Abstract
The DNA entry and exit points on the nucleosome core regulate the initial invasion of the nucleosome by factors requiring access to the underlying DNA. Here we describe in vivo consequences of eliminating a single protein–DNA interaction at this position through mutagenesis of histone H3 Lys 42 to alanine. This substitution has a dramatic effect on the Saccharomyces cerevisiae transcriptome in both the transcriptional output and landscape of mRNA species produced. We attribute this in part to decreased histone H3 occupancy at transcriptionally active loci, leading to enhanced elongation. Additionally we show that this lysine is methylated in vivo, and genetic studies of methyl-lysine mimics suggest that this modification may be crucial in attenuating gene expression. Interestingly, this site of methylation is unique to Ascomycota, suggesting a recent evolutionary innovation that highlights the evolvability of post-translational modifications of chromatin.
Keywords: evolution, histone methylation, transcription elongation, transcriptional output, yeast
Nucleosomes, the octameric histone protein complex wrapped by 147 base pairs (bp) of DNA, are core components of chromatin in eukaryotes. At a molecular level, the nucleosome crystal structure shows that there are 14 DNA–histone interactions within the core particle, positioned at each minor groove (Luger et al. 1997). Twelve of these interactions are mediated by histone fold motifs of all four histone proteins, and the remaining two involve residues in the histone H3 αN helix at the DNA entry and exit points. Although the majority of these interactions are mediated by nonspecific electrostatics between the DNA and protein backbones, important positively charged histone side chains have also been shown to play a role (Luger and Richmond 1998). Recently, it has been shown in vitro that not all histone–DNA interactions within the nucleosome are of equal strength. Using an optical trapping method, Hall et al. (2009) mapped the strongest interactions to three precise locations on the nucleosome core, including the dyad axis, and ±40 bp on either side of this.
The different classes of intranucleosomal interactions illustrate how the cell can tune the stability of the nucleosome by reinforcing or destabilizing these interactions, thus controlling the timing and extent of accessibility of the underlying DNA. Various proteins and complexes are recruited to chromatin to alter nucleosome stability either temporally or spatially throughout the genome, including ATP-dependent chromatin remodelers, histone variant proteins, and enzymes that chemically modify histone amino acid side chains. (for review, see Hartzog er al. 2002; Li et al. 2007; Clapier and Cairns 2009). The question then arises as to which structurally important interactions defining nucleosome stability at a molecular level are targeted by these processes.
First, it is known that modification of histone residues is quite complex. Modification of residues lying both at the N termini of histone proteins and within the globular core perturbs the formation of higher-order chromatin structure by influencing electrostatic interactions, and through interactions with specific information “readers” (Watanabe et al. 2010). Second, it has been suggested that the residues at the dyad axis are targeted by chromatin remodeling enzymes, because introducing histone mutations in this region (so-called “SIN alleles”) facilitated transcription of genes that typically require chromatin remodeling in the absence of the ATP-dependent factors (Kruger et al. 1995; Recht and Osley 1999) and lead to nucleosome instability in vitro (Kurumizaka and Wolffe 1997). Furthermore, residues at/close to the dyad axis in the nucleosome do not tolerate mutagenesis well, as these positions are overrepresented in collections of lethal histone mutations (Dai et al. 2008; Nakanishi et al. 2008). Third, it has been shown that certain mutations in histone H3 resulted in the formation of spurious transcription from intragenic locations (Cheung et al. 2008), suggesting that the role of the wild-type residue may be to prevent nucleosome destabilization at the 3′ end of ORFs. These residues are located close to the dyad axis as well as at the base of the histone H3 αN helix.
In this study, we sought to understand the contribution of the DNA entry and exit points to nucleosome function in vivo and ascertain whether it plays a dynamic role in controlling the accessibility of nucleosomal DNA. In the crystal structure, the DNA at this region is organized exclusively by the histone H3 αN helix, and a single residue, Lys 42, within this helix interacts directly with DNA by inserting its side chain between the two DNA gyres. In spite of the fact that it has been shown that this interaction is comparatively weak within the nucleosome (Hall et al. 2009), we know from in vitro studies that mutating this residue makes purified nucleosomes inherently more mobile (Somers and Owen-Hughes 2009). We studied the H3-K42A mutation, and, although not a lethal substitution in vivo, yeast cells harboring this histone mutant as the sole source of histone H3 grow slowly and display a variety of phenotypes. The most striking observation is vastly increased transcriptional output of H3-K42A-expressing cells. Surprisingly, this residue is a substrate for methylation, and our data support a role for this modification in transcriptional repression. We therefore identified a critical interaction within the nucleosome that is targeted to fine-tune gene expression, and we propose that K42, and possibly its modification, serves as a structural “gatekeeper” of transcription. Remarkably, K42 is a “young” lysine evolutionarily; in most other eukaryotes, this residue is an arginine, suggesting recent evolution of its thus-far unidentified methylation machinery.
Results
Histone H3-K42A mutants are pleiotropic in Saccharomyces cerevisiae
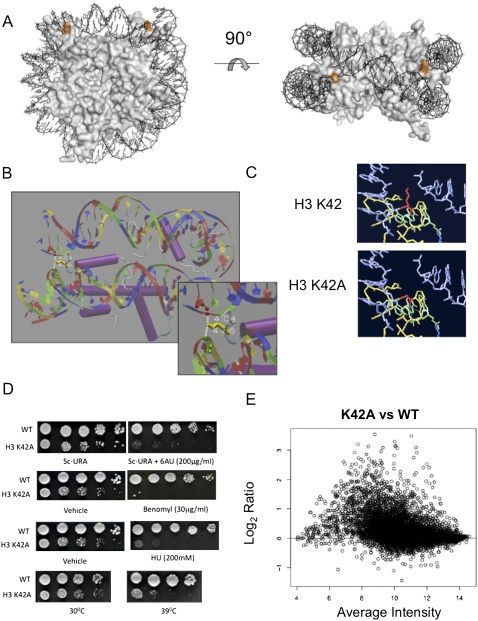
Histone H3-K42 is a crucial residue that mediates the sole DNA contact of the nucleosome at the DNA entry and exit positions. (Fig. 1A). In the crystal structure (White et al. 2001), the K42 side chain is positioned in the solvent-exposed space between the two DNA helices. Its ɛ amino group points away from the nucleosome core and forms two hydrogen bonds with the phosphodiester backbone (Fig. 1B). To assess the contribution of these interactions to nucleosome function, we mutated this lysine residue to alanine, eliminating hydrogen bond potential (Fig. 1C), and performed phenotypic analyses.
Figure 1.
Histone H3-K42A is a pleiotropic mutation, which disrupts a structurally important nucleosome surface. (A) The crystal structure of the S. cerevisiae nucleosome highlighting histone H3 Lys 42 (orange) positioned at the DNA entry and exit points. The figure was generated using PYMOL. (B) Nucleosome crystal structure surrounding H3-K42 (yellow), indicating hydrogen bonds between the lysine side chain and DNA. (C) Predicted structures of H3-K42A mutation in its nucleosomal context. (Red) Residue 42; (blue) DNA; (green/yellow) histone H3. Shown is the single rotamer that exists for this mutation. Graphics were generated using Swiss PDB viewer. (D) Growth assays were undertaken for the indicated strains as described in Materials and Methods to detect defects in transcription elongation, cell cycle, and DNA repair, and to monitor temperature sensitivity at 39°C. (E) An MVA plot indicating gene expression changes versus spot intensity of normalized microarray data of K42A versus wild-type (WT) histone H3-expressing cells.
