Abstract
Many cancers escape host immunity without losing tumor-specific rejection antigens or MHC class I expression. This study tracks the evolution of one such cancer that developed in a mouse following exposure to ultraviolet light. The primary autochthonous tumor was not highly malignant and was rejected when transplanted into naïve immunocompetent mice. Neoplastic cells isolated from the primary tumor were susceptible to the growth-inhibitory effects of IFNγ in vitro, but expressed very low levels of MHC I antigen and were resistant to tumor-specific T cells unless they were first exposed to IFNγ. Serial passage of the primary tumor cells in vivo led to a highly aggressive variant that caused fast-growing tumors in normal mice. In vitro, the variant tumor cells showed increased resistance to the growth-inhibitory effects of IFNγ but expressed high levels of immunoproteasomes and MHC I molecules and were susceptible to tumor-specific T cells even without prior exposure to IFNγ.
Keywords: mice, cultured tumor cells, melanoma, MHC, IFN-gamma, cytotoxic T lymphocytes, immunoproteasome
Introduction
Tumor progression is the continuous increase in malignant behavior and is typical for cancers of mice and men (1-3). Clear and most dramatic examples are the development of progressor variants from regressor tumors (4, 5). Regressor tumor cells at any challenge dose are rejected by normal naïve hosts (6), except for infrequent variants which arise from the original tumor (4, 5). These variants have always been found to be due to heritable changes in the cancer cells within the same lineage (7-9). Aggressively growing cancers may lack T cell-recognized target antigens, show altered proteasomal processing and/or have reduced expression of MHC I molecules (for review see 10), suggesting the power of adaptive T cell immunity in cancer surveillance. This notion is reinforced by experiments showing that a mixture of anti-CD8, -CD4 and -IFNγ antibodies promoted the outgrowth of tumors in euthymic mice injected subcutaneously (s.c.) with methylcholanthrene (11). However, detailed analyses of progressor variants developing from UV-induced regressor tumors cells have shown that the majority of variants evolved without loss of target antigen or MHC expression. Understanding principle mechanisms involved in the progression of these relatively frequent antigen-retention variants should help develop better, mechanistically sound approaches for eradicating either earlier or advanced stages of cancers. Improving this understanding was the goal of the current study.
Results
Derivation of 4102 primary and variant cell lines
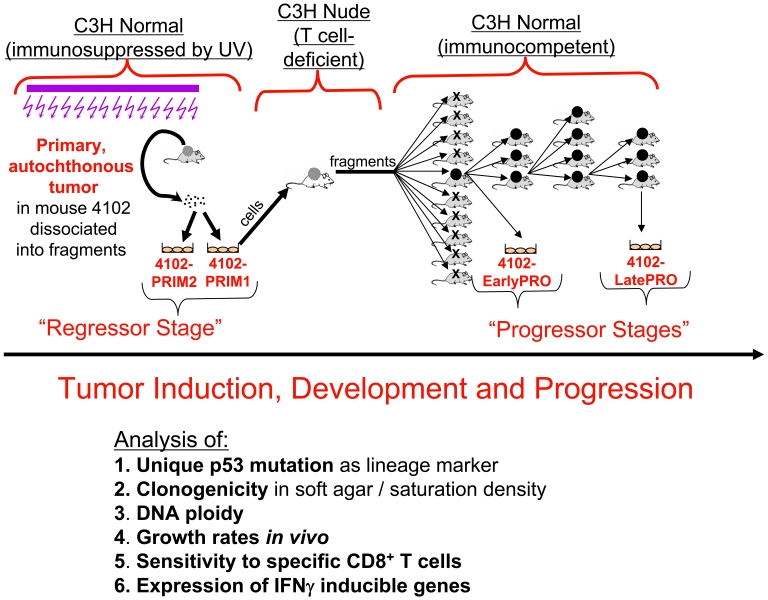
The primary 4102 tumor was induced in a C3H mouse by chronic UV irradiation (Figure 1). Two fragments of the primary tumor were cultured briefly (for less than 20 days) to establish the PRIM1 and PRIM2 cell cultures and aliquots of these cell populations were stored in liquid nitrogen for subsequent analyses. A suspension of PRIM1 cells caused a tumor in a nude C3H mouse. When fragments of this tumor were subsequently transplanted, nine of ten normal C3H mice rejected the tumor challenge. The only tumor that developed in a normal mouse was adapted to culture as EarlyPRO cells and was serially transplanted to normal mice for three additional passages. In each transplant generation, all of the normal mice developed progressive lethal tumors, indicating that the EarlyPRO cells had already acquired a heritably stable progressor phenotype. After three successive transplantations, the growing tumor was adapted to culture and cloned to generate LatePRO. In a repeat experiment, the same frequency of progressor variant developed when normal mice were challenged with tumor fragments generated in nude mice by injection of PRIM1 cancer cells: in 5 mice challenged bilaterally with tumor fragments from the nude mouse, only 1 of the 10 inocula developed a tumor.
Figure 1.
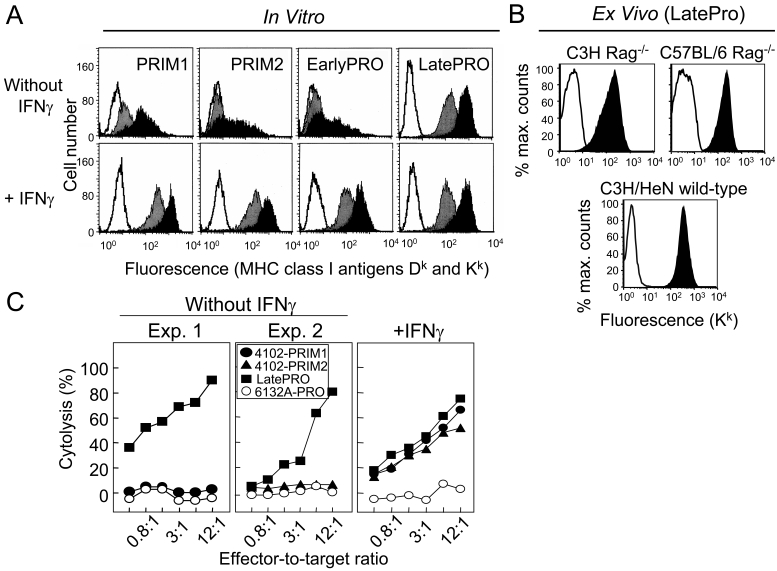
Derivation of the 4102 tumor cell lines. The primary 4102 tumor arose in a C3H mouse chronically exposed to UV light. Two separate fragments were adapted to culture to generate the PRIM1 and PRIM2 cell lines. The PRIM1 cell line was used to generate more progressive tumors by serial passage first in a C3H nude mouse and then in immunocompetent mice. In the first transplant generation, the tumor grew progressively in only one of ten normal mice; in subsequent transplant generations, the tumor grew progressively in every mouse. The LatePRO cell line was established after the fourth transplant generation.
The 4102 cell lines share a unique tumor-specific p53 mutation
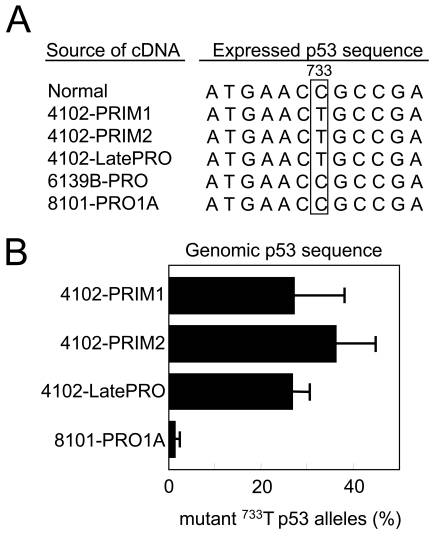
Mutations in p53 are one of the earliest genetic changes associated with human UV-induced skin carcinogenesis and are found in the earliest premalignant UV-induced lesions (12). We have shown that unique p53 mutations are convenient and reliable markers to track the origin of tumor cell lines (9). To confirm that the 4102 tumor cell lines are derived from a common precursor, the expressed p53 sequences from PRIM1, PRIM2, and LatePRO were analyzed by RT-PCR followed by direct sequencing. PCR primers were designed to span exons 5 through 8 to include the majority of p53 mutation hot spots. All of the sequences derived from the 4102 tumor cell lines contained a C to T transition mutation that changed amino acid 245 from arginine to cysteine (Figure 2A). Arg245 in the mouse p53 protein is equivalent to Arg248 in the human p53 protein, which is one of the most commonly mutated residues associated with familial Li-Fraumeni syndrome (13). This mutation was neither found in the heart lung fibroblasts from the mouse that developed the primary 4102 tumor nor in other independently derived tumor cell lines. No other mutation was detected within this region (data not shown).
Figure 2.
