Abstract
Properties of a mutant bacteriophage T2 DNA [N6-adenine] methyltransferase (T2 Dam MTase) have been investigated for its potential utilization in RecA-assisted restriction endonuclease (RARE) cleavage. Steady-state kinetic analyses with oligonucleotide duplexes revealed that, compared to wild-type T4 Dam, both wild-type T2 Dam and mutant T2 Dam P126S had a 1.5-fold higher kcat in methylating canonical GATC sites. Additionally, T2 Dam P126S showed increased efficiencies in methylation of non-canonical GAY sites relative to the wild-type enzymes. In agreement with these steady-state kinetic data, when bacteriophage λ DNA was used as a substrate, maximal protection from restriction nuclease cleavage in vitro was achieved on the sequences GATC, GATN and GACY, while protection of GACR sequences was less efficient. Collectively, our data suggest that T2 Dam P126S can modify 28 recognition sequences. The feasibility of using the mutant enzyme in RARE cleavage with BclI and EcoRV endonucleases has been shown on phage λ DNA and with BclI and DpnII endonucleases on yeast chromosomal DNA embedded in agarose.
INTRODUCTION
The mapping and subcloning of large genomes could be greatly simplified and accelerated by the development of methods to catalyze sequence-specific cleavage of DNA at a limited number of all the possible site-specific locations within a genome. As currently developed, most methods for sequence-specific cleavage of large DNAs are severely limited because of the high frequency of cleavage sites. However, several excellent proposals have been suggested for decreasing the frequency of restriction and predetermining the sites of cutting by sequence-specific endonucleases (ENases) using cognate DNA methyltransferases (MTases) (1,2).
One such method, called RecA-assisted restriction endonuclease (RARE) cleavage by Ferrin and Camerini-Otero (3) and RecA Achilles cleavage (RecA-AC) by Koob et al. (4), is a simple general method that allows selective DNA cleavage at a single predetermined restriction site. This technique is based on the ability of the RecA protein to bind to an oligonucleotide in the presence of a non-hydrolyzable ATP analog, which results in formation of a stable complex. This preformed nucleoprotein complex binds specifically to homologous duplex DNA to form a triple-stranded DNA–protein complex. The triple helix complex protects DNA from methylation by the cognate MTase, but not the remaining double-helical regions. After complex dissociation and MTase inactivation, DNA cleavage by the cognate ENase is limited to the site that was precluded from methylation.
Originally, RARE cleavage was proposed as a method to simplify and accelerate mapping of large genomes (3,4). Later this approach was used for human genome mapping (5–7) and was also applied to experiments on transgenic expression (8) and site-directed mutagenesis (9,10). However, in practice there are several problems limiting use of RARE cleavage as a general method: (i) the number of commercially available MTases is far lower than that of ENases; (ii) MTases must have a very high activity in order to modify all potential cleavage sites; (iii) MTases should be free of non-specific nuclease contamination, especially for modification of long DNA fragments. In this regard, it was noted that many commercially available MTases are not suitable for RARE cleavage (5,6). At present two adenine-specific MTases associated with cognate ENases (M.EcoRI/R.EcoRI and M.HhaII/R.HinfI) are applicable to RARE cleavage, but EcoRI is the only system that has been used (11).
In order to expand usage of the RARE method, this investigation explores the advantages of using the MTases from the closely related T-even bacteriophages T4 and T2. These MTases, which methylate the tetranucleotide palindrome GATC, differ in only three amino acids (12) and have previously been overexpressed, purified and characterized (13,14). The choice to use adenine-specific MTases in RARE technologies, instead of cytosine-specific MTases, was made because vertebrate DNAs have no methylated adenines (15). This avoids difficulties in interpretation of RARE cleavage results, which can be created using cytosine-specific MTases. At present five recognition sequences overlapping GATC are known in which adenine methylation blocks DNA cleavage by the corresponding ENases: CGATCG (XorII), GATC (MboI), GGATC(4/5) (AlwI), R/GATCY (MflI) and T/GATCA (BclI) (2). T4 and T2 Dam MTases also modify some non-canonical sites containing the sequence GAY (GAT or GAC) (16,17), which in turn protects DNA against cleavage not only by ENases having GATC in the target recognition site but also by those that have GAY, e.g. ClaI, EcoRV and FokI (14,18). There are 16 ENases known to have a GAY sequence within their recognition site that can be protected against cleavage by adenine methylation (2,19). Additionally, the T2 Dam MTase has been shown to have a 2-fold higher kcat for canonical and non-canonical sites relative to T4 Dam (14).
In order to further expand the utility of the T2 and T4 MTases, strategies were designed to utilize mutants of these enzymes for the RARE technology. In this regard, Revel and Hattman (20) obtained mutants of T4 and T2 phage designated damh that are able to grow on P1 lysogens, suggesting that methylation protects phage DNA against P1 restriction of the non-canonical sequence AGACC. The nature of the damh mutation has been elucidated in both T2 and T4; it is a change of Pro126 to Ser (12). In addition, the P126S mutant has an increased ability to protect phage DNA against HinfI ENase (GANTC) at enzyme levels at which the parental Dam+ enzymes could not (21).
