Abstract
The ATM-related TRRAP protein is a component of several different histone acetyltransferase (HAT) complexes but lacks the kinase activity characteristic of other ATM family members. We identified a novel function for this evolutionarily conserved domain in its requirement for the assembly of a functional HAT complex. Ectopic expression of TRRAP protein with a mutation in the ATM-related domain inhibits Myc-mediated oncogenic transformation. The Myc-binding region of TRRAP maps to a separable domain, and ectopic expression of this domain inhibits cell growth. These findings demonstrate that the ATM-related domain of TRRAP forms a structural core for the assembly and recruitment of HAT complexes by transcriptional activators.
Keywords: TRRAP, histone acetyltransferase, Myc, ATM domain, oncogenesis
Members of the ATM family of proteins function in humans, yeast, and other organisms as critical sensors of genomic integrity or growth conditions. In humans, the ATM family includes ATM, ATR, FRAP, and DNA-PKcs, and homologs of these proteins are found in yeast, flies, and other organisms (e.g., Mec1p, Tel1p, RAD3p, TOR1p, and Mei-41; Keith and Schreiber 1995). The defining features of the proteins in the ATM family are their general large size (2500–4000 amino acids) and a 300-amino-acid motif in their C terminus that resembles the catalytic domain of PI3-kinases. However, most of these proteins have been shown to phosphorylate other proteins rather than lipids. More extended alignment of the ATM family members identifies two other conserved domains in addition to the PI3-kinase-like domain (Bosotti et al. 2000). The PI3-kinase-like domain is flanked by the FATC domain of 35 amino acids at the extreme C terminus and the FAT domain, which comprises ∼500 amino acids amino terminal of the catalytic domain. All existing evidence suggests that the integrity of the kinase domain is essential for the function of ATM family proteins, but the functions of the FATC and FAT domains remain unknown. However, our recent identification of the novel ATM-related TRRAP proteins has raised new questions about the function of these conserved domains. The TRRAP protein family was identified as transcriptional cofactors that mediate the recruitment of large multiprotein histone acetyltransferase (HAT) complexes to sequence-specific activators (Grant et al. 1998; McMahon et al. 1998; Saleh et al. 1998; Vassilev et al. 1998). We initially identified TRRAP orthologs in humans, Saccharomyces cerevisiae, Schizosaccharomyces pombe, and Caenorhabditis elegans, and more recently in Arabidopsis thaliana and Drosophila melanogaster. The TRRAP proteins are true members of the ATM family because they possess the FAT, kinase-related, and FATC domains. However, the kinase-related domain is conserved in sequence but not in function as the specific residues mediating phosphate transfer are absent (McMahon et al. 1998). This lack of catalytic activity has been supported by biochemical studies on both the human and yeast TRRAP proteins (Saleh et al. 1998). The paradox of sequence conservation in the absence of kinase activity prompted us to explore a noncatalytic role for this conserved domain in the recruitment of HAT complexes to transcriptional activation domains.
Results
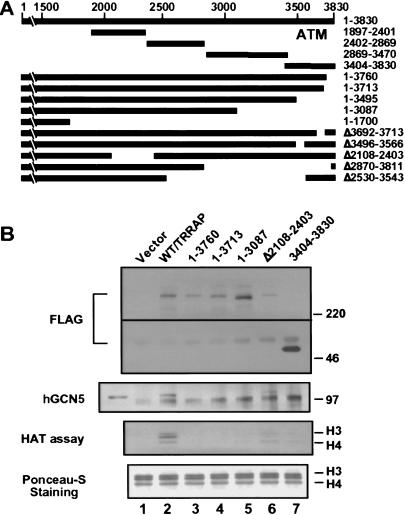
To investigate the function of the ATM-related region of TRRAP, a progressive series of C-terminal deletions (1–3760, 1–3713, and 1–3087) was constructed from a FLAG epitope-tagged full-length (1–3830) TRRAP cDNA expression vector (Fig. 1A). An internal deletion (Δ2108–2403) within the TRRAP cDNA was also constructed. Each protein was transiently expressed in HEK293 cells, immunoprecipitated with anti-FLAG antibodies, eluted from the beads with an excess of FLAG peptide, and subjected to histone acetyltransferase assays using core histones as substrates. The reaction products were resolved by graded porosity SDS-PAGE and transferred onto nitrocellulose membranes. The upper section of the membrane was probed with antibodies for FLAG and hGCN5 and the bottom section was subjected to fluorography after Ponceau-S staining (Fig. 1B). We found that the full-length TRRAP protein bound to the endogenous hGCN5 protein and that the immunoprecipitates had readily assayed histone acetyltransferase activity. In contrast, all of the C-terminal deletion mutations abolished TRRAP binding to hGCN5 and the recruitment of histone acetyltransferase activity, including the TRRAP(1–3760) protein with only a 70-amino-acid deletion including the FATC