H3-K42A cells grow almost twice as slowly as wild-type yeast (data not shown). We scored the viability of this mutant under different conditions and found it was sensitive to (1) the DNA-damaging agents hydroxyurea (HU) and methyl-methanesulfonate; (2) benomyl, the microtubule-destabilizing drug; (3) 6-azauracil (6AU), a drug that interferes with transcription; and (4) temperatures >39°C (Fig. 1D). It did not display growth defects in the presence of camptothecin, a topoisomerase II inhibitor, or cold temperatures (data not shown). The pleiotropic behavior of H3-K42A indicates that this residue either is directly critical for numerous cellular processes, or its affect on nucleosome structure indirectly influences multiple pathways, possibly by influencing target gene expression. To test this, we analyzed the transcript profile of cells harboring H3-K42A. Strikingly, in these cells, we found that the expression of ∼42% of genes was altered in comparison with wild-type cells, with the majority of these (90%) up-regulated (Fig. 1E). The average fold increase for affected genes was two; the expression of a subset of loci was altered up to 12-fold above wild-type levels. No specific gene classes (based on Gene Ontology [GO] terms) were preferentially perturbed by histone H3-K42A (Supplemental Fig. S1). Additionally, no correlation was observed between chromosome location and transcriptional response to the H3-K42A substitution (data not shown).
We next examined whether these effects were specific to K42A or whether mutating other residues close to this nucleosome location exhibited similar phenotypes. We know that this region is important for faithful mitotic chromosome segregation, serving as a docking site for the recruitment of Sgo1p to pericentromeric regions during mitosis (Luo et al. 2010). This explains why mutations in this region, including K42A, are benomyl-sensitive, which is suppressed by overexpressing SGO1 (data not shown). We found that, although the H3-K42 region mutants do not confer 6AU sensitivity, we did observe increased expression of certain genes in these strains (Supplemental Fig. S2). However, the extent of this effect is nonuniform, and K42A exhibits the most severe transcriptional phenotype. Taken together, these observations highlight the structural importance and functionality of this nucleosome location and suggest that H3-K42 is a key residue within this region in transcriptional regulation.
Histone H3-K42A profoundly alters the S. cerevisiae transcriptome
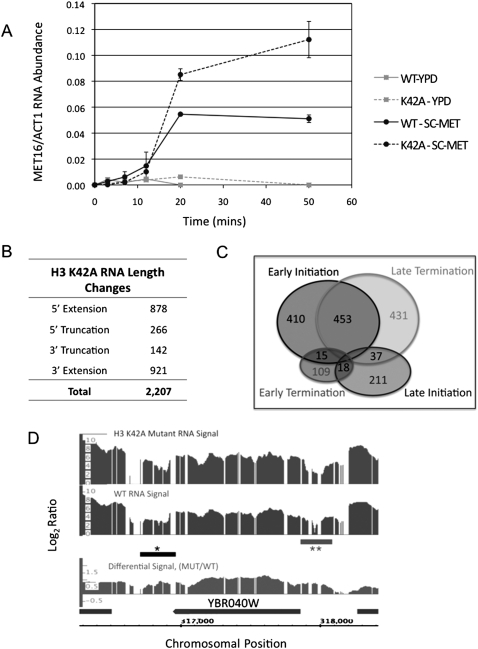
To probe mechanisms underlying the potential role of this nucleosome surface during transcription, we sought to understand the hypertranscription phenotype of H3-K42A. The microarray analysis indicated that H3-K42A does not lead to erroneous derepression of transcription genome-wide, as the majority of genes typically repressed under the conditions tested were unaffected by H3-K42A. Therefore, we hypothesized that H3-K42A influences an active transcription cycle, and thus analyzed the dynamic expression of a single inducible locus: MET16. Our results indicate that the initial appearance of MET16 mRNA under inducing conditions was unaltered in H3-K42A cells, suggesting that regulation of the first round of transcription initiation is unaffected. However, MET16 transcript overaccumulates with time in the mutant cells (Fig. 2A), consistent with a defect in establishing a proper set point for transcription rate or inefficient transcription termination (Fig. 2A). MET16 expression level is not affected under repressive conditions, supporting the hypothesis that the effect of K42A depends on transcriptional activation (Fig. 2A).
Figure 2.
Histone H3-K42A alters the S. cerevisiae transcriptome. (A) Kinetics of appearance of MET16 mRNA in cells expressing wild-type (WT) or K42A histone H3, as described in the Materials and Methods. Error bars represent the standard deviation between two biological replicates of each sample, each assayed in triplicate. (B) Table summarizing RNA length changes detected in H3-K42A cells using yeast genomic tiling array as described in the Materials and Methods. Four classes of mRNA length changes were analyzed, and the number of transcripts in K42A cells that fall into each class is indicated. (C) Venn diagram representing the number of genes within each class of RNA length change. The overlap regions indicate the counts of loci showing aberrant RNA length changes that can be classified by two distinct transcriptional events. (D) Integrated Genome Browser screen shot of log2 wild-type (WT) and H3-K42A RNA levels, as well as the differential RNA levels (log2 K42A/WT) for a segment of chromosome 2 spanning YBR040W. The horizontal bars marked with asterisks represent aberrant length changes, indicating both 5′ extension (**) and 3′ extension (*) events at this locus.
To gain a higher resolution of the hypertranscription phenotype, we examined the length of RNA species produced in K42A cells transcriptome-wide. We hybridized total RNA extracted from mutant cells to the Affymetrix genomic tiling array, which interrogates the yeast genome at 5-bp resolution. A methodology was employed that identifies stretches of RNA that extend beyond the length of wild-type transcripts as well as differential RNA lengths within an ORF (Poorey et al. 2010). Strikingly, we found that ∼30% of all transcripts analyzed deviated from their expected length in H3-K42A cells (Supplemental Table S1). The majority (61%) of these transcripts also displayed increased abundance in the K42A microarray data set. The distribution of actual base pair length change was broad, ranging from 101 bp to 19,196 bp (median = 902 bp). Altered-length transcripts are categorized relative to the ORF of the affected locus as 5′ extensions, 5′ truncations, 3′ truncations, and 3′ extensions. Figure 2B shows that the K42A data set was significantly enriched for both 5′ and 3′ extensions, although truncation events were also detected. Significant overlap was seen between the 5′ and 3′ extension events, indicating that both events occur at the same locus with high frequency in K42A cells (Fig. 2C). A representative example of a transcript length change is shown in Figure 2D for YBR040W, a gene up-regulated 12-fold by H3-K42A. At this single locus, the transcript in K42A cells displayed both 5′ and 3′ extensions. This analysis indicates that H3-K42A drastically alters not only the amplitude, but also the “quality” of the yeast transcriptome.
Hypertranscription phenotype depends on elongation factors
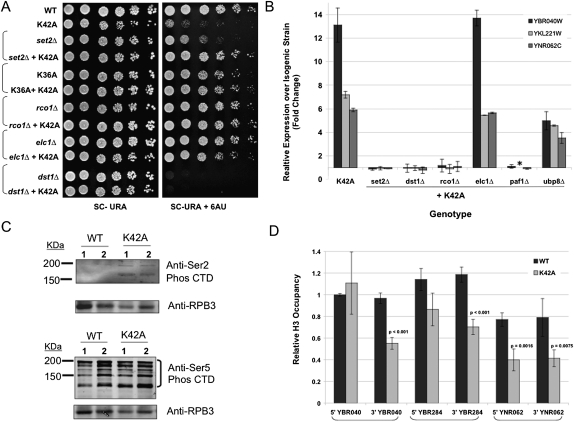
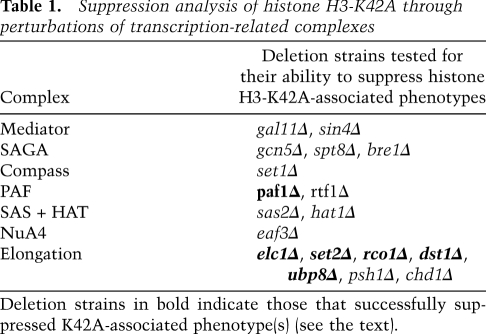
Chromatin negatively impacts many aspects of transcription, and a wide variety of complex mechanisms exist to overcome this obstacle. Based on our analysis of MET16 expression, we hypothesized that histone H3-K42A influences the function of one or more such protein(s) in an event downstream from transcription initiation. We studied transcription component knockouts to probe how K42A elicits its transcriptional effects. We chose 20 nonessential gene mutants known to play roles in the initiation and elongation phases and initially scored their ability to influence K42A 6AU sensitivity. Our results indicate that the K42A 6AU phenotype was specifically suppressed by the deletion of the elongation factors SET2, ELC1, or RCO1 (Fig. 3A). In contrast, we found no genetic interactions between K42A and a series of genes involved primarily in transcription initiation, such as SET1, GCN5, GAL11, and BRE1 (data not shown). Table 1 lists all of the genes tested in this analysis.
