Abstract
We describe a novel mammalian DNA binding activity that requires at least two symmetrically methylated CpG dinucleotides in its recognition sequence, preferably within the sequence 5′CGCG. A key component of the activity is Kaiso, a protein with POZ and zinc-finger domains that is known to associate with p120 catenin. We find that Kaiso behaves as a methylation-dependent transcriptional repressor in transient transfection assays. Kaiso is a constituent of one of two methyl–CpG binding complexes originally designated as MeCP1. The data suggest that zinc-finger motifs are responsible for DNA binding, and may therefore target repression to specific methylated regions of the genome. As Kaiso associates with p120 catenin, Kaiso may link events at the cell surface with DNA methylation-dependent gene silencing.
Keywords: DNA methylation, methy-CpG binding protein, Kaiso, transcriptional repression, POZ/BTB zinc-finger protein
Methylation of cytosine in cytidine–guanidine (CpG) dinucleotide pairs causes repression of transcription, and has been implicated in the stable modifications of gene expression during development (Walsh and Bestor 1999). Mouse embryos, frog embryos, and cultured cells with reduced CpG methylation all misexpress a number of genes that may normally be repressed by DNA methylation (Stancheva and Meehan 2000; Jackson-Grusby et al. 2001). A family of five mammalian proteins, MeCP2, MBD1, MBD2, MBD3, and MBD4, are of likely importance in the interpretation of the DNA methylation signal (Hendrich and Bird 1998). Each family member has a related stretch of 60–80 residues termed the methyl–CpG binding domain (MBD) (Nan et al. 1993). This domain is essential for MeCP2 binding to chromosomes (Nan et al. 1996), and is required for the specific binding of MBD1, MBD2, and MBD4 to methylated DNA in vitro and in vivo (Hendrich and Bird 1998; Ng et al. 2000). The histone deacetylase inhibitor trichostatin A (TSA) can relieve transcriptional repression mediated by MeCP2 (Jones et al. 1998; Nan et al. 1998), MBD1 (Ng et al. 2000), and MBD2 (Ng et al. 1999), indicating that all three behave as transcriptional repressors that involve histone deacetylases. Correspondingly, MeCP2 associates with the Sin3A/HDAC corepressor complex and MBD2 copurifies with the histone deacetylase-containing NuRD complex as part of the methyl–CpG binding complex originally known as MeCP1 (Meehan et al. 1989; Jones et al. 1998; Nan et al. 1998; Ng et al. 1999; Wade et al. 1999; Feng and Zhang 2001).
All of the above proteins bind methyl–CpG sites with little apparent specificity for flanking nucleotide sequence. Here, we describe characterization of a novel DNA binding activity that requires an appropriately organized cluster of methyl–CpG moieties. A key component of the activity is the nuclear protein Kaiso, which belongs to the BTB/POZ family of zinc-finger (ZF) proteins (POZ-ZF proteins; for review, see Collins et al. 2001). We find that Kaiso acts as a transcriptional repressor that requires cytosine methylation at its consensus DNA binding sequence. Kaiso was first identified in a yeast two-hybrid screen with human p120 catenin as bait (Daniel and Reynolds 1999), and was subsequently shown to associate with p120 catenin in vivo. Its additional involvement in methylation-dependent gene silencing raises the possibility that it may link cell surface signaling with the control of gene expression.
Results and Discussion
A novel methyl–CpG-specific DNA binding activity in CSML-0 nuclear extracts
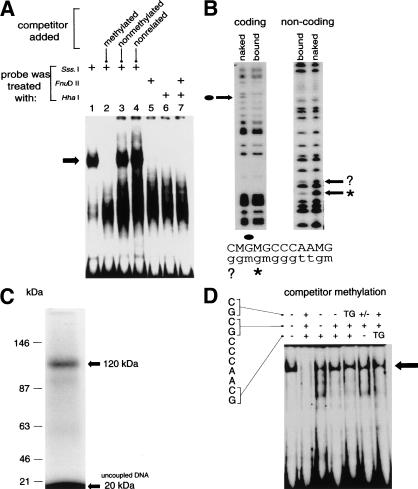
A methyl–CpG-specific DNA binding complex was detected using a DNA probe from an intron of the murine S100A4 gene in electrophoretic mobility shift assays (EMSA) with nuclear extracts from the CSML-0 adenocarcinoma cell line (Fig. 1A, lane 1). The probe coincides with a DNase 1 hypersensitive site in the gene (Prokhortchouk et al. 1998), and contains three closely spaced CpGs in the sequence 5′-MGMGCCCAAMG-3′ (where M is 5-methylcytosine). The complex was abolished by adding a 100-fold molar excess of unlabeled methylated fragment (lane 2), but neither an excess nonmethylated probe (lane 3) nor an unrelated nonmethylated sequence (lane 4) was able to compete for binding. Similar methylation-dependent complexes were detected in various mammalian nuclear extracts, but not in extracts from HeLa cells (see below). Methylation interference footprinting localized binding of the complex to the methylated CpG cluster, and showed specific contacts with G residues within the MGMG motif (Fig. 1B). To estimate the molecular weight of the bound protein, an excised DNA–protein complex was cross-linked by ultraviolet light. A labeled band of ∼120 kD was observed, which corrected to ∼100 kD when the weight of bound DNA (∼20 kD) was subtracted (Fig. 1C).