The 4102 cell lines share a common point mutation in the p53 gene. (A) p53 was sequenced from cDNA generated by RT-PCR using primers spanning exons 5 through 8, which contain the majority of mutation hot spots. The expressed p53 sequence in the 4102 cell lines shares a common C to T point mutation in exon 7, resulting in an amino acid substitution from Arg to Cys at position 245. All of the p53 cDNA sequences examined contained this point mutation, suggesting that the wild type sequence was not expressed. The independently generated tumors 6139B-PRO and 8101-PRO do not have this mutation. (B) The primary 4102 cell lines and the 4102-LatePRO clone contained a similar percentage of p53 alleles with the lineage-specific mutation. The number of wild type and mutant p53 sequences in each cell line was quantified by real-time PCR using PCR primers specific for the wild type or the mutant p53 sequence. The results were expressed as a percentage of mutant p53 out of total genomic p53. The specificity of the assay is reflected by the near-absence of mutant p53 sequence in the unrelated 8101-PRO1A tumor cell line. These results suggest that the primary 4102 cell lines are composed homogenously of tumor cells.
To determine whether all or only some of the primary tumor cells have the 4102-specific p53 mutation, genomic DNA from the tumor cell lines was analyzed by real-time PCR. The analysis was performed using antisense primers specific for either the wild-type (C) or mutated (T) base in exon 7 with a sense primer specific for intron 6, chosen to prevent amplification of the p53 pseudogene. The wild type- and mutant-specific primer combinations specifically amplified p53 cDNAs with cytosine or thymine at position 733, respectively (data not shown). In addition, the mutation-specific primer failed to amplify the p53 sequence from the unrelated 8101-PRO1A tumor cell line lacking the C to T mutation. Approximately 25% of the amplified p53 sequences from PRIM1, PRIM2, and LatePRO were mutated (Figure 2B). As shown in Figure 3A, these tumor cells are near tetraploid and therefore likely contain 3 non-mutant alleles in addition to the mutant allele, although the wild type sequences were not expressed (Figure 2A). This result and the fact that only one p53 mutation was found suggest that all of the 4102 tumor cells developed within a single lineage.
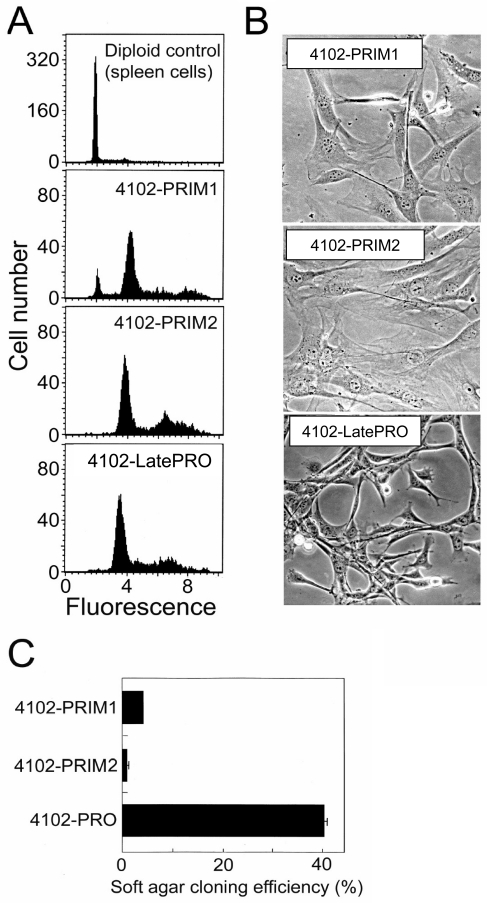
Figure 3.
Phenotype of the primary 4102 tumor cells in vitro. (A) 4102 cell lines have abnormal DNA content. Cells fixed and permeabilized with ethanol were stained with propidium iodide and analyzed for DNA content by flow cytometry. Normal spleen cells were used as a diploid control. The PRIM2 and LatePRO cell lines were composed mainly of tetraploid cells and the PRIM1 cell line contained in addition 8% near diploid cells. (B) Morphology of 4102 tumor cells in culture. The primary tumor cells were very similar in appearance to normal fibroblasts in culture, and were easily distinguished from the LatePRO cells by their large size, flatness, and low refractivity. (C) The PRIM1 and PRIM2 cell lines failed to form colonies in soft agar. Clonogenicity of the tumor cell lines was determined by overlaying 100 tumor cells in 0.21% agarose atop a 0.26% agarose base layer and culturing for 10 days. The results are represented as the average cloning efficiency in four wells, calculated using the formula: cloning efficiency = (colonies greater than or equal to 0.05 mm in diameter / total colonies) x 100.
The 4102 primary cell lines are poorly tumorigenic
PRIM1 and PRIM2 were characterized for DNA content, morphology in culture, clonogenicity in soft agar, and growth in nude and normal mice. The 4102 cell lines were composed mainly of tetraploid cells; PRIM1 contained in addition 8% near-diploid cells (Figure 3A) that are most likely tumor cells that had not duplicated their genome but could also be contaminating fibroblasts. PRIM1 and PRIM2 tumor cells appeared very similar to normal fibroblasts in their large size, low refractivity, flat cytoplasm and tight attachment to plastic (Figure 3B). The primary tumor cells also failed to form full size colonies in soft agar, demonstrating their strong anchorage dependence (Figure 3C); after 10 days in culture, most cells appeared viable but had undergone only two or three cell divisions. In contrast, LatePRO grew as small, highly refractive cells with arched-up cytoplasm, often piling up on each other (Figure 3B). They were also much less adherent to plastic and formed colonies effectively in soft agar, demonstrating that LatePRO had acquired anchorage-independent growth.
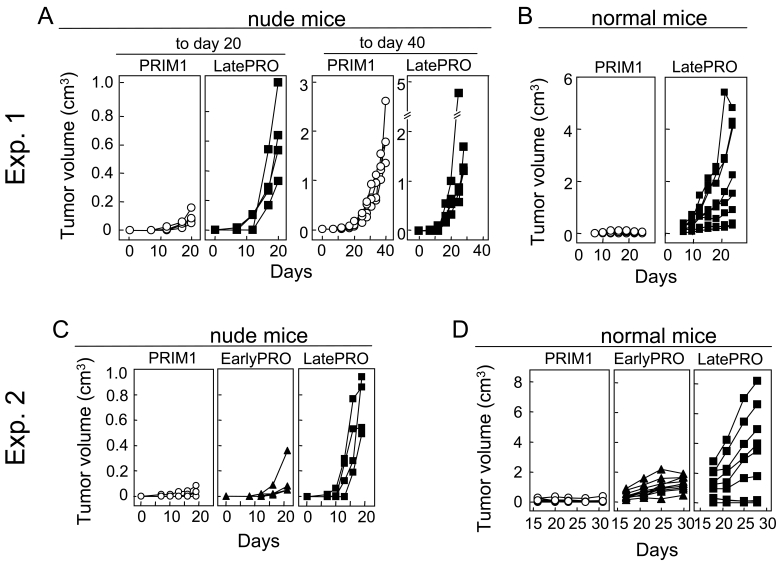
A suspension of PRIM1 cells injected s.c. caused tumors in nude mice that grew slowly but progressively to 1-3 cm3 after 40 days (Figure 4, panels A and C). When fragments of these tumors were transplanted, 6 of 7 normal mice rejected the tumor challenge (Figure 4, panels B and D). EarlyPRO tumors, which represent the next step in tumor progression, grew at a similar slow rate in nude mice (Figure 4C) but none of the transplanted tumor fragments were rejected by normal mice (Figure 4D). The LatePRO tumors were the most aggressive overall; they were not only the fastest growing tumors in nude mice (Figure 4, panels A and C), but nearly all of the normal mice transplanted with tumor fragments developed progressive tumors that were 2-3 times larger than the EarlyPRO tumors over the same time frame (Figure 4D). Together, our results show that 4102 primary tumor cells are indeed an early neoplastic population that still retained some of the morphological and functional characteristic of nontransformed cells and showed a long latency before outgrowth even in T cell-deficient mice.
Figure 4.
Primary 4102 tumor cells are tumorigenic in immunodeficient nude mice but not in normal mice. (A and C) Nude C3H/HeN mice were injected with 105 PRIM1, EarlyPRO or LatePRO tumor cells. The PRIM1 and EarlyPRO tumors developed slowly and the mean tumor volume 20 days after injection was only one fifth of the LatePRO tumors. However, they continued to grow progressively to reach a similar size as the LatePRO. (B and D) Normal C3H mice were injected with approximately 0.3 cm3 of PRIM1, EarlyPRO and LatePRO tumor fragments isolated from the nude C3H/HeN mice used in panel A and C. Fragments of the PRIM1 tumor were rejected by normal C3H mice while EarlyPRO and LatePRO tumor fragments grew progressively and killed the recipients.
The 4102 primary cells express low constitutive levels of MHC I molecules and are resistant to cytolysis by tumor-specific CTLs
PRIM1 and PRIM2 tumor cells expressed low levels of MHC I molecules H-2Kk and H-2Dk (Figure 5A, upper panels). In order to compare MHC I expression between the 4102 primary tumor cells and normal keratinocytes, a suspension of PRIM2 was injected s.c. into C3H Rag-2-/- mice to induce a tumor with an overlying layer of normal keratinocytes. Immunohistochemical analyses showed that H-2Kk was undetectable on PRIM2 tumor cells, but expressed abundantly on the adjacent normal keratinocytes (Supplementary Figure 1). A similar approach was used to examine MHC I expression on the LatePRO tumor cells in vivo. LatePRO tumors were removed from tumor-bearing mice more than 2 weeks after inoculation when the tumors were >1 cm in average diameter. Single cell suspensions were prepared and analyzed immediately for MHC I expression by ex vivo flow cytometry. Figure 5B shows that the high level MHC I expression on the LatePRO tumor cells in vitro (Figure 5A) was maintained in growing tumors in both immunodeficient and immunocompetent hosts.
