Abstract
Pax-6 genes, known to be essential for eye development, encode an evolutionarily conserved transcription factor with two DNA-binding domains. To corroborate the contribution of each DNA-binding domain to eye formation, we generated truncated forms of the Drosophila Pax-6 gene eyeless and tested their capacity to rescue the ey2 mutant. Surprisingly, EY deleted of the homeodomain rescued the ey2 mutant and triggered ectopic eyes morphogenesis. In contrast, EY lacking the paired domain failed to rescue the ey2 mutant, led to truncation of appendages, and repressed Distal-less when misexpressed. This result suggests distinct functions mediated differentially by the two DNA-binding domains of eyeless.
Keywords: eyeless, Pax-6, eye development, paired domain, homeodomain
The animal body plan is set up during embryogenesis by a combinatorial genetic interaction between selector genes. The Hox gene family is responsible for the anterior-posterior segmentation pattern of the embryo. First, the segmentation genes establish a repetitive pattern of body segments. Then, the Hox genes specify the identity of each segment and induce another class of selector genes that determine the different appendages or organs within a given segment. The exclusive expression of those genes gives each organ its specific identity. Recently, several Drosophila genes have been identified as being capable of inducing organogenesis when ectopically expressed. Vestigial is essential for wing and haltere identity (Kim et al. 1996), Distal-less (Dll) for leg identity, and in combination with extradenticale and homothorax for antenna determination (Casares and Mann 1998; Gonzalez-Crespo et al. 1998). The selector gene for eye morphogenesis is the Pax-6 gene (Halder et al. 1995).
Pax genes encode nuclear transcription factors that play a key role in organogenesis (Dahl et al. 1997). They are characterized by a structurally conserved DNA-binding domain known as the paired domain (PD). The Pax family is subdivided into different subgroups, according to the presence or absence of additional conserved domains, namely, a paired-like homeodomain (HD) or a truncated paired-like homeodomain and an octapeptide (Strachan and Read 1994). The paired domain is a bipartite DNA-binding domain, subdivided into a N- and C-terminal part referred to as the PAI and RED domain, respectively (Jun and Desplan 1996). PAI, RED, and HD consist of three α-helices each, with the third helix contacting the bases in the major groove of the DNA (Xu et al. 1999).
Pax-6 contains a paired domain, and a paired-like homeodomain, but lacks the octapeptide (Ton et al. 1991). The role of Pax-6 appears to be evolutionarily conserved during eye development in both mammals and flies. An important function for Pax-6 in mammalian eye development was deduced from the disruption of eye development in homozygous Small eye (Sey) mice carrying a Pax-6 mutation (Hill et al. 1991). In humans, heterozygous mutations of Pax-6 are known to cause various forms of congenital eye abnormalities, such as Aniridia or Peter's anomaly, and a complete absence of eyes in homozygous mutants. Further analysis in mice revealed an early expression in most structures of the developing eye (Walther and Gruss 1991; Grindley et al. 1995). Beside its role in eye morphogenesis, Pax-6 has important functions in the development of the brain and the spinal cord.
In contrast to vertebrates, in which multiple protein isoforms derived from a single Pax-6 gene are found, in Drosophila two Pax-6 genes, ey (Quiring et al. 1994) and twin of eyeless (toy) (Czerny et al. 1999), and a related gene called eye gone (eyg) have been identified. Like toy and ey, eyg is a Pax class transcription factor and has a RED domain and a paired-class HD, but lacks the N-terminal arm of the PAI domain (Jun et al. 1998). High-affinity binding assays with the RED domain revealed a binding specificity similar to the one described for the Pax-6 5a isoform (Epstein et al. 1994; Jun et al. 1998). This splice form contains a 14-amino-acid insertion in the PD found in all vertebrate Pax-6 genes analyzed so far. No Pax-6 splice forms have been reported in Drosophila, suggesting that this organism solves the complex regulation of development by gene duplication and modification rather than by differential splicing.
Two hypomorphic mutants of eyeless, ey2 and eyR, both inactivating the eye-specific enhancer, result in partial to complete loss of compound eyes (Quiring et al. 1994). The early expression pattern of toy and ey and the mutant phenotypes of ey suggest that Pax-6 is also a crucial regulator of the development of the insect eye. Gain-of-function experiments in which toy, ey, or Sey are ectopically expressed, lead to the formation of ectopic eyes on Drosophila appendages (Halder et al. 1995; Czerny et al. 1999). Misexpression of Pax-6 in Xenopus leads to the formation of ectopic eye structures (Altmann et al. 1997; Chow et al. 1999). The conservation of Pax-6 genes in the animal kingdom, their ability to induce ectopic eyes, and their mutant phenotypes puts them high up in the genetic hierarchy of eye development.
The three genes sine oculis (so), eyes absent (eya), and dachshund (dac) encode evolutionarily conserved nuclear proteins that are essential for Drosophila eye development (Bonini et al. 1993; Cheyette et al. 1994; Mardon et al. 1994). All three genes are activated by EY and are required for EY-induced formation of ectopic eyes, which suggests that these genes act downstream of ey in the eye development pathway (Bonini et al. 1997; Shen and Mardon 1997; Halder et al. 1998). This hypothesis was confirmed for so, which is a direct target gene of ey (Niimi et al. 1999). In contrast, toy acts upstream of ey, because ectopic expression of toy induces ey, but not vice versa (Czerny et al. 1999). Whereas the crosstalk between toy, ey, and their downstream targets has been shown, the relationship between eyg, ey, and toy remains unclear.
