Abstract
When directed to the nucleus by TGF-β or BMP signals, Smad proteins undergo cyclin-dependent kinase 8/9 (CDK8/9) and glycogen synthase kinase-3 (GSK3) phosphorylations that mediate the binding of YAP and Pin1 for transcriptional action, and of ubiquitin ligases Smurf1 and Nedd4L for Smad destruction. Here we demonstrate that there is an order of events—Smad activation first and destruction later—and that it is controlled by a switch in the recognition of Smad phosphoserines by WW domains in their binding partners. In the BMP pathway, Smad1 phosphorylation by CDK8/9 creates binding sites for the WW domains of YAP, and subsequent phosphorylation by GSK3 switches off YAP binding and adds binding sites for Smurf1 WW domains. Similarly, in the TGF-β pathway, Smad3 phosphorylation by CDK8/9 creates binding sites for Pin1 and GSK3, then adds sites to enhance Nedd4L binding. Thus, a Smad phosphoserine code and a set of WW domain code readers provide an efficient solution to the problem of coupling TGF-β signal delivery to turnover of the Smad signal transducers.
Keywords: signal transduction, transcription, Smad, BMP, TGF-β, WW domains
An important gap in the current understanding of cytokine-driven transcriptional control is about the processes that remove mediator molecules that have participated in gene regulation. A case in point is the Smad transcription factors, central mediators of the TGF-β and BMP pathways (Li and Flavell 2008; Massagué 2008; Wu and Hill 2009). Fundamental aspects of metazoan embryo development and tissue homeostasis are controlled by TGF-β and BMP through Smad-mediated transcription of master regulator genes. In the course of this action in the nucleus, Smad proteins undergo certain phosphorylation events that enable peak transcriptional activity but also mark the proteins for destruction (Alarcon et al. 2009; Gao et al. 2009). These findings presented a paradox, but also an opportunity to define how the delivery of TGF-β and BMP signals is coupled to the turnover of the Smad signal transducers.
Several key phosphorylations drive the Smad signaling process. The ligand cytokines activate receptor serine/threonine protein kinases that phosphorylate Smad proteins at the C terminus. The BMP receptors act on Smad1, Smad5, and Smad8, and the receptors for the TGF-β/nodal/activin /myostatin group of ligands act mainly on Smad2 and Smad3 (Shi and Massagué 2003). The phosphorylated C terminus provides a binding site for Smad4, which is an essential component in the assembly of target-specific transcriptional complexes. These phosphorylations are reversed by protein phosphatases that limit the general pool of activated Smad molecules (Inman et al. 2002; Xu et al. 2002; Lin et al. 2006; Schmierer et al. 2008).
Receptor-activated Smad proteins that associate with Smad4 and bind to target genes undergo a second set of phosphorylations; these are catalyzed by the transcriptional cyclin-dependent kinases CDK8 and CDK9 (Alarcon et al. 2009; Gao et al. 2009) and glycogen synthase kinase-3 (GSK3) (Fuentealba et al. 2007; Sapkota et al. 2007; Alarcon et al. 2009). CDK8 and CDK9 are part of the transcriptional Mediator and Elongation complexes, respectively (Komarnitsky et al. 2000; Malik and Roeder 2000; Durand et al. 2005). GSK3 is a Wnt- and PI3K-regulated kinase (Cohen and Frame 2001; Wu and Pan 2010). CDK8/9 phosphorylation of Smad serves as the priming event for phosphorylation by GSK3. These phosphorylations are clustered in an interdomain linker region and enable peak activation of Smads, but also mark the proteins for polyubiquitination and proteasome-mediated degradation (Alarcon et al. 2009; Gao et al. 2009). Degradation of agonist-activated Smads (Lo and Massagué 1999; Alarcon et al. 2009) occurs alongside dephosphorylation of the linker (Wrighton et al. 2006; Sapkota et al. 2007). Whereas dephosphorylation recycles the Smad proteins for repeated rounds of signaling, action-coupled destruction of Smad depletes the pool of signal transducer. In a different context, the Smad linker region is phosphorylated by MAP kinases and cell division CDKs in response to mitogens and stresses to constrain TGF-β and BMP signaling (Kretzschmar et al. 1997, 1999; Matsuura et al. 2009).
Four proteins are known to bind specifically to linker phosphorylated Smads during BMP and TGF-β signal transduction. The HECT domain ubiquitin ligase Smurf1 (Sapkota et al. 2007) and the transcriptional effector of the Hippo pathway YAP bind to linker phosphorylated Smad1/5 (Alarcon et al. 2009), whereas the Smurf1-related protein Nedd4L (Gao et al. 2009) and the peptidyl-prolyl cis/trans isomerase Pin1 (Matsuura et al. 2009) bind to linker phosphorylated Smad2/3. YAP cooperates with Smad1 to activate ID genes that suppress neural differentiation in mouse embryonic stem cells in response to BMP signals (Alarcon et al. 2009). Pin1 cooperates with Smad2/3 to stimulate cancer cell migration in response to TGF-β (Matsuura et al. 2009). Smurf1 and Nedd4L target activated Smad1/5 and Smad2/3, respectively, for polyubiquitination and proteasome-dependent degradation. Common to this set of Smad-binding proteins is the presence of WW domains: one in Pin1, two in Smurf1 and YAP, and four in Nedd4L. WW domains are 38- to 40-amino-acid residue units characterized by two highly conserved tryptophans and folded as a three-strand β sheet that typically binds proline-rich sequences (e.g., PPxY or “PY box”) or, in the case of Pin1, phospho-SP motifs (Macias et al. 2002). A PY box is located near the CDK/GSK3 phosphorylation sites in the linker region of Smad proteins.
These lines of evidence present a scenario in which different nuclear protein kinases phosphorylate agonist-activated Smads to create docking sites for competing transcriptional cofactors and ubiquitin ligases. The outcome of these interactions governs Smad function, and is therefore important in BMP and TGF-β signal transduction. However, the convergence of activation and turnover functions on a clustered set of Smad modifications raises questions about how Smads get to act before undergoing disposal. We postulated that a mechanism must exist that ensures the orderly sequence of events in this process by somehow switching Smad proteins from binding transcriptional cofactors to binding ubiquitin ligases. Combining the power of functional and structural approaches, we uncovered such a switch mechanism and defined the basis for its operation and specificity in the BMP and TGF-β pathways.
Results
GSK3 switches the Smad1 binding preference from YAP to Smurf1
Smad proteins consist of a globular N-terminal MH1 (Mad Homology 1) domain with DNA-binding activity, a C-terminal MH2 domain that mediates key protein–protein interactions, and an interdomain linker region with a conserved cluster of phosphorylation sites adjacent to a PY motif (Fig. 1A,B; Shi and Massagué 2003). Phosphorylation of these sites follows TGF-β- and BMP-driven C-terminal phosphorylation and nuclear translocation of Smads, as seen in human cell lines, mouse embryonic stem cells, the mouse embryo, and the Xenopus embryo (Supplemental Fig. 1A–C; Fuentealba et al. 2007; Sapkota et al. 2007; Alarcon et al. 2009). In Smad1, CDK8/9 phosphorylate S206 and S214, which prime T202 and S210, respectively, for phosphorylation by GSK3. To dissect this process, we tested the effect of pharmacological inhibitors of CDK8/9 and GSK3 in human embryonic kidney 293 (HEK293) cells expressing epitope-tagged Smurf1 or YAP constructs. A catalytically inactive Smurf1 mutant (Smurf1DD) (Ebisawa et al. 2001) was used in order to avoid confounding the effects of Smurf1-dependent Smad degradation. The BMP inhibitor noggin was added to the culture medium in order to block endogenous BMP and thus set a basal state. Incubation of the cells with BMP rapidly induced the formation of Smad1–YAP and Smad1–Smurf1 complexes (Fig. 1C,D). The CDK8/9 inhibitor flavopiridol, which inhibits all BMP-induced linker phosphorylations (Alarcon et al. 2009), prevented the formation of both complexes (Fig. 1C,D). Addition of LiCl, which inhibits GSK3 site phosphorylation (Fuentealba et al. 2007), also prevented the Smad1–Smurf1 interaction (Fig. 1C). Interestingly, LiCl did not inhibit, but rather increased, the level of Smad1–YAP complex (Fig. 1D). These results suggested that the formation of the YAP–Smad1 complex in response to BMP requires CDK8/9 but not GSK3, whereas the formation of the Smurf1–Smad1 complex requires both kinase activities.
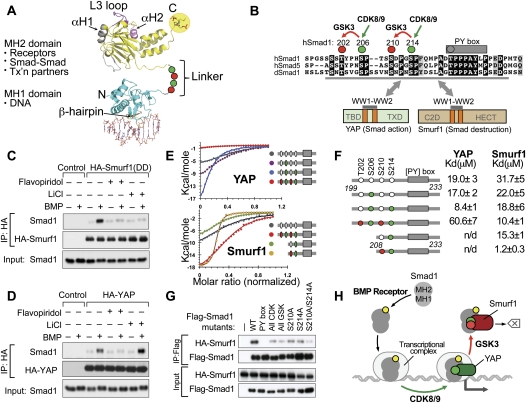
Figure 1.