Figure 1.
A methyl–CpG-specific DNA binding activity in nuclear extracts from CSML-0 cells. (A) Nuclear extracts from CSML-0 cells form a DNA methylation-specific complex (arrow) when mixed with a labeled 210-bp probe containing three CpG motifs in the sequence 5′CGCGCCCAACG. A 100-fold molar excess of unlabeled competitor DNA was added to the mixtures shown in lanes 2–4 consisting of a methylated 210-bp probe fragment, an unmethylated 210-bp fragment, or an unrelated nonmethylated 220-bp fragment from the S100A4 gene, respectively. (B) Methylation interference analysis of the methyl–CpG-specific complex. Lanes “bound” correspond to DNA from the excised complex. “Naked” DNA was treated with DMS as a control. G residues that are protected in the complex are arrowed and marked correspondingly on the sequence shown below the panels. An apparently protected G on the noncoding strand (?) showed protection that was five times weaker than of the neighboring clearly protected residue (asterisk). (C) Cross-linking of a ∼100-kD protein to DNA. The methylated probe was BrdU-substituted, 32P-labeled, and subjected to EMSA. The complex was then cross-linked with UV light in situ and analyzed by electrophoresis by SDS-PAGE. The cross-linked complex (∼120 kD) and unbound DNA (∼20 kD) are arrowed. (D) Methylation specificity of the DNA–protein complex (arrow) formed by incubating CSML-0 nuclear extracts with methylated Sm probe in the presence of variably methylated nonlabeled Sm competitors (100-fold molar excess). The sequence of the DNA binding site is shown. The methylation status (+ or −) of each CpG in the competitor is shown above the panel. TG indicates replacement of CG by TG. A hemimethylated site is marked by +/−.
The dependence of binding on multiple methylated CpGs was first tested by partially methylating the probe using CpG modifying enzymes (Fig. 1A, lanes 5–7). Probes that were fully methylated by M.HhaI, M.FnuDII, or both enzymes failed to form the complex, indicating a requirement for multiple symmetrically methylated CpG sites. This conclusion was extended by EMSA experiments using a variably methylated 25-bp duplex containing the 5′CGCGCCCAACG core (probe “Sm”) as competitor. The normal complex (Fig. 1D, lane 1) was abolished by a 100-fold molar excess of fully CpG-methylated Sm (lane 2), but none of the competitors with fewer than three MGs (lanes 3–7), or with thymine in place of M (lanes 5 and 7), reduced Sm binding significantly. All three methyl–CpG pairs are evidently required, and their arrangement is important. Insertion of A between CpG1 and CpG2 (MGAMGCCCAAMG) in the Sm sequence abolished binding to the complex (data not shown), suggesting a stringent requirement for these methylated moieties to be adjacent. On the other hand, the distance between CpG2 and CpG3 could be changed from 5 nucleotides to 4 (MGMGCCAAMG) or 6 (MG MGCCCCAAMG or MGMGCCAAAAMG) nucleotides without reducing the affinity for the complex (data not shown).
Kaiso copurifies with the methyl–CpG binding activity
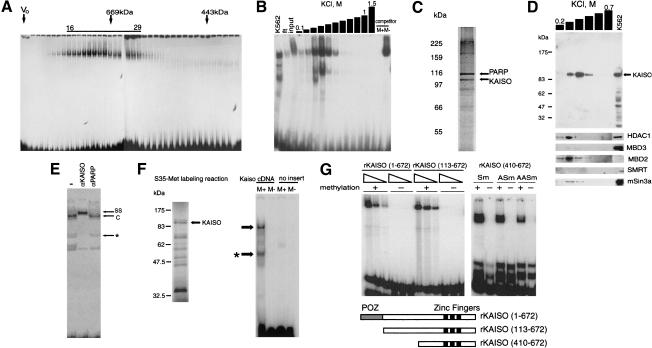
To enrich the methylation-dependent complex, nuclear extracts from K562 erythroleukemia cells were initially fractionated by size exclusion (Fig. 2A). The peak at ∼700 kD was pooled and further fractionated by affinity for immobilized methylated-Sm DNA (Fig. 2B). The peak eluting above 0.3 M KCl (Fig. 2B) gave two silver-stained bands close to the size of cross-linked proteins (∼100 kD) on SDS-polyacrylamide gels (Fig. 2C), and these were found by mass-spectrometric analysis to contain polyADP-ribose polymerase (PARP) and the BTB/POZ zinc finger protein Kaiso (GenBank no. AF097416). Subsequent analysis cast doubt on the involvement of PARP in the complex. In particular, an identical purification scheme using HeLa extracts, which lack the methyl–CpG-specific complex, again enriched for PARP, but Kaiso was absent (data not shown). PARP is a common contaminant in DNA affinity-purified protein fractions (Kadonaga 1996).