Figure 3.
Histone H3-K42 is an important residue during transcription elongation. (A) Strains with the indicated genotypes were plated as described in the Materials and Methods to detect transcriptional elongation defects. (B) Quantitative RT–PCR analysis of the YBR040W, YKL221W, and YNR062C transcripts using RNA prepared from cells of the indicated genotype. Expression levels in the deletion strains expressing wild-type (WT) histones were set to 1 and were compared with those from the same deletion strain harboring H3-K42A. Data are presented as fold change over wild type and represent three individual colonies of each sample analyzed in triplicate and internally normalized to ACT1 transcript levels. (*) The expression of YNR062C was not determined in the paf1Δ strain. (C) Western blot analysis of yeast whole-cell extracts from strains of the indicated genotype detecting Ser 2 phosphorylation of RNAPII CTD (top panel) and Ser 5 phosphorylation of RNAP II CTD (bottom panel). Rpb3p, a subunit of the RNAPII complex, was detected and served as a loading control. (D) ChIP analysis of H3 occupancy at both the promoter region (5′) and coding region (3′) of the loci indicated in wild-type and K42A-expressing cells. Data represent two individual colonies of each sample, each of which underwent two parallel immunoprecipitations. Quantitative PCR was used to determine the amount of DNA in each immunoprecipitation. Data were normalized internally to the levels of histone H3 at the ACT1 locus, expression of which is not affected by histone H3-K42A. Significant differences between wild type and K42A are indicated with the corresponding probability value.
Table 1.
Suppression analysis of histone H3-K42A through perturbations of transcription-related complexes
As certain elongation factor deletions are themselves sensitive to 6AU—such as dst1Δ and paf1Δ, for example—we sought additional evidence for the ability of elongation factors to suppress K42A-mediated hypertranscription. We employed quantitative RT–PCR to monitor the expression of three specific loci up-regulated by H3-K42A: YBR040W, YNR062C, and YKL221W. We found that deletions of SET2, DST1, RCO1, or PAF1 completely reversed the ability of H3-K42A to increase transcription of these genes (Fig. 3B). These interactions are consistent with the idea that K42A hypertranscription reflects hyperelongation. If we explain the 6AU sensitivity of K42A as the inability of these cells to generate essential transcripts when nucleotide pools are decreased, perhaps a direct consequence of the extent of genome-wide hypertranscription, then mutations that decrease elongation rates will allow those nucleotides to be used more effectively and restore growth in the presence of 6AU. In contrast, although elc1Δ suppressed the 6AU phenotype of K42A, this strain did not alter the expression pattern of the three test genes. It was also noted that ubp8Δ, a gene involved in the transition between transcription initiation and elongation, had a slight but reproducible suppressive effect in both assays tested (Fig. 3B; data not shown).
Since SET2 is the histone methyltransferase for H3-K36, a residue in close proximity to H3-K42, we tested whether substitutions at H3-K36 would also suppress K42A and found that H3-K36A suppressed K42A slow growth and hypertranscription phenotypes as well as its 6AU sensitivity both in cis and in trans (Fig. 3A; data not shown).
These genetic data show a striking pattern: The H3-K42A phenotypes are specifically suppressed by a subset of elongation factor mutants, suggesting that the mode of action of this allele may be to induce hyperelongation or uncontrolled elongation. The altered elongation rate might in turn explain the altered 3′ end formation observed in >1000 different genes. To test this hypothesis, we determined the levels of RNA polymerase II (RNAPII) actively involved in elongation as opposed to initiation by analyzing the phosphorylation status of its C-terminal domain (CTD). Ser 2 phosphorylation of the CTD is a hallmark of the elongating RNAPII; immunoblot analysis indicated that the levels of this modification were increased in K42A-expressing cells compared with the wild type. In contrast, CTD phosphorylation at Ser 5 is important during transcription initiation; K42A did not affect this modification significantly (Fig. 3C).
Histone H3-K42A alters chromatin structure at actively transcribed genes
Data presented thus far suggest a potential role for H3-K42 in negatively regulating transcription elongation and support the removal of this inhibitory effect by the K42A substitution. Given that nucleosomes harboring a mutation at this site display increased mobility in vitro, we hypothesized that the K42A phenotype may be due to a direct effect on chromatin structure. Figure 2A shows that H3-K42A increases the levels of MET16 expression under inducing conditions specifically, indicating that activated transcription is necessary to reveal the K42A phenotype. Therefore, we hypothesized that K42A-containing nucleosomes give rise to an altered chromatin template that is more permissive to transcription once initiated. We determined the occupancy of histone H3 at genes up-regulated by the H3-K42A mutation by chromatin immunoprecipitation (ChIP), as a decrease in nucleosome density across a gene would promote transcription. Additionally, the generation of 5′ truncated transcripts, as seen in K42A cells, has been accounted for previously by the lack of restoration of a repressive chromatin state in the wake of RNAPII elongation (Carrozza et al. 2005; Joshi and Struhl 2005; Keogh et al. 2005). Figure 3D indicates that there is a significant loss of histone H3 at these loci in K42A cells. Interestingly, in two out of the three genes tested, this difference was evident only at the 3′ end of the ORF. There was no detectable difference in H3 occupancy at genes that are unresponsive to K42A; namely, ACT1, inactivated MET16, and an intergenic noncoding locus in K42A cells (data not shown).
During transcription, nucleosomes are continuously disassembled and reassembled by histone chaperones to facilitate the passage of RNAPII along a gene while maintaining a closed state to prevent aberrant initiation (Schwabish and Struhl 2004; Kulaeva et al. 2007). Spt6p is an essential histone chaperone that binds histone H3 and is important for replacing histone H3/H4 tetramers behind RNAPII. Cells expressing a temperature-sensitive (ts) allele of SPT6, spt6-1004, display phenotypes very similar to that of H3-K42A at semipermissive conditions with respect to transcription up-regulation, decreased nucleosome density in gene coding regions, intragenic transcription, and temperature sensitivity (Kaplan et al. 2003; Cheung et al. 2008). Based on these similarities, we hypothesized that the phenotypes associated with K42A result from defects in the SPT6 pathway, and, accordingly, we expected no exacerbation of the common phenotypes when both alleles are combined. We integrated the spt6-1004 allele into our histone knockout strain background and observed no enhancement of spt6-1004 temperature sensitivity by H3-K42A. Furthermore, mRNA length changes in spt6-1004 cells were not altered in the presence of K42A and at either permissive or semipermissive temperatures (Supplemental Fig. S4).
Histone H3-K42 is dimethylated in S. cerevisiae.
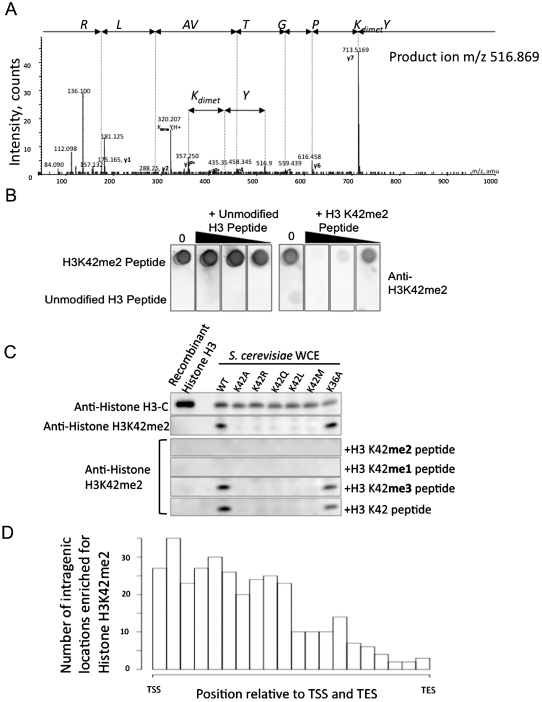
During the course of this work, our ongoing proteomic effort to identify novel post-translational modifications (PTMs) in the globular core of nucleosomes revealed, to our surprise, that histone H3-K42 can be methylated in yeast. We detected both monomethyl and dimethyl forms of K42 using electrospray tandem mass spectrometry in three independent samples (Fig. 4A).