GSK3 switches the binding preference of Smad1 from YAP to Smurf1. (A) Schematic representation of the Smad protein domains and their main functions. The MH1 domain (cyan) contains a β hairpin that mediates binding to dsDNA (orange) (PDB code: 1MHD) (Shi et al. 1998). The MH2 domain (yellow) binds to the type I TGF-β receptor, which involves the L3 loop (magenta); to Smad4 via the phosphorylated C terminus (highlighted) and the α-helix 1 (gray); and to various DNA-binding cofactors and histone-modifying enzymes (PDB code: 1KHX) (Wu et al. 2001). The interdomain linker region (dotted line) contains CDK8/9 and GSK3 phosphorylation sites, represented by green and red circles, respectively. (B) Sequence alignment of the linker region of human Smad1 and Smad5 and Drosophila MAD (dSmad1) proteins, with conserved residues highlighted. The conserved CDK8/9 sites (green) and CDK8/9-primed GSK3 sites (red) and the PY box are shown. The Smad1 (199–232) segment used in this study is underlined. The domain composition of Smurf1 and YAP proteins and the regions that mediate binding to linker phosphorylated Smad1 are indicated. (C) BMP-dependent formation of a complex between HA-Smurf1(DD) and endogenous Smad1 in HEK293 cells, and effects of flavopiridol and LiCl on the formation of this complex. (D) BMP-dependent formation of a complex between HA-YAP and endogenous Smad1 in HEK293 cells, and effects of flavopiridol and LiCl on the formation of this complex. (E) ITC curves for the binding of Smurf1 and YAP WW1–WW2 segments to Smad1 synthetic peptides. (F) Synthetic Smad1 (phospho-)peptides and their affinity for recombinant WW1–WW2 segments of YAP and Smurf1. Colored circles denote phosphorylation of the residues. (G) Effect of alanine mutations in the PY box and the indicated phosphorylation sites on the ability of Flag-tagged Smad1 constructs to bind HA-Smurf1(DD) in HEK293 cells. (H) Schematic summary of the Smad action turnover switch operated by CDK8/9 and GSK3 in combination with YAP and Smurf1.
We performed isothermal titration calorimetry (ITC) binding assays to investigate the interaction between recombinant WW1–WW2 segments and Smad1 linker phosphopeptides (Fig. 1E,F). We tested versions of the Smad1 199–233 linker region with no phosphorylation, with phosphorylation at CDK8/9 sites S206 and S214, or with additional phosphorylation at GSK3 sites T202 and S210 (Fig. 1F). The YAP WW1–WW2 segment bound the unphosphorylated Smad1 peptide with K = 19.0 ± 3 μM and the CDK8/9-phosphorylated peptide with KD = 8.4 ± 1 μM. Notably, this gain in affinity was fully erased by phosphorylation at the GSK3 sites (KD = 60.6 ± 7) (Fig. 1E,F). In contrast, the affinity of the Smurf1 WW1–WW2 segment for the Smad1 peptide was increased by phosphorylation at the CDK8/9 sites, and was further increased by phosphorylation at the GSK3 sites (Fig. 1E,F).
Further refinement of the Smurf1–Smad1 interaction revealed a strong preference of Smurf1 for pS214 over pS206, achieving the highest affinity (KD = 1.2 ± 0.3 μM) with a Smad1 208–233 peptide containing pS210 and pS214 (Fig. 1E,F). Moreover, mutation of S210 or S214 to alanine inhibited the Smad1–Smurf1 interaction in HEK293 cells, and mutation of both residues further decreased binding (Fig. 1G). Collectively, these results suggest that CDK8/9-mediated phosphorylation of the Smad1 linker creates binding sites for competing YAP and Smurf1 WW1–WW2 domains, and GSK3 switches this balance in favor of Smurf1 binding and at the expense of the YAP interaction (Fig. 1H).
Structure of the Smurf1 WW pair bound to the Smad1 linker
We used NMR spectroscopy to calculate the structure of the Smurf1 WW1–WW2 segment bound to the Smad1 linker peptide (208–233) diphosphorylated at S210 and S214 in solution. Triple-resonance NMR spectroscopy was applied to assign the WW1–WW2 pair in this complex, whereas homonuclear and half-filter spectra were used to assign the Smad1 peptide and its contacts with Smurf1 (Fig. 2A; Supplemental Fig. 2A,B). In the complex, each of the WW domains adopts the typical triple-stranded anti-parallel β-sheet fold, even in the case of WW1 that lacks the first highly conserved tryptophan. The WW domains do not contact each other, but each contacts a portion of the linker. This arrangement provides enough freedom for the WW domains to adopt an anti-parallel orientation, forming a continuous binding surface that smoothly cradles the phosphorylated Smad1 linker. The Smad1 linker adopts an extended conformation, with the diphosphorylated T208–P215 segment bound to the WW1 domain and the PY motif bound to the WW2 domain (Fig. 2A). The segment between P215 and the PY motif forms a turn defined by interactions between the F217 and A220 backbone atoms. The 10-residue segment connecting the two Smurf1 WW domains adopts a helical structure in its first half (Fig. 2A). This configuration allows access to CKIP1 (casein kinase 2-interacting protein-1), a protein that binds to this region to enhance the Smurf1–Smad1 interaction (Lu et al. 2008).
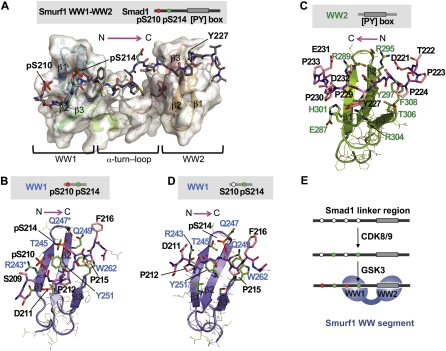
Figure 2.
Structure of the Smurf1 WW1–WW2 segment bound to the Smad1 linker. (A) NMR model of the complex between the human Smurf1 WW1–WW2 pair (residues 232–314) and the 208–233 segment of the Smad1 linker diphosphorylated at S210 and S214. Smurf1 is shown as a semitransparent surface, with all elements of secondary structure represented. The Smad1 peptide is shown with a stick representation, with the backbone colored in gray. There are several relative orientations of the WW domains that satisfy all experimental NMR restraints (shown in Supplemental Fig. 2), and, due to this, we call this complex the NMR model. (B) Detailed view of the refined structure of the Smurf1 WW1 domain (slate) bound to the diphosphorylated pS210/pS214 region of the Smad1 linker. Key residues in Smad1 (black) and Smurf1 (blue) are indicated. (Asterisks) Three residues that, when jointly mutated to alanine, decreased the binding affinity of the complex by ∼25-fold. (C) Detailed view of the refined structure of the Smurf1 WW2 domain (green) bound to the PY motif of Smad1. Key residues in Smad1 (black) and Smurf1 (green) are indicated. (D) Detailed view of the refined structure of the Smurf1 WW1 domain (slate) bound to the monophosphorylated pS214 region of the Smad1 linker. (E) Schematic representation of the mode of binding of Smurf1 to the Smad1 linker region.
To facilitate the presentation in the text, we use the one-letter amino acid notation for Smad residues and the three-letter notation for residues in its binding partners. The Smurf1 WW1 domain binds the Smad1 pS210 residue through contacts with Tyr251, Arg243, and Leu253 side chains. The Tyr251 hydroxyl and the Arg243 guanidinium groups jointly coordinate the phosphate group of pS210. pS214 also contacts Tyr251, and the phosphate group is coordinated by the hydroxyl of Thr245 and the side chains of Gln247 and Gln249. P215 is packed parallel to the aromatic ring of Trp262, and P212 is sandwiched between the Leu253 and Ser260 side chains in a cavity perpendicular to the β sheet (Fig. 2B). The pS210–D211–P212–G213 segment forms a turn favored by a D211–P212 cis bond, whereas pS214–P215 is in trans. Single-alanine mutations of Arg243, Gln247, or Gln249 decreased the affinity to KD values of ∼30 μM, confirming the importance of these residues in the interaction of WW1 with the pS210 and pS214 phosphate groups.
The Smurf1 WW2 domain binds to the PY motif in a manner similar to canonical group 1 WW complexes (Macias et al. 2002). P224 and P225 contact Tyr297 and Phe308, respectively, and Y227 binds between His301 and Arg304 (Fig. 2C; Supplemental Fig. 2B). The six residues after the tyrosine in the PY motif fold over the first strand of WW2. Abundant contacts are observed between P229 and P230 and His301 and Glu287, respectively. The side chain of E231 points toward the Tyr297 hydroxyl and shows contacts with the Arg289 side chain.
We also solved the structure of the Smurf1 WW1–WW2 segment bound to the Smad1 linker monophosphorylated at pS214 (Supplemental Fig. 2B). Most of the contacts between the two molecules are like those in Smurf1 WW1–WW2 bound to the pS210/pS214 diphosphorylated peptide, except that both the D211–P212 and pS214–P215 bonds are in trans and are bound differently (Fig 2, cf. D and B). S210 is less ordered than in the phosphorylated state, and only weak contacts are observed between D211 and Arg243 side chains (Fig. 2D). These weaker contacts explain the intermediate affinity of the WW1–WW2 domain for the Smad1 linker monophosphorylated at S214. In conclusion, formation of the Smurf1–Smad1 complex involves recognition of the Smad1 PY motif by the Smurf1 WW2 domain, and of the Smad1 GSK3 and CDK phosphorylated sites by the WW1 domain (Fig. 2E).