Figure 2.
Kaiso copurifies with the methyl–CpG binding activity. (A) EMSA of fractions from a S-300 Sephacryl gel filtration column using methylated Sm duplex as a probe. The void volume is marked (V0), and molecular weight standards are depicted by arrows. Fractions 16–29 were pooled. (B) EMSA with fractions eluted from the Sm DNA affinity column. Complexes detected with crude K562 nuclear extracts (K562), flow-through fractions from the column (ft), and pooled gel filtration fractions (input) are shown. The methyl–CpG-dependent complex formed with fractions eluting at ∼0.4 M KCl resisted competition by nonmethylated Sm competitor (M−), but was lost in the presence of methylated Sm (M+). (C) The fraction eluting at 0.4 M KCl was analyzed by denaturing SDS gels followed by silver staining. Bands that were characterized by mass-spectrometry are arrowed. (D) Cofractionation of Kaiso and methyl–CpG binding activities by Western blot analysis with anti-Kaiso monoclonal (6F) antibodies of DNA affinity column fractions from 0.2 to 0.7 M KCl. The presence of Kaiso in 30 μg of the starting K562 nuclear extract is shown. Below are shown blots of the same fractions reacted with antibodies to HDAC1, MBD3, MBD2, Smrt, and mSin3a. These proteins do not appear to cofractionate with the Kaiso-containing peak. (E) The methyl–CpG-specific complex (C) is supershifted (SS) by anti-Kaiso antibodies (αKaiso), but not by anti-polyADPribose polymerase polyclonal antibodies (αPARP). The faster migrating complex (*) was not reproducible, and could be due to a degradation product. (F) In vitro translated Kaiso exhibits methyl–CpG-specific DNA binding. Translation of pcDNA3.1-Kaiso (containing a Kaiso cDNA insert) generated apparently full-length Kaiso plus shorter products (left panel). Polyacrylamide EMSA with the products of in vitro transcription/translation of pcDNA3.1-Kaiso and pcDNA3.1 (no insert). Methyl–CpG-specific complexes are arrowed. Asterisk marks shifts presumably produced by truncated DNA binding competent parts of Kaiso. (G) Recombinant Kaiso binds to variants of the Sm probe in a methylation-dependent manner. (Left panel) EMSA using 1, 3, and 10 ng of full-length rKaiso(1–672) or rKaiso(113–672) gave a complex with the methylated probe (+) but not the nonmethylated probe (−). (Right panel) Methyl–CpG-dependent (lanes +) binding by the zinc-finger domain of Kaiso (410–672) is reduced by insertion of A residues between the CpG motifs 1 and 2 (see Fig. 1D; Table 1). No complex is detected in the presence of the nonmethylated probe (lanes −).
Several lines of evidence confirm that Kaiso, a protein of 70 kD that migrates with an apparent size of ∼100 kD on SDS-polyacrylamide gels (Daniel and Reynolds 1999), is a key component of the novel methyl–CpG binding complex. First, Western blots probed with three independent anti-Kaiso antibodies confirmed the presence of the 100-kD Kaiso band comigrating with DNA affinity fractions that formed the methylation-specific complex (e.g., Fig. 2D). Components of known corepressor complexes (HDAC1, MBD2, MBD3, SMRT, and mSin3a) did not coelute with the methyl–CpG-specific complex (Fig. 2D). Second, monoclonal antibodies against mouse Kaiso (12H; Daniel and Reynolds 1999) supershifted the methyl–CpG-specific EMSA complex (Fig. 2E), whereas antibodies against PARP, which was also identified in the DNA-affinity fractions, did not. Third, DNA binding studies showed that Kaiso is a methylation-specific DNA binding protein in its own right. Like the native complex, in vitro translated Kaiso showed strong specificity for methylated DNA (Fig. 2F). Moreover, recombinant GST-fusion protein containing intact Kaiso (amino acids 1–672), Kaiso minus the POZ domain (amino acids 113–672), or the zinc-finger domain alone (amino acids 410–672) absolutely required CpG methylation for DNA binding (Fig. 2G; Table 1). The DNA binding zinc-finger domain preferred two adjacent MGs in the probe, as one, two, or three intervening nucleotides reduced binding moderately (MGAMG) or severely (MGAAMG and MGAAAMG). The third MG of the Sm probe had a minimal effect on binding of recombinant Kaiso, as probes lacking this modification bound efficiently (e.g., Table 1, probe TG3Sm). Thus, Kaiso shows methyl–CpG-dependent binding via its zinc-finger domain with a DNA sequence specificity that is somewhat relaxed compared to the endogenous complex in crude extracts. Relaxed specificity may be due to competition for binding with other methyl–CpG binding activities present in nuclear extracts, or to the presence of Kaiso-associated factors that modulate its DNA binding specificity. The dissociation constant of the recombinant Kaiso–Sm probe complex was measured in EMSA assays using full-length Kaiso as ∼1 nanomolar and ∼1 micromolar for the methylated and nonmethylated sites, respectively (data not shown). For comparison, MBD binding to a single methylated CpG falls within the 10 nanomolar range (Free et al. 2001). Thus, Kaiso exhibits the highest reported affinity for DNA methylated at CpG. Its binding affinity is increased ∼1000-fold by CpG methylation.
