Abstract
Previous work has established a role for p53 in triggering apoptosis in response to DNA damage; p53 also induces apoptosis in response to deregulation of the Rb cell cycle pathway. The latter event is consistent with a role for the Rb-regulated E2F1 protein as a specific inducer of apoptosis and p53 accumulation. We now show that DNA damage leads to a specific induction of E2F1 accumulation, dependent on ATM kinase activity and that the specificity of E2F1 induction reflects a specificity in the phosphorylation of E2F1 by ATM as well as the related kinase ATR. We identify a site for ATM/ATR phosphorylation in the amino terminus of E2F1 and we show that this site is required for ATM-mediated stabilization of E2F1. Finally, we also show that E2F1 is required for DNA damaged induced apoptosis in mouse thymocytes. We conclude that the cellular response to DNA damage makes use of signals from the Rb/E2F cell cycle pathway.
Keywords: E2F1, DNA damage, ATM, ATR, p53, apoptosis
The role of E2F transcription activity in the regulation of cell cycle progression, particularly the G1/S transition, is now well established (Dyson 1998; Nevins 1998). Overexpression of E2F, in the absence of other growth signals, is sufficient to induce quiescent cells to enter S phase (Johnson et al. 1993). The growth suppression activity of the Rb tumor suppressor protein is dependent on its ability to bind and repress E2F activity, although associations with other proteins are likely also important for other pRb function.
The tumorigenesis resulting from loss of pRb function is in part mediated through the action of E2F1 as a loss of E2F1 reduces the oncogenic potential of Rb (+/−) mice (Yamasaki et al. 1998). A variety of experiments have suggested distinct roles for individual members of the E2F family in the control of cell growth including distinct roles for E2F proteins in transcription activation and transcription repression (Dyson 1998; Nevins 1998). This includes an apparent unique ability of E2F1 to induce apoptosis in addition to an ability to trigger S phase entry (Qin et al. 1994; Shan and Lee 1994; Wu and Levine 1994; Kowalik et al. 1995, 1998). Although overexpression of each E2F protein, except for E2F5, is able to induce S phase entry, only E2F1 overexpression induces apoptosis in serum-starved quiescent fibroblasts (DeGregori et al. 1997). A physiological role for the E2F1-induced death is indicated by the observation that E2F1−/− mice develop thymic hyperplasia attributable to a defect in thymocyte apoptosis (Field et al. 1996). In addition, homozygous Rb mutations lead to embryonic lethality in mice attributable to abnormalities in CNS and hematopoietic development (Jacks et al. 1992; Lee et al. 1992). Excessive apoptosis was observed in the developing neurons, lens, and myocytes of these embryos, which strongly argues for a specific role of Rb in preventing apoptosis. At least part of the Rb−/− phenotype is attributable to E2F1 deregulation (Pan et al. 1998; Tsai et al. 1998; Yamasaki et al. 1998), but it is also true that other studies have provided evidence for a role for E2F3 in the Rb-dependent apoptosis pathway (Ziebold et al. 2001). Moreover, overexpression of E2F2 and E2F3 were also reported to induce apoptosis in another experimental setting (Vigo et al. 1999) although it is not clear from these studies whether the observed E2F2 or E2F3-induced apoptosis is dependent on E2F1. In any event, it seems clear from these studies that E2F1 plays a significant role in apoptosis signaling and that additional E2Fs, particularly E2F3, also contribute to the induction of apoptosis.
Various studies have demonstrated that E2F1-induced apoptosis can be both dependent (Wu and Levine 1994) and independent of p53 (Phillips et al. 1997). The p53 dependence coincides with our previous work that has demonstrated an induction of p53 protein accumulation following expression of E2F1 (Kowalik et al. 1998). As with the induction of apoptosis, the stimulation of p53 accumulation is an event unique to E2F1. A mechanism by which E2F1 could induce p53 accumulation has now been revealed by studies of the role of the p19ARF protein in controlling Mdm2 function. Mdm2 has been shown to control p53 protein levels by targeting p53 for ubiquitin mediated degradation (Honda et al. 1997). ARF interacts with Mdm2 and blocks the ability of Mdm2 to target p53 for destruction (Weber et al. 1999). The link with E2F1 is now seen from the observation that E2F induces p19ARF expression, likely a direct transcription activation via E2F sites in the ARF promoter, and with an apparent specificity for E2F1 (DeGregori et al. 1997; Bates et al. 1998).
DNA damaging agents, such as γ irradiation or chemotherapeutic drugs, can cause cell cycle arrest as well as apoptosis, largely dependent on p53. Several recent reports have described an increase in E2F1 protein, resulting from stabilization of the E2F1 protein, following the treatment of a variety of tumor cells with DNA damaging agents (Huang et al. 1997; Blattner et al. 1999; Hofferer et al. 1999; Meng et al. 1999). We have now further examined the response of E2F1 to treatment of cells with various chemotherapeutic agents. We find that the induction is specific to E2F1 and is not observed for the other E2F family proteins. We also show that this induction is dependent on ATM/ATR, a kinase known to activate p53 in response to DNA damage, and that specificity in E2F1 induction reflects a specificity of phosphorylation by ATM/ATR. Finally, we show that the induction of E2F1 is functionally important for DNA damage-induced apoptosis in thymocytes, establishing a role for E2F1 in the response to DNA damage.
Results
Induction of E2F1 accumulation in tumor cells following DNA damage
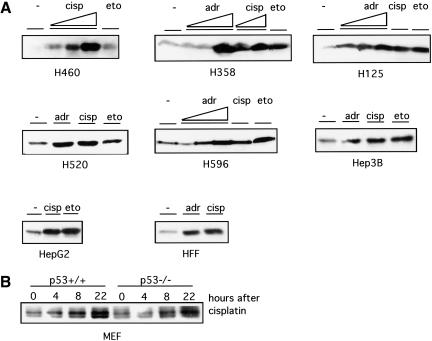
We examined the accumulation of E2F1 protein following treatment of a panel of tumor cell lines with a variety of chemotherapeutic drugs. These cell lines were derived from lung cancer (H460, H358, H125, H520, H596) or hepatoma (Hep3B, HepG2) and reflect a variation in the genetic status of p53 and Rb. Although the kinetics of induction varies depending on the type of cell lines and the drugs being used, E2F1 is induced in each of these cells following treatment with the drugs (Fig. 1A). The magnitude of the induction of E2F1 protein is correlated roughly to the sensitivity of the particular cell line to a particular drug, as assayed by propidium iodide for the percentage of apoptosis induced by the drug (data not shown). This result is thus consistent with several recent reports describing the induction of E2F1 in various other types of cancer cell lines in response to a variety of forms of DNA damage including γ-irradiation, UV-irradiation, and chemotherapeutic drugs (Huang et al. 1997; Blattner et al. 1999; Hofferer et al. 1999; Meng et al. 1999). As such, it would appear that the induction of E2F1 protein may be a general response to DNA damage. In addition, the induction is independent of p53 status as seen by the analysis of E2F1 following treatment of either wild-type or p53 null mouse embryo fibroblasts with cisplatin (Fig. 1B).
Figure 1.
Induction of E2F1 accumulation following DNA damage. (A) Western blot analysis of E2F1 among a panel of human cell lines after 24 h treatment of adriamycin (adr), cisplatin (cisp), or etoposide (eto) shows induction of E2F1 protein regardless of their p53 or Rb status. The concentration of cisplatin was from 5 to 50 μM, adriamycin 50 to 500 nM, and etoposide 5 to 50 μM. H460, H358, H125, H520, and H596 are derived from lung cancer. Hep3B and HepG2 are hepatoma cell lines. HFF is a nontransformed human foreskin fibroblast cell line. H460 carries wild-type p53. H358 and Hep3B are p53-null. H125, H520, and H596 carry mutated p53. H596 and Hep3B have no Rb protein expression, whereas H460, H358, H125, and HepG2 carry normal Rb. Equal protein loading of each lane was confirmed by Ponceau-S staining. (B) Mouse embryo fibroblasts prepared from p53−/− embryos or wild-type littermates were treated with cisplatin (20 μM) for the indicated time. E2F1 in the lysates was detected by immunoblotting.