Figure 5.
4102 primary cells express low constitutive levels of MHC I molecules and are resistant to cytolysis by tumor-specific CTLs. (A) Expression of MHC I molecules in vitro by 4102 cancer cells. Primary tumor cells grown in vitro were incubated with or without 100 U/ml recombinant IFNγ for 2 days. The cells were analyzed by flow cytometry after being incubated with supernatants from hybridomas specific for H-2Dk (grey) or H-2Kk (black) followed by goat anti-mouse Ig-FITC. Control cells were stained with only the second-step antibody (unshaded). (B). Expression of MHC I Kk molecules on LatePRO tumor cells in vivo. LatePRO tumor cells were isolated from growing tumors more than 14 days after inoculation and greater than or equal to 1 cm in diameter. Top panels: EGFP-transduced LatePRO tumor cells recovered from immunodeficient C3H Rag-2-/- mice and C57BL/6 Rag-1-/- mice. The tumor cells were identified as the EGFP-positive population. Bottom panel: Untransduced LatePRO tumor cells recovered from normal immunocompetent C3H mice. The tumor cells were identified as the CD45-negative population. 4102 tumor cell suspensions were directly stained with anti-Kk (black) or isotype control (unshaded) antibodies and analyzed for MHC I expression by ex vivo flow cytometry. (C) Primary 4102 tumor cells that express low levels of MHC I are resistant to cytolysis by tumor-specific CTLs unless first exposed to IFNγ. Tumor cells were incubated with or without 100 U/ml recombinant IFNγ for 2 days prior to use as 51Cr-labeled targets in a 4.5 h cytolysis assay. The CTL line was previously shown to recognize an unique tumor antigen expressed only on tumors of the 4102 lineage. Without IFNγ treatment, PRIM1 and PRIM2 were poorly lysed (left panel). However, after IFNγ treatment, these cell lines were lysed as effectively as LatePRO by the antigen-specific CTL (right panel), indicating the presence of the unique tumor antigen. 6132A-PRO is an independently generated UV-induced tumor that does not express the 4102 tumor-specific antigen.
To determine whether the low level of MHC I expression on the primary tumor cells prevents killing by a tumor-specific CTL line, PRIM1 and PRIM2 were used as targets in 4.5 h chromium release assays. We had previously generated a CTL line that recognizes an unique tumor antigen expressed on LatePRO (5). This cloned CTL line very effectively lysed LatePRO but not PRIM1, PRIM2, EarlyPRO and the independently generated H-2k tumor cells 6132A-PRO (Figure 5C, left and middle panels). IFNγ treatment increased MHC I expression on the primary tumor cells by 50-fold (Figure 5A, lower panels) as well as CTL-mediated cytolysis (Figure 5C, right panel), suggesting that the primary tumor cells expressed the lineage-specific tumor antigen but were not recognized by the CTL due to insufficient antigen processing and/or presentation. Since this assay was done in vitro with a cloned T cell line, we can exclude an influence of tumor-infiltrating regulatory T cells, myeloid-derived suppressor cells (MDSC), immunosuppressive effects of the tumor stroma, or other mechanisms that may interfere with the effector phase in vivo and could have caused resistance of the PRIM cells.
Given the lack of antigen presentation, rejection of the transplanted PRIM cells by naïve T cell-competent mice (Figure 4, panels B and D) cannot be easily explained. It is possible that s.c. transplantation induced an artificial inflammatory response and adjuvant effect that transiently upregulated presentation of the rejection antigen; this in turn induced a primary T cell response that eliminated the PRIM inoculum. Induction of memory response is known to require a more sustained antigen exposure. Therefore, we determined next whether rejection of PRIM cells protected against a subsequent lethal challenge with LatePRO. Interestingly, we found that rejection of PRIM cells failed to induce protective immunity against a lethal LatePRO challenge in 3 out of 4 cases (data not shown). Thus, mice injected with early stage tumor cells may not be primed sufficiently to generate a protective memory response against LatePRO.
IFNγ suppresses proliferation and colony formation of 4102 primary but not late-stage variant cells
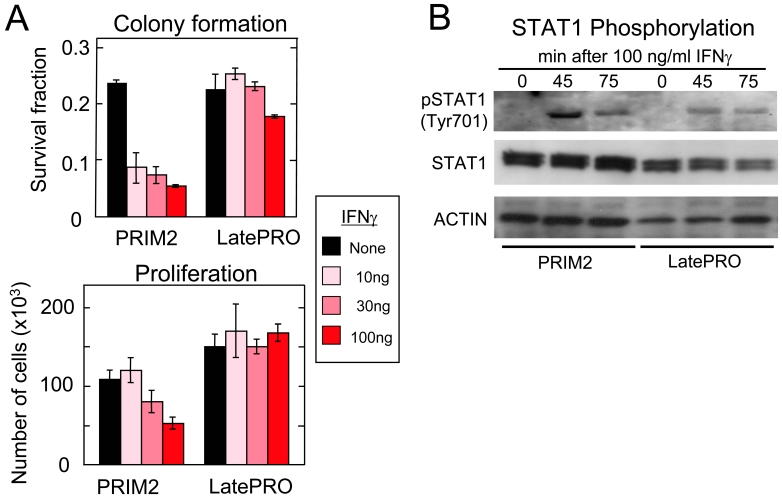
Since IFNγ enhanced MHC I expression and CTL-mediated cytotoxicity of the early but not the late 4102 lineage cancer cells, we investigated the effect of IFNγ on the growth of the cancer cells in vitro. Figure 6A shows that IFNγ inhibited both colony formation and proliferation of the PRIM2 primary tumor cells but not of the LatePRO tumor cells. The failure of IFNγ to inhibit growth or further increase MHC I expression on the LatePRO cancer cells (Figure 5A) could be due to failure of LatePRO tumor cells to signal through IFNγR/Stat1 after exposure to IFNγ. However, Figure 6B shows that LatePRO cells expressed similar levels of STAT1 and pSTAT1 phosphorylated at tyrosine residue 701 as those observed in the primary cancer cells exposed to IFNγ.
Figure 6.
IFNγ suppresses colony formation and proliferation of primary 4102 tumor cells but not of the late stage variant. (A) Analysis of the effects of IFNγ on colony formation and proliferation of various 4102 lineage cells in vitro. (B) Analysis of the phosphorylation of STAT1 of various 4102 lineage cells. Cells were treated by IFNγ and lysed after 0, 45 and 75 min of treatment. Proteins were separated in two parallel gels and transferred onto PVDF membranes for detection of phosphorylated STAT1 (pSTAT1) and total STAT1 (STAT1). Actin was used as internal loading control. Data show that administration of IFNγ leads to Tyr701 phosphorylation of STAT1 in both PRIM2 and LatePRO, indicating intact IFNγR/Jak1 signaling system in both primary and late progressor tumors.
Changes in TAP expression and proteasome composition and specificity
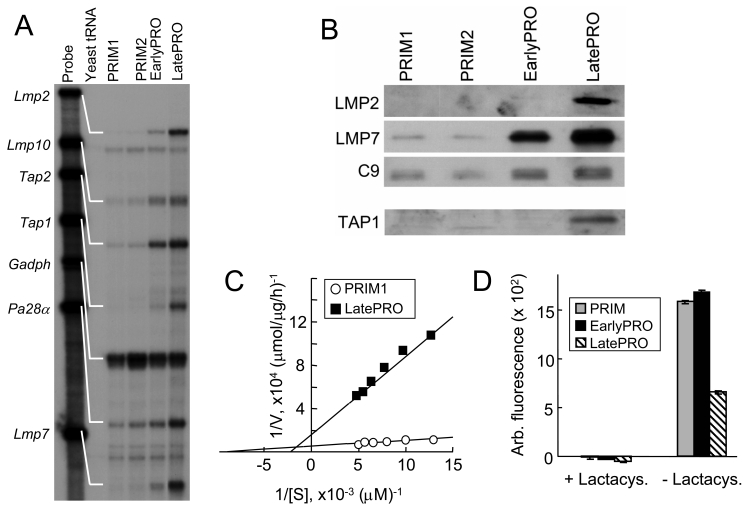
MHC I expression can be increased by the coordinated upregulation of the transporter associated with antigen presentation TAP-1 and -2, the proteasome subunits LMP2, LMP7, and LMP10 and the proteasome activator PA28 α and β subunits. Constitutively elevated expression of these genes in the LatePRO tumor cells may have led to the increased MHC I expression. Indeed, RNase protection assays showed that these genes were transcribed at low levels in the PRIM tumor cells and increased as the tumor progressed to EarlyPRO and then to LatePRO (Figure 7A). Consistent with the mRNA quantification, the amount of LMP2 and LMP7 proteins incorporated into functional proteasomes and the amount of TAP-1 protein also increased with tumor progression (Figure 7B).
Figure 7.
