Abstract
Exit from mitosis requires the degradation of regulatory proteins including the mitotic cyclins and securin through ubiquitination by the anaphase promoting complex (APC) bound to Cdc20 or Cdh1. Cdc20–APC is regulated through inhibition by the spindle assembly checkpoint protein MAD2. Knowledge of Cdh1–APC regulation is limited to the phosphorylation-dependent dissociation of Cdh1 from APC. We report a novel means of regulating Cdh1 by the MAD2-related gene, MAD2L2. MAD2L2 specifically binds and inhibits Cdh1–APC, paralleling the effect of MAD2 on Cdc20. We suggest that MAD2L2 and MAD2 inhibit the release of substrates from APC and propose a mechanism of inhibition.
Keywords: Cdh1, APC, MAD2, MAD2L2, ubiquitin, checkpoint
Ubiquitin-mediated protein degradation plays a key role in the cell cycle by targeting key regulatory proteins for degradation. A set of enzymes ending with a ubiquitin protein ligase (E3) attaches ubiquitin to a target protein. Additional ubiquitin molecules are added to form a chain. Poly-ubiquitinated substrates are degraded by the proteasome (for review, see Hershko and Ciechanover 1998).
The anaphase promoting complex (APC) is an E3 required for progression from metaphase to anaphase, and exit from mitosis by targeting the inhibitor of sister chromatid separation (securin) as well as the mitotic cyclins and the chromokinesin Xkid for degradation (Irniger and Nasmyth 1997; Fang et al. 1998a; for review, see Glotzer et al. 1991; Zachariae and Nasmyth 1999; Zou et al. 1999; Funabiki and Murray 2000). APC remains active in G1, and is present in nondividing tissues such as brain (Gieffers et al. 1999).
APC activity is observed during mitosis and G1, but not during S or G2, although the core complex of the APC persists throughout the cell cycle. APC is controlled largely by cell cycle-dependent association with Cdc20 or Cdh1 (Schwab et al. 1997; Visintin et al. 1997; Fang et al. 1998a). Cdc20 levels fluctuate, peaking in mitosis when it associates with APC (Weinstein 1997; Fang et al., 1998a) and declining rapidly as cells exit mitosis and enter G1 (Prinz et al. 1998; Shirayama et al. 1998).
The Cdc20-related protein, Cdh1 (Fizzy-related in Drosophila) is stable (Schwab et al. 1997; Visintin et al. 1997; Fang et al. 1998a), although a recent report suggests it undergoes cell cycle-regulated degradation (Kramer et al. 2000). Despite its presence throughout the cell cycle, Cdh1 only associates with APC late in mitosis and during G1. In its phosphorylated state during S and G2, Cdh1 loses affinity for the APC and cannot bind or activate it. At the end of mitosis, the Cdc14 phosphatase removes the inhibitory phosphorylation, and Cdh1 associates with APC (Jaspersen et al. 1999).
Cdc20–APC and Cdh1–APC have shared and specific substrates (Schwab et al. 1997; Visintin et al. 1997; Fang et al., 1998a; Pfleger and Kirschner 2000). Both Cdc20 and Cdh1 target destruction box (D box) containing substrates like the mitotic cyclins. In Drosophila, the Cdc20 homolog, fizzy, targets the mitotic cyclins in mitosis, and fizzy-related (which appears later in development) maintains the low level of mitotic cyclins once a G1 phase appears (Sigrist and Lehner 1997). The two activators also target different substrates. In the budding yeast, Cdc20 targets securin and Clb5 (Shirayama et al. 1999); both Cdc20 and Cdh1 target Clb2 (Baumer et al. 2000); Cdh1 targets the other mitotic cyclins and Ase1 (Visintin et al. 1997). Cdh1 also targets substrates not recognized by Cdc20 (for review, see Fang et al. 1998a; Zachariae and Nasmyth 1999; Pfleger and Kirschner 2000). Cdh1-specific substrates contain either a D box (conserved or less well-defined) or the Cdh1-specific targeting signal, the KEN box (Pfleger and Kirschner 2000).
Cdc20–APC is regulated by the checkpoint protein MAD2 (Mitotic Arrest Deficient) (Li and Murray 1991; Fang et al. 1998b). MAD2 monitors attachment of the kinetochore to the spindle, causing arrest if chromosomes are not aligned and kinetochores are unattached (for review, see Glotzer 1996). Sequence searching reveals one MAD2 gene in S. cerevisiae and two MAD2-like genes in higher eukaryotes, MAD2L1 (MAD2-Like1) and MAD2L2 (also referred to as MAD2β and hRev7) (Nelson et al. 1999; Cahill et al. 1999; Murakumo et al. 2000). MAD2L1 displays a homologous function to yeast MAD2, and will be referred to here simply as MAD2. MAD2 arrests cells in metaphase by inhibiting Cdc20–APC (Fang et al. 1998b).