The PD and the HD are the most conserved regions within the Pax-6 protein, pointing out evolutionary constraints imposed to maintain specific binding to target genes. It has been suggested that the protein might activate target genes either through the PD, the HD, or both. Alternatively, both domains could work in a cooperative manner to regulate their target genes (Jun and Desplan 1996). Furthermore, recent evidence indicates that these two DNA-binding domains are also involved in protein–protein interactions (Plaza et al. 2001). Despite the importance of Pax-6 for eye development, the respective functions of the two DNA-binding domains are unknown. Thus, understanding their contribution in vivo is crucial for the correlation of the different mutations to their respective phenotypes. To unravel the functional role of the PD and the HD in EY, we generated different truncated forms and tested their capacity to rescue an ey mutant eye phenotype. We show that the PD within the EY protein is essential and sufficient for the induction of eye development and that the HD within the EY protein is sufficient to repress the selector gene Dll. Thus, Pax-6 can exert a dual function as an activator and a repressor via its two different DNA-binding domains.
Results
The eyeless homeodomain is not required to rescue ey2 mutants and to induce ectopic eyes
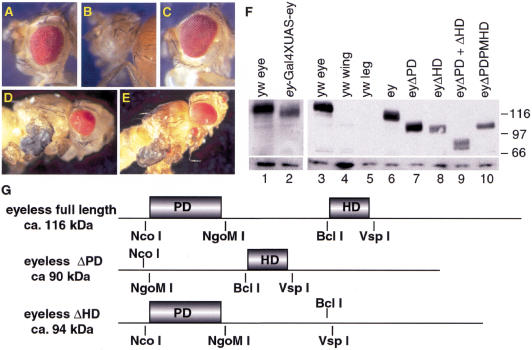
It has been shown previously that full-length ey cDNA was able to efficiently rescue the eye phenotype of ey2 mutants when expressed in the eye disc under the control of the eye-specific ey enhancer (Halder et al. 1998). To assess the contribution of the EY DNA-binding domains to eye development, we expressed mutant ey cDNAs with ey-enhancer Gal4 in an ey mutant background. We generated ey cDNA under the control of the UAS promoter (Brand et al. 1994), which lacked either the paired box (eyΔPD) or the homeobox (eyΔHD) (Fig. 1G). When expressed in the eye disc, eyΔPD was not able to rescue the eye phenotype of ey2, but enhanced the eye reduction, leading to a complete loss of compound eye in 64% of the flies (Fig. 1B). Unexpectedly, eyΔHD rescued the eye phenotype at an even higher efficiency than the full-length ey-cDNA (Fig. 1A,C), suggesting that the HD of ey was dispensable for eye formation in the ey2 mutant background. Similar results were obtained in the ey null mutant eyJ5.71 (data not shown) isolated recently in our laboratory by an EMS mutagenesis screen. This mutant is RNA and protein null (see also Figs. 2F and 3C, below) due to a 9-kb deletion in the 5′ region of the gene (eyJ5.71; S. Flister, U. Kloter, and W.J. Gehring, unpubl.). To corroborate these findings, we performed ectopic expression experiments in a wild-type background. We found that misexpression of eyΔPD by dppblink–Gal4 in various imaginal discs did not lead to the formation of any eye structures, but generated severely truncated appendages (Fig. 1D). In contrast, misexpression of eyΔHD resulted in ectopic eye formation at the same efficiency as full-length ey (Fig. 1E). These results confirmed our finding that the PD was sufficient to induce the eye developmental pathway and suggested that the HD might act as a repressor for a gene involved in leg development.
Figure 1.
Rescue and ectopic expression of the different ey constructs. (A–C) Different UAS constructs were crossed to ey enhancer Gal4 in an ey2 mutant background. Three independent crosses were done with two independent ey–Gal4 driver lines. Percentages given relate to an eye size that is >80% of the wild-type eye size. They represent the average of six measurements, in which at least 70 flies were analyzed. (A) Rescue with full-length ey (50%). (B) No rescue with eyΔPD. (C) Rescue with eyΔHD (79%). (D,E) Ectopic expression of UAS constructs with dppblink–Gal4 in wild-type background. (D) Misexpression of eyΔPD leads to truncation of appendages. (E) Misexpression of eyΔHD leads to formation of ectopic eyes. (F) Western blot analysis on third instar discs of EY proteins expressed during the rescue (lanes 1,2) and the ectopic expression (lanes 3–10) experiments. (Lanes 1,3) yw control eye discs; (lane 2) eye discs expressing ey with ey enhancer Gal4 in an ey2 mutant background; (lanes 4,5) yw control wing and leg discs, respectively; (lanes 6–10) leg discs expressing the various ey constructs by dppblink Gal4; (lane 6) misexpression of full-length ey; (lane 7) misexpression of eyΔPD; (lane 8) misexpression of eyΔHD; (lane 9) misexpression of eyΔPD+ΔHD; (lane 10) misexpression of eyΔPDPMHD. Molecular weight marker is indicated at right. The Western blot below indicates the loading control with an anti-β-Tubulin antibody. (G) Schematic representation of the ey constructs. The ey-cDNA was used to generate the different deletions by use of the indicated restriction sites. The size of the new proteins is indicated in kilodaltons next to the schematically drawn cDNA.
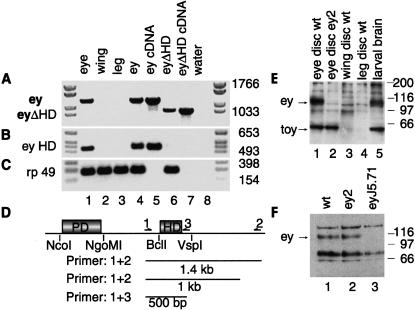
Figure 2.