LatePRO tumor cells express higher levels of IFNγ-inducible genes involved in MHC I antigen processing and presentation and show cleavage specificity consistent with immunoproteasome activity. (A) RNase protection assay showed that transcription of Tap1, Tap2, Lmp2, Lmp7, Lmp10 and Pa28α is up-regulated in EarlyPRO and further increased in the LatePRO cancer cells. (B, rows 1-3) Proteasomes isolated from LatePRO tumor cells by differential centrifugation contain higher abundance of immunoproteasome subunits LMP2 and LMP7. C9 is a structural subunit found in all proteasomes and was therefore used to equalize the amount of proteasomes from different cell preparations for Western blot analyses. (B, row 4) LatePRO tumor cells also express higher level of TAP-1 protein. (C) LatePRO tumor cells exhibit increased immunoproteasome activity. Hydrolysis of the chymotrypsin-like fluorescent substrate LLVY-MCA was used to measure immunoproteasome activity. Proteasomes were enriched from PRIM1 and LatePRO tumor cell lysates by differential centrifugation. 1 µg of each preparation was incubated with 25-200 µM LLVY-MCA for 1 h. The reduced hydrolysis of LLVY-MCA suggested that proteasomes isolated from LatePRO tumor cells were predominately immunoproteasomes. (D) To confirm that the hydrolysis was mediated by proteasomes, proteasomes were incubated with 10 µM lactacystin, a proteasome-specific inhibitor, for 1 h before the hydrolysis assay was performed.
There are two types of proteasomes which are characterized by either the presence of LMP2, LMP7, and LMP10 (immunoproteasomes) or the presence of homologous, constitutive subunits Y, X, and Z (constitutive proteasomes). The results described above suggested that progression of the 4102 tumor lineage is associated with a shift in proteasome composition toward increasing amounts of immunoproteasomes. Since the proteasome composition can affect cleavage site specificity (14-16), proteasomes were prepared by differential centrifugation and incubated with the tetrapeptide substrate LLVY-MCA to measure the chymotrypsin-like hydrolytic activity typically associated with the constitutive proteasome. As expected, proteasomes isolated from PRIM1 tumor cells exhibited higher hydrolysis of LLVY-MCA compared to LatePRO proteasomes (Figure 7C). The observed hydrolytic activity was mediated by proteasomes because it was completely abrogated by pretreatment with the proteasome-specific inhibitor lactacystin (Figure 7D). The measured Km for proteasomes from PRIM1 and LatePRO were 104 µM and 453 µM, respectively (Figure 7C). Similar results were observed with PRIM2 and EarlyPRO (data not shown).
Discussion
With increased exposure to UV light linked to ozone depletion of the atmosphere, the incidence of skin cancer has reached epidemic proportions (17) with now one in five Americans developing skin cancer in their life times (18). In addition, immunosuppressed solid organ transplant recipients have a 65- to 250-fold increased risk of developing skin cancer (19). These cancers are often more aggressive in these patients and kill 7-9% of them due to metastases (for review see 20). The increased incidence in transplant patients is linked to suppressed T cell function, and two thirds of these patients experience reduced skin cancer development after immunosuppressive medications are stopped (21). It appears therefore that many of these human UV-induced cancers are actually "regressor" tumors and have arisen only because host defenses had been damaged. Developing specific T cell probes and longitudinal analysis of the stepwise progression of a skin tumor in patients is not feasible. Fortunately, there is a striking homology with skin cancers induced experimentally in mice by UV irradiation, since these tumors are often rejected when transplanted into normal immunocompetent mice (5, 22). The study of these UV-induced regressor tumors provides important clinically relevant insight into what happens in humans, and how to prevent or reverse the process.
Premalignant or early neoplastic cells, even when isolated from the earliest detectable lesion, may still represent a rather heterogeneous population (23) that undergoes rapid changes in their properties once expanded in culture or transplanted in vivo. The act of observing may therefore change the phenomenon being observed, also referred to as "observer effect". Our current study has attempted to minimize these problems by isolating neoplastic cells directly from the original autochthonous UV-induced tumor and using a lineage-specific p53 mutation and a CTL-recognized tumor antigen to establish that they were the precursors of the highly malignant cells.
We expected that the early neoplastic cells would be highly susceptible to killing by tumor-specific T cells. Surprisingly, they expressed very low levels of MHC I molecules and were not recognized by tumor-specific T cells in vitro unless pre-exposed to IFNγ. These results suggest that the autochthonous host may never have generated antigen-specific CTLs against the 4102 primary tumor because there was not enough IFNγ produced at the tumor site to induce sufficient presentation of the tumor antigen. Indeed, UV-irradiation induces various immune alterations (24, 25). Inhibition of IFNγ production (26, 27) and IFNγ responsiveness by the cells targeted by the transforming agent (27) appears to be critical for promoting primary UV-induced tumor development. In addition, responses of the host stroma to IFNγ and IFNγ production by innate and adoptive immunity have also been reported to have major effects in preventing tumor induction by certain carcinogens such as 3-methylcholanthrene (28-30).
The rejection of transplanted 4102 primary tumor cells by normal mice suggests tumor recognition can be improved by an inflammatory response such as that induced artificially by tumor transplantation (31), which is invariably associated with local production of type I and type II interferons. Consistent with this idea, topical application of Imiquimod, a TLR7 agonist, can induce immunological destruction of premalignant actinic keratoses and basal and squamous carcinomas (32, 33). Imiquimod also enhances IFNγ production and T cell effector function when topically applied to UV-induced skin cancers in transplant recipients (34). Furthermore, cervical intraepithelial neoplasia (CIN), which is associated with a high risk of developing cervical cancer, consistently have an allele-specific or general downregulation of MHC I molecules (35, 36). In addition, intracervical injections of IFNγ caused upregulation of MHC I alleles and correlated with clearance of the cervical intraepithelial lesions in patients (37).
Since the genes encoding LMP2, LMP7, TAP-1 and TAP-2 are all located in the MHC locus on chromosome 17, we tested whether translocation involving chromosome 17 could explain the upregulation of the immunoproteasome components LMP2 and LMP7 and was therefore directly responsible for, or contributed to, the increased MHC I expression on the LatePRO tumor cells. Indeed, 16 of 17 metaphase spreads of the LatePRO cells contained translocations involving chromosome 17 fused to chromosome 4 and to other as yet unidentified chromosomes, while no translocations were found in metaphase spreads of PRIM1 and PRIM2, even though these cancer cells were also near tetraploid and contained 3 to 4 copies of chromosome 17. Whether these translocations increased expression of genes within the MHC gene cluster remains unclear. However, upregulation of Lmp10 and Pa28α, which are localized to chromosome 8 (38) and 14 (39), respectively, point to a more global effect such as cellular oxidative stress response (40, 41) or a constitutively activated IFNγ signaling pathway. The latter is consistent with the resistance of LatePRO tumor cells to the growth inhibitory effects of IFNγ in vitro. We showed previously that tumor clones overexpressing IFNγ/Stat1-dependent genes both in immunocompromised (42) and immunocompetent mice (43) were resistant to IFNγ and genotoxic insult without disruption of the upstream signaling system. These effects were Stat1-dependent because enforced expression of Stat1 led to acquisition of IFNγ resistance and suppression of Stat1 led to increased sensitivity to genotoxic stress and suppression of aggressive tumor growth.
A challenge for future investigations will be the elimination of late stage aggressive cancers such as the LatePRO. LatePRO tumor cells exhibited a phenotype that should have led to tumor rejection: they constitutively expressed high levels of MHC I molecules and were highly susceptible to cytolysis by tumor-specific T cells in vitro without prior exposure to IFNγ. At present, we do not know whether LatePRO tumors grow because of a priming and/or effector phase defect. As antigen is key (44), we are working to express a defined tumor-specific antigen in LatePRO that will allow us to measure the anti-tumor T cell response and the influence of the tumor stroma on antigen priming and the effector phase. We previously reported that the aggressive behavior of LatePRO is associated with a paracrine stimulatory loop that recruits inflammatory cells and induces tumor stroma which, when inhibited, converts the progressor to a regressor phenotype in euthymic mice (45, 46). We believe that signals from the tumor cells are critical to initiate and maintain the paracrine stimulatory loop. For example, LatePRO expresses high levels of the potent monocyte chemoattractant MCP-1 (45) which can enhance tumor progression and angiogenesis (47, 48). Production of these critical cytokines and chemokines by LatePRO may well require the immunoproteasome. Our hypothesis originates from the facts that (i) in an experimental colitis model, lmp7-/- mice have severely reduced production of IL-6, RANTES, MCP-1 (CCL-2), KC and other proteins that may promote tumor growth (49); (ii) in response to LPS-stimulation, primary macrophages from lmp7 and mecl-1 double knockout mice have reduced transcription of IL-1 and IL-6 (50), with the latter having been implicated as a tumor promoter in colitis-associated cancer development (51); and (iii) selective inhibition of LMP7 reduced cytokine and chemokine production and progression of experimental arthritis (52). Thus, we plan to test this hypothesis using non-specific proteasome inhibitors such as bortezomib, as well as less toxic inhibitors that specifically target LMP7 in immunoproteasomes (53). We expect that inhibiting LMP7 activity in LatePRO cancer cells will reduce the recruitment of stromal progenitors, paracrine stimulation and intratumoral immune suppression. Furthermore, selective inhibition of LMP7 has been shown to induce apoptosis of primary multiple myeloma cells and cell lines (54). Another potential therapeutic target is the proteasome activator PA28γ, also known as gankyrin. This protein was initially identified as the third member of the PA28 family, but it is restricted to the nucleus and not co-regulated with the PA28 α and β subunits (55). Gankyrin is commonly upregulated in many important human cancers including hepatocellular, pancreatic and colorectal cancer (56) and plays an important role in tumorigenesis by accelerating proteasomal degradation of p53 and Rb tumor suppressor genes (56). Suppression of gankyrin reduced the oncogenic potential of several types of tumors in vitro and in vivo (57).