Table 1.
Binding of recombinant zinc fingers of Kaiso to methylated DNA
| Probe
|
Sequence
|
Binding
|
||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Sm | C | A | G | C | A | G | C | M | G | M | G | C | C | C | A | A | M | G | C | T | G | G | G | A | ++++ | |||
| Sm-A | C | A | G | C | A | G | C | M | G | A | M | G | C | C | C | A | A | M | G | C | T | G | G | G | A | +++ | ||
| Sm-AA | C | A | G | C | A | G | C | M | G | A | A | M | G | C | C | C | A | A | M | G | C | T | G | G | G | A | ++ | |
| VIRTUAL | C | A | G | C | A | G | C | M | G | A | A | A | M | G | A | A | A | M | G | C | T | G | G | G | A | + | ||
| Asm | C | A | G | C | A | G | C | M | G | M | G | C | C | C | A | A | A | M | G | C | T | G | G | G | A | +++ | ||
| AASm | C | A | G | C | A | G | C | M | G | M | G | C | C | C | A | A | A | A | A | M | G | C | T | G | G | G | A | +++ |
| CCSm | C | A | G | C | A | G | C | M | G | M | G | C | A | A | M | G | C | T | G | G | G | A | +++ | |||||
| CG3Sm | C | A | G | C | A | G | C | M | G | M | G | C | C | C | A | A | C | T | G | G | G | A | +++ | |||||
| 1A2Sm | C | A | G | C | A | G | C | M | G | A | M | G | C | C | C | A | A | C | T | G | G | G | A | +++ | ||||
| 2A2Sm | C | A | G | C | A | G | C | M | G | A | A | M | G | C | C | C | A | A | C | T | G | G | G | A | + | |||
| Sm-TG | C | A | G | C | A | G | C | T | G | T | G | C | C | C | A | A | T | G | C | T | G | G | G | A | − | |||
| AACG-Sm | C | A | G | C | A | G | C | A | A | M | G | C | C | C | A | A | C | T | G | G | G | A | +/− | |||||
| TGCG-Sm | C | A | G | C | A | G | C | T | G | M | G | C | C | C | A | A | C | T | G | G | G | A | + | |||||
| CGTG-Sm | C | A | G | C | A | G | C | M | G | T | G | C | C | C | A | A | C | T | G | G | G | A | − | |||||
| CGCA-Sm | C | A | G | C | A | G | C | M | G | C | A | C | C | C | A | A | C | T | G | G | G | A | − | |||||
| CACG-Sm | C | A | G | C | A | G | C | C | A | M | G | C | C | C | A | A | C | T | G | G | G | A | − | |||||
A motif based on C2H2 zinc fingers (three Kaiso zinc fingers belong to C2H2 class) that is capable of binding preferentially to a methylated site (GMGC) was selected previously by phage display (Choo 1998). Although Kaiso's zinc fingers belong to the C2H2 class, the amino acid sequence of this artificial protein does not resemble Kaiso. Two studies have detected natural complexes with a binding specificity similar to Kaiso. A methyl–CpG-dependent DNA binding complex was identified via its affinity for a domain of the methylated Xist promoter (Huntriss et al. 1997). The complex required a motif containing the sequence MGMGGMG for full binding, and cross-linking studies detected a 100-kD protein close to the size of Kaiso. A methyl–CpG-dependent complex formed on the sequence MGMGGGAAAAA MG from the promoter of the retinoblastoma gene (Rb) (De Fiore et al. 1999), has subsequently been shown by EMSA supershifts to include Kaiso (A. Prokhortchouk, unpubl.). The common feature of both Xist and Rb promoters is the requirement for MGMG motifs, but the distance between MG2 and MG3 is one nucleotide and seven nucleotides, respectively. These data suggest further flexibility in this aspect of the DNA recognition sequence. There is as yet no evidence that any of the five gene fragments used to detect Kaiso binding in this and previous studies (S100A4, Rb, Xist; E-cadherin, and Pgk, see below) is an in vivo target.