RT–PCR analysis to test for the presence of ey HD and molecular analysis of the ey mutants. (A–C) RT–PCR on leg discs of third instar larval stage in which ey and eyΔHD were misexpressed. (Lanes 1–3) yw controls. (Lane 1) Eye; (lane 2) wing; (lane 3) leg discs; (lane 4) misexpression of full-length ey; (lane 5) ey cDNA control; (lane 6) misexpression of eyΔHD; (lane 7) eyΔHD cDNA control; (lane 8) water control. (A) Detection of full-length ey, primer 1 + 2 of D. (B) Detection of ey HD fragment, primer 1 + 3 of D. (C) rp49 control. In lane 6, in which eyΔHD was misexpressed, we failed to detect full-length ey (A) and the HD fragment (B), whereas rp49 was expressed normally. (D) Schematic drawing of ey cDNA and the respectively used primers for the RT–PCR in A and B. (E,F) Molecular analysis of ey mutants. (E, lanes 1–5) Western blot analysis of ey2 mutants of third instar larval stage with an anti PD antibody; (lanes 1,2) eye discs of wild-type and ey2, respectively. EY is only detectable in wild-type in contrast to TOY. (Lanes 3,4) Wing and leg discs control of wild type; (lane 5) larval brain of wild type, both proteins are expressed. (F, lanes 1–3): Western blot analysis of adult head with an anti EY antibody. (Lane 1) Wild type; (lane 2) ey2; (lane 3) eyJ5.71. EY could be detected in wild type and ey2 , but not in eyJ5.71.
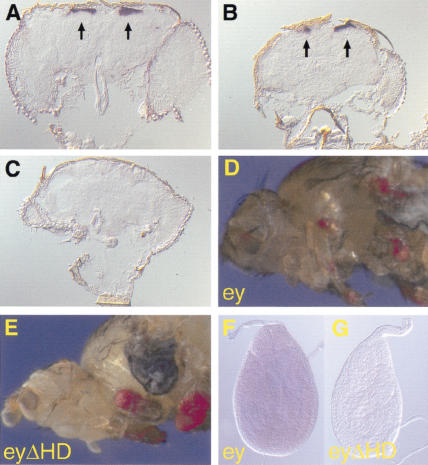
Figure 3.
Expression analysis of ey and ectopic eyes in ey2 mutants. (A–C) In situ hybridization on cryosections of adult heads with an anti-sense Dig ey probe. Expression was only detected in the brain (arrows), but not in the retina. (A) Wild type; (B) ey2; (C) eyJ5.71. (D,E) Ectopic eyes induced by ey and eyΔHD, respectively with dppblink–Gal4 in an ey2 mutant background. (F,G) In situ hybridization with an anti-sense Dig toy probe on third instar leg discs of ey2 mutants in which ey and eyΔHD were misexpressed with dppblink–Gal4. In both cases, toy could not be detected.
To further characterize the function of the HD, we generated the two following constructs: one lacking PD and HD (eyΔPD+ΔHD) and one lacking the PD and carrying two point mutations of amino acids directly involved in DNA binding of the HD (point mutations, S50A and N51A) (eyΔPDPMHD). Misexpression of both eyΔPD+ΔHD and eyΔPDPMHD, by dppblink–Gal4 did not induce any appendage truncation (data not shown). Previously published results had shown that the nuclear localization signals are contained within the DNA-binding domains of Pax-6 (Carriere et al. 1995). To ensure that our point-mutated construct was still localized in the nucleus, we performed immunofluorecent analysis. We found that eyΔPD+ΔHD was no longer transported into the nucleus, whereas eyΔPDPMHD was nuclear (data not shown). Therefore, we concluded that the truncated appendages we observed by misexpression of eyΔPD are due to DNA binding of the HD.
To ensure that the Gal4 system used during our rescue assay does not over produce the ey constructs and that the ectopically induced proteins were correctly expressed, we performed Western analysis using an α-EY antibody. We analyzed eye discs of ey2 mutants that expressed full-length ey by ey-enhancer Gal4, and leg discs of third instar larvae that misexpressed the various EY proteins in the dppblink domain and compared them with endogenous EY levels. We found that the Gal4 system does not over express ey in our rescue experiment (Fig. 1F, lanes 1 and 2) and that our ectopically induced proteins were expressed with the expected molecular weight and at comparable levels (Fig. 1F, lanes 3–10). Therefore, we concluded that the phenotypes obtained were due to the misexpression of the different mutated ey constructs.
eyΔHD is sufficient to trigger eye development in the absence of endogenous ey
Regulatory feedback loops between ey and its downstream genes so, dac, and eya have been demonstrated in an ectopic situation (Chen et al. 1997; Pignoni et al. 1997). Therefore, we tested whether during eyΔHD-induced ectopic eye development, the endogenous intact ey gene might get activated and in turn be responsible for the ectopic eye formation. We tested for the presence of endogenous ey by RT–PCR analysis in leg imaginal discs in which ey and eyΔHD were misexpressed in a wild-type background. First, we used a set of primers able to detect both intact ey and eyΔHD (Fig. 2A) (primer 1 + 2; Fig. 2D). Full-length ey was detected only in yw eye discs (Fig. 2A, lane 1), and in leg discs in which it had been misexpressed (Fig. 2A, lane 4). It was absent in yw control wing (Fig. 2A, lane 2) and leg discs (Fig. 2A, lane 3), and in leg discs in which eyΔHD had been misexpressed (Fig. 2A, lane 6). To exclude template competition, we repeated the RT–PCR experiment using an additional set of primers able to prime only the intact ey and not the ΔHD transcript (Fig. 2B) (primer 1 + 3; Fig. 2D). Again, we failed to detect any full-length ey (Fig. 2B, lane 6) upon misexpression of eyΔHD. This experiment shows that eyΔHD is not able to induce full-length ey, suggesting that in an ectopic situation, the homeodomain is dispensable during the larval stages of eye development.
