Abstract
Despite major advances in characterizing the eukaryotic transcriptional machinery, the function of promoter-specific transcriptional activators (activators) is still not understood. For example, in no case have the direct in vivo targets of a transcriptional activator been unambiguously identified, nor has it been resolved whether activators have a single essential target or multiple redundant targets. Here we address these issues for the prototype acidic activator yeast Gal4p. Gal4p binds to the upstream activating sequence (UAS) of GAL1 and several other GAL genes and stimulates transcription in the presence of galactose. Previous studies have shown that GAL1 transcription is dependent on the yeast SAGA (Spt/Ada/GCN5/acetyltransferase) complex. Using formaldehyde-based in vivo cross-linking, we show that the Gal4p activation domain recruits SAGA to the GAL1 UAS. If SAGA is not recruited to the UAS, the preinitiation complex (PIC) fails to assemble at the GAL1 core promoter, and transcription does not occur. SAGA, but not other transcription components, is also recruited by the Gal4p activation domain to a plasmid containing minimal Gal4p-binding sites. Recruitment of SAGA by Gal4p and stimulation of PIC assembly is dependent on several SAGA subunits but not the SAGA histone acetyl-transferase (HAT) GCN5. Based on these and other results, we conclude that SAGA is an essential target of Gal4p that, following recruitment to the UAS, facilitates PIC assembly and transcription.
Keywords: Transcription, GAL1, SAGA, Gal4p, TAF, TBP
Transcription of eukaryotic structural genes often involves a cis-acting element that functions at a distance from the core promoter, termed an enhancer in mammalian cells (McKnight and Tjian 1986) and a UAS (upstream activating sequences) in yeast (Stargell and Struhl 1996). Enhancers and UASs provide binding sites for transcriptional activator proteins (activators), which contain two functional domains: one that directs DNA binding and another that activates transcription (Ptashne 1988). A central question in the field is how different promoter-specific activators function through a common set of general transcription factors (GTFs) that assemble on the core promoter to form the preinitiation complex (PIC). Although it is clear that activators function, at least in part, by stimulating PIC assembly (Orphanides et al. 1996; Roeder 1996; Ptashne and Gann 1997; Lee and Young 2000), the detailed mechanism by which this occurs has not been clearly elucidated.
Activator-mediated stimulation of PIC assembly is believed to result from a direct interaction between the activation domain and one or more components of the transcription machinery, termed the target. Based primarily on in vitro protein-protein interaction experiments, a variety of factors have been proposed to be the direct targets of activators (Orphanides et al. 1996; Ptashne and Gann 1997; Zaman et al. 1998). However, whether any of these are bona fide in vivo targets required for stimulation of PIC assembly and transcription remains to be determined. An important prediction is that a true target may be directly recruited to the promoter by the activator in vivo and in a manner not dependent on other transcription components.
The well-characterized acidic activator Gal4p is responsible for the transcriptional induction of GAL genes, such as GAL1, which contain Gal4p-binding sites in their promoters (Johnston 1987; Johnston and Carlson 1992; Dudley et al. 1999). In this study, we use formaldehyde-based in vivo cross-linking (Orlando and Paro 1993; Strahl-Bolsinger et al. 1997) in conjunction with transcriptional, mutational, and kinetic analyses to identify the target of Gal4p.
Results
Recruitment of SAGA to the GAL1 UAS
Yeast SAGA is a 1.8-MD complex, which contains ≥14 subunits: Ada1p, Ada2p, Ada3p, Ada5p/Spt20p, Spt3p, Spt7p, Spt8p, Gcn5p, Tra1p, TAFII17, TAFII25, TAFII60, TAFII68, and TAFII90 (Hampsey 1997; Grant et al. 1998; Brown et al. 2000). Several of these subunits (e.g., Spt3p, Spt7p, Spt8p, and Spt20p) are present exclusively in SAGA and thus can be used to study the intact complex. Because previous studies have shown that transcription of GAL1 (Roberts and Winston 1997; Dudley et al. 1999; Sterner et al. 1999) as well as other genes (Horiuchi et al. 1997) is dependent on SAGA, we analyzed recruitment of SAGA to the GAL1 promoter using a formaldehyde-based in vivo cross-linking/immunoprecipitation assay. For these experiments, we used either a c-myc mouse monoclonal antibody and yeast strains expressing a C-terminal epitope-tagged SAGA subunit (Spt3p, Spt20p, Gcn5p, or Ada2p) or polyclonal antibodies to various SAGA TAFIIs. Two sets of promoter-specific primer-pairs were used that, as shown below, could distinguish binding to the GAL1 UAS or core promoter.