In this study we have investigated the relative abilities of the wild-type T4 and T2 MTases and mutant T2 Dam P126S MTase to methylate synthetic oligonucleotide duplexes containing non-canonical GAY sites. In addition, we have investigated the ability of the mutant MTases to afford protection to phage λ DNA against cleavage by ENases with GAY-containing recognition sequences. Based on the results of these experiments we tested the applicability of T2 Dam P126S for RARE cleavage of λ phage DNA in solution and dissection of yeast chromosomes embedded in agarose.
MATERIALS AND METHODS
Materials
Strains of Escherichia coli GM 2971 containing plasmids pSLRT4 dam+, pSLRT2 dam+ and pINT2damP126S (13,14) were used for enzyme purification.
Oligonucleotides were synthesized by Integrated DNA Technologies (Coraville, IA). RecA, ENases and N6-methyladenine-free λ DNA were purchased from New England Biolabs (Beverly, MA). S-adenosyl-l-methionine (AdoMet) and adenosine 5′-O-(3-thiotriphosphate) (ATP-γ-S) were obtained from Sigma. [Methyl-3H]AdoMet was purchased from NEN Life Science Products.
T2 Dam P126S purification
The T4 Dam purification protocol developed by Kossykh et al. (13) was modified to obtain T4 Dam, T2 Dam and T2 Dam P126S. Frozen cells (10 g) were suspended in 50 ml of PEM buffer (20 mM phosphate buffer pH 7.4, 1 mM EDTA, 7 mM 2-mercaptoethanol) with 0.1% Nonidet P-40 and 0.4 M NaCl. Lysozyme was added to a concentration of 500 µg/ml and after a 2 h incubation at 4°C the cells were disrupted by sonication. Insoluble debris was removed by centrifugation at 100 000 g for 1 h. The supernatant was diluted 2-fold in PEM buffer and passed through a DEAE–cellulose (DE52) column connected to a P-11 phosphocellulose column. Proteins bound to the P11 phosphocellulose were eluted in a 120 ml NaCl gradient (200–800 mM) in PEM buffer. Appropriate fractions were pooled, diluted 2-fold in PEM buffer and applied to a 5 ml hydroxyapatite column equilibrated in column buffer (PEM with 0.2 M NaCl). Proteins bound to the matrix were eluted with a 60 ml gradient of 20–500 mM potassium phosphate (pH 7.4) in column buffer. In order to further increase the purity of the enzyme, one additional step in the purification was added. After elution of the proteins from the hydroxyapatite column, appropriate fractions were collected, dialyzed against PEM buffer with 0.1 M NaCl and applied to a 5 ml High S column (Bio-Rad). Proteins that bound to the matrix were eluted with a 40 ml gradient of 0.1–1.0 M NaCl in PEM buffer. The enzyme was dialyzed against PEM with 0.1 M NaCl and 50% glycerol. Protein concentration was measured with the Bio-Rad Dye Reagent and purity of preparations was tested by SDS–PAGE (22).
Steady-state studies
Methyl transfer assays were carried out as described (13). Assay mixtures (50 µl) contained 20 mM Tris–HCl pH 8.0, 1 mM EDTA, 1 mM DDT, 0.2 mg/ml BSA and 0.55 µM [3H-CH3]AdoMet. For determination of steady-state kinetic parameters 3–13 nM MTase and various concentrations of substrate were used. All 24mer oligonucleotide duplexes studied had the same general structure
5′-CGCGGGCGCCG(GATC)CGGGCGGGC-3′
3′-GCGCCCGCCGC(CTAG)GCCCGCCCG-5′
except for individual base substitutions in the target GATC site (Table 1, indicated by italics). Oligonucleotide concentrations were determined spectrophotometrically from molar extinction coefficients of individual oligonucleotides and their sequences. Oligonucleotide duplexes were obtained by heating and annealing mixtures of equimolar amounts of complementary individual oligonucleotides. Kinetic parameters were calculated using the Enzyme Kinetics program from Trinity Software.
Table 1. T4 Dam, T2 Dam and T2 Dam P126S steady-state kinetic parameters determined with synthetic oligonucleotides.