To test whether the HD would also be dispensable during the pupal stages of development, we ectopically misexpressed eyΔHD in ey2 mutants. There, we expect no endogenous EY to be present in the tissue that gives rise to the eye, because the eye-specific enhancer of the gene is disrupted (Quiring et al. 1994). All ey mutants characterized so far show a partial to complete loss of the compound eye, although to a highly variable degree. As an additional control, we used the eyeless null mutant eyJ5.71. As revealed by in situ hybridization, ey is not expressed in the embryonic eye-anlagen and in the larval eye discs of ey2 mutants, whereas toy expression remains unaffected (Quiring et al. 1994; Czerny et al. 1999). Western blot analysis on ey2 eye discs confirmed that EY is not detectable, in contrast to TOY (Fig. 2E, lanes 1–5). Western blot analysis of adult heads revealed that EY is absent in eyJ5.71 mutants, but present in ey2 mutants (Fig. 2F, lanes 1–3). To elucidate whether ey is expressed at all in the adult eye, we performed in situ hybridization on cryosections of adult heads. We found that ey expression in the adult head is restricted to the brain area (Fig. 3A–C). Therefore, the residual expression of EY in ey2 adult heads is due to the expression of the gene in the brain, ey2 being a mutation affecting only the eye-specific enhancer of the gene (Quiring et al. 1994). This result allowed us to analyze the role of the ey homeodomain during the late stages of eye development. Misexpression of eyΔHD in ey2 induced ectopic eyes at the same efficiency as full-length ey (Fig. 3D,E). The same result was obtained in eyJ5.71 mutants (data not shown), showing that the HD of ey is not required during ectopic eye development.
Because Drosophila has two Pax-6 genes, we wanted to exclude the possibility that eyΔHD would activate toy and that the TOY–HD would be responsible for the ectopic eye development. Although it has been demonstrated that in an ectopic situation ey does not activate toy, we performed in situ hybridization on leg discs of ey2 mutants in which eyΔHD was misexpressed. However, we could not detect ectopically induced toy transcripts (Fig. 3F,G), ruling out the possibility that the TOY–HD may functionally replace EY–HD during ectopic eye development. Thus, these results show that eyΔHD induces ectopic eyes independently of endogenous toy and ey.
ey target genes are induced via the paired domain
Our data predict that ey downstream target genes required for eye development are activated by the paired domain. Therefore, we tested the expression of so as a direct target gene of ey (Niimi et al. 1999). The various ey constructs were misexpressed by dppblink–Gal4. Induction of so expression was detected by lac-Z staining of an enhancer trap line (Cheyette et al. 1994). so was ectopically activated by the full-length (Fig. 4B) as well as the EYΔHD (Fig. 4D) protein. It was not ectopically activated when the paired domain was missing (Fig. 4C), showing that the PD within the EY protein is sufficient to induce its direct target so.
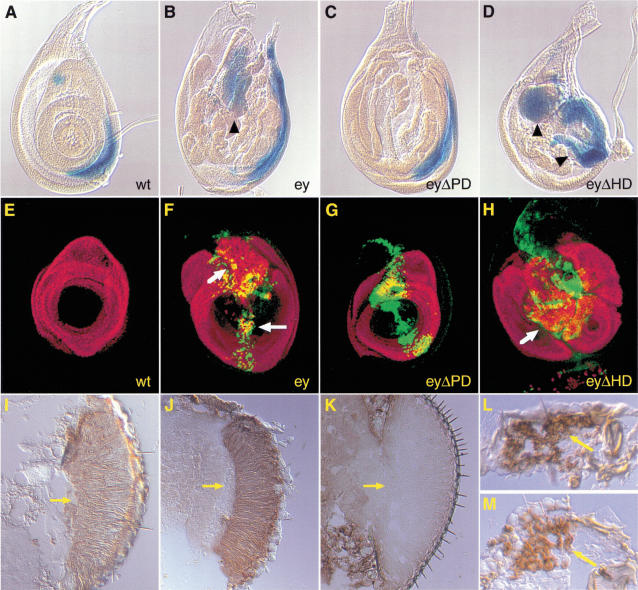
Figure 4.
Activation of downstream genes of ey required during eye development. (A–D) Induction of so enhancer trap line by misexpression of the various ey constructs. (E–H) Confocal sections showing the induction of DAC by misexpression of the various ey constructs (DAC, red; EY, green). All panels show late third instar larval leg discs in which the various constructs were misexpressed by dppblink–Gal4. (A,E) Wild-type expression pattern; (B,F) misexpression of ey (arrowhead and arrow highlight regions of induction); (C,G) misexpression of eyΔPD (so and DAC are both not induced); (D,H) misexpression of eyΔHD leads, in both cases, to a stronger induction than with full-length ey; (I–M) Rhodopsin-1 expression in various mutants and in ectopic eyes. The expression of Rhodopsin was monitored with an α-Rhodopsin-1 antibody on cryosections; (I,J) Rhodopsin-1 expression in the retina in ey2 (I) and eyJ5.71 (J) mutants; (K) no staining in the rhodopsin-1 mutant ninaE; (L,M) Rhodopsin-1 expression in ectopic eyes induced ey (L) and eyΔHD (M) in an ey2 mutant. Arrows indicate the retina.
dac is an indirect target gene of ey and misexpression of dac has been shown to be in part responsible for appendage truncation and to be able to induce ectopic eye development (Shen and Mardon 1997). We repeated the misexpression experiments done for so and used antibodies to detect both EY and DAC. Like so, DAC is only ectopically activated if the PD in EY is present (Fig. 4F–H), as expected for an essential gene in eye development. This suggests that in our case, the truncated appendages are not due to DAC misexpression, because it does not get ectopically activated by eyΔPD.