domain. In addition, an internal 20-amino-acid deletion (Δ3692–3713) of a highly conserved sequence within the ATM-related domain also abolished the interaction with hGCN5 (data not shown). An internal deletion (Δ2108–2403), distant from the ATM-related domain, showed little or no difference in hGCN5 binding or HAT activity compared with full-length TRRAP. These data establish that the ATM-related domain of TRRAP is necessary for HAT recruitment. To determine whether the ATM-related domain of TRRAP was sufficient for HAT recruitment, the C terminus of TRRAP (3404–3830) was expressed with a FLAG epitope tag (Fig. 1B, lane 7). The smaller C-terminal domain was expressed at a much higher level than full-length TRRAP, but this segment was inactive in binding to hGCN5 or in the recruitment of histone acetyltransferase activity. Thus, the ATM/PI-3 kinase homology domain is necessary but not sufficient for the assembly of a functional HAT enzyme complex with TRRAP.
Figure 1.
The ATM/PI-3 kinase domain of TRRAP is critical for the recruitment of histone acetyltransferase activity. (A) Schematic representation of wild-type, full-length TRRAP (top) with the ∼300 C-terminal amino acids homologous to the ATM/PI-3 kinase family members indicated as ATM. Below is shown a series of TRRAP internal fragments, C-terminal truncation mutants, and internal deletion mutants with their respective sizes at the right. (B) ATM-domain dependence for HAT recruitment. Complexes containing transiently expressed FLAG-tagged TRRAP or TRRAP mutants were isolated from HEK293 cells (McMahon et al. 1998). The FLAG–TRRAP or TRRAP mutants were eluted from beads and assayed for HAT activity on calf thymus core histones. Reaction mixtures were resolved on an SDS-PAGE gradient gel (4%–15%) and transferred to a nitrocellulose membrane. The upper section was probed with anti-FLAG and anti-hGCN5 antibodies. Cell lysate containing 100 μg of protein was included as a positive control for hGCN5 (lane left of lane 1). The bottom portion of the membrane below the 30-kD molecular mass marker was stained with Ponceau-S and subjected to fluorography for HAT activities. The 97-kD band common to lanes 1–7 in the hGCN5 panel is a nonspecific band that arises from the anti-FLAG beads, but is not found with cell lysates (lane left of lane 1).
The recruitment of hGCN5 by TRRAP is expected to create an enzyme complex with predominantly H3-specific HAT activity (Yang et al. 1996; Wang et al. 1997), but the TRRAP immunoprecipitates showed an equal preference for H3 and H4 acetylation. The identity of the H4-specific histone acetyltransferase in association with TRRAP requires further characterization but could derive from human MYST family proteins TIP60, MOZ, or MORF (Sterner and Berger 2000).
Functional TRRAP and hGCN5 proteins are required for cellular transformation by the c-Myc oncoprotein (McMahon et al. 1998, 2000). The involvement of the TRRAP ATM-related homology domain in the recruitment of histone acetyltransferase activities prompted us to investigate its physiological role in the cell cycle and cellular transformation. Expression vectors for full-length TRRAP or the TRRAP(1–3760) truncation mutant were cotransfected with expression vectors for c-Myc and/or oncogenic H-rasG12V into early-passage rat embryo fibroblasts (Fig. 2A). Neither TRRAP itself nor the truncation mutant had intrinsic transforming activity in conjunction with the H-rasG12V oncogene. Cotransfection of c-myc and H-rasG12V leads to focus formation, and the addition of the wild-type TRRAP expression vector had no effect on this activity. In contrast, cotransfection of c-myc and H-rasG12V with the TRRAP(1–3760) mutant that is defective for HAT-recruitment resulted in a 50% inhibition of focus formation. The finding of only a partial dominant-negative phenotype with the TRRAP(1–3760) mutant may be due to the large size of the TRRAP protein and relatively low level of ectopic expression, which may not allow effective competition with the endogenous wild-type TRRAP. In parallel with the REF transformation assay, we tested for transfection efficiency or any toxicity from the TRRAP expression constructs by scoring for puromycin-resistant colonies after cotransfection with the pSG5-PuromycinR plasmid (Fig. 2B). Approximately equal numbers of puromycin-resistant colonies were obtained with all plasmid combinations tested, indicating that the inhibitory effect of the TRRAP(1–3760) truncation mutant on oncogenic transformation did not result from cellular toxicity or general growth inhibition.