Cdc20 is regulated both by MAD2 and proteolysis, but regulation of Cdh1 relies on its phosphorylation-dependent dissociation of Cdh1 from APC. Because Cdh1 levels do not vary significantly, additional controls on Cdh1 seem likely. Evidence for a MAD2-like mechanism controlling Cdh1–APC has been lacking. We hereby demonstrate that the MAD2-related protein MAD2L2 negatively regulates Cdh1 by direct binding to Cdh1–APC.
Results
Cloning of the Xenopus homolog of MAD2L2
To identify new developmentally important genes by expression cloning, we tested the ability of small pools of injected RNA from a Xenopus laevis blastula stage (64 to 128 cell embryos) cDNA library to generate developmental phenotypes (Salic et al. 1997). One of the positive pools caused low-level duplications of the trunk. Injection of individual RNAs from a subpool of 48 clones resulted in two positive clones that both encoded the Xenopus homolog of MAD2L2. Subsequent experiments failed to reproduce the original phenotype, suggesting that xMAD2L2 had effects that were dependent on the batch of injected embryos. Although the specific developmental phenotype was not reproducible, injections of 2 ng of xMAD2L2 into one blastomere at the two-cell stage typically causes other developmental abnormalities such as gastrulation arrest and embryos with reduced heads and shortened body axis. By in situ hybridization, the xMAD2L2 mRNA is present maternally and zygotically, and appears uniformly distributed in embryos (data not shown) consistent with observations of MAD2L2 in human tissues (Nelson et al. 1999; Murakumo et al. 2000). xMAD2L2 protein levels do not vary with the cell cycle; in vitro-translated protein is stable in interphase extracts, mitotic extracts, and Cdh1-supplemented interphase extracts (data not shown).
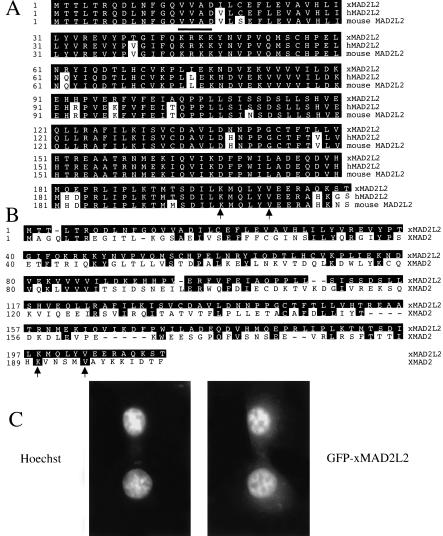
xMAD2L2 shows significant homology to human MAD2L2, and is highly conserved in vertebrates (Fig. 1A). The amino acid sequence of MAD2L2 reveals a weak, putative, nuclear localization signal, and a C-terminal lysine and valine (indicated by arrows) conserved between MAD2L2 and MAD2 (Fig. 1B). When eGFP-xMAD2L2 is injected into embryos or transfected into BHK cells, in interphase it is localized principally in the nucleus in a pattern complementary to the DNA (Fig. 1C); in mitotic cells, the protein disperses throughout the cytoplasm, showing no obvious localization (data not shown).
Figure 1.
MAD2L2 sequence conservation. (A) Comparison of xMAD2L2 with MAD2L2 sequences from human and mouse. Shading indicates residues in common with xMAD2L2; a bold line indicates a putative nuclear localization signal. (B) Sequence comparison of xMAD2L2 with XMAD2. Arrows indicate a conserved C-terminal lysine and valine (A,B). (C) Hoechst staining of eGFP-xMAD2L2 transfected BHK cells (left); eGFP-xMAD2L2 signal (right).
xMAD2L2 interacts with Cdh1–APC
We tested the ability of xMAD2L2 to interact with APC complexes. Cdc20 and Cdh1 bind to APC on αCdc27 beads but not to control beads (data not shown). When in vitro-translated 35S-labeled xMAD2L2 was incubated with interphase APC (iAPC), or APC on αCdc27 beads prebound with equal amounts of hCdc20 or hCdh1 (Fig. 2A), significant xMAD2L2 bound hCdh1–APC but not hCdc20–APC, iAPC, or control beads (Fig. 2B). We cannot rule out lower affinity interaction between xMAD2L2 and Cdc20–APC, or in vivo modifications of either xMAD2L2 or Cdc20–APC, which would promote interaction, but under our conditions we have not seen interaction between xMAD2L2 and Cdc20–APC. We have been unsuccessful in showing binding between xMAD2L2 and Cdh1 in the absence of APC (data not shown). This might indicate that MAD2L2 binds preferentially to Cdh1–APC, or that the antibody beads used in the pull-down assays displace MAD2L2 and Cdh1 from each other.
Figure 2.