It is important to point out that most studies with proteasome inhibitors focused on their pro-apoptotic or anti-inflammatory effects. However, it is unlikely that the LatePRO tumor can be eradicated without the participation of tumor-specific T cells. Thus, it is important to consider that inhibition of proteasomes may also affect the repertoire of peptides presented by the tumor cells and the quality of the host immune response (58). On one hand, it is possible that the 4102 tumor antigen is generated independently of immunoproteasomes and selective inhibition may sensitize the tumor cells to cytolysis by CTLs (59); on the other hand, inhibiting components of the immunoproteasome could have an undesirable impact on the repertoire of tumor-derived peptides that are dependent on immunoproteasome. Thus, it is possible that selective inhibition of immunoproteasomes results in loss of the 4102 rejection tumor antigen and poor tumor cell recognition (60, 61). Therefore, even if the proteasome inhibitors delay tumor progression in T cell-deficient mice, it will be necessary to determine whether they negatively influence T cell-mediated destruction of the cancer cells and eventually prevent eradication of the cancer.
In summary, our studies characterized the stepwise progression of an antigenic tumor and showed that different mechanisms work to prevent tumor cell recognition and elimination throughout this process. This can be a major obstacle to effective immunotherapy against established tumors which contain a heterogeneous population of tumor cells, all at different stages of progression. Therefore, we propose that a multi-pronged approach, such as one combining induction of IFNγ at the tumor site with inhibition of immunoproteasome activity, may render the tumor cells and stroma more susceptible for destruction by tumor-specific T cells.
Acknowledgements
This work was supported by National Institute of Health grants R01-CA37516, P01-CA74182 and R01-CA2267, by University of Chicago Cancer Center Grant CA-14599, and by the Ludwig Institute for Cancer Research.
References
- 1.Rous P, Beard JW. The progression to carcinoma of virus-induced rabbit papillomas (Shope). J Exp Med. 1935;62:523–528. doi: 10.1084/jem.62.4.523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Foulds L. The experimental study of tumor progression. A review. Cancer Res. 1954;14:327–339. [PubMed] [Google Scholar]
- 3.Nowell PC. The clonal evolution of tumor cell populations. Science. 1976;194:23–29. doi: 10.1126/science.959840. [DOI] [PubMed] [Google Scholar]
- 4.Urban JL, Burton RC, Holland JM, Kripke ML, Schreiber H. Mechanisms of syngeneic tumor rejection. Susceptibility of host-selected progressor variants to various immunological effector cells. J Exp Med. 1982;155:557–573. doi: 10.1084/jem.155.2.557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ward PL, Koeppen H, Hurteau T, Schreiber H. Tumor antigens defined by cloned immunological probes are highly polymorphic and are not detected on autologous normal cells. J Exp Med. 1989;170:217–232. doi: 10.1084/jem.170.1.217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kripke ML. Antigenicity of murine skin tumors induced by ultraviolet light. J Natl Cancer Inst. 1974;53:1333–1336. doi: 10.1093/jnci/53.5.1333. [DOI] [PubMed] [Google Scholar]
- 7.Dubey P, Hendrickson RC, Meredith SC, Siegel CT, Shabanowitz J, Skipper JC, Engelhard VH, Hunt DF, Schreiber H. The immunodominant antigen of an ultraviolet-induced regressor tumor is generated by a somatic point mutation in the DEAD box helicase p68. J Exp Med. 1997;185:695–705. doi: 10.1084/jem.185.4.695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Beck-Engeser GB, Monach PA, Mumberg D, Yang F, Wanderling S, Schreiber K, Espinosa R 3rd, Le Beau MM, Meredith SC, Schreiber H. Point mutation in essential genes with loss or mutation of the second allele: relevance to the retention of tumor-specific antigens. J Exp Med. 2001;194:285–300. doi: 10.1084/jem.194.3.285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Schreiber K, Wu TH, Kast WM, Schreiber H. Tracking the common ancestry of antigenically distinct cancer variants. Clin Cancer Res. 2001;7:871s–875s. [PubMed] [Google Scholar]
- 10.Marincola FM, Jaffee EM, Hicklin DJ, Ferrone S. Escape of human solid tumors from T-cell recognition: molecular mechanisms and functional significance. Adv Immunol. 2000;74:181–273. doi: 10.1016/s0065-2776(08)60911-6. [DOI] [PubMed] [Google Scholar]
- 11.Koebel CM, Vermi W, Swann JB, Zerafa N, Rodig SJ, Old LJ, Smyth MJ, Schreiber RD. Adaptive immunity maintains occult cancer in an equilibrium state. Nature. 2007;450:903–907. doi: 10.1038/nature06309. [DOI] [PubMed] [Google Scholar]
- 12.Ziegler A, Jonason AS, Leffell DJ, Simon JA, Sharma HW, Kimmelman J, Remington L, Jacks T, Brash DE. Sunburn and p53 in the onset of skin cancer. Nature. 1994;372:773–776. doi: 10.1038/372773a0. [DOI] [PubMed] [Google Scholar]
- 13.Hollstein M, Sidransky D, Vogelstein B, Harris CC. p53 mutations in human cancers. Science. 1991;253:49–53. doi: 10.1126/science.1905840. [DOI] [PubMed] [Google Scholar]
- 14.Driscoll J, Brown MG, Finley D, Monaco JJ. MHC-linked LMP gene products specifically alter peptidase activities of the proteasome. Nature. 1993;365:262–264. doi: 10.1038/365262a0. [DOI] [PubMed] [Google Scholar]
- 15.Gaczynska M, Rock KL, Goldberg AL. Gamma-interferon and expression of MHC genes regulate peptide hydrolysis by proteasomes. Nature. 1993;365:264–267. doi: 10.1038/365264a0. [DOI] [PubMed] [Google Scholar]
- 16.Boes B, Hengel H, Ruppert T, Multhaup G, Koszinowski UH, Kloetzel PM. Interferon gamma stimulation modulates the proteolytic activity and cleavage site preference of 20S mouse proteasomes. J Exp Med. 1994;179:901–909. doi: 10.1084/jem.179.3.901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cancer Facts & Figures 2009. http://www.cancer.org/Research/CancerFactsFigures/CancerFactsFigures/cancer-facts-figures-2009
- 18.Robinson JK. Sun exposure, sun protection, and vitamin D. JAMA. 2005;294:1541–1543. doi: 10.1001/jama.294.12.1541. [DOI] [PubMed] [Google Scholar]
- 19.Bouwes Bavinck JN, Hardie DR, Green A, Cutmore S, MacNaught A, O'Sullivan B, Siskind V, Van Der Woude FJ, Hardie IR. The risk of skin cancer in renal transplant recipients in Queensland, Australia. A follow-up study. Transplantation. 1996;61:715–721. doi: 10.1097/00007890-199603150-00008. [DOI] [PubMed] [Google Scholar]
- 20.Berg D, Otley CC. Skin cancer in organ transplant recipients: Epidemiology, pathogenesis, and management. J Am Acad Dermatol. 2002;47:1–20. doi: 10.1067/mjd.2002.125579. quiz 18-20. [DOI] [PubMed] [Google Scholar]
- 21.Otley CC, Coldiron BM, Stasko T, Goldman GD. Decreased skin cancer after cessation of therapy with transplant-associated immunosuppressants. Arch Dermatol. 2001;137:459–463. [PubMed] [Google Scholar]
- 22.Kripke ML. Latency, histology, and antigenicity of tumors induced by ultraviolet light in three inbred mouse strains. Cancer Res. 1977;37:1395–1400. [PubMed] [Google Scholar]
- 23.Klein G, Klein E. Immune surveillance against virus-induced tumors and nonrejectability of spontaneous tumors: contrasting consequences of host versus tumor evolution. Proc Natl Acad Sci U S A. 1977;74:2121–2125. doi: 10.1073/pnas.74.5.2121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fisher MS, Kripke ML. Systemic alteration induced in mice by ultraviolet light irradiation and its relationship to ultraviolet carcinogenesis. Proc Natl Acad Sci U S A. 1977;74:1688–1692. doi: 10.1073/pnas.74.4.1688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schwarz T. The dark and the sunny sides of UVR-induced immunosuppression: photoimmunology revisited. J Invest Dermatol. 2010;130:49–54. doi: 10.1038/jid.2009.217. [DOI] [PubMed] [Google Scholar]
- 26.Gensler HL, Simpson J, Gerrish K, Gilmore J. Reduction of interferon-gamma as a critical mechanism by which ultraviolet radiation prevents tumor rejection. Photochem Photobiol. 1995;62:862–868. doi: 10.1111/j.1751-1097.1995.tb09148.x. [DOI] [PubMed] [Google Scholar]