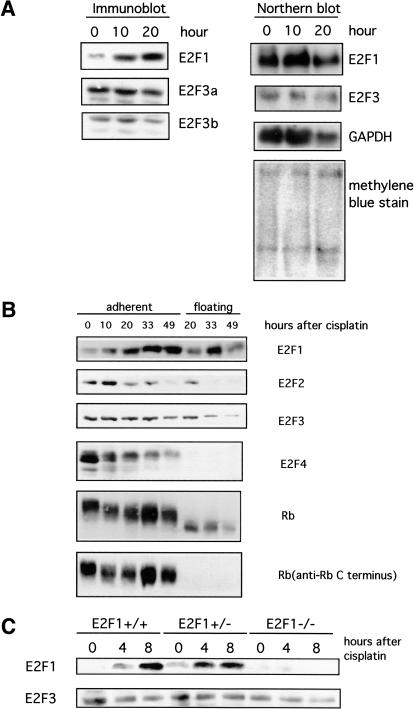
Consistent with recent work, it is apparent that the induced accumulation of E2F1 protein does not result from an activation of E2F1 transcription. Although there is a marked increase in the level of E2F1 protein in H460 tumor cells treated with DNA damaging drugs, there was little or no change in the level of E2F1 mRNA (Fig. 2A). It was also clear from this experiment that the levels of E2F3 transcripts were not altered in response to DNA damage.
Figure 2.
Specificity in posttranscriptional induction of E2F1 accumulation. (A) Measurements of E2F protein accumulation (immunoblot) and RNA accumulation (Northern blot) following treatment of H460 cells with cisplatin (50 μM). Two species of E2F3 protein are detected (E2F3a and E2F3b) that derive from alternate transcription initiation sites within the E2F3 locus (Leone et al. 2000). The two E2F3 transcripts are not resolved in this Northern analysis. (B) Western blot analysis of E2F and Rb proteins in the H460 human lung cancer cell line following treatment with cisplatin (50 μM). Rb protein is seen to be degraded to a truncated product that lacks the carboxyl terminus in the cells undergoing apoptosis, likely a result of caspase action (Tan and Wang 1998). (C) Western blot analysis of E2F in primary mouse embryonic fibroblasts (MEF). Primary MEF cultures were prepared from embryos with the indicated genotypes and then treated with cisplatin (20 μM).
Specific induction of E2F1 accumulation following DNA damage
Given the fact that the accumulation of E2F1 is tightly regulated during the cell cycle, including the control of E2F1 protein levels, the DNA damage-induced accumulation could simply reflect an indirect consequence of a cell cycle inhibition, with some fraction of the cells accumulating at G1/S, the time at which E2F1 normally accumulates. Previous work has shown that the E2F1, E2F2, and E2F3 family members are commonly regulated with respect to the cell cycle. Each gene is induced in response to growth stimulation with peak accumulation at G1/S. The activities then continue to oscillate in the cell cycle, again peaking at G1/S (Leone et al. 1998). In contrast, the E2F4 and E2F5 gene products are constitutively expressed, with little or no fluctuation as a function of cell cycle progression. Thus, if the accumulation of E2F1 in response to DNA damage was the result of cell cycle arrest, we would expect to see an accumulation of E2F2 and E2F3 as well. However, as shown by the data in Figure 2, it is apparent that the induction of E2F1 protein accumulation is specific for E2F1. H460 cells that are undergoing apoptosis in response to DNA-damaging drugs detach from the tissue culture plate whereas the remaining, adherent cells are enriched for cell cycle-arrested cells, as confirmed by propidium iodide staining. The lysates from the two H460 cell populations were analyzed by Western blot with antibodies specific for E2F1, E2F2, E2F3, E2F4, and Rb. As seen in previous assays, there was an induction of E2F1 protein following treatment with cisplatin. We also confirm the specificity of E2F1 induction in mouse embryonic fibroblast (Fig. 2C), in which we observed rapid induction of E2F1 within 4 h of cisplatin treatment, but no change in E2F3 protein. This data also clearly shows that the response was specific for E2F1 with no increase seen in the levels of the other E2F proteins; indeed, there was a substantial decline in the E2F4 protein. It was also evident from this experiment that the Rb protein is dephosphorylated during DNA damage. The specific induction of E2F1, but not the E2F2 or E2F3 proteins, strongly suggests that the induction is not a consequence of enrichment for G1/S population by DNA damage-induced G1/S arrest. Rather, this result suggests a distinct regulatory mechanism controlling E2F1 accumulation.
Induction of E2F1 accumulation in response to DNA damage requires ATM
Other work has demonstrated a role for the ATM/ATR class of protein kinases in the DNA damage response. For instance, the role of these kinases in the phosphorylation of p53 following DNA damage, both directly (Banin et al. 1998; Canman et al. 1998) and indirectly (Chehab et al. 2000; Shieh et al. 2000) is now well established. We thus considered the possibility that the induction of E2F1 accumulation following DNA damage might involve the same pathways.
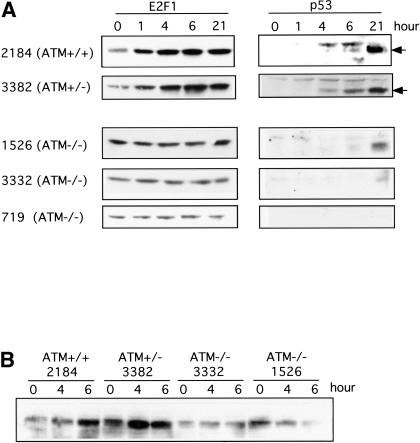
We made use of various cell lines deficient in ATM gene function to explore the role of the kinase in the induction of E2F1. We examine the induction of E2F1 and p53 in the lymphoblastoid cell lines established from AT patients or normal subjects. As shown in Figure 3A, adriamycin treatment induces a rapid accumulation of E2F1 protein in the cells with wild-type ATM (GM2184) as well as in cells that are heterozygous for the ATM mutation (GM3382). Similar results are seen for the induction of p53 protein in these cell lines. In contrast, E2F1 is not induced in the cell lines bearing ATM homozygous mutation (GM1526, GM3332, and GM719). In addition, the induction is not limited to adriamycin because E2F1 is also induced in cells with either a wild-type or heterozygous ATM genotype following treatment with etoposide but was not induced in the ATM null cells by etoposide (Fig. 3B). Based on this result, we conclude that the induction of E2F1 in response to DNA damaging agents involves ATM.
Figure 3.
Role of ATM in the induction of E2F1 following DNA damage. (A) Various human lymphoblastoid cell lines with the indicated ATM genotypes were treated with adriamycin (1 μM). E2F1 protein accumulation was assayed by Western blot analysis. For comparison, p53 protein accumulation was measured in the same samples. (B) Induction of E2F1 by etoposide. The lymphoblastoid cell lines were treated with etoposide (50 μM) for the indicated time and E2F1 was detected by immunoblotting.
Selective phosphorylation of E2F1 by ATM/ATR
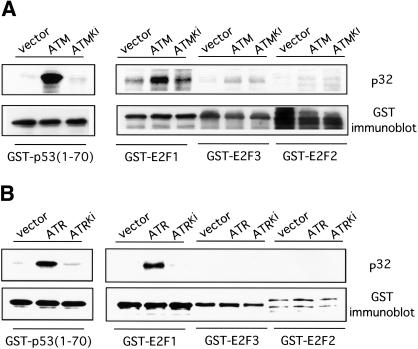
To further explore the mechanism for the involvement of ATM in E2F1 stabilization, we examined the possibility that E2F1 might be a direct substrate for ATM/ATR kinase activity. We used ATM or ATR kinase immunoprecipitated from 293T cell extracts following transfection with either ATM- or ATR-expressing plasmids as well as plasmids encoding kinase inactive versions of each. The immunoprecipitated kinase was then used to phosphorylate GST–E2F purified from bacterial lysate. As shown by the data in Figure 4, the active ATM and ATR kinases can phosphorylate E2F1 whereas the kinase-inactive ATM or ATR fails to do so. Thus, by this in vitro phosphorylation assay, E2F1 appears to be a substrate for the action of ATM and ATR.
Figure 4.