MAD2L2 binds to Cdh1 and not Cdc20. (A) 35S-labeled in vitro-translated Cdc20 and Cdh1 prebound to APC on αCdc27 beads. (B) Binding of in vitro-translated 35S-labeled xMAD2L2 to Cdh1–APC on αCdc27 beads but not to Cdc20–APC on beads, iAPC on beads, or αCdc27 beads alone.
xMAD2L2 inhibits Cdh1–APC activity
Xenopus eggs and early embryos lack detectable Cdh1 protein (Lorca et al. 1998); however, interphase extracts supplemented with recombinant Cdh1 actively degrade Cdh1–APC substrates (Pfleger and Kirschner 2000). Interphase extracts do not degrade mB99, a p53 inducible gene (Utrera et al. 1998), which contains a KEN box, nor do they degrade Xkid (Funabiki and Murray 2000), securin, or the N terminus of Xenopus cyclin B (cyc B), all of which contain the D box. All four substrates are degraded in interphase extracts supplemented with Cdh1 (Fig. 3A). Cyc B, securin, and Xkid disappear with a half-life (t1/2) of 10 to 20 min, and mB99 shows a t1/2 of 30 min in Cdh1 extracts. Addition of recombinant his-tagged xMAD2L2 (Fig. 3A) or recombinant MBP-MAD2L2 (data not shown) stabilizes these proteins.
Figure 3.
xMAD2L2 inhibits Cdh1 but not Cdc20 activity. (A) Degradation assays in interphase extracts, Cdh1-supplemented interphase extracts, and Cdh1 extracts in the presence of recombinant His–MAD2L2 for mB99, Xkid, securin, and cyc B. Phosphorylation of mB99 in interphase extracts causes a shift; mB99 samples were CIP treated to clarify the difference between phosphorylation and degradation. (B) Gel filtration profiles of recombinant MBP-MAD2L2 on a superdex 200 column using the SMART system (upper panel) and tetramer, trimer, and monomer peaks rerun over the same column (lower panel). (C) t1/2 of Xkid in Cdh1 extracts (upper panel) or Δ90 extracts (lower panel) with increasing concentrations of MAD2M, MAD2O, MBP-xMAD2L2M or MBP-xMAD2L2O. Graph reflects typical results from one extract; t1/2 varies between extracts, but the effect on t1/2 is consistent. (D) Ubiquitination assays of cyc B using Cdh1-activated APC in the presence of 5 μM MAD2O or 5 μM MBP–xMAD2L2.
Bacterially expressed MAD2 forms monomers (MAD2M), dimers, and tetramers (MAD2O). Injection of MAD2O but not MAD2M into Xenopus embryos causes cells to arrest in mitosis. Both MAD2O and MAD2M bind Cdc20, but only MAD2O inhibits APC in mitotic Xenopus extracts; MAD2M has no measurable effect (Fang et al. 1998b). By gel filtration, MBP-MAD2L2 exists as tetrameric, trimeric, dimeric, and monomeric protein (Fig. 3B, upper panel). In our experience, his-tagged proteins occasionally form dimers, but MBP is strictly monomeric, so the oligomerization we saw was most likely due to MAD2L2 and not the tagging epitope. Repeated gel filtration of the MBP-MAD2L2 tetramer, trimer, and monomer peaks revealed that tetramer and monomer populations did not substantially interconvert, but the trimer readily interconverted into tetramer, dimer, and monomer (Fig. 3B, lower panel).
Neither MAD2O nor MAD2M inhibit degradation of Xkid (Fig. 3C) or cyc B (data not shown) in Cdh1-supplemented interphase extracts. However, all species of MAD2L2 stabilized cyc B and Xkid, securin, and mB99 (shown for Xkid in Fig. 3C for monomeric MAD2L2 [MAD2L2M], and tetrameric MAD2L2 [MAD2L2O]). At higher concentrations, His-MAD2L2 forms aggregates and precipitates, but at lower concentrations, His-MAD2L2 forms predominantly monomer by gel filtration (although a small amount of dimer is observed; data not shown); monomer is fully active (Fig. 3A). This suggests no requirement for a stable oligomer of MAD2L2, but we cannot rule out dynamic assembly into higher molecular weight forms.
Ubiquitination of APC substrates can be reconstituted with purified components (Fang et al. 1998a). Cdh1–APC quickly ubiquitinates cyc B, and completely converts all of the original unmodified polypeptide into higher molecular weight conjugates by 15 min. Addition of MAD2O has little effect on Cdh1–APC activity, but addition of MBP–MAD2L2 significantly reduces poly-ubiquitination of substrates (although cyc B is still converted to the three major mono-ubiquitinated species) (Fig. 3D).
xMAD2L2 is selective for Cdh1–APC
Because in vitro-translated xMAD2L2 does not bind Cdc20–APC (Fig. 2), we surmised it would not affect Cdc20–APC specific degradation processes. To assay Cdc20–APC under physiological conditions, we used a Xenopus extract stably activated in mitosis by Δ90 cyclin where the active form of APC is Cdc20–APC. In these extracts, addition of MAD2O stabilizes cyclin (Fang et al. 1998b). Because of the tendency of His–MAD2L2 to precipitate and aggregate at higher concentrations and the difficulty in purifying large amounts of clean, soluble protein, we focused our characterization on MBP–MAD2L2. cyc B, securin (data not shown) and Xkid degradation (Fig. 3C) are not inhibited by MBP–MAD2L2M or MBP–MAD2L2O (tested from multiple preparations of the protein), but are inhibited by MAD2O at similar molar ratios. At higher concentrations, we see an inhibitory effect of MBP–MAD2L2 on Cdc20–APC-mediated degradation; this effect is small compared to MAD2O (Fig. 3C). His–MAD2L2 shows lower selectivity for Cdh1–APC, and greater variability between preparations, extending the t1/2 to between 20 min and 40 min at 5 μM (data not shown); because of the greater variability, the effects on Cdc20–APC may be nonspecific, possibly caused by contaminants or aggregation.