Figure 4.
Histone H3-K42 is dimethylated in S. cerevisiae. (A) Electrospray tandem mass spectrum (MS/MS) of the doubly charged dimethylated peptide: YK(dimet.)PGTVALR, m/z 516.869, recorded using a quadrupole time-of-flight mass spectrometer in an LC-MS/MS experiment (Agilent 1200 series CapLC coupled to QSTAR Pulsar). It was possible to confirm the site of modification using the doubly charged y ions y7 and y8. The spectrum was further confirmed by comparison with the corresponding unmodified peptide, which was also subjected to MS/MS (data not shown). (B) Dot blot analysis of anti-H3-K42me2 serum. The unmodified and K42me2-modified H3 peptides were spotted onto the PVDF membrane and probed with anti-H3-K42me2 serum in the presence of increasing amounts of either the competing modified or unmodified peptide. (C) Western blot analysis. Recombinant Escherichia coli expressed histone H3 or whole-cell extracts (WCE) prepared from yeast expressing wild-type (WT) or various substituted histone H3 were run on a gel and probed with either anti-H3-K42me2 serum or anti-histone H3 C terminus antibody. Peptide competition of the H3-K42me2 band was done in the presence of K42 mono-, di-, or trimethylated synthetic peptides. (D) Genome-wide histone H3-K42me2 ChIP in wild-type yeast was undertaken as described in the Materials and Methods and analyzed on yeast tiling array. The abundance of this modification across a “standard” yeast gene is indicated relative to the transcription start site (TSS) and transcription end site (TES).
Residue 42 is an arginine in all other eukaryotes except Ascomycota (Supplemental Fig. S5), suggesting that, if this lysine is indeed modified, the methylation machinery must be newly evolved. With considerable skepticism, we raised an antibody against a K42 dimethyl-containing peptide. With such an antiserum in hand, experiments to probe for this modification in S. cerevisiae by an independent method became possible. The dot blot in Figure 4B indicates the specificity of this antibody in recognizing the dimethyl-modified histone peptide over the unmodified using competition assays with specific and control unmodified peptides. Immunoblot analyses (Fig. 4C) supported the specificity of the antibody and the existence in vivo of dimethylation of H3-K42 (H3-K42me2). These data illustrate that the serum detects its antigen in both purified histones and whole-cell protein extracts from yeast expressing wild-type histone H3, but fails to detect histone H3 from strains harboring mutations at residue 42 or unmodified bacterially expressed yeast histone H3. Additionally, the modified band in the immunoblot could be competed away by addition of either a monomethyl or dimethylated histone peptide, but not trimethyl or unmodified control peptides.
Histone H3-K42 methylation is enriched at the 5′ end of genes
In our efforts to characterize histone H3-K42me2, we undertook ChIP-on-chip analysis using the methyl-specific antibody to probe for the localization of this modification across the yeast genome. Histone H3-K42 dimethylation was significantly enriched at 491 genome locations distributed seemingly randomly across all 16 S. cerevisiae chromosomes (at a false discovery rate [FDR] threshold of <0.1). The modification was enriched in intragenic regions (65%); however, a substantial proportion was also located in intergenic regions (35%). Figure 4D shows the distribution of H3-K42me2 averaged genome-wide across all genes, indicating enrichment in the 5′ half of genes.
Establishing a genetic system to probe the possible function of histone H3-K42me2
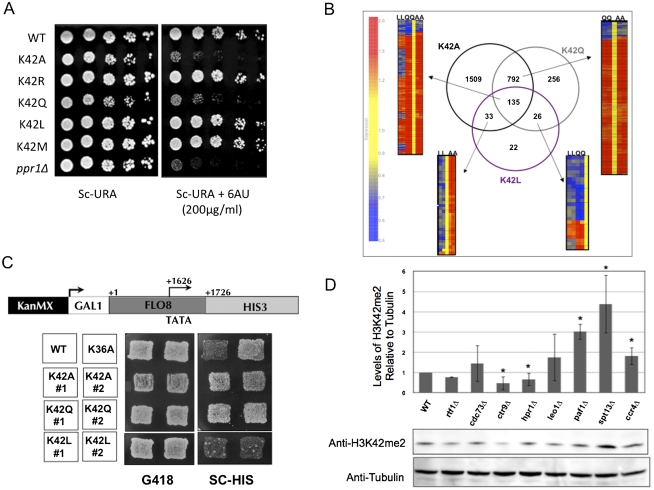
We next sought in vivo evidence for the functionality of this modification in yeast. The H3-K42A mutation eliminates this modification in cells, and therefore the phenotypes associated with this substitution give us some insight into the effects of losing H3-K42me2. However, alanine is not a conservative substitution and, as stated previously, in addition to preventing K42 methylation, it also eliminates potential K42 hydrogen bonding with DNA. We therefore mutated K42 to arginine to mimic a constitutively nonmethylated form of K42 while maintaining hydrogen-bonding potential. Unfortunately, this allele displayed no detectable phenotypes in any assay tested (Fig. 5A; data not shown). We rationalized that perhaps arginine can act a substrate for methylation at this site, supported by the presence of arginine at this position in all other eukaryotes. However, we were unsuccessful in detecting methylation at this site in cells by mass spectrometry. Therefore, we chose to use K42Q as a surrogate for a demethylated version of K42, as it cannot be methylated but is predicted to retain the H bond to DNA (Supplemental Fig. S6).
Figure 5.
Histone H3-K42me2 may play a role in transcription. (A) Strains expressing the indicated histone H3 allele were assayed for their sensitivity to 6AU to detect transcription elongation defects. ppr1Δ acts as a positive control for comparison. (B) A diagram of microarray data showing the overlap of results obtained from the hybridization of total RNA from cells expressing either K42A, K42Q, or K42L histone H3 alleles to the yeast Affymetrix chip. The expression of the genes in common between the microarray experiments are visualized with heat maps based on the clustering using Genespring software. (C) Schematic of the intragenic transcription reporter whereby HIS3 was placed downstream from the cryptic promoter in the FLO8 ORF. Plating assay to detect the ability of strains expressing the indicated histone H3 allele to initiate transcription at the FLO8 cryptic promoter. (D) Western blot analysis detecting H3-K42me2 in whole-cell extracts from strains with the indicated deletions of PAF1 complex components. Tubulin detection served as a loading control. The graph displays the quantification of H3-K42me2 band intensity as compared with tubulin levels; error bars represent the standard deviation between two biological replicates. (*) Statistically significant values.
A complementary approach that is widely used to determine the contribution of a PTM in vivo is to generate a mutation at the site that mimics a constitutively modified state. The closest natural mimics to a methylated lysine are leucine and methionine, which represent monomethyl and dimethyl side chains, respectively. Structures of nucleosomes predicted by energy minimization (Guex and Peitsch 1997) harboring these substitutions are illustrated in Supplemental Figure S6. Although the length of the side chains of the substituted amino acids do not extend as far as lysine, their solvent exposure seems unaltered, and it could be argued that the recognition of their functional groups is similar to that of the modified and unmodified native residues. In particular, the solvent-exposed side chain terminal dimethyl of leucine has a contour strikingly similar to that of dimethyl lysine.
Histone H3-K42me2 may regulate transcription
We generated strains that expressed either H3-K42Q, H3-K42L, or H3-K42M as their sole source of histone H3 and monitored their phenotypes. Initially we determined each strains' sensitivity to 6AU, and Figure 5A shows that our data are consistent with the modification status dictating the sensitivity to this drug. Cells expressing the unmodified mimic K42Q behaved like K42A and were sensitive to the drug, whereas cells expressing the modified mimics K42L and K42M were unaffected by 6AU.