Figure 2.
Expression of a TRRAP mutant defective in HAT recruitment inhibits c-Myc oncogenic activity. (A) Primary rat embryo fibroblasts were transfected with expression vectors for c-Myc, H-RasG12V, wild-type full-length TRRAP, and the HAT-defective TRRAP(1–3760) truncation mutant in the different combinations indicated. (y axis) Number of transformed foci. Three plates were assayed for each bar. (B) TRRAP and TRRAP mutants do not inhibit colony growth. Primary rat embryo fibroblasts were transfected as indicated along with pSG5 puromycinR plasmid. PuromycinR colonies were counted after 14 d. Two plates were assayed for each bar in the graph.
In addition to TRRAP–Tra1p and GCN5–PCAF, the SAGA–PCAF complexes contain SPT3, which has been suggested to regulate transcription through interaction with TBP (Dudley et al. 1999; Belotserkovskaya et al. 2000). SPT3 protein has been found in all TRRAP complexes that contain hGCN5 (Brand et al. 1999). Because mutations in the ATM-homology region abolished binding to hGCN5, we were interested in determining what other components of the complex were dependent on this domain. To assess whether hSPT3 binding to TRRAP was dependent on the ATM-related domain, immunoprecipitates of FLAG-tagged full-length or a C-terminal deletion of TRRAP were probed with anti-SPT3 antibodies (Fig. 3). Full-length TRRAP bound to hSPT3, but binding was abolished by a small deletion in the ATM-related domain.
Figure 3.
The ATM/PI-3 kinase domain of TRRAP is critical for interaction with hSPT3. HEK293 cells were transiently transfected with expression vectors for FLAG-tagged TRRAP fragment or its deletion mutant, TRRAP(1–3713). Cell lysates were prepared and immunoprecipitated with anti-FLAG antibodies. Each precipitate was resolved by SDS-PAGE and analyzed for the extent of its coprecipitation with endogenous hSPT3. To check the relative expression levels of TRRAP fragments and hGCN5 recruitment, the same membrane was reprobed with anti-FLAG antibodies (top) and anti-hGCN5 (middle).
The exclusive enrichment of TRRAP by the c-Myc N terminus in the initial affinity purification strategy suggested that the interaction between these two nuclear proteins is direct and unlikely to require any associated cofactors (McMahon et al. 1998). We were interested in determining which domain or motif in TRRAP is important for c-Myc binding. Progressive deletions and several internal fragments in the TRRAP C terminus were used to map the sequence for Myc binding (Fig. 4). Plasmids expressing FLAG-tagged TRRAP (or mutants thereof) were transiently cotransfected into HEK293 cells with an expression vector for c-Myc, and then anti-FLAG immunoprecipitates isolated under nondenaturing conditions were probed by Western blot analysis for c-Myc protein (Fig. 4A, bottom). The expression levels and recovery of the TRRAP proteins were tested by anti-FLAG Western blotting (Fig. 4A, top), showing that smaller TRRAP mutants were generally expressed better than the full-length protein. In the initial experiments, deletions from the C terminus up to amino acid 3087 retained the ability to coprecipitate with c-Myc, in contrast with the hGCN5 binding described above. Expression of only the N-terminal half of TRRAP, 1–1700, coprecipitated a negligible amount of c-Myc despite the fact that the expression level was significantly higher than the larger TRRAP proteins. These data indicate that the Myc-binding site maps between amino acids 1700 and 3087. To refine the mapping further, three FLAG-tagged internal fragments of TRRAP (1899–2401, 2402–2869, and 2869–3470) were transiently expressed, immunoprecipitated with anti-FLAG antibody, and assayed for binding to endogenous c-Myc by Western blot (Fig. 4B, top). In parallel, immunoprecipitates by anti-FLAG antibodies were analyzed by Western blotting to assess the expression level of FLAG-tagged TRRAP fragments (Fig. 4B, bottom). Only the TRRAP(1899–2401) fragment coprecipitates with endogenous c-Myc, suggesting that it contains the primary c-Myc interaction domain in the context of intact TRRAP.