xMAD2L2 causes cell cycle arrest in Xenopus embryos
Injection of 2.5 ng of purified MBP–xMAD2L2O (Fig. 4) or MBP–xMAD2L2M (data not shown) into one cell of a two-cell Xenopus embryo results in a dramatic gastrulation arrest (after MBT). These concentrations of protein show no effect initially and embryos develop normally (in terms of cell size and nuclear staining) until after MBT. The injected halves of the embryos fail to gastrulate normally, with cells on the injected side larger than on the uninjected side. Hoechst or propidium iodide staining at MBT showed that cells on the injected side either lack nuclei or have abnormally larger, more intensely staining nuclei (Fig. 4F,G) than cells on the uninjected side (Fig. 4E,G). The more intense staining most likely results from an increase in the amount of DNA. In some cases, cells on the injected side show a postanaphase state with two separated nuclei in which the cell fails to completely divide, leaving a cytoplasmic bridge between the nuclei (data not shown). The gastrulation arrest phenotype upon MAD2L2 overexpression is consistent with inhibition of endogenous Cdh1–APC once it appears post-MBT. Furthermore, coinjection of 2.5 ng purified Cdh1 protein with MAD2L2 reverses the arrest (Fig. 4C), resulting in embryos that develop completely normally, as do embryos injected with Cdh1 protein. The number of embryos rescued (Fig. 4I) is typical of coinjection rescue experiments in the Xenopus system. Unlike protein injections, RNA injections of Cdh1 alone in one cell of two cell embryos result in an arrest near MBT with nuclei that stain diffusely for DNA (Fig. 4H), suggesting an inability to enter mitosis. The discrepancy is likely due to the fact that Cdh1 protein accumulates during the course of development following RNA injection. Failure to enter mitosis may reflect continued activation of APC by Cdh1, consistent with the effect seen by overexpression of Cdh1 in tissue culture cells (Sorensen et al. 2000).
Figure 4.
xMAD2L2-mediated inhibition of gastrulation is rescued by coinjection with Cdh1. Vegetal views of stage 11 embryos injected (left half of each embryo) at the two-cell stage with (A) 2.5 ng MBP–MAD2L2, (B) 2.5 ng His6–Cdh1, (C) 2.5 ng MBP–MAD2L2 plus 2.5 ng His6–Cdh1, or (D) buffer. Injections of MBD–MAD2L2 result in cells that have enlarged and enhanced nuclear staining properties. One cell of two-cell stage embryos injected with 2.5 ng of MBP–MAD2L2, allowed to develop to stages 8–9, fixed, and stained with propidium iodide (E,F) or Hoechst (G). (E) Uninjected side and (F) injected side of the same embryo. (G) A view showing an injected region (left) next to an uninjected region. (H) Injections of Cdh1 RNA result in enlarged cells that give diffuse nuclear staining. One cell of two-cell embryos were injected with 500 pg of Cdh1 (left side), allowed to develop to stages 10.5, fixed, and stained with Hoechst. (I) Summary of the gastrulation arrest and rescue of the embryos shown in A–D.
MAD2L2 and MAD2 inhibit substrate release from Cdh1 and Cdc20, respectively
Inhibition by MAD2 is not due to competition for Cdc20 but by formation of an inhibitory MAD2–Cdc20–APC ternary complex (Fang et al. 1998b). Similarly, MAD2L2 does not prevent binding of Cdh1 to APC. When in vitro-translated 35S-labeled Cdh1 was incubated with APC on αCdc27-coated beads or control beads, similar levels of Cdh1 were recovered in the presence or absence of MAD2L2 (data not shown).
