Abstract
Capping, splicing, and cleavage/polyadenylation of pre-mRNAs are interdependent events that are all stimulated in vivo by the carboxy-terminal domain (CTD) of RNA Pol II. We show that the CTD independently enhances splicing and 3′ processing and that stimulation of splicing by enhancers is facilitated by the CTD. We provide evidence that stimulation of 3′ processing by the CTD requires contact with the 50-kD subunit of the cleavage stimulation factor, CstF. Overexpression of the CTD-binding domain of CstF p50 had a dominant-negative effect on 3′ processing without disrupting the CstF complex. The CTD comprises 52 heptad repeats. The CTD carboxyl terminus including heptads 27–52 supported capping, splicing, and 3′ processing but the amino terminus supported only capping. We conclude that the CTD independently stimulates all three major pre-mRNA processing steps and that different regions of the CTD can serve distinct functions in pre-mRNA processing.
Keywords: mRNA processing, RNA polymerase II, CstF, carboxy-terminal domain, splicing
Messenger RNA production requires synthesis of a pre-mRNA by RNA Pol II and processing of the nascent precursor by 5′ capping, splicing of introns, and 3′ cleavage/polyadenylation to make mature mRNA. In vivo, mRNA processing occurs cotranscriptionally (Beyer and Osheim 1988; Bauren et al. 1998) and is directed to RNAs made by Pol II and not other RNA polymerases (Smale and Tjian 1985; Gunnery and Mathews 1995; McCracken et al. 1998). The details of how processing is specifically coupled to Pol II transcription are poorly understood; however, it is known that the carboxy-terminal domain (CTD) of the Pol II large subunit is required for all three major mRNA processing steps in vivo (McCracken et al. 1997a,b). The CTD is a unique feature of RNA Pol II that is essential for cell viability and is thought to function as a landing pad for Pol II holoenzyme subunits and RNA processing factors (Bentley 1999; Hirose and Manley 2000; Lee and Young 2000). The mammalian Pol II CTD contains 52 tandem heptads whose consensus, YSPTSPS, is absolutely conserved among most eukaryotes. Carboxy-terminal of the last heptad are 10 residues that are not essential (Bartolomei et al. 1988) and are less well conserved than the heptad repeats. The serines at positions 2 and 5 of the heptads are targets for phosphorylation (Dahmus 1996) and dephosphorylation (Komarnitsky et al. 2000; Schroeder et al. 2000) during transcription. Eighteen of the amino-terminal 26 heptads in the human and mouse CTDs are identical to the consensus whereas only three of the carboxy-terminal 26 heptads conform to this sequence. A deletion of heptads 23–52 is lethal (Bartolomei et al. 1988) whereas a deletion of heptads 23–36 is viable (Litingtung et al. 1999). Most carboxy-terminal heptads diverge from the consensus at position 7. The significance of this sequence variation is not fully understood; however heptads with Lys at position 7 are preferred over consensus heptads for phosphorylation of Ser 5 by the TFIIH-associated kinase CDK7 (Rickert et al. 1999).
Precisely how the CTD contributes to efficient processing of pre-mRNAs remains a major unanswered question. Because capping, splicing, and cleavage/polyadenylation are interdependent, it is not obvious which effects of the CTD on processing are direct and which are indirect. The 5′ cap can enhance splicing of the first intron (Inoue et al. 1989; Lewis et al. 1996) as well as 3′ processing (Hart et al. 1985; Cooke and Alwine 1996; Flaherty et al. 1997). 3′ Processing depends on splicing of the last intron (Niwa et al. 1990; Bauren et al. 1998; Dye and Proudfoot 1999), and conversely splicing of the last intron depends on recognition of the poly(A) site, which defines the 3′ end of the last exon (Niwa and Berget 1991; Bauren et al. 1998; Vagner et al. 2000).
The strongest evidence for a direct role of the CTD in a processing step is for capping, which occurs when the nascent RNA is about 25 bases long (Coppola et al. 1983; Rasmussen and Lis 1993). Capping enzymes bind to the phosphorylated CTD in vitro (Cho et al. 1997; McCracken et al. 1997a; Yue et al. 1997), and CTD phosphorylation is required for recruitment of capping enzymes to sites of transcription in vivo (Komarnitsky et al. 2000; Schroeder et al. 2000). The mammalian guanylyltransferase recognizes and is activated by as few as two phosphorylated heptads (Ho and Shuman 1999).
A direct role of the CTD in 3′ processing is suggested by the fact that the CTD can stimulate the cleavage reaction in vitro in the absence of ongoing transcription (Hirose and Manley 1998). Moreover, yeast polyadenylation factors Pcf11 and Pta1 (Barilla et al. 2001; Rodriguez et al. 2000) as well as mammalian cleavage/polyadenylation specificity factor (CPSF) and cleavage stimulation factor (CstF; McCracken et al. 1997b) bind to CTD affinity resins. CPSF and CstF also copurify with HeLa Pol II holoenzyme and are displaced from it by anti-CTD monoclonal antibody (Yankulov et al. 1999). CstF is composed of three subunits p77, p64, and p50, held together by interactions between p77 and the other two subunits (Takagaki and Manley 2000). When translated in rabbit reticulocyte lysate, the p50, but not the p64 or p77, subunits bound to the CTD (McCracken et al. 1997b). These studies with crude lysates do not address whether the binding of polyadenylation factors to the CTD is direct or indirect. Nor is it known whether protein–protein interactions between 3′ processing factors and the CTD are actually required for efficient cleavage/polyadenylation in vivo or in vitro.
The role of the CTD in splicing is least well understood of the three major pre-mRNA processing events. Hyperphosphorylated Pol II coimmunoprecipitates with splicing factors including SR proteins as originally postulated by Greenleaf (1993), and the CTD is required for recruitment of these factors to sites of transcription in the nucleus (Mortillaro et al. 1996; Vincent et al. 1996; Yuryev et al. 1996; Kim et al. 1997; Misteli and Spector 1999). No direct interaction between a mammalian splicing factor and the CTD has yet been documented, however. A variety of effects have been reported when the CTD is added to in vitro splicing reactions that are not coupled to transcription. Short CTD peptides inhibited splicing (Yuryev et al. 1996), whereas full-length recombinant CTD either had no effect (Hirose et al. 1999) or specifically stimulated splicing of substrates that permit exon definition (Zeng and Berget 2000). Exon definition involves bridging interactions across exons that are mediated in part by SR proteins binding to exonic splicing enhancer elements (Blencowe 2000). Interestingly, intact Pol II with phosphorylated CTD could substitute for the SR protein ASF/SF2 during in vitro splicing of HIV-1 tat intron 2 (Hirose et al. 1999). These in vitro studies suggest that the CTD could be directly involved in splicing; however, the question of whether it influences splicing independently of other processing events in vivo remains open.
Here, we document the protein–protein interaction between the amino terminus of CstF p50 and the carboxy-terminal variant heptads of the CTD and provide evidence that this interaction is important for pre-mRNA 3′ processing in vivo. We also demonstrate that the CTD independently stimulates each of the three major pre-mRNA processing events in vivo. In addition, we report that the amino terminus of the CTD can support capping without efficient splicing or 3′ processing whereas the carboxyl terminus supports all of the pre-mRNA processing steps. Therefore, different segments of the CTD can fulfill different functions in enhancing pre-mRNA processing.
Results
The CTD is required for splicing independent of its effect on 3′ processing
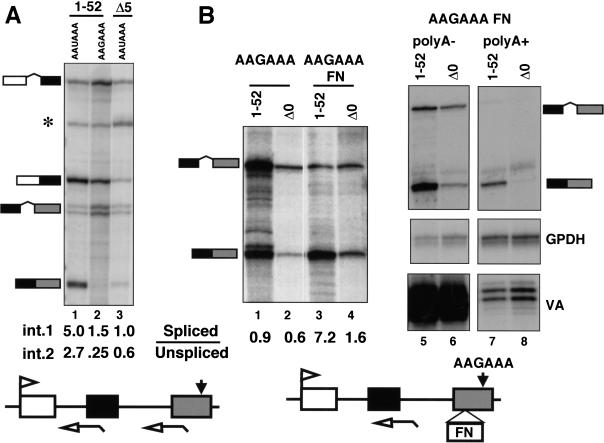
It was shown previously that transcripts made by Pol II with only five amino-terminal heptad repeats are defective in splicing of a 3′ intron (McCracken et al. 1997b). If the primary effect of CTD truncation were to inhibit 3′ processing, it could reduce splicing of adjacent introns indirectly. To test this possibility, we compared the effect of mutating the poly(A) site with the effect of deleting the CTD on splicing of β-globin transcripts. Reporters containing either the wild-type (AATAAA) or mutant (AAGAAA) poly(A) sites were transfected into 293 cells along with expression vectors for α-amanitin-resistant full-length (1–52) or truncated Pol II large subunit with only five amino-terminal heptad repeats (Δ5; Gerber et al. 1995). Approximately 16 h after transfection, α-amanitin was added to inhibit endogenous Pol II; mRNA made after this time is synthesized by Pol II that has incorporated the resistant large subunit. RNA harvested after 36–48 h of α-amanitin treatment was assayed by RNase protection with antisense probes that span the 3′ splice sites of introns 1 and 2 (Fig. 1A). The protection products were quantified by PhosphorImager and corrected for [32P]uridine content. Mutation of the poly(A) site did, in fact, inhibit splicing of both introns 1 and 2 when the gene was transcribed by full-length Pol II. The ratios of spliced to unspliced transcripts for introns 1 and 2 were 5.0 and 2.7, respectively, for AAUAAA and 1.5 and 0.25 for the AAGAAA mutant (Fig. 1A, lanes 1,2). Splicing of intron 2 was inhibited more than intron 1 consistent with in vitro results (Niwa and Berget 1991). The spliced to unspliced ratios for introns 1 and 2 with the Δ5 CTD truncation were 1.0 and 0.6, respectively, compared with 5.0 and 2.7 for full-length Pol II (Fig. 1A, lanes 1,3). Although absolute values of processed to unprocessed RNAs at a particular intron or poly(A) site varied between experiments (Fig. 1A, lane 2; Fig. 1B, lane 1), the differences between samples in a given experiment were reproducible. We conclude that mutation of the poly(A) site inhibited splicing in a way that resembles the effect of truncating the Pol II CTD. Therefore, it is important to establish whether CTD truncation affects splicing independently of 3′ processing.