Structure of the YAP WW pair bound to the Smad1 linker
Alanine mutations in either the WW1 domain or WW2 domain in YAP almost completely abolished the interaction of overexpressed YAP and Smad1 in transiently transfected human cells (Supplemental Fig. 3A), suggesting that both domains are essential for this interaction. Given the high affinity of the YAP WW1–WW2 module for a Smad1 (199–233) linker peptide phosphorylated at the CDK8/9 sites pS206 and pS214 (see Fig. 1F), we solved the structure of this complex first. We used double- and triple-labeled WW1–WW2 samples (YAP 163–266 segment) to assign the protein resonances in combination with filtered and homonuclear experiments to obtain the chemical shifts of the bound peptide and the contacts between both molecules. We also used independent domains to assist in the assignment. The 25-residue connector between the two WW domains adopts a helix–loop–helix structure, as determined on the basis of the detected nuclear Overhauser effects (NOEs) and carbon chemical shift analysis. Several contacts are present between each WW domain and this connector, but these contacts do not prevent the WW domains from adopting an optimal orientation for interactions with the Smad1 linker. No contacts were observed between the WW domains.
In the complex between the WW1–WW2 pair and the Smad1 34-residue peptide, both WW domains adopt the canonical fold and participate in the interaction with Smad1 (Fig. 3A; Supplemental Fig. 3B). The WW1 domain contacts the pS206 region and the WW2 domain contacts the PY motif. The Smad1 pS206 and P207 side chains are accommodated in the aromatic cavity formed by Tyr188 and Trp199 in the YAP WW1 domain (Fig. 3B). The pS206 phosphate group is at a hydrogen bond distance from the hydroxyl groups of Thr182 and Tyr188 and the Gln186 side chain. Trp199 is also involved in a network of contacts with residues comprised between P207 and S209. The structure of the WW2 domain bound to the Smad1 PY motif resembles that of Smurf1 WW2 bound to this region. The interaction is well defined, involving eight Smad1 residues between D221 and D232 and nine out of 13 residues on the WW2 domain surface (Fig. 3C). We also observed NOEs from the E231 side chain with T241 and Tyr247 and from D232 to Gln242, suggesting the presence of intermolecular salt bridges between these residues. Thus, formation of the YAP–Smad1 complex involves recognition of the Smad1 PY and the CDK phosphorylated site pS206 by the WW2 and WW1 domains, respectively.
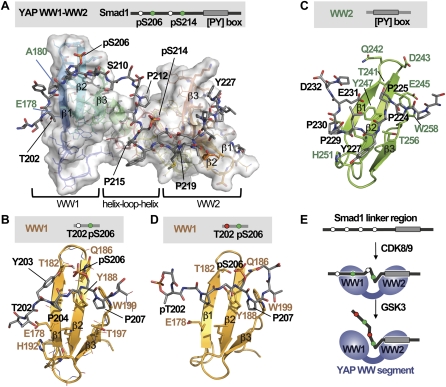
Figure 3.
Structure of the YAP WW1–WW2 pair bound to the Smad1 linker. (A) NMR model of the complex between the human YAP WW1–WW2 pair (residues 163–266) and the 199–233 segment of the Smad1 linker diphosphorylated at S206 and S214. YAP is shown as a semitransparent surface, and Smad1 is shown as gray sticks. (B) Detailed view of the refined structure of the YAP WW1 domain (gold) bound to the mono-pS206 phosphorylation site of Smad1 (gray). Key residues in Smad1 (black) and YAP (brown) are indicated. (C) Detailed view of the refined structure of the YAP WW2 domain (green) bound to the PY motif region of Smad1 (gray), with the key residues indicated. (D) Detailed view of the refined structure of the YAP WW1 domain (gold) bound to the diphosphorylated pT202, pS206 region of the Smad1 linker. (E) Schematic representation of the mode of binding of YAP to the Smad1 linker region.
The GSK3 phosphorylated Smad1 linker avoids YAP binding
Next we analyzed the interactions between the YAP WW1–WW2 pair and Smad1 linker peptides containing GSK3 site phosphorylations. NMR-based titrations with a peptide containing pT202, pS206, pS210, and pS214 require a fourfold to fivefold peptide excess to induce chemical shift changes in the YAP WW1–WW2 pair (Supplemental Fig. 5F), corroborating the weak interaction measured by ITC and precluding the determination of the complex structure. In a complex of YAP WW1–WW2 domains with a peptide containing pT202, pS206, pS214, and the PY site, but no phosphorylation at S210, the NOEs detected from the N-terminal end of the peptide to the WW1 domain of YAP were weak and the structure is defined only for the P204–pS206–P207 site (Fig. 3D; Supplemental Fig. 3B). In particular, the Glu178 side chain and the His192 ring in the WW1 domain fail to contact the methyl of T202. Y203 and P204 are only partially ordered. Thus, the presence of a phosphate group in T202 destabilizes the interaction of Smad1 with the YAP WW1 domain, and the presence of phosphates at both T202 and at S210 drastically reduces the interaction between YAP and the Smad1 linker.
To better illuminate the different binding preferences of the Smurf1 and YAP complexes, we compared the charge distribution on the surfaces of the Smurf1 and YAP WW1 domains. Both WW1 domains contain Gln residues in the surroundings of pS214 and pS206, respectively, but the positively charged patch of Smurf1WW1 that interacts with pS210 (Fig. 4A,B) is absent in YAP. Instead, YAP contains a negatively charged region suited to interact with T202 but incompatible with the presence of a phosphate group at T202 (Fig. 4C,D).
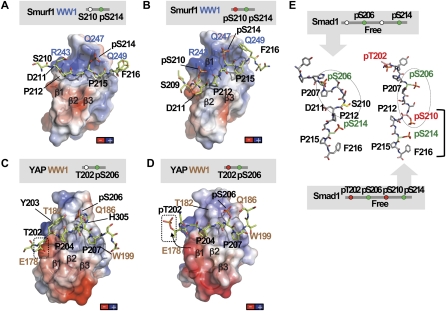
Figure 4.
The GSK3 phosphorylated Smad1 linker prevents YAP binding. (A,B) Charge distribution on the surface of the Smurf1WW1 domain in complex with the Smad1 linker monophosphorylated at S214 (A) or diphosphorylated at S210 and S214 (B). Negatively charged patches are shown in red, and positively charged patches are shown in dark blue. Smurf1 WW1 is shown as a semitransparent surface, and Smad1 is shown as green sticks. Key residues in Smad1 (black) and Smurf1 WW1 (blue) are shown. The complex is shown in the same orientation as that of Figure 2. (C,D) Charge distribution on the surface of the YAP WW1 domain in complex with the Smad1 linker monophosphorylated at S206 (C) or diphosphorylated at T202 and S206 (D). The YAP WW1 domain is shown as a semitransparent surface and with the same orientation as in Figure 3. The position of T202 is shown in a box. The conformational change observed in pT202 is represented with an arrow. (E) Molecular simulations performed on two peptides corresponding to Smad1 phosphorylated at S206 and S214 (left) or at T202, S206, S210, and S214 (right). Key residues are labeled.
Notably, in the complex of YAP WW1–WW2, the Smad1 linker segment between residues S210 and D221 runs across the inter-WW connector with three prolines (P212, P215, and P219) in trans (see Fig. 3A). The trans configuration of the D211P and pS214P bonds favors the formation of two β turns that facilitate the interaction of the Smad1 pS206 site with YAP. This feature likely explains the higher affinity of YAP for the pS206/pS214 diphosphorylated peptide (8.4 ± 1 μM) compared with its affinity for the pS206 monophosphorylated peptide (17.0 ± 2 μM). The negative effect of pS210 on the YAP–Smad1 interaction observed by ITC and NMR titration experiments could arise from a conformational change in the Smad1 fragment forced by electrostatic repulsion between the pS210 phosphate group and the negatively charged D211. To test the potential impact of this phosphate, we performed NMR experiments and molecular dynamic simulations of peptides containing pS214, pS210, and pS206. The simulations revealed that pS210 favors an extended conformation without the β turn centered at D211–P212 (Fig. 4E) that would decrease the likelihood of WW1 interacting with the pS206 site.
Collectively these observations suggest that phosphorylation of Smad1 by CDK8/9 creates a binding site for the YAP WW1 domain in pS206, and the downstream all-trans configuration imposed by D211P and pS214P favors this binding interaction. GSK3 phosphorylation of the Smad1 linker at T202 and, particularly, at S210 creates a conformation that avoids recognition by the YAP WW1 domain (Fig. 3E) while favoring recognition by the Smurf1 WW1 domain.