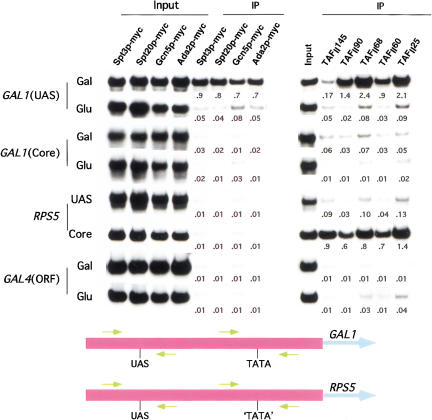
Figure 1 shows that under conditions in which the Gal4p activation domain was active (galactose media), the SAGA subunits Spt3p, Spt20p, Gcn5p, and Ada2p were present at the GAL1 UAS but not the core promoter. Significantly, these same SAGA subunits were not associated with the GAL1 UAS when the Gal4p activation domain was inactive (glucose [Fig. 1] or raffinose [see below] media). In addition, SAGA was not associated with an irrelevant DNA sequence (GAL4 ORF) or with the UAS or core of the RPS5 promoter, the transcription of which does not require SAGA (Lee et al. 2000; see below).
Figure 1.
Recruitment of SAGA to the GAL1 upstream activating sequence (UAS). Yeast strains were grown at 30°C in 1% yeast extract containing 2% peptone plus 2% glucose or galactose as indicated. Formaldehyde-based in vivo cross-linking/immunoprecipitation was performed as previously described (Li et al. 2000). Primer-pairs located in the UAS or core promoter of GAL1 and RPS5 (see Materials and Methods) were used for PCR analysis of the immunoprecipitated DNA samples. A PCR fragment corresponding to the GAL4 ORF was used as a control for background binding. Immunoprecipitation was performed using a mouse monoclonal antibody against the c-myc epitope-tag (9E10; Santa Cruz) or a polyclonal antibody against the indicated TAFII. The promoter, primer-pair, and media are indicated on the left. The ratio of immunoprecipite over the input is indicated below each band. Ratios ≤0.01 are designated .01. IP, immunoprecipitate.
We also analyzed the association of the TAFIIs with the GAL1 promoter. Previous studies have shown that TAFIIs are not required for GAL1 transcription and that TBP binds the TATA box in a form not associated with TAFIIs (Li et al. 2000). Figure 1 shows that following growth in galactose, the SAGA TAFIIs (TAFII90, TAFII68, TAFII60, and TAFII25), like the other SAGA components, were associated with the GAL1 UAS but not the core promoter. TAFII145, a component of TFIID but not SAGA, was not associated with the GAL1 UAS. These results indicate that the TAFIIs are recruited to the GAL1 UAS as part of the SAGA complex. In contrast, on the RPS5 promoter, the transcription of which requires TAFIIs (Shen and Green, 1997; Li et al. 2000) but not SAGA, the TFIID-specific TAFII145 and the other TAFIIs are associated with the core promoter but not the UAS. These results indicate that TFIID is recruited to the RPS5 core promoter during PIC assembly.
Gal4p mediates SAGA recruitment
Figure 1 indicates that in the presence of galactose, SAGA was specifically recruited to the UAS, which harbors the Gal4p-binding sites. These results strongly suggest that recruitment of SAGA to the UAS requires Gal4p. To confirm this supposition, we performed in vivo cross-linking in strains lacking Gal4p. Figure 2A shows that in a GAL4 deletion strain, SAGA was not recruited to the UAS.
Figure 2.
Gal4p mediates recruitment of SAGA to the GAL1 upstream activating sequence (UAS). Wild-type and GAL4 deletion strains were first grown in glucose-containing (YPD) and then shifted to galactose-containing (YPG) media 5 h before treatment with formaldehyde. Primer-pairs located in the (A) GAL1 UAS or (B) core promoter were used for PCR analysis of the immunoprecipitated DNA samples. Immunoprecipitations were performed using polyclonal antibodies against a TAFII or TBP, the mouse monoclonal antibody 9E10 (Santa Cruz) against the c-myc epitope-tag, or the mouse monoclonal antibody 8WG16 (Covance) against the CTD domain of the RNA polymerase II large subunit (RPB1).