| Structure of | T4 Dam | T2 Dam | T2 Dam P126S | ||||||
| recognition site |
Km (nM) |
kcat (s–1 10–3) |
Relative kcat (%) |
Km (nM) |
kcat (s–1 10–3) |
Relative kcat (%) |
Km (nM) |
kcat (s–1 10–3) |
Relative kcat (%) |
| GATC | 10.3 (0.6) | 24.80 (4.6) | 100 | 13.9 (1.5) | 41.90 (4.3) | 169 | 11.0 (1.6) | 39.30 (3.5) | 158 |
| GATT | 4.4 (1.8) | 5.70 (0.4) | 23 | 8.6 (2.4) | 9.10 (1.2) | 37 | 16.3 (2.6) | 13.90 (1.1) | 56 |
| GATA | 5.7 (0.9) | 5.80 (0.7) | 23 | 7.1 (0.5) | 9.10 (0.4) | 37 | 12.2 (0.8) | 15.30 (1.8) | 62 |
| GATG | 5.0 (0.8) | 12.40 (0.4) | 50 | 11.7 (4.1) | 19.70 (0.3) | 79 | 17.5 (0.6) | 26.10 (1.3) | 105 |
| GACC | 24.6 (4.4) | 7.50 (1.0) | 30 | 14.6 (3.9) | 7.90 (0.6) | 32 | 13.2 (3.6) | 7.30 (2.0) | 29 |
| GACT | 21.0 (1.3) | 1.24 (0.29) | 5 | 61.9 (13.9) | 2.10 (0.6) | 8 | 15.5 (2.4) | 8.50 (1.5) | 34 |
| GACA | 3.2 (0.6) | 0.13 (0.07) | 0.5 | 15.6 (1.7) | 0.11 (0.06) | 0.4 | 10.5 (0.1) | 0.46 (0.09) | 1.9 |
| GACG | 3.2 (0.4) | 0.10 (0.02) | 0.4 | 5.9 (1.9) | 0.14 (0.03) | 0.6 | 10.0 (2.3) | 0.36 (0.04) | 1.5 |
Standard error values are in parentheses.
Testing the effect of adenine methylation on cleavage by restriction ENases
The methylation mixture (20 µl) contained 20 mM Tris–HCl pH 8.0, 50 mM NaCl, 7 mM 2-mercaptoethanol, 10 mM EDTA, 0.1 mg/ml BSA, 1 µg N6-methyladenine-free λ DNA, T2 Dam P126S MTase and 50 µM AdoMet. One unit of MTase was defined as that amount of enzyme required to protect all GATC sites in 1 µg λ DNA in 1 h at 37°C. The amount of enzyme added depended on the DNA sequence to be protected; further details will be addressed in Results. After 1 h incubation at 37°C the reaction was stopped by heat inactivation of the MTase at 65°C for 15 min. Five to ten units of ENase were added. The concentrations of Tris, MgCl2, NaCl and BSA were pre-adjusted for optimum cleavage conditions and the reaction mixture incubated for an additional 1 h at the temperature recommended by the enzyme supplier. The reaction products were separated by electrophoresis through an agarose gel and visualized by ethidium bromide staining. Control samples of λ DNA were subjected to the same manipulations, but with no MTase added. Unmethylated and undigested λ DNA was used as a molecular length marker. Methylated, undigested λ DNA was used to assay for possible degradation during the methylation reaction with T2 Dam P126S.
RARE cleavage of λ DNA
The RARE cleavage protocol was a combination of the original procedure by Ferrin and Camerini-Otero (3) and that developed by Koob et al. (4). The oligonucleotide was a 40mer corresponding to the phage λ DNA sequence positions 8816–8856: 5′-ATGAGCGATATCCCGGCACTGTCAGATTTGATCACCAGTA-3′. It contains recognition sites (underlined) for BclI (TGATCA) and EcoRV (GATATC). The solution for formation of the oligonucleotide:RecA complex (9 µl) contained 25 mM Tris–acetate pH 8.0, 1 mM Mg acetate, 60 ng oligonucleotide and 1.5 µg RecA. [The working ratio of oligonucleotide:RecA was experimentally determined using the approach described by Koob et al. (4).] After incubation of the mixture at 37°C for 1 min, an aliquot of 1 µl of 0.4 mM ATP-γ-S was added and the reaction was allowed to proceed for another 10 min. To form the oligonucleotide:RecA:λ DNA complex the buffer was adjusted to 25 mM Tris–acetate pH 8.0, 4 mM Mg acetate, 10 mM DDT, 0.1 mg/ml BSA, 0.5 mM spermidine and 1 µl (0.5 µg) of λ DNA was added, bringing the final volume of the mixture to 28 µl. After 30 min at 37°C an aliquot of 1 µl of T2 Dam P126S and 1 µl of 1.5 mM AdoMet were added. Conditions of methylation and subsequent ENase cleavage are described above.