Specific phosphorylation of E2F1 by ATM/ATR. (A) Wild-type ATM or catalytically inactive ATM (ATMki) was immunoprecipitated from transfected cells 293T cells and used to phosphorylate GST–p53 (aa 1–70), or GST–E2F1, GST–E2F2, and GST–E2F3 proteins in in vitro reactions. The upper panel depicts the 32P autoradiograph and the lower panel shows an immunoblot with GST antibody. (B) Similar assays as described in A were performed with wild-type ATR or catalytically inactive ATR (ATRki).
We also assayed the ability of ATM or ATR to phosphorylate the E2F2 or E2F3 proteins. As shown by the date in Figure 4, neither ATM nor ATR kinase was able to phosphorylate E2F2 or E2F3 in vitro. This result thus provides good evidence for specificity in the phosphorylation of E2Fs by ATM or ATR and as such, provides a potential explanation for the specificity in induction of E2F1 protein following DNA damage. Taken together with the observation that E2F1 induction requires ATM activity, as seen in the assays using the AT cell lines result described above, this result strongly suggests that ATM/ATR is responsible for the specific induction of E2F1 during DNA damage.
A basis for selective phosphorylation and stabilization of E2F1
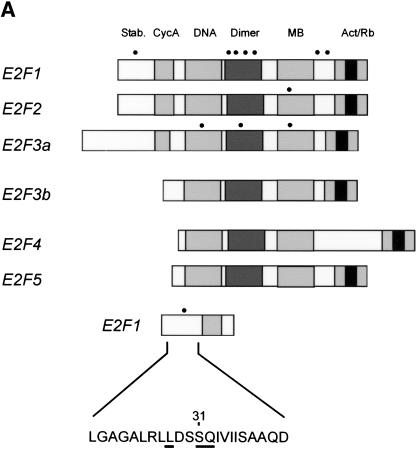
Previous work has detailed the recognition properties of ATM and ATR as requiring a serine or threonine adjacent to a glutamine (Kim et al. 1999; O'Neill et al. 2000). Additional amino acids neighboring these residues, and in particular the presence of hydrophobic residues at the N-1 and N-3 positions, also contribute to specificity, but it is the strict requirement of an SQ or TQ that defines specificity for ATM or ATR. An examination of the amino acid sequence of the three E2F proteins with respect to the presence of ATM/ATR consensus sequence reveals a number of potential sites in the three proteins. As summarized in Figure 5A, there are seven potential sites in E2F1, one site in E2F2, and three sites in E2F3. Interestingly, one of the sites in E2F1, a serine residue at position 31, lies within the amino-terminal domain that has been shown to be required for degradation of the E2F1 protein (Marti et al. 1999). This site also contains a hydrophobic residue at the N-3 position. In contrast, there are no potential ATM/ATR sites in the amino-terminal domain of either E2F2 or E2F3.
Figure 5.
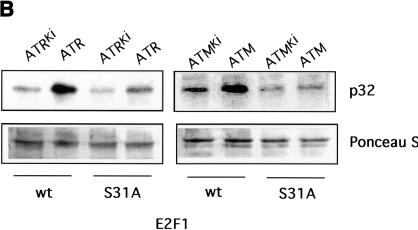
A basis for the specific phosphorylation of E2F1 by ATM and ATR. (A) Schematic depiction of the E2F family with potential sites for ATM/ATR phosphorylation. ATM/ATR consensus sites (S/T-Q) found within the E2F1–3 subfamily are indicated by the dots. The amino acid sequence containing the unique amino-terminal site in E2F1 is indicated at the bottom. (B) In vitro phosphorylation of E2F1 at serine 31. Wild-type E2F1 protein or the E2F1S31A mutant was prepared as described in Materials and Methods and used as a substrate for ATM/ATR kinase assays in vitro. The upper panel depicts the 32P autoradiograph after the kinase reaction whereas the lower panel depicts staining of the input proteins.
The in vitro phosphorylation of E2F1 by ATM/ATR occurred mainly at serine according to phosphoamino acid analysis, and likely localizes to a single AspN digested peptide (data not shown). Furthermore, immunoblotting of phosphorylated E2F1 proteolytic fragments shows that the site of phosphorylation by ATM/ATR lies in the amino terminus of E2F1 (data not shown). To explore the possible role of the serine 31 residue in E2F1 as a ATM/ATR phosphorylation site, we created a mutant protein in which this residue was changed to alanine. The protein was produced and purified from bacterial lysate and assayed as a substrate for ATM/ATR phosphorylation. As shown by the data in Figure 5B, both ATM and ATR readily phosphorylated the wild-type protein but the phosphorylation was largely abolished in the mutant protein. These results thus suggest that this serine 31 residue is indeed an ATM/ATR phosphorylation site and in fact is the major site for ATM/ATR-mediated phosphorylation within E2F1.
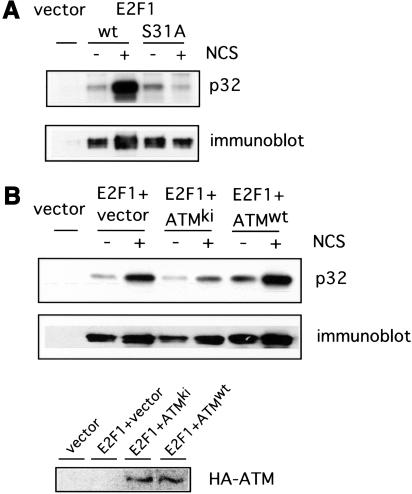
We have also assayed the role of the Ser 31 site in the phosphorylation of E2F1 in vivo, in response to DNA damage. Treatment of cells with the DNA damaging agent neocarzinostatin (NCS) leads to double strand DNA breaks and induction of ATM kinase activity (Banin et al 1998). Indeed, many experiments suggest that this agent is a selective inducer of ATM. We thus transfected cells with plasmids encoding either wild-type E2F1 or the Ser 31 mutant of E2F1 and then treated the cells with NCS. The cells were then labeled with 32P orthophosphate, extracts prepared, and E2F1 proteins were isolated by immunoprecipitation and then analyzed in an SDS acrylamide gel. As depicted in Figure 6A, the wild-type E2F1 protein was strongly phosphorylated in response to NCS (>12-fold increase in 32P signal by PhosphorImager quantitation). In sharp contrast, the Ser 31 E2F1 mutant protein was not phosphorylated despite the equivalent level of production of the two proteins. Based on these results, we conclude that E2F1 is indeed phosphorylated in vivo in response to DNA damage and that this involves a specific phosphorylation of Ser 31.
Figure 6.
DNA damage induced phosphorylation of E2F1. (A) 293T cells were transfected with HA-E2F1 or HA-E2F1S31A and labeled with 32P orthophosphate in the absence or presence of neocarzinostatin (NCS) at 300 ng/ml. E2F1 protein was then immunoprecipitated with HA beads and phosphorylation was detected by autoradiography (top). Also shown is the protein level as seen by immunoblotting (bottom). (B) A role for ATM in DNA damage induced phosphorylation of E2F1. 293T cells were transfected with HA-E2F1 along with vector, HA-ATMwt, or HA-ATMki. The cells were then labeled with 32P orthophosphate in the absence or presence of NCS and E2F1 protein was immunoprecipitated and detected as in A. Expression of transfected HA-ATM was confirmed by immunoblotting the HA bead precipitate with ATM antibody (Ab-3).
To explore the role of ATM in the in vivo phosphorylation of E2F1 in response to DNA damage, we have taken advantage of the ability of the kinase dead mutant of ATM (ATMki) to act as a dominant-negative inhibitor of ATM activity (Lim et al. 2000). Cells were transfected with the E2F1 expression vector either alone or together with wild-type ATM or the ATMki mutant. Cells were treated with NCS and labeled with 32P. Once again, DNA damage induced the phosphorylation of E2F1 which was enhanced by coexpression of wild-type ATM (Fig. 6B). The basal level of E2F1 phosphorylation in untreated cells after cotransfection with wild-type ATM was slightly higher than that seen with vector controls. This is consistent with a partial activation of overexpressed ATM as reported (Lim et al. 2000). In contrast, coexpression of the ATMki mutant abolished the phosphorylation of E2F1. Based on these results, together with the in vitro phosphorylation data, we conclude that E2F1 is indeed a substrate for ATM and that the DNA damage induced phosphorylation is mediated by ATM.