We have shown previously that in the presence of galactose, Gal4p stimulates PIC assembly as evidenced by recruitment of GTFs and RNA polymerase II to the core promoter (Li et al. 1999, 2000). Consistent with these previous results, in a GAL4 deletion strain, TBP and RNA polymerase II were not recruited to the core promoter (Fig. 2B).
Role of specific SAGA subunits
We next analyzed various yeast deletion strains to determine the role of individual SAGA subunits in SAGA recruitment, PIC assembly, and transcription. In yeast strains lacking SPT20, SAGA was not recruited to the GAL1 UAS (Fig. 3A), and PIC assembly (Fig. 3B) and transcription (Fig. 3C) were reduced substantially. Thus, Spt20p is required for recruitment of SAGA to the GAL1 promoter, consistent with previous studies showing that Spt20p is required for integrity of the SAGA complex (Grant et al. 1997; Sterner et al. 1999). Figure 3D shows that strains lacking SPT20 were also defective for transcription of the Gal4p-dependent GAL2, GAL7, and GAL10 genes. Thus, the requirement for SAGA is not specific to GAL1 but rather is general to Gal4p-dependent genes.
Figure 3.
Requirement of SAGA subunits for GAL1 complex assembly and transcription. Wild-type and SAGA subunit deletion mutants were grown as in Fig. 2 before treatment with formaldehyde. Primer-pairs located in the (A) GAL1 upstream activating sequence (UAS) or (B) core promoter were used for PCR analysis of the immunoprecipitated DNA samples. (C) Transcription. Total cellular RNA was prepared from the wild type or deletion mutants (top), and transcription from the indicated gene (left) was quantitated by primer-extension. The percentage transcription relative to wild type is indicated below. (D) Other Gal4p-dependent genes (as in panel C). (E) Requirement of SAGA following artificial activation of Gal4p (as in panel C).
In strains lacking SPT3, recruitment of SAGA to the UAS was reduced only modestly (Fig. 3A). Significantly, however, PIC assembly (Fig. 3B) and transcription (Fig. 3C) were reduced substantially. Thus, Spt3p appears to function by facilitating PIC assembly following recruitment of SAGA to the UAS.
Finally, in strains lacking GCN5, SAGA recruitment, PIC assembly, and transcription occurred at near wild-type levels (Fig. 3A–C). Collectively, these data reveal that individual SAGA components are differentially required for SAGA recruitment, PIC assembly, and transcription.
Requirement of SAGA following GAL80 deletion
The Gal4p activation domain is controlled through binding of the negative regulator Gal80p. In the presence of galactose, Gal4p is activated through a complex pathway that ultimately counteracts Gal80p (Johnston 1987; Johnston and Carlson 1992; Ostergaard et al. 2001). It was therefore possible that the inability of Gal4p to activate transcription in SAGA deletion strains was caused by an inability to counteract Gal80p and not by the absence of an essential target of the Gal4p activation domain.
To distinguish between these possibilities, we artificially activated Gal4p by deleting the gene encoding the negative regulator Gal80p and analyzed the requirement for SAGA (Fig. 3E). As expected, in raffinose media GAL1 was transcribed at high levels in a GAL80 deletion but not a wild-type strain. Significantly, inactivation of SAGA by deletion of SPT20 again dramatically reduced GAL1 transcription, even under these artificial activation conditions. As expected, transcription of TUB2 and RPS5, which do not require SAGA (Dudley et al. 1999; Lee et al. 2000), was unaffected by deletion of SPT20 (or GAL80). Thus, the requirement of SAGA for GAL1 transcription is at a step subsequent to establishment of a functional Gal4p activation domain.
Recruitment of SAGA to minimal Gal4p-binding sites
The above experiments indicate that in the presence of active Gal4p, SAGA is recruited to the UAS, the PIC assembles at the core promoter, and transcription occurs. These observations raise the possibility that SAGA is a direct target of the Gal4p activation domain. However, because these experiments were performed with the intact GAL1 promoter under conditions permissive for transcription, it remained possible that other transcription components, such as GTFs, played a role in SAGA recruitment to the UAS. To address this issue, we asked whether SAGA could be recruited by Gal4p to a plasmid bearing only Gal4p-binding sites and not other promoter elements. Figure 4A shows that similar to the results with the GAL1 UAS, SAGA was recruited to a plasmid bearing Gal4p-binding sites in galactose but not glucose media.