Kaiso represses transcription in vivo in a methyl–CpG-dependent manner
Several BTB/POZ zinc-finger proteins are transcriptional repressors (Huynh and Bardwell 1998). We asked whether Kaiso acts as a DNA methylation-dependent transcriptional repressor by assaying its effects when coexpressed in mammalian cells with methylated or nonmethylated reporter genes (Fig. 3A). To avoid repression of the methylated reporter due to endogenous MeCP1, we used Mbd2(−/−) cells, which are defective in repression of a transfected methylated gene (Hendrich et al. 2001). One methylated reporter construct contained the luciferase gene under the control of an SV40 promoter, and contained 11 dispersed Kaiso consensus binding sites. As expected, transcription of a fully CpG-methylated reporter gene in Mbd2−/− cells was 30–35% of nonmethylated control transcription. Expression of intact Kaiso, however, reduced transcription to 5–7% of nonmethylated controls. Enhanced repression of the methylated reporter was not achieved by expression of truncated Kaiso lacking the POZ domain (KaisoΔPOZ) or zinc fingers (KaisoΔZF) or both (KaisoΔPOZ+ZF). Therefore, Kaiso can restore DNA methylation-dependent repression to silencing-defective Mbd2−/− cells, and this effect requires both POZ and zinc-finger domains. To test the effect of Kaiso on a naturally methylated promoter, we chose the mouse phosphoglycerate kinase (Pgk) gene promoter, which is naturally silenced and methylated upon X chromosome inactivation (Singer-Sam et al. 1990). The promoter contains consensus Kaiso binding sites, and binds Kaiso in vitro in a methylation-dependent manner (see below; Fig. 3D). Coexpression of Kaiso markedly decreased transcription from a methylated Pgk promoter in Mbd2−/− cells from 7% of nonmethylated control to 2% (Fig. 3A). Comparable restoration of methyl–CpG-specific transcriptional repression of SV40 or Pgk promoters has been shown for MBD2 and MeCP2 when overexpressed in this system (Hendrich et al. 2001; Guy et al. 2001). Transcriptional silencing by Kaiso was not diminished in the presence of the histone deacetylase inhibitor TSA (data not shown). MBD2-mediated repression has been shown to be sensitive to the presence of TSA in some, but not all, instances (Ng et al. 1999). Further work is, therefore, required to test for involvement of histone deacetylation in Kaiso-mediated repression.
Figure 3.
Kaiso represses transcription in vivo in a methyl–CpG-dependent manner, and is a component of the MeCP1 doublet complex. (A) Methyl–CpG-dependent repression by Kaiso and its truncated derivatives (diagram, top) in a transient transfection assay. Kaiso expression constructs were cotransfected with an SV40-luciferase reporter into mouse cells that are compromised in methyl–CpG-dependent transcriptional repression (Mbd2−/−; shaded bars) or into their normal counterparts (Mbd2+/−, unshaded bars). Relative reporter activity is presented as a percentage (methylated reporter expression/nonmethylated reporter expression × 100). Results are the average of at least three experiments. (B) Kaiso is responsible for the faster migrating component of the MeCP1 complex that is seen in many cell types. Complexes between HeLa and NIH3T3 nuclear extracts and the methylated (M+CG11) or nonmethylated (CG11) probe were detected by EMSA using agarose gels. Increasing amounts (up to 1 μL) of ZFH6 anti-Kaiso antibodies (αKaiso) or preimmune serum (P.I.) were added to determine which complexes were supershifted. The KGB complex is arrowed. (C) Kaiso is responsible for formation of a methyl–CpG-specific complex on a fragment of the E-cadherin promoter. Nuclear extracts from NIH3T3 cells and either methylated (M+Ecad) or nonmethylated Ecad probe were used in EMSA using agarose gels. Increasing amounts (up to 1 μL) of ZFH6 anti-Kaiso antibodies (αKaiso) or anti-MBD2 (αMBD2) antibodies were added to determine which complexes were supershifted. The KGB complex is arrowed. (D) Kaiso forms a methyl–CpG-specific complex on a fragment of the Pgk (−430; +74 bp from transcriptional start of the gene) promoter. Nuclear extracts from NIH3T3 cells and either methylated (M+Pgk) or nonmethylated (Pgk) probes were used in EMSA. The slight supershift on lane 5 is nonspecific due to the high concentration of rabbit serum in this lane. (E) Kaiso comigrates with a 700-kD protein complex on a Superose 6 column as assayed by EMSA in an agarose gel. Molecular weight markers are arrowed. The same fractions (pooled in pairs) were analyzed by Western blot hybridization with anti-Kaiso and anti-MBD2 antibodies. Input lanes represent mouse liver nuclear extract. The anti-Kaiso cross-reacting band seen in fractions 25–31 has a different electrophoretic mobility from Kaiso.
Kaiso is a component of the MeCP1 doublet complex
The methyl–CpG binding activity MeCP1 has been detected in several mammalian cell types as a doublet of DNA–protein bands on agarose gels (Meehan et al. 1989). HeLa cells, however, invariably displayed one MeCP1 complex on agarose gels (Boyes and Bird 1991; Ng et al. 1999). As HeLa cells also lack the methyl–CpG-specific complex detected in this study (data not shown; Fig. 3B), we asked whether the missing MeCP1 complex might include Kaiso. EMSA on agarose gels using the MeCG11 probe (Meehan et al. 1989), which contains four copies of a consensus Kaiso complex binding site (MGMGN5MG), confirmed that NIH3T3 fibroblast cell extracts gave two major DNA–protein complexes on agarose gels, whereas HeLa extracts gave one complex (Fig. 3B). Addition of ZFH6 anti-Kaiso antibodies led to loss of the lower NIH3T3 complex. Neither the upper NIH3T3 complex nor the sole HeLa complex was affected by the Kaiso antibody (Fig. 3B).