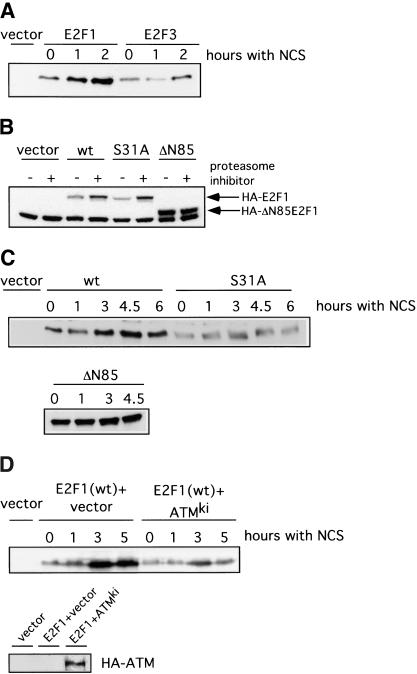
To investigate the relationship between ATM-dependent phosphorylation of E2F1 at Ser 31 and the DNA damage-induced accumulation of E2F1 protein, we have assayed the induction of either wild-type E2F1 or the Ser 31 mutant in response to DNA damage. Our approach was to employ transient transfection of plasmids encoding either the wild-type protein or the Ser 31 mutant and then treatment of the transfected cells with a DNA damaging drug. Following treatment, cells are harvested and assayed for the tagged E2F1 protein by Western blot. As shown by the data in Figure 7A, treatment of cells transfected with wild-type E2F1 with NCS led to an induction of E2F1 protein equivalent to that seen for the endogenous protein in other assays. In contrast, there was no effect on the accumulation of wild-type E2F3 protein used as a control like wild-type E2F1. Similar to the wild-type E2F1 protein, the Ser 31 mutant is also degraded by the proteasome as seen by the ability of the proteasome inhibitor MG-132 to promote an increase in E2F1 (Fig. 7B). In contrast, an amino-terminal deletion of E2F1 was already stable and not affected by treatment with MG-132, consistent with previous work demonstrating a requirement for these amino-terminal sequences for targeted degradation. A comparison of the wild-type E2F1 protein versus the Ser 31 mutant for a response to NCS is shown in Figure 7C. The wild-type protein was again induced by DNA damage but in contrast, the Ser 31 was not affected. As a further control, the amino-terminal deletion was also not affected by treatment with NCS.
Figure 7.
The ATM phosphorylation site in E2F1 is required for DNA damaged induction. (A) 293T cells were transfected with HA-E2F1 or HA-E2F3 and then treated with neocarzinostatin (NCS) for the indicated time. HA-tagged E2F proteins were detected by immunoblotting with an HA-specific antibody. (B) Degradation of E2F1 by the proteasome. 293T cells were transfected with HA-E2F1, HA-E2F1S31A, or HA-E2F1ΔN85 and then treated with MG-132 at 20 μM for 5 h. Proteins were detected by immunoblotting with an HA specific antibody. (C) 293T cells were transfected with HA-E2F1, HA-E2F1S31A, or HA-E2F1ΔN85 and then treated with NCS for the indicated time. Proteins were detected by immunoblotting with an HA specific antibody. (D) Induction of E2F1 in response to DNA damage requires ATM. 293T cells were transfected with HA-E2F1 along with HA-ATMKi or vector. The cells were then treated with NCS for the indicated time. Transfected E2F1 was detected with an HA-specific antibody (top). Expression of transfected ATMki protein was confirmed by immunoprecipitation with HA beads followed by immunoblotting with ATM antibody (Ab-3) (bottom).
To explore the role of ATM in the DNA damage-mediated induction of E2F1, we again made use of the ATMki mutant to block ATM activity. Cells were transfected with the E2F1-expressing plasmid alone or together with the ATMki mutant, treated with NCS, and then assayed for E2F1 protein accumulation. As shown in Figure 7D, NCS again induced E2F1 protein accumulation, which was blocked by the ATMki mutant. Based on these results, we conclude that the induction of E2F1 protein accumulation that follows a DNA damage event is dependent on ATM and involves the specific phosphorylation of E2F1 at a unique amino-terminal site.
A role for E2F1 in the DNA damage response in mouse thymocytes
The specific induction of E2F1 protein following DNA damage, together with the past work that has established a role for E2F1 as a signal for p53-dependent apoptosis, suggests the possibility of a functional role for E2F1 in the DNA damage response. We have thus investigated whether there is a role for E2F1 in cell cycle repression or apoptosis following DNA damage using mice in which the E2F1 gene has been disrupted by homologous recombination.
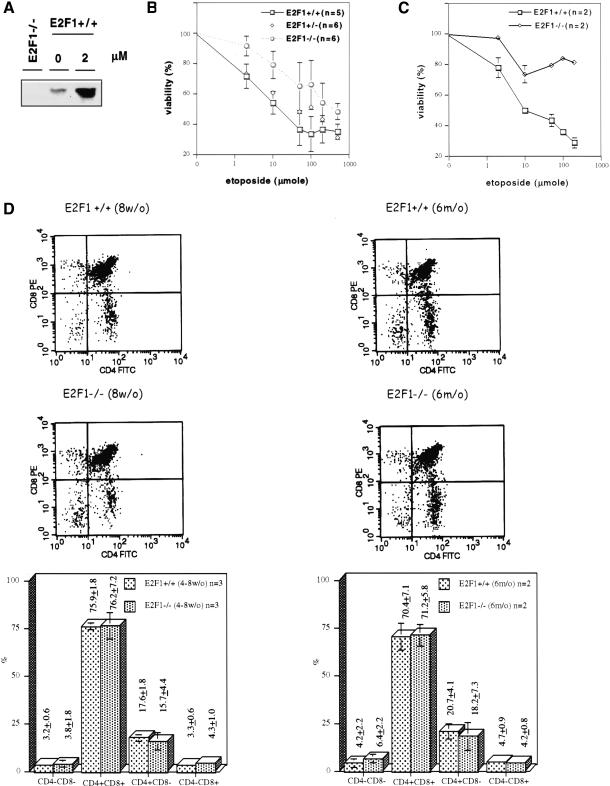
Previous work has shown that DNA damage-induced apoptosis in thymocytes is p53-dependent (Clarke et al. 1993; Lowe et al. 1993). Moreover, other work has shown a role for E2F1 in thymocyte cell death associated with negative selection (Field et al. 1996). As such, we have examined the role of E2F1 in thymocyte apoptosis following treatment with DNA damaging drugs. Consistent with the results seen in the tumor cells and mouse fibroblasts, E2F1 protein accumulation was induced by treatment with etoposide (Fig. 8A). We have not detected an induction of either E2F2 or E2F3 protein under these conditions (data not shown) although we also cannot detect basal levels of these proteins to confirm that the induction is indeed specific. Although there was no apparent effect of E2F1 status on the G1/S arrest as measured by 3H-thymidine incorporation (data not shown), we did find that the thymocytes from E2F1−/− mice were more resistant to etoposide-induced cell death as compared with thymocytes from their wild-type siblings (Fig. 8B). The sensitivity of the E2F1+/− heterozygotes fell between wild-type and E2F1−/− counterparts. In our short-term overnight culture, the viability of thymocytes without etoposide treatment is around 80% (E2F1+/+, 84.5 ± 8%; E2F1+/−, 81.9 ± 5%; E2F1−/−, 86.6% ± 5%). The viability of thymocytes after etoposide treatment is normalized to untreated for each animal. The assay was carried out in five or six 4- to 8-week-old animals from each genotype and they are siblings from several different pairs of parents. When the experiment was carried out in six-month-old mice, we also observe an attenuation of etoposide-induced apoptosis in thymocytes from E2F1−/− mice (Fig. 8C). A similar difference in drug sensitivity was observed when cisplatin or adriamycin was used (data not shown).
Figure 8.