Figure 4.
Recruitment of SAGA to minimal Gal4p-binding sites. (A) Gal4p recruits SAGA but not other transcription factors to minimal Gal4p-binding sites. Yeast strains were grown, and in vivo cross-linking analysis was performed as described in Fig. 1. Immunoprecipitation was performed using polyclonal antibodies against the indicated TAFII or TBP, or a mouse monoclonal antibody against the c-myc epitope-tag, HA epitope-tag, or TFIIB. The primers used for the PCR analysis are adjacent to the Gal4p-binding sites in the plasmid. (B) Dependence of SAGA recruitment on the Gal4p activation domain. As in panel A except strains were used that expressed the Gal4p derivative indicated on the left. (C) In vitro interaction between the Gal4p activation domain and SAGA. Whole-cell extracts were prepared from a wild-type strain or an SPT20 deletion mutant and incubated with immobilized GST or GST-34 in buffer A for 30 min at 4°C. The eluate was analyzed by immunoblotting using the c-myc mouse monoclonal antibody against Spt3p-myc. The position of Spt3p-myc is indicated.
The requirement of galactose for SAGA recruitment, suggested an essential role for the Gal4p activation domain. Consistent with this idea, Figure 4B shows that SAGA was not recruited by Gal4p(1–147), which contains an intact DNA-binding domain but lacks the activation domain. In vivo cross-linking indicated that Gal4p(1–147) bound to the Gal4p-binding sites efficiently; in fact, the level of Gal4p(1–147) binding was actually higher than that of Gal4p. Thus, the inability of Gal4p(1–147) to recruit SAGA was not because of a failure to bind DNA.
Previous studies have shown that residues 840–881 of Gal4p comprise a functional galactose-dependent transcriptional activation domain (Wu et al. 1996). Figure 4B shows that addition of this 41 amino acid activation domain to a minimal Gal4p DNA-binding domain, Gal4p(1–100), also resulted in SAGA recruitment. Thus, SAGA is also targeted by the Gal4p 41 amino acid C-terminal activation domain.
Finally, we performed an in vitro protein affinity-chromatography experiment to detect interactions between the Gal4p activation domain and SAGA. Figure 4C shows that in a yeast whole-cell extract, SAGA bound to a glutathione-S-transferase (GST) fusion-protein containing a 34-residue Gal4p activation domain (841–875) but not to a control GST protein. Consistent with the in vivo cross-linking data of Figure 3A, the Gal4p activation domain–SAGA interaction did not occur in a yeast whole-cell extract prepared from an SPT20 deletion mutant.
We also used the in vivo cross-linking assay to look for interactions between Gal4p and several other transcription components, some of which are proposed Gal4p targets on the basis of in vitro protein-protein interaction experiments (Melcher and Johnston 1995; Wu et al. 1996; Ansari et al. 1998; Koh et al. 1998; Xie et al. 2000a,b). Figure 4A shows that although SAGA was recruited by Gal4p relatively efficiently (cross-linking signal within twofold of Gal4p itself), cross-linking of TFIID components (TAFII145 and TBP), TFIIB, RNA polymerase II, and Srb4p were substantially lower, which in some cases approximated the background level of the assay. Although these data do not rule out these other factors as possible Gal4p targets, the low level of cross-linking argues that these other factors are not required to mediate (in particular bridge) the Gal4p–SAGA interaction. Collectively, these data indicate that SAGA is a direct target of the Gal4p activation domain.
A stepwise pathway of transcription complex assembly on the GAL1 promoter
Next, we analyzed the kinetics of Gal4p and SAGA recruitment to the UAS and several GTFs to the core promoter. The analysis was performed in three carbon sources: glucose (repressing and noninducing), raffinose (noninducing), and galactose (inducing). Consistent with previous studies (Johnston 1987; Johnston and Carlson 1992; Lohr et al. 1995) and the results presented above, Figure 5A shows that in glucose, Gal4p was bound at a low level, SAGA was not associated with the UAS, and GTFs were not associated with the core promoter. In raffinose, Gal4p was bound to the UAS, but neither SAGA (UAS) nor GTFs (core) were recruited. Finally, in galactose, in which GAL1 is transcriptionally active, Gal4p and SAGA were associated with the UAS, and the GTFs were associated with the core promoter.