Next, we asked whether the ectopic eyes generated by eyΔHD also express late marker genes of eye development. rhodopsin-1 has been proposed to be directly regulated by the homeodomain of ey (Sheng et al. 1997). We therefore analyzed the presence of rhodopsin-1 in ectopic eyes generated by eyΔHD in ey2 mutants, in which we showed that neither EY–HD nor TOY–HD are required for ectopic eye development. Immunostainings on cryosections by use of an α-Rhodopsin-1 antibody revealed that Rhodopsin-1 is expressed in the retina of ectopic eyes generated by ey and eyΔHD in ey2 mutants (Fig. 4L,M). Rhodopsin-1 expression was also detected in the eyes of both ey2 and eyJ5.71 mutants, but not in the eyes of the rhodopsin-1 mutant ninaE (Fig. 4I–K). This indicates that the expression of Rhodopsin-1 is independent of the homeodomain of ey and does not require the presence of EY in the adult eye. It strengthens the hypothesis that rhodopsin-1 is likely to be activated by another paired type HD containing a gene other than ey.
The ey homeodomain is able to repress distal-less
Because the homeodomain of the Pax-6 proteins is highly conserved, we wondered about its function during development. It has been shown previously that ey is able to repress Dll in an ectopic situation (Kurata et al. 2000). So far, our experiments have suggested that the HD may confer gene repression. Therefore, we tested whether Dll repression by ey is mediated by the homeodomain. We ectopically expressed ey, eyΔPD, eyΔHD, and eyΔPDPMHD on all appendages with dppblink–Gal4 and monitored EY and DLL expression by antibody detection. Ectopic expression of ey (data not shown) and eyΔPD (Fig. 5A) were able to repress Dll expression in the respective areas of overlap, in contrast to ectopic expression of eyΔHD (Fig. 5B) and eyΔPDPMHD (data not shown). This result shows that the HD of ey mediates Dll repression by DNA binding. Therefore, we conclude that the truncated appendages are due in part to the repression of Dll.
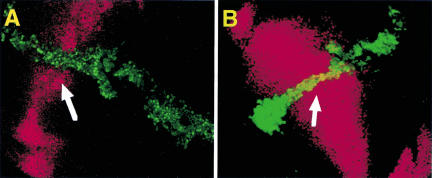
Figure 5.
DLL repression by ectopic expression of eyΔPD. (A,B) Confocal sections of antibody staining on wing discs in which the different ey constructs were misexpressed with dppblink–Gal4 (DLL, red; EY, green). (A) Misexpression of eyΔPD leads to a repression of DLL in the respective areas of overlap (arrow: yellow staining), whereas misexpression of eyΔHD does not (B).
Discussion
Pax-6 is one of the most important transcription factors controlling eye development. It has two evolutionarily conserved DNA-binding domains and, therefore, we considered it a priori unlikely that one of the two domains would be sufficient to induce all genes required for eye development. However, the rescue of the ey2 mutant by eyΔHD led us to the conclusion that the HD of ey could be dispensable for eye formation. To corroborate this finding, we switched to ectopic expression experiments. We showed that eyΔHD induces ectopic eyes in wild-type and in an ey mutant background. All target genes tested are only activated in the presence of the PD. In addition, misexpression of eyΔHD was not able to induce endogenous ey in wild-type or endogenous toy in an ey mutant background. To corroborate our hypothesis that the HD of ey was dispensable, we further characterized the ey2 mutant, showing that endogenous full-length ey is not expressed in cells that give rise to the eye. Therefore, together with the rescue experiment, we conclude that the expression of a PD containing EY protein is sufficient to induce eye development and that the HD of ey is dispensable for target gene activation and EY-mediated eye morphogenesis. Sequence analysis of Pax-6 mutations in patients with Aniridia or Peter's anomaly showed that point mutations of Pax-6 affecting eye development are mostly located in the PD (Glaser et al. 1992; Hanson et al. 1994; Martha et al. 1994). This raises the question of the function of the homeodomain being so highly conserved among the Pax-6 genes. Obviously, the HD might have an essential function in the development of the brain and the ventral nerve cord. We were able to show in an ectopic situation that the repression of Dll (at the RNA level) is mediated by EY–HD. This may explain why we observed truncated appendages when eyΔPD was misexpressed, and it suggests that the EY–HD can confer repression during development. The repression might be achieved either directly or by activation of a repressor. We do not rule out the possibility that the HD might be able to activate gene expression in organs other than the eye. In vertebrates, different splice forms of Pax-6 have been characterized, some of them lacking the paired domain (Carriere et al. 1993). In Caenorhabditis elegans, one splice form without the paired domain was found to be important for the development of the peripheral nervous system (Zhang and Emmons 1995). Therefore, the homeodomain of Pax-6 might play a major role during nervous system development in Drosophila also. Thus, eye development could provide the selective pressure directed toward the conservation of the PD, whereas the nervous system put constrains on the HD. This would connect one protein to the development of two different organ systems, both of which are required for vision.