A role for E2F1 in DNA damage-induced thymocyte death. (A) Western blot analysis of E2F1 protein induction in thymocytes analyzed 22 h following treatment with etoposide (2 μM). (B) Viability assays. Thymocytes were prepared from 4–8-week-old mice and cell viability was analyzed 20 h following treatment with etoposide at concentrations ranging from 2 to 500 μM. (C) Viability assays. Thymocytes were prepared from six month-old mice and cell viability was analyzed 20 h following treatment with etoposide at concentrations ranging from 2 to 200 μM. (D) Flow cytometry profiles following CD4 and CD8 immunostaining of thymocytes from E2F1+/+ or E2F1−/− mice at either eight wk of age or six mo of age.
One possible explanation for the differential sensitivity of wild-type and E2F1−/− thymocytes could be an alteration in the thymocyte population, and in particular an increase in the proportion of single positive cells (CD4+CD8− or CD4−CD8+) because these single positive cells have been shown to be more resistant to apoptosis as compared with the double positive thymocytes (Lowe et al. 1993). To address this possibility, we have analyzed the population of thymocytes, by CD4 and CD8 immunostaining, from the wild-type and E2F1−/− mice. As seen by the analysis in Figure 8D, the proportion of each population is quite similar between E2F1+/+ and E2F1−/− mice either in age of 4- to 8-weeks or six months. To further examine the population of thymocytes, cells were stained with an antibody to CD3ɛ, a component of the T cell receptor expressed at high levels in mature cells. The percent of CD3ɛhi thymocytes was essentially the same between E2F1+/+ and E2F1−/− mice, either at 4–8 weeks of age (E2F1+/+ 24.4 ± 3.7; E2F1−/− 22.7 ± 6.0) or at six months of age (E2F1+/+ 27.7 ± 5.3; E2F1−/− 28.3 ± 10). Thus, based on these results, we conclude that it is the absence of E2F1 activity in thymocytes derived from E2F1−/− mice, rather than a shift in the thymocyte population, that attenuates the DNA damage-induced apoptosis response. This finding, together with the observations that demonstrate an induction of E2F1 accumulation following DNA damage, strongly suggests a role for the E2F1 pathway in the DNA damage response.
Discussion
An understanding of the mechanisms involved in the DNA damage response has developed rapidly over the past several years, placing control of p53 accumulation at a critical juncture in the decision to arrest cell growth or initiate programmed cell death. These studies have led to the realization that part of the DNA damage response involves the activation of protein kinases of the PI3-K family, including ATM and ATR, that can specifically target p53 for phosphorylation, either directly or indirectly through the chk2 kinase (Hirao et al. 2000). In part, these phosphorylation events are believed to block the ability of Mdm2 to inhibit p53 transcriptional activity as well as to target p53 for degradation. As such, the level of active p53 protein rises, leading to enhanced expression of key p53 target genes that function as cell cycle inhibitors as well as inducers of apoptosis.
Likewise, an understanding of the role of p53 as a cell cycle checkpoint, responding to alterations in either the Rb pathway or the Myc protein, has also rapidly advanced. In particular, the role of Mdm2 as a p53-specific ubiquitin ligase, the role of the recently identified p19ARF protein in the control of Mdm2 activity, and the role of the E2F1 protein as a signal for apoptosis and inducer of ARF expression, now defines a pathway that leads from the Rb/E2F cell cycle pathway to p53 accumulation. Moreover, recent data has also shown that at least part of the action of Myc as an inducer of apoptosis and p53 accumulation is dependent on E2F1 (Leone et al. 2001). The results presented here now suggest a role for E2F1 in signaling apoptosis not only as a consequence of deregulation of the Rb cell cycle pathway or in response to Myc, but also in response to DNA damage, involving the same ATM/ATR pathway that controls p53 accumulation.
E2F1 protein is specifically induced by DNA damage
Consistent with reports by others (Huang et al. 1997; Blattner et al. 1999; Hofferer et al. 1999; Meng et al. 1999), we observe an induction of E2F1 by several DNA-damaging chemotherapeutic drugs in a variety of tumor cell lines independent of p53 or Rb status. The data we present here now demonstrates a similar induction of E2F1 in normal fibroblasts and thymocytes, indicating that DNA damage-induced E2F1 is a general phenomenon. This induction is rapid (within 4 h of drug treatment) and is mediated through a posttranscriptional mechanism that likely involves protein stabilization (Blattner et al. 1999). More importantly, we also show that E2F1, unique among the E2F family, is specifically induced, which makes it unlikely that its induction is simply a secondary phenomenon caused by an enrichment of G1/S arrested population. Taken together, these observations demonstrate a specific response of E2F1 following a DNA damage event.
The mechanisms that control the accumulation of E2F1 activity in normally growing cells are complex and involve transcriptional control of the E2F1 promoter (Hsiao et al. 1994; Johnson et al. 1994; Sellers et al. 1995), control of E2F1 DNA-binding activity through the action of cyclin A/cdk2 (Krek et al. 1994), and control of E2F1 protein stability through ubiquitin-mediated degradation by the proteasome (Hateboer et al. 1996). Two activities have been implicated in the control of E2F1 degradation—the binding of Rb to the carboxyl terminus of E2F1 (Hateboer et al. 1996; Hofmann et al. 1996) and the binding of p45Skp2 to the amino-terminal sequences of the protein (Marti et al. 1999). The binding of unphosphorylated Rb to the carboxyl terminus of E2F1 may be important for the stability of Rb–E2F1-repressor complexes during an exit from cell cycle progression to quiescence but likely cannot account for the specific induction of E2F1 in response to DNA damage because cell lines lacking Rb expression still maintain the inducibility of E2F1 protein.
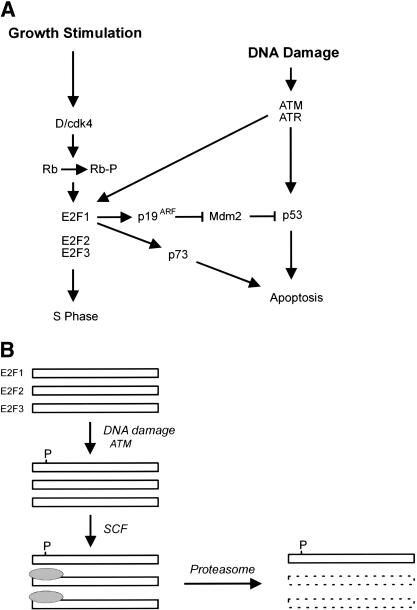
A more likely mechanism controlling E2F1 stability in response to DNA damage would be the role of the ubiquitin-protein ligase SCFSkp2 and E2F1 (Marti et al. 1999). p45Skp2 is a member of a rapidly expanding family of F box protein components of SKP1-CDC53 (cullin)-F-box (SCF) complexes that function as E3 ligases (Winston et al. 1999). E3 ubiquitin-protein ligases mediate a critical step in substrate recognition and are essential for ubiquitination reaction. The SCF complexes utilize different F-box proteins as a substrate specific receptor and target a specific protein to degradation. Recently, p45Skp2 was reported to be the F-box protein that can bind to the amino terminus of E2F1 and target E2F1 for degradation in S/G2 phase (Marti et al. 1999). We can only speculate as to how ATM-mediated phosphorylation might alter the degradation of E2F1 but we do note that the site of phosphorylation (serine 31) lies within the domain recognized by Skp2. Thus, it is possible that the ATM/ATR-mediated phosphorylation inhibits the binding and function of Skp2 and thus prevents the normal degradation of E2F1 (Fig. 9).
Figure 9.
Role of E2F1 in the DNA damage response. (A) A link between the Rb/E2F pathway and the DNA damage response. In addition to the role of ATM and ATR in inducing p53 accumulation, the results presented here identify E2F1 as a target for ATM and ATR in response to DNA damage. The consequence of the E2F1 induction could be seen as a further augmentation of p53 accumulation through an activation of p19ARF. In addition, given the ability of E2F1 to induce the p73 gene, the action through E2F1 could lead to a p53-independent effect on apoptosis. (B) Potential mechanism for ATM/ATR-mediated stabilization of E2F1. Previous work has documented the role of an SCF complex in targeting an amino-terminal domain of E2F1, leading to ubiquitin-dependent proteasome degradation. The site for ATM/ATR phosphorylation lies within this domain; as such, we speculate that the phosphorylation might prevent the interaction of the SCF complex with E2F1 and thus prevent degradation.