Figure 1.
The CTD is required for enhancer-dependent splicing independent of 3′ processing. (A) Mutation of the poly(A) site and deletion of the CTD inhibit splicing of β-globin introns 1 and 2. RNase protection of RNA from α-amanitin-treated 293 cells transfected with pSVβ128 AAUAAA or pSVβ128 AAGAAA β-globin with wild-type or mutant poly(A) sites (closed arrow) and expression vectors for α-amanitin-resistant full-length, 1–52 [HA-WT, (Gerber et al. 1995)] or CTD-truncated large subunit with 5 heptads (HA-Δ5). (*) Undigested probe; (open arrows) RNase protection probes. Ratios of spliced to unspliced transcripts for introns 1 and 2 were calculated from PhosphorImager data corrected for the [32P]uridine content of the protected fragments. (B) CTD-dependent splicing driven by the FN EDI enhancer. (Lanes 1–4) RNase protection of pSVβ128 AAGAAA or pSVβ128 AAGAAA-FN transcripts made by α-amanitin-resistant Pol II (1–52) or Δ0. Spliced to unspliced ratios were calculated as in A. (Lanes 5–8) poly(A)+ and poly(A)− RNAs analyzed with β-globin intron2, GPDH and VA probes. Note that most spliced β-globin transcripts are poly(A)−.
To study splicing independently of 3′ processing, we tested whether efficient splicing of β-globin intron 2 could be restored to the AAGAAA mutant by inserting a splicing enhancer element into exon 3. The 73-base enhancer element from the alternatively spliced fibronectin exon ED I (FN EDI; Lavigueur et al. 1993) was inserted in the forward orientation into exon 3 of the AAGAAA mutant. The AAGAAA FN construct was cotransfected into 293 cells with the adenovirus VA gene and cDNA expression vectors (Nguyen et al. 1996) for α-amanitin resistant full-length (1–52) or CTD-deleted (Δ0) large subunit. Complete deletion of the CTD has the same effect as the truncation with five heptads used previously. The FN EDI enhancer in the forward orientation but not in the reverse orientation (data not shown) restored efficient splicing of intron 2 in the AAGAAA mutant when it was transcribed by full-length Pol II regardless of whether or not it was α-amanitin resistant (Fig. 1B, lanes 1,3; data not shown). Spliced β-globin RNA was predominantly in the poly(A)− fraction (Fig. 1B, cf. lanes 5 and 7) whereas endogenous GPDH mRNA was predominantly poly(A)+ and Pol III VA transcripts were poly(A)− (Fig. 1B, lanes 5–8). The small amount of spliced β-globin RNA in the poly(A)+ fraction (lane 7) was presumably processed at a cryptic poly(A) site. These results show that the terminal exon definition signal that is normally provided by a poly(A) site can be substituted by a splicing enhancer element.
Next, we tested whether the CTD was required for enhancer-dependent splicing without a functional poly(A) site by comparing transcripts made by full-length Pol II with those made by Pol IIΔCTD. As we observed previously, fewer reporter gene transcripts accumulate with Pol IIΔCTD than with full-length Pol II (Fig. 1B, lanes 1,2) probably because of reduced transcription (Gerber et al. 1995) and reduced stability of unprocessed RNA. The experiment in Figure 1B showed that splicing of intron 2 driven by the FN EDI enhancer element was substantially inhibited by CTD deletion (Fig. 1B, lane 3,4). The spliced to unspliced ratio declined from 7.2 to 1.6 when the gene was transcribed by Pol II ΔCTD relative to wild type. Similar results were obtained for two other splicing enhancer elements (see Fig. 8B, below). We conclude that the CTD is important for enhancer-dependent splicing, independently of 3′ processing.
Figure 8.
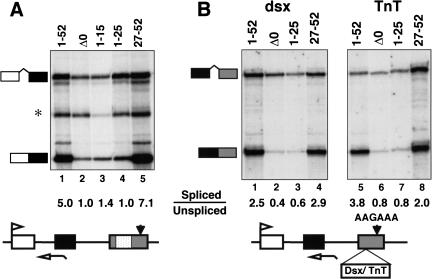
The carboxyl terminus of the CTD is necessary and sufficient to enhance splicing. (A) Efficient splicing of β-globin intron 1 is supported by heptads 27–52 but not by heptads 1–25. RNAs are the same as those analyzed in Fig. 7B. (*) Irrelevant undigested probe. (B) Enhancer-dependent splicing, independent of 3′ processing, is supported by heptads 27–52 but not by heptads 1–25 or 1–15. RNase protection of transcripts from pSVβ128 AAGAAA-dsx and pSVβ128 AAGAAA-TnT, which have mutant poly(A) sites and splicing enhancers inserted into exon 3 as diagrammed.
CTD deletion inhibited 3′ processing independently of splicing
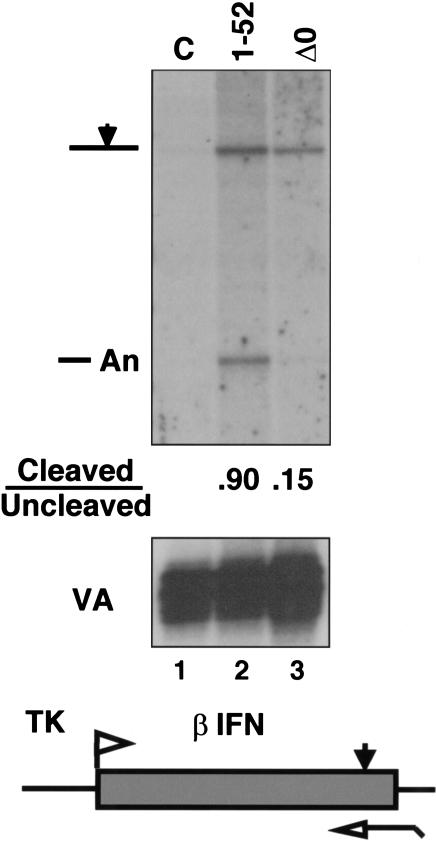
Because splicing and 3′ processing are closely coupled events, it is also possible that CTD deletion inhibits the 3′ cleavage reaction indirectly by inhibiting splicing of the adjacent intron. Therefore, we tested whether CTD deletion affected 3′ cleavage of transcripts from the human β-interferon (β-IFN) gene, which lacks introns. 293 Cells were cotransfected with the β-IFN gene driven by the HSV TK promoter and expression vectors for α-amanitin-resistant Pol II large subunits (1–52 or Δ0) or an irrelevant expression vector, CMV-neo (C). Detectable β-IFN RNA was made only when α-amanitin-resistant large subunit was expressed (Fig. 2, cf. lane 1 with lanes 2 and 3). RNase protection analysis showed that the fraction of transcripts cleaved at the poly(A) site was diminished by sixfold when the β-IFN gene was transcribed by Pol II ΔCTD relative to full-length Pol II (Fig. 2, lanes 2,3). Therefore, this observation confirmed that the CTD enhances 3′ processing independent of any effects it may have on splicing.
Figure 2.
CTD-dependent 3′ processing independent of splicing. RNase protection of TK–βIFN transcripts from α-amanitin-treated cells cotransfected with a CMV-neo control (C) or cDNA expression vectors for the full-length (1–52) or CTD-deleted (Δ0) Pol II large subunit. VA transcripts served as a control for transfection efficiency.