RARE cleavage of yeast genomic DNA embedded in agarose
Saccharomyces cerevisiae (strainYPH80, obtained from Dr P.Goldfarb, New England Biolabs) was used to prepare yeast genomic DNA embedded in agarose. For preparation of yeast genomic DNA embedded in 0.7% agarose plugs we used the kit and protocol from Bio-Rad. The oligonucleotide used for RARE cleavage at the overlapping BclI and DpnII sites (underlined) was 60 nt in length: 5′-GCAAGGAGTTATATTTTGGAAACGATTGATCAATATTTAAAGATTACTTCAAAGGAAGAT-3′. RARE cleavage was performed on DNA in 50 µl plugs according to detailed protocols and tips for troubleshooting (23,24). Immediately prior to use the agarose plugs were equilibrated at room temperature in RARE cleavage buffer (25 mM Tris–acetate, pH 8.0, 4 mM Mg acetate, 0.4 mM DTT and 5 mM spermidine). The components added were 15 µl of 5× RARE buffer, 30 µg RecA protein, 10 µl of 10 mM ATP-γ-S and oligonucleotide (10–100 ng oligonucleotide/µg RecA protein were used to optimize the concentration for efficient cleavage) and water to a total volume of 200 µl. After 15 min at 37°C, 50 µl of agarose plug and 10 µl of 2 mg/ml acetylated BSA were added and the mixture incubated further at 37°C for 30 min. Then 2 µl of 4.6 mM AdoMet and 10–50 U MTase were added and incubation continued for 120 min at 37°C. Methylation was stopped by adding 200 µl of 50 mM EDTA and 2% SDS. After 15 min at 50°C the plugs were washed and equilibrated in restriction buffer with 200 µl final exchange, then 20 U restriction endonuclease was added for digestion for 1 h at 37°C. The reaction was stopped and the products were examined by pulsed field gel electrophoresis (PFGE), performed in a CHEF-DR II apparatus (Bio-Rad) at 15°C with 200 V applied, switch times of 60 s for 15 h and then 90 s for 9 h. The buffer used was 0.5× TBE.
RESULTS AND DISCUSSION
Steady-state kinetics
Standard analyses of the dependence of substrate concentration on velocity were used to obtain the catalytic rate and Michaelis constants and to calculate specific coefficient values from steady-state assays. In preliminary experiments using the canonical duplex the Km for AdoMet was calculated to be 0.30 µM. Thus, to determine kinetic parameters for different duplexes (summarized in Table 1) AdoMet was used at a concentration of 0.55 µM and the kinetics of methyl group transfer were linear under steady-state conditions for T4 Dam, T2 Dam and T2 Dam P126S. As expected, methylation was most efficient on canonical GATC substrates for all enzymes; additionally, sequences containing GATN and GACC were efficiently methylated by all enzymes. In contrast, T2 Dam and T4 Dam poorly methylated GACT, while T2 Dam P126S was very efficient on this sequence. Sequences containing GATR were somewhat more efficient for T2 Dam P126S, but kcat was still reduced ∼100-fold from the canonical site.
Compared with T4 Dam, the T2 Dam enzyme had an ∼2-fold higher kcat for most of the DNA substrates, as well as a slightly higher Km. With the canonical GATC-containing substrate, T2 Dam P126S had similar kinetic parameters to the wild-type T2 Dam. In contrast, with most non-canonical substrates T2 Dam P126S had a 2- to 4-fold higher kcat and Km compared to T4 Dam. Based on these data, T2 Dam P126S was chosen for protection of GAY-containing sites against cleavage by restriction ENases and for RARE cleavage.
Methylation by T2 Dam P126 protects GAY-containing sequences against cleavage by restriction ENases
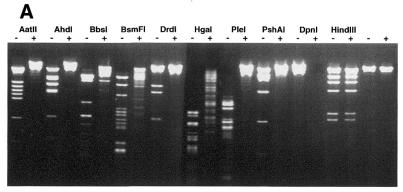
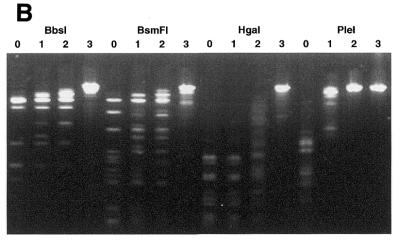
It is known that methylation of N6-adenine in 16 different GAY-containing recognition sequences inhibits DNA cleavage by certain ENases (2,19). A list of these sequences is presented in Table 2. Additionally, a search of the Rebase Enzyme Data Base (New England Biolabs) revealed 12 commercially available enzymes with recognition sequences that overlap GAY and different from those cited above. Using T2 Dam P126S we tested the sensitivity to adenine methylation of this group, namely AatII, AhdI, BbsI, BciVI, BsaI, BsmFI, DrdI, HgaI, PflFI, PleI, PshAI and Tth111I (an isoschizomer of PflFI). To verify that the conditions of T2 Dam P126S methylation could afford protection, ENases BclI, BstF51, EcoRV and FokI were included, since they have been shown previously to be inhibited by the presence of N6-methyladenine in the cleavage site. The results in Figure 1 are representative of the analyses for methylation protection against cleavage