Our results show that each domain of the Drosophila Pax-6 gene ey can function separately, and that simultaneous binding of both domains, on the same regulatory element, is not required. Similar deletion analysis have also been done for other homeodomain containing proteins (Fitzpatrick et al. 1992; Ananthan et al. 1993; Bertuccioli et al. 1996). In contrast to this analysis, in which the lack of the HD was not able to fully rescue endogenous protein function, in the case of ey, the HD is dispensable to rescue an ey mutant eye phenotype. Thus, these results might reflect different ways of actions for the different homeodomain-containing proteins. Therefore, we would like to propose a model for ey in which the same transcription factor can act as a repressor and an activator via its two different DNA-binding domains in the context of different organs. The question of how the protein exerts its different functions might be explained by recruiting different cofactors as has been shown recently for Pax-5 (Eberhard et al. 2000). We are currently searching for such interacting factors.
Materials and methods
Molecular methods
Western blot and cloning procedure were done according to the standard protocol described in Sambrook et al. (1989). The embryonic ey cDNA (E10) was deleted between S19-GT-A181 for eyΔPD by use of the NcoI–NgoMI sites. The amino acids GT were inserted in the linker to connect the two restriction sites. The eyΔHD was deleted between L405–T523 using the sites BclI–VspI and also by connecting them with a linker. The double deletion contains the same deletion regions. The point mutations S50 to A50 (TCA to GCA); N51 to A51 (ACC to GCC) were done by standard PCR experiments. Each construct generated was confirmed by sequencing. Western blot experiments were done with a rabbit α-EY antibody at a dilution of 1:200, in which the antibody was preabsorbed with larval tissue or with a rabbit α-PD of squid Pax-6 antibody at a dilution of 1:500. Each lane was loaded with extracts from 10 discs, 5 adult heads, or 5 larval brains, respectively. Extracts of all Western were boiled for 6 min. Correct transfer was tested by ponceau red staining. Additionally, equal loading was tested with an α-β-Tubulin antibody at a dilution of 1:10. The secondary antibody for detection of the signal was used at a dilution of 1:2000 (HRP-coupled swine α-rabbit antibody from DAKO A/S), and the signal was revealed by use of a chemoluminescence kit (Amersham). The RT–PCR was performed as follows: total RNA was extracted from discs (with Trizol; Life Technologies) and then reverse transcribed (80 leg discs per 800 μL of Trizol). The single-stranded cDNA was amplified by SMART PCR cDNA Synthesis Kit (Clontech). The PCR was further performed as follows: 28 cycles of, 20 sec at 95°C, 20 sec at 57°C, 13 sec at 72°C for rp49; 20 sec at 95°C, 30 sec at 55°C, 1 min 40 sec at 72°C for primer 1 + 2; 20 sec at 95°C, 20 sec at 58°C, 13 sec at 72°C for primer 1 + 3 by use of the Taq polymerase from Pharmacia. Primer sequences. Primer 1, 5′-AGTCCGATGAAACGGG-3′; Primer 2, 5′-CCTAGACCCACGGTGAG-3′; Primer 3, 5′-GG GACCCCCAGCTGATCCGG-3′; rp49 sens, 5′-CGAACAAGC GCACCCGC-3′; rp49 antisens, 5′-CGCAGGCGACCGTTGG GG-3′.
Histology
In situ hybridization on sections was performed as described in Janssens and Gehring (1999). In situ hybridization on discs, β-Galactosidase staining and antibody stainings on cryosections were performed as described in Ashburner (1989). Antibody staining on discs were performed as described in Halder et al. (1998). The concentration of the antibodies was as follows: rat α-EY 1:500 (Haldes et al. 1998); mAb α-DLL 1:20 (Wu and Cohen 1999); mAb α-DAC 1:100 (Mardón et al. 1994).
Acknowledgments
We thank S. Tomarev, U. Walldorf, S. Cohen, and G. Mardon for the rabbit α-Pax-6 PD, the EY antibodies, the DLL antibody, and the DAC antibody, respectively. The mAb α-Rhodopsin-1 and the mAb α-β-Tubulin were obtained from the Developmental Studies Hybridoma Bank, developed under the auspices of the NICHD and maintained by the University of Iowa, Department of Biological Sciences, Iowa City. We also thank S. Flister, L. Michaut, S. Plaza, M. Seimiya, N. Grieder, B. Gafford, B. Dichtl, and A. Grötzinger for critical reading of the manuscript. This work was supported by the Swiss National Science Foundation and the Kantons of Basel-Stadt and Baselland. The last stages of this work were supported by Grant-in-Aid from the Ministry of Education, Science, Sports, and Culture, Japan, the Mitsubishi Foundation and the Sumitomo Foundation to S.K.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL Walter.Gehring@unibas.ch; FAX 41-61-267-20-78.