- 27.Aragane Y, Schwarz A, Luger TA, Ariizumi K, Takashima A, Schwarz T. Ultraviolet light suppresses IFN-gamma-induced IL-7 gene expression in murine keratinocytes by interfering with IFN regulatory factors. J Immunol. 1997;158:5393–5399. [PubMed] [Google Scholar]
- 28.Ibe S, Qin Z, Schuler T, Preiss S, Blankenstein T. Tumor rejection by disturbing tumor stroma cell interactions. J Exp Med. 2001;194:1549–1559. doi: 10.1084/jem.194.11.1549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shankaran V, Ikeda H, Bruce AT, White JM, Swanson PE, Old LJ, Schreiber RD. IFN gamma and lymphocytes prevent primary tumour development and shape tumour immunogenicity. Nature. 2001;410:1107–1111. doi: 10.1038/35074122. [DOI] [PubMed] [Google Scholar]
- 30.Qin Z, Kim HJ, Hemme J, Blankenstein T. Inhibition of methylcholanthrene-induced carcinogenesis by an interferon gamma receptor-dependent foreign body reaction. J Exp Med. 2002;195:1479–1490. doi: 10.1084/jem.20011887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schreiber K, Rowley DA, Riethmuller G, Schreiber H. Cancer immunotherapy and preclinical studies: why we are not wasting our time with animal experiments. Hematol Oncol Clin North Am. 2006;20:567–584. doi: 10.1016/j.hoc.2006.03.001. [DOI] [PubMed] [Google Scholar]
- 32.Hurwitz DJ, Pincus L, Kupper TS. Imiquimod: a topically applied link between innate and acquired immunity. Arch Dermatol. 2003;139:1347–1350. doi: 10.1001/archderm.139.10.1347. [DOI] [PubMed] [Google Scholar]
- 33.Peris K, Micantonio T, Fargnoli MC, Lozzi GP, Chimenti S. Imiquimod 5% cream in the treatment of Bowen's disease and invasive squamous cell carcinoma. J Am Acad Dermatol. 2006;55:324–327. doi: 10.1016/j.jaad.2006.04.004. [DOI] [PubMed] [Google Scholar]
- 34.Huang SJ, Hijnen D, Murphy GF, Kupper TS, Calarese AW, Mollet IG, Schanbacher CF, Miller DM, Schmults CD, Clark RA. Imiquimod enhances IFN-gamma production and effector function of T cells infiltrating human squamous cell carcinomas of the skin. J Invest Dermatol. 2009;129:2676–2685. doi: 10.1038/jid.2009.151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Aptsiauri N, Cabrera T, Mendez R, Garcia-Lora A, Ruiz-Cabello F, Garrido F. Role of altered expression of HLA class I molecules in cancer progression. Adv Exp Med Biol. 2007;601:123–131. doi: 10.1007/978-0-387-72005-0_13. [DOI] [PubMed] [Google Scholar]
- 36.Garrido F, Cabrera T, Aptsiauri N. "Hard" and "soft" lesions underlying the HLA class I alterations in cancer cells: implications for immunotherapy. Int J Cancer. 2010;127:249–256. doi: 10.1002/ijc.25270. [DOI] [PubMed] [Google Scholar]
- 37.Sikorski M, Bobek M, Zrubek H, Marcinkiewicz J. Dynamics of selected MHC class I and II molecule expression in the course of HPV positive CIN treatment with the use of human recombinant IFN-gamma. Acta Obstet Gynecol Scand. 2004;83:299–307. [PubMed] [Google Scholar]
- 38.Cruz M, Elenich LA, Smolarek TA, Menon AG, Monaco JJ. DNA sequence, chromosomal localization, and tissue expression of the mouse proteasome subunit lmp10 (Psmb10) gene. Genomics. 1997;45:618–622. doi: 10.1006/geno.1997.4977. [DOI] [PubMed] [Google Scholar]
- 39.Kandil E, Kohda K, Ishibashi T, Tanaka K, Kasahara M. PA28 subunits of the mouse proteasome: primary structures and chromosomal localization of the genes. Immunogenetics. 1997;46:337–344. doi: 10.1007/s002510050281. [DOI] [PubMed] [Google Scholar]
- 40.Callahan MK, Wohlfert EA, Menoret A, Srivastava PK. Heat shock up-regulates lmp2 and lmp7 and enhances presentation of immunoproteasome-dependent epitopes. J Immunol. 2006;177:8393–8399. doi: 10.4049/jimmunol.177.12.8393. [DOI] [PubMed] [Google Scholar]
- 41.Seifert U, Bialy LP, Ebstein F, Bech-Otschir D, Voigt A, Schröter F, Prozorovski T, Lange N, Steffen J, Rieger M, Kuckelkorn U, Aktas O, Kloetzel PM, Krüger E. Immunoproteasomes preserve protein homeostasis upon interferon-induced oxidative stress. Cell. 2010;142:613–624. doi: 10.1016/j.cell.2010.07.036. [DOI] [PubMed] [Google Scholar]
- 42.Khodarev NN, Minn AJ, Efimova EV, Darga TE, Labay E, Beckett M, Mauceri HJ, Roizman B, Weichselbaum RR. Signal transducer and activator of transcription 1 regulates both cytotoxic and prosurvival functions in tumor cells. Cancer Res. 2007;67:9214–9220. doi: 10.1158/0008-5472.CAN-07-1019. [DOI] [PubMed] [Google Scholar]
- 43.Khodarev NN, Roach P, Pitroda SP, Golden DW, Bhayani M, Shao MY, Darga TE, Beveridge MG, Sood RF, Sutton HG, Beckett MA, Mauceri HJ, Posner MC, Weichselbaum RR. STAT1 pathway mediates amplification of metastatic potential and resistance to therapy. PLoS One. 2009;4:e5821. doi: 10.1371/journal.pone.0005821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Buckwalter MR, Srivastava PK. "It is the antigen(s), stupid" and other lessons from over a decade of vaccitherapy of human cancer. Semin Immunol. 2008;20:296–300. doi: 10.1016/j.smim.2008.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pekarek LA, Starr BA, Toledano AY, Schreiber H. Inhibition of tumor growth by elimination of granulocytes. J Exp Med. 1995;181:435–440. doi: 10.1084/jem.181.1.435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Seung LP, Seung SK, Schreiber H. Antigenic cancer cells that escape immune destruction are stimulated by host cells. Cancer Res. 1995;55:5094–5100. [PubMed] [Google Scholar]
- 47.Ueno T, Toi M, Saji H, Muta M, Bando H, Kuroi K, Koike M, Inadera H, Matsushima K. Significance of macrophage chemoattractant protein-1 in macrophage recruitment, angiogenesis, and survival in human breast cancer. Clin Cancer Res. 2000;6:3282–3289. [PubMed] [Google Scholar]
- 48.Neumark E, Anavi R, Witz IP, Ben-Baruch A. MCP-1 expression as a potential contributor to the high malignancy phenotype of murine mammary adenocarcinoma cells. Immunol Lett. 1999;68:141–146. doi: 10.1016/s0165-2478(99)00043-7. [DOI] [PubMed] [Google Scholar]
- 49.Schmidt N, Gonzalez E, Visekruna A, Kühl AA, Loddenkemper C, Mollenkopf H, Kaufmann SH, Steinhoff U, Joeris T. Targeting the proteasome: partial inhibition of the proteasome by bortezomib or deletion of the immunosubunit LMP7 attenuates experimental colitis. Gut. 2010;59:896–906. doi: 10.1136/gut.2009.203554. [DOI] [PubMed] [Google Scholar]
- 50.Reis J, Hassan F, Guan XQ, Shen J, Monaco JJ, Papasian CJ, Qureshi AA, Van Way CW 3rd, Vogel SN, Morrison DC, Qureshi N. The Immunoproteasomes Regulate LPS-Induced TRIF/TRAM Signaling Pathway in Murine Macrophages. Cell Biochem Biophys. 2011;60:119–126. doi: 10.1007/s12013-011-9183-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Grivennikov S, Karin E, Terzic J, Mucida D, Yu GY, Vallabhapurapu S, Scheller J, Rose-John S, Cheroutre H, Eckmann L, Karin M. IL-6 and Stat3 are required for survival of intestinal epithelial cells and development of colitis-associated cancer. Cancer Cell. 2009;15:103–113. doi: 10.1016/j.ccr.2009.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Muchamuel T, Basler M, Aujay MA, Suzuki E, Kalim KW, Lauer C, Sylvain C, Ring ER, Shields J, Jiang J, Shwonek P, Parlati F, Demo SD, Bennett MK, Kirk CJ, Groettrup M. A selective inhibitor of the immunoproteasome subunit LMP7 blocks cytokine production and attenuates progression of experimental arthritis. Nat Med. 2009;15:781–787. doi: 10.1038/nm.1978. [DOI] [PubMed] [Google Scholar]
- 53.Kuhn DJ, Hunsucker SA, Chen Q, Voorhees PM, Orlowski M, Orlowski RZ. Targeted inhibition of the immunoproteasome is a potent strategy against models of multiple myeloma that overcomes resistance to conventional drugs and nonspecific proteasome inhibitors. Blood. 2009;113:4667–4676. doi: 10.1182/blood-2008-07-171637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Singh AV, Bandi M, Aujay MA, Kirk CJ, Hark DE, Raje N, Chauhan D, Anderson KC. PR-924, a selective inhibitor of the immunoproteasome subunit LMP-7, blocks multiple myeloma cell growth both in vitro and in vivo. Br J Haematol. 2011;152:155–163. doi: 10.1111/j.1365-2141.2010.08491.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Jiang H, Monaco JJ. Sequence and expression of mouse proteasome activator PA28 and the related autoantigen Ki. Immunogenetics. 1997;46:93–98. doi: 10.1007/s002510050246. [DOI] [PubMed] [Google Scholar]
