Abstract
Ikaros is a unique regulator of lymphopoiesis that associates with pericentromeric heterochromatin and has been implicated in heritable gene inactivation. Binding and competition experiments demonstrate that Ikaros dimers compete with an Ets activator for occupancy of the lymphocyte-specific TdT promoter. Mutations that selectively disrupt Ikaros binding to an integrated TdT promoter had no effect on promoter function in a CD4+CD8+ thymocyte line. However, these mutations abolished down-regulation on differentiation, providing evidence that Ikaros plays a direct role in repression. Reduced access to restriction enzyme cleavage suggested that chromatin alterations accompany down-regulation. The Ikaros-dependent down-regulation event and the observed chromatin alterations appear to precede pericentromeric repositioning. Current models propose that the functions of Ikaros should be disrupted by a small isoform that retains the dimerization domain and lacks the DNA-binding domain. Surprisingly, in the CD4+CD8+ thymocyte line, overexpression of a small Ikaros isoform had no effect on differentiation or on the pericentromeric targeting and DNA-binding properties of Ikaros. Rather, the small isoform assembled into multimeric complexes with DNA-bound Ikaros at the pericentromeric foci. The capacity for in vivo multimer formation suggests that interactions between Ikaros dimers bound to the TdT promoter and those bound to pericentromeric repeat sequences may contribute to the pericentromeric repositioning of the inactive gene.
Keywords: Ikaros, TdT, lymphocytes, transcription
Lymphocyte development is regulated by extracellular and intracellular signals that drive sequential changes in the gene expression program within the nucleus. Our understanding of the molecular events that control lymphopoiesis is limited, but has greatly benefited from the identification of proteins that bind control regions for lymphocyte-specific genes (Glimcher and Singh 1999). Ikaros and its family members, Aiolos and Helios, are intriguing examples of DNA-binding proteins that contribute to the development of all lymphocyte lineages (for review, see Cortes et al. 1999). Members of the Ikaros family influence developmental events at least in part by regulating signaling thresholds (for review, see Cortes et al. 1999).
Although Ikaros contains a classical C2H2 zinc finger DNA-binding domain, the functions of this protein appear to be atypical. Two methods, immunogold electron microscopy and immunoFISH, have demonstrated predominant localization to pericentromeric heterochromatin in progenitor and mature lymphocytes (Brown et al. 1997; Klug et al. 1998). The immunoFISH method revealed a striking correlation between the physical location of Ikaros and that of a variety of inactive, developmentally regulated genes (Brown et al. 1997, 1999), providing evidence that genes migrate to pericentromeric foci during heritable inactivation. This notion gained support from a recent study of stable transgenes, which revealed that an active enhancer is required for transgene escape from centromeric foci (Francastel et al. 1999).
The colocalization of Ikaros with inactive genes suggests that Ikaros contributes to heritable gene inactivation, possibly by recruiting genes to foci of pericentromeric heterochromatin. Ikaros and inactive genes migrate towards these foci with similar kinetics as B cells enter the cell cycle, thus providing support for this hypothesis (Brown et al. 1999). Further support for a role in repression was provided by the finding that Ikaros can interact with histone deacetylase complexes, and that GAL4–Ikaros fusion proteins repress transcription from some artificial promoters on transient overexpression (Kim et al. 1999; Koipally et al. 1999).
Although the above results suggest that Ikaros contributes to heritable gene inactivation, this function remains hypothetical. The need for a more extensive examination is highlighted by evidence that Ikaros (1) colocalizes with DNA replication forks (Avitahl et al. 1999), (2) interacts with SWI/SNF ATP-dependent nucleosome remodeling complexes in addition to histone deacetylases (Kim et al. 1999; O'Neill et al. 2000), (3) contains an activation domain (Molnár and Georgopoulos 1994), and (4) fails, in its native state, to repress artificial reporter genes in transient and stable transfection assays (B.S. Cobb, unpubl.). Furthermore, the hypothesis that Ikaros directly recruits genes to centromeric foci must be reconciled with our recent finding that Ikaros is targeted to centromeric foci via direct DNA binding, most likely to pericentromeric repeat sequences (Cobb et al. 2000). Unless Ikaros forms multimeric complexes in vivo, the DNA-binding domains of Ikaros dimers bound to pericentromeric repeats would not be available for interactions with target genes.
Our understanding of Ikaros has been limited in part by the absence of primary target genes. Several genes contain Ikaros binding sites, but their functional relevance has not been established. One candidate for a primary target of Ikaros is the gene-encoding terminal transferase (TdT), a template-independent DNA polymerase expressed in immature lymphocytes that increases junctional diversity of immunoglobulin (Ig) and TCR genes (for review, see Ernst et al. 1999). In double-positive thymocytes, the TdT gene, along with the RAG-1 and RAG-2 genes, is rapidly down-regulated by TCR cross-linking or treatment with PMA plus ionomycin. Heritable inactivation of the TdT gene is accompanied by an increased propensity for pericentromeric localization (Brown et al. 1999). In a transformed cell line in which the TdT gene is only transiently down-regulated, pericentromeric repositioning was not observed (Brown et al. 1999). Ikaros binds an element in the TdT promoter that is required for transcription, but this element also binds members of the Ets family (Ernst et al. 1993, 1996; Hahm et al. 1994, 1998). An analysis of promoter point mutations in a transient transfection assay revealed that the nucleotides required for Ets protein binding correlate closely with those required for promoter activity (Ernst et al. 1993, 1996). In contrast, mutations that selectively disrupt Ikaros binding without altering Ets binding had either no effect or slightly enhanced promoter activity. These findings strongly suggest that an Ets protein is a relevant activator of TdT transcription (Ernst et al. 1996). Additional experiments provided evidence that the relevant Ets family member is Elf-1 (Ernst et al. 1996).
Although the TdT promoter analysis failed to identify a function for Ikaros, the presence of an Ikaros binding site overlapping the Elf-1 site suggests that Ikaros may cooperate with Elf-1 during TdT activation or, alternatively, contribute to down-regulation by competing with Elf-1. The results presented here demonstrate that intact Ikaros binding sites are essential for transcriptional down-regulation of the TdT gene in double-positive thymocytes, strongly suggesting that Ikaros makes a direct contribution to down-regulation. The results also suggest that the chromatin structure of the TdT promoter is altered on down-regulation and that some chromatin alterations precede pericentromeric repositioning. Finally, evidence is presented that Ikaros forms multimeric structures in vivo, helping to reconcile the binding of Ikaros to both target genes and pericentromeric repeats. Together, these findings support the hypothesis that Ikaros plays a direct role in heritable gene inactivation.
Results
Delineation of tandem Ikaros binding sites within the TdT D‘ element
The ability of Ikaros to bind the TdT D‘ element raises the possibility that it cooperates with an Ets protein during transcriptional activation or competes with an Ets protein during inactivation. To examine these possibilities in greater depth, the binding sites within the D‘ element were defined using gel shift and methylation-interference assays. These detailed experiments were essential for elucidating the potential for simultaneous or competitive binding of Ets and Ikaros proteins. More importantly, these experiments were a prerequisite for the functional studies presented in Figure 4 (see below).
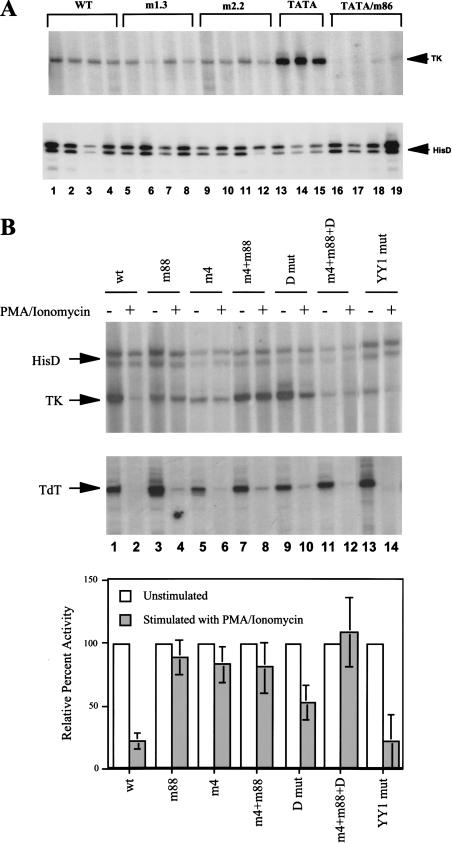
Figure 4.
Ikaros recognition sequences are required for TdT promoter down-regulation. (A) Stable transfection experiments were performed in RLm11 cells, using the wild-type and mutant TdT promoters fused to an HSV–TK reporter gene (see Materials and Methods). TdT promoter activity was monitored by primer extension in three or four independent clones from each plasmid (top). The promoters analyzed included the wild-type promoter (lanes 1–4), two different mutants that disrupt sequences between −25 and −35 (m1.3, lanes 5–8; m2.2 lanes 9–12), a promoter variant containing a consensus TATA box (lanes 13–15), and a promoter variant containing the consensus TATA box and mutant D‘ element (lanes 16–19). HisD transcription was monitored by primer extension as a control (bottom). (B) Stable transfection experiments were performed in VL3–3M2 cells using the wild-type TdT(TATA) promoter and the promoters containing mutations in the D, D‘, and YY1 elements, as indicated at the top of each lane, with all promoters containing the TATA box. Cells were left untreated (odd-numbered lanes) or were treated with PMA-ionomycin (even-numbered lanes). HSV–TK, HisD, and endogenous TdT mRNAs were monitored by primer extension. The graph at the bottom shows the activity of each promoter, obtained by averaging results from three independent cell clones. HSV–TK signals were normalized to the HisD signals and are presented as a percentage of the signal obtained in the absence of PMA-ionomycin stimulation (set at 100%).