Figure 5.
Kinetic analysis of transcription complex assembly on the GAL1 promoter. (A) Association of Gal4p, SAGA, and GTFs with the GAL1 promoter in different carbon sources. The carbon source is indicated on the left and association of transcription factors (top) is given with the upstream activating sequence (UAS) or core region of the GAL1 promoter analyzed by formaldehyde-mediated cross-linking/immunoprecipitation. (B) Kinetics of Gal4p binding on switch from glucose to raffinose. Cells were grown in glucose and shifted to raffinose for the times (includes 15 min cross-linking) indicated on the left. Binding of Gal4p to the GAL1 UAS was analyzed by formaldehyde-mediated cross-linking/immunoprecipitation. (C) Kinetics of complex assembly on switch from raffinose to galactose. Cells were grown in raffinose and shifted to galactose for the times indicated on the left. Association of the indicated transcription factor (top) with the UAS or core region of the GAL1 promoter was analyzed by formaldehyde-mediated cross-linking/ immunoprecipitation. (D) Kinetics of complex assembly on switch from glucose to galactose. As in panel C except that cells were grown in glucose and shifted to galactose. (E) Plot of TBP and RNA polymerase II association with the GAL1 core promoter upon switch from glucose to galactose. The data from an independent experiment (inset) was quantitated and plotted. Each point was normalized to the maximum cross-linking signal (assigned value of 100%).
Figure 5B shows that on switch from glucose to raffinose media, Gal4p binding was first detectable in ∼2 h, reflecting the time required for relief of glucose repression (Johnston 1987; Johnston and Carlson 1992; Lohr et al. 1995; Ostergaard et al. 2001). Figure 5C shows that on switch from raffinose to galactose media, recruitment of SAGA to the UAS was first detectable between 30 and 45 min, and concomitantly, binding of TBP to the TATA box became evident. Shortly thereafter (60 min), recruitment of RNA polymerase II to the core promoter was first detectable. Figure 5D shows that on switch from glucose to galactose, association of Gal4p was first detectable at 90 min, SAGA and TBP at 130 min, and RNA polymerase II at 140 min. Figure 5E plots the kinetics of TBP and RNA polymerase II association with the GAL1 core promoter following shift from glucose to galactose and again demonstrates entry of TBP shortly before RNA polymerase II. Collectively, the data of Figure 5 reveal a kinetic pathway of complex assembly on the GAL1 promoter initiated by binding of Gal4p, followed by recruitment of SAGA, the apparent simultaneous association of TBP, and finally entry of RNA polymerase II.
To determine whether this kinetic pathway reflects a series of obligatory steps, we analyzed complex assembly in yeast strains bearing mutations in Gal4p, SAGA, TBP, or RNA polymerase II. As described above, strains deleted of GAL4 failed to recruit SAGA to the UAS (Fig. 2A) and GTFs to the core promoter (Fig. 2B). In the SPT20 deletion strain, Gal4p binding was normal (Fig. 6A), but SAGA was not recruited to the UAS (Fig. 3A) and PIC assembly did not occur (Fig. 3B). Temperature-sensitive inactivation of TBP did not interfere with recruitment of Gal4p and SAGA to the UAS (Fig. 6A,B), but the PIC failed to assemble (Fig. 6C). Finally, temperature-sensitive inactivation of RNA polymerase II did not interfere with recruitment of Gal4p or SAGA to the UAS (Fig. 6A,B) and had only a modest effect on recruitment of TBP to the TATA box (Fig. 6C). Thus, the kinetic assembly pathway reflects a stepwise series of obligatory interactions shown schematically in Figure 7 and discussed below.
Figure 6.
A stepwise pathway of transcription complex assembly on the GAL1 promoter. Deletion strains and their isogenic wild-type counterparts were first grown in glucose, switched to galactose for 5 h, and treated with formaldehyde. Temperature-sensitive mutant stains and their isogenic wild-type versions were grown in galactose to an OD600 of 0.85 at 23°C, switched to 37°C for 1 h, and treated with formaldehyde. Association of Gal4p (A) and SAGA (TAFII68; B) with the GAL1 upstream activating sequence (UAS) or TBP and RNA polymerase II (C) with the GAL1 core promoter was analyzed by formaldehyde-mediated cross-linking/immunoprecipitation.