Interaction between the CTD and the amino terminus of CstF p50
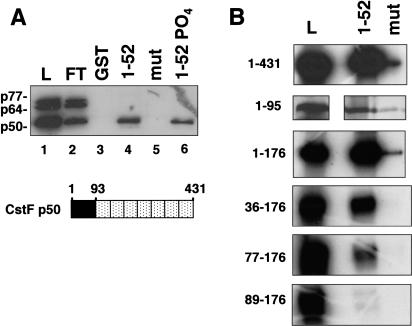
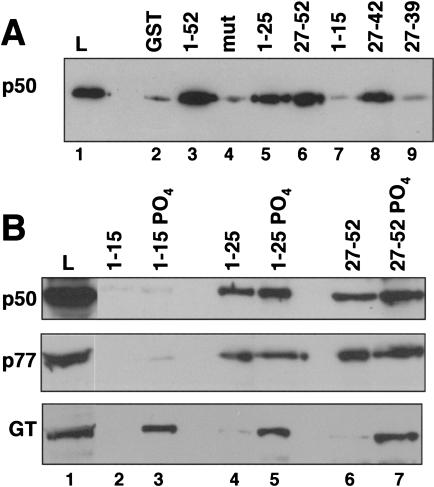
The simplest model for how the CTD affects 3′ processing is that it directly contacts one or more subunits of the cleavage/polyadenylation machinery. Our preliminary studies showed that purified CPSF bound poorly to the CTD in affinity chromatography experiments (S. McCracken, unpubl.). Therefore, we tested whether any of the subunits of CstF make direct contacts with the CTD. Polyhistidine-tagged p77, p64, and p50 subunits of CstF were expressed in Sf9 cells infected with recombinant baculoviruses and partially purified by Ni2+ affinity chromatography. The p50 and p64 subunits were at least 50% pure and the p77 was at least 10% pure as judged by silver staining (data not shown). The CstF subunits were incubated individually (data not shown) or as a mixture with beads containing GST fused to full-length mouse CTD either phosphorylated (see Materials and Methods) or unphosphorylated. CstF p50, but not p64 or p77, showed significant binding over background to both phosphorylated and unphosphorylated CTD resins (Fig. 3A, lanes 4,6). The slightly higher binding of p50 to unphosphorylated CTD (Fig. 3A, lanes 4,6) was not reproducibly observed (see Fig. 6B, below). The recombinant CstF subunits did not associate with one another in our experiments probably because of interference by the epitope tags. As negative controls, GST and GST–mutant CTD with 15 heptad repeats containing a Ser to Ala substitution at position 5 (West and Corden 1995) were used (Fig. 3A, lanes 3,5). In principle, contaminating insect proteins could contribute to CTD binding; however, this possibility is made unlikely by the observation that binding is completely specific to p50. In summary, this experiment suggests that a direct protein–protein contact occurs between the 50-kD subunit of CstF and the CTD.
Figure 3.
Interaction between the CTD and the amino terminus of CstF p50. (A) A mix of partially purified baculoviral His-tagged CstF p50, p64, and p77 was incubated with glutathione–Sepharose beads containing immobilized GST or fusions with wild-type (1–52) or mutant CTD (mut) with 15 repeats of YSPTAPS. High-salt eluates were immunoblotted with anti-Xpress antibody. 20% of the load (L) and 10% of the flowthrough (FT) and the eluates were loaded. Diagram of CstF p50 with its amino-terminal CTD-binding domain (black) and WD40 repeats (stippled). (B) The CTD-binding domain of CstF p50 maps to the amino-terminal 95 residues. [35S]methionine-labeled fragments of rat p50 were made by in vitro translation and incubated with wild-type (1–52) or mutant (mut) GST–CTD as in A. 10% of the load (L) and 50% of the high-salt eluates were loaded.
Figure 6.
Binding of CstF and capping enzyme to different segments of the CTD. (A) Binding of partially purified baculoviral CstF p50 to GST, GST–mut CTD (Fig. 3A), and fusions with segments of the CTD. 2.5% of the load (L) and 50% of the eluates were immunoblotted with anti-Xpress antibody. Note binding to heptads 27–42 but not to 1–15 or 27–39. (B) Binding of HeLa CstF (p50, p77) and capping enzyme guanylyltransferase (GT) to phosphorylated and unphosphorylated fragments of the CTD. HeLa nuclear extract was chromatographed on columns of immobilized GST fusion proteins. 1% of the load (L) and 8% of the eluates were analyzed by Western blotting. Note GT binds heptads 1–15-PO4 but CstF does not.
Deletions of CstF p50 were used to define the CTD contact site. CstF p50 contains a unique amino-terminal region of 92 amino acids followed by seven WD40 repeats (Fig. 3A; Takagaki and Manley 1992). In vitro-translated fragments comprising amino acids 1–176 or 1–95 of rat CstF p50 bound to the CTD approximately as well as the full-length protein (1–431) with minimal binding to the mutant CTD resin (Fig. 3B). Deletion of the first 36 or 77 residues (36–176 and 77–176) caused a progressive reduction in CTD binding and deletion of 89 amino-terminal residues (89–176) eliminated all binding (Fig. 3B). We conclude that the CTD interaction domain of CstF p50 lies within the amino-terminal 95 amino acids.
Dominant-negative effect of CstF p50 amino terminus on 3′ processing and capping
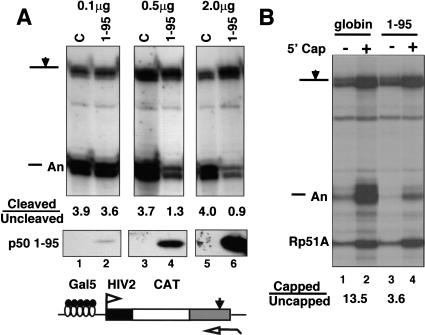
To investigate the functional significance of the interaction between the CstFp50 amino terminus and the CTD, we adopted a dominant-negative strategy. The rationale is that an excess of the CTD-binding domain of CstF p50 in the nucleus might compete with intact CstF for binding to the CTD and thereby inhibit 3′ processing. We transiently overexpressed the CTD-binding domain of CstF p50 with an SV40 nuclear localization signal (NLS) and asked whether it affected cleavage at the poly(A) site of a cotransfected reporter gene. We cotransfected 293 cells with constant amounts of Gal5HIV2CATΔt reporter and GAL4–VP16 expression vector and different amounts of expression plasmid for CstF p50 (1–95) or empty vector (C). Western blotting with antibody against the amino terminus of CstF p50 showed that transfected CTD-binding domain was greatly overexpressed relative to endogenous full-length protein (data not shown) and that its level increased with the amount of transfected expression plasmid (Fig. 4A, lower panel). Cleavage at the SV40 late poly(A) site in the reporter gene was quantified as in Figure 1. The ratio of cleaved to uncleaved RNAs decreased from 3.6 to 0.9 as the amount of CstF p50 (1–95) expression vector was increased from 0.1 to 2.0 μg (Fig. 4A, lanes 2,4,6). Increasing the amount of empty vector (C, Fig. 4A, lanes 1,3,5) had no effect. Overexpression of CstF p50 (1–176) had a similar dominant-negative effect (data not shown). We conclude that the CstF p50 amino terminus acts as a dominant-negative inhibitor of 3′ processing.
Figure 4.
Dose-dependent dominant-negative effect of overexpressed CstF p50 amino terminus on 3′ processing and capping. (A) (Top) RNase protection of Gal5HIV2CATΔt reporter transcripts activated by GAL4–VP16 (see diagram). Cells were cotransfected with 0.1, 0.5, or 2.0 μg of EFpLinkCstFp50(1–95) or EFpLink vector (C). Ratios of cleaved to uncleaved RNA at the SV40 late poly(A) site are given. (Bottom) Western blotting of p50 (1–95) with antibody against the amino terminus. (B) RNase protection as in A of Gal5HIV2CATΔt transcripts that were separated into capped and uncapped fractions by binding to GST–eIF4E. Cells were cotransfected with 40 μg of either EFpLinkTag globin control or EFpLinkTagCstFp50(1–95). Yeast Rp51A RNA served as a control for the GST–eIF4E selection procedure. Capped to uncapped ratios are given for the sum of cleaved plus uncleaved transcripts. CstF p50 1–95 overexpression inhibited both cleavage at the poly(A) site and the extent of capping.
If the CstF p50 amino terminus competes with CstF for binding sites on the CTD, then it might also compete with capping enzymes and thereby inhibit capping. This idea was tested by cotransfection of the Gal5HIV2CATΔt reporter and GAL4–VP16 expression vectors with a large excess of expression plasmid for CstF p50 (1–95) or a fragment of β-globin with an SV40 NLS, as a control (Fig. 4B). Capped and uncapped RNAs were separated by binding to GST–eIF4E. As a control for the efficiency of this procedure, samples were spiked with total yeast RNA and assayed for the Rp51A mRNA in the capped and uncapped fractions. The sums of the cleaved and uncleaved HIV2CAT transcripts in the capped and uncapped fractions were determined and normalized to Rp51A. Overexpression of CstF p50 (1–95) reduced the ratio of capped to uncapped transcripts (cleaved plus uncleaved) from 13.5 to 3.6 (Fig. 4B). As we observed previously (McCracken et al. 1997a), the uncapped fractions were enriched for uncleaved precursors. The fact that overexpression of the CTD-binding domain of CstF p50 affects the extent of capping as well as 3′ processing suggests that it binds to the CTD in vivo and competes with endogenous processing factors.