Previous DNase I footprinting studies revealed that partially purified Ikaros protects a 30-bp region (−48 to −77), suggesting that the D‘ element may contain two or more Ikaros binding sites (Lo et al. 1991; Ernst et al. 1993). Indeed, two sites that resemble core recognition motifs for Ikaros are present within the D‘ element (Fig. 1A, bold; Hahm et al. 1998). Recombinant GST–Ikaros containing the amino-terminal half of the protein (including the DNA-binding zinc fingers) yielded a single abundant gel shift complex with a probe containing the wild-type D‘ element (Fig. 1B, lane 3). This complex was unaffected by five different mutations spanning the element (Fig. 1B, lanes 4–8), consistent with the possibility that recombinant Ikaros binds independently to two sites within the D‘ element, with each mutation disrupting only one of the two sites. The absence of a slow-mobility complex containing GST–Ikaros bound simultaneously to both sites within the wild-type probe suggests that binding of the recombinant protein was not cooperative. (In fact, competition experiments suggested a slight negative cooperativity [data not shown], possibly caused by steric effects on binding of the recombinant fusion protein.) Notably, the complex observed with GST–Ikaros migrates much more rapidly than that observed with native Ikaros from lymphocyte extracts (Fig. 1B, lane 1; complex a), despite the fact that the molecular weight of recombinant GST–Ikaros (containing Ikaros amino acids 1–304) is comparable to that of full-length Ikaros. This difference suggests that GST–Ikaros binds as a monomer and native Ikaros, as a dimer, as predicted previously (Hahm et al. 1994; Molnár and Georgopoulos 1994). Monomeric binding by GST–Ikaros was not surprising because it lacks the carboxy-terminal dimerization domain (Sun et al. 1996). We have had difficulty expressing recombinant, full-length proteins that retain this domain, as it appears to be removed by proteolysis.
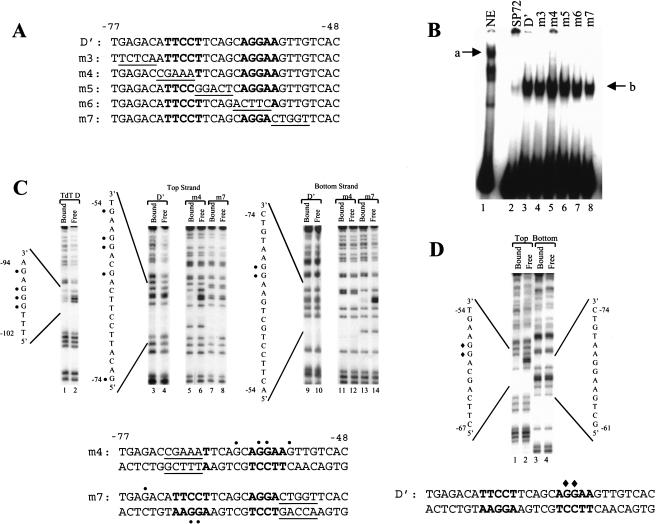
Figure 1.
Interactions between Ikaros and Elf-1 at the TdT D‘ element. (A) The sequence of the TdT D‘ element is shown (−48 to −77), along with the sequences of five mutant elements (mutant sequences underlined). Potential recognition sites for Ikaros are indicated in bold. (B) A gel shift experiment was performed with GST–Ikaros using probes containing the wild-type or mutant D‘ elements (lanes 3–8; complex b). Lane 2 shows background binding of GST–Ikaros to a probe derived from vector sequences. Lane 1 shows the Ikaros-containing complex (a) formed with nuclear extracts from RLm11 cells and the D‘ probe. (C) Methylation interference experiments were performed with GST–Ikaros and probes containing the D (lanes 1,2), D‘ (lanes 3,4,9,10), D‘m4 (lanes 5,6,11,12), and D‘m7 (lanes 7,8,13,14) elements. Reaction products from bound and free probe samples are shown, as indicated at the top. D‘ probes were labeled on the sense (lanes 3–8) or antisense (lanes 9–14) strands. The guanines that inhibit Ikaros binding when methylated are depicted on the DNA sequences (circles). (D) A methylation interference experiment was performed with GST–Elf-1. Guanines that inhibit binding when methylated are depicted (diamonds).
Next, methylation interference experiments were performed. As an initial control, an isolated Ikaros binding site located between −102 and −94 of the TdT promoter was examined. Ikaros can bind this element (the D element; Hahm et al. 1994), but its in vivo relevance remains uncertain because substitution mutations had no effect on promoter activity in transient assays (Lo et al. 1991; see below). As shown in Figure 1C (lanes 1 and 2), the Ikaros–D interaction was abolished by methylation of three guanines (−97 to −99) and weakened by methylation of a fourth guanine (−95). In contrast, the Ikaros–D‘ interaction was not abolished by methylation of any of the guanines, although some weak effects were observed (Fig. 1C, lanes 3,4,9,10). This result supports the hypothesis that recombinant Ikaros recognizes two sites independently, as each site would be occupied on only ∼50% of the probe molecules.
The two recognition sites for Ikaros within the wild-type D‘ element were revealed by analysis of mutants m4 and m7, which disrupt the distal and proximal consensus sites, respectively. When the distal site was disrupted, two essential guanine contacts were observed at the proximal end of the D‘ element (Fig. 1C, lanes 5 and 6, nucleotides −58 and −59). Two other guanines made modest contributions to binding affinity (Fig. 1C, lanes 5 and 6, nucleotides −55 and −62). When the proximal site was disrupted, three guanines at the distal end of D‘ were critical for binding, one on the top strand (Fig. 1C, lanes 7 and 8, nucleotide −74) and two on the bottom (Fig. 1C, lanes 13 and 14, nucleotides −67 and −68). These data confirm the existence of two tandem Ikaros binding sites.
Evidence of mutually exclusive binding by Ikaros and Ets proteins
The nucleotides found to be critical for Ikaros binding at the proximal end of the D‘ element (−58 and −59) are within the core Ets recognition sequence. Previous methylation interference and X-ray crystallographic studies revealed that these guanines are critical for Ets-protein binding (Graves and Peterson 1998). As expected, methylation of these same guanines prevented binding of recombinant Elf-1 (Fig. 1D), the putative activator of TdT transcription (Ernst et al. 1996). Because a methyl group on either of these guanines disrupts binding of both Elf-1 and Ikaros, the two proteins appear to occupy the same physical space when bound to the proximal end of D‘, strongly suggesting that they cannot bind simultaneously. It therefore is highly unlikely that Elf-1 and Ikaros act in concert with one another at the proximal D‘ site during TdT activation.
Competition between Ets and Ikaros proteins
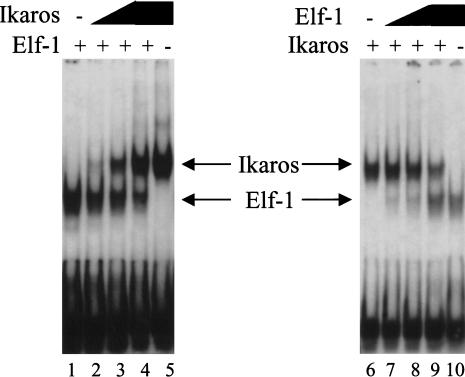
As an independent method of comparing Ets and Ikaros interactions at the D‘ element, competition experiments were performed using a gel shift assay. Addition of increasing concentrations of GST–Ikaros to binding reactions containing the D‘ probe and constant GST–Elf-1 led to a gradual reduction in the abundance of the Elf-1 complex, with no appearance of a slower-mobility complex that would suggest simultaneous binding of Ikaros and Elf-1 (Fig. 2, lanes 1–4). Similarly, addition of increasing concentrations of GST–Elf-1 in the presence of constant GST–Ikaros resulted in a gradual reduction in the Ikaros complex, again with no appearance of a slower-mobility complex (Fig. 2, lanes 6–9). Because these results were obtained in vitro with recombinant proteins, they do not demonstrate that native Ikaros and Elf-1 compete for D‘ occupancy in vivo under physiological conditions. However, they agree with the methylation interference results and lend further credence to the hypothesis that Ikaros and Elf-1 cannot bind simultaneously to the proximal end of the D‘ element.
Figure 2.
Competition between Ikaros and Ets proteins for D‘ occupancy. Gel shift assays were performed with constant GST–Elf-1 and increasing concentrations of GST–Ikaros (lanes 2–4), or constant GST–Ikaros with increasing GST–Elf-1 (lanes 7–9). Reactions with GST–Elf-1 alone (lanes 1,10) and GST–Ikaros alone (lanes 5,6) are also shown.
Binding of a native Ikaros dimer or multimer to the TdT D‘ element
To characterize the DNA-binding properties of native Ikaros, gel shift experiments were performed with nuclear extracts from the TdT-expressing thymocyte line, RLm11. The predominant complex obtained with the D‘ element migrated slowly (Fig. 3A, lane 1). As demonstrated previously by its sensitivity to antibodies (Hahm et al. 1994), this complex contains Ikaros (Fig. 3A, lane 3). A faster migrating Elf-1 complex was detected with different gel conditions (Ernst et al. 1996), but not with the conditions used in this study.