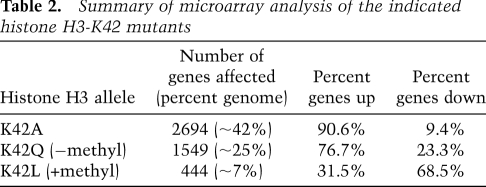
To ask whether the effect of K42A on transcription might be related to H3-K42 modification state, we analyzed K42Q and K42L transcript profiles. Histone H3-K42Q altered expression of ∼25% of the genome and, as in K42A-expressing cells, the majority (∼77%) of these genes were up-regulated. K42L, however, had a more modest phenotype, resulting in dysregulation of only 7% of the genome. Strikingly, however, the major effect of K42L was to down-regulate gene expression (Table 2). We next determined the number and identity of target genes common to the three profiling experiments (Fig. 5B). There was high concordance between genes affected by both K42A and K42Q and the direction of change in expression of these genes. One-hundred-thirty-five genes were affected by all three alleles: K42A, K42Q, and K42L. Whereas the direction of change in these genes was concordant in K42A and K42Q, ∼40% of the genes were oppositely regulated by K42L. Additionally, almost all genes common to the K42A and K42L data sets were differentially regulated by each mutation. Collectively, these data support a model in which K42 methylation is a repressive mark for transcription.
Table 2.
Summary of microarray analysis of the indicated histone H3-K42 mutants
A more direct test of this model is to compare the loci that are enriched by K42 methylation, as determined by our ChIP-on-chip experiment, and those genes whose expression was dysregulated by K42 alleles. Consistent with K42 methylation acting to inhibit transcription, we found significant overlap between genomic regions containing this modification and the loci up-regulated by both K42A (P-value = 1.6 × 10−4) and K42Q (P-value = 3.1 × 10−6) substitutions, both of which mimic a lack of K42me2. Consistently, the relatively few genes that showed decreased expression in the presence of K42A or K42Q did not show significant enrichment of H3-K42me2, consistent with these representing secondary effects. Furthermore, we found that loci down-regulated by the constitutively methylated mimic K42L were significantly enriched for nucleosomes containing H3-K42me2 (P-value = 1.3 × 10−4).
Given our knowledge that H3-K42A causes mRNA length changes, we asked whether other K42 alleles displayed a similar effect. Specifically, we determined the ability of K42 mutations to initiate transcription at a site downstream from the natural start site. We used a reporter in which the HIS3 ORF was driven by the intragenic cryptic promoter in the FLO8 gene (Fig. 5C; Cheung et al. 2008) and assayed the ability of K42 mutants to grow in the absence of histidine. Our results indicate that, similar to K42A, the demethylated H3-K42 mimic K42Q was capable of inducing cryptic promoter–HIS3 expression, whereas K42L, the constitutively methylated mimic, was not—consistent with a potential role for H3-K42me2 in preventing erroneous transcription (Fig. 5C).
Components of the PAF1 complex influence histone H3-K42 dimethylation
The transcript-profiling studies of the different K42 alleles suggested that methylation of this site may be important in the regulation of transcription, and our genetic investigations into the K42A allele implied a role for the modification during transcription elongation. To directly test this hypothesis, we determined whether strains with defects in transcription elongation might signal through H3-K42me2 and perturb the abundance of this modification. We quantified the levels of H3-K42me2 in 87 deletion strains that either lacked a known nonessential transcription elongation factor or were known to be sensitive to 6AU (Supplemental Table S2). Strikingly, the only strains that showed any effect on the levels of H3-K42me2 were those lacking a component of the PAF1 elongation complex. ctr9Δ and hpr1Δ showed statistically significant decreases in H3-K42me2 and rtf1Δ displayed a modest defect in this modification. ccr4Δ, paf1Δ, and spt13Δ displayed elevated levels (up to threefold that of wild type) of this methyl mark (Fig. 5D).
Discussion
Comprehensive histone mutagenesis in yeast has shown that the DNA entry/exit point of the nucleosome does not tolerate substitutions. We identified a group of histone H3 residues in this location that influence transcription, the most critical among them being Lys 42. We showed that the integrity of K42 is essential to maintain the correct transcriptional output in a yeast cell, potentially explaining why certain mutations at this site are pleiotropic. This is a novel site of methylation in yeast: We used a K42-methyl-specific antibody in conjunction with a genetic approach to probe the function and biological significance of this modification. Given its nucleosomal location and the results of our studies, we believe that K42, and possibly its methylation, acts as a transcriptional “gatekeeper” by limiting access to nucleosomal DNA during transcriptional elongation.
The histone H3-K42A substitution results in the abundance of many transcripts, as well as substantial effects on the lengths of many mRNAs. Considering how the elongating RNAPII is influenced by many negative factors, not least the chromatin template, and that it is not uncommon for this massive complex to halt, stall, or become completely arrested (for review, see Shilatifard 2004), we believe that chromatin containing K42A is intrinsically less restrictive to the elongating RNAPII protein complex because the “gate is constitutively open,” promoting more efficient elongation. The two most prominent changes in mRNA length that we detected in K42A cells are those that generate 5′ and 3′ extensions, indicating that the effect of K42A-containing nucleosomes extends beyond canonical transcriptional units. The inability of transcription to be terminated may reflect the permissiveness of K42A chromatin to elongating RNAPII. We hypothesize that, once RNAPII is engaged in transcription, the chromatin landscape in K42A cells is sufficient to promote elongation through normal stop signals even in the presence of transcription-terminating factors. In support of this, our data show that, in K42A cells, we detected an increase in elongation-competent RNAPII, as monitored by increased Ser 2 phosphorylation of the CTD. Furthermore, we found that, by deleting positive elongation factors, the hypertranscription phenotype of K42A was suppressed. For example, DST1 is required to re-engage stalled RNAPII (Christie et al. 1994; Kulish, Struhl 2001); by eliminating DST1, we decreased the number of RNAPII complexes that complete transcripts, thereby masking K42A phenotypes. Similar results were seen for mutants of PAF1, an RNAPII-associated protein required to recruit proelongation factors to actively transcribed genes, including Rad6p (Wood et al. 2003), Dot1p (Krogan et al. 2003), and the FACT complex (for review, see Jaehning 2010). Since our tiling array analysis does not assign strand specificity, ambiguity exists when transcripts span multiple loci and, consequently, ∼30% of 5′ extensions could be classified as 3′ extensions of an upstream locus resulting from hyperelongation (Supplemental Fig. S7). Additionally, our data indicate that K42A cells do not induce expression of repressed genes, illustrating that K42A chromatin does not generically promote ectopic recruitment of initiation complexes genome-wide. These data, taken together with the fact that deleting nonessential initiation factors did not alter K42A phenotypes, suggest that aberrant transcription elongation efficiency specifically underlies K42A-mediated transcriptional phenotypes.
What are the mechanisms behind the “open gate” state of nucleosomes in K42A cells? Our ChIP analyses reveal that there is a loss of histone H3 in K42A cells at certain actively transcribed genes, which is more apparent at the 3′ end of the ORF. We also provided genetic evidence suggesting that both K42A and spt6-1004, a Ts− mutation in the histone chaperone Spt6p, feed into the same pathway and are mechanistically similar. This raises the hypothesis that the histone H3-containing K42A mutation may not be redeposited efficiently in the wake of RNAPII elongation. However, this explanation alone is insufficient to account for the greater number of transcripts affected by K42A cells than in spt6-1004. We speculate that the permissiveness of elongation in K42A cells may arise from inefficient redeposition of histone H3 in conjunction with increased mobility of K42A nucleosomes, as evidenced in vitro.
Our discovery that H3-K42 is methylated in yeast corroborates the importance of this nucleosome residue, providing us with a mechanism by which a cell regulates this transcriptional gatekeeper. The contrasting transcript profiles of methyl (K42L) and nonmethyl (K42Q) mimics of K42 provide indirect evidence to support such a mechanism. Furthermore, we saw a direct effect on methylation levels in mutants with a defective Paf1 complex. This suggests that perturbations to elongation, specifically through the Paf1 complex pathway, influence K42me2 abundance, placing this modification in the Paf1 pathway. The exact mechanism underlying the pattern of H3-K42me2 variations in the individual Paf1 complex subunit deletions is likely to be complex, although it is attractive to speculate that the Paf1 complex is necessary for the recruitment of the elusive K42-specific methyltransferase. In support of this notion, deletion of PAF1 leads to both initiation of transcription at intragenic cryptic promoters (Chu et al. 2007) and 3′ extensions of certain mRNAs (Penheiter et al. 2005; Sheldon et al. 2005), similar to H3-K42A.