by some ENases. All the ENases tested in this particular experiment had a restricted cleavage of DNA that had been methylated by T2 Dam P126S. After a series of experiments varying the MTase concentration it was determined that the ENases tested could be divided into five distinct groups according to the level of MTase required to block cleavage completely. (I) This group has GATC in its recognition sequence, so it was not surprising that loss of DNA cleavage capability was achieved at a relatively low concentration (1–5 U) of MTase for DpnII and BclI. (II) For non-canonical sequences GATA or GACY ∼40 U T2 Dam P126S were required to protect against cleavage by BciVI, BsaI, EcoRV and PleI. These results are also consistent with steady-state kinetic analyses of T2 Dam P126S methylation of synthetic duplexes, although the kcat values with the GATA- and GACY-containing substrates were 2–5 times lower than with the canonical GATC-containing substrate (Table 1). (III) BstF51 and FokI. About three times more T2 Dam P126S (∼120 U) was needed to fully protect non-canonical GATG sites in λ DNA relative to the amount needed to protect GATA sites. According to steady-state kinetic analysis the kcat for the GATG-containing duplex was comparable with the kcat for the GATA-containing substrate. However, the apparent difference can be explained by the fact that these two sequences are present in λ DNA at different frequencies. (IV) This group of enzymes contain GACX in their recognition sequence. Full protection of GACR or GACC sequences against AatII, AhdI, DrdI, PflFI, PshAI and Tth111I required ∼400 U T2 Dam P126S. Poor protection against cleavage by AatII suggests that GACR sites were poor substrates for T2 Dam P126S methylation. This notion was confirmed by steady-state kinetic analysis of GACR-containing synthetic duplexes, namely the kcat values were ∼100 times lower than kcat for a duplex with a canonical GATC site. Five ENases (AhdI, DrdI, PflFI, PshAI and Tth111I) mentioned in this group have a GACX sequence in their recognition site. Obviously, efficacy of DNA protection by T2 Dam P126S against cleavage by these ENases depends on which base, Y or R, is in the fourth position. (V) BbsI, BsmFI and HgaI. Methylation of λ DNA with up to 1000 U T2 Dam P126S was sufficient to fully block the action of BsmFI and HgaI ENases, which recognize sequences containing GACG or some subset of GACX.
Table 2. Recognition sequences containing GAY triplets and blocked by overlapping Dam methylation.
| ENase group |
No. |
Recognition sequence |
Restriction enzymes |
| I | 1 | /GATCa | Bsp143Ib, BstEIIIb, DpnIIb,c, MboIb, MmeIIb, NdeIIb |
| 2 | CGAT/CGa | XorIIb | |
| 3 | T/GATCAd | AtuCIb, BclIb,c, BspXIIb, BstGIb, CpeIb | |
| 4 | GGATC(4/5)a | AlwIb,c, BinIb | |
| 5 | R/GATCYa | MflIb | |
| II | 6 | (5/6)GGATACd | BciVIe |
| 7 | GAT/ATCd | EcoRVb,c | |
| 8 | (5/4)GACTCd | PleIe | |
| 9 | (5/1)GAGACCd | BsaIe | |
| 10 | AGACCa | EcoPIb | |
| 11 | GACCGA(11/9)a | TagIIb | |
| III | 12 | GGATG(2/0)d | BstF5If |
| 13 | GGATG(9/13)d | FokIb,c | |
| 14 | AT/CGATa | BanIIIb, BseCIb, BsiXIb, Bsp106Ib, BspDIb,c, BspXIb, Bsu15Ib, ClaIb,c | |
| 15 | (9/5)GATGCa | SfaNIb,e | |
| 16 | GATNN/NNATCa | BsaBIb,c, Bsh1365Ib, BsiBIb, BsrBRIg, MamIb | |
| 17 | GGATG(10/14)a | StsIb | |
| IV-A | 18 | GACGT/Cd | AatIIe |
| 19 | GACN/NNGTCd | PflFIe, Tth111Ie | |
| 20 | GACNN/NNGTCd | PshAIe | |
| 21 | GACNNN/NNGTCd | AhdIe | |
| 22 | GACNNNN/NNGTCd | DrdIe | |
| IV-B | 23 | GAAGAC(2/6)d | BbsIe |
| 24 | GACGC(5/10)d | HgaIe | |
| 25 | GGGAC(10/14)d | BsmFIe | |
| IV-C | 26 | (5/1)GAGACa | Alw26Ib |
| 27 | (5/1)GAGACGa | Esp3Ib | |
| 28 | G/TCGACa | Rrh4273Ib, SalIb |
aSequences that are predicted to belong to the mentioned group.
bMcClelland et al. (2)
cSensitivity to adenine methylation was shown using T4 Dam (13).
dSequences that were tested in this paper.
eSensitivity to adenine methylation was shown in this paper.
fDegtyarev et al. (19).
gPromega catalog.
Figure 1.
Methylation by T2 Dam P126S MTase alters the sensitivity of phage λ DNA to restriction ENase. Methylation and the cleavage assay were performed under the conditions described in Materials and Methods. ENases are designated at the top. (A) Phage λ DNA was methylated (+) or not (–) with 400 U T2 Dam P126S MTase and then incubated with the designated ENases. (B) The amounts of T2 Dam P126S MTase in the incubation mixture were as follows: lane 0, 0 U; lane 1, 6 U; lane 2, 60 U; lane 3, 600 U.