A Smad action turnover switch in the TGF-β pathway
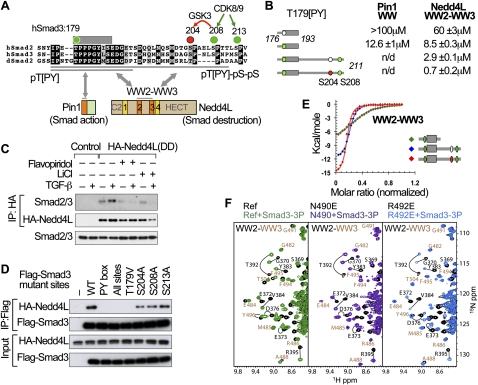
Several clues suggested that a similar Smad action turnover switch, with its own phospho-amino acid code and set of WW domain code readers, may operate in the TGF-β/nodal version of the pathway. The linker regions of Smad2 and Smad3 contain a conserved PY motif, three agonist-dependent CDK phosphorylation sites, and one CDK-primed GSK3 site (Fig. 5A; Alarcon et al. 2009; Wang et al. 2009). Significant differences exist in the configuration of these elements compared with Smad1. In the Smad3 linker, a T179 immediately preceding the PY motif is rapidly phosphorylated by CDK8/9 in response to TGF-β. This is followed by phosphorylation of two CDK8/9 sites (S208 and S213) and a single GSK3 site (S204), all of which are located downstream from the PY motif in this case. The pT179[PY] motif is recognized by the single WW domain of Pin1 (Fig. 5A; Matsuura et al. 2009). Using ITC titrations, we corroborated that pT179 within a Smad3 176–193 peptide is the preferred binding site for the Pin1 WW domain (KD = 12.6 ± 1 μM) (Fig. 5B), as compared with pS204/pS208 within a Smad3 202–211 peptide (KD = 23.4 ± 7 μM) or pS208/pS213 within a Smad3 206–215 peptide (KD = 49 ± 11 μM).
Figure 5.
Elements of a Smad action turnover switch in the TGF-β pathway. (A) Sequence alignment of the linker regions of human Smad2 and Smad3 and Drosophila Smad2, with the conserved residues highlighted. The conserved CDK8/9 sites (green) and CDK8/9-primed GSK3 site (red) and the PY box are shown. Two Smad3 segments (176–193 and 176–211) used in this study are underlined. The domain composition of Pin1 and Nedd4L proteins and the regions that mediate binding to linker phosphorylated Smad3 are indicated. (B) Synthetic Smad3 (phospho-)peptides and their affinity for the recombinant Pin1 WW domain and Nedd4L WW2–WW3 pair. Colored circles denote phosphorylation of the indicated residues. (n.d.) Not determined. (C) TGF-β-dependent formation of a complex between HA-Nedd4L(DD) and endogenous Smad3, and effects of flavopiridol and LiCl on the formation of this complex. (D) Effect of alanine mutations in the PY box and the indicated phosphorylation sites on the ability of Flag-tagged Smad3 constructs to bind HA-Nedd4L(DD) in HEK293 cells. (E) ITC curves and corresponding fitting to pairs of Nedd4L WW domains and the indicated Smad3 (phospho-)peptides. (F) NMR titrations of WW2–WW3 pairs (wild type in green) with point mutations introduced in two residues that coordinate the pS204pS208 site (violet and royal blue). Residues that belong to WW2 and WW3 are labeled in black and camel, respectively.
The pT179[PY] motif is also recognized by the Nedd4L WW2 domain (Gao et al. 2009). A recombinant WW2 domain bound this motif with KD = 8.5 ± 0.3 μM (Fig. 5B). However, formation of the TGF-β-dependent Nedd4L–Smad3 complex was inhibited not only by the CDK inhibitor flavopiridol, but also by the GSK3 inhibitor LiCl (Fig. 5C). Furthermore, alanine mutation of the GSK3 site S204 diminished the binding of Smad3 to Nedd4L in HEK293 cells, as did mutation of the GSK3-priming CDK site S208 (Fig. 5D). Mutation of S213 had no discernible effect on binding. These results suggest that the pS208-primed, GSK3-mediated phosphorylation of S204 further augments the affinity of Nedd4L for Smad3. Thus, we postulated that another WW domain in Nedd4L must recognize the pS204–pS208 site and collaborate with the binding of WW2 to the pT179[PY] motif.
To test this hypothesis, we determined whether pairs of Nedd4L WW domains could bind to an extended region of Smad3, including the pT179[PY] motif as well as the phosphorylated S208 site or the phosphorylated S204 and S208 sites (Fig. 5B). Of the three consecutive pairs present in Nedd4L, only the WW2–WW3 pair showed high affinity for the diphosphorylated peptide (KD = 2.9 ± 0.1 μM) and a further gain in affinity for the triphosphorylated peptide (KD = 0.7 ± 0.2 μM) (Fig. 5B,E). This affinity is 15-fold stronger than the affinity (KD = 10.1 ± 0.7 μM) of the same peptide for the WW2–WW3 region of another family member, Smurf2. NMR titrations with increasing concentrations of the pT[PY]-pS-pS peptide corroborated chemical shift changes in both domains of a recombinant WW2–WW3 pair. The residues affected in WW2 were the same as those affected in titrations with the pT179[PY] peptide, whereas residues affected in WW3 were located in and around the first loop and in the third strand (Fig. 5F). The effects of point mutations in WW2 (Leu384 to Tyr) or in WW3 (Ile496 to Tyr) provided further evidence that WW2 recognizes the pT179[PY] motif and WW3 recognizes the pS204–pS208 sites (Supplemental Fig. 4A), as summarized in Figure 5A.
Basis for Nedd4L and Pin1 recognition of the Smad3 linker phosphorylation code
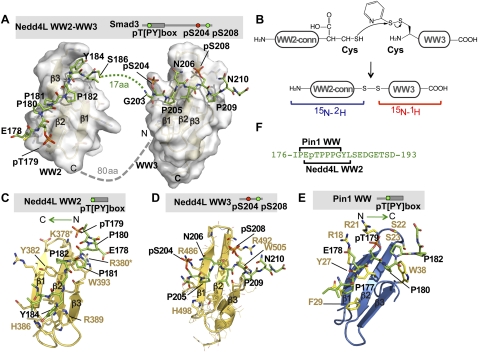
Determination of the solution structure of the Nedd4L WW2–WW3 module (Nedd4L 364–512 segment) bound to the Smad3 linker peptide pT[PY]-pS-pS (Smad3 176–211 segment triphosphorylated at T179, S204, and S208) (Fig. 6A) posed a challenge owing to poor NMR signal dispersion of the 80-amino-acid region connecting WW2 and WW3. To simplify the interpretation of the NMR data, we prepared the WW2–WW3 module as two separate fragments, which allowed the use of sequential isotope labeling. A disulfide bond protein ligation strategy was used to connect these two fragments (Fig. 6B; Baca et al. 1995; Nair and Burley 2003). Applying a stepwise protocol to solve the structure of the complex (see the Materials and Methods), we were able to fully assign the WW2 and WW3 domains and ∼80% of the residues in the linker (Fig. 6A). The unassigned residues are located in a proline-rich segment immediately upstream of the WW3 domain. The α and β 13C chemical shifts analysis revealed that the segment connecting the WW2–WW3 pair (residues 400–480) lacks elements of a secondary structure in both the free and bound conformations, and is unaffected by binding of the Smad3 peptide.
Figure 6.
Basis for Nedd4L and Pin1 recognition of the phosphorylated Smad3 linker. (A) Model of the complex structure between the human Nedd4L WW2–WW3 pair (residues 364–512) and the 176–211 segment of the Smad3 linker triphosphorylated at T179, S204, and S208. Nedd4L is shown as a semitransparent surface, and Smad3 is shown as green sticks. Smad3 residues involved in the interaction with the Nedd4L WW2–WW3 pair are indicated. The 80-amino-acid region connecting the WW2 and WW3 domains (dotted line) does not adopt a defined secondary or tertiary structure, as indicated by near-random 13C chemical shifts. Due to the complexity that this long unstructured part adds to the calculation of the complex, the model has been calculated using three independent molecules (WW2, WW3, and the Smad3 peptide) without the 80-amino-acid region. Three possible orientations of the WW2 and WW3 pair were obtained using a set of RDC experiments. The orientation that yields the best view of the bound Smad3 peptide is shown here. In the bound Smad3, the segment between S186 and G203 does not contact either WW2 or WW3 and is not represented. (B) Schematic of the protein ligation strategy employed to prepare the WW2–WW3 module as two separate fragments for sequential isotope labeling. A fully deuterated WW2 and connector (WW2-conn) segment of Nedd4L was ligated to the protonated WW3 domain, as shown. Using this strategy, the signals from the first part of the protein were filtered, and the analysis of data corresponding to the WW3 domain bound to Smad3 and of that of Smad3 itself was simplified. In the calculation of the complex, this information was combined with that of the WW2 in complex with the pT[PY] peptide and with residual dipolar coupling data obtained from the entire WW2–WW3 segment. (C) Detailed view of the Nedd4L WW2 domain (gold) bound to the phosphorylated PY motif (pT179[PY]) of Smad3 (green). Key residues in Smad3 (black) and Nedd4L (brown) are indicated. This complex has been refined using the data of the WW2–WW3 pair that corresponds to the second WW and the fragment of 176–190 of Smad3. (Asterisks) Two residues that, when jointly mutated to alanine, decreased the binding affinity of the complex by ∼11-fold. (D) Detailed view of the Nedd4L WW3 domain (gold) bound to the diphosphorylated pS204–pS208 sites of Smad3 (green). Key residues in Smad3 (black) and Nedd4L (brown) are indicated. This complex has been refined using the data of the WW2–WW3 that corresponds to the WW3 site and the fragment of 203–211 of Smad3. (E) Solution structure of the Pin1 WW domain bound to the Smad3 pT179[PY] motif. The WW domain is shown as a ribbon representation, shown in marine. Key residues in Smad3 (black) and Pin1 (light brown) are indicated. This complex is displayed using the same orientation as that of the Nedd4L WW2 complex (shown in C) to highlight that these WW domains bind to the pT179[PY] site in opposite orientations. (F) Representation of the distinct portions of the pT179[PY] motif of Smad3 that provide contacts with Pin1 and Nedd4L.