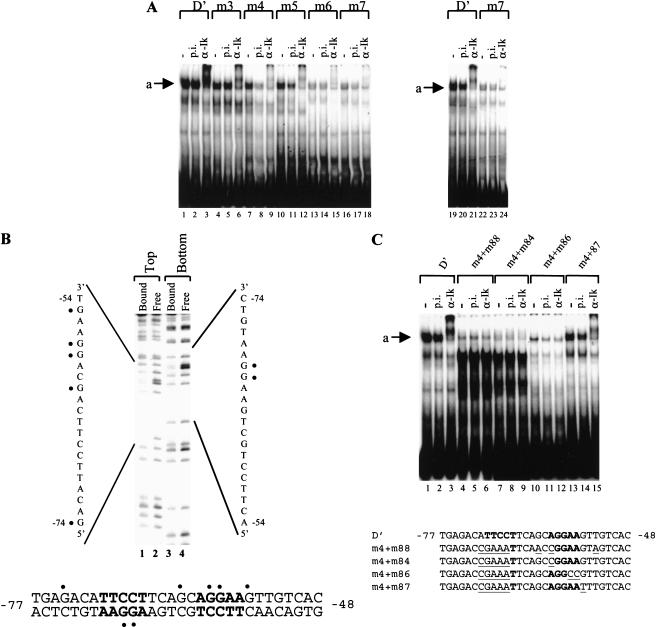
Figure 3.
Interactions between native Ikaros and the TdT D‘ element. (A) Gel shift experiments were performed with nuclear extracts from RLm11 cells, using the wild-type and mutant D‘ probes, as indicated (top). Reactions contained no antibody, preimmune (p.i.), or anti-Ikaros serum (α-Ik). The Ikaros-containing complex is indicated (a). (B) A methylation interference experiment was performed with complex “a” obtained with the wild-type D‘ probe. Pools of bound and free probe molecules were analyzed. Probes were labeled on the top or bottom strand as indicated. Guanines that inhibit complex formation when methylated are indicated on the sequence (circles). (C) Gel shift experiments were performed with nuclear extracts from RLm11 cells, using wild-type and mutant D‘ elements, as indicated at the top and in the diagram at the bottom (mutant sequences underlined; Ikaros recognition sites in bold). The presence of p.i. or α-Ik antibodies is indicated at the top.
Analysis of probes containing the 5-bp mutations revealed that the abundance of the slow-mobility Ikaros complex was reduced significantly by all of the mutations (Fig. 3A, lanes 4–18), suggesting that the stability of the endogenous complex depends on both Ikaros consensus sites within the D‘ element. Disruption of the distal Ikaros recognition site (m3, m4, and m5) reduced complex abundance by only a few-fold, but disruption of the proximal site (m6 and m7) reduced complex abundance dramatically. Because complex abundance in lanes 1–18 (Fig. 3A) was gradually diminished from left to right, the difference between the wild-type and m7 probes was confirmed in a side-by-side comparison (Fig. 3A, lanes 19–24). The substantial reduction in complex formation when either the proximal site or distal site was mutated suggests that native Ikaros binds cooperatively to the two sites. Most importantly, because the native complex binds poorly to the distal site in the absence of the proximal site, Ikaros is unlikely to occupy the distal site while an Ets protein occupies the proximal site. It is interesting to note that preferential binding to the proximal site was not observed with recombinant Ikaros in either direct binding or competition experiments (Fig. 1; data not shown). Because the recombinant protein contained the isolated DNA-binding domain of Ikaros, preferential binding to the proximal site does not appear to be an intrinsic property of the DNA-binding domain, but rather a unique property of full-length native Ikaros.
The simplest interpretation of the above results is that the slow-mobility complex contains an Ikaros dimer bound to both halves of D‘. Alternatively, the native complex may contain an Ikaros subunit and an Ets subunit bound cooperatively to the distal and proximal sites, respectively. Although the slow-mobility complex was unaffected by Elf-1 antibodies (Ernst et al. 1996), it remained possible that this complex contains a related Ets protein. To examine this possibility, two experiments were performed. First, a methylation interference experiment with the native complex demonstrated that precisely the same seven guanines involved in binding of recombinant Ikaros contributed to binding of the native complex (Fig. 3B), suggesting that Ikaros proteins are responsible for both interactions. Second, gel shift experiments were performed with probes containing mutations that selectively disrupt Ikaros- or Ets-protein binding. This experiment was based on previous evidence that different sets of nucleotides are required for Ikaros and Ets binding (Ernst et al. 1996). Specifically, mutations m84 and m88, both of which alter an adenosine at −60 (see Fig. 3C, bottom), disrupted binding of recombinant Ikaros, with no effect on binding of Ets-1, Fli-1, or Elf-1 (Ernst et al. 1996). In contrast, mutation m87, which alters a guanine at −55, disrupted binding of Ets proteins, with no effect on Ikaros binding. A fourth mutation, m86, alters nucleotides −56 and −57 and disrupted binding of both proteins.
By themselves, none of these mutations had a significant effect on the slow-mobility complex (data not shown), presumably because sufficient binding energy was provided by the distal site in combination with the remaining wild-type nucleotides at the proximal site. The mutations were therefore coupled to the m4 mutation, which disrupts the distal site. Gel shift analyses revealed that the complex was strongly reduced by the m4+m84 and m4+m88 mutations, as well as by m4+m86 (Fig. 3C, lanes 4–12). In contrast, the m4+m87 mutation reduced complex formation by only ∼threefold, comparable to the effect of the m4 mutation by itself (Fig. 3C, lanes 13–15; see Fig. 3A). (The abundant complexes that appear in Figure 3A lanes 4–9 do not appear to be either Ets or Ikaros proteins on the basis of antibody supershift experiments [data not shown].) These results further confirm that, in the native complex, Ikaros proteins contact both the proximal and distal ends of D‘.
An Ikaros–D‘ interaction is required for TdT down-regulation
The above results suggest that Ikaros complexes may compete for TdT promoter occupancy and therefore may be involved in TdT down-regulation. Because Ikaros proteins appear to be essential for the survival of developing lymphocytes (Georgopoulos et al. 1994), Ikaros-deficient cells most likely cannot be used to assess the importance of Ikaros for TdT down-regulation. (The survival of some T cells in Ikaros null mice is most likely attributable to compensation by Helios and Aiolos [Cortes et al. 1999].) Therefore, to examine the dynamic relationship between Ikaros and Ets proteins at the TdT promoter, a stable transfection assay was designed in which the dynamics could be recapitulated and altered. This system involved the stable integration of well-characterized TdT promoter mutants into cells that can be stimulated to undergo a physiologically relevant developmental transition that is accompanied by the down-regulation of TdT transcription. The detailed binding studies described above were a prerequisite for this analysis.
To develop a stable transfection assay, a 1700-bp TdT promoter fragment (−1700 to +58) was inserted into a vector containing a herpes simplex virus thymidine kinase (HSV-TK) reporter gene and a hisD dominant selectable marker (under the control of an SV40 promoter). This plasmid was initially transfected into the TdT-expressing thymocyte line, RLm11. Clones that had integrated the plasmid were selected and expanded in the presence of histidinol. TdT promoter activity was then monitored by primer extension using a primer complementary to HSV-TK sequences. Figure 4A (lanes 1–4, top) shows the primer extension signal obtained with four independent clones. Weak, but consistent TdT promoter activity was detected, with transcription initiating at the correct start site. HisD transcripts from the SV40 promoter were also monitored (Fig. 4A, bottom).
Although the TdT promoter was active in this assay, promoter activity was weak in the RLm11 line and even weaker in other cell lines that were analyzed (data not shown). In an attempt to enhance promoter activity without losing physiological regulation, a consensus TATA box was introduced into the −30 region of the promoter. Previous studies demonstrated that a consensus TATA box enhanced TdT promoter activity in transient transfection and in vitro transcription assays while retaining D‘-dependent transcription (Garraway et al. 1996). The results in Figure 4A reveal a similar effect in the stable transfection assay. In this assay, insertion of a consensus TATA box at the −30 region of the TdT promoter enhanced promoter activity 5–10-fold (Fig. 4A, TATA, lanes 13–15). In contrast, two different 5-bp mutations at the −30 region that did not introduce a TATA box were found to have no significant effect on promoter activity (Fig. 4A, m1.3 and m2.2, lanes 5–12), demonstrating that the −30 region of the wild-type TdT promoter does not contain an important control element. Importantly, when a D‘ mutation that disrupts Ets protein binding (and also Ikaros binding) was introduced into the TdT promoter containing a TATA box, promoter activity was abolished (Fig. 4A, TATA/m86, lanes 16–19). This result demonstrates that D‘-dependence was retained in the presence of the TATA box.
To test the relevance of Ikaros for TdT down-regulation, the stable transfection assay was used in the VL3–3M2 murine thymocyte line (Groves et al. 1995). This line exhibits properties of double-positive thymocytes and can be induced with PMA plus ionomycin to undergo the early stages of positive selection, which include the rapid down-regulation of TdT transcription. Chromosomal integration of the TdT(TATA)-TK reporter into the VL3–3M2 line resulted in efficient promoter activity (Fig. 4B, lane 1), consistent with the RLm11 results. Importantly, treatment with PMA plus ionomycin for 24 h resulted in the down-regulation of promoter activity, demonstrating that the 1700-bp promoter fragment is sufficient for down-regulation. Figure 4B (lanes 1 and 2) shows the primer extension results obtained with a representative clone. The bar graph at the bottom shows the quantified results from three independent cell clones following normalization to the respective hisD primer extension signals. As an additional control, endogenous TdT mRNA levels were monitored by primer extension (Fig. 4B). The results reveal that steady-state HSV-TK mRNA was reduced an average of fivefold. A similar fivefold reduction in steady-state TK mRNA was observed when the cells were treated with Actinomycin D instead of PMA plus ionomycin (data not shown), suggesting that the residual TK mRNA may represent incomplete degradation rather than residual, ongoing transcription.
It is interesting to note that normalization of the TK signal to the hisD signal was possible because hisD transcription was not significantly down-regulated on addition of PMA plus ionomycin, despite the expected close linkage of the hisD gene and TK reporter gene. (The slight reductions in the hisD signals in Figure 4, lanes 2 and 4 of the gel image were not reproducible.) The maintenance of hisD transcription suggests that the down-regulation of TdT promoter activity in the stable transfection assay was not accompanied by long-range chromatin modifications (see below).