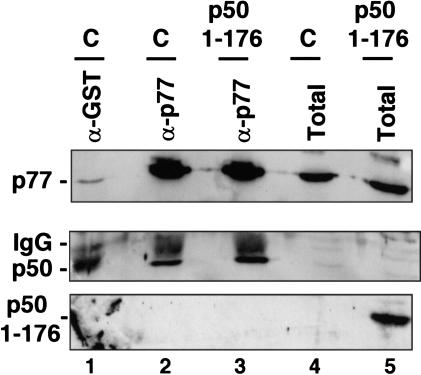
It remains possible that an excess of the p50 amino terminus could also inhibit 3′ processing by disrupting the association between p50 and p77 in the CstF complex (Takagaki and Manley 2000). To control for this possibility, we overexpressed the CstF p50 amino terminus and asked whether CstF remained intact. We cotransfected 293 cells with the expression vector for c-Myc-tagged CstF p50 (1–176), the Gal5HIV2CATΔt reporter, GAL4–VP16, and GFP expression plasmids. GFP-expressing cells were selected by FACS. RNA was prepared from a fraction of the cells to confirm that 3′ processing had been inhibited (data not shown). Protein extracts from the sorted cells and untransfected controls were immunoprecipitated with anti-CstF p77 antibody, and the precipitates were probed for p77 and p50 by Western blotting. The amount of CstF p50 coprecipitating with p77 was unaffected by overexpression of the p50 CTD-binding domain (Fig. 5, middle, cf. lanes 2 and 3). Furthermore, the CTD-binding domain (1–176) did not coprecipitate with CstF p77, indicating that these proteins do not form a stable complex in vivo (Fig. 5, lower panel, lane 3). In summary, overexpression of the CTD-binding domain of CstF p50 has a dominant-negative effect that resembles the effect of deleting the CTD; it inhibits both 3′ processing and capping without disrupting the CstF complex.
Figure 5.
Overexpression CstF p50 amino terminus does not disrupt the CstF complex. Cells were cotransfected with EFpLinkTag p50(1–176) (1μg/plate). Gal5HIV2CATΔt reporter, GAL4–VP16, and GFP expression vectors. Transfected cells were selected by FACS sorting for GFP expression. Extracts from the transfected cells (p50 1–176) and untransfected controls (C) were immunoprecipitated with anti-CstFp77 (lanes 2,3) or anti-GST as a negative control (lane 1). Immunoprecipitates and total extracts were immunoblotted with anti-CstF p77, anti-p50, and anti-Myc, which recognizes p50 (1–176). Note that overexpression of p50 (1–176) does not disrupt the association of p77 with p50 and that p50 (1–176) does not form a stable complex with p77 (lane 3).
Variant heptads in the carboxy-terminal region of the CTD enhance binding to CstF p50
Although the CTD is a highly repeated structure, it is not uniform in sequence (see Fig. 10, below). Variant heptad repeats with residues other than Ser at position 7 are clustered in the carboxy-terminal part of the CTD. We tested GST fusions of several fragments of the CTD, for their ability to bind recombinant CstF p50 (Fig. 6A). Heptads 1–15 (Fig. 6A, lane 7) did not bind to CstF p50 significantly above background (Fig. 6A, lanes 2,4) whereas heptads 27–42 bound almost as well as full-length CTD (1–52; Fig. 6A, lanes 3,8). Deletion of heptads 40–42 with Lys or Thr at position 7 to make GST–CTD 27–39 eliminated binding of CstF p50 (Fig. 6A, lane 9). Heptads 1–15 of the CTD include 11 consensus heptads, YSPTSPS, whereas heptads 27–42 include 13 variants with Asn, Arg, Thr, Lys or Glu at position 7 (see Fig. 10, below). Increasing the length of the CTD ligand from 15 to 25 repeats in GST–CTD 1–25 adds eight consensus and two variant heptads with Asn at position 7 (heptads 22, 23) and permitted binding to CstF p50 (Fig 6A, lane 5). CstF p50 also bound strongly to heptads 27–52 (Fig. 6A, lane 6). We conclude that, although they are not necessarily essential, certain variant heptads including those with Lys or Thr at position 7, enhance binding to CstF p50 and permit association with a segment of only 15 repeats.
Figure 10.
Amino acid sequence of the human CTD with heptad repeats numbered.
Binding of the CstF complex and the capping enzyme guanylyltransferase (GT) to different segments of the CTD was compared by chromatography of HeLa nuclear extract on CTD affinity columns. Because GT requires CTD phosphorylation for binding, we phosphorylated GST fusions of heptads 1–15, 1–25, and 27–52 as in Figure 3A. Consistent with the results for recombinant p50, intact CstF detected with anti-p50 and anti-p77 antibodies bound to phosphorylated and unphosphorylated heptads 1–25 and 27–52 approximately equally, but did not bind to heptads 1–15 (Fig. 6B). In contrast, the capping enzyme (GT) bound to heptads 1–15 as well as 1–25 and 27–52 (Fig. 6B lower panel) provided they were phosphorylated. Therefore, the capping enzyme and the polyadenylation factor CstF differ in their specificities for different segments of the CTD.
The carboxy-terminal half of the CTD is sufficient for 3′ processing
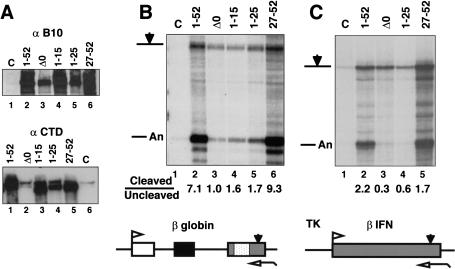
We tested whether the differences in binding of CstF to different fragments of the CTD in vitro correlated with different functional properties in vivo. Full-length (1–52) and CTD truncated mutants comprising heptads 27 to the carboxyl terminus (27–52), 1–15, 1–25, and Δ0 were expressed in transiently transfected cells as shown by Western blotting with antibody against the amino-terminal B10 epitope (Fig. 7A). Immunoprecipitation with an anti-B10 antibody followed by Western blotting with an anti-CTD antibody confirmed that the 1–15, 1–25, and 27–52 truncations did indeed express CTD sequences (Fig. 7A, bottom). The signal present in lanes 2 and 6 (Fig. 7A, bottom) is due to contaminating endogenous Pol II in the precipitates.
Figure 7.
The carboxyl terminus of the CTD is necessary and sufficient to enhance 3′ processing. (A) Expression of B10 epitope-tagged CTD deletion mutants of α-amanitin-resistant Pol II large subunit. (Top) Western blot of extracts from transfected 293 cells and untransfected control (C) with anti-B10 antibody. (Bottom) Western blot of anti-B10 immunoprecipitates probed with rabbit anti-CTD antibody. (B) Cleavage at the β-globin poly(A) site is supported by heptads 27–52 but not by heptads 1–25. RNase protection of pSVβ128Rpbex5 transcripts from α-amanitin-treated cells cotransfected with CMV-neo (C), or expression vectors for α-amanitin-resistant Pol II large subunits with full-length (1–52) or truncated CTDs. Ratios of cleaved to uncleaved transcripts are given. (C) Cleavage at the β-IFN poly(A) site is supported by heptads 27–52 but not by heptads 1–25. RNase protection analysis of intronless TK-βIFN transcripts made by full-length and CTD-deleted Pol II. Ratios of cleaved to uncleaved transcripts are given.
The effects of these CTD deletions on 3′ processing of a cotransfected β-globin reporter gene (pSVβ128Rpbex5) are shown in Figure 7B. Remarkably, the CTD carboxyl terminus (27–52) functioned as well as the full-length (1–52) in enhancing 3′ processing (Fig. 7B, lanes 2,6) whereas the amino-terminal heptads 1–25 were nearly equivalent to complete deletion of the CTD, Δ0 (Fig. 7B, lanes 3,5). Heptads 1–15 also failed to support efficient 3′ processing (Fig. 7B, lane 4). Transcription by Pol II (27–52) consistently yielded greater amounts of RNA than Pol II (1–25). This difference may be due in part to better expression of the 27–52 construct (Fig. 7A) but it may also indicate that transcripts made by Pol II (27–52) are more stable or that heptads 27–52 are better than 1–25 at carrying out activated transcription.
To test whether the amino- and carboxy-terminal regions of the CTD also affect 3′ processing differently in the absence of splicing, we examined the effects of CTD deletions on cleavage at the poly(A) site of the intronless TK-β-IFN gene (Fig. 7C). Heptads 27–52 support 3′ processing at the β-IFN poly(A) site almost as well as full-length CTD, 1–52; however heptads 1–25 are not much more effective than the total deletion, Δ0 (Fig. 7C, lanes 2–5). In summary, the carboxy-terminal half of the CTD is sufficient to enhance 3′ processing independently of splicing but the amino-terminal half is not.