The Nedd4L WW2 domain recognizes the EpTPPPGY segment in this complex as well as in a complex with a shorter Smad3 peptide (pT[PY] peptide, Smad3 176–193 segment) that we also solved (Fig. 6C). P181 and P182 bind in a cavity formed by the Tyr382 and Trp393 aromatic rings, whereas Y184 binds to His386, Arg389, and backbone atoms in loop 2. The Lys378 and Arg380 side chains coordinate the phosphate group of pT179, while the Trp393 aromatic ring contacts the methyl and β protons. The electrostatic interactions of Lys378 and Arg380 with the phosphate group explain the high affinity of this interaction and compensate for an absence of contacts with residues downstream from the PY motif observed in YAP and Smurf1 complexes. Versions of the Nedd4L WW2 domain with Glu mutations in Lys378 or Arg380 decreased the affinity for the pT[PY] peptide from KD = 4.1 ± 0.3 μM to KD = 20–23 μM, and mutation of both residues caused a further decrease (KD = 44.6 ± 15 μM), confirming the importance of these residues in binding (Fig. 6C). We solved the structures of the mutant proteins to verify that the mutations do not affect the overall fold of the WW domain, but that they alter the charge distribution of loop 1, involved in the phosphate recognition of the pT179 site (Supplemental Fig. 5C–F). Negatively charged residues in and around loop1 are highly unusual in WW sequences (SMART database of protein domains). Interestingly, the wild-type sequence of the YAP and TAZ WW2 domains are unique for having these negatively charged residues. The weak affinity of the YAP WW1 domain for PY motifs (Macias et al. 1996; Pires et al. 2001; Toepert et al. 2001) and the presence of a negatively charged patch in the YAP WW2 domain that prevents binding to a pT[PY] motif explain our previous result that YAP does not interact efficiently with linker phosphorylated Smad3 (Supplemental Fig. 5F–H; Alarcon et al. 2009).
The Nedd4L WW3 domain binds pS208 between the Asn490 side chain and the guanidinium group of Arg492, and also contacts the aromatic ring of Phe494 (Fig. 6D). Additional contacts of P209 and P211 to the aromatic ring of Trp505 help position pS208. pS204 is bound by the Arg486 guanidinium group and the hydrophobic part of Glu484, and the ring of P205 is are accommodated between the side chains of Glu484, Arg486, and Tyr496. Indeed, alanine mutation of either Arg486 or Arg492 caused a fourfold reduction in the affinity of the WW2–WW3 construct for the pT[PY]-pSpS peptide. Overall, the GSK3-CDK8/9 diphosphorylated motifs of Smad3 and Smad1 are recognized by the corresponding WW domains of Nedd4L and Smurf1 in a structurally similar manner, with the phospho-serines positioned close to arginine or glutamine side chains.
The structure of the Pin1 WW domain bound to a Smad3 pT[PY] peptide (Fig. 6E) revealed similarities but also significant differences compared with how Nedd4L binds to this motif (Fig. 6F). The Pin1 WW domain recognizes the PEpTPPP motif of Smad3, with P177 bound between the Tyr27 and Phe29 aromatic rings, and P180, P181, and P182 all in trans and contacting Trp38. The proline aromatic stacking used by Pin1 provides a different binding strategy compared with the network of van der Waals interactions used by the Nedd4L WW2 domain to bind these prolines (Fig. 6, cf. C and E). The pT179 phosphate is coordinated by arginine side chains (Arg18 and Arg21) in β strand 1 and loop 1 of the Pin1 WW domain, which also differs from the coordination of this phosphate group by β strands 2 and 3 in the case of Nedd4L WW2. These different modes of recognition of the Smad3 pT179-PY motif by Pin1 and Nedd4L may reflect the different outcomes of these two interactions: peak transcriptional action in the case of Pin1, and Smad3 polyubiquitination in the case of Nedd4L.
Discussion
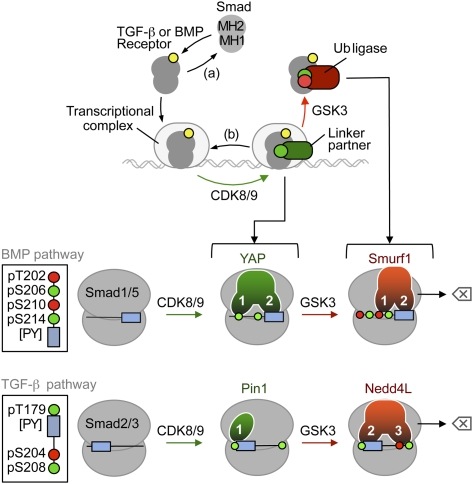
We show here that tandem WW domains in Smad-binding proteins function as readers of a phospho-serine code that dictates Smad peak transcriptional action as well as the subsequent elimination of Smad molecules that participate in transcription. The code of this action turnover switch is written by kinases CDK8/9 and GSK3 acting on the linker region of activated, transcriptionally poised Smad proteins. In our model (Fig. 7), CDK8/9 create binding sites that are preferentially recognized by WW-containing Smad transcriptional cofactors. These phosphorylations additionally prime Smads for subsequent GSK3-mediated phosphorylation, which creates sites for ubiquitin ligase binding at the expense of transcriptional cofactor-binding sites. Thus, GSK3 switches the phosphorylation code in the Smad linker region from one that favors Smad action to one that favors Smad destruction. As a result, TGF-β/BMP signal delivery becomes coupled to Smad turnover.
Figure 7.
The Smad action turnover switch in the BMP and TGF-β pathways: pSer codes and WW domain code readers. (Top panel) Schematic summary of the Smad action turnover switch in the BMP and TGF-β pathways. Following receptor-mediated phosphorylation (yellow circle), Smad proteins translocate to the nucleus and assemble transcriptional complexes, which are phosphorylated at CDK8/9 sites (green circle) in the MH1–MH2 interdomain linker region. This phosphorylation creates high-affinity binding sites for transcriptional partners (such as YAP in the case of the BMP mediator Smad1, Pin1 in the case of the TGF-β mediator Smad3, and probably others), thus achieving peak transcriptional action. Phosphorylation by CDK8/9 also primes the Smads for GSK3-mediated phosphorylation (red symbol) at the −4 position, which favors the binding of ubiquitin ligases Smurf1 (BMP pathway) and Nedd4L (TGF-β pathway), leading to proteasome-dependent degradation of Smad molecules that participate in transcription (erase symbol). Alternatively, C-terminal Smad phosphatases (a) and linker phosphatases (b) reverse these phosphorylation states. See the text for details and citations. (Bottom panels) Schematic of the Smad linker phospho-amino acid codes (insets) and WW domain code readers. The conserved CDK8/9 phosphorylation sites (green circles) and GSK3 sites (red circles) are located at the indicated positions relative to the PY box (slate box). Amino acid positions correspond to Smad1 and Smad3. In the BMP pathway, the YAP WW1 domain binds to pS206 in Smad1, as long as p210 is not phosphorylated. The Smurf1 WW1 domain binds with higher affinity to the pS210–pS214 motif. The WW2 domains bind the [PY] motif. In the TGF-β pathway, the sole WW domain of Pin1 binds the pT179[PY] motif, as does the WW2 domain of Nedd4L. However, the Nedd4L WW3 domain increases the binding affinity by recognizing the pS204–pS208 motif. See the text for additional details and citations on the known roles of these WW domain proteins in Smad signal transduction.
We propose that degradation is a price that Smad molecules pay for participating in transcription. CDK8 and CDK9 are components of the CDK8/CyclinC/Med12/Med13 transcriptional mediator complex and P-TEFb Cdk9/CyclinT elongation complex that regulate RNA polymerase II during transcription (Komarnitsky et al. 2000; Malik and Roeder 2000; Durand et al. 2005). CDK8/9 have access to Smad molecules on the chromatin but not to receptor-activated Smad molecules that fail to engage in transcriptional complexes (Alarcon et al. 2009). Another fate for activated Smads is C-terminal dephosphorylation, which mediates Smad recycling for new rounds of signaling and thereby links the duration of signaling to the presence of activated TGF-β or BMP receptors (Batut et al. 2008; Schmierer et al. 2008). However, Smad C-terminal dephosphorylation does not require prior participation of the molecule in transcription. Smad C-terminal dephosphorylation may modulate the global pool of activated Smads, whereas the action-coupled turnover process elucidated here eliminates Smad molecules as a function of their exposure to CDK8/9 during target gene regulation.