Residue 42 in histone H3 is arginine in most other braches of eukaryotes, including Basidiomycetes (Supplemental Fig. S6), indicating that the lysine substitution appeared within the last 400 million years, after Ascomycota and Basidiomycota diverged. Therefore, H3-K42me2 is not a conserved modification, and, strikingly, the ability to methylate this residue is a very recent innovation in evolution, possibly through neo- or subfunctionalization of a conserved enzyme. To explore what protein might have evolved such a function, we searched for a K42-specific methyltransferase in yeast. Unfortunately, to date, our efforts have proved futile, as individual deletion of all 62 known annotated methyltransferases in yeast, including all SET domain proteins and Dot1p, failed to abolish H3-K42me2, as detected by immunoblotting (data not shown). From this analysis, we assume that Ascomycetes have co-opted a noncanonical methyltransferase to methylate H3-K42, or, alternatively, redundant enzymes may modify K42.
Although the prevalence of K42me2 histones throughout the genome is limited, as illustrated by our ChIP-on-chip data, this might indicate that there was a selective advantage to yeast in acquiring this novel modification at those particular sites. It is attractive to interpret the K42A and K42Q mutant data as reflecting the loss of H3-K42me2, and that the ability of the cell to regulate the transcriptional output may be the selective pressure driving the evolution of the modification. In support of this idea, there is a correlation between genes affected by nonmethyl mimics K42A and K42Q and those enriched with the modification. Additionally, these alleles permit the production of intragenic transcripts, whereas the methyl-mimic K42L suppresses it. Although it has been shown that additional nucleosome core mutations promote intragenic transcription, our study illustrates how this phenomenon may be regulated within the cell by PTM. One tempting model might be that H3-K43me2 regulates the interaction between Spt6p and histone H3.
To uncover alternative sources of selective pressure promoting histone H3-K42me2 evolution, we took a bioinformatic approach and looked for differences in specific chromatin-related proteins between the Ascomycota and Basidiomycota phyla, the latter lacking H3-K42. One potentially relevant candidate is the linker histone, H1, which binds DNA at the nucleosome entry and exit points (Thoma et al. 1979), stabilizing the nucleosome and restricting transcription in most eukaryotes. However, S. cerevisiae and other Ascomycetes species lack a canonical H1 protein. Given that histone H3-K42 is positioned in the same nucleosomal region, and our data suggest a role for H3-K42me in transcriptional inhibition, it is plausible that Ascomycota have evolved this modification to serve the function of canonical histone H1. In support of this, our sequence analysis showed that histone H1 divergence largely conforms to phylogeny; H1 sequences from different Basidiomycetes species are more similar than those from Ascomycetes. Similarly, we found that the fungal distribution of a second protein, HP-1, and, presumably, the histone H3-K9 methylation system that generates the repressive chromatin mark that it recognizes follow a similar pattern. Interestingly, in this example, Ascomycetes species lack any detectable homolog of HP-1. These preliminary findings suggest that the lack of canonical histone H1 and/or H3-K9 methylation in Ascomycota could have driven the evolution of a novel repressive chromatin mark, H3-K42me2. Future experiments will be required to directly test this hypothesis.
If we assume that the lack of conservation of this site among higher eukaryotes implies that K42 methylation is a recent acquisition of Ascomycota, we conclude that its function in attenuating transcriptional output provides an evolutionary advantage to these species. We know that, under certain growth conditions, wild-type yeast actively produce cryptic transcripts that play physiological roles (Ono et al. 2005; Bickel and Morris 2006), and that certain intragenic transcripts can be translated into functional proteins, increasing the complexity of the yeast proteome, perhaps providing the cell with additional capabilities and functions. Given that up to 17% of genes within the yeast genome contain intragenic cryptic initiation sites, regulatory mechanisms must be in place to modulate the extent of this form of pervasive transcription, perhaps limiting it temporally or under certain conditions. Methylation of the gate to the globular core of the nucleosome at histone H3-K42 is one such mechanism.
Materials and methods
Strains and media
Standard laboratory techniques for budding yeast manipulations were used. The previously described yeast strain JPY12 (Park et al. 2002) was the parental strain employed for all genetic manipulations unless otherwise noted. A uracil auxotrophy derivative of JPY12, EMHY510, was generated. EMHY510 used in 6AU growth assay was transformed with the URA3 CEN plasmid pRS316. Deletion strains of JPY12 were made using a PCR product amplified from the appropriate BY4742 strain of the KanMX deletion collection. Colony PCR was used to verify the correct integration of the knockout cassette in JPY12. Supplemental Table S3 contains a complete list of strains generated. The Ts− allele of SPT6, spt6-1004, was integrated in EMHY510 by transformation with the SphI-linearized form of the integrating plasmid pCK125 and selecting on synthetic complete (SC)–Ura. Ura+ transformants were streaked to YPD, replica-plated to 5FOA, and subsequently screened for a Ts− phenotype at 39°C. Colony PCR was used to verify the correct targeting of the Ts− alleles.
All media were supplemented with 0.64 mM adenine. SC medium contained 0.67% (w/v) yeast nitrogen base without amino acids (YNB, Difco), 2% (w/v) glucose, and 2% (w/v) bacto-agar, supplemented with 2% (w/v) “Hopkins mix” amino acids. HU medium was prepared by supplementing SC with 200 mM HU. Benomyl was added to YPD-rich medium at a concentration of 30 μg/mL. 6AU-containing medium was SC–Ura containing 200 μg/mL 6AU.
Plasmids
Plasmids pDM18 (Park et al. 2002) and pEMH7 (Hyland et al. 2005) carrying HHT2 and HHF2 and pJP11 encoding HHT1 and HHF1 were used in this study. pEMH17 was generated whereby the NaeI/SacI fragment from pEMH7 was excised and subcloned into pRS315. Histone H3 mutants were engineered into pEMH7, as described previously (Hyland et al. 2005). Plasmids were sequenced with oligonucleotides JB6505 and JB6504. Plasmid pBS4 harboring histone H3-K36A mutation was obtained from B. Strahl. The integrating plasmid pCK125 harboring spt6-1004 was provided by F. Winston.
Growth assays
Yeast strains used in growth assays were grown overnight in either YPD or selective medium where appropriate. The next day, cultures were diluted to an OD A600 of 0.5, and fivefold serial dilutions were plated on both growth control plates and experimental plates. Plates were photographed after 2 d of incubation at 30°C.
Intragenic transcript detection
The FLO8 intragenic reporter was generated by V. Cheng and F. Winston. The reporter construct (4.5 kb) was amplified from strain VC-377-1B using primers JB11738 and JB11739, and was used to transform EMHY510 harboring pBS4 (H3-K36A). Transformants were selected on YPD + G418 and were screened by colony PCR for the correct integration, generating strain EMHY578. This strain was transformed with pEMH17 harboring histone alleles, and pBS4 was shuffled. Transformants were patched onto YPD + G418, incubated for 2 d at 30°C, replica-plated to SC–His, and further incubated for 2 d at 30°C. A His+ phenotype in this assay reports inappropriate intragenic transcription initiation.
Transcriptional profiling; data acquisition and normalization
RNA was extracted from log phase cultures of JPY12 strains expressing histone H3-K42 mutations using the acid phenol method. Ten micrograms of total RNA of two replicate samples was used to create labeled cRNA cocktails, as outlined in the Affymetrix protocol. Fifteen micrograms each of labeled cRNA was hybridized on the Affymetrix GeneChip Yeast 2.0 arrays. Data analysis was conducted on the chips' CEL file probe signal values at the Affymetrix probe pair level using the statistical technique Robust Multiarray Analysis (Irizarry et al. 2003) with the bioconductor package Affy. Contrast normalization was carried out as described in Bolstad et al. (2003) using the following control probes: “Bacillus subtilis,” “Bacteriophage,” “Escherichia coli,” “synthetic insert,” and other non-S. cerevisiae “control” probes, excluding all Schizosaccharomyces pombe and S. cerevisiae “control” probes on the microarray. Between-condition and between-replicate variations were examined with pairwise MvA plots, where the base 2 log ratios (M) between two samples are plotted against their averaged base 2 log signals (A). An empirical Bayes method with lognormal–normal modeling, as implemented in the bioconductor package EBarrays, was used to estimate the posterior probabilities of the differential expression of genes between the sample conditions (Kendziorski et al. 2003; Newton et al. 2001). The criterion of a posterior probability >0.5 was used to produce the differentially expressed gene list. Bioconductor packages are found at http://www.bioconductor.org, and all computations were performed under the R environment (http://www.r-project.org). The raw data were deposited at Gene Expression Omnibus (GEO), accession number GSE13889.