The Kaiso-containing complex also interacted in vitro with the natural E-cadherin CpG island promoter (Fig. 3C), which is often silenced by methylation in malignant epithelial cancers (Hirohashi 1998; Graff et al. 2000). The methylated probe (199 bp) contains a consensus binding site for the Kaiso complex (MGMGTCTAGMG) ∼150 bp upstream of the transcription start site, and formed a slow-migrating complex that could be efficiently supershifted by anti-Kaiso antibodies, but not anti-MBD2 antibodies (Fig. 3C). No equivalent complex formed with the nonmethylated probe. Two faster migrating, methylation-independent complexes were not affected by either antibody. The naturally methylated Pgk promoter also gave a methylation-dependent complex whose migration was retarded by low concentrations of anti-Kaiso antibodies but not by preimmune serum (Fig. 3D).
Size-exclusion chromatography of a mouse liver nuclear extract confirmed Kaiso as a component of the faster migrating complex formed on methylated CG11 DNA. The prominent methyl–CpG-dependent complex chromatographed with an apparent molecular weight of ∼700 kD (Fig. 3E, fractions 20–25; compare polyacrylamide gel EMSA, Fig. 2A), colocalized with the peak of Kaiso on Western blots (Fig. 3E), and was supershifted by anti-Kaiso antibodies (data not shown). MBD2, which is a key component of HeLa MeCP1 (Ng et al. 1999; Feng and Zhang. 2001), did not coincide with the Kaiso-containing complex (Fig. 3E; see also Fig. 2D), and did not supershift the EMSA complex (Fig. 3C), indicating that it is not part of the Kaiso-containing complex. The data distinguish a Kaiso complex or KGB (for Kaiso Generated Band) from the MBD2-containing MeCP1 complex seen in HeLa cells. This view is supported by the finding that Mbd2−/− mouse cells lack one component of the MeCP1 complex, but retain other components, including KGB (Hendrich et al. 2001).
Does Kaiso receive signals from the cytoplasm?
Our work identifies Kaiso as a highly specific methylated DNA binding protein within the KGB complex that binds to DNA containing the core recognition sequence MGMG. Kaiso was originally identified as a binding partner of p120 catenin (Daniel and Reynolds 1999), a protein that also associates with the cytoplasmic domain of the cell surface protein E-cadherin (for review, see Anastasiadis and Reynolds 2000). Recent studies suggest that p120 catenin signals between cell–cell junctions and the cell motility machinery of the cytoplasm (Anastasiadis et al. 2000), but there is also evidence for its presence in the nucleus (van Hengel et al. 1999), where Kaiso is found (Daniel and Reynolds 1999). A speculative possibility is that E-cadherin, p120 catenin, and Kaiso are components of a signal transduction pathway that transmits signals from the cell surface to the nucleus, leading to the shutdown of specific methylated genes. A pathway of this kind would have important implications for the study of development and carcinogenesis.
Materials and methods
Plasmids and primers
The 210-bp probe from intron 1 of the murine S100A4 gene (Prokhortchouk et al. 1998) and E-cadherin promoter (−169; +8 bp from transcriptional start) fragment were amplified by PCR. The pGL2-Pgk plasmid contains mouse Pgk gene promoter (−430; +74 bp from transcriptional start) in the pGL2-Basic vector. The CG11 and MeCG11 probes were prepared and labeled as described (Meehan et al. 1989). Full-length human Kaiso cDNA (2–672 aa), POZ-deleted (113–672 aa), zinc fingers-deleted (2–490 aa), and both POZ- and zinc fingers-deleted (113–490) were into the pFLAG-CMV2 (Kodak) mammalian expression vector at NotI–EcoRI sites. Plasmid pcDNA3.1-Kaiso contained mouse Kaiso cDNA in the pcDNA3.1 (Invitrogen) vector.
Repression test
Mbd2−/− and Mbd2+/− cells were transfected using Lipofectamine according to manufacturer's instructions (Life Technologies). Each well of a six-well plate was transfected with 2 μg of either M.SssI methylated or unmethylated pGL2-Promoter plasmid and 50 ng of the pRL-SV40 control plasmid (Promega), and 100 ng of a CMV–Kaiso expression construct. Luciferase levels were measured after approximately 40 h using the Dual Luciferase Assay Kit (Promega). Sample values were obtained according to the following formula: (luciferase sample − luciferase control)/(renilla sample − renilla control) where control values were obtained from untransfected cells. Relative luciferase values are defined as the sample value obtained using a methylated pGL-Promoter plasmid divided by the sample value obtained using the unmethylated pGL-Promoter plasmid.
Chromatography
For purification of the methyl–CpG binding activity, nuclear extracts (300 mg protein; Dignam et al. 1983) from K562 cells were fractionated on a S-300 Sephacryl (Amersham-Pharmacia) column (XK 50/60) followed by DNA-affinity chromatography (Kadonaga 1996).