Next, cell lines were generated with TdT(TATA)-TK reporter genes containing promoter mutations. With a promoter mutant that disrupts binding of both Elf-1 and Ikaros, no drug-resistant cells lines were obtained that yielded a detectable TK mRNA signal in either unstimulated or stimulated cells (data not shown), consistent with the results obtained in RLm11 cells. In contrast, the m88 mutation, which disrupts the proximal Ikaros recognition site without affecting Ets binding, retained strong promoter activity in unstimulated cells (Fig. 4B, lane 3). The absolute activity varied from clone to clone, as would be expected for differences in copy number and chromosomal integration site (data not shown). Importantly, the promoter activity observed in three independent cell clones was reduced only slightly following treatment with PMA plus ionomycin (Fig. 4B, lane 4). Similar results were obtained with the m4 and m4+m88 mutants (Fig. 4B, lanes 5–8). Although the m88 mutation by itself does not lead to an appreciable effect on the binding of native Ikaros in a gel shift assay (see above), the hypothesis that Ikaros and Ets proteins compete for promoter occupancy implies that relatively small differences in binding affinity could alter the competition.
A mutation was also tested in the Ikaros binding site at −100 (the D element). This mutation, like the m4 and m88 mutations, had no consistent effect on promoter activity in unstimulated cells, but down-regulation by PMA plus ionomycin was limited to an average of twofold (Fig. 4B, lanes 9 and 10). Down-regulation was abolished when the m4, m88, and D mutations were combined in the same plasmid (Fig. 4B, lanes 11 and 12). As a final control, a mutation was introduced into a recognition site for YY1, located immediately adjacent to the D element: This mutant promoter was down-regulated an average of fivefold following PMA plus ionomycin treatment (Fig. 4B, lanes 13 and 14), comparable to the results obtained with the wild-type promoter. Taken together, these results provide strong evidence that Ikaros binding at the D‘ element, perhaps with some contribution from the D element, is essential for the down-regulation of TdT promoter activity.
TdT down-regulation is accompanied by an altered chromatin structure
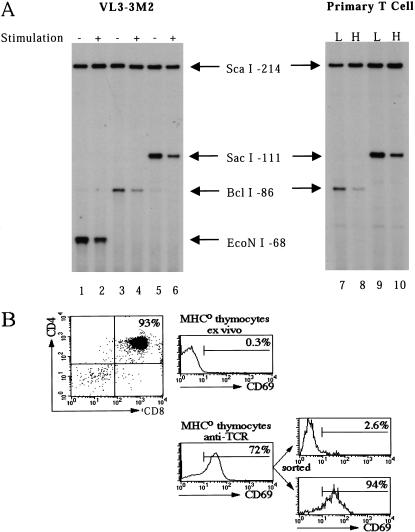
To determine whether chromatin alterations occur at the TdT promoter during down-regulation in VL3–3M2 cells, a restriction enzyme accessibility assay was employed (Weinmann et al. 1999). Briefly, nuclei from unstimulated and stimulated cells were treated with a limiting concentration of a restriction enzyme that recognizes a site within the TdT promoter. The cleaved genomic DNA was purified and digested in vitro to completion with a different restriction enzyme that recognizes a site located farther upstream. The genomic DNA cleavage products were then analyzed by LM–PCR. The in vitro cleavage step is critical for normalization of the results, which are typically presented as the ratio of nuclear cleavage to in vitro cleavage. A decrease in this ratio on cell stimulation would suggest that the locus had assumed a tighter chromatin configuration.
The restriction enzyme analysis was performed with the endogenous TdT promoter instead of the stably integrated promoters because the latter are constitutively accessible to restriction enzyme cleavage (Weinmann et al. 2001; data not shown). The results in Figure 5A reveal a substantial (three- to fivefold) decrease in nuclear cleavage on stimulation of VL3–3M2 cells when any of three enzymes that cleave within the endogenous TdT promoter were tested (Fig. 5A, lanes 1–6). These results suggest that TdT down-regulation is accompanied by chromatin alterations.
Figure 5.
Restriction enzyme accessibility analysis of the endogenous TdT promoter. (A) The restriction enzyme accessibility assay (see text) was performed with nuclei from unstimulated (lanes 1,3,5) or stimulated (lanes 2,4,6) VL3–3M2 cells, using three different restriction enzymes for nuclear cleavage (EcoNI, BclI, and SacI). The in vitro normalization cleavage was performed with ScaI. Cleavage products were analyzed by LM–PCR, using primers complementary to the TdT promoter sequences. CD69loCD5lo (lanes 7,9) and CD69hiCD5hi (lanes 8,10) populations were analyzed following nuclear cleavage with two different restriction enzymes. (B) A profile of the unstimulated MHC0 thymocyte population is shown, indicating that 93% of the cells were CD4+CD8+. Before and after stimulation by TCR cross-linking, 0.3 and 72% of the cells, respectively, expressed CD69. The stimulated cells were sorted into CD69lo and CD69hi populations, which were used for the restriction enzyme assay.
Previous studies have shown that the heritable down-regulation of TdT transcription in primary double-positive thymocytes is accompanied by an increased propensity for pericentromeric positioning of the TdT locus (Brown et al. 1999). However, on stimulation of VL3–3M2 cells, repositioning was not observed and the TdT gene was only transiently down-regulated; removal of the PMA-ionomycin resulted in the rapid restoration of transcription (Brown et al. 1999). These results suggest that VL3–3M2 cells support some, but not all of the steps involved in pericentromeric repositioning and heritable inactivation.
To determine whether pericentromeric repositioning and heritable inactivation of the TdT locus are accompanied by more dramatic changes in chromatin structure than were observed in VL3–3M2 cells, primary double-positive thymocytes were analyzed using the restriction enzyme assay. Double-positive (CD4+CD8+) thymocytes were isolated from MHC-deficient (MHC0) mice as described previously (Brown et al. 1999). After stimulation by TCR cross-linking, 72% of the cells up-regulated expression of CD69, which serves as an activation marker (Fig. 5B). This population was sorted into CD69hi and CD69lo subpopulations, which, as demonstrated previously, are TdT− and TdT+, respectively (Fig. 5B; Brown et al. 1999). In CD69hi cells, about 75% of the TdT loci are positioned at pericentromeric foci (Brown et al. 1999).
Analysis of the CD69hi and CD69lo populations by restriction enzyme accessibility revealed substantially reduced accessibility in the CD69hi population (Fig. 5A, lanes 7–10), consistent with an altered chromatin structure. Interestingly, the magnitude of this reduced accessibility relative to the CD69lo population was similar to that observed in stimulated versus unstimulated VL3–3M2 cells, despite the different propensities for centromeric localization of the TdT loci in the primary and transformed cells. Similar results were obtained when unstimulated and stimulated primary thymocytes were compared in the absence of sorting (data not shown).
One potential explanation for the similar results in VL3–3M2 cells and primary cells is that the magnitude of the reduction may be underestimated in the primary cells, because the TdT gene may be active in only a fraction of the CD69lo cells (resulting in an artificially low signal in this population). However, an alternative and more intriguing possibility is that the inactive TdT locus in VL3–3M2 cells may have acquired a subset of the chromatin changes that occur during heritable down-regulation (i.e., those changes that diminish restriction enzyme access), despite the fact that the locus is only transiently inactive and has not been repositioned to pericentromeric heterochromatin.
In vivo evidence that Ikaros proteins form multimeric complexes at pericentromeric foci
The finding that Ikaros contact sites within the TdT promoter are essential for transcriptional down-regulation suggests that Ikaros may help recruit the TdT locus to foci of pericentromeric heterochromatin, where Ikaros molecules are concentrated. Our previous results demonstrated, however, that Ikaros dimers are themselves targeted to the pericentromeric foci by direct DNA binding, presumably to pericentromeric repeats (Cobb et al. 2000). If Ikaros dimers are bound to pericentromeric repeats, their DNA-binding domains will not be available for binding to target genes.
One hypothesis that would be consistent with both of the above results is that Ikaros molecules bound to inactive genes (e.g., the TdT gene) form multimeric or lattice structures with Ikaros molecules bound to pericentromeric repeats, thereby recruiting the inactive genes to pericentromeric foci. We demonstrated previously that Ikaros migrates in a broad, high molecular weight peak when analyzed by gel filtration chromatography (Hahm et al. 1998), providing some support for this hypothesis. However, it was not clear from these experiments whether Ikaros multimers exist in vivo. It also was not clear whether the complexes observed in vitro contained Ikaros multimers or an Ikaros dimer associated with other proteins, as suggested previously (Kim et al. 1999). Furthermore, our attempts to determine the size of Ikaros complexes using other approaches (e.g., chemical cross-linking and glycerol gradient analyses) have revealed that the predominant stable species in nuclear extracts is a dimer (Hahm et al. 1998; A.S. McCarty, B.S. Cobb, and S.T. Smale, unpubl.). It therefore is not known whether Ikaros molecules exist as dimers or as multimeric structures at pericentromeric foci in vivo.
If Ikaros can indeed form multimeric structures at pericentromeric foci, one prediction is that Ikaros molecules lacking the DNA-binding domain would be capable of associating with endogenous Ikaros dimers that are directly bound to the pericentromeric foci. The experiments described below serve as a critical test of this prediction. It is important to note, however, that these experiments were originally designed with an alternative goal in mind. The original intent was to overexpress a dominant negative isoform of Ikaros that would disrupt high-affinity DNA-binding by endogenous Ikaros proteins. If our current models are correct, disruption of high-affinity DNA-binding should disrupt the centromeric targeting of endogenous Ikaros, as well as TdT down-regulation.