Figure 4.
TRRAP amino acids 1899–2026 mediate Myc binding. (A) In vivo coimmunoprecipitation. HEK293 cells were transiently cotransfected with CMV-driven expression vectors for c-Myc and FLAG-tagged full-length TRRAP or a series of TRRAP C-terminal truncation mutants. Cell lysates were prepared and the FLAG-tagged TRRAP protein or mutants were immunoprecipitated with anti-FLAG antibodies. Precipitates were resolved by SDS-PAGE and analyzed by Western blotting for anti-c-Myc antibodies (bottom). The same membrane was also probed with anti-FLAG antibodies to detect TRRAP expression (top). Apparent molecular masses in kilodaltons of protein markers are indicated at the left of fluorogram. (B) Localization of the c-Myc-binding domain. HEK 293 cells were transiently transfected with expression vectors for FLAG-tagged TRRAP fragment. Cell lysates were prepared and immunoprecipitated with anti-FLAG antibodies. Each precipitate was resolved by SDS-PAGE and analyzed for the extent of its coprecipitation with endogenous c-Myc (top). Cell lysate from the c-Myc transfection was included as a positive control (lane left of lane 1). To check the relative expression levels of TRRAP fragments, the same membrane was reprobed with anti-FLAG antibodies (bottom). (C) An internal region of TRRAP (spanning amino acids 1591–2026) interacts with N-Myc. GST fusion proteins containing the indicated TRRAP regions were expressed in E. coli and bound to glutathione beads. Equal amounts of bead-bound fusion proteins were incubated with affinity-purified FLAG-tagged N-Myc protein. After washing, interacting proteins were subjected to SDS-PAGE analysis and immunoblotted with anti-FLAG antibody. The FLAG-N-Myc starting material (20% of input) was run with the bound material.
Coprecipitation of c-Myc and TRRAP from whole cell lysates could be mediated by another component of SAGA or other intermediate protein. A more direct assay of protein–protein interactions using the GST-pulldown technique was used to map the Myc-binding domain on TRRAP (Fig. 4C). To broaden the analysis, we tested the binding of N-Myc to TRRAP as N-Myc binds with an affinity equal to that of c-Myc (Wood et al. 2000). A series of TRRAP protein fragments was expressed to equal levels in Escherichia coli and purified on beads. The beads were then incubated with affinity-purified N-Myc protein to assess binding. Five segments spanning 50% of the 3830-amino-acid TRRAP protein were tested for N-Myc binding, and only one segment (1591–2026) exhibited significant binding compared with GST alone. This segment overlaps the binding domain predicted from mammalian expression studies above, suggesting that the major site of Myc interaction with TRRAP maps between amino acids 1899 and 2026.
To provide an in vivo test of the N-Myc–TRRAP interaction domain, two FLAG–TRRAP protein segments (1261–1579 and 1899–2401) were stably expressed at equal levels in human neuroblastoma cells, which contain an amplified N-myc gene (data not shown). Because Myc is required for cell cycle progression, competition between endogenous N-Myc and a critical cofactor would be expected to impede cell growth. Consistent with this model, expression of FLAG–TRRAP(1899–2401) containing the Myc-binding domain increased the doubling time of IMR5 cells from 28 to 63 h, or 2.25-fold (Table 1). Expression of another segment of TRRAP (1261–1579) had no impact on growth rate compared with control cells, even though the latter can inhibit c-Myc and N-Myc transforming activity in primary fibroblasts (McMahon et al. 1998).
Table 1.
Myc-interacting region of TRRAP has a dominant negative effect on cell growth
| Cell line
|
TRRAP segment
|
Doubling time (h)
|
|---|---|---|
| IMR-5 | — | 26.7 ± 6.4 |
| IMR-5 | 1261–1579 | 27.9 ± 5.3 |
| IMR-5 | 1899–2401 | 63.1 ± 12.2 |
IMR-5 human neuroblastoma cells were stably transfected with expression vectors for FLAG-tagged TRRAP fragments as indicated. Cell lysates were immunoprecipitated with anti-FLAG antibody followed by immunoblot analysis with the same antibody (data not shown). Both fragments were comparably expressed. Doubling times were calculated from three individual plates from each line (± SE).