Our study provides a structural and functional basis for the involvement of tandem WW domains in these protein–protein interactions. On the one hand, the different WW domains of Smurf1, Nedd4L, and YAP proteins achieve overall high specificity in target recognition by acting in pairs, extending the interacting surface to recognize not only the canonical PY site, but also the adjacent pS/pTP motifs. The interactions with the pS sites were unexpected from predictions based on sequence conservation, since only the WW domain of Pin1 has been described as a pS/pTP-binding motif, and the Pin1 residues involved in the phosphate interaction are not strictly conserved in YAP, Smurf1, and Nedd4L WW domains. On the other hand, since the linker region of Smads contains a set of CDK phosphorylation sites and a set of CDK-primed GSK3 sites, the phospho-Ser motifs can be tuned to optimally bind either transcriptional cofactors (monophosphorylated) or ubiquitin ligases (diphosphorylated). The WW2 domains of Smurf1 and YAP recognize a canonical PY motif in Smad1, whereas the WW2 domain of Nedd4L and the Pin1 WW domain recognize a phospho-threonine-PY motif in Smad3. The PY-independent, monophospho-serine motif of Smad1 is recognized by the WW1 domain of YAP, whereas the atypical WW1 domain of Smurf1 and the Nedd4L WW3 domain recognize diphospho-serine motifs in Smad1 and Smad3, respectively. The ability of WW domains to recognize both pT[PY] and pS(-4)pS motifs was previously unknown. The interaction of these proteins with Smads is determined not only by the binding specificity of individual WW domains, but also by the configuration of the two domains. For example, Smurf1 cannot bind Smad3 because the N-terminal position of the pT[PY] motif relative to the pSer cluster in Smad3 is opposite to the orientation required by the WW1 and WW2 domains of Smurf1. The present findings expand the known structural and functional versatility of WW domains as protein–protein interaction modules (Macias et al. 2002).
The recognition of distinct Smad phosphorylation codes by different WW domain proteins provides ample opportunities for regulation. As an adaptor that binds to the WW1–WW2 connector of Smurf1 (Lu et al. 2008), CKIP1 may enforce an optimal orientation of these WW domains for contact with their cognate sites on the Smad1 linker. A Smurf1 isoform with a longer WW1–WW2 connector exists that may differ in this regulatory function (Schultz et al. 2000). The ability of serum/glucocorticoid-regulated kinase 1 (SGK1) to phosphorylate the WW2–WW3 connector and inhibit Nedd4L binding to Smad3 (Gao et al. 2009) may similarly be based on a change in the binding ability of these WW domains. In response to mitogens and stresses, MAPKs primarily phosphorylate Smad3 at S208/S213, not at T179 (Gao et al. 2009). The poor ability of Nedd4L to recognize such MAPK phosphorylated Smad3 species (Gao et al. 2009) can be explained by the key role of the pT179 phosphate group in contacting the Nedd4L WW2 domain. Furthermore, the signals that inhibit GSK3 and increase the half-life of Smad proteins (Fuentealba et al. 2007; Guo et al. 2008; Wang et al. 2009) can now be regarded as inputs that extend the period of CDK8/9-dependent Smad peak performance. In mammalian cells and Drosophila, YAP enhances certain BMP responses but not others (Alarcon et al. 2009). Similarly, Pin1 enhances certain effects of TGF-β on mammalian cell migration but not other effects (Matsuura et al. 2009). Different factors may fulfill these roles in other contexts or on other target genes. A larger repertoire of Smad linker code-reading factors than presently known may therefore exist.
The action turnover switch delineated here involves a remarkable concentration of opposing protein-binding functions in a discrete region of the Smad proteins. The fulfillment of dual roles under a phosphorylation-dependent switch is also characteristic of another key component of this pathway: the regulatory GS region of type I TGF-β receptor kinase (Huse et al. 2001). The core TGF-β/Smad pathway is therefore characterized by the economical use that it makes of the structural elements that switch key pathway components from one activation state into another.
Materials and methods
Mammalian cell expression vectors
The plasmids encoding Smad1, Smad3, Smurf1(DD), and Nedd4L(DD) are described elsewhere (Sapkota et al. 2007; Alarcon et al. 2009; Gao et al. 2009). The linker phosphorylation site mutant Smad3 (all sites) was denoted previously as Smad3 (EPSM) (Kretzschmar et al. 1999). PCR-based site-directed mutagenesis was employed to generate all of the other mutants of the linker phosphorylation sites of Smad1 and Smad3 using the primers containing the desired mutation. The YAP construct was obtained from M.B. Yaffe (Massachusetts Institute of Technology). The YAP mutants with WW domain mutations were generated by PCR-based site-directed mutagenesis. The primers are YAPWW1AA-F (CAGACAACAACAGCGCAGGACGCCAGGAAGGCCATG), YAPWW1AA-R (CATGGCCTTCCTGGCGTCCTGCGCTGTTGTTGTCTG), YAPWW2AA-F (AAGACCACCTCTGCGCTAGACGCAAGGCTTGACCCT), and YAPWW2AA-R (AGGGTCAAGCCTTGCGTCTAGCGCAGAGGTGGTCTT).
The human PIN1 cDNA was from Open Biosystems, and the XhoI, EcoRI fragment was cloned into the PCI vector (Promega). Sequences of all constructs generated and used in this study were verified by DNA sequencing.
Transfection, immunoprecipitation, and immunoblotting
Transfections of the indicated plasmids were performed as described previously (Alarcon et al. 2009; Gao et al. 2009). Cells were used 36 h later. HEK293T cells were incubated as indicated with BMP2 (25 ng/mL; R&D Systems), TGF-β1 (100 pM; R&D Systems), noggin (50 ng/mL; R&D Systems), or SB431542 (10 μM; TOCRIS). When used, flavopiridol (0.6 mM; National Cancer Institute) and LiCl (15 mM) were added to the cells for 1 h prior to the addition of growth factors. Immunoprecipitations and Western immunoblotting were done as described previously (Sapkota et al. 2007). Antibodies raised against full-length Smad1 were from R&D Systems or were generated in-house. Antibodies against phospho-tail Smad1 were raised against Smad1 (pS463/pS465) (Cell Signaling). Antibodies against Smad2/3, Smad1pS206 were produced in-house (Sapkota et al. 2007), and Smad1 pS214 was produced in-house. Antibodies against Smad1 pS210 were a gift from E. De Robertis (Fuentealba et al. 2007). Other antibodies used include rat monoclonal anti-HA-peroxidase (Roche), mouse mononclonal anti-Flag M2-peroxidase antibody (Sigma), agarose-conjugated mouse monoclonal anti-HA clone 7 (Sigma), and anti-Flag M2 affinity gel (Sigma).
Recombinant products
The three consecutive WW domain pairs in human Nedd4L (WW1–WW2 193–400, WW2–WW3 364–512, and WW3–WW4 476–563) and the WW domains of human Smurf1 (WW1 232–267, WW2 277–314, and WW1–WW2 232–314) were cloned into a pETM11 vector. The WW domains of human YAP (WW1 163–206, WW2 227–266, and WW1–WW2 163–266) and Pin1 (1–40) were cloned into a pETM30 vector (a gift from the EMBL-Heidelberg Protein Expression Facility) using NcoI and HindIII sites. All point mutations described in the text were introduced using the QuickChange site-directed mutagenesis kit (Stratagene) with the appropriate complementary mutagenic primers. All wild-type and mutant constructs were confirmed by sequencing.
Generation of Nedd4L WW2–WW3 segment by protein ligation
We selected a segmental labeling approach that requires the protein to be expressed as two fragments, each produced with a different labeling pattern, and then combined by chemical ligation. For the ligation, we used the formation of a disulfide bridge built from the side chains of two cysteine residues—one at the C terminus of fragment 1 (WW2-connector-C471) and the other at the N terminus of fragment 2 (C472-WW3)—using mutants I515Y and H517A as templates. In order to guide the reaction toward the formation of the hetero-disulfide WW2–WW3 product, the (C472-WW3) site was activated prior to the ligation reaction (Baca et al. 1995). The pyridylsulfenyl-cysteine WW3 product was identified as a new peak with a higher retention time in the reverse-phase high-performance liquid chromatography (RP-HPLC) profile of the reaction, and its mass was corroborated by MALDI-TOF mass spectroscopy. The product was purified by semipreparative RP-HPLC and lyophilized. 2H,15N-labeled Nedd4L WW2-linker-C471 (0.4 μmol) and 1H,15N-labeled Nedd4L PyS-(C472)-WW3 domain (0.3 μmol) were dissolved in 600 μL of 6 M guanidine–HCl and sodium acetate buffer (20 mM, 100 mM NaCl at pH 4.5) and stirred overnight at 4°C. The ligation product was identified by RP-HPLC/MALDI-TOF mass spectroscopy. After purification, the product was lyophilized.