Thus, all the ENases were inhibited by T2 Dam P126S methylation of their DNA recognition sequences. Based on these results, it is possible to add 12 more members to the list of the GAY-containing recognition sequences in which adenine methylation blocks ENase cleavage. These data also show that any GAY site is an appropriate in vitro substrate for T2 Dam P126S methylation, although GACR sites are methylated very slowly. Finally, these results allow one to select ENases for RARE cleavage experiments.
RARE cleavage of phage λ DNA
In order to demonstrate the utility of T2 Dam P126S for use in a RARE cleavage assay, a 40mer synthetic oligonucleotide (corresponding to the phage λ DNA sequence 8816–8856) was used for strand invasion. This DNA contains both a BclI site and an EcoRV site, thus it provides an opportunity for a direct comparison of two modification systems, the canonical T2 Dam P126S/BclI and non-canonical T2Dam-P126S/EcoRV.
To develop the T2 Dam P126S/BclI RARE cleavage protocol, a number of parameters were tested: concentrations of MTase and RecA proteins, the RecA:oligonucleotide ratio and duration of the methylation reaction. In our hands the optimal amount of RecA was shown to be 1.5 µg/60 ng oligonucleotide. Increasing the RecA concentration resulted in incomplete methylation (data not shown), presumably because of non-specific RecA binding to λ DNA. Determination of the optimal RecA:oligonucleotide ratio is a standard procedure required for optimal RARE cleavage. To determine this ratio we carried out the titration reaction according to Koob et al. (4) with 1.5 µg RecA protein and 30, 60, 120, 180 and 240 ng oligonucleotide. Sixty nanograms of the oligonucleotide was shown to be optimal both to provide protection of the targeted restriction site and to avoid non-specific protection of other sites (data not shown). Experiments to determine the amount of T2 Dam P126S sufficient for full methylation under RARE cleavage conditions showed that the MTase had the same high activity and stability as under standard optimal conditions (data not shown). Three units of the enzyme and a 30 min reaction time is recommended for methylation of canonical GATC sites. These results clearly demonstrate that T2 Dam P126S, in combination with methyladenine-sensitive, GATC-recognizing ENases, was perfectly suited for the RARE cleavage technique.
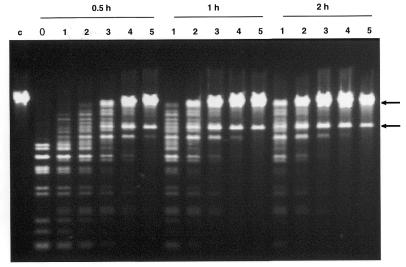
Figure 2 shows a test of the applicability of T2 Dam P126S to RARE cleavage using EcoRV, which has a non-canonical Dam methylation site within the ENase cleavage sequence. Having experimentally determined the optimal RecA:oligonucleotide ratio for the canonical GATC sequence, a time course T2 Dam P126S/EcoRV RARE cleavage experiment was performed with varying MTase concentrations (Fig. 2). Again, the amount of T2 Dam P126S sufficient for complete protection was close to that found in preliminary experiments on methylation (namely 40 U in a 60 min reaction). These data indicate that a combination of T2 Dam P126S with the non-canonical site-recognizing ENase EcoRV can be successfully applied to RARE cleavage.
Figure 2.
RARE cleavage of phage λ DNA with EcoRV. The conditions are described in Materials and Methods. The RecA:oligonucleotide (40mer) duplex ratio was 1.5 µg:60 ng. Duration of the methylation reactions was 0.5, 1 or 2 h as shown. The amounts of T2 Dam P126S MTase in the incubation mixture were as follows: lane 0, no MTase; lane 1, 2.5 U; lane 2, 5 U; lane 3, 10 U; lane 4, 20 U; lane 5, 40 U. Lane c, λ DNA. Arrows show localization of products of RARE cleavage of λ DNA.