A specific role of E2F1 induction following DNA damage
Our finding that a DNA damage response is attenuated in thymocytes lacking E2F1 provides evidence that the induction of E2F1 protein is in fact important. Although there was little or no evidence for any significant difference in the response of primary mouse embryonic fibroblasts (MEFs) with different E2F1 genotypes to various chemotherapeutic drugs, as measured by the extent and kinetics of G1/S arrest or apoptosis (data not shown), and there was no significant difference of G1/S arrest in thymocytes from these mice, we found that the DNA damage-induced apoptosis is significantly attenuated in thymocytes from E2F1−/− mice. This tissue-dependent differential function for one gene is not without precedent. Although p53 induction is important for DNA damage-induced apoptosis in thymocytes (Clarke et al. 1993; Lowe et al. 1993), the role for p53 in fibroblasts is mainly for G1/S arrest (Kastan et al. 1992). Although the etoposide-induced apoptosis is attenuated but not completely abrogated in E2F1−/− thymocytes (Fig. 8), the etoposide-induced apoptosis in p53−/− thymocytes was shown to be decreased rather than completely abolished (Clarke et al. 1993). As shown by our analysis of thymocyte populations (Fig. 8D), the attenuation of apoptosis in E2F1−/− thymocytes is attributable to a lack of E2F1 induction rather than a secondary phenomenon from a shift in thymocyte differentiation because there is no significant difference in the subpopulation (defined by CD3ɛ, CD4, and CD8 surface marker expression) among the mice we used. The reason that we failed to observe a similar difference on thymocyte subpopulation in E2F1−/− mice as reported previously (Field et al. 1996) may be related to a different genetic background as we have bred the mice to C57/BL6 background for at least eight generations.
Our finding that E2F1 is induced in response to DNA damage, dependent on ATM/ATR kinase activity, and the fact that ATM/ATR can specifically phosphorylate E2F1, now expands the events involving p53 that respond to DNA damage. The consequence of this E2F1 induction could be twofold (Fig. 9). Given the role of E2F1 in inducing p53 accumulation through an activation of p19ARF, the effect of ATM/ATR on E2F1 could be to simply amplify the accumulation of p53. That is, ATM/ATR induces p53 both by preventing Mdm2 action and by reducing Mdm2 levels. Alternatively, E2F1 may activate additional genes such as p73 (Irwin et al. 2000; Lissy et al. 2000; Stiewe and Putzer 2000), in addition to p19ARF, that further contribute to the apoptosis response.
Materials and methods
Cells
H460, H358, H125, H596, H520 cell lines were gifts from Gerold Bepler (Roswell Park Memorial Institute) and were maintained in RPMI-1640 supplemented with 10% fetal bovine serum (FBS). HepG2 and Hep3B were purchased from ATCC and HFF (human foreskin fibroblast) were obtained from Clonetics. Primary cultured MEFs of different E2F1 genotypes were prepared from 12.5- to 14.5-day-old embryos as described previously (Kamijo et al. 1997). p53−/− and wild-type littermate MEFs have been described previously (Kowalik et al. 1995). MEF, Hep3B, HepG2, and HFF cultures were maintained in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% heat inactivated FBS. GM2184, 3382, 1526, 3332, and 719 were purchased from Coriell Cell Repositories and were maintained in RPMI 1640 supplemented with 15% heat inactivated FBS. To obtain thymocytes, thymuses from mice with different E2F1 genotypes were crushed between two glass slides. Connective tissue was removed by passage through nylon mesh, and thymocytes were purified over a Ficoll-Paque gradient and cultured at a density of 1 × 106 cells/mL in RPMI 1640 supplemented with 15% heat inactivated FBS and 50 μM 2-mercapto ethanol. Cells were obtained in pups derived from E2F1+/− × E2F1+/− parents, and the E2F1−/− cells were compared with the cells derived from their sibling littermates. 293T cells were a gift from Dr. Robert Abraham.
Mice
The E2F1−/− mice have been described previously (Field et al. 1996) and were kindly provided by Dr. M. Greenberg (Harvard Medical School). The mice were bred to C57BL6.
Immunoblotting
Cell lysates (100 μg of protein) were fractionated by SDS-PAGE and electrotransferred to Imobilon-P membrane. Equal protein loading was confirmed with Ponceau-S staining. E2F1 antibody (C-20 and KH-95), E2F2 antibody (C-20), E2F3 antibodies (C-18 and N-20), E2F4 antibody (C-20), and Rb-carboxy-terminal-specific antibody (C-15) were purchased from Santa Cruz, and the immunoblot was performed with enhanced chemiluminescence assay (Amersham) according to the manufacturer's instructions. Rb antibody (G3-245), which recognizes an epitope between amino acid 300 and 380 of human Rb, and p53 antibody (Pab 240) were purchased from Pharmingen. GST antibody (B-14) and HA antibody (Y-11) were purchased from Santa Cruz. ATM antibody (Ab-3) was purchased from Oncogene Research Products.
RNA analysis
The isolation of mRNA and Northern blot with E2F1-specific and E2F3-specific probes were performed as described previously (Leone et al. 1998).
Drug sensitivity assay
Cisplatin, etoposide, and adriamycin were obtained from Sigma. Cells were treated with drugs as indicated and cell viability was assayed by trypan blue exclusion. Drug-induced apoptosis was also confirmed with annexin V-FITC/propidium iodide staining (Pharmingen) and propidium iodide DNA staining followed by flow cytometry (Kowalik et al. 1995).
ATM/ATR kinase assays
293T cells were transiently transfected with either pBJ-HA-ATM, pBJ-HA-ATMki, pcDNA3-Flag-ATR, or pcDNA3-Flag-ATRki by the Fugene6 method (Roche). The ATM and ATR kinases were prepared as described previously (Tibbetts et al. 1999) with 12CA5 antibody (Boehringer-Mannheim) for HA-ATM and M2 antibody (Sigma) for Flag-ATR. GST–p53 (aa 1–70), GST–E2F1, GST–E2F2, and GST–E2F3 were produced and purified from Escherichia coli.
Plasmid constructs and transfections
ATR and ATM expression vectors were kind gifts from Dr. Robert Abraham (Tibbetts et al. 1999). E2F1 mutant S31A was generated using GeneEditor in vitro site-directed Mutagenesis System (Promega) with primer 5′-CGGCTGCTCGACTCTGCGC AGATCGTCATC-3′. ΔN85 deletional mutant was generated by PCR with primers: 5′-CTCGGATCCGCCAGCGCCCC GCCGGTGAAG-3′ and 5′-CCGAGTGGCTCAGCAGCTC CAGGAAGCG-3′. The PCR product was then digested with BamHI and BlpI and cloned into BamHI–BlpI fragment of pcDNA3HAE2F1 (a gift from Dr. Wilhelm Krek, Friedrich Miescher Institute, Basel, Switzerland). The mutation and deletion were verified by sequencing. For transfection, 293T cells were transiently transfected with plasmids by the Fugene6 method. To ensure equal transfection efficiency, one transfected master plate was then split equally to generate plates for individual samples.
In vivo labeling
Two days after transfection, 293T cells were starved for 30 min in 2 mL of phosphate-free DMEM and 32P-orthophosphate (2 mCi) was added for additional 2.5 h. Cells were washed and lysed in RIPA buffer supplemented with protease inhibitors and phosphatase inhibitors. Lysates were clarified by centrifugation and HA-E2F1 was immunoprecipitated with HA.11 affinity matrix (BAbCO).
Thymocyte surface marker staining
Thymocytes were suspended in PBS with 2% FBS and incubated with anti-mouse CD16/CD32 (FCγIII/II receptor) for 5 min. The cells were then stained with allophycocyanin (APC)-conjugated anti-CD3ɛ (clone 145–2C11), fluorescein isothiocyanate (FITC)-conjugated anti-CD4 (clone H129.19), and R-phycoerythrin (PE)-conjugated anti-CD8 (clone 53–6.7) (all obtained from Pharmingen) for 15 min. The cells were then washed with PBS/2% FBS twice and fixed in fresh 1% paraformaldehyde for flow cytometry.