Figure 7.
Summary of transcription complex assembly on the GAL1 promoter. The entry of SAGA and TBP were not kinetically resolvable, but the results of mutational experiments (Fig. 6) indicate that SAGA can associate with GAL1 in the absence of TBP. Although not analyzed here, it has been shown previously that other GTFs are associated with the transcriptionally active GAL1 promoter (Li et al. 1999, 2000). The model shows the other general transcription factors (GTFs) entering subsequent to TBP, but it is possible that some GTFs may be recruited to the promoter simultaneously with TBP (see Li et al. 1999, 2000).
Discussion
The major conclusion of this present study and the study of Larschan and Winston (2001) is that SAGA is an essential and likely direct target of the prototype acidic activator Gal4p. This conclusion is supported by several lines of evidence. First, SAGA is required for Gal4p to stimulate PIC assembly and transcription, indicating that SAGA performs an essential and nonredundant function. Second, in vivo Gal4p recruits SAGA to the GAL1 UAS as well as to minimal Gal4p-binding sites. Recruitment of SAGA is dependent on a functional Gal4p activation domain. In this same assay, other transcription components, including some previously proposed Gal4p targets, were not recruited by Gal4p, strongly suggesting that they are not involved in SAGA recruitment. Finally, the proposed pathway of transcription complex assembly on GAL1 (Fig. 7), based on kinetic and mutational analyses, is also consistent with the direct targeting of SAGA by Gal4p.
Our work suggests a model in which Gal4p first recruits SAGA to the UAS, and then UAS-bound SAGA facilitates PIC assembly and thus transcription (Fig. 7). The most likely mechanism by which UAS-bound SAGA functions is by serving as an adaptor that directly contacts one or more components of the PIC. This model fits in very well with several other studies. For example, it has been previously shown that SAGA is required for GAL1 transcription (Roberts and Winston 1997; Dudley et al. 1999; Sterner et al. 1999), acts following Gal4p binding (Dudley et al. 1999), and facilitates the TBP–TATA box interaction (Dudley et al. 1999).
The detailed mechanism by which UAS-bound SAGA interacts with the transcriptional machinery and stimulates PIC assembly remains to be determined. However, our results suggest that the SAGA subunit Spt3p may be involved: Spt3p was dispensable for SAGA recruitment but required for Gal4p-mediated PIC assembly (Fig. 3). Significantly, previous studies have provided genetic and physical evidence for interactions between Spt3p and TBP (Eisenmann et al. 1992, 1994). Thus, an attractive idea would be that UAS-bound SAGA facilitates PIC assembly by interacting with TBP through Spt3p. The manuscript by Larschan and Winston (2001) also provides strong, independent support for this possibility.
Several experimental observations have suggested that activators, in particular acidic activators, have multiple, functionally redundant targets. For example, in vitro protein-protein interaction experiments have shown that a single activator can interact with multiple transcription components (Struhl 1996; Ptashne and Gann 1997). Moreover, in some instances a single activator can stimulate transcription synergistically on a promoter bearing multiple activator-binding sites (see Carey et al. 1990). The interpretation of this synergistic effect has been that each of the bound activators contacts a different component of the PIC.
An important conclusion of our studies is that SAGA is a nonredundant target of Gal4p. It is possible that Gal4p is atypical and that most activators will, as generally believed, have multiple, redundant targets. However, the previous experimental observations suggesting target redundancy require re-examination. For example, although an activator might interact in vitro with more than one transcription component, these interactions may not be involved in transcriptional activation in vivo. With regard to transcriptional synergy, not all activators can synergize with themselves (see Davidson et al. 1988; Fromental et al. 1988), suggesting distinct classes of activators, which likely function through different targets. Even for activators that can synergize with themselves, the activator could interact with a single target, such as SAGA, which in turn makes multiple contacts with the PIC. According to this idea, it is the adaptor, rather than the activator, that has redundant targets.
In several instances, a HAT-containing component or complex has been shown to be required for transcription of or recruited to a particular gene (e.g., p300; Sterner and Berger 2000; Roth et al. 2001). This result has in general been interpreted as a requirement for the HAT activity. Here we find that a HAT-containing complex, SAGA, is recruited to the GAL1 promoter and required for transcription. Because SAGA is a multi-subunit complex, we have been able to show that the HAT activity is dispensable for recruitment to the promoter, stimulation of PIC assembly, and transcription, consistent with previous results (Dudley et al. 1999). Our results raise the possibility that in these other instances, the major function of the HAT-containing component or complex may not as generally believed be to provide a HAT activity.