- 56.Higashitsuji H, Liu Y, Mayer RJ, Fujita J. The oncoprotein gankyrin negatively regulates both p53 and RB by enhancing proteasomal degradation. Cell Cycle. 2005;4:1335–1337. doi: 10.4161/cc.4.10.2107. [DOI] [PubMed] [Google Scholar]
- 57.Li H, Fu X, Chen Y, Hong Y, Tan Y, Cao H, Wu M, Wang H. Use of adenovirus-delivered siRNA to target oncoprotein p28GANK in hepatocellular carcinoma. Gastroenterology. 2005;128:2029–2041. doi: 10.1053/j.gastro.2005.03.001. [DOI] [PubMed] [Google Scholar]
- 58.van Hall T, Sijts A, Camps M, Offringa R, Melief C, Kloetzel PM, Ossendorp F. Differential influence on cytotoxic T lymphocyte epitope presentation by controlled expression of either proteasome immunosubunits or PA28. J Exp Med. 2000;192:483–494. doi: 10.1084/jem.192.4.483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Seeger JM, Schmidt P, Brinkmann K, Hombach AA, Coutelle O, Zigrino P, Wagner-Stippich D, Mauch C, Abken H, Krönke M, Kashkar H. The proteasome inhibitor bortezomib sensitizes melanoma cells toward adoptive CTL attack. Cancer Res. 2010;70:1825–1834. doi: 10.1158/0008-5472.CAN-09-3175. [DOI] [PubMed] [Google Scholar]
- 60.Sijts AJ, Standera S, Toes RE, Ruppert T, Beekman NJ, van Veelen PA, Ossendorp FA, Melief CJ, Kloetzel PM. MHC class I antigen processing of an adenovirus CTL epitope is linked to the levels of immunoproteasomes in infected cells. J Immunol. 2000;164:4500–4506. doi: 10.4049/jimmunol.164.9.4500. [DOI] [PubMed] [Google Scholar]
- 61.Schwarz K, van Den Broek M, Kostka S, Kraft R, Soza A, Schmidtke G, Kloetzel PM, Groettrup M. Overexpression of the proteasome subunits LMP2, LMP7, and MECL-1, but not PA28 alpha/beta, enhances the presentation of an immunodominant lymphocytic choriomeningitis virus T cell epitope. J Immunol. 2000;165:768–778. doi: 10.4049/jimmunol.165.2.768. [DOI] [PubMed] [Google Scholar]
- 62.Gaczynska M, Rock KL, Spies T, Goldberg AL. Peptidase activities of proteasomes are differentially regulated by the major histocompatibility complex-encoded genes for LMP2 and LMP7. Proc Natl Acad Sci U S A. 1994;91:9213–9217. doi: 10.1073/pnas.91.20.9213. [DOI] [PMC free article] [PubMed] [Google Scholar]
Materials and methods
Mice and tumor cell lines
Normal and nude MMTV- C3H/HeN mice were purchased from the National Cancer Institute, Frederick Cancer Research Facility (Frederick, MD). All mice were maintained in a specific pathogen-free barrier facility at The University of Chicago. All animal procedures have been approved by the Institutional Animal Care and Use Committee (IACUC) at The University of Chicago. The standard method used to induce the C3H tumors 4102, 6132A, and 6139B and the C57BL/6 tumors 8101 and 8101-PRO1A have been described in detail (5, 7, 9). Briefly, the shaved backs of 4-5 month old mice were exposed three times per week to a bank of six FS40 sunlamps mounted in parallel 10.5 cm apart and 20 cm above the bottom of the cage. The autochthonous skin tumors were removed, adapted to culture, and aliquots of early (less than 20 days) passage of the tumor cells were stored in liquid nitrogen for future analyses. Tumor cells were also passaged in vivo as described in Results to generate progressor variants. Autologous non-malignant cells and DNA were also isolated from each mouse of tumor origin. All cell lines were maintained in Dulbecco's Modification of Eagle's medium (DMEM) supplemented with 5% heat-inactivated fetal bovine serum in a 37˚C incubator with 10% CO2. Cells were tested for mycoplasma contamination by staining with Hoechst #33258 followed by fluorescence microscopy.
Analysis of p53
Total RNA (10 µg) from tumor cells was reverse transcribed into cDNA with M-MLV reverse transcriptase (Life Technologies, Rockville, MD) in a 50 µl reaction mixture containing 0.4 mM dNTP, 4 mg/ml oligo (dT)10, 0.5 mg/ml random hexamer, and 0.6 U/ml RNasin (Promega, Madison, WI). A partial p53 sequence was amplified from cDNA using the primers 5'-ATG CGA ATT CTC CTC CCC TCA ATA AGC T-3' and 5'-TAT AGG AAT TCA GGG CAA AGG ACT TCC T-3' and the cycling conditions: 5 s at 94˚C, 1 min at 68˚C, and 1 min at 72˚C for 35 cycles. PCR products were cloned into pGEM-T Easy vector (Promega, Madison, WI) and submitted to the DNA sequencing facility at The University of Chicago. Sequences were aligned using the Vector NTI DNA analysis software (Informax, Bethesda, MD).
The amount of p53 alleles with the 4102-specific mutation was quantified by real-time PCR using the SYBR Green PCR core reagents and the GeneAmp 5700 sequence detection system (Applied Biosystems, Foster City, CA). The PCR primers were designed using Primer Express software (Applied Biosystems, Foster City, CA) to specifically amplify the wild-type or the mutant sequence. The mutant allele was amplified using the sense primer 5'-ACC TGG ATC CTG TGT CTT CCC-3' (Integrated DNA Technologies, Coralville, IA) and the antisense primer 5'-TGA TGG TAA GGA TAG GTC GGC A-3' (Life Technologies, Rockville, MD). The wild-type allele was amplified using the same sense primer together with the antisense primer 5'-GAT GGT AAG GAT AGG TCG GCG-3' (Integrated DNA Technologies, Coralville, IA). The PCR mixture contained 1.5 mM MgCl2 and 0.3 mM of each primer. The PCR conditions used were: 2 min at 50˚C, 10 min at 95˚C, 15 s at 95˚C, and 1 min at 65˚C, the last two steps being repeated 39 times.
Analysis of DNA content
Tumor cells (3 x 106) were washed twice and resuspended in 1.2 ml PBS. The cells were permeabilized with the addition of 3 ml of 100% ethanol while mixing and incubated at room temperature for 30 min. The cells were centrifuged at 650 x g for 10 min, washed with PBS, and incubated in 0.1 mg/ml boiled RNase A in PBS at 37˚C for 30 min. The cells were washed again and incubated with 100 mg/ml propidium iodide (Sigma, St Louis, MO) for 30 min at 4˚C before analysis by flow cytometry.
Clonogenicity of tumor cells in soft agar
A solution of 5% cloning agarose in pyrogen-free water was autoclaved, diluted to 0.26% with fresh DMEM supplemented with 10% FBS, glutamine, and non-essential amino acids, and maintained at 41.5˚C. For the base layer, 0.4 ml of the 0.26% agarose solution was added with a preheated glass pipet to each well of a 24-well plate and allowed to solidify over a stack of wet paper towels at 4˚C. The plates were transferred to a 37˚C, humidified incubator and equilibrated for 1 h. Tumor cells were resuspended to 104 cells/ml and then mixed with 0.26% agarose at a ratio of 1:4 (v:v) to make a 0.21% agarose solution containing 2 x 103 cells/ml. The cell-agarose solution was added dropwise to the base layer and allowed to solidify before returning to the incubator. After 10 days, colonies equal to or greater than 0.05 mm in diameter were counted.
Effect of IFNγ on colony formation or proliferation of cancer cells in vitro
Cells were maintained in low glucose DMEM medium (GIBCO, Carlsbad, CA) supplemented with 10% FBS and 1% penicillin/streptomycin at 37˚C and 5% CO2. Cells were collected by mild trypsinization, diluted with fresh media to appropriate concentrations to produce between 64 and 640 cells/plate in 5 ml media and transferred to T35 culture plates. Three independent plates were used for each measurement. After 18 h, mouse IFNγ was added to final concentrations of 10, 30 and 100 ng/ml and the plates were incubated for 7 days. The culture media were discarded and the surviving colonies were rinsed with 0.85% NaCl and stained with 20% crystal violet in ethanol/water mixture (1:1). After drying at room temperature, colonies containing more than 50 cells were counted. All data were normalized to the plating efficiency (fraction of colony-forming cells in untreated plates). Differences between the surviving fraction of each cell line at each dose of IFNγ were estimated by a two-tailed unpaired t test with cut off P < 0.05. Error bars indicate standard deviations. For growth curves, cells were plated in 24-well plates at a density of 20,000 cells/well. After 24 h, IFNγ was added to final concentrations of 10, 30 and 100 ng/ml and cells were counted 24 hours after IFNγ addition.