Real-time quantitative PCR
Strains were grown in rich medium supplemented with 0.64 mM adenine and were harvested at log phase. RNA was prepared from these cells and treated with DNase, and 500 ng of this RNA was reverse-transcribed using SuperScript III (Invitrogen). Following RNase H digestion, cDNA equivalent to 25 ng of input RNA was added to a real-time PCR reaction using the Applied Biosystems SYBR Green RT–PCR system. Reactions were run in a 96-well plate using the Applied Biosystems Prism 7900HT fast real-time PCR system. The CT values for expression of gene of interest were compared with that of an internal ACT1 control. The primers used for ACT1 amplification were JB9639 and JB9640 for YBR040W; JB10085 and JB10086 for YNR062C; JB10091 and JB10092 for YKL221W; JB10112 and JB10113 for MCD1; and JB12292 and JB12293.
Analysis of dynamic MET16 transcription
Overnight cultures of the indicated strains were diluted 1:20 in rich medium and grown to log phase. Cells were pelleted at room temperature, washed once in water, and added to either SC–Met or SC prewarmed medium, a procedure that took up to 4 min. Aliquots of cells were taken at regular intervals, added to equal volume of ice-cold 100% ethanol, and centrifuged immediately at 4°C. Cell pellets were frozen on dry ice. RNA was extracted from cells and the MET16 transcript was analyzed by quantitative RT–PCR as above using primers JB12026 and JB12027. ACT1 served as the internal control.
Immunoblotting
Whole-cell lysates were prepared from 2.5 A600 units of cells for SDS-PAGE using the alkaline lysis method as described previously (Kushnirov 2000). For histone analyses, lysates were resolved on 4%–20% SDS polyacrylamide gels and transferred to nitrocellulose. Blots were probed with polyclonal antibodies against the C terminus of histone H3 (1:1000 dilution) (Abcam, ab1971), and histone H3-K42me-specific (1:500 dilution) followed by horseradish peroxidase (HRP)-conjugated antibodies against rabbit IgGs (Amersham Pharmacia) and chemiluminescence detection (Pierce). Polyclonal anti-H3-K42me antibodies, generated by Upstate Biotechnologies, were raised in rabbits upon exposure to the H3 peptide PPEMH2, 39-HRYKme2PGTVAL-48. Antibodies were affinity-purified from the rabbit serum using PPEMH2 and eluted at pH 1.7. Peptide competition of Western blots was undertaken by incubating the H3-K42me2-specific antibody with 0.5 μg/mL appropriate peptide (PPEMH1, 39-HRYKPGTVAL-48; PPEMH2, 39-HRYKme2PGTVAL-48; PPEMH3, 39-HRYKmePGTVAL-48; and PPEMH4, 39-HRYKme3PGTVAL-48 ) for 1 h at room temperature prior to probing the blot. Analysis of the phosphorylation state of the C terminus of RNAPII was undertaken as described previously (Patturajan et al. 1998).
Histone H3 ChIP
The protocol employed was described previously (Dedon et al. 1991) using the histone H3 C-terminal antibody (Abcam, ab1971). Immunoprecipitated DNA was quantitated using quantitative RT–PCR as outlined in the preceding section. The following primer pairs were used: JB10085 and JB10086 for 3′ YBR040W, JB12360 and JB12361 for 5′ YBR040W, JB12450 and JB12451 for 5′ YBR284, JB10114 and JB10115 for 3′ YBR284, JB12452 and JB12453 for 5′ YNR062C, and JB12454 and JB12455 for 3′ YNR062C.
mRNA length change detection
RNA was extracted from log phase cultures of JPY12 strains expressing histone H3-K42 mutations using the acid phenol method. The analysis of genomic tiling array data was carried out by methods described in Poorey et al. (2010). TAS (Tiling Analysis software version 1.1) was used to analyze the raw array data to generate graph files containing log2 (fold change) or differential signals. Differential signals were calculated using the Hodges-Leman estimator, which is associated with the Wicoxon rank-sum test, over a sliding window of 500 bp for treatment versus control. We applied a 0.3 cutoff to the absolute value of log2 (fold change)—i.e., the differential signal data—to identify differentially expressed segments. By comparing these segments to annotated 5′ and 3′ ends, we identified RNA length changes in mutants relative to control. These changes were categorized as 5′ extensions, 5′ truncations, 3′ truncations, or 5′ extensions. Putative length changes that did not satisfy certain criteria—including minimum overlap with the annotations, minimum length change, and minimum signal difference—were filtered out (Poorey et al. 2010).
Acknowledgments
We thank Min-Hao Kuo for communicating results prior to publication. We thank Vanessa Cheung and Fred Winston for providing the intragenic transcription reporter, and Steve Desiderio for the CTR9 haploid deletion strain. We thank Sean Taverna for helpful advice on modification-specific antibody characterization, and Jeffry Corden for providing the RNAPII CTD phosphorylation site-specific antibodies. We are grateful to Pamela Meluh and Melissa Wells for helpful discussions. This work was supported in part by NIH Roadmap grant U54 RR020839 to J.D.B., and NIH grant GM55763 to D.T.A.
Footnotes
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.2050311.
Supplemental material is available for this article.