Acknowledgments
We thank M. E. Greenberg for providing the E2F1−/− mice. We thank Bob Abraham, Randy Tibbetts, and Kathy Brumbaugh for ATM and ATR constructs and for advice on kinase assays. We thank Mike Cook and Lynn Martinek of the Duke Comprehensive Cancer Center Flow Cytometry Facility for the FACS analyses. We also thank laboratory members for discussions and Kaye Culler for help with the preparation of the manuscript. W.C.L. is supported by the Cancer Research Fund of the Damon Runyon Walter Winchell Foundation (fellowship DRG 093). J.R.N. is an Investigator in the Howard Hughes Medical Institute.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL J.Nevins@duke.edu; FAX (919) 681-8973.
References
- Banin S, Moyal L, Shieh S, Taya Y, Anderson CW, Chessa L, Smorodinsky NI, Prives C, Reiss Y, Shiloh Y, et al. Enhanced phosphorylation of p53 by ATM in response to DNA damage. Science. 1998;281:1674–1677. doi: 10.1126/science.281.5383.1674. [DOI] [PubMed] [Google Scholar]
- Bates S, Phillips AC, Clark PA, Stott F, Peters G, Ludwig RL, Vousden KH. p14ARF links the tumour suppressors RB and p53. Nature. 1998;395:124–125. doi: 10.1038/25867. [DOI] [PubMed] [Google Scholar]
- Blattner C, Sparks A, Lane D. Transcription factor E2F-1 is upregulated in response to DNA damage in a manner analogous to that of p53. Mol Cell Biol. 1999;19:3704–3713. doi: 10.1128/mcb.19.5.3704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canman CE, Lim DS, Cimprich KA, Taya Y, Tamai K, Sakaguchi K, Appella E, Kastan MB, Siliciano JD. Activation of the ATM kinase by ionizing radiation and phosphorylation of p53. Science. 1998;281:1677–1679. doi: 10.1126/science.281.5383.1677. [DOI] [PubMed] [Google Scholar]
- Chehab NH, Malikzay A, Appel M, Halazonetis TD. Chk2/hCds1 functions as a DNA damage checkpoint in G(1) by stabilizing p53. Genes & Dev. 2000;14:278–288. [PMC free article] [PubMed] [Google Scholar]
- Clarke AR, Purdie CA, Harrison DJ, Morris RG, Bird CC, Hooper ML, Wyllie H. Thymocyte apoptosis induced by p53-dependent and independent pathways. Nature. 1993;362:849–852. doi: 10.1038/362849a0. [DOI] [PubMed] [Google Scholar]
- DeGregori J, Leone G, Miron A, Jakoi L, Nevins JR. Distinct roles for E2F proteins in cell growth control and apoptosis. Proc Natl Acad Sci. 1997;94:7245–7250. doi: 10.1073/pnas.94.14.7245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dyson N. The regulation of E2F by pRB-family proteins. Genes & Dev. 1998;12:2245–2262. doi: 10.1101/gad.12.15.2245. [DOI] [PubMed] [Google Scholar]
- Field SJ, Tsai F-Y, Kuo F, Zubiaga AM, Kaelin WG, Jr, Livingston DM, Orkin SH, Greenberg ME. E2F-1 functions in mice to promote apoptosis and suppress proliferation. Cell. 1996;85:549–561. doi: 10.1016/s0092-8674(00)81255-6. [DOI] [PubMed] [Google Scholar]
- Hateboer G, Kerkhoven RM, Shvarts A, Bernards R, Beijersbergen RL. Degradation of E2F by the ubiquitin-proteasome pathway: Regulation by retinoblastoma family proteins and adenovirus transforming proteins. Genes & Dev. 1996;10:2960–2970. doi: 10.1101/gad.10.23.2960. [DOI] [PubMed] [Google Scholar]
- Hirao A, Kong YY, Matsuoka S, Wakeham A, Ruland J, Yoshida H, Liu D, Elledge SJ, Mak TW. DNA damage-induced activation of p53 by the checkpoint kinase Chk2. Science. 2000;287:1824–1827. doi: 10.1126/science.287.5459.1824. [DOI] [PubMed] [Google Scholar]
- Hofferer M, Wirbelauer C, Humar B, Krek W. Increased levels of E2F-1 dependent DNA binding activity after UV- or gamma-irradiation. Nucleic Acids Res. 1999;27:491–495. doi: 10.1093/nar/27.2.491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofmann F, Martelli F, Livingston DM, Wang Z. The retinoblastoma gene product protects E2F-1 from degradation by the ubiquitin-proteasome pathway. Genes & Dev. 1996;10:2949–2959. doi: 10.1101/gad.10.23.2949. [DOI] [PubMed] [Google Scholar]
- Honda R, Tanaka H, Yasuda H. Oncoprotein Mdm2 is a ubiquitin ligase E3 for tumor suppressor p53. FEBS Lett. 1997;420:25–27. doi: 10.1016/s0014-5793(97)01480-4. [DOI] [PubMed] [Google Scholar]
- Hsiao K-M, McMahon SL, Farnham PJ. Multiple DNA elements are required for the growth regulation of the mouse E2F1 promoter. Genes & Dev. 1994;8:1526–1537. doi: 10.1101/gad.8.13.1526. [DOI] [PubMed] [Google Scholar]
- Huang Y, Ishiko T, Nakada S, Utsugisawa T, Kato T, Yuan ZM. Role for E2F in DNA damage-induced entry of cells into S phase. Cancer Res. 1997;57:3640–3643. [PubMed] [Google Scholar]
- Irwin M, Martin MC, Phillips AC, Seelan RS, Smith DI, Liu W, Flores ER, Tsai KY, Jacks T, Vousden KH, et al. Role for the p53 homologue p73 in E2F-1-induced apoptosis. Nature. 2000;407:645–648. doi: 10.1038/35036614. [DOI] [PubMed] [Google Scholar]
- Jacks T, Fazeli A, Schmitt EM, Bronson RT, Goodell MA, Weinberg RA. Effects of an Rb mutation in the mouse. Nature. 1992;359:295–300. doi: 10.1038/359295a0. [DOI] [PubMed] [Google Scholar]
- Johnson DG, Schwarz JK, Cress WD, Nevins JR. Expression of transcription factor E2F1 induces quiescent cells to enter S phase. Nature. 1993;365:349–352. doi: 10.1038/365349a0. [DOI] [PubMed] [Google Scholar]
- Johnson DG, Ohtani K, Nevins JR. Autoregulatory control of E2F1 expression in response to positive and negative regulators of cell cycle progression. Genes & Dev. 1994;8:1514–1525. doi: 10.1101/gad.8.13.1514. [DOI] [PubMed] [Google Scholar]
- Kamijo T, Zindy F, Roussel MF, Quelle DE, Downing JR, Ashmun RA, Grosveld G, Sherr CJ. Tumor suppression at the mouse INK4a locus mediated by the alternative reading frame product p19ARF. Cell. 1997;91:649–659. doi: 10.1016/s0092-8674(00)80452-3. [DOI] [PubMed] [Google Scholar]
- Kastan MB, Zhan Q, El-Deiry WS, Carrier F, Jacks T, Walsh WV, Plunkett BS, Vogelstein B, Fornace AJ., Jr A mammalian cell cycle checkpoint pathway utilizing p52 and GADD45 is defective in ataxia-telangiectasia. Cell. 1992;71:587–597. doi: 10.1016/0092-8674(92)90593-2. [DOI] [PubMed] [Google Scholar]
- Kim ST, Lim DS, Canman CE, Kastan MB. Substrate specificities and identification of putative substrates of ATM kinase family members. J Biol Chem. 1999;274:37538–37543. doi: 10.1074/jbc.274.53.37538. [DOI] [PubMed] [Google Scholar]
- Kowalik TF, DeGregori J, Schwarz JK, Nevins JR. E2F1 overexpression in quiescent fibroblasts leads to induction of cellular DNA synthesis and apoptosis. J Virol. 1995;69:2491–2500. doi: 10.1128/jvi.69.4.2491-2500.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kowalik TF, DeGregori J, Leone G, Nevins JR. E2F1-specific induction of apoptosis and p53 accumulation is modulated by mdm2. Cell Growth & Diff. 1998;9:113–118. [PubMed] [Google Scholar]