The carboxy-terminal half of the CTD is sufficient to enhance splicing
We asked whether the amino- and carboxy-terminal regions of the CTD also differ in their ability to support splicing. The effect of CTD truncations on splicing of β-globin intron 1 is shown in Figure 8A (lanes 1–5). Heptads 27–52 supported a level of splicing equivalent to full-length Pol II, 1–52 (Fig. 8A, lanes 1,5) but heptads 1–15, 1–25 (lanes 3,4), and 27–42 (data not shown) had little activity above the complete deletion, Δ0 (lane 2).
To investigate the role of the amino- and carboxy-terminal halves of the CTD in splicing independent of 3′ processing, we examined transcripts from β-globin AAGAAA reporter genes containing the Drosophila doublesex (dsx) exon 4 or cardiac Troponin T (cTnT) exon 5 enhancer (Coulter et al. 1997) inserted into exon 3. Both of these elements, like the FN EDI enhancer (Fig. 1B), stimulated splicing of β-globin intron 2 in the context of a mutant poly(A) site. Heptads 1–25 did not support any splicing of β-globin intron 2 above that observed with a complete deletion of the CTD (Fig. 8B, cf. lanes 2 and 3 with lanes 6 and 7). In contrast, heptads 27–52 retained significantly more splicing function than the complete deletion, Δ0 (Fig. 8B, cf. lane 2 with 4 and lane 6 with 8). We conclude that the carboxy-terminal half but not the amino-terminal half of the CTD is sufficient to stimulate enhancer-dependent splicing, independent of 3′ processing.
The amino and carboxyl termini of the CTD support capping
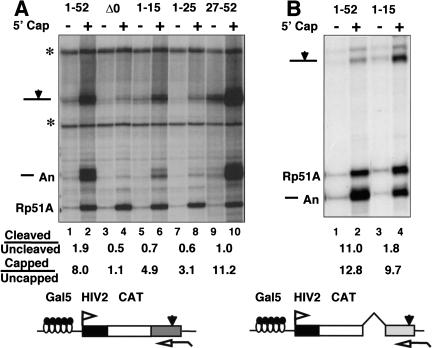
Heptads 1–15 did not support efficient splicing or 3′ processing nor did they bind CstF in vitro; however, when phosphorylated, they did bind to capping enzyme (Fig. 6B). Therefore, we tested whether heptads 1–15 retained the ability to enhance capping in vivo independent of splicing and 3′ processing. RNA made by α-amanitin-resistant Pol II CTD truncations was fractionated by binding to GST–eIF4E as in Figure 4B, and transcripts in the capped and uncapped populations were quantified with a probe complementary to the SV40 late poly(A) site. Cleavage efficiency for full-length Pol II, 1–52 and the amino-terminal truncation, 27–52, was reduced in this experiment relative to most others (see Fig. 4A, lanes 1,3,5). The results in Figure 9A showed that both amino- and carboxy-terminal segments of the CTD support more capping than the complete CTD deletion, Δ0. The capped to uncapped ratios normalized to Rp51A were 8.0, 4.9, 3.1, and 11.2, respectively, for 1–52, 1–15, 1–25, and 27–52 compared with 1.1 for Δ0.
Figure 9.
Either the amino or carboxyl terminus of the CTD can enhance capping. (A) The Gal5HIV2CATΔt reporter activated by GAL4–VP16 was transcribed by full-length (1–52) and CTD-truncated α-amanitin-resistant mutants of Pol II. Capped and uncapped RNA was analyzed with probe complementary to the SV40 late poly(A) site. (*) Undigested probes. Ratios of cleaved to uncleaved and capped to uncapped transcripts are shown. Capped to uncapped ratios are for the sum of cleaved plus uncleaved transcripts and were normalized to Rp51A as in Fig. 4B. (B) Heptads 1–15 support efficient capping but not 3′ processing. Gal5HIV2CAT transcription was activated by GAL4–SW6 and HIV-1 Tat. Capped and uncapped RNA was analyzed with a probe complementary to the SV40 early poly(A) site. Capped to uncapped ratios are for the sum of cleaved plus uncleaved transcripts as in A.
The effects of heptads 1–15 on capping of transcripts from the Gal5HIV2CAT gene were compared with full-length CTD (1–52) in Figure 9B. Transcription was activated by HIV-1 Tat and the GAL4–VP16 mutant activator SW6 (Walker et al. 1993). Capped and uncapped RNA was assayed with a probe spanning the SV40 early poly(A) site. The extent of capping was only modestly reduced (capped to uncapped ratios: 12.8 and 9.7) when heptads 16–52 were deleted (Fig. 9B) whereas cleavage at the poly(A) site was substantially inhibited (cleaved to uncleaved ratios: 11.0 and 1.8). In summary, the results in Figure 9 show that both the amino- and carboxy-terminal segments of the CTD are able to support capping.
Discussion
Independent stimulation of capping, splicing, and 3′ processing by the CTD
The CTD stimulates three interdependent pre-mRNA processing events: capping, splicing, and 3′ processing. In this paper, we have begun to dissect the role of the CTD in each of these processing steps. The CTD is required for 3′ processing independent of splicing as shown by examination of transcripts from an intronless gene (Fig. 2). The CTD is also required for splicing independent of 3′ processing as shown by examination of enhancer-dependent splicing of a gene with a mutant poly(A) site (Figs. 1B and 8). By testing CTD truncations in the context of α-amanitin-resistant Pol II in vivo, we found a striking difference between the amino- and carboxy-terminal halves of the CTD in the ability to stimulate different processing steps. Whereas the carboxyl terminus supports all three major pre-mRNA processing steps, the amino terminus (heptads 1–15 and 1–25, Fig. 9) supports only capping. This observation shows that capping can be enhanced by the CTD independently of splicing and 3′ processing. Therefore, defects in capping cannot fully explain the CTD dependence of splicing and 3′ processing. We conclude that the CTD independently stimulates each of the three major pre-mRNA processing steps.
CstF p50 binding to the CTD and 3′ processing
The 3′ processing factor CstF p50 binds the CTD but does not bind equally well to all segments of the CTD. CstF p50 can bind to either heptads 1–25 or 27–52 in vitro; however, variant heptads strengthen the interaction. CstF p50 did not bind to heptads 1–15 but did bind to heptads 27–42, which are enriched in variant repeats (Figs. 6A and 10). Deletion of three repeats with Lys or Thr at position 7 (heptads 27–42 vs. 27–39) abolished binding (Fig. 6A, lanes 8,9).
The CTD-binding domain of CstF p50 was mapped to the amino-terminal 95 amino acids (Fig. 3B). This amino-terminal region was found previously to bind full-length CstF p50 whereas the carboxy-terminal WD40 repeats interact with CstF p77, PCNA, and BARD1 (Kleiman and Manley 1999; Takagaki and Manley 2000). When overexpressed, the p50 CTD-binding domain had a dose-dependent dominant-negative effect on cleavage at a poly(A) site without disrupting the CstF complex (Figs. 4A and 5). That the CstF p50 amino terminus binds to the CTD in vivo is supported by the fact that it also inhibited capping (Fig. 4B) probably by competing with capping enzymes for binding sites on the CTD.
Amino- and carboxy-terminal segments of the CTD bind guanylyltranferase and support capping
In contrast to CstF, the capping enzyme guanylyltransferase bound to the phosphorylated form of heptads 1–15 (Fig. 6B) in accord with the fact that heptads 1–15 support capping but not polyadenylation (Figs. 7–9). Phosphorylated heptads 27–52 also bound the capping enzyme in vitro (Fig. 6B) and supported capping in vivo (Fig. 9). Therefore, it is apparent that the guanylyltransferase need not bind a unique site within the CTD to carry out efficient capping.
The CTD stimulates enhancer-dependent splicing independent of 3′ processing
The details of how the CTD influences splicing in vivo are unclear; however, both positive and negative effects have been reported in vitro (Yuryev et al. 1996; Hirose et al. 1999; Zeng and Berget 2000). We show that the FN EDI, Drosophila dsx and cTnT splicing enhancers restored splicing of β-globin intron 2 after it was inhibited by mutation of the poly(A) site (Figs. 1B and 8B). In all cases, the CTD was required for this enhancer-dependent splicing, independent of 3′ processing. Splicing enhancers function by binding to SR proteins. The dsx exon 4 enhancer binds the SR-like Tra2 proteins (Dauwalder et al. 1996; Tacke et al. 1998); the cTnT exon 5 enhancer binds to ASF/SF2, SRp40, SRp55, and SRp75 (Ramchatesingh et al. 1995) and the FN EDI enhancer binds ASF/SF2 and 9G8 (Cramer et al. 1999; Lavigueur et al. 1993). Whether or not direct interactions between the CTD and SR proteins occur has not been fully explored. Although we do not know at what step(s) in the splicing reaction the CTD is required, one possibility that is consistent with our results and those of Zeng and Berget (2000) is that the CTD is needed for the function of at least some splicing enhancers.