Several of the critical SAGA subunits (e.g., Spt3p, Spt20p) are not essential for yeast viability (Roberts and Winston 1996), and whole-genome expression analysis indicates that SAGA is required for expression of only a small subset of yeast genes (Lee et al. 2000). Therefore, the activator targets for the vast majority of yeast genes must be components other than SAGA. The experimental approaches described here may be generally useful to identify the targets of other activators.
Materials and methods
Plasmids
Plasmids pRJR182 (Gal4p 1–100+840–881; Wu et al. 1996), pRJR217 (Gal4p 1–100; Wu et al. 1996), and pMA241 (Gal4p 1–147; Ma and Ptashne 1987) were obtained from Mark Ptashne (Memorial Sloan Kettering Cancer Center, NY). Plasmids pGEX2T (GST) and pGEXCSNGal4AD (GST-34) (Melcher and Johnston 1995) were obtained from Stephen A. Johnston (University of Texas Southwestern Medical Center, TX). Plasmid SGP4 (three Gal4p-binding sites) was generated by cloning a DNA fragment containing three Gal4p-binding sites into the low copy number plasmid pRS416.
Yeast strains and media
Yeast strains harboring null mutations in SPT3 (FY294), SPT20 (FY1097), and GCN5 (FY1370) and their isogenic wild-type equivalents, FY631, FY67, and FY1369, respectively, were obtained from Fred Winston (Harvard Medical School, Boston, MA) (Roberts and Winston 1996, 1997; Sterner et al. 1999). Strains FY1992 (MATa his4–917δ lys2–173R2 ura3–52 leu2Δ1), FY1993 (MATa his4–917δ lys2–173R2 ura3–52 leu2Δ1 gal80Δ::LEU2), and FY1994 (MATa his4–917δ lys2–173R2 ura3–52 leu2Δ1 gal80Δ::LEU2 spt20Δ::URA3) were obtained from Erica Larschan and Fred Winston (Harvard Medical School, Boston, MA). The myc-tagged strains, SGY1 (Spt3p–myc; TRP1), SGY2 (Spt20p–myc; TRP1), SGY3 (Ada2p–myc; TRP1), and SGY4 (Gcn5p–myc; TRP1) were generated by insertion of multiple myc-epitope tags at the original chromosomal loci of SPT3, SPT20, ADA2, and GCN5, respectively, in FY631 (Longtine et al. 1998). SGY96 (Spt20p–myc; TRP1) was generated by multiple myc-epitope tags at the original chromosomal locus of SPT20 in FY294. SGY97 (Spt3p–myc; HIS3) and SGY98 (Spt3p–myc; Kan) were generated by multiple myc-epitope tags at the original chromosomal locus of SPT3 in IPY36 and IPY37, respectively. The strains SGY7 and SGY8 were generated by multiple myc tags at the chromosomal locus of SPT3 in FY67 and FY1097. The endogenous GAL4 gene of SGY1 and SGY2 was disrupted using the PCR method (Brachmann et al. 1998) to generate SGY14 (Spt3p–myc; gal4Δ::URA3) and SGY15 (Spt20p–myc; gal4Δ::URA3), respectively. The plasmid SGP4, carrying three Gal4p-binding sites, was transformed into SGY27 (multiple myc-epitope tags with TRP1 at the original chromosomal locus of SPT3 in w303a) to generate SGY30. The GAL4 gene of SGY27 was disrupted to generate SGY28 (gal4Δ::URA3), which was then streaked on a 5FOA plate to pop out URA3. URA3–popped-out SGY28 was transformed by the plasmids pMA241 (Gal4p 1–147) and SGP4 or pRJR216 (Gal4p 1–100) and SGP4 or pRJR182 (Gal4p 1–100 + 840–881) and SGP4 to generate SGY71, SGY72, and SGY73, respectively. The plasmid SGP4 was also transformed into SGY2 and SGY81 (HA3 epitope tag with TRP1 at the chromosomal locus of SRB4 in w303a) to generate SGY99 and SGY82, respectively.