RARE cleavage of yeast genomic DNA
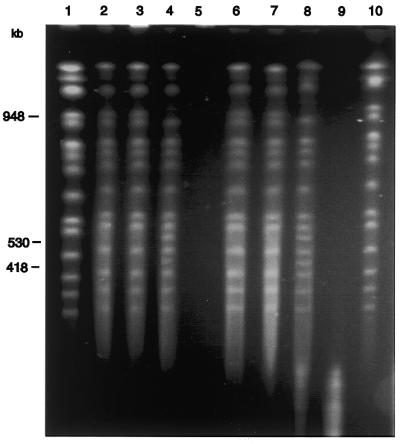
To determine whether large DNA molecules methylated with T2 Dam 126S MTase can be effectively analyzed by RARE cleavage whole chromosomes of the yeast S.cerevisiae (16 linear chromosomes and a total genome size of 11 Mb) were used as a model system. To protect a specific site on chromosome XVI we added a 60mer oligonucleotide (containing BclI and DpnII sites) that was homologous to the sequence 531716–531776. Methylation of yeast chromosomes in the presence of oligonucleotide and RecA, and subsequent digestion with BclI or with DpnII, resulted in cleavage of chromosome XVI at a single site to produce two fragments (Fig. 3). Preliminary RARE experiments were performed to optimize the oligo:RecA ratio to form a complex, to protect the targeted restriction sites and to eliminate non-specific protection of other sites. We found that efficient cleavage occurred at a molar stoichiometry of 3–5 oligonucleotide bases per RecA protein. This is close to what we had observed for RARE cleavage of phage λ DNA. In these experiments the ratio of RecA to oligonucleotides is very important. RecA has to saturate the oligonucleotides, but there should be no excess RecA remaining, as that could promote non-specific protection against MTase, leading to non-specific cuts. Non-specific binding of RecA to DNA can be eliminated either by titrating the oligonucleotide with RecA protein (23,24) or by adding a non-specific oligonucleotide, such as oligo(dT) (4). These aspects of the procedure were stressed by Szybalski in his review (25). In this regard, Koob et al. (4) found that an additional back titration with oligo(dT) can be a more predictable and convenient procedure to remove excess RecA, especially where spurious DNA bands appeared after electrophoresis.
Figure 3.
RARE cleavage of yeast genomic DNA. DNA in agarose plugs from yeast strain YHP80 was subjected to RARE cleavage at BclI and DpnII sites corresponding to positions 531742 and 531741 on yeast chromosome XVI. Details are given in Materials and Methods. Aliquots of 30 µg RecA and 1150 ng 60mer oligonucleotide were used in these reactions. Treated chromosomes were analyzed by PFGE. Sizes of yeast chromosomes were from public databases (NCBI). The two RARE cleavage products have molecular sizes of 530 and 418 kb. The yeast chromosomes and their products were visualized by staining with ethidium bromide. Lanes 1 and 10 show control chromosomes without treatment. Chromosomes were incubated with the following enzyme(s)/oligo: lane 2, RecA/oligo; lane 3, RecA/oligo, T2 Dam P126S MTase; lane 4, RecA/oligo, T2 Dam P126S MTase and DpnII; lane 5, DpnII; lane 6, RecA/oligo; lane 7, RecA/oligo, T2 Dam P126S MTase; lane 8, T2 Dam P126S MTase and BclI; lane 9, BclI.
In this study we have exploited the ability of T2 Dam P126S to methylate non-canonical GAY sites and investigated the sensitivity to adenine methylation of several ENases using λ DNA as a substrate. We also tested the applicability of T2 Dam P126S MTase to RARE cleavage, modifying the canonical sequence GATC, as well as non-canonical GAY sites. Additionally, we showed the feasibility of performing RARE cleavage, exploiting the advantage of methylation by bacteriophage T2 Dam P126S MTase. The approach of utilization of mutant MTases with extended methylation capabilities allows use of a broader range of restriction enzymes in the RARE method and can create new tools for precise physical dissection of small and large DNA molecules.
Acknowledgments
ACKNOWLEDGEMENTS
We thank Dr S.L.Schlagman for constructing some of the plasmids used in this study and Dr V.Perepnikhatka for help in performing the PFGE. This work was supported by the Texas Advanced Technology Program under grant no. 003658-0127 from the Texas Higher Education Coordinating Board (to V.K.), NIEHS grant P30 ES06676 (to R.S.L.) and a US Public Health Service grant GM29227 from the National Institutes of Health (to S.H.). R.S.L. is the holder of the Mary Gibbs Jones Distinguished Chair in Environmental Toxicology from the Houston Endowment.