We anticipated that neutralization of the dimerization domain of Ikaros would be the preferred method for disrupting high-affinity DNA-binding. This prediction was based on our knowledge that the zinc finger dimerization domain of Ikaros isoform VI (or a leucine zipper engineered in its place) is required for centromeric targeting and for high-affinity binding to most sites, including the binding sites within the TdT promoter and pericentromeric repeats (Cobb et al. 2000). To neutralize the dimerization domain of endogenous Ikaros proteins, Ikaros isoform I was chosen because it lacks the amino-terminal DNA-binding domain but retains the carboxy-terminal zinc finger dimerization domain (Hahm et al. 1994; Sun et al. 1996). In fact, using recombinant proteins expressed in Escherichia coli, this isoform, which is naturally expressed in some primary and transformed cells (Hahm et al. 1994; Klug et al. 1998), was found previously to disrupt DNA binding by large Ikaros isoforms (Sun et al. 1996). The results presented below dramatically alter our view of the dominant negative potential of small Ikaros isoforms. At the same time, the results provide strong support for the hypothesis that Ikaros molecules form multimeric structures at pericentromeric foci.
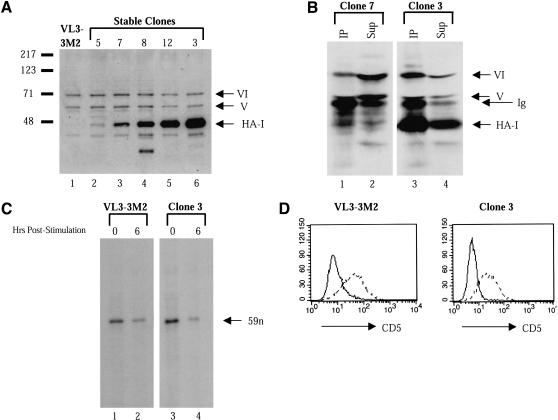
For these experiments, an HA epitope-tagged version of Ikaros isoform I (HA-I; Hahm et al. 1994) was overexpressed in VL3–3M2 cells using the pEBB expression vector (Mayer et al. 1995). Drug-resistant clones that had stably integrated the plasmid were isolated and screened for HA-I expression by Western blot using Ikaros antibodies. Figure 6A (lanes 2–6) shows the expression level of HA-I relative to that of the endogenous Ikaros isoforms (V and VI) in extracts from five independent clones. HA-I was expressed at variable levels, with the highest expression observed in clones 12 and 3 (Fig. 6A, lanes 5 and 6).
Figure 6.
Overexpression of HA-tagged Ikaros isoform I (HA-I) in VL3–3M2 cells. (A) A Western blot was performed with extracts from untransfected VL3–3M2 cells (lane 1) and five HA-I-expressing stable lines (lanes 2–6). The membrane was probed with Ikaros (CTS) antibodies that recognize all isoforms. Bands corresponding to HA-I and endogenous isoforms V and VI are indicated. (B) Immunoprecipitations were performed with HA antibodies and extracts from clones 7 (lanes 1,2) and 3 (lanes 3,4). Equivalent fractions of the pellets (lanes 1,3) and supernatants (lanes 2,4) were then analyzed by Western blot using CTS antibodies. (C) Untransfected VL3–3M2 cells (lanes 1,2) and clone 3 cells (lanes 3,4) were stimulated with PMA (20 ng/mL) plus ionomycin (700 ng/mL) for 6 h (lanes 2,4) or were left unstimulated (lanes 1,3). TdT mRNA levels were monitored by RNase protection. The specific, 59-nucleotide product is indicated. (D) Untransfected VL3–3M2 cells (left) and clone 3 cells (right) were stimulated with PMA plus ionomycin (dashed lines) or were left unstimulated (solid lines). CD5 up-regulation was monitored by flow cytometry.
To examine the efficiency with which HA-I associates with the endogenous Ikaros isoforms in extracts from the HA-I-expressing clones, coimmunoprecipitation experiments were performed with antibodies directed against the HA epitope. The proteins in the pellet and supernatant were then compared by Western blot, using Ikaros antibodies. Only a small fraction of the endogenous Ikaros isoforms coprecipitated with HA-I in extracts from clone 7 (Fig. 6B, lanes 1 and 2), consistent with the low expression of HA-I in this clone. In contrast, isoforms V and VI appeared to coimmunoprecipitate with HA-I in a quantitative manner in clones 3 (Fig. 6B, lanes 3 and 4) and 12 (data not shown), which express the highest concentrations of HA-I. That is, the HA antibodies precipitated comparable fractions of all three isoforms from the extracts. We were unable to precipitate all of the HA-I from the extracts, presumably because of its high concentration.
The anticipated effect of HA-I overexpression was that it would inhibit the functions of Ikaros by acting in a dominant negative manner. Surprisingly, no effect on the growth or viability of the cells was observed (data not shown). In addition, in multiple experiments, we observed no effect on the down-regulation of TdT transcription following PMA-ionomycin treatment, as determined by RNase protection (Fig. 6C). By flow cytometry, only a modest (twofold) effect on the up-regulation of CD5 (Fig. 6D) or CD69 (data not shown) was observed. These results suggested that the functions of Ikaros may not be essential for the growth and/or differentiation of VL3–3M2 cells. Alternatively, the properties of endogenous Ikaros may not be disrupted by overexpressed HA-I. The following experiments support the latter possibility.
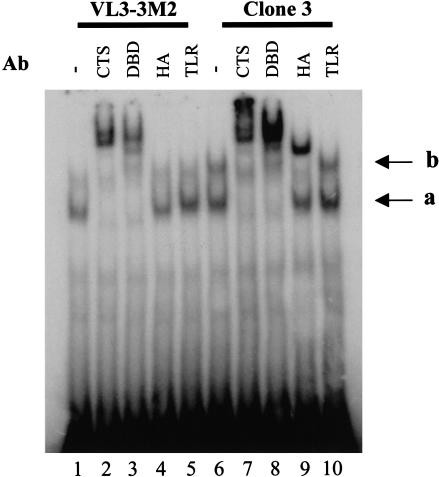
To examine the effect of overexpressed HA-I on the properties of the endogenous Ikaros isoforms, gel shift experiments were performed with nuclear extracts from VL3–3M2 cells and clone 3 cells (Fig. 7). The results revealed that overexpression of HA-I had no effect on the abundance of the Ikaros gel shift complex observed with a probe containing a consensus recognition site that strongly prefers an Ikaros dimer (Fig. 7, cf. lanes 1 and 6; Molnár and Georgopoulos 1994; Cobb et al. 2000). Furthermore, this complex contained only the large Ikaros isoforms, as its migration was altered by polyclonal antibodies against the common carboxy-terminal sequence (CTS) or the DNA-binding domain (DBD) of Ikaros (Fig. 7, lanes 7 and 8), but not by the HA antibody (Fig. 7, lane 9). Similar results were obtained with extracts from HEK 293 cells containing overexpressed HA-I and Ikaros isoform VI (data not shown). As demonstrated previously (Cobb et al. 2000; see above), this complex is likely to contain dimers of Ikaros isoforms V and VI. Most likely, the absence of HA-I in this complex and the absence of an effect of HA-I overexpression are attributable to the fact that the dimerization domain allows for rapid exchange between Ikaros subunits. This property has been well-documented in an independent biochemical and structural analysis of the carboxy-terminal zinc finger domain (A.S. McCarty and S.T. Smale, unpubl.). Although Ikaros proteins efficiently dimerize in solution through their carboxy-terminal domains, this rapid exchange facilitates efficient and stable DNA-binding by dimers of endogenous Ikaros isoforms, even in the presence of high concentrations of HA-I.
Figure 7.
Overexpressed HA-I does not disrupt the DNA-binding activity of large Ikaros isoforms. Gel shift experiments were performed with an Ikbs4 probe (Molnár and Georgopoulos 1994; Cobb et al. 2000) and extracts from untransfected VL3–3M2 cells (lanes 1–5) and clone 3 cells (lanes 6–10). Reactions contained no antibody (lanes 1,6) or antibodies directed against the common carboxy-terminal sequence of Ikaros (CTS, lanes 2,7), DNA-binding domain of Ikaros (DBD, lanes 3,8), HA epitope found on HA-I (lanes 4,9), or Toll-like receptor 4 as a negative control (lanes 5,10). The faster migrating complex (a) contains dimers of the endogenous Ikaros isoforms (V and VI). The slower migrating complex (b) is likely to contain multimers that include endogenous Ikaros dimers and one or more HA-I molecules.
Although overexpressed HA-I had no effect on the gel shift complex containing dimers of the large Ikaros isoforms, it enhanced a slower mobility complex (Fig. 7, cf. lanes 1 and 6). Interestingly, the mobility of this complex was altered by HA antibodies, as well as by the antibody against the DNA-binding domain of Ikaros (Fig. 7, lanes 8 and 9). This complex therefore contains HA-I in addition to endogenous Ikaros isoforms. Similar results were obtained with HEK 293 extracts containing overexpressed Ikaros proteins (data not shown). Given the slow migration of this complex relative to the predominant dimeric complex, it is likely to contain one or more HA-I molecules associated with a dimer of endogenous Ikaros isoforms. Importantly, a complex with a similar mobility is often observed in gel shift experiments with lymphocyte extracts in the absence of HA-I overexpression (e.g., Hahm et al. 1994). The abundance of this complex varies widely from experiment to experiment, suggesting that it is relatively unstable in vitro. Our ability to detect this complex in the absence of HA-I overexpression demonstrates that it does not depend on overexpression.