We recently have shown that both Cdc20 and Cdh1 bind substrates independent of APC (C. Pfleger, E. Lee, and M. Kirschner, in prep.). Myc-tagged hCdc20 or myc-tagged hCdh1 on anti-myc beads binds at least 10% of in vitro-translated 35S-labeled Xkid. Addition of MAD2O to the Cdc20–Xkid binding assay or MBP–MAD2L2 to the Cdh1–Xkid binding assay did not inhibit substrate binding, but rather increased the amount bound (data not shown). Because these may or may not have been equilibrium conditions, the increase of substrate bound could reflect changes in the affinity of the activator for substrate or a specific decrease in the on- and off-rates for substrate bound to activator. To test for an effect on the release of substrate, we separated Xkid bound to Cdc20 into separate fractions to which we added buffer, 5 μM MAD2O or 5 μM MAD2M. We also separated Xkid bound to Cdh1 into fractions to which we added buffer or 5 μM MAD2L2. After 20 min, we added an excess of cold Xkid. In the presence of buffer, little hot Xkid was bound after 4 h, showing that cold Xkid completely replaced hot Xkid bound to Cdc20 or Cdh1. In the presence of MAD2O, however, substantial hot Xkid was bound (Fig. 5) even after 4 h, indicating that the original Xkid was not released. In the presence of MAD2M, we saw retention of more Xkid than with the buffer control (roughly 20% after 4 h) but not as much as with MAD2O (data not shown). Similarly, addition of MAD2L2 but not buffer resulted in Cdh1-retention of hot Xkid (Fig. 5). MAD2O (5 μM) did not cause Cdh1 retention of Xkid, and 5 μM MAD2L2 caused Cdc20 retention of Xkid similar to MAD2M (data not shown).
Figure 5.
MAD2 and MAD2L2 prevent release of substrate bound to Cdc20 or Cdh1, respectively. Graph of substrate binding assay showing prebound Xkid retained by Cdc20 or Cdh1 beads over time after addition of excess cold Xkid in the presence of 5 μM MAD2O or MAD2L2, respectively, compared to buffer alone.
Discussion
In APC-dependent degradation, the major point of regulation is APC itself rather than the individual substrates, allowing several degradation events to be coordinately regulated. APC is regulated by Cdc20 and Cdh1, which in turn, are regulated by other proteins or by phosphorylation. The spindle assembly checkpoint responds to unattached or unstably attached chromosomes through MAD2, and arrests the cell cycle in metaphase (for review, see Glotzer 1996). Phosphorylation of Cdh1 also regulates APC activity (Jaspersen et al. 1999; Kotani et al. 1999). Degradation of Cdc20 at the end of mitosis causes a loss of Cdc20–APC activity, while at the same time Cdc14 dephosphorylation of Cdh1 allows for Cdh1–APC activity to arise. In the S-phase, CDK-dependent phosphorylation of Cdh1 causes Cdh1 to dissociate from the APC, which is thus kept inactive during S phase and G2 until the following mitosis when it partners with Cdc20 again (Fang et al. 1998a; Jasperson et al. 1999).
We have cloned the Xenopus homolog of MAD2L2. In extracts and ubiquitination assays, xMAD2L2 inhibits Cdh1–APC in a manner similar to the MAD2 inhibition of Cdc20–APC. MBP–xMAD2L2 forms monomers and oligomers, all of which inhibit Cdh1–APC. xMAD2L2 is selective for Cdh1–APC; there is very little inhibition of Cdc20 (and no effect on SCF-mediated β-catenin degradation; A. Salic, unpubl.). We have made several forms of MAD2L2 including bacterially expressed His- and MBP–MAD2L2. The MBP-tagged protein is soluble, mono-disperse, and displays high selectivity for Cdh1, although selectivity is decreased for His–MAD2L2. Further evidence for selectivity comes from binding experiments. Nontagged in vitro-translated xMAD2L2 selectively binds Cdh1–APC and not Cdc20–APC (Fig. 2). However, as a cautionary note, the capacity of MAD2 to exist in several different conformations (Luo et al. 2000) suggests that we cannot rule out that another conformation of MAD2L2 with inhibitory activity for Cdc20 could exist.
MAD2 inhibition of Cdc20 does not disrupt formation of a Cdc20–APC complex (Fang et al. 1998b). Similarly, we observe no effect of recombinant MAD2L2 on Cdh1 binding to APC. We have, therefore, suggested previously that the activity of MAD2 must occur in the ternary complex with APC. How might MAD2 or MAD2L2 inhibit APC? First, MAD2 and MAD2L2 might interfere with substrate binding. Using a recently developed a substrate-binding assay (C. Pfleger, E. Lee, and M. Kirschner, in prep.), we show that neither MAD2 nor MAD2L2 inhibits the binding of Xkid to Cdc20 or Cdh1, respectively. Surprisingly, MAD2O and MAD2L2 increase the binding of substrate by preventing the release of substrate once bound. We have not determined if substrate retained in this manner is ubiquitinated, however, MAD2L2 inhibits poly-ubiquitination of cyc B. This suggests two related mechanisms. In the presence of MAD2 and MAD2L2, respectively, Cdc20–APC and Cdh1–APC might bind substrates nonproductively; alternatively, if ubiquitination occurs but release is prevented, turnover of the enzymatic activity of APC would be prevented.
MAD2O injection into Xenopus embryos causes an almost immediate arrest in mitosis. In contrast, purified xMAD2L2 protein injection causes a post-MBT gastrulation arrest. Nuclei are large and intensely staining, or are arrested late in nuclear division. This is consistent with xMAD2L2 inhibiting endogenous Cdh1–APC activity in vivo once it appears post-MBT. The latter may result from a failure to completely exit mitosis, while the former may represent higher amounts of DNA from extra rounds of replication. Additional rounds of replication could result from stabilization of the Cdh1–APC substrate Cdc6 (Peterson et al. 2000) and other substrates postulated to be involved in replication (Sorensen et al. 2000).