Discussion
This study provides the first functional mapping of domains within the large ATM-related TRRAP protein that is implicated in transcriptional regulation and chromatin modification from yeast to man. We find that the c-Myc-binding region is distinct from the ATM/PI-3 kinase domain, and this region is conserved among TRRAP orthologs from other species, but not conserved in other ATM family members. Interestingly, this region overlaps the direct binding site for transcriptional activators on the yeast ortholog of TRRAP (Tra1p; J. Workman, pers. comm.), suggesting that the function of TRRAP–Tra1p as a mediator of HAT-activator interactions has been highly conserved in evolution. Furthermore, this same domain of Tra1p binds to Yng2p in yeast HAT complexes and hence may play a major functional role in all TRRAP–Tra1p complexes (Loewith et al. 2000).
Although the ATM-related C-terminal domain of TRRAP lacks kinase activity, we show here that this domain is essential for the recruitment of HAT activity by the complex, and similar findings have been made for the yeast Tra1p (J. Workman, pers. comm.). Generally protein–protein interactions are mediated by relatively small motifs, and, therefore, it was unexpected that mutations in the ATM-related domain abolished HAT recruitment whereas the isolated ATM-related domain is defective for binding. These results suggest that TRRAP may adopt a complex three-dimensional structure that is dependent on the ATM homology domain for the recruitment of HATs and most other components of the complex. A parallel can be drawn with the ATM-related Rad3p in fission yeast, which requires an N-terminal protein domain for the activity of the C-terminal kinase domain (Chapman et al. 1999). ATM/PI3 kinase family proteins are frequently involved in checkpoint or sensing functions within the cell, suggesting that the activity of TRRAP itself may also be regulated.
A common mechanism of chromatin modification is the acetylation of N-terminal lysine residues on histones, which reduces histone–DNA interactions to promote a more open chromatin configuration (Grant et al. 1997). A key feature of the multiprotein HAT complexes is the ability to acetylate nucleosomal histones, whereas the HAT enzyme alone can only acetylate free histones (Kuo et al. 1996; Grant et al. 1997). Therefore, one or more components of these complexes must modify HAT activity to facilitate nucleosome recognition, as well as mediate binding to transcriptional activators. TRRAP is remarkably conserved in evolution as a 3800 ± 100-amino-acid polypeptide, implying that size and the orientation of functional elements are critical. TRRAP–Tra1p appears to be a direct, global mediator of transcriptional activator–HAT interactions, and further elucidation of its structure and function may provide important insights into the dynamics of transcriptional activation and chromatin modification.
Materials and methods
Plasmid constructs and transfections
Expression plasmids were created by standard methods in a CMV-promoter driven vector containing a FLAG-epitope tag and verified by sequence analysis (McMahon et al. 1998). Details of individual constructs are available on request.
HAT assays
FLAG-tagged TRRAP protein was transiently expressed in HEK293 cells and isolated from whole-cell lysates by binding to anti-FLAG antibody covalently conjugated to beads (Sigma). The complexes containing FLAG–TRRAP or its mutants were released by addition of excess FLAG peptide in HAT assay buffer (50 mM Tris at pH 8.0, 10% glycerol, 50 mM KCl, 0.1 mM EDTA, 10 mM butyric acid, 1 mM DTT, 1 mM PMSF). Calf thymus core histones (3 μg) along with 14C-acetyl-CoA were incubated with each eluate, and reaction mixtures were resolved on a SDS-PAGE gradient gel (4%–15%; Ogryzko et al. 1996). Proteins were transfered onto a nitrocellulose membrane, and the section above the 30-kD molecular mass marker was subjected to Western blot analysis and probed with anti-FLAG and anti-hGCN5 antibodies. The bottom section of the membrane was stained with Ponceau-S and subjected to fluorography for HAT activities.
GST pulldown assay
TRRAP fragments were PCR amplified, cloned into pGXT4T1 (Amersham Pharmacia Biotech) and expressed as fusion proteins in E. coli. For large-scale purification, bacterial pellets were sonicated in STE buffer (10 mM Tris at pH 8.0, 150 mM NaCl, 1 mM EDTA, 5 mM DTT, 1.5% Sarkosyl, PMSF–aprotinin). After addition of Triton X-100 to a final concentration of 2%, lysates were cleared by centrifugation, and the fusion proteins were bound to glutathione–Sepharose 4B beads (Amersham Pharmacia Biotech). Beads were analyzed for the fusion protein by SDS-PAGE followed by Western analysis for GST.