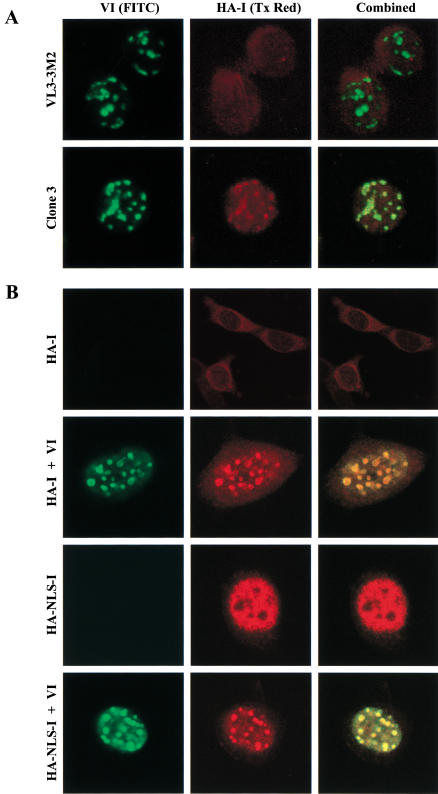
Because overexpressed HA-I had no effect on DNA binding by endogenous Ikaros proteins, one would predict that it would have no effect on the pericentromeric targeting of endogenous Ikaros. To examine endogenous Ikaros in the presence of overexpressed HA-I, confocal microscopy was performed with the antibodies that interact only with endogenous isoforms through the DNA-binding domain (Fig. 8A, green). The results revealed efficient targeting of endogenous Ikaros to centromeric foci in both wild-type VL3–3M2 cells and in clone 3 cells. (Centromeric targeting was more efficient in these experiments than in previous experiments performed with VL3–3M2 cells [Hahm et al. 1998], perhaps due to the use of different fixation procedures or variable properties of the cultured cells.) Given the above gel shift data and our previous evidence that the zinc finger dimerization domain (or a leucine zipper dimerization domain in its place) is essential for centromeric targeting and high-affinity DNA-binding to pericentromeric sequences (Cobb et al. 2000), the complexes bound at the centromeric foci must contain dimers of large Ikaros isoforms.
Figure 8.
Formation of multimeric Ikaros structures at pericentromeric foci. (A) Confocal immunofluorescence was performed with untransfected VL3–3M2 cells (top) and clone 3 cells (bottom). Fixed cells were costained with Ikaros antibodies that specifically bind the endogenous Ikaros isoforms (DBD, green, left) and HA antibodies that bind HA-I (red, center). Combined images are also shown. (B) Confocal immunofluorescence was performed with NIH 3T3 cells transduced with retroviruses expressing HA-I (HA-I, top row), HA-I plus isoform VI (HA-I+ VI, second row), an HA-I variant containing a nuclear localization signal (HA–NLS-I, third row), or HA–NLS-I plus isoform VI (HA–NLS-I+ VI, bottom row). Cells were costained as described above.
When considered in combination with the results in Figures 6 and 7, the most striking finding was that a significant fraction of the HA-I protein in clone 3 cells colocalized with the endogenous Ikaros isoforms to the centromeric foci (Fig. 8A, red). Similar results were obtained in other VL3–3M2 clones that express lower concentrations of HA-I (data not shown). Similar results were also obtained in 3T3 fibroblasts (Fig. 8B), in which ectopically expressed Ikaros isoform VI is known to localize to pericentromeric foci (Cobb et al. 2000). In the absence of isoform VI, HA-I localized primarily to the cytoplasm of 3T3 cells (Fig. 8B, top row). A variant of HA-I containing a nuclear localization signal (HA–NLS-I) localized to the nucleus, but was not concentrated at the centromeric foci (Fig. 8B, third row). In contrast, coexpression of either HA-I or HA–NLS-I with isoform VI revealed colocalization to centromeric foci (Fig. 8B, second and fourth rows). These results are consistent with previous evidence that large and small Ikaros isoforms colocalize to punctate nuclear spots (Sun et al. 1996). However, the current data lead to different conclusions due to documentation of the extent of overexpression and the effects on DNA-binding and endogenous gene expression (Figs. 6 and 7).
Because dimerization is absolutely essential for pericentromeric targeting (Cobb et al. 2000) and because HA-I overexpression had no effect on DNA-binding by endogenous Ikaros dimers (Fig. 7), it is highly unlikely that the HA-I-containing species positioned at the pericentromeric foci are heterodimers between HA-I and an endogenous Ikaros isoform. Therefore, the only reasonable explanation for these results is that HA-I can assemble into multimeric complexes with endogenous Ikaros dimers that are bound to pericentromeric DNA sequences. Thus, these results provide strong in vivo support for the existence of multimeric Ikaros structures at centromeric foci.
Discussion
This study used detailed knowledge of Ikaros contacts within the TdT D‘ element to assess the importance of Ikaros for TdT promoter function. The results provide evidence that an Ikaros–D‘ interaction is critical for TdT down-regulation following stimulation of double-positive thymocytes. The results also suggest that down-regulation is accompanied by changes in restriction enzyme access of the TdT locus. Because the TdT locus is not repositioned on differentiation of VL3–3M2 cells (Brown et al. 1999), transcriptional down-regulation by Ikaros and the chromatin alterations that accompany down-regulation are likely to precede the repositioning event in primary thymocytes. Of particular significance is the in vivo evidence that Ikaros forms multimeric structures at pericentromeric foci. These data may help to reconcile our evidence that Ikaros binds directly to centromeric foci (Cobb et al. 2000) with our evidence that Ikaros binding sites are essential for TdT down-regulation.
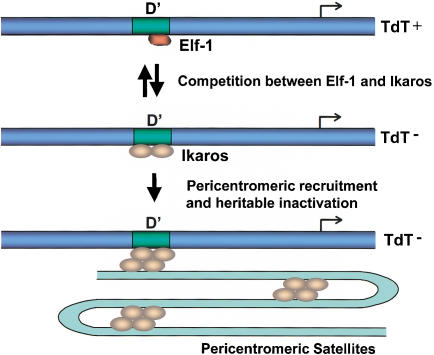
The current working model is shown in Figure 9. Following the stimulation of double-positive thymocytes, Ikaros dimers compete for occupancy of the TdT D‘ element, thereby displacing Elf-1 and suppressing transcription. This event is readily reversible (Brown et al. 1999; data not shown) and is accompanied by chromatin alterations that are apparent in restriction enzyme accessibility experiments. In primary cells, the Ikaros dimer bound to the D‘ element then becomes competent for association with multimeric Ikaros complexes positioned at the centromeric foci, thereby recruiting the TdT genes to these foci. In VL3–3M2 cells, centromeric repositioning does not occur, most likely because of their transformed properties. Although these hypotheses must be tested much more extensively, the results provide initial evidence using physiologically relevant assays that Ikaros is indeed a direct participant in gene repression and has the potential to assemble into multimeric structures at centromeric foci.
Figure 9.
Working model for the down-regulation and heritable inactivation of the TdT gene on differentiation of double-positive thymocytes.
The inability of HA-I to disrupt centromeric targeting and DNA-binding of endogenous Ikaros isoforms may explain why no effect was observed on cell growth, differentiation, or gene expression. However, one might still expect to observe an effect, as the incorporation of HA-I into multimeric complexes may reduce the number of endogenous Ikaros molecules that can enter into these complexes. There are two possible explanations for these negative results. First, the incorporation of HA-I into the complex may not significantly reduce the capacity for endogenous Ikaros molecules to associate with the complexes. Consistent with this hypothesis, the pericentromeric localization of endogenous Ikaros isoforms was completely unaffected by the overexpressed HA-I (Fig. 8). The second possibility is that these complexes may not be important for the growth and differentiation of VL3–3M2 cells. This possibility would be consistent with the fact that the TdT loci do not migrate toward centromeric foci in these cells (Brown et al. 1999; see above). It will be interesting to examine the effect of overexpressed HA-I in primary double-positive thymocytes that support the centromeric repositioning of inactive loci. The severe phenotype observed in Ikaros-deficient mice that retain expression of small Ikaros isoforms (Georgopoulos et al. 1994) suggests that HA-I will indeed act as a strong dominant negative in primary cells. However, the dominant negative effect may be observed only when the concentrations of the large Ikaros isoforms are reduced. Indeed, transgenic mice overexpressing small Ikaros isoforms exhibit phenotypes that are less severe than anticipated on the basis of the gene disruption phenotypes (C. Wilson, pers. comm.; C. Klug, pers. comm.).
The use of the native TdT promoter appears to have been critical for detecting transcriptional repression by Ikaros proteins, as we have been unable to detect repression of artificial reporter constructs containing multiple Ikaros binding sites (B.S. Cobb, unpubl.). To further confirm that Ikaros is involved in TdT down-regulation, the Ikaros binding sites within the endogenous TdT promoter will need to be disrupted using a knock-in strategy. The results of such an analysis may confirm the results shown here, although it remains possible that redundant mechanisms contribute to down-regulation of the endogenous gene when analyzed in the context of its full complement of control regions. Additional support could be provided by an examination of Ikaros binding to the endogenous TdT promoter using a chromatin immunoprecipitation (ChIP) assay.
The competition model supported by these data requires a switch from Elf-1 to Ikaros occupancy of the D‘ element following thymocyte stimulation. One mechanism that may allow Ikaros to compete for D‘ occupancy is the simple up-regulation of Ikaros protein levels. Ikaros protein levels are up-regulated to a modest extent (∼fourfold) on stimulation of VL3–3M2 cells, but the down-regulation of TdT transcription occurs more rapidly than the Ikaros up-regulation (R. Ferrini, unpubl.). A second potential mechanism is via a posttranslational modification of Ikaros. Two-dimensional phosphopeptide mapping experiments have revealed changes in the phosphorylation pattern of Ikaros following VL3–3M2 stimulation (S. Dovat, unpubl.). Additional experiments will be required to determine whether these changes contribute to the putative switch. Alternative mechanisms that may regulate the switch include a posttranslational modification of Helios or Elf-1, or modification of a hypothetical DNA-binding protein, coactivator, or corepressor, which may stabilize or destabilize Ikaros or Elf-1 binding. Further examination of the competition mechanism and further tests of the model shown in Figure 9 should advance our knowledge of the regulation of lymphopoiesis.