Information on the biological role of Cdh1 is limited to the cell cycle and by our knowledge of its substrates. No role outside of the cell cycle has been reported. Cdh1 has an expanded set of substrates compared to Cdc20 (Fang et al. 1998a; Pfleger and Kirschner 2000). Furthermore, Cdh1 activity exists in nondividing cells (Gieffers et al. 1999), suggesting a physiological role outside of cell cycle events. Recently, Y. Wan and M. Kirschner (in prep.) have found several Cdh1 homologs, suggesting diversified functions in cell physiology. MAD2 plays a clearly defined role in the spindle assembly checkpoint by inhibiting Cdc20–APC. We expect that MAD2L2 may play a related role at other stages of the cell cycle, such as an exit-from-mitosis checkpoint, ensuring that DNA is replicated only once per cell cycle, or have other developmental roles. Like cyclin-dependent kinase inhibitors, MAD2L2 may link external signals to cell cycle progression or cell differentiation. Given the phenotype of MAD2L2 overexpression in early Xenopus development, we think that Cdh1 may target substrates involved in important developmental processes.
Materials and methods
Purification of recombinant xMAD2L2
xMAD2L2 was cloned into a pET 28 bacterial expression vector (N-terminal His tag) and a pMAL vector (N-terminal MBP tag). After expression in bacteria, soluble protein was purified on nickel beads and eluted with high imidazole or on amylose beads and eluted with high maltose, respectively. His-tagged protein expressed poorly, but low levels of soluble protein could be obtained by induction at an OD of 0.3. Substantial soluble protein was produced by the MBP fusion.
Pull-down assays using immunopurified APC
Immunopurified APC on αCdc27 beads was prebound with equal amounts of in vitro-translated hCdc20 or hCdh1 by incubation for 1 h on a TOMY shaker followed by addition of 35S-labeled in vitro-translated xMAD2L2 and incubation for 1 h. Beads were washed on minicolumns on a vacuum manifold with XB containing 500 mM KCl and 0.5% NP40, XB, and eluted with sample buffer. Parallel experiments using iAPC on αCdc27 beads and αCdc27 control beads were subjected to identical binding and washing conditions.
Preparation of low-speed supernatant interphase extracts and degradation assays
Extracts were prepared as described previously (Murray 1991; Pfleger and Kirschner 2000). Degradation assays were described previously (Pfleger and Kirschner 2000). Low-speed extracts were supplemented with ubiquitin, energy, and cycloheximide and buffer, Cdh1, or Δ90. After a 20-min preincubation with inhibitory protein (or buffer), in vitro-translated 35S-labeled substrate was added.
In vitro ubiquitination assays
Ubiquitination assays were described previously (Fang et al. 1998a; Pfleger and Kirschner 2000).
Substrate binding assays
Cold in vitro-translated myc-tagged hCdc20 or hCdh1 was prebound to 9E10 anti-myc coupled beads from Santa Cruz. Half the volume of 35S-methionine-labeled Xkid was added, in addition to 100 μg/mL cycloheximide 1mg/mL BSA in binding buffer (50 mM HEPES at pH 7.7, 100 mM NaCl, 1 mM MgCl2, 1 mM EDTA, and 0.2% Tween 20). After 2 h, samples were split and supplemented with buffer or protein, as indicated, and incubated for 20 min, followed by the addition of 10-fold excess of cold in vitro-translated Xkid. Samples were loaded onto minicolumns on a vacuum assembly and washed four times with 1 mL of binding buffer and eluted with sample buffer.
RNA and protein injection of Xenopus embryos and DNA staining
The indicated amounts of RNA or protein were injected into one cell of two-cell Xenopus embryos. To control for the Cdh1 rescue of MAD2L2 protein-injected embryos, boiled Cdh1 protein was coinjected with MAD2L2, showing no rescue. Capped CDH1 RNA was synthesized from linearized plasmid DNA templates. As the cap analog (GpppG) is a potent inhibitor of translation, care was taken to remove all unincorporated cap. Messenger RNAs were resuspended in water to 100–500 ng/μL. Embryos were fixed in MEMFA (0.1 M MOPS at pH 7.4, 2 mM EGTA, 1 mM MgSO4, and 3.7% formaldehyde) for 2 h at room temperature followed by three washes in PBS plus 0.2% Tween 20 (PBT). Embryos were either incubated with Hoechst (10 μg/mL) for 1 h at room temperature or Propidium Iodide (10 μg/mL) plus 20 μg/mL RNAse A for 1 h at 37°C. Propidium Iodide stained embryos were washed three times with PBT, dehydrated for 5 min in methanol, and mounted in 2:1 benyzl benzoate/benzyl alcohol.