Article and publication are at http://www.genesdev.org/cgi/doi/10.1101/gad.196401
References
- Altmann CR, Chow RL, Lang RA, Hemmati-Brivanlou A. Lens induction by Pax-6 Xenopus laevis. Dev Biol. 1997;185:119–123. doi: 10.1006/dbio.1997.8573. [DOI] [PubMed] [Google Scholar]
- Ananthan J, Baler R, Morrissey D, Zuo J, Lan Y, Weir M, Voellmy R. Synergistic activation of transcription is mediated by the N-terminal domain of Drosophila fushi tarazu homeoprotein and can occur without DNA binding by the protein. Mol Cell Biol. 1993;13:1599–1609. doi: 10.1128/mcb.13.3.1599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashburner M. Drosophila: A laboratory manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- Bertuccioli C, Fasano L, Jun S, Wang S, Sheng G, Desplan C. In vivo requirement for the paired domain and homeodomain of the paired segmentation gene product. Development. 1996;122:2673–2685. doi: 10.1242/dev.122.9.2673. [DOI] [PubMed] [Google Scholar]
- Bonini NM, Leiserson WM, Benzer S. The eyes absent gene: Genetic control of cell survival and differentiation in the developing Drosophila eye. Cell. 1993;72:379–395. doi: 10.1016/0092-8674(93)90115-7. [DOI] [PubMed] [Google Scholar]
- Bonini NM, Bui QT, Gray-Board GL, Warrick JM. The Drosophila eyes absent gene directs ectopic eye formation in a pathway conserved between flies and vertebrates. Development. 1997;124:4819–4826. doi: 10.1242/dev.124.23.4819. [DOI] [PubMed] [Google Scholar]
- Brand AH, Manoukian AS, Perrimon N. Ectopic expression in Drosophila. Methods Cell Biol. 1994;44:635–654. doi: 10.1016/s0091-679x(08)60936-x. [DOI] [PubMed] [Google Scholar]
- Carriere C, Plaza S, Martin P, Quantannens B, Bailly M, Stehelin D, Saule S. Characterization of quail Pax-6 (Pax-QNR) proteins expressed in the neuroretina. Mol Cell Biol. 1993;13:7257–7266. doi: 10.1128/mcb.13.12.7257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carriere C, Plaza S, Caboche J, Dozier C, Bailly M, Martin P, Saule S. Nuclear localization signals, DNA binding, and transactivation properties of quail Pax-6 (pax-QNR) isoforms. Cell Growth Differ. 1995;6:1531–1540. [PubMed] [Google Scholar]
- Casares F, Mann RS. Control of antennal versus leg development in Drosophila. Nature. 1998;392:723–726. doi: 10.1038/33706. [DOI] [PubMed] [Google Scholar]
- Chen R, Amoui M, Zhang Z, Mardon G. Dachshund and eyes absent proteins form a complex and function synergistically to induce ectopic eye development in Drosophila. Cell. 1997;91:893–903. doi: 10.1016/s0092-8674(00)80481-x. [DOI] [PubMed] [Google Scholar]
- Cheyette BN, Green PJ, Martin K, Garren H, Hartenstein V, Zipursky SL. The Drosophila sine oculis locus encodes a homeodomain-containing protein required for the development of the entire visual system. Neuron. 1994;12:977–996. doi: 10.1016/0896-6273(94)90308-5. [DOI] [PubMed] [Google Scholar]
- Chow RL, Altmann CR, Lang RA, Hemmati-Brivanlou A. Pax6 induces ectopic eyes in a vertebrate. Development. 1999;126:4213–4222. doi: 10.1242/dev.126.19.4213. [DOI] [PubMed] [Google Scholar]
- Czerny T, Halder G, Kloter U, Souabni A, Gehring WJ, Busslinger M. Twin of eyeless, a second Pax-6 gene of Drosophila, acts upstream of eyeless in the control of eye development. Mol Cell. 1999;3:297–307. doi: 10.1016/s1097-2765(00)80457-8. [DOI] [PubMed] [Google Scholar]
- Dahl E, Koseki H, Balling R. Pax genes and organogenesis. BioEssays. 1997;19:755–765. doi: 10.1002/bies.950190905. [DOI] [PubMed] [Google Scholar]
- Eberhard D, Jimenez G, Heavey B, Busslinger M. Transcriptional repression by Pax5 (BSAP) through interaction with corepressors of the Groucho family. EMBO J. 2000;19:2292–2303. doi: 10.1093/emboj/19.10.2292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Epstein JA, Glaser T, Cai J, Jepeal L, Walton DS, Maas RL. Two independent and interactive DNA-binding subdomains of the Pax6 paired domain are regulated by alternative splicing. Genes & Dev. 1994;8:2022–2034. doi: 10.1101/gad.8.17.2022. [DOI] [PubMed] [Google Scholar]
- Fitzpatrick VD, Percival-Smith A, Ingles CJ, Krause HM. Homeodomain-independent activity of the fushi tarazu polypeptide in Drosophila embryos. Nature. 1992;356:610–612. doi: 10.1038/356610a0. [DOI] [PubMed] [Google Scholar]
- Glaser T, Walton DS, Maas RL. Genomic structure, evolutionary conservation and aniridia mutations in the human PAX6 gene. Nat Genet. 1992;2:232–239. doi: 10.1038/ng1192-232. [DOI] [PubMed] [Google Scholar]
- Gonzalez-Crespo S, Abu-Shaar M, Torres M, Martinez AC, Mann RS, Morata G. Antagonism between extradenticle function and Hedgehog signalling in the developing limb. Nature. 1998;394:196–200. doi: 10.1038/28197. [DOI] [PubMed] [Google Scholar]
- Grindley JC, Davidson DR, Hill RE. The role of Pax-6 in eye and nasal development. Development. 1995;121:1433–442. doi: 10.1242/dev.121.5.1433. [DOI] [PubMed] [Google Scholar]
- Halder G, Callaerts P, Gehring WJ. Induction of ectopic eyes by targeted expression of the eyeless gene in Drosophila. Science. 1995;267:1788–1792. doi: 10.1126/science.7892602. [DOI] [PubMed] [Google Scholar]
- Halder G, Callaerts P, Flister S, Walldorf U, Kloter U, Gehring WJ. Eyeless initiates the expression of both sine oculis and eyes absent during Drosophila compound eye development. Development. 1998;125:2181–2191. doi: 10.1242/dev.125.12.2181. [DOI] [PubMed] [Google Scholar]