References
- Bickel KS, Morris DR 2006. Role of the transcription activator Ste12p as a repressor of PRY3 expression. Mol Cell Biol 26: 7901–7912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolstad BM, Irizarry RA, Astrand M, Speed TP 2003. A comparison of normalization methods for high density oligonucleotide array data based on variance and bias. Bioinformatics 19: 185–193 [DOI] [PubMed] [Google Scholar]
- Carrozza MJ, Li B, Florens L, Suganuma T, Swanson SK, Lee KK, Shia WJ, Anderson S, Yates J, Washburn MP, et al. 2005. Histone H3 methylation by Set2 directs deacetylation of coding regions by Rpd3S to suppress spurious intragenic transcription. Cell 123: 581–592 [DOI] [PubMed] [Google Scholar]
- Cheung V, Chua G, Batada NN, Landry CR, Michnick SW, Hughes TR, Winston F 2008. Chromatin- and transcription-related factors repress transcription from within coding regions throughout the saccharomyces cerevisiae genome. PLoS Biol 6: e277 doi: 10.1371/journal.pbio.0060277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christie KR, Awrey DE, Edwards AM, Kane CM 1994. Purified yeast RNA polymerase II reads through intrinsic blocks to elongation in response to the yeast TFIIS analogue, P37. J Biol Chem 269: 936–943 [PubMed] [Google Scholar]
- Chu Y, Simic R, Warner MH, Arndt KM, Prelich G 2007. Regulation of histone modification and cryptic transcription by the Bur1 and Paf1 complexes. EMBO J 26: 4646–4656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clapier CR, Cairns BR 2009. The biology of chromatin remodeling complexes. Annu Rev Biochem 78: 273–304 [DOI] [PubMed] [Google Scholar]
- Dai J, Hyland EM, Yuan DS, Huang H, Bader JS, Boeke JD 2008. Probing nucleosome function: a highly versatile library of synthetic histone H3 and H4 mutants. Cell 134: 1066–1078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dedon PC, Soults JA, Allis CD, Gorovsky MA 1991. A simplified formaldehyde fixation and immunoprecipitation technique for studying protein–DNA interactions. Anal Biochem 197: 83–90 [DOI] [PubMed] [Google Scholar]
- Guex N, Peitsch MC 1997. SWISS-MODEL and the swiss-PdbViewer: an environment for comparative protein modeling. Electrophoresis 18: 2714–2723 [DOI] [PubMed] [Google Scholar]
- Hall MA, Shundrovsky A, Bai L, Fulbright RM, Lis JT, Wang MD 2009. High-resolution dynamic mapping of histone–DNA interactions in a nucleosome. Nat Struct Mol Biol 16: 124–129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartzog GA, Speer JL, Lindstrom DL 2002. Transcript elongation on a nucleoprotein template. Biochim Biophys Acta 1577: 276–286 [DOI] [PubMed] [Google Scholar]
- Hyland EM, Cosgrove MS, Molina H, Wang D, Pandey A, Cottee RJ, Boeke JD 2005. Insights into the role of histone H3 and histone H4 core modifiable residues in Saccharomyces cerevisiae. Mol Cell Biol 25: 10060–10070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irizarry RA, Hobbs B, Collin F, Beazer-Barclay YD, Antonellis KJ, Scherf U, Speed TP 2003. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics 4: 249–264 [DOI] [PubMed] [Google Scholar]
- Jaehning JA 2010. The Paf1 complex: platform or player in RNA polymerase II transcription? Biochim Biophys Acta 1799: 379–388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joshi AA, Struhl K 2005. Eaf3 chromodomain interaction with methylated H3-K36 links histone deacetylation to pol II elongation. Mol Cell 20: 971–978 [DOI] [PubMed] [Google Scholar]
- Kaplan CD, Laprade L, Winston F 2003. Transcription elongation factors repress transcription initiation from cryptic sites. Science 301: 1096–1099 [DOI] [PubMed] [Google Scholar]
- Kendziorski CM, Newton MA, Lan H, Gould MN 2003. On parametric empirical bayes methods for comparing multiple groups using replicated gene expression profiles. Stat Med 22: 3899–3914 [DOI] [PubMed] [Google Scholar]
- Keogh MC, Kurdistani SK, Morris SA, Ahn SH, Podolny V, Collins SR, Schuldiner M, Chin K, Punna T, Thompson NJ, et al. 2005. Cotranscriptional set2 methylation of histone H3 lysine 36 recruits a repressive Rpd3 complex. Cell 123: 593–605 [DOI] [PubMed] [Google Scholar]
- Krogan NJ, Dover J, Wood A, Schneider J, Heidt J, Boateng MA, Dean K, Ryan OW, Golshani A, Johnston M, et al. 2003. The Paf1 complex is required for histone H3 methylation by COMPASS and Dot1p: linking transcriptional elongation to histone methylation. Mol Cell 11: 721–729 [DOI] [PubMed] [Google Scholar]
- Kruger W, Peterson CL, Sil A, Coburn C, Arents G, Moudrianakis EN, Herskowitz I 1995. Amino acid substitutions in the structured domains of histones H3 and H4 partially relieve the requirement of the yeast SWI/SNF complex for transcription. Genes Dev 9: 2770–2779 [DOI] [PubMed] [Google Scholar]
- Kulaeva OI, Gaykalova DA, Studitsky VM 2007. Transcription through chromatin by RNA polymerase II: histone displacement and exchange. Mutat Res 618: 116–129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kulish D, Struhl K 2001. TFIIS enhances transcriptional elongation through an artificial arrest site in vivo. Mol Cell Biol 21: 4162–4168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurumizaka H, Wolffe AP 1997. Sin mutations of histone H3: influence on nucleosome core structure and function. Mol Cell Biol 17: 6953–6969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kushnirov VV 2000. Rapid and reliable protein extraction from yeast. Yeast 16: 857–860 [DOI] [PubMed] [Google Scholar]
- Li B, Carey M, Workman JL 2007. The role of chromatin during transcription. Cell 128: 707–719 [DOI] [PubMed] [Google Scholar]
- Luger K, Richmond TJ 1998. DNA binding within the nucleosome core. Curr Opin Struct Biol 8: 33–40 [DOI] [PubMed] [Google Scholar]
- Luger K, Mader AW, Richmond RK, Sargent DF, Richmond TJ 1997. Crystal structure of the nucleosome core particle at 2.8 A resolution. Nature 389: 251–260 [DOI] [PubMed] [Google Scholar]
- Luo J, Xu X, Hall H, Hyland EM, Boeke JD, Hazbun T, Kuo MH 2010. Histone h3 exerts a key function in mitotic checkpoint control. Mol Cell Biol 30: 537–549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakanishi S, Sanderson BW, Delventhal KM, Bradford WD, Staehling-Hampton K, Shilatifard A 2008. A comprehensive library of histone mutants identifies nucleosomal residues required for H3K4 methylation. Nat Struct Mol Biol 15: 881–888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newton MA, Kendziorski CM, Richmond CS, Blattner FR, Tsui KW 2001. On differential variability of expression ratios: improving statistical inference about gene expression changes from microarray data. J Comput Biol 8: 37–52 [DOI] [PubMed] [Google Scholar]
- Ono B, Futase T, Honda W, Yoshida R, Nakano K, Yamamoto T, Nakajima E, Noskov VN, Negishi K, Chen B, et al. 2005. The Saccharomyces cerevisiae ESU1 gene, which is responsible for enhancement of termination suppression, corresponds to the 3′-terminal half of GAL11. Yeast 22: 895–906 [DOI] [PubMed] [Google Scholar]
- Park JH, Cosgrove MS, Youngman E, Wolberger C, Boeke JD 2002. A core nucleosome surface crucial for transcriptional silencing. Nat Genet 32: 273–279 [DOI] [PubMed] [Google Scholar]
- Patturajan M, Schulte RJ, Sefton BM, Berezney R, Vincent M, Bensaude O, Warren SL, Corden JL 1998. Growth-related changes in phosphorylation of yeast RNA polymerase II. J Biol Chem 273: 4689–4694 [DOI] [PubMed] [Google Scholar]
- Penheiter KL, Washburn TM, Porter SE, Hoffman MG, Jaehning JA 2005. A posttranscriptional role for the yeast Paf1–RNA polymerase II complex is revealed by identification of primary targets. Mol Cell 20: 213–223 [DOI] [PubMed] [Google Scholar]
- Poorey K, Sprouse RO, Wells MN, Viswanathan R, Bekiranov S, Auble DT 2010. RNA synthesis precision is regulated by preinitiation complex turnover. Genome Res 20: 1679–1688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Recht J, Osley MA 1999. Mutations in both the structured domain and N-terminus of histone H2B bypass the requirement for swi–snf in yeast. EMBO J 18: 229–240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwabish MA, Struhl K 2004. Evidence for eviction and rapid deposition of histones upon transcriptional elongation by RNA polymerase II. Mol Cell Biol 24: 10111–10117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheldon KE, Mauger DM, Arndt KM 2005. A requirement for the Saccharomyces cerevisiae Paf1 complex in snoRNA 3′ end formation. Mol Cell 20: 225–236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shilatifard A 2004. Transcriptional elongation control by RNA polymerase II: a new frontier. Biochim Biophys Acta 1677: 79–86 [DOI] [PubMed] [Google Scholar]
- Somers J, Owen-Hughes T 2009. Mutations to the histone H3 α N region selectively alter the outcome of ATP-dependent nucleosome-remodelling reactions. Nucleic Acids Res 37: 2504–2513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thoma F, Koller T, Klug A 1979. Involvement of histone H1 in the organization of the nucleosome and of the salt-dependent superstructures of chromatin. J Cell Biol 83: 403–427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe S, Resch M, Lilyestrom W, Clark N, Hansen JC, Peterson C, Luger K 2010. Structural characterization of H3K56Q nucleosomes and nucleosomal arrays. Biochim Biophys Acta 1799: 480–486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- White CL, Suto RK, Luger K 2001. Structure of the yeast nucleosome core particle reveals fundamental changes in internucleosome interactions. EMBO J 20: 5207–5218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood A, Schneider J, Dover J, Johnston M, Shilatifard A 2003. The Paf1 complex is essential for histone monoubiquitination by the Rad6–Bre1 complex, which signals for histone methylation by COMPASS and Dot1p. J Biol Chem 278: 34739–34742 [DOI] [PubMed] [Google Scholar]