References
- 1.Koob M., Grimes,E. and Szybalski,W. (1988) Conferring operator specificity on restriction endonucleases. Science, 241, 1084–1086. [DOI] [PubMed] [Google Scholar]
- 2.McClelland M., Nelson,M. and Raschke,E. (1994) Effect of site-specific modification on restriction endonucleases and DNA modification methyltransferases. Nucleic Acids Res., 22, 3640–3659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ferrin L.J. and Camerini-Otero,R.D. (1991) Selective cleavage of human DNA: Rec-A-assisted restriction endonuclease (RARE) cleavage. Science, 254, 1494–1497. [DOI] [PubMed] [Google Scholar]
- 4.Koob M., Burkiewicz,A., Kur,J. and Szybalski,W. (1992) RecA-AC: single-site cleavage of plasmids and chromosomes at any predetermined restriction site. Nucleic Acids Res., 20, 5831–5836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gnirke A., Iadonato,S.P., Kwok,P.-Y. and Olson,M.V. (1994) Physical calibration of yeast artificial chromosome contiguous maps by RecA-assisted restriction endonuclease (RARE) cleavage. Genomics, 24, 199–210. [DOI] [PubMed] [Google Scholar]
- 6.Lauer P., Schneider,S.S. and Gnirke,A. (1998) Construction and validation of yeast artificial chromosome contig maps by RecA-assisted restriction endonuclease cleavage. Proc. Natl Acad. Sci. USA, 95, 11318–11323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Xiang Z., Hu,X.L., Flint,J. and Riethman,H.C. (1999) A sequence-ready map of the human chromosome 17p telomere. Genomics, 58, 207–210. [DOI] [PubMed] [Google Scholar]
- 8.Nielsen L.B., Kahn,D., Duell,T., Weier,H.U., Taylor,S. and Young,S.C. (1998) Apolipoprotein B gene expression in a series of human apolipoprotein B transgenic mice generated with RecA-assisted restriction endonuclease cleavage-modified bacterial artificial chromosomes. An intestine-specific enhancer element is located between 54 and 62 kilobases 5′ to the structural gene. J. Biol. Chem., 273, 21800–21807. [DOI] [PubMed] [Google Scholar]
- 9.Weiner M.P., Felts,K.A., Simcox,T.G. and Braman,J.C. (1993) A method for the site-directed mono- and multi-mutagenesis of double-stranded DNA. Gene, 126, 35–41. [DOI] [PubMed] [Google Scholar]
- 10.Boren J., Lee,I., Callow,M.J., Rubin,E.M. and Innerarity,T.L. (1996) A simple and efficient method for making site-directed mutants, deletions and fusions of large DNA such as P1 and BAC clones. Genome Res., 6, 1123–1130. [DOI] [PubMed] [Google Scholar]
- 11.Ferrin L.J. (1995) Manipulating and mapping DNA with RecA-assisted restriction endonuclease (RARE) cleavage. Genet. Eng., 17, 21–30. [PubMed] [Google Scholar]
- 12.Miner Z. and Hattman,S. (1988) Molecular cloning, sequencing and mapping of the bacteriophage T2 dam gene. J. Bacteriol., 170, 5177–5184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kossykh V.G., Schlagman,S.L. and Hattman,S. (1995) Phage T2 [N6-adenine] methyltrasferase. Overexpression, purification and characterization. J. Biol. Chem., 270, 14389–14393. [DOI] [PubMed] [Google Scholar]
- 14.Kossykh V.G., Schlagman,S.L. and Hattman,S. (1997) Comparative studies of the phage T2 and T4 DNA (N6-adenine) methyltransferases: amino acid changes that affect catalytic activity. J. Bacteriol., 179, 3239–3243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Martienssen R.A. and Richards,E.J. (1995) DNA methylation in eukaryotes. Curr. Opin. Genet. Dev., 5, 234–242. [DOI] [PubMed] [Google Scholar]
- 16.Brooks J.E. and Hattman,S. (1978) In vitro methylation of phage λ DNA by wild-type (dam+) and mutant (damh) forms of the phage T2 DNA-adenine methylase. J. Mol. Biol., 126, 381–394. [DOI] [PubMed] [Google Scholar]
- 17.Hattman S., van Ormondt,H. and de Waard,A. (1978) Sequence specificity of wild-type dam+ and mutant (damh) forms of bacteriophage T2 DNA adenine methylase. J. Mol. Biol., 119, 361–376. [DOI] [PubMed] [Google Scholar]
- 18.Schlagman S.L. and Hattman,S. (1989) The bacteriophage T2 and T4 DNA-[N6-adenine] methyltransferases (Dam) sequence specificities are not identical. Nucleic Acids Res., 17, 109–112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Degtyarev S.K., Netesova,N.A., Abdurashitov,M.A. and Shevchenko,A.V. (1997) Primary structure and strand specificity of BstF51-1 DNA methyltransferase which recognizes 5′-GGATG-3′. Gene, 187, 217–219. [DOI] [PubMed] [Google Scholar]
- 20.Revel H.R. and Hattman,S. (1971) Mutants of T2gt with altered DNA methylase activity: relation to restriction by prophage P1. Virology, 45, 484–495. [DOI] [PubMed] [Google Scholar]
- 21.Miner Z., Schlagman,S.L. and Hattman,S. (1989) Single amino acid changes that alter DNA sequence specificity of the dna [N6-adenine] methyltransferase of bacteriophage T4. Nucleic Acids Res., 17, 8149–8157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Laemmli U.K. (1970) Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature, 227, 680–685. [DOI] [PubMed] [Google Scholar]
- 23.Iadonato S.P. and Gnirke,A. (1996) RARE-cleavage analysis of YACs. Methods Mol. Biol., 54, 75–85. [DOI] [PubMed] [Google Scholar]
- 24.Ferrin L.J. (1995) Manipulation and mapping DNA with RecA-assisted restriction endonuclease (RARE) cleavage. In Setlow J.K. (ed.), Genetic Engineering: Principle and Methods. Plenum Press, New York, Vol. XVII, pp. 21–30. [PubMed]
- 25.Szybalski W. (1997) RecA-mediated Achilles’ cleavage. Curr. Opin. Biotechnol., 8, 75–81. [DOI] [PubMed] [Google Scholar]