai.68.10.5710-5715.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montzka KA, Steitz JA. Additional low-abundance human small nuclear ribonucleoproteins: U11, U12, etc. Proc Natl Acad Sci. 1988;85:8885–8889. doi: 10.1073/pnas.85.23.8885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muffler A, Fischer D, Hengge-Aronis R. The RNA-binding protein HF-I, known as a host factor for phage Qβ RNA replication, is essential for rpoS translation in Escherichia coli. Genes & Dev. 1996;10:1143–1151. doi: 10.1101/gad.10.9.1143. [DOI] [PubMed] [Google Scholar]
- Okamoto K, Freundlich M. Mechanism for the autogenous control of the crpoperon: Transcriptional inhibition by a divergent RNA transcript. Proc Natl Acad Sci. 1986;83:5000–5004. doi: 10.1073/pnas.83.14.5000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pepe CM, Suzuki C, Laurie C, Simons RW. Regulation of the “tetCD” genes of transposon Tn10. J Mol Biol. 1997;270:14–25. doi: 10.1006/jmbi.1997.1094. [DOI] [PubMed] [Google Scholar]
- Rudd KE. Linkage map of Escherichia coliK-12, edition 10: The physical map. Microbiol Mol Biol Rev. 1998;62:985–1019. doi: 10.1128/mmbr.62.3.985-1019.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ————— Novel intergenic repeats of Escherichia coliK-12. Res Microbiol. 1999;150:653–664. doi: 10.1016/s0923-2508(99)00126-6. [DOI] [PubMed] [Google Scholar]
- Seoane AS, Levy SB. Identification of new genes regulated by the marRAB operon in Escherichia coli. J Bacteriol. 1995;177:530–535. doi: 10.1128/jb.177.3.530-535.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silhavy TJ, Berman ML, Enquist LW. Experiments with gene fusions. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory; 1984. [Google Scholar]
- Sledjeski DD, Gupta A, Gottesman S. The small RNA, DsrA, is essential for the low temperature expression of RpoS during exponential growth in Escherichia coli. EMBO J. 1996;15:3993–4000. [PMC free article] [PubMed] [Google Scholar]
- Sledjeski DD, Whitman C, Zhang A. Hfq is necessary for regulation by the untranslated RNA DsrA. J Bacteriol. 2001;183:1997–2005. doi: 10.1128/JB.183.6.1997-2005.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsui H-CT, Feng G, Winkler M. Negative regulation of mutS and mutH repair gene expression by the Hfq and RpoS global regulators of Escherichia coliK-12. J Bacteriol. 1997;179:7476–7487. doi: 10.1128/jb.179.23.7476-7487.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyc K, Steitz JA. U3, U8 and U13 comprise a new class of mammalian snRNPs localized in the cell nucleolus. EMBO J. 1989;8:3113–3119. doi: 10.1002/j.1460-2075.1989.tb08463.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urbanowski ML, Stauffer LT, Stauffer GV. The gcvB gene encodes a small untranslated RNA involved in expression of the dipeptide and oligopeptide transport systems in Escherichia coli. Mol Microbiol. 2000;37:856–868. doi: 10.1046/j.1365-2958.2000.02051.x. [DOI] [PubMed] [Google Scholar]
- Vytvytska O, Jakobsen J, Balcunaite G, Andersen J, Baccarini M, von Gabain A. Host Factor I, Hfq, binds to Escherichia coli ompAmRNA in a growth-rate dependent fashion and regulates its stability. Proc Natl Acad Sci. 1998;95:14118–14123. doi: 10.1073/pnas.95.24.14118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vytvytska O, Moll I, Kaberdin VR, von Gabain A, Blasi U. Hfq (HF1) stimulates ompAmRNA decay by interfering with ribosome binding. Genes & Dev. 2000;14:1109–1118. [PMC free article] [PubMed] [Google Scholar]
- Wassarman KM, Steitz JA. The low abundance U11 and U12 snRNAs interact to form a two snRNP complex. Mol Cell Biol. 1992;12:1276–1285. doi: 10.1128/mcb.12.3.1276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wassarman KM, Storz G. 6S RNA regulates E. coliRNA polymerase activity. Cell. 2000;101:613–623. doi: 10.1016/s0092-8674(00)80873-9. [DOI] [PubMed] [Google Scholar]
- Wassarman KM, Zhang A, Storz G. Small RNAs in Escherichia coli. Trends Microbiol. 1999;7:37–45. doi: 10.1016/s0966-842x(98)01379-1. [DOI] [PubMed] [Google Scholar]
- Zhang A, Altuvia S, Tiwari A, Argaman L, Hengge-Aronis R, Storz G. The oxyS regulatory RNA represses rpoStranslation by binding Hfq (HF-1) protein. EMBO J. 1998;17:6061–6068. doi: 10.1093/emboj/17.20.6061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y-N, Gottesman S. Regulation of proteolysis of the stationary-phase ς factor RpoS. J Bacteriol. 1998;180:1154–1158. doi: 10.1128/jb.180.5.1154-1158.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]