FLAG–N-Myc was stably expressed from a CMV promoter in IMR-5 neuroblastoma cells after neo cotransfection and G418 selection. Cells from three semiconfluent 15-cm plates were lysed in E1A lysis buffer (McMahon et al. 1998), followed by overnight immunoprecipitation with anti-FLAG antibody (Sigma) and Protein A/G beads (Santa Cruz). Bound FLAG–N-Myc was eluted with FLAG peptide (Sigma). Equal amounts of GST–TRRAP or control beads (based on the Western), were incubated with 30–50 μL of the FLAG–N-Myc proteins in binding buffer for 3 h in 4°C. Beads were washed three times in the same buffer, and then bound proteins were resolved by SDS-PAGE. N-Myc interaction was detected with anti-FLAG antibody, and the same blot was reprobed with anti-GST to assess the amount of protein on the beads.
Cell culture
Rat embryo fibroblasts were prepared by trypsinization of 15-d embryos (Land 1995), and focus formation was assessed as described (McMahon et al. 1998). Foci were scored after 18 d. Growth properties after transfection of the same DNAs were assessed on duplicate plates of primary rat embryo fibroblasts cotransfected as indicated along with 1 μg of pSG5 puromycinR plasmid. Transfected cells were selected in 2.5 μg/mL puromycin for 14 d, at which time the number of colonies per plate was determined.
FLAG–TRRAP fragments were stably expressed in IMR-5 neuroblastoma cells by cotransfection with neo and selection with G418. Growth rates and protein expression were assessed at early passage of G418-resistant polyclonal populations. Equal numbers of viable cells were plated in parallel for the parental IMR-5 line and the two TRRAP fragment-expressing lines. Cells were counted 48 and 72 h after plating, and the doubling time was calculated.
Acknowledgments
We thank John Maris for providing IMR-5 cells, and Christine Brown and Jerry Workman for communicating results in advance of publication. This work was supported by a grant from the National Cancer Institute to MDC. This work was also supported by a Leukemia Society Special Fellow award and grants from the Edward Mallinckrodt, Jr. Foundation, the Mary L. Smith Charitable Lead Trust, the Gustavus and Louise Pfeiffer Research Foundation, and the American Association for Cancer Research to S.B.M.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL mcole@molbio.princeton.edu; FAX (609) 258-4575.
Article and publication are at http://www.genesdev.org/cgi/doi/10.1101/gad.900101.
References
- Belotserkovskaya R, Sterner DE, Deng M, Sayre MH, Lieberman PM, Berger SL. Inhibition of TATA-binding protein function by SAGA subunits Spt3 and Spt8 at Gcn4-activated promoters. Mol Cell Biol. 2000;20:634–647. doi: 10.1128/mcb.20.2.634-647.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bosotti R, Isacchi A, Sonnhammer EL. FAT: A novel domain in PIK-related kinases. Trends Biochem Sci. 2000;25:225–227. doi: 10.1016/s0968-0004(00)01563-2. [DOI] [PubMed] [Google Scholar]
- Brand M, Yamamoto K, Staub A, Tora L. Identification of TATA-binding protein-freeTAFII-containing complex subunits suggests a role in nucleosome acetylation and signal transduction. J Biol Chem. 1999;274:18285–18289. doi: 10.1074/jbc.274.26.18285. [DOI] [PubMed] [Google Scholar]
- Chapman CR, Evans ST, Carr AM, Enoch T. Requirement of sequences outside the conserved kinase domain of fission yeast Rad3p for checkpoint control. Mol Biol Cell. 1999;10:3223–3238. doi: 10.1091/mbc.10.10.3223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dudley AM, Rougeulle C, Winston F. The Spt components of SAGA facilitate TBP binding to a promoter at a post-activator-binding step in vivo. Genes & Dev. 1999;13:2940–2945. doi: 10.1101/gad.13.22.2940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grant PA, Duggan L, Cote J, Roberts SM, Brownell JE, Candau R, Ohba R, Owen-Hughes T, Allis CD, Winston F, et al. Yeast Gcn5 functions in two multisubunit complexes to acetylate nucleosomal histones: Characterization of an Ada complex and the SAGA (Spt/Ada) complex. Genes & Dev. 1997;11:1640–1650. doi: 10.1101/gad.11.13.1640. [DOI] [PubMed] [Google Scholar]