Acknowledgments
We thank Louise Evans for technical help and Hui Zou for helpful suggestions in the preparation of this work. E. Lee is a Merck fellow supported by the Helen Hay Whitney Foundation. We thank the National Institute of General Medical Science (GM26875 and GM39023) for their support.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL marc@hms.harvard.edu; FAX (617) 432-0420.
Article and publication are at http://www.genesdev.org/cgi/doi/10.1101/gad.897901.
References
- Baumer M, Braus GH, Irniger S. Two different modes of cyclin Clb2 proteolysis during mitosis in Saccharomyces cerevisiae. FEBS Lett. 2000;468:142–148. doi: 10.1016/s0014-5793(00)01208-4. [DOI] [PubMed] [Google Scholar]
- Cahill DP, da Costa LT, Carson-Walter EB, Kinzler KW, Vogelstein B, Lengauer C. Characterization of MAD2B and other mitotic spindle checkpoint genes. Genomics. 1999;58:181–187. doi: 10.1006/geno.1999.5831. [DOI] [PubMed] [Google Scholar]
- Fang G, Yu H, Kirschner MW. Direct binding of CDC20 protein family members activates the anaphase-promoting complex in Mitosis and G1. Mol Cell. 1998a;2:163–171. doi: 10.1016/s1097-2765(00)80126-4. [DOI] [PubMed] [Google Scholar]
- Fang G, Yu H, Kirschner MW. The checkpoint protein MAD2 and the mitotic regulator Cdc20 form a ternary complex with the anaphase-promoting complex to control anaphase initiation. Genes & Dev. 1998b;12:1871–1883. doi: 10.1101/gad.12.12.1871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Funabiki H, Murray AW. The Xenopus Chromokinesin Xkid is essential for metaphase chromosome alignment and must be degraded to allow anaphase chromosome movement. Cell. 2000;102:411–424. doi: 10.1016/s0092-8674(00)00047-7. [DOI] [PubMed] [Google Scholar]
- Gieffers C, Peters BH, Kramer ER, Dotti CG, Peters JM. Expression of the CDH1-associated form of the anaphase-promoting complex in postmitotic neurons. Proc Natl Acad Sci. 1999;96:11317–11322. doi: 10.1073/pnas.96.20.11317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glotzer M. Mitosis: Don't get mad, get even. Curr Biol. 1996;6:1592–1594. doi: 10.1016/s0960-9822(02)70781-1. [DOI] [PubMed] [Google Scholar]
- Glotzer M, Murray AW, Kirschner MW. Cyclin is degraded by the ubiquitin pathway. Nature. 1991;349:132–137. doi: 10.1038/349132a0. [DOI] [PubMed] [Google Scholar]
- Hershko A, Ciechanover A. The ubiquitin system. Annu Rev Biochem. 1998;67:425–479. doi: 10.1146/annurev.biochem.67.1.425. [DOI] [PubMed] [Google Scholar]
- Irniger S, Nasmyth K. The anaphase-promoting complex is required in G1 arrested yeast cells to inhibit B-type cyclin accumulation and to prevent uncontrolled entry into S-phase. J Cell Sci. 1997;110:1523–1531. doi: 10.1242/jcs.110.13.1523. [DOI] [PubMed] [Google Scholar]
- Jaspersen SL, Charles JF, Morgan DO. Inhibitory phosphorylation of the APC regulator Hct1 is controlled by the kinase Cdc28 and the phosphatase Cdc14. Curr Biol. 1999;9:227–236. doi: 10.1016/s0960-9822(99)80111-0. [DOI] [PubMed] [Google Scholar]
- Kotani S, Tanaka H, Yasuda H, Todokoro K. Regulation of APC activity by phosphorylation and regulatory factors. J Cell Biol. 1999;146:791–800. doi: 10.1083/jcb.146.4.791. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Kramer ER, Scheuringer N, Podtelejnikov AV, Mann M, Peters JM. Mitotic regulation of the APC activator proteins CDC20 and CDH1. Mol Biol Cell. 2000;11:1555–1569. doi: 10.1091/mbc.11.5.1555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li R, Murray AW. Feedback control of mitosis in budding yeast. Cell. 1991;66:519–531. doi: 10.1016/0092-8674(81)90015-5. [DOI] [PubMed] [Google Scholar]
- Lorca T, Castro A, Martinez A-M, Vigneron S, Morin N, Sigrist S, Lehner S, Dorée M, Labbé JC. Fizzy is required for activation of the APC/cyclosome in Xenopus egg extracts. EMBO J. 1998;17:3565–3575. doi: 10.1093/emboj/17.13.3565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo X, Fang G, Coldiron M, Lin Y, Yu H, Kirschner MW, Wagner G. Structure of the Mad2 spindle assembly checkpoint protein and its interaction with Cdc20. Nat Struct Biol. 2000;7:224–229. doi: 10.1038/73338. [DOI] [PubMed] [Google Scholar]