Proteins
Unlabeled; 15N-labeled; 13C, 15N-labeled; and 2H, 13C, 15N-labeled proteins were expressed in Escherichia coli BL21 (DE3) in LB medium or minimal medium (M9) using either H2O or D2O (99.89%, CortecNet) enriched with 15NH4Cl and/or D-[13C] glucose as the sole sources of carbon and nitrogen, respectively (Marley et al. 2001). E. coli extracts were lysed using an EmulsiFlex-C5 (Avestin) cell disrupter equipped with an in-house-developed Peltier temperature controller system. Soluble fusion proteins were purified by nickel-affinity chromatography (HiTrap Chelating HP column, GE Healthcare Life Science), and samples were eluted using buffer A with EDTA. Smurf1 WW1–WW2 protein was mostly in the insoluble fraction after centrifugation of E. coli lysates, and was solubilized with 6 M guanidine–hydrochloride and then purified using the HiTrap HP column. After buffer exchange, fusion tags were removed by overnight TEV protease digestion at 4°C followed by a second nickel-affinity binding step. All proteins were further purified with an additional gel filtration chromatography step using HiLoad Superdex 30, 75, or 200 16/60 prep-grade columns (GE Healthcare), depending on the protein size. Fractions containing the purified proteins were concentrated to 1–2 mM for NMR experiments. To ensure the presence of a 1:1 protein:peptide ratio, and to avoid formation of aggregates or misfolded samples, Smurf1 and YAP proteins were concentrated in the presence of the Smad1 peptides prior to NMR experiments. The NMR buffer was 20 mM deuterated Tris-HCl (pH 7.2–7.4), 100 mM NaCl, 0.01% NaN3, and 2.5 mM deuterated dithiothreitol in the presence of 10% D2O.
Peptides
All peptides were prepared using Fmoc solid-phase peptide synthesis with 0.10–0.15 mmol FastMoc protocols as described (Alarcon et al. 2009; Gao et al. 2009). The syntheses of Smad3 3P and Smad1 3P and 4P peptides were optimized by combining manual and automated strategies. Crude peptides were purified by RP-HPLC using a Vydac C18 or C4 Sephasil preparative columns and an ÄKTApurifier10 (GE Healthcare) or in a Waters HPLC delta 600 system using a high preparative Waters SunFire C18 column. Fractions containing the desired peptides were identified by MALDI-TOF mass spectrometry.
ITC
ITC experiments were performed using a VP-ITC MicroCalorimeter (MicroCal) at 10°C and 25°C essentially as described (Alarcon et al. 2009; Gao et al. 2009). ITC isotherms were fit to the simplest model with MicroCal's ORIGIN software.
NMR spectroscopy
All experiments were recorded on a Bruker Avance III 600-MHz spectrometer equipped with a Z pulse field gradient unit and either triple (1H, 13C, 15N) or quadruple (plus 31P) resonance probe heads. Double- and/or triple-labeled samples were prepared to obtain sequence-specific [HNCACB/HN(CO)CACB or CBCA(CO)NH/CBCANH] experiments. Side chain resonance assignments were obtained using standard triple-resonance experiments [15N-TOCSY, CC(CO)NH, HCCH-TOCSY (12-msec mixing time) and 15N-, 13C NOESY experiments (with 100- and 150-msec mixing times depending on the protein size)]. Intramolecular proton distance restraints were obtained from peaks assigned in 2D-NOESY, 15N-NOESY, 13C-NOESY, and half-filtered experiments. All spectra were processed with the NMRPipe/NMRDraw (Delaglio et al. 1995) software and were analyzed with CARA (Bartels et al. 1995). Spectra used for the calculation were integrated with the batch integration method of the XEASY package. 3JHN-Hα scalar couplings were obtained from HNHA experiments. One-bond N–HN RDCs were determined by using the IPAP 15N HSQC sequence. Hydrogen bonds were obtained by acquiring a set of 1H15N-HSQC experiments after dissolving the lyophilized protein in D2O. Double- and half-filtered experiments were run to assign the peptides in the bound state (Sattler et al. 1999).
NMR titration experiments with peptides
For the 15N-HSQC experiments, 15N-labeled protein domains were prepared at 0.25 mM concentration in the same buffer as described above, and unlabeled ligand was added to the 15N-labeled sample up to a final molar ratio of 3:1. Measurements were performed at 285 K or 295 K on a Bruker Avance III 600 MHz.
Structure calculation
For the structure calculation, distance restraints derived from NOESY experiments, 3J(HN, HA) obtained from HNHA spectra, and hydrogen bond restraints determined by D2O exchange were used. The structures were calculated using CNS (Brünger et al. 1998) with an in-house-modified protocol of Aria 1.2 (Nilges et al. 1997). Since only unambiguously assigned restraints were used, the protocol was reduced to two iterations of one and 120 structures, respectively, using 100,000 cooling steps. All calculated structures were submitted to water refinement, and were ranked based on minimum values of energy terms and violations. The water refinement protocol was also modified by weighing the value of unambiguous NOEs, hydrogen bonds, and dihedral restraints by a factor of 10. In this way, all experimental restraints are used during the refinement process, and the obtained structures are in better agreement with the experimental data while retaining good Ramachandran values. The Nedd4L WW2–WW3 complex was refined using residual dipolar couplings obtained from aligned samples in different alkyl-poly(ethylene glycol) mixtures (Ruckert and Otting 2000).
Analysis of the quality of the lowest-energy structures was performed using PROCHECK-NMR (Laskowski et al. 1996). The statistics from the analysis are shown in Supplemental Table S1.
Molecular dynamic simulations
Molecular dynamic simulations were performed with the Gromacs package (Hess et al. 2008). Prior to the simulations, we generated an extended model of each molecule with CNS, surrounded by a charged-equilibrated, periodic cubic water box. Then, the system was energy-minimized and short position-restrained molecular dynamics was performed to equilibrate the water molecules. Finally, a 40-nsec molecular dynamics in explicit solvent with Particle Mesh Ewald electrostatics was carried out. Calculated structures and the results of the molecular dynamic simulations were analyzed with PyMOL (http://www.pymol.org). Sequence alignments were performed using ClustalX (Thompson et al. 1997) and BoxShade 3.21 (http://www.ch.embnet.org/software/BOX_form.html).
Accession numbers
For each of the nine complexes (short names in bold, as in Supplemental Table S1), we deposited 20 structures in the Protein Data Bank (PDB), and the list of restraints and chemical shifts in the BioMagResBank (BMRB) database. For the Smurf1–Smad1 complexes, the corresponding PDB and BMRB codes are, respectively, WW1–pS214: 21az, 17541; WW1–pS210pS214: 21b0, 17542; and WW2–PY: 21b1, 17543. For the YAP–Smad1 complexes, the corresponding PDB and BMRB codes are, respectively, WW1–pS206: 21ay, 17540; WW1–pT202pS206: 21ax, 17539; and WW2–PY: 21aw, 17538. For the Nedd4L–Smad3 complexes, the corresponding PDB and BMRB codes are, respectively, WW2–pTPY: 21b2, 17544; and WW3–pS204pS208: 2laj, 17529. For the PIN1–Smad3 complex, the corresponding PDB and BMRB codes are, respectivey, WW1–pT: 21b3, 17545.
Acknowledgments
We thank M. Royo (Parc Cientific de Barcelona) for advice on protein chemical ligation and peptide synthesis. N.G. and A.E have an IRB and FPU-AP2009-3242 PhD fellowship, respectively. This work was supported by SMSI-BFU2008-02795 grant (to M.J.M.) and by NIH grant CA34610 (to J.M.). M.J.M. is an ICREA Programme Investigator. J.M. is an Investigator of the Howard Hughes Medical Institute.
Footnotes
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.2060811.
Supplemental material is available for this article.