For analytical gel filtration 1.2 mg of mouse liver nuclear protein was fractionated on a Superose 6 column.
Recombinant proteins
A region of cDNA that encoded the Kaiso zinc fingers (amino acids 481–672) was cloned into the pQE30 vector. Recombinant protein was used for production of ZFH6 antibodies in rabbits. To obtain GST fusion proteins, we cloned either full-length Kaiso cDNA PCR product (1–672 aa) or fragments encoding amino acids 113–672 and 410–672 into the pGEX-2T vector. We synthesized Kaiso in vitro using an Ambion PROTEINscript-PRO T7 transcription-translation coupled kit on the template of pcDNA3.1-Kaiso plasmid.
Acknowledgments
We thank Dr. Albert Reynolds for providing anti-Kaiso monoclonal antibodies and a Kaiso plasmid, and M.D. Gusev for the help with antibody production. This work was supported by grant RB1-2041 from CRDF, and by a Progamme Grant from the Wellcome Trust. E.P. is presently supported by an EMBO Fellowship.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL egorbp@holyrood.ed.ac.uk; FAX 44-131-650-5379.
Article and publication are at http://www.genesdev.org/cgi/doi/10.1101/gad.198501.
References
- Anastasiadis PZ, Reynolds AB. The p120 catenin family: Complex roles in adhesion, signaling and cancer. J Cell Sci. 2000;113:1319–1334. doi: 10.1242/jcs.113.8.1319. [DOI] [PubMed] [Google Scholar]
- Anastasiadis PZ, Moon SY, Thoreson MA, Mariner DJ, Crawford DJ, Zheng Y, Reynolds AB. Inhibition of RhoA by p120 catenin. Nat Cell Biol. 2000;2:637–644. doi: 10.1038/35023588. [DOI] [PubMed] [Google Scholar]
- Boyes J, Bird A. DNA methylation inhibits transcription indirectly via a methyl–CpG binding protein. Cell. 1991;64:1123–1134. doi: 10.1016/0092-8674(91)90267-3. [DOI] [PubMed] [Google Scholar]
- Choo Y. Recognition of DNA methylation by zinc fingers. Nat Struct Biol. 1998;5:264–265. doi: 10.1038/nsb0498-264. [DOI] [PubMed] [Google Scholar]
- Collins T, Stone JR, Williams AJ. All in the family: The BTB/POZ, KRAB, and SCAN domains. Mol Cell Biol. 2001;21:3609–3615. doi: 10.1128/MCB.21.11.3609-3615.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniel JM, Reynolds AB. The catenin p120(ctn) interacts with Kaiso, a novel BTB/POZ domain zinc finger transcription factor. Mol Cell Biol. 1999;19:3614–3623. doi: 10.1128/mcb.19.5.3614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Fiore B, Palena A, Felsani A, Palitti F, Caruso M, Lavia P. Cytosine methylation transforms an E2F site in the retinoblastoma gene promotor into a binding site for the general repressor methylcytosine-binding protein (MeCP2) Nucleic Acids Res. 1999;27:2852–2859. doi: 10.1093/nar/27.14.2852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dignam JD, Lebovitz RM, Roeder RG. Accurate transcription initiation by RNA polymerase II in a soluble extract from isolated mammalian nuclei. Nucleic Acids Res. 1983;5:1475–1489. doi: 10.1093/nar/11.5.1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng Q, Zhang Y. The MeCP1 complex represses transcription through preferential binding, remodeling, and deacetylating methylated nucleosomes. Genes & Dev. 2001;15:837–832. doi: 10.1101/gad.876201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Free A, Wakefield RID, Smith BO, Dryden DTF, Barlow PN, Bird A. DNA recognition by the methyl–CpG binding domain of MeCP2. J Biol Chem. 2001;276:3353–3360. doi: 10.1074/jbc.M007224200. [DOI] [PubMed] [Google Scholar]
- Graff JR, Gabrielson E, Fujii H, Baylin SB, Herman JG. Methylation patterns of the E-cadherin expression. J Biol Chem. 2000;275:2727–2732. doi: 10.1074/jbc.275.4.2727. [DOI] [PubMed] [Google Scholar]
- Guy J, Hendrich B, Holmes M, Martin J, Bird A. A mouse Mecp2-null mutation causes neurological symptoms that mimic Rett syndrome. Nat Genet. 2001;27:322–326. doi: 10.1038/85899. [DOI] [PubMed] [Google Scholar]
- Hendrich B, Bird A. Identification and characterization of a family of mammalian methyl–CpG binding proteins. Mol Cell Biol. 1998;18:6538–6547. doi: 10.1128/mcb.18.11.6538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendrich B, Guy J, Ramsahoye B, Wilson V, Bird A. Closely related proteins MBD2 and MBD3 play distinctive but interacting roles in mouse development. Genes & Dev. 2001;15:710–723. doi: 10.1101/gad.194101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirohashi S. Inactivation of the E-cadherin-mediated cell adhesion system in human cancers. Am J Pathol. 1998;153:333–339. doi: 10.1016/S0002-9440(10)65575-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huntriss J, Lorenzi R, Purewal A, Monk M. A methylation-dependent DNA-binding activity recognising the methylated promoter region of the mouse Xist gene. Biochem Biophys Res Commun. 1997;235:730–738. doi: 10.1006/bbrc.1997.6876. [DOI] [PubMed] [Google Scholar]