- Hanson IM, Fletcher JM, Jordan T, Brown A, Taylor D, Adams RJ, Punnett HH, van Heyningen V. Mutations at the PAX6 locus are found in heterogeneous anterior segment malformations including Peters' anomaly. Nat Genet. 1994;6:168–173. doi: 10.1038/ng0294-168. [DOI] [PubMed] [Google Scholar]
- Hill RE, Favor J, Hogan BL, Ton CC, Saunders GF, Hanson IM, Prosser J, Jordan T, Hastie ND, van Heyningen V. Mouse small eye results from mutations in a paired-like homeobox-containing gene. Nature. 1991;354:522–525. doi: 10.1038/354522a0. [DOI] [PubMed] [Google Scholar]
- Janssens H, Gehring WJ. Isolation and characterization of drosocrystallin, a lens crystallin gene of Drosophila melanogaster. Dev Biol. 1999;207:204–214. doi: 10.1006/dbio.1998.9170. [DOI] [PubMed] [Google Scholar]
- Jun S, Desplan C. Cooperative interactions between paired domain and homeodomain. Development. 1996;122:2639–2650. doi: 10.1242/dev.122.9.2639. [DOI] [PubMed] [Google Scholar]
- Jun S, Wallen RV, Goriely A, Kalionis B, Desplan C. Lune/eye gone, a Pax-like protein, uses a partial paired domain and a homeodomain for DNA recognition. Proc Natl Acad Sci. 1998;95:13720–13725. doi: 10.1073/pnas.95.23.13720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, Sebring A, Esch JJ, Kraus ME, Vorwerk K, Magee J, Carroll SB. Integration of positional signals and regulation of wing formation and identity by Drosophila vestigial gene. Nature. 1996;382:133–138. doi: 10.1038/382133a0. [DOI] [PubMed] [Google Scholar]
- Kurata S, Go MJ, Artavanis-Tsakonas S, Gehring WJ. Notch signaling and the determination of appendage identity. Proc Natl Acad Sci. 2000;97:2117–2122. doi: 10.1073/pnas.040556497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mardon G, Solomon NM, Rubin GM. dachshund encodes a nuclear protein required for normal eye and leg development in Drosophila. Development. 1994;120:3473–3486. doi: 10.1242/dev.120.12.3473. [DOI] [PubMed] [Google Scholar]
- Martha A, Ferrell RE, Mintz-Hittner H, Lyons LA, Saunders GF. Paired box mutations in familial and sporadic aniridia predicts truncated aniridia proteins. Am J Hum Genet. 1994;54:801–811. [PMC free article] [PubMed] [Google Scholar]
- Niimi T, Seimiya M, Kloter U, Flister S, Gehring WJ. Direct regulatory interaction of the eyeless protein with an eye- specific enhancer in the sine oculis gene during eye induction in Drosophila. Development. 1999;126:2253–2260. doi: 10.1242/dev.126.10.2253. [DOI] [PubMed] [Google Scholar]
- Pignoni F, Hu B, Zavitz KH, Xiao J, Garrity PA, Zipursky SL. The eye-specification proteins So and Eya form a complex and regulate multiple steps in Drosophila eye development. Cell. 1997;91:881–891. doi: 10.1016/s0092-8674(00)80480-8. [DOI] [PubMed] [Google Scholar]
- Plaza S, Prince F, Jaeger J, Kloter U, Flister S, Benassayag C, Cribbs D, Gehring WJ. Molecular basis for the inhibition of Drosophila eye development by Antennapedia. EMBO J. 2001;20:802–811. doi: 10.1093/emboj/20.4.802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quiring R, Walldorf U, Kloter U, Gehring WJ. Homology of the eyeless gene of Drosophila to the Small eye gene in mice and Aniridia in humans. Science. 1994;265:785–789. doi: 10.1126/science.7914031. [DOI] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T. Molecular cloning: A laboratory manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- Shen W, Mardon G. Ectopic eye development in Drosophila induced by directed dachshund expression. Development. 1997;124:45–52. doi: 10.1242/dev.124.1.45. [DOI] [PubMed] [Google Scholar]
- Sheng G, Thouvenot E, Schmucker D, Wilson DS, Desplan C. Direct regulation of rhodopsin 1 by Pax-6/eyeless in Drosophila: Evidence for a conserved function in photoreceptors. Genes & Dev. 1997;11:1122–1131. doi: 10.1101/gad.11.9.1122. [DOI] [PubMed] [Google Scholar]
- Strachan T, Read AP. PAX genes. Curr Opin Genet Dev. 1994;4:427–438. doi: 10.1016/0959-437x(94)90032-9. [DOI] [PubMed] [Google Scholar]
- Ton CC, Hirvonen H, Miwa H, Weil MM, Monaghan P, Jordan T, van Heyningen V, Hastie ND, Meijers-Heijboer H, Drechsler M, et al. Positional cloning and characterization of a paired box- and homeobox- containing gene from the aniridia region. Cell. 1991;67:1059–1074. doi: 10.1016/0092-8674(91)90284-6. [DOI] [PubMed] [Google Scholar]
- Walther C, Gruss P. Pax-6, a murine paired box gene, is expressed in the developing CNS. Development. 1991;113:1435–1449. doi: 10.1242/dev.113.4.1435. [DOI] [PubMed] [Google Scholar]
- Wu J, Cohen SM. Proximodistal axis formation in the Drosophila leg: Subdivision into proximal and distal domains by Homothorax and Distal-less. Development. 1999;126:109–117. doi: 10.1242/dev.126.1.109. [DOI] [PubMed] [Google Scholar]
- Xu HE, Rould MA, Xu W, Epstein JA, Maas RL, Pabo CO. Crystal structure of the human Pax6 paired domain-DNA complex reveals specific roles for the linker region and carboxy-terminal subdomain in DNA binding. Genes & Dev. 1999;13:1263–1275. doi: 10.1101/gad.13.10.1263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Emmons SW. Specification of sense-organ identity by a Caenorhabditis elegans Pax-6 homologue. Nature. 1995;377:55–59. doi: 10.1038/377055a0. [DOI] [PubMed] [Google Scholar]