References
- Alarcon C, Zaromytidou AI, Xi Q, Gao S, Yu J, Fujisawa S, Barlas A, Miller AN, Manova-Todorova K, Macias MJ, et al. 2009. Nuclear CDKs drive Smad transcriptional activation and turnover in BMP and TGF-β pathways. Cell 139: 757–769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baca M, Muir TW, Schnölzer M, Kent SBH 1995. Chemical ligation of cysteine- containing peptides: synthesis of a 22 kDa tethered dimer of HIV-1 protease. J Am Chem Soc 116: 10797–10798 [Google Scholar]
- Bartels C, Xia T-H, Billeter M, Güntert P, Wüthrich K 1995. The program XEASY for computer-supported NMR spectral analysis of biological macromolecules. J Biol NMR 5: 1–10 [DOI] [PubMed] [Google Scholar]
- Batut J, Schmierer B, Cao J, Raftery LA, Hill CS, Howell M 2008. Two highly related regulatory subunits of PP2A exert opposite effects on TGF-β/Activin/Nodal signalling. Development 135: 2927–2937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brünger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, Jiang JS, Kuszewski J, Nilges M, Pannu NS, et al. 1998. Crystallography and NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr D Biol Crystallogr 54: 905–921 [DOI] [PubMed] [Google Scholar]
- Cohen P, Frame S 2001. The renaissance of GSK3. Nat Rev Mol Cell Biol 2: 769–776 [DOI] [PubMed] [Google Scholar]
- Delaglio F, Grzesiek S, Vuister GW, Zhu G, Pfeifer J, Bax A 1995. NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J Biol NMR 6: 277–293 [DOI] [PubMed] [Google Scholar]
- Durand LO, Advani SJ, Poon AP, Roizman B 2005. The carboxyl-terminal domain of RNA polymerase II is phosphorylated by a complex containing cdk9 and infected-cell protein 22 of herpes simplex virus 1. J Virol 79: 6757–6762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebisawa T, Fukuchi M, Murakami G, Chiba T, Tanaka K, Imamura T, Miyazono K 2001. Smurf1 interacts with transforming growth factor-β type I receptor through Smad7 and induces receptor degradation. J Biol Chem 276: 12477–12480 [DOI] [PubMed] [Google Scholar]
- Fuentealba LC, Eivers E, Ikeda A, Hurtado C, Kuroda H, Pera EM, De Robertis EM 2007. Integrating patterning signals: Wnt/GSK3 regulates the duration of the BMP/Smad1 signal. Cell 131: 980–993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao S, Alarcon C, Sapkota G, Rahman S, Chen PY, Goerner N, Macias MJ, Erdjument-Bromage H, Tempst P, Massagué J 2009. Ubiquitin ligase Nedd4L targets activated Smad2/3 to limit TGF-β signaling. Mol Cell 36: 457–468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo X, Waddell DS, Wang W, Wang Z, Liberati NT, Yong S, Liu X, Wang XF 2008. Ligand-dependent ubiquitination of Smad3 is regulated by casein kinase 1 γ 2, an inhibitor of TGF-β signaling. Oncogene 27: 7235–7247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hess B, Kutzner C, van der Spoel D, Lindahl E 2008. GROMACS 4: algorithms for highly efficient, load-balanced, and scalable molecular simulation. J Chem Theory Comput 4: 435–447 [DOI] [PubMed] [Google Scholar]
- Huse M, Muir TW, Xu L, Chen YG, Kuriyan J, Massagué J 2001. The TGF β receptor activation process: an inhibitor- to substrate-binding switch. Mol Cell 8: 671–682 [DOI] [PubMed] [Google Scholar]
- Inman GJ, Nicolas FJ, Hill CS 2002. Nucleocytoplasmic shuttling of Smads 2, 3, and 4 permits sensing of TGF-β receptor activity. Mol Cell 10: 283–294 [DOI] [PubMed] [Google Scholar]
- Komarnitsky P, Cho EJ, Buratowski S 2000. Different phosphorylated forms of RNA polymerase II and associated mRNA processing factors during transcription. Genes Dev 14: 2452–2460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kretzschmar M, Doody J, Massagué J 1997. Opposing BMP and EGF signalling pathways converge on the TGF-β family mediator Smad1. Nature 389: 618–622 [DOI] [PubMed] [Google Scholar]
- Kretzschmar M, Doody J, Timokhina I, Massagué J 1999. A mechanism of repression of TGFβ/ Smad signaling by oncogenic Ras. Genes Dev 13: 804–816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laskowski RA, Rullmannn JA, MacArthur MW, Kaptein R, Thornton JM 1996. AQUA and PROCHECK-NMR: programs for checking the quality of protein structures solved by NMR. J Biol NMR 8: 477–486 [DOI] [PubMed] [Google Scholar]
- Li MO, Flavell RA 2008. TGF-β: a master of all T cell trades. Cell 134: 392–404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin X, Duan X, Liang YY, Su Y, Wrighton KH, Long J, Hu M, Davis CM, Wang J, Brunicardi FC, et al. 2006. PPM1A functions as a Smad phosphatase to terminate TGFβ signaling. Cell 125: 915–928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lo RS, Massagué J 1999. Ubiquitin-dependent degradation of TGF-β-activated smad2. Nat Cell Biol 1: 472–478 [DOI] [PubMed] [Google Scholar]
- Lu K, Yin X, Weng T, Xi S, Li L, Xing G, Cheng X, Yang X, Zhang L, He F 2008. Targeting WW domains linker of HECT-type ubiquitin ligase Smurf1 for activation by CKIP-1. Nat Cell Biol 10: 994–1002 [DOI] [PubMed] [Google Scholar]
- Macias MJ, Hyvonen M, Baraldi E, Schultz J, Sudol M, Saraste M, Oschkinat H 1996. Structure of the WW domain of a kinase-associated protein complexed with a proline-rich peptide. Nature 382: 646–649 [DOI] [PubMed] [Google Scholar]
- Macias MJ, Wiesner S, Sudol M 2002. WW and SH3 domains, two different scaffolds to recognize proline-rich ligands. FEBS Lett 513: 30–37 [DOI] [PubMed] [Google Scholar]
- Malik S, Roeder RG 2000. Transcriptional regulation through Mediator-like coactivators in yeast and metazoan cells. Trends Biochem Sci 25: 277–283 [DOI] [PubMed] [Google Scholar]
- Marley J, Lu M, Bracken C 2001. A method for efficient isotopic labelling of recombinant proteins. J Biol NMR 20: 71–75 [DOI] [PubMed] [Google Scholar]
- Massagué J 2008. TGFβ in cancer. Cell 134: 215–230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuura I, Chiang KN, Lai CY, He D, Wang G, Ramkumar R, Uchida T, Ryo A, Lu K, Liu F 2009. Pin1 promotes transforming growth factor-β-induced migration and invasion. J Biol Chem 285: 1754–1764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nair SK, Burley SK 2003. X-ray structures of Myc-Max and Mad-Max recognizing DNA. Molecular bases of regulation by proto-oncogenic transcription factors. Cell 112: 193–205 [DOI] [PubMed] [Google Scholar]
- Nilges M, Macias MJ, O'Donoghue SI, Oschkinat H 1997. Automated NOESY interpretation with ambiguous distance restraints: the refined NMR solution structure of the pleckstrin homology domain from β-spectrin. J Mol Biol 269: 408–422 [DOI] [PubMed] [Google Scholar]
- Pires JR, Taha-Nejad F, Toepert F, Ast T, Hoffmuller U, Schneider-Mergener J, Kuhne R, Macias MJ, Oschkinat H 2001. Solution structures of the YAP65 WW domain and the variant L30K in complex with the peptides GTPPPPYTVG, N-(n-octyl)-GPPPY and PLPPY and the application of peptide libraries reveal a minimal binding epitope. J Mol Biol 314: 1147–1156 [DOI] [PubMed] [Google Scholar]
- Ruckert M, Otting G 2000. Alignment of biological macromolecules in novel nonionic liquid crystalline media for NMR experiments. J Am Chem Soc 122: 7793–7797 [Google Scholar]
- Sapkota G, Alarcon C, Spagnoli FM, Brivanlou AH, Massagué J 2007. Balancing BMP signaling through integrated inputs into the Smad1 linker. Mol Cell 25: 441–454 [DOI] [PubMed] [Google Scholar]
- Sattler M, Schleucher J, Griesinger C 1999. Heteronuclear multidimensional NMR experiments for the structure determination of proteins in solution employing pulsed field gradients. Prog Nucl Magn Reson Spectrosc 34: 93–158 [Google Scholar]
- Schmierer B, Tournier AL, Bates PA, Hill CS 2008. Mathematical modeling identifies Smad nucleocytoplasmic shuttling as a dynamic signal-interpreting system. Proc Natl Acad Sci 105: 6608–6613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schultz J, Copley RR, Doerks T, Ponting CP, Bork P 2000. SMART: a Web-based tool for the study of genetically mobile domains. Nucleic Acids Res 28: 231–234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Y, Massagué J 2003. Mechanisms of TGF-β signaling from cell membrane to the nucleus. Cell 113: 685–700 [DOI] [PubMed] [Google Scholar]
- Shi Y, Wang YF, Jayaraman L, Yang H, Massagué J, Pavletich NP 1998. Crystal structure of a Smad MH1 domain bound to DNA: insights on DNA binding in TGF-β signaling. Cell 94: 585–594 [DOI] [PubMed] [Google Scholar]
- Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG 1997. The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res 25: 4876–4882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toepert F, Pires JR, Landgraf C, Oschkinat H, Schneider-Mergener J 2001. Synthesis of an array comprising 837 variants of the hYAP WW protein domain. Angew Chem Int Ed Engl 40: 897–900 [DOI] [PubMed] [Google Scholar]
- Wang G, Matsuura I, He D, Liu F 2009. Transforming growth factor-β-inducible phosphorylation of Smad3. J Biol Chem 284: 9663–9673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wrighton KH, Willis D, Long J, Liu F, Lin X, Feng XH 2006. Small C-terminal domain phosphatases dephosphorylate the regulatory linker regions of Smad2 and Smad3 to enhance transforming growth factor-β signaling. J Biol Chem 281: 38365–38375 [DOI] [PubMed] [Google Scholar]
- Wu MY, Hill CS 2009. TGF-β superfamily signaling in embryonic development and homeostasis. Dev Cell 16: 329–343 [DOI] [PubMed] [Google Scholar]
- Wu D, Pan W 2010. GSK3: a multifaceted kinase in Wnt signaling. Trends Biochem Sci 35: 161–168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu JW, Hu M, Chai J, Seoane J, Huse M, Li C, Rigotti DJ, Kyin S, Muir TW, Fairman R, et al. 2001. Crystal structure of a phosphorylated Smad2. Recognition of phosphoserine by the MH2 domain and insights on Smad function in TGF-β signaling. Mol Cell 8: 1277–1289 [DOI] [PubMed] [Google Scholar]
- Xu L, Kang Y, Col S, Massagué J 2002. Smad2 nucleocytoplasmic shuttling by nucleoporins CAN/Nup214 and Nup153 feeds TGFβ signaling complexes in the cytoplasm and nucleus. Mol Cell 10: 271–282 [DOI] [PubMed] [Google Scholar]