- Huynh KD, Bardwell VJ. The BCL-6 POZ domain and other POZ domains interact with the co-repressors N-CoR and SMRT. Oncogene. 1998;17:2473–2484. doi: 10.1038/sj.onc.1202197. [DOI] [PubMed] [Google Scholar]
- Jackson-Grusby L, Beard C, Possemat R, Tudor M, Fambrough D, Csankovszki G, Dausman J, Lee P, Wilson C, Lander E, et al. Loss of genomic methylation causes p53-dependent apoptosis and epigenetic deregulation. Nat Genet. 2001;27:31–39. doi: 10.1038/83730. [DOI] [PubMed] [Google Scholar]
- Jones PL, Veenstra GJC, Wade PA, Vermaak D, Kass SU, Landsberger N, Strouboulis J, Wolffe AP. Methylated DNA and MeCP2 recruit histone deacetylase to repress transcription. Nat Genet. 1998;19:187–191. doi: 10.1038/561. [DOI] [PubMed] [Google Scholar]
- Kadonaga JT. Purification of transcription factor AP-1 from HeLa cells. In: Marshak D, et al., editors. Strategies for protein purification and characterization. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1996. pp. 127–204. [Google Scholar]
- Meehan RR, Lewis JD, McKay S, Kleiner EL, Bird AP. Identification of a mammalian protein that binds specifically to DNA containing methylated CpGs. Cell. 1989;58:499–507. doi: 10.1016/0092-8674(89)90430-3. [DOI] [PubMed] [Google Scholar]
- Nan X, Meehan RR, Bird A. Dissection of the methyl–CpG binding domain from the chromosomal protein MeCP2. Nucleic Acids Res. 1993;21:4886–4892. doi: 10.1093/nar/21.21.4886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nan X, Ng H-H, Johnson CA, Laherty CD, Turner BM, Eisenman RN, Bird A. Transcriptional repression by the methyl–CpG-binding protein MeCP2 involves a histone deacetylase complex. Nature. 1998;393:386–389. doi: 10.1038/30764. [DOI] [PubMed] [Google Scholar]
- Nan X, Tate P, Li E, Bird AP. DNA methylation specifies chromosomal localization of MeCP2. Mol Cell Biol. 1996;16:414–421. doi: 10.1128/mcb.16.1.414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng H-H, Zhang Y, Hendrich B, Johnson CA, Burner BM, Erdjument-Bromage H, Tempst P, Reinberg D, Bird A. MBD2 is a transcriptional repressor belonging to the MeCP1 histone deacetylase complex. Nat Genet. 1999;23:58–61. doi: 10.1038/12659. [DOI] [PubMed] [Google Scholar]
- Ng H-H, Jeppesen P, Bird A. Active repression of methylated genes by the chromosomal protein MBD1. Mol Cell Biol. 2000;20:1394–1406. doi: 10.1128/mcb.20.4.1394-1406.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prokhortchouk E, Prokhortchouk A, Rouzov A, Kiselev S, Lukanidin E, Georgiev G. A minisatellite “core” element constitutes a novel, chromatin-specific activator of mts1 gene transcription. J Mol Biol. 1998;280:227–236. doi: 10.1006/jmbi.1998.1857. [DOI] [PubMed] [Google Scholar]
- Singer-Sam J, Grant M, LeBon JM, Okuyama K, Chapman V, Monk M, Riggs AD. Use of a HpaII-polymerase chain reaction assay to study DNA methylation in the Pgk-1 CpG island of mouse embryos at the time of X-chromosome inactivation. Mol Cell Biol. 1990;10:4987–4989. doi: 10.1128/mcb.10.9.4987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stancheva I, Meehan RR. Transient depletion of xDnmt1 leads to premature gene activation in Xenopus embryos. Genes & Dev. 2000;14:313–327. [PMC free article] [PubMed] [Google Scholar] [Retracted]
- van Hengel J, Vanhoenacker P, Staes K, van Roy F. Nuclear localization of the p120(ctn) Armadillo-like catenin is counteracted by a nuclear export signal and by E-cadherin expression. Proc Natl Acad Sci. 1999;96:7980–7985. doi: 10.1073/pnas.96.14.7980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wade PA, Gegonne A, Jones PL, Ballestar E, Aubry F, Wolffe AP. Mi-2 complex couples DNA methylation to chromatin remodelling and histone deacetylation. Nat Genet. 1999;23:62–66. doi: 10.1038/12664. [DOI] [PubMed] [Google Scholar]
- Walsh CP, Bestor TH. Cytosine methylation and mammalian development. Genes & Dev. 1999;13:26–34. doi: 10.1101/gad.13.1.26. [DOI] [PMC free article] [PubMed] [Google Scholar]