The carboxyl terminus of the CTD is sufficient for 3′ processing and splicing
In the context of α-amanitin-resistant Pol II large subunit, the carboxyl terminus of the CTD (heptads 27–52) enhanced 3′ processing whereas heptads 1–25 or 1–15 had no significant activity above the complete CTD deletion (Figs. 7B,C). Note that our experiments do not address whether the 10 residues carboxy-terminal of the heptad 52 contribute to the processing function of the carboxy-terminal half of the CTD. The ability of heptads 27–52 and the failure of heptads 1–15 to support 3′ processing are consistent with the fact that 27–52 binds CstF in vitro whereas 1–15 does not (Fig. 6). On the other hand, heptads 1–25 and 27–42, which bind CstF in vitro, do not support efficient 3′ processing in vivo possibly because they do not recruit some unknown factor(s) that is required in addition to CstF. Alternatively, the binding sites for CstF on these CTD fragments may be obscured in the context of Pol II holoenzyme complexes in vivo. These results are consistent with the hypothesis that binding of CstF to the carboxyl terminus of the CTD is important but not necessarily sufficient for the stimulation of transcription-coupled 3′ processing.
The CTD carboxyl terminus (27–52) was also sufficient to support splicing even in the absence of a poly(A) site, whereas heptads 1–25 and 1–15 were not (Fig. 8). Fragments of the carboxy-terminal region comprising heptads 27–42 and 40–52 did not support either 3′ processing or splicing (data not shown). These results are consistent with the finding that overexpression of full-length CTD was more effective than overexpression of the amino-terminal 13 heptads in inhibiting splicing of β-globin intron 1 (Du and Warren 1997). We conclude that although the CTD is a highly repeated structure, there is functional specialization of different segments within it.
Materials and methods
Transfections and RNA analysis
293 Cells (150 mm plates) were transiently transfected with 5 μg of reporter plasmid, 0.5 μg of activator expression plasmid, 0.5 μg of pSPVA, and 2.5 μg of Pol II expression vectors or CMV-neo control plasmid by calcium phosphate precipitation. α-Amanitin (2.5 μg/mL) was added 12–16 h after transfection, and cells were harvested after 65 h. CMV-neo and VA controls are shown only in some figures. Fractionation of capped and uncapped RNAs by binding to GST–eIF4E was as described (McCracken et al. 1997a). Poly(A)+ selection was done with BioMag oligodT(20) (Polysciences) according to the manufacturer's directions. RNase protection was as described (McCracken et al. 1997a). Fixed, dried gels were quantified by PhosphorImager by use of Imagequant software. In all cases, signal volume over background was as least 5000 units.
Plasmids
pGal5HIV2CAT, pGal5HIV2CATΔt, pSPVA, pSVβ128, and GAL4–VP16 plasmids and RNase protection probes were described previously (McCracken et al. 1997a,b).
pSVβ128-AAGAAA contains the human β-globin gene with SV40 enhancer and mutant poly(A) site made by mismatch oligonucleotide–directed PCR.
pSVβ128Rpbex5 contains the human β-globin gene with a 280-bp in-frame insertion from the mouse Pol II large subunit exon 5 into the BstXI site. This insertion modestly increased the efficiency of 3′ processing and had no effect on the splicing of intron 2.
pBSKS-β5′int2 used to make an RNase protection probe for the β-globin intron 2 5′ splice site contains a PCR fragment extending 218 bases 5′ and 92 bases 3′ of the splice site.
pEGFP2-C2 (Clontech) was used for GFP expression (Fig. 5).
pVZ GPDH used to make an RNase protection probe for human GPDH contains a 246 base AluI cDNA fragment.
pBS Rp51A was used to make an RNase protection probe complementary to bases −67 to +188 relative to the 3′ splice site of yeast Rp51A.
pSVβ128-AAGAAA-FN+, pSVβ128-AAGAAA-dsx, and pSVβ128-AAGAAA-TnT were made by insertion of the respective splice enhancer elements into the BstXI site in the β-globin exon 340 bp from the 3′ splice site. The FN enhancer was the 73-bp blunt-ended XhoI–StuI fragment from pSVEDA-HIV (Cramer et al. 1999). The cTnT enhancer element was the N mutant 5′-CA AGAGGAAGAAGAAGAAGAGGAAGACGACGA-3′ (Ramchatesingh et al. 1995) and the dsx element was 5′-GTTT CTTCAATCAACAGAAG-3′ (Coulter et al. 1997). Both were inserted in frame.
EFpLinkTag p50(1–176) contains codons 1–176 of rat CstF p50 cDNA with an amino-terminal c-Myc tag and carboxy-terminal SV40 NLS in the EF1α promoter-driven expression vector EFpLinkTag (a gift of R. Treisman, ICRF, London, UK)
EFpLinkTag p50(1–95) contains codons 1–95 of rat CstF p50 with a carboxy-terminal SV40 NLS.
EFpLinkTag globin contains human β-globin exons 1 and 2 with a c-Myc amino-terminal epitope tag and carboxy-terminal SV40 NLS.
pIFpTK2 has a human β-IFN genomic fragment from +72 to +1302 driven by the HSV TK promoter (−105 to +57) and was a gift of S. Goodbourn (University of London, UK).
pBSKS-IFNpoly(A) containing the ClaI–AccI (+638 to +948) fragment spanning the poly(A) site was used to make an RNase protection probe.
pAT7RpbwtAmr expresses a cDNA of the human full-length Pol II large subunit with an amino-terminal B10 epitope tag and a carboxy-terminal His6 tag and was a gift of M. Vigneron (Nguyen et al. 1996).
CTD deletions of pAT7RpbwtAmr were made initially by replacing the CTD with a short polylinker 5′-GTTTAAACGC GGCCGCTCTAGA-3′ in three frames. Fragments corresponding to mouse CTD heptads 1–15, 27–42, and 27–52 (including the natural carboxyl terminus) were inserted in-frame into this polylinker. pAT7Rpb1–25Amr was derived from pAT7RpbwtAmr by deletion of the sequences between the SpeI site in heptad 26 of the human CTD and the carboxyl terminus. The mouse and human CTDs have identical amino acid sequences except for one substitution of Ala for Thr in mouse CTD at position 4 of heptad 38.
Recombinant proteins
Baculoviral rat CstF p50, p64, and p77 were expressed in the pBlueBacHis2B vector (Invitrogen) with amino-terminal Xpress and His6 tags. Extracts from Sf9 cells were chromatographed on Ni2+ affinity columns for purification. The p64 and p50 subunits did not coimmunoprecipitate with p77 either when Sf9 cells were coinfected or when extracts from singly infected cells were mixed. Failure to associate could be due to interference by the epitope tags.
[35S]methionine-labeled p50 (Fig. 3B) was made by in vitro transcription/translation (TNT, Promega). Rat p50 1–431 and 1–176 were made from the pet21dCstFp50 template; p50 (1–95) was made from pcDNA3CstF p50 (1–95); p50 (36–176) and (77–176) were made from pet21dCstFp50Δ36 and Δ77, respectively, which were made by PCR-mediated deletion of the amino terminus; and p50 (89–176) was made from pet21d CstFp50 Aat1–StuI (McCracken et al. 1997b).
Affinity chromatography
Full-length wild-type murine GST–CTD (1–52), GST–CTD 1–15, and GST–mut CTD (A5)15 have been described (McCracken et al. 1997a). GST–CTD(27–52) was expressed from pet21aGST–TEVmCTD(27–52) made by cloning of a PCR fragment of the murine CTD that extends to the carboxyl terminus. GST–CTD(27–42) and GST–CTD(27–39) were made by deletion of pet21aGSTmCTD(27–52) at two SspI sites. GST–CTD(1–25) was made by subcloning of a SmaI–SpeI fragment of human Rpb1 cDNA into pet21aGSTTEV. This fragment includes 20 residues amino-terminal of the first heptad repeat. Phosphorylated CTD was prepared by incubation in HeLa nuclear extract as described (McCracken et al. 1997a) followed by extensive high salt washing. This material (1–52, 1–15, 1–25, and 27–52) reacted strongly with monoclonal antibody H14, which is specific for phosphorylation on Ser 5, and weakly with monoclonal antibody H5 (Patturajan et al. 1998), which is specific for phosphorylation on Ser 2.
For small-scale batch chromatography of in vitro-translated proteins (diluted fivefold) and baculoviral CstF (Figs. 3,6A), 25 μL of the resins was incubated with 100 μL of extract in binding buffer (20 mM Hepes at pH 7.9, 0.1 mM EDTA, 2 mM DTT, 20% glycerol, 0.1 M NaCl, 0.1% NP-40, 0.05% nonfat dried milk, 400 μg/mL ethidium bromide) for 1 h at 25°C. Beads were washed three times in 0.5 mL of binding buffer without milk or ethidium bromide, then eluted with 40 μL of the same buffer plus 0.9 M NaCl. Chromatography of HeLa nuclear extract (Fig. 6B, 1.25 mg in 250 μL) was at 4°C on 200-μL columns (0.5 mg/mL immobilized GST–CTD 1–15 and 27–42, 3 mg/mL 1–52, 1–25, and 27–39 and >6 mg/mL GST, GST-mutCTD and 27–52) in binding buffer plus 0.5 μM microcystin and 1 mM β-glycerophosphate. Columns were washed five times in 1 mL as above and eluted with 3 × 0.25 mL of 1 M NaCl elution buffer.