Materials and methods
Plasmids
Most plasmids used for gel shift and methylation interference studies were reported previously (Ernst et al. 1993). To generate plasmids containing the D‘ element with mutations in both the proximal and distal ends, the following oligonucleotides were annealed with their complementary sequences, phosphorylated with T4 polynucleotide kinase, and inserted into the BamHI site of pSP72 (Promega): m4+m88 (5′-AGTGAGACCGAAAT TCAACCGGAAGTAGTC-3′), m4+m84 (5′-AGTGAGACCGA AATTCAGCCGGAAGTTGTC-3′), m4+m86 (5′-AGTGAGAC CGAAATTCAGCAGGCCGTTGTC-3′), and m4+m87 (5′-AGT GAGACCGAAATTCAGCAGGAATTTGTC-3′).
TdT reporter constructs containing the −30 mutations were described previously (Garraway et al. 1996). For stable transfection of RLm11 cells, the cells were cotransfected with these constructs and pSV2his (Hartman and Mulligan 1988). For stable transfection of VL3–3M2 cells, a vector, pHis-TK, was created that contained the S. typhimurium hisD gene, the HSV-TK coding sequence, and an upstream multiple cloning sequence. First, the following oligonucleotide and its complement, containing the multiple cloning sequence (MCS), was inserted into the BamHI site of pSV2his (Hartman and Mulligan 1988): 5′-GATCGGATCCAAGCTTGTCGACTCTAGACTCG AGAGATCTGATC-3′. Next, a 1.2 kb PCR fragment containing the HSV-TK gene was inserted into the HindIII and BamHI sites of the MCS. Reporter constructs were generated by PCR site-directed mutagenesis and subcloning. First, a promoter variant containing a consensus TATA box between −25 and −30 was prepared using the Stratagene QuikChange Site-Directed Mutagenesis method and a plasmid, pIG (Ernst et al. 1993), as template. pIG contains TdT promoter sequences from −1700 to +58. The following oligonucleotide and its complement were used as primers for this procedure: 5′-GGGTGGTACCTATGTATAAAATGGTGAGAGGACTCAGAGCC-3′. The resulting plasmid, pIG/T, was used as the template for generating all subsequent promoter mutants. The mutant promoters were then excised with BamHI and BglII and inserted into the BglII site of pHis-TK.
For stable overexpression of HA-I, a cDNA carrying an amino-terminal HA epitope tag was cloned into the pEBB expression vector (Mayer et al. 1995).
Proteins and DNA-binding assays
GST fusion proteins containing an amino-terminal fragment of Ikaros isoform VI were produced in E. coli as described previously (Hahm et al. 1994). GST–Elf-1 and GST–Fli-1 fusion proteins were prepared in E. coli and purified as described previously (Ernst et al. 1996). RmL11 nuclear extracts for gel shift and methylation interference assays were prepared as described previously (Hahm et al. 1994).
All gel shift probes, except Ikbs4, were prepared by cleaving the plasmids described above with XbaI and EcoRI. Restriction fragments were labeled with [γ-32P]ATP. The labeled fragments were gel-purified and normalized to 10,000 cpm/μL. The Ikbs4 gel shift probe was prepared by annealing complementary synthetic oligonucleotides (5′-TGACAGGGAATACACATTCCC AAAAGC-3′). The oligonucleotides were designed with 5′ overhangs that were filled in with Klenow and α-32P-labeled nucleotides, followed by purification over a G-50 spin column. Methylation interference probes were prepared either from restriction fragments cleaved at the BglII and XhoI sites or by PCR using T7 and SP6 primers. One of the two PCR primers was phosphorylated with [γ-32P]ATP and T4 polynucleotide kinase. PCR products were then gel purified and quantified.
Gel shift assays with nuclear extracts in the presence of antibodies were performed as follows: RLm11 nuclear extracts (2–4 μg) were incubated with preimmune or α-Ikaros IgG (Hahm et al. 1994) in HGED.1 (20 mM HEPES at pH 7.9, 0.2 mM EDTA, 20% glycerol, 100 mM KCl, 1 mM dithiothreitol) containing 2 μg poly (dI-dC), 1 μg bovine serum albumin (BSA) and 10 μM ZnCl2 for 15 min before addition of probe (10,000 cpm). The total volume of the binding reactions was 25 μL. Control reactions lacking antibody were also incubated for 15 min prior to probe addition. Reaction mixtures were then incubated at ambient temperature for 30 min and applied to a gel containing 4% polyacrylamide and 0.25× TBE (22 mM Tris-borate at pH 8.3, 0.5 mM EDTA). Gel shift assays with recombinant proteins were performed as above, except lower concentrations of poly(dI-dC) were employed. Gel shift assays with VL3–3M2 nuclear extracts (4.5 μg) were performed similarly except 1 μg poly(dI-dC) and 20,000 cpm of probe was used. Also, preincubation with antibodies was extended to 30 min.
Purified probes for methylation interference were modified with 1 μM dimethyl sulfate as described in Ausubel et al. (1994). Binding reactions were identical to those described above except reactions were increased 10-fold. Piperdine cleavage was performed as described previously (Ausubel et al. 1994) with the products analyzed on a 12% denaturing polyacrylamide gel.
Cell culture and stable transfections
RLm11 and VL3–3M2 T cells were grown in RPMI 1640 medium supplemented with 10% newborn calf serum or 5% fetal calf serum, respectively, and 100 μg/mL streptomycin. Stable transfections were performed with 1.0 × 107 cells by electroporation (300 mV, 960 μF, Bio-Rad Gene Pulser) in RPMI 1640 containing 16.6% fetal calf serum and 60 μg DNA. HA-I stable clones were generated by transfecting 2.0 × 107 cells in RPMI containing 30 μg expression plasmid and 3 μg cotransfected selection marker plasmid (pBABE puro; Morgenstern et al. 1990). After electroporation, cells were plated onto 96-well plates at 100 μL per well. After 1 d, selection with 1 mM histidinol or 1 μg/mL puromycin was initiated. Clones containing integrated reporter plasmids were identified by PCR. Transcription from the wild-type and mutant TdT promoters was monitored by primer extension using a 26-nucleotide primer complementary to HSV-TK sequences (Smale and Baltimore 1989). For TdT down-regulation experiments, stable transfectants were passaged to a density of 2 × 105 cells/mL, allowed to grow for 8–10 h, and then treated with 20 ng/mL phorbol-12-myristate-13 acetate (PMA, Calbiochem) and 700 ng/mL ionomycin (Calbiochem). Cytoplasmic RNA was isolated 24 h after stimulation and analyzed by primer extension using 30 μg of RNA (Smale and Baltimore 1989). Transcripts from the TdT and HisD genes were also monitored by primer extension (Smale and Baltimore 1989; Ernst et al. 1996). Expression of isoforms VI and HA-I in 3T3 cells was achieved by retroviral transduction as described previously (Cobb et al. 2000). Double infections were performed sequentially for 2.5 h each.
Restriction enzyme accessibility
Restriction enzyme accessibility assays were performed as described previously (Weinmann et al. 1999), using the enzymes shown in Figure 5. Primary double-positive thymocytes were isolated from MHC-deficient mice and were stimulated and sorted as described previously (Brown et al. 1999).
Western blots and immunoprecipitations
Western blot analysis of stable HA-I pEBB clones was performed with nuclear extracts prepared by osmotic swelling and Dounce homogenization as described previously (Lo et al. 1991). Protein concentrations were quantified by Bradford assay and 2.5 μg of total protein of each sample was loaded onto a 10% denaturing polyacrylamide gel. After transfer to nitrocellulose the blots were probed with rabbit antisera raised against the carboxyl terminus of Ikaros.
For immunoprecipitations, 50 μL (normalized to 1 μg/μL) of nuclear extract was incubated with mouse monoclonal α-HA (12CA5) in a total volume of 100 μL. Total volume was raised to 100 μL with NTN (100 mM NaCl, 20 mM Tris at pH 8.0, 0.5% NP-40). After a 1 h incubation at 4°C, 100 μL of a 50% protein A-Sepharose slurry was added and the mixture was allowed to incubate for one additional hour at 4°C on a rocker. The resin was pelleted and washed four times with NTN buffer and then analyzed by SDS-PAGE as described above.
Flow cytometry
CD5 was detected by immunochemical staining with FITC rat anti-mouse CD5 (Pharmingen). Cells were incubated at 4°C for 30 min in 100 μL of PBS containing 1% BSA. After washing, cells were fixed in 4% paraformaldehyde at 4°C for 20 min and analyzed using a Beckton Dickenson FACSCalibur within 24 h.
RNase protection
RNase protections were carried out with the RPA II RNase protection kit (Ambion). TdT template DNA was described previously (Lo et al. 1991). From these templates, RNA probes were produced using the MAXIscript in vitro transcription kit (Ambion).
Confocal microscopy
VL3–3M2 cells were adhered by centrifugation (2500 rpm) onto poly-L-lysine coated coverslips and fixed immediately. 3T3 cells were plated on uncoated coverslips and allowed to adhere for several hours. Cells were washed once with PBS and then fixed for 1 min at −20°C. Cells were immediately washed three times in PBS, permeabilized for 10 min in staining buffer (PBS with 0.1% saponin and 10% FCS), and then stained with rabbit polyclonal antibody against the DNA binding domain of Ikaros, and mouse monoclonal α-HA (12CA5). Staining was performed for 1 h at 4°C before cells were washed three times in PBS with 0.1% saponin (3 min/wash). Cells were then incubated with secondary antibodies, FITC goat α-rabbit (Jackson Immunoresearch) and Texas Red α-mouse (Jackson Immunoresearch) for 30 min in staining buffer followed by three more 3-min washes in PBS with 0.1% saponin and a final rinse in water. The coverslips were then mounted onto slides.