Analysis of phosphorylation of STAT1
Proteins were extracted in modified radio-immunoprecipitation assay (RIPA) buffer supplemented with protease inhibitors (1× PBS, 1% NP40, 0.1% SDS, 1 mmol/l Na3VO4, 2 µg/ml aprotinin, 1 mmol/l PMSF). Cell lysates were centrifuged at 15,000 x g and 4˚C to remove cell debris. All samples were normalized by protein concentration using Bradford reagent. Concentration of all samples was adjusted to 1 mg/ml and equal amounts of protein lysates (10 µg for total Stat1 and 20-25 µg for pStat1) were loaded in each well. Proteins were separated on 7.5% SDS-PAGE (Bio-Rad/Life Science Research, Hercules, CA) and transferred to polyvinylidene difluoride (PVDF) membranes. Total Stat1 was detected using rabbit polyclonal IgG antibodies specific for Stat1 p84/p91 (E-23) and phosphorylated Stat1 was detected using goat polyclonal IgG antibodies specific for p-Stat1 (Tyr 701). For internal loading control, goat polyclonal IgG specific for actin (I-19) was used. All antibodies were purchased from Santa Cruz Biotechnology (Santa Cruz, CA). Membranes were developed with SuperSignal West Pico Chemiluminescent Substrate (PIERCE/Thermo Scientific, Rockford, IL).
Tumor inoculation and growth measurement
Tumor cells (105) were washed twice with ice-cold, serum-free DMEM before being injected s.c. into the flanks of nude C3H mice. Tumors isolated from the nude mice were chopped into small pieces and injected using a trocar (0.3 cm3 per mouse) into the flanks of normal C3H mice; tumor fragments were used because a suspension of 4102-PRO tumor cells does not cause tumors in normal mice. Tumors were measured along the three orthogonal axes (a, b and c) and tumor volume was calculated using the formula: tumor volume = abc/2.
Tumor-specific T cells and 51Cr-release assay
Standard 4.5 h 51Cr-release assays were performed with a cloned 4102-specific T cell line as described previously (5). Tumor cells were incubated with or without 100 U/ml recombinant IFNγ (Genentech, San Francisco, CA) for 2 d to induce surface MHC I molecule expression. 51Cr-labeled tumor cells (5 x 103) were incubated with T cells at the indicated effector-to-target ratio in complete RPMI 1640 in 96-well V-bottom plates in a humidified incubator. After 4.5 h, 51Cr released into the medium was measured in a gamma counter (Micromedic Systems, Horsham, PA). The results were represented as percentage lysis, calculated using the formula: cytolysis (%) = [(experimental release-spontaneous release) / (maximum release-spontaneous release)] x100.
Analysis of MHC I expression
For in vitro MHC I analyses, tumor cells were stained with mouse hybridoma supernatants 15.5.5 (anti-H-2Dk) or 16.1.11N (anti-H-2Kk) followed with FITC-conjugated goat anti-mouse Ig antibody (American Qualex, San Clemente, CA). Analysis was performed on a FACScan II cell sorter (Becton Dickinson, Mountain View, CA). For ex vivo MHC I analyses, tumors were minced into 1-2 mm3 pieces and digested in RPMI containing 1 mg/ml collagenase D and 0.25 mg/ml DNase I (Roche) for 30 min at 37˚C, 5% CO2. Trypsin (1 mg/ml) was then added and the suspension was mixed by pipetting up and down for 2 min to enhance tissue disruption. DMEM with 10% FCS was added to the cell suspension to stop enzymatic activity. The cells were centrifuged, resuspended in cold PBS and filtered through a 70-µm nylon filter mesh. Single-cell suspensions were incubated with 2.4G2 hybridoma supernatant to block antibody binding to the Fc receptors, stained with antibodies specific for CD45 (30-F11, eBioscience), Kk (36-7-5, BD Pharmingen) or mouse IgG2a isotype control (BD Pharmingen) for 15 min at 4˚C, and washed twice with cold PBS, 0.1% sodium azide, 1% BSA. Dead cells were identified by 7AAD (BD Pharmingen) staining and excluded by electronic gating. Data were acquired on a FACSCalibur and analyzed with FlowJo software. For immunohistochemical analyses, a tumor and the overlying skin were resected, embedded in OCT compound, and frozen in liquid nitrogen. Sections (5 µm) were stained with biotin-conjugated mouse anti-H-2Kk antibody (Pharmingen, San Diego, CA), followed with horseradish peroxidase-conjugated streptavidin (Vector, Burlingame, CA). Bound antibody was detected using 3,3',4,4'-tetraaminobiphenyl tetrahydochloride (DAB) as a substrate and the sections were counterstained with hematoxylin. The images were captured with an Axioskop microscope (Zeiss, Thornwood, NY) and an Axiocam camera (Zeiss, Thornwood, NY) at the Digital Light Microscopy Facility at The University of Chicago.
RNase protection assay
Complementary DNA encoding LMP2, LMP10, TAP-1, TAP-2, PA28α, and GAPDH were amplified by PCR and cloned into pGEM-T Easy vector (Promega, Madison, WI); the LMP7 PCR product was cloned into pcDNA 3.0 vector (Invitrogen, Carlsbad, CA). The identity and orientation of the PCR products in the vectors was verified by restriction digestion. The plasmids were linearized and used as templates to produce [α-32P]rUTP-labeled RNA probes using Riboprobe in vitro Transcription System-SP6 (Promega, Madison, WI). The transcription reactions were separated in 6% acrylamide/8 M urea denaturing polyacrymide gel and exposed to autoradiographic film (Eastman Kodak, Rochester, NY) to locate the full-length RNA probes. Thin gel fragments containing the full-length RNA probes were excised and the probes were eluted into probe elution buffer (0.5 M ammonium acetate, 1 mM EDTA, and 0.2% SDS) overnight at 37˚C. A cocktail containing all 7 probes (104 cpm each) was mixed with 5 µg of total RNA isolated from each tumor cell line and RNase protection assays performed using the RPA III kit (Ambion, Austin, TX) following the manufacturer's protocol. The protected fragments were separated in a 6% acrylamide/8 M urea denaturing polyacrylamide gel and detected by autoradiography. The sizes of the full-length probe and protected fragments (in parentheses) are as follows: LMP2 622 nt (545 nt); LMP7 195 nt (181 nt); LMP10 511 nt (494 nt); Tap-1 398 nt (321 nt); Tap-2 454 nt (377 nt); Gapdh 340 nt (263 nt); Pa28α 291 nt (214 nt).
Analysis of proteasome composition and activity
Proteasomes were enriched by differential centrifugation as described (62). Cells were lysed and homogenized in 50 mM Tris-HCl pH 7.4, 5 mM MgCl2, 1 mM DTT, 2 mM ATP, 250 mM sucrose. After an initial centrifugation at 10,000 x g for 20 min, the supernatants were transferred to clean centrifuge tubes (Du Pont, Wilmington, DE) and centrifuged at 100,000 x g for 1 h. After the supernatants were centrifuged again at 100,000 x g for 5 h, the pellet was resuspended in 50 mM Tris-HCl, pH 7.4, 5 mM MgCl2, 1 mM DTT, 2 mM ATP, 20% glycerol, aliquoted, and stored at -80˚C until used. Proteasome subunits were separated by 3% SDS-PAGE, transferred onto Trans-Blot nitrocellulose membrane (BIO-RAD, Hercules, CA), and detected using rabbit antisera against LMP2, LMP7, and C9 (kindly provided by Dr. J. J. Monaco, University of Cincinnati, OH), followed with horseradish peroxidase (HRP)-labeled goat anti-rabbit serum (Promega, Madison, WI). Western blots were developed by chemiluminescence using ECL Plus Western Blotting Reagent Pack (Amersham Biosciences, Piscataway, NJ). For proteasome hydrolysis assays, proteasome preparations from the cell lines were equalized according to the amount of the proteasome structural subunit C9. The amount of C9 in each preparation was determined by Western blot analysis followed by densitometry analysis using the Nucleotech gel documentation station (NucleoTech, San Mateo, CA) and the GelExpert software (NucleoTech, San Mateo, CA). Proteasomes were incubated with LLVY-MCA (Sigma, St. Louis, MO) at 37˚C in 50 mM Tris HCl, pH 7.5, 25 mM KCl, 10 mM NaCl, 0.2 mM EDTA, 1 mM DTT, 5 units/ml apyrase. Fluorescence was measured in a CytoFluor II microplate reader (Applied Biosystems, Foster City, CA) at an excitation wavelength of 360 nm and emission wavelength of 460 nm. For Western blot analysis of TAP-1 expression, cells were lysed and incubated for 30 min at 4˚C in 10 mM Tris-HCl, pH 7.4, 130 mM NaCl, 5 mM CaCl2, 0.5% (v/v) Nonidet P (NP)-40, and 1 mM PMSF. After cell debris was removed by centrifugation, cell lysates (40 µg) were subjected to 10% SDS-PAGE, transferred onto Trans-Blot nitrocellulose membrane (Bio-Rad, Hercules, CA), and detected with rabbit antisera against TAP-1 (kindly provided by Dr. K. Früh, Oregon Health Sciences University, OR) followed with HRP-labeled goat anti-rabbit antiserum (Promega, Madison, WI).
Supplemental data
Download from http://www.cancerimmunity.org/v11p2/110602_suppl_fig1.pdf (735 KB PDF file).
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Download from http://www.cancerimmunity.org/v11p2/110602_suppl_fig1.pdf (735 KB PDF file).