Antibodies and immunoprecipitation
Transfected cells were lysed in 50 mM Tris HCl (pH 8.0), 500 mM NaCl, 1 mM EDTA, 10 mM β-glycerophosphate, 1% NP40, 0.5μM microcystin, and protease inhibitors for 30 min on ice then cleared by microcentrifuging for 15 min. Immunoprecipitation was for 3 h at 4°C in lysis buffer plus 19 mM EDTA and 10 mM EGTA with anti-B10 antibody bound to protein A–Sepharose. Precipitates were washed four times in 1 mL of 50 mM Tris-HCl (pH 7.4), 1% Triton-X100, 300 mM NaCl, and protease inhibitors.
Anti-p50, anti-p77, anti-CTD, and anti-capping enzyme antibodies were raised in rabbits. Anti-Myc 9E10, anti-B10, and anti-Xpress epitope antibodies were purchased from Roche, Chemicon, and Invitrogen, respectively.
Acknowledgments
We thank A. Kornblihtt, S. Goodbourn, M. Vigneron, D. Licatalosi, and R. Treisman for plasmids and antibodies, D. Zorio for help with the capping assay, the UCHSC Cancer Center Sequencing and Flow Cytometry Facilities. We are grateful to D. Zorio, S. Schroeder, J. Jaehning, M. Huang, J. Tyler, P. McGee, T. Evans, and T. Blumenthal for valuable criticisms and suggestions and to T. Boudreau for secretarial help. This work was funded by NIH grant GM58613 to D.B.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL david.bentley@UCHSC.edu; FAX (303) 315-8215.
Article and publication are at http://www.genesdev.org/cgi/doi/10.1101/gad.889101.
References
- Barilla D, Lee BA, Proudfoot NJ. Cleavage/polyadenylation factor IA associates with the carboxyl-terminal domain of RNA polymerase II in Saccharomyces cerevisiae. Proc Natl Acad Sci. 2001;98:445–450. doi: 10.1073/pnas.98.2.445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartolomei MS, Halden NF, Cullen CR, Corden JL. Genetic analysis of the repetitive carboxyl-terminal domain of the largest subunit of mouse RNA polymerase II. Mol Cell Biol. 1988;8:330–339. doi: 10.1128/mcb.8.1.330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bauren G, Belikov S, Wieslander L. Transcriptional termination in the Balbiani ring 1 gene is closely coupled to 3′-end formation and excision of the 3′-terminal intron. Genes & Dev. 1998;12:2759–2769. doi: 10.1101/gad.12.17.2759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bentley D. Coupling RNA polymerase II transcription with pre-mRNA processing. Curr Opin Cell Biol. 1999;11:347–351. doi: 10.1016/S0955-0674(99)80048-9. [DOI] [PubMed] [Google Scholar]
- Beyer AL, Osheim YN. Splice site selection, rate of splicing, and alternative splicing on nascent transcripts. Genes & Dev. 1988;2:754–765. doi: 10.1101/gad.2.6.754. [DOI] [PubMed] [Google Scholar]
- Blencowe BJ. Exonic splicing enhancers: Mechanism of action, diversity and role in human genetic diseases. Trends Biochem Sci. 2000;25:106–110. doi: 10.1016/s0968-0004(00)01549-8. [DOI] [PubMed] [Google Scholar]
- Cho EJ, Takagi T, Moore CR, Buratowski S. mRNA capping enzyme is recruited to the transcription complex by phosphorylation of the RNA polymerase II carboxy-terminal domain. Genes & Dev. 1997;11:3319–3326. doi: 10.1101/gad.11.24.3319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooke C, Alwine JC. The cap and the 3′ splice site similarly affect polyadenylation efficiency. Mol Cell Biol. 1996;16:2579–2584. doi: 10.1128/mcb.16.6.2579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coppola JA, Field AS, Luse DS. Promoter-proximal pausing by RNA polymerase II in vitro: Transcripts shorter than 20 nucleotides are not capped. Proc Natl Acad Sci. 1983;80:1251–1255. doi: 10.1073/pnas.80.5.1251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coulter LR, Landree MA, Cooper TA. Identification of a new class of exonic splicing enhancers by in vivo selection. Mol Cell Biol. 1997;17:2143–2150. doi: 10.1128/mcb.17.4.2143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cramer P, Caceres JF, Cazalla D, Kadener S, Muro AF, Baralle FE, Kornblihtt AR. Coupling of transcription with alternative splicing: RNA Pol II promoters modulate SF2/ASF and 9G8 effects on an exonic splicing enhancer. Mol Cell. 1999;4:251–258. doi: 10.1016/s1097-2765(00)80372-x. [DOI] [PubMed] [Google Scholar]
- Dahmus ME. Reversible phosphorylation of the C-terminal domain of RNA polymerase II. J Biol Chem. 1996;271:19009–19012. doi: 10.1074/jbc.271.32.19009. [DOI] [PubMed] [Google Scholar]
- Dauwalder B, Amaya-Manzanares F, Mattox W. A human homologue of the Drosophila sex determination factor transformer-2 has conserved splicing regulatory functions. Proc Natl Acad Sci. 1996;93:9004–9009. doi: 10.1073/pnas.93.17.9004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du L, Warren SL. Functional interaction between the carboxy-terminal domain of RNA polymerase II and pre-messenger RNA splicing. J Cell Biol. 1997;136:5–18. doi: 10.1083/jcb.136.1.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dye MJ, Proudfoot NJ. Terminal exon definition occurs cotranscriptionally and promotes termination of RNA polymerase II. Mol Cell. 1999;3:371–378. doi: 10.1016/s1097-2765(00)80464-5. [DOI] [PubMed] [Google Scholar]
- Flaherty SM, Fortes P, Izaurralde E, Mattaj IW, Gilmartin GM. Participation of the nuclear cap binding complex in pre-mRNA 3′ processing. Proc Natl Acad Sci. 1997;94:11893–11898. doi: 10.1073/pnas.94.22.11893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerber HP, Hagmann M, Seipel K, Georgiev O, West MA, Litingtung Y, Schaffner W, Corden JL. RNA polymerase II C-terminal domain required for enhancer-driven transcription. Nature. 1995;374:660–662. doi: 10.1038/374660a0. [DOI] [PubMed] [Google Scholar]
- Greenleaf AL. Positive patches and negative noodles: Linking RNA processing to transcription? Trends Biochem Sci. 1993;18:117–119. doi: 10.1016/0968-0004(93)90016-g. [DOI] [PubMed] [Google Scholar]
- Gunnery S, Mathews MB. Functional mRNA can be generated by RNA polymerase III. Mol Cell Biol. 1995;15:3597–3607. doi: 10.1128/mcb.15.7.3597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hart RP, McDevitt MA, Nevins JR. Poly(A) site cleavage in a HeLa nuclear extract is dependent on downstream sequences. Cell. 1985;43:677–683. doi: 10.1016/0092-8674(85)90240-5. [DOI] [PubMed] [Google Scholar]
- Hirose Y, Manley JL. RNA polymerase II is an essential mRNA polyadenylation factor. Nature. 1998;395:93–96. doi: 10.1038/25786. [DOI] [PubMed] [Google Scholar]
- ————— RNA polymerase II and the integration of nuclear events. Genes & Dev. 2000;14:1415–1429. [PubMed] [Google Scholar]
- Hirose Y, Tacke R, Manley JL. Phosphorylated RNA polymerase II stimulates pre-mRNA splicing. Genes & Dev. 1999;13:1234–1239. doi: 10.1101/gad.13.10.1234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho C, Shuman S. Distinct effector roles for Ser2 and Ser5 phosphorylation of the RNA polymerase II CTD in the recruitment and allosteric activation of mammalian capping enzyme. Mol Cell. 1999;3:405–411. doi: 10.1016/s1097-2765(00)80468-2. [DOI] [PubMed] [Google Scholar]
- Inoue K, Ohno M, Sakamoto H, Shimura Y. Effect of the cap structure on pre-mRNA splicing in Xenopus oocyte nuclei. Genes & Dev. 1989;3:1472–1479. doi: 10.1101/gad.3.9.1472. [DOI] [PubMed] [Google Scholar]
- Kim E, Du L, Bregman DB, Warren SL. Splicing factors associate with hyperphosphorylated RNA polymerase II in the absence of pre-messenger RNA. J Cell Biol. 1997;136:19–28. doi: 10.1083/jcb.136.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleiman FE, Manley JL. Functional interaction of BRCA1-associated BARD1 with polyadenylation factor CstF-50. Science. 1999;285:1576–1579. doi: 10.1126/science.285.5433.1576. [DOI] [PubMed] [Google Scholar]