Acknowledgments
We thank Amanda Fisher and Prim Singh for valuable discussions and Cynthia Guidos for providing the VL3–3M2 cell line. This work was supported by U.S. Public Health Service Grant R01 DK43726 (S.T.S.), by National Research Service Awards GM07185 (A.S.W. and P.E.), GM07104 (K.H.), HG00117 (I.P.G.), CA09120 (P.E.), by University of California Dissertation-Year (A.S.W.) and Office of the President (P.E.) Fellowships, and by the Medical Research Council, U.K. (M.M.). S.T.S. is an Investigator with the Howard Hughes Medical Institute.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL steves@hhmi.ucla.edu; FAX (310) 206-8623.
Article and publication are at http://www.genesdev.org/cgi/doi/10.1101/gad.905601.
References
- Ausubel FM, Brent R, Kingston RE, Moore DD, Seidman JG, Smith JA, Struhl K. Current protocols in molecular biology. NY: John Wiley & Sons; 1994. [Google Scholar]
- Avitahl N, Winandy S, Friedrich C, Jones B, Ge Y, Georgopoulos K. Ikaros sets thresholds for T cell activation and regulates chromosome propagation. Immunity. 1999;10:333–343. doi: 10.1016/s1074-7613(00)80033-3. [DOI] [PubMed] [Google Scholar]
- Brown KE, Guest SS, Smale ST, Hahm K, Merkenschlager M, Fisher AG. Association of transcriptionally silent genes with Ikaros complexes at centromeric heterochromatin. Cell. 1997;91:845–854. doi: 10.1016/s0092-8674(00)80472-9. [DOI] [PubMed] [Google Scholar]
- Brown KE, Baxter J, Graf D, Merkenschlager M, Fisher AG. Dynamic repositioning of genes in the nucleus of lymphocytes preparing for cell division. Mol Cell. 1999;3:207–217. doi: 10.1016/s1097-2765(00)80311-1. [DOI] [PubMed] [Google Scholar]
- Cobb BS, Morales-Alcelay S, Kleiger G, Brown KE, Fisher AG, Smale ST. Targeting of Ikaros to pericentromeric heterochromatin by direct DNA binding. Genes & Dev. 2000;14:2146–2160. doi: 10.1101/gad.816400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cortes M, Wong E, Koipally J, Georgopoulos K. Control of lymphocyte development by the Ikaros gene family. Curr Op Immunol. 1999;11:167–171. doi: 10.1016/s0952-7915(99)80028-4. [DOI] [PubMed] [Google Scholar]
- Ernst P, Hahm K, Smale ST. Both LyF-1 and an Ets protein interact with a critical promoter element in the murine terminal transferase gene. Mol Cell Biol. 1993;13:2982–2992. doi: 10.1128/mcb.13.5.2982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ernst P, Hahm K, Trinh L, Davis JN, Roussel MF, Turck CW, Smale ST. A potential role for Elf-1 in terminal transferase gene regulation. Mol Cell Biol. 1996;16:6121–6131. doi: 10.1128/mcb.16.11.6121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ernst P, Hahm K, Cobb BS, Brown KE, Trinh LA, McCarty AS, Merkenschlager M, Klug CA, Fisher AG, Smale ST. Mechanisms of transcriptional regulation in lymphocyte progenitors: Insight from an analysis of the terminal transferase promoter. Cold Spring Harb Symp Quant Biol. 1999;64:87–97. doi: 10.1101/sqb.1999.64.87. [DOI] [PubMed] [Google Scholar]
- Francastel C, Walters MC, Groudine M, Martin DI. A functional enhancer suppresses silencing of a transgene and prevents its localization close to centrometric heterochromatin. Cell. 1999;99:259–269. doi: 10.1016/s0092-8674(00)81657-8. [DOI] [PubMed] [Google Scholar]
- Garraway IP, Semple K, Smale ST. Transcription of the lymphocyte-specific terminal deoxynucleotidyltransferase gene requires a specific core promoter structure. Proc Natl Acad Sci. 1996;93:4336–4341. doi: 10.1073/pnas.93.9.4336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Georgopoulos K, Bigby M, Wang JH, Molnar A, Wu P, Winandy S, Sharpe A. The Ikaros gene is required for the development of all lymphoid lineages. Cell. 1994;79:143–156. doi: 10.1016/0092-8674(94)90407-3. [DOI] [PubMed] [Google Scholar]
- Glimcher LH, Singh H. Transcription factors in lymphocyte development—T and B cells get together. Cell. 1999;96:13–23. doi: 10.1016/s0092-8674(00)80955-1. [DOI] [PubMed] [Google Scholar]
- Graves BJ, Petersen JM. Specificity within the ets family of transcription factors. Adv Cancer Res. 1998;75:1–55. doi: 10.1016/s0065-230x(08)60738-1. [DOI] [PubMed] [Google Scholar]
- Groves T, Katis P, Madden Z, Manickam K, Ramsden D, Wu G, Guidos CJ. In vitro maturation of clonal CD4+CD8+ cell lines in response to TCR engagement. J Immunol. 1995;154:5011–5022. [PubMed] [Google Scholar]
- Hahm K, Ernst P, Lo K, Kim G, Turck C, Smale ST. The lymphoid transcription factor LyF-1 is encoded by a specific, alternatively spliced RNA derived from the Ikaros gene. Mol Cell Biol. 1994;114:7111–7123. doi: 10.1128/mcb.14.11.7111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hahm K, Cobb BS, McCarty AS, Brown KE, Klug CA, Lee R, Akashi K, Weissman IL, Fisher AG, Smale ST. Helios, a T cell-restricted Ikaros family member that quantitatively associates with Ikaros at centromeric heterochromatin. Genes & Dev. 1998;12:782–796. doi: 10.1101/gad.12.6.782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartman SC, Mulligan RC. Two dominant-acting selectable markers for gene transfer studies in mammalian cells. Proc Natl Acad Sci. 1988;85:8047–8051. doi: 10.1073/pnas.85.21.8047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, Sif S, Jones B, Jackson A, Koipally J, Heller E, Winandy S, Viel A, Sawyer A, Ikeda T, et al. Ikaros DNA-binding proteins direct formation of chromatin remodeling complexes in lymphocytes. Immunity. 1999;10:345–355. doi: 10.1016/s1074-7613(00)80034-5. [DOI] [PubMed] [Google Scholar]
- Klug CA, Morrison SJ, Masek M, Hahm K, Smale ST, Weissman IL. Hematopoietic stem cells and lymphoid progenitors express different Ikaros isoforms, and Ikaros is localized to heterochromatin in immature lymphocytes. Proc Natl Acad Sci. 1998;95:657–662. doi: 10.1073/pnas.95.2.657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koipally J, Renold A, Kim J, Georgopoulos K. Repression by Ikaros and Aiolos is mediated through histone deacetylase complexes. EMBO J. 1999;18:3090–3100. doi: 10.1093/emboj/18.11.3090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lo K, Landau NR, Smale ST. LyF-1, a transcriptional regulator that interacts with a novel class of promoters for lymphocyte-specific genes. Mol Cell Biol. 1991;11:5229–5243. doi: 10.1128/mcb.11.10.5229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayer BJ, Hirai H, Sakai R. Evidence that SH2 domains promote processive phosphorylation by protein-tyrosine kinases. Curr Biol. 1995;5:296–305. doi: 10.1016/s0960-9822(95)00060-1. [DOI] [PubMed] [Google Scholar]
- Molnár A, Georgopoulos K. The Ikaros gene encodes a family of functionally diverse zinc finger DNA binding proteins. Mol Cell Biol. 1994;83:785–794. doi: 10.1128/mcb.14.12.8292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgenstern JP, Land H. Advanced mammalian gene transfer: High titre retroviral vectors with multiple drug selection markers and a complementary helper-free packaging cell line. Nucleic Acids Res. 1990;18:3587–3596. doi: 10.1093/nar/18.12.3587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Neill DW, Schoetz SS, Lopez RA, Castle M, Rabinowitz L, Shor E, Krawchuk D, Goll MG, Renz M, Seelig HP, et al. An ikaros-containing chromatin-remodeling complex in adult-type erythroid cells. Mol Cell Biol. 2000;20:7572–7582. doi: 10.1128/mcb.20.20.7572-7582.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smale ST, Baltimore D. The “initiator” as a transcription control element. Cell. 1989;57:103–113. doi: 10.1016/0092-8674(89)90176-1. [DOI] [PubMed] [Google Scholar]
- Sun L, Liu A, Georgopoulos K. Zinc finger mediated Ikaros protein interactions modulate their activity in transcription: a putative on/off switch for lymphocyte proliferation. EMBO J. 1996;15:5358–5369. [PMC free article] [PubMed] [Google Scholar]
- Weinmann AS, Plevy SE, Smale ST. Rapid and selective remodeling of a positioned nucleosome during the induction of IL-12 p40 transcription. Immunity. 1999;11:665–675. doi: 10.1016/s1074-7613(00)80141-7. [DOI] [PubMed] [Google Scholar]
- Weinmann AS, Mitchell DM, Sanjabi S, Bradley MN, Hoffmann A, Liou HC, Smale ST. Nucleosome remodeling at the IL-12 p40 promoter is a TLR-dependent, Rel-independent event. Nature Immunol. 2001;2:51–57. doi: 10.1038/83168. [DOI] [PubMed] [Google Scholar]