- Grant PA, Schieltz D, Pray-Grant MG, Yates JRR, Workman JL. The ATM-related cofactor Tra1 is a component of the purified SAGA complex. Mol Cell. 1998;2:863–867. doi: 10.1016/s1097-2765(00)80300-7. [DOI] [PubMed] [Google Scholar]
- Keith CT, Schreiber SL. PIK-related kinases: DNA repair, recombination, and cell cycle checkpoints. Science. 1995;270:50–51. doi: 10.1126/science.270.5233.50. [DOI] [PubMed] [Google Scholar]
- Kuo M-H, Brownell JE, Sobel RE, Ranalli TA, Cook RG, Edmonson DG, Roth SY, Allis CD. GCN5p, a yeast nuclear histone acetyltransferase, acetylates specific lysines in histones H3 and H4 that differ from deposition-related acetylation sites. Nature. 1996;383:269–272. doi: 10.1038/383269a0. [DOI] [PubMed] [Google Scholar]
- Land H. Transformation of primary rat embryo cells. Methods Enzymol. 1995;254:37–41. doi: 10.1016/0076-6879(95)54005-9. [DOI] [PubMed] [Google Scholar]
- Loewith R, Meijer M, Lees-Miller SP, Riabowol K, Young D. Three yeast proteins related to the human candidate tumor suppressor p33(ING1) are associated with histone acetyltransferase activities. Mol Cell Biol. 2000;20:3807–3816. doi: 10.1128/mcb.20.11.3807-3816.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMahon SB, Van Buskirk HA, Dugan KA, Copeland TD, Cole MD. The novel ATM-related protein TRRAP is an essential cofactor for the c-Myc and E2F oncoproteins. Cell. 1998;94:363–374. doi: 10.1016/s0092-8674(00)81479-8. [DOI] [PubMed] [Google Scholar]
- McMahon SB, Wood MA, Cole MD. The c-Myc cofactor TRRAP recruits the histone acetylase hGCN5. Mol Cell Biol. 2000;20:556–562. doi: 10.1128/mcb.20.2.556-562.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogryzko VV, Schiltz RL, Russanova V, Howard BH, Nakatani Y. The transcriptional coactivators p300 and CBP are histone acetyltransferases. Cell. 1996;87:953–959. doi: 10.1016/s0092-8674(00)82001-2. [DOI] [PubMed] [Google Scholar]
- Saleh A, Schieltz D, Ting N, McMahon SB, Litchfield DW, Yates JR, III, Lees-Miller SP, Cole MD, Brandl CJ. Tra1p is a component of the yeast ADA/SPT transcriptional regulatory complexes. J Biol Chem. 1998;273:26559–26570. doi: 10.1074/jbc.273.41.26559. [DOI] [PubMed] [Google Scholar]
- Sterner DE, Berger SL. Acetylation of histones and transcription-related factors. Microbiol Mol Biol Rev. 2000;64:435–459. doi: 10.1128/mmbr.64.2.435-459.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vassilev A, Yamauchi J, Kotani T, Prives C, Avantaggiati ML, Qin J, Nakatani Y. The 400 kDa subunit of the PCAF histone acetylase complex belongs to the ATM superfamily. Mol Cell. 1998;2:869–875. doi: 10.1016/s1097-2765(00)80301-9. [DOI] [PubMed] [Google Scholar]
- Wang L, Mizzen C, Ying R, Candau R, Barlev N, Brownell J, Allis CD, Berger S. Histone acetyltransferase activity is conserved between yeast and human GCN5 and required for complementation of growth and transcriptional activation. Mol Cell Biol. 1997;17:519–527. doi: 10.1128/mcb.17.1.519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood MA, McMahon SB, Cole MD. An ATPase/helicase complex is an essential cofactor for oncogenic transformation by c-Myc. Mol Cell. 2000;5:321–330. doi: 10.1016/s1097-2765(00)80427-x. [DOI] [PubMed] [Google Scholar]
- Yang X-J, Ogryzko VV, Nishikawa J, Howard BH, Nakatani Y. A p300/CBP-associated factor that competes with the adenoviral oncoprotein E1A. Nature. 1996;382:319–324. doi: 10.1038/382319a0. [DOI] [PubMed] [Google Scholar]