- Murakumo Y, Roth T, Ishii H, Rasio R, Numata S-I, Croce CM, Fishel R. A human REV7 homolog that interacts with the polymerase ζ catalytic subunit hREV3 and the spindle assembly checkpoint protein MAD2. J Biol Chem. 2000;275:4391–4397. doi: 10.1074/jbc.275.6.4391. [DOI] [PubMed] [Google Scholar]
- Murray AW. Cell-cycle extracts. Methods Cell Biol. 1991;36:581–605. [PubMed] [Google Scholar]
- Nelson K, Schlondorff J, Blobel CP. Evidence for an interaction of the metalloprotease-disintegrin tumour necrosis factor alpha convertase (TACE) with mitotic arrest deficient 2 (MAD2), and of the metalloprotease-disintegrin MDC9 with a novel MAD2-related protein, MAD2β. Biochem J. 1999;343:673–680. [PMC free article] [PubMed] [Google Scholar]
- Petersen BO, Wagener C, Marinoni F, Kramer ER, Melixetian M, Denchi EL, Gieffers C, Matteucci C, Peters JM, Helin K. Cell cycle- and cell growth-regulated proteolysis of mammalian CDC6 is dependent on APC-CDH1. Genes & Dev. 2000;14:2330–2343. doi: 10.1101/gad.832500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfleger CM, Kirschner MW. The KEN box: An APC recognition signal distinct from the D box targeted by Cdh1. Genes & Dev. 2000;14:655–665. [PMC free article] [PubMed] [Google Scholar]
- Prinz S, Hwang ES, Visintin R, Amon A. The regulation of Cdc20 proteolysis reveals a role for the APC components Cdc23 and Cdc27 during S phase and early mitosis. Curr Biol. 1998;8:750–760. doi: 10.1016/s0960-9822(98)70298-2. [DOI] [PubMed] [Google Scholar]
- Salic AN, Kroll KL, Evans LM, Kirschner MW. Sizzled: A secreted Xwnt8 antagonist expressed in the ventral marginal zone of Xenopus embryos. Development. 1997;124:4739–4748. doi: 10.1242/dev.124.23.4739. [DOI] [PubMed] [Google Scholar]
- Schwab M, Lutum AS, Seufert W. Yeast Hct1 is a regulator of Clb2 cyclin proteolysis. Cell. 1997;90:683–693. doi: 10.1016/s0092-8674(00)80529-2. [DOI] [PubMed] [Google Scholar]
- Shirayama M, Zachariae W, Ciosk R, Nasmyth K. The Polo-like kinase Cdc5p and the WD-repeat protein Cdc20p/fizzy are regulators and substrates of the anaphase promoting complex in Saccharomyces cerevisiae. EMBO J. 1998;17:1336–1349. doi: 10.1093/emboj/17.5.1336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shirayama M, Toth A, Galova M, Nasmyth K. APCCdc20 promotes exit from mitosis by destroying the anaphase inhibitors Pds1 and cyclin Clb5. Nature. 1999;402:203–207. doi: 10.1038/46080. [DOI] [PubMed] [Google Scholar]
- Sigrist SJ, Lehner CF. Drosophila fizzy-related down-regulates mitotic cyclins and is required for cell proliferation arrest and entry into endocycles. Cell. 1997;90:671–681. doi: 10.1016/s0092-8674(00)80528-0. [DOI] [PubMed] [Google Scholar]
- Sorensen CS, Lukas C, Kramer ER, Peters JM, Bartek J, Lukas J. Non-periodic activity of the human Anaphase-Promoting Complex-Cdh1 ubiquitin ligase results in continuous DNA synthesis uncoupled from mitosis. Mol Cell Biol. 2000;20:7613–7623. doi: 10.1128/mcb.20.20.7613-7623.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Utrera R, Collavin L, Lazarevic D, Delia D, Schneider C. A novel p53-inducible gene coding for a microtubule-localized protein with G2-phase-specific expression. EMBO J. 1998;17:5015–5025. doi: 10.1093/emboj/17.17.5015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visintin R, Prinz S, Amon A. Cdc20 and Cdh1: A family of substrate-specific activators of APC-dependent proteolysis. Science. 1997;278:460–463. doi: 10.1126/science.278.5337.460. [DOI] [PubMed] [Google Scholar]
- Weinstein J. Cell cycle-regulated expression, phosphorylation, and degradation of p55Cdc. A mammalian homolog of CDC20/Fizzy/slp1. J Biol Chem. 1997;272:28501–28511. doi: 10.1074/jbc.272.45.28501. [DOI] [PubMed] [Google Scholar]
- Zachariae W, Nasmyth K. Whose end is destruction: Cell division and the anaphase-promoting complex. Genes & Dev. 1999;13:2039–2058. doi: 10.1101/gad.13.16.2039. [DOI] [PubMed] [Google Scholar]
- Zou H, McGarry TJ, Bernal T, Kirschner MW. Identification of a vertebrate sister-chromatid separation inhibitor involved in transformation and tumorigenesis. Science. 1999;285:418–422. doi: 10.1126/science.285.5426.418. [DOI] [PubMed] [Google Scholar]