- Komarnitsky P, Cho EJ, Buratowski S. Different phosphorylated forms of RNA polymerase II and associated mRNA processing factors during transcription. Genes & Dev. 2000;14:2452–2460. doi: 10.1101/gad.824700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavigueur A, La Branche H, Kornblihtt AR, Chabot B. A splicing enhancer in the human fibronectin alternate ED1 exon interacts with SR proteins and stimulates U2 snRNP binding. Genes & Dev. 1993;7:2405–2417. doi: 10.1101/gad.7.12a.2405. [DOI] [PubMed] [Google Scholar]
- Lee TI, Young RA. Transcription of eukaryotic protein-coding genes. Annu Rev Genet. 2000;34:77–137. doi: 10.1146/annurev.genet.34.1.77. [DOI] [PubMed] [Google Scholar]
- Lewis JD, Izaurralde E, Jarmolowski A, Mcguigan C, Mattaj IW. A nuclear cap-binding complex facilitates association of U1-snRNP with the cap-proximal 5′ splice-site. Genes & Dev. 1996;10:1683–1698. doi: 10.1101/gad.10.13.1683. [DOI] [PubMed] [Google Scholar]
- Litingtung Y, Lawler AM, Sebald SM, Lee E, Gearhart JD, Westphal H, Corden JL. Growth retardation and neonatal lethality in mice with a homozygous deletion in the C-terminal domain of RNA polymerase II. Mol Gen Genet. 1999;261:100–105. doi: 10.1007/s004380050946. [DOI] [PubMed] [Google Scholar]
- McCracken S, Fong N, Rosonina E, Yankulov K, Brothers G, Siderovski D, Hessel A, Foster S, Shuman S, et al. Amgen EST Program. 5′-Capping enzymes are targeted to pre-mRNA by binding to the phosphorylated carboxy-terminal domain of RNA polymerase II. Genes & Dev. 1997a;11:3306–3318. doi: 10.1101/gad.11.24.3306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCracken S, Fong N, Yankulov K, Ballantyne S, Pan GH, Greenblatt J, Patterson SD, Wickens M, Bentley DL. The C-terminal domain of RNA polymerase II couples messenger RNA processing to transcription. Nature. 1997b;385:357–361. doi: 10.1038/385357a0. [DOI] [PubMed] [Google Scholar]
- McCracken S, Rosonina E, Fong N, Sikes M, Beyer A, O'Hare K, Shuman S, Bentley D. Role of RNA polymerase II carboxy-terminal domain in coordinating transcription with RNA processing. Cold Spring Harbor Symp Quant Biol. 1998;63:301–309. doi: 10.1101/sqb.1998.63.301. [DOI] [PubMed] [Google Scholar]
- Misteli T, Spector DL. RNA polymerase II targets pre-mRNA splicing factors to transcription sites in vivo. Mol Cell. 1999;3:697–705. doi: 10.1016/s1097-2765(01)80002-2. [DOI] [PubMed] [Google Scholar]
- Mortillaro MJ, Blencowe BJ, Wei XY, Nakayasu H, Du L, Warren SL, Sharp PA, Berezney R. A hyperphosphorylated form of the large subunit of RNA polymerase II is associated with splicing complexes and the nuclear matrix. Proc Natl Acad Sci. 1996;93:8253–8257. doi: 10.1073/pnas.93.16.8253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen VT, Giannoni F, Dubois MF, Seo SJ, Vigneron M, Kedinger C, Bensaude O. In vivo degradation of RNA polymerase II largest subunit triggered by alpha-amanitin. Nucleic Acids Res. 1996;24:2924–2929. doi: 10.1093/nar/24.15.2924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niwa M, Berget SM. Mutation of the AAUAAA polyadenylation signal depresses in vitro splicing of proximal but not distal introns. Genes & Dev. 1991;5:2086–2095. doi: 10.1101/gad.5.11.2086. [DOI] [PubMed] [Google Scholar]
- Niwa M, Rose SD, Berget SM. In vitro polyadenylation is stimulated by the presence of an upstream intron. Genes & Dev. 1990;4:1552–1559. doi: 10.1101/gad.4.9.1552. [DOI] [PubMed] [Google Scholar]
- Patturajan M, Schulte RJ, Sefton BM, Berezney R, Vincent M, Bensaude O, Warren SL, Corden JL. Growth-related changes in phosphorylation of yeast RNA polymerase II. J Biol Chem. 1998;273:4689–4694. doi: 10.1074/jbc.273.8.4689. [DOI] [PubMed] [Google Scholar]
- Ramchatesingh J, Zahler AM, Neugebauer KM, Roth MB, Cooper TA. A subset of SR proteins activates splicing of the cardiac troponin T alternative exon by direct interactions with an exonic enhancer. Mol Cell Biol. 1995;15:4898–4907. doi: 10.1128/mcb.15.9.4898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasmussen EB, Lis JT. In-vivo transcriptional pausing and cap formation on three Drosophila heat-shock genes. Proc Natl Acad Sci. 1993;90:7923–7927. doi: 10.1073/pnas.90.17.7923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rickert P, Corden JL, Lees E. Cyclin C/CDK8 and cyclin H/CDK7/p36 are biochemically distinct CTD kinases. Oncogene. 1999;18:1093–1102. doi: 10.1038/sj.onc.1202399. [DOI] [PubMed] [Google Scholar]
- Rodriguez CR, Cho EJ, Keogh MC, Moore CL, Greenleaf AL, Buratowski S. Kin28, the TFIIH-associated carboxy-terminal domain kinase, facilitates the recruitment of mRNA processing machinery to RNA polymerase II. Mol Cell Biol. 2000;20:104–112. doi: 10.1128/mcb.20.1.104-112.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schroeder S, Schwer B, Shuman S, Bentley D. Dynamic association of capping enzymes with transcribing RNA polymerase II. Genes & Dev. 2000;14:2435–2440. doi: 10.1101/gad.836300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smale ST, Tjian R. Transcription of herpes simplex virus tk sequences under the control of wild-type and mutant human RNA polymerase I promoters. Mol Cell Biol. 1985;5:352–362. doi: 10.1128/mcb.5.2.352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tacke R, Tohyama M, Ogawa S, Manley JL. Human Tra2 proteins are sequence-specific activators of pre-mRNA splicing. Cell. 1998;93:139–148. doi: 10.1016/s0092-8674(00)81153-8. [DOI] [PubMed] [Google Scholar]
- Takagaki Y, Manley JL. A human polyadenylation factor is a G protein beta-subunit homologue. J Biol Chem. 1992;267:23471–23474. [PubMed] [Google Scholar]
- ————— Complex protein interactions within the human polyadenylation machinery identify a novel component. Mol Cell Biol. 2000;20:1515–1525. doi: 10.1128/mcb.20.5.1515-1525.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vagner S, Vagner C, Mattaj IW. The carboxyl terminus of vertebrate poly(A) polymerase interacts with U2AF 65 to couple 3′-end processing and splicing. Genes & Dev. 2000;14:403–413. [PMC free article] [PubMed] [Google Scholar]
- Vincent M, Lauriault P, Dubois MF, Lavoie S, Bensaude O, Chabot B. The nuclear matrix protein p255 is a highly phosphorylated form of RNA polymerase II largest subunit which associates with spliceosomes. Nucleic Acids Res. 1996;24:4649–4652. doi: 10.1093/nar/24.23.4649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker S, Greaves R, O'Hare P. Transcriptional activation by the acidic domain of VMW65 requires the integrity of the domain and involves additional determinants distinct from those necessary for TFIIB binding. Mol Cell Biol. 1993;13:5233–5244. doi: 10.1128/mcb.13.9.5233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- West M, Corden J. Construction and analysis of yeast RNA polymerase II CTD deletion and substitution mutations. Genetics. 1995;140:1223–1233. doi: 10.1093/genetics/140.4.1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yankulov K, Todorov I, Romanowski P, Licatalosi D, Cilli K, McCracken S, Laskey R, Bentley DL. MCM proteins are associated with RNA polymerase II holoenzyme. Mol Cell Biol. 1999;19:6154–6163. doi: 10.1128/mcb.19.9.6154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yue Z, Maldonado E, Pilluta R, Cho H, Reinberg D, Shatkin A. Mammalian capping enzyme complements mutant Saccharomyces cerevisiae lacking mRNA guanylyltransferase and selectively binds the elongating form of RNA Pol II. Proc Natl Acad Sci. 1997;94:12898–12903. doi: 10.1073/pnas.94.24.12898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuryev A, Patturajan M, Litingtung Y, Joshi R, Gentile C, Gebara M, Corden J. The CTD of RNA polymerase II interacts with a novel set of SR-like proteins. Proc Natl Acad Sci. 1996;93:6975–6980. doi: 10.1073/pnas.93.14.6975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng C, Berget SM. Participation of the C-terminal domain of RNA polymerase II in exon definition during pre-mRNA splicing. Mol Cell Biol. 2000;20:8290–8301. doi: 10.1128/mcb.20.21.8290-8301.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]