- Krek W, Ewen ME, Shirodkar S, Arany Z, Kaelin WG, Livingston DM. Negative regulation of the growth-promoting transcription factor E2F-1 by a stably bound cyclin A-dependent protein kinase. Cell. 1994;78:161–172. doi: 10.1016/0092-8674(94)90582-7. [DOI] [PubMed] [Google Scholar]
- Lee EYHP, Chang C-Y, Hu N, Wang Y-CJ, Lai C-C, Herrup K, Lee W-H, Bradley A. Mice deficient for Rb are nonviable and show defects in neurogenesis and haematopoiesis. Nature. 1992;359:288–294. doi: 10.1038/359288a0. [DOI] [PubMed] [Google Scholar]
- Leone G, DeGregori J, Yan Z, Jakoi L, Ishida S, Williams RS, Nevins JR. E2F3 activity is regulated during the cell cycle and is required for the induction of S phase. Genes & Dev. 1998;12:2120–2130. doi: 10.1101/gad.12.14.2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leone G, Nuckolls F, Ishida S, Adams M, Sears R, Jakoi L, Miron A, Nevins JR. Identification of a novel E2F3 product suggests a mechanism for determining specificity of repression by Rb proteins. Mol Cell Biol. 2000;20:3626–3632. doi: 10.1128/mcb.20.10.3626-3632.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leone, G., Sears, R., Huang, E., Rempel, R., Nuckolls, F., Park, C.-H., Giangrande, P., Wu, L., Saavedra, H.I., Field, S.J., et al. 2001. Myc requires distinct E2F activities to induce S phase and apoptosis. Mol. Cell (in press). [DOI] [PubMed]
- Lim D-S, Kim S-T, Xu B, Maser RS, Lin J, Petrini JHJ, Kastan MB. ATM phosphorylates p95/nbs1 in an S-phase checkpoint pathway. Nature. 2000;404:613–617. doi: 10.1038/35007091. [DOI] [PubMed] [Google Scholar]
- Lissy NA, Davis PK, Irwin M, Kaelin WG, Dowdy SF. A common E2F-1 and p73 pathway mediates cell death induced by TCR activation. Nature. 2000;407:642–644. doi: 10.1038/35036608. [DOI] [PubMed] [Google Scholar]
- Lowe SW, Schmitt EM, Smith SW, Osborne BA, Jacks T. p53 is required for radiation-induced apoptosis in mouse thymocytes. Nature. 1993;362:847–849. doi: 10.1038/362847a0. [DOI] [PubMed] [Google Scholar]
- Marti A, Wirbelauer C, Scheffner M, Krek W. Interaction between SCFSKP2 ubiquitin protein ligase and E2F-1 underlies regulation of E2F-1 degradation. Nat Cell Biol. 1999;1:14–19. doi: 10.1038/8984. [DOI] [PubMed] [Google Scholar]
- Meng RD, Phillips P, El-Deiry WS. p53-independent increase in E2F-1 expression enhances the cytotoxic effects of etoposide and of adriamycin. Int J Oncol. 1999;14:5–14. [PubMed] [Google Scholar]
- Nevins JR. Toward an understanding of the functional complexity of the E2F and Retinoblastoma families. Cell Growth & Diff. 1998;9:585–593. [PubMed] [Google Scholar]
- O'Neill T, Dwyer AJ, Ziv Y, Chan DW, Lees-Miller SP, Abraham RH, Lai JH, Hill D, Shiloh Y, Cantley LC, et al. Utilization of oriented peptide libraries to identify substrate motifs selected by ATM. J Biol Chem. 2000;275:22719–22727. doi: 10.1074/jbc.M001002200. [DOI] [PubMed] [Google Scholar]
- Pan H, Yin C, Dyson NJ, Harlow E, Yamasaki L, van Dyke T. Key roles for E2F1 in signaling p53-dependent apoptosis and in cell division within developing tumors. Mol Cell. 1998;2:283–292. doi: 10.1016/s1097-2765(00)80273-7. [DOI] [PubMed] [Google Scholar]
- Phillips AC, Bates S, Ryan KM, Helin K, Vousden KH. Induction of DNA synthesis and apoptosis are separable functions of E2F-1. Genes & Dev. 1997;11:1853–1863. doi: 10.1101/gad.11.14.1853. [DOI] [PubMed] [Google Scholar]
- Qin X-Q, Livingston DM, Kaelin WG, Adams PD. Deregulated transcription factor E2F-1 expression leads to S-phase entry and p53-mediated apoptosis. Proc Natl Acad Sci. 1994;91:10918–10922. doi: 10.1073/pnas.91.23.10918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sellers WR, Rodgers JW, Kaelin WG., Jr A potent transrepression domain in the retinoblastoma protein induces a cell cycle arrest when bound to E2F sites. Proc Natl Acad Sci. 1995;92:11544–11548. doi: 10.1073/pnas.92.25.11544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shan B, Lee W-H. Deregulated expression of E2F-1 induces S-phase entry and leads to apoptosis. Mol Cell Biol. 1994;14:8166–8173. doi: 10.1128/mcb.14.12.8166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shieh SY, Ahn J, Tamai K, Taya Y, Prives C. The human homologs of checkpoint kinases Chk1 and Cds1 (Chk2) phosphorylate p53 at multiple DNA damage-inducible sites. Genes & Dev. 2000;14:289–300. [PMC free article] [PubMed] [Google Scholar]
- Stiewe T, Putzer BM. Role of the p53 homologue p73 in E2F1-induced apoptosis. Nat Genet. 2000;26:391–392. doi: 10.1038/82617. [DOI] [PubMed] [Google Scholar]
- Tan X, Wang JYJ. The caspase-Rb connection in cell death. Trends Cell Biol. 1998;8:116–120. doi: 10.1016/s0962-8924(97)01208-7. [DOI] [PubMed] [Google Scholar]
- Tibbetts RS, Brumbaugh KM, Williams JM, Sarkaria JN, Cliby WA, Shieh S-Y, Taya Y, Prives C, Abraham RT. A role for ATR in the DNA damage-induced phosphorylation of p53. Genes & Dev. 1999;13:152–157. doi: 10.1101/gad.13.2.152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai KY, Hu Y, Macleod KF, Crowley D, Yamasaki L, Jacks T. Mutation of E2f-1 suppresses apoptosis and inappropriate S phase entry and extends survival of Rb-deficient mouse embryos. Mol Cell. 1998;2:293–304. doi: 10.1016/s1097-2765(00)80274-9. [DOI] [PubMed] [Google Scholar]
- Vigo E, Muller H, Prosperini E, Hateboer G, Cartwright P, Moroni MC, Helin K. CDC25A phosphatase is a target of E2F and is required for efficient E2f-induced S phase. Mol Cell Biol. 1999;19:6379–6395. doi: 10.1128/mcb.19.9.6379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber JD, Taylor LJ, Roussel MF, Sherr CJ, Bar-Sagi D. Nucleolar Arf sequesters Mdm2 and activates p53. Nat Cell Biol. 1999;1:20–26. doi: 10.1038/8991. [DOI] [PubMed] [Google Scholar]
- Winston JT, Koepp DM, Zhu C, Elledge SJ, Harper JW. A family of mammalian F-box proteins. Curr Biol. 1999;9:1180–1182. doi: 10.1016/S0960-9822(00)80021-4. [DOI] [PubMed] [Google Scholar]
- Wu X, Levine AJ. p53 and E2F-1 cooperate to mediate apoptosis. Proc Natl Acad Sci. 1994;91:3602–3606. doi: 10.1073/pnas.91.9.3602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamasaki L, Bronson R, Williams BO, Dyson NJ, Harlow E, Jacks T. Loss of E2F-1 reduces tumorigenesis and extends the lifespan of Rb1(+/−) mice. Nat Genet. 1998;18:360–364. doi: 10.1038/ng0498-360. [DOI] [PubMed] [Google Scholar]
- Ziebold U, Reza T, Caron A, Lees JA. E2F3 contributes both to the inappropriate proliferation and to the apoptosis arising in Rb mutant embryos. Genes & Dev. 2001;15:386–391. doi: 10.1101/gad.858801. [DOI] [PMC free article] [PubMed] [Google Scholar]