Glucose-repressed strains were grown in YPD (YP + 2% glucose) to an OD600 of 1. Galactose-induced strains were grown in YPG (YP + 2% galactose) to an OD600 of 1. Raffinose noninduced strains were grown in YPR (YP + 2% Raffinose). For gal4Δ, spt3Δ, spt20Δ, and gcn5Δ strains, cells were first grown in YPD to an OD600 of 0.8 and then tranferred to YPG for 5 h. Minimal media containing either 2% glucose or galactose were used for strains with reporter plasmid, protein expression plasmid, or both.
GST protein affinity chromatography
GST and GST-34 were overexpressed in 100 mL of Escherichia coli culture by IPTG induction for 3.5 h. The overexpressed proteins were immobilized on 0.2 mL of a 50% slurry of glutathione beads for 30 min at 4°C with gentle agitation. The beads were washed four times with phosphate-buffered saline with 1% triton X-100. Finally, the beads were washed twice with buffer A (50 mM HEPES at pH 7.5, 150 mM NaCl, 10% glycerol, 0.1% tween 20, 0.5 mM DTT, 1 mM PMSF, 2 μg/μL leupeptin, and 2 μg/μL pepstatin A). Yeast whole cell extract (WCE) was prepared from Saccharomyces cerevesiae SGY7 and SGY8 as described by Woontner et al. (1991).
To 50 μL GST or GST-34 immobilized beads, 20 μL (20 μg/μL) yeast WCE and 180 μL buffer A were added and gently agitated at 4°C for 30 min. Beads were washed four times with buffer A and boiled in 1× SDS-PAGE buffer for 5 min at 95°C. The eluate was analyzed by immunoblotting using the c-myc monoclonal antibody.
Primer-extension analysis
Primer-extension analysis was performed as described by Li et al. (2000). The primers used for analysis of GAL1, GAL2, GAL7, GAL10, RPS5, and TUB2 mRNA are as follows: GAL1, 5′-CCT TGACGTTAAAGTATAGAGG-3′; GAL2, 5′-GCTTGGGGTT GCTGTGAAACA-3′; GAL7, 5′-CGGTTAGTGGATTGTAAC GTCT-3′; GAL10, 5′-CAATGTATCCAGCACCACCTGT-3′; RPS5, 5′-GACTGGGGTGAATTCTTCAACAACTTC-3′; and TUB2, 5′-CCAATTTGGTTACCACACTGACCT-3′
Formaldehyde-based in vivo cross-linking
Formaldehyde-based in vivo cross-linking and chromatin immunoprecipitation was performed as described by Li et al. (2000). The immunoprecipitated DNA was amplified by PCR. Primer-pairs used for PCR analysis are as follows: GAL1 (UAS), 5′-CGCTTAACTGCTCATTGCTATATTG-3′ and 5′-TTGTT CGGAGCAGTGCGGCGC-3′; GAL1 (Core), 5′-ATAGGATGA TAATGCGATTAGTTTTTTAGCCTT-3′ and 5′-GAAAATGT TGAAAGTATTAGTTAAAGTGGTTATGCA-3′; RPS5 (UAS), 5′-AGAAACAATGAACAGCCTTGAGTTCTC-3′ and 5′-GCA GGGCCATTCTCATCTGA-3′; RPS5 (Core), 5′-GGCCAACTT CTACGCTCACGTTAG-3′ and 5′-CGGTGTCAGACATCTT TGGAATGGTC-3′; and GAL4 (ORF), 5′-CTTGTTCAATGCA GTCCTAGTACCC-3′ and 5′-CACAAGTCTGGATTTTAAAA GTGGCC-3′.
Primer-pairs flanking Gal4p-binding sites in the plasmid SGP4 are 5′-GGTGGCGGCCGCTCTAGAACTAGT-3′ and 5′-TTGACCGTAATGGGATAGGTCACG-3′.
Autoradiograms were scanned and quantitated by the National Institutes of Health image 1.62 program. Immunoprecipitated (IP) DNAs were quantitated and presented as the ratio of IP to input.
Acknowledgments
We thank Mark Ptashne for plasmids and helpful discussions, Erica Larschan and Fred Winston for yeast strains, Danny Reinberg for the monoclonal yTFIIB antibody, and Stephen A. Johnston for plasmids. We are especially grateful to Erica Larschan and Fred Winston for discussions and communication of results before publication. This work was supported in part by a grant from the National Institutes of Health to M.R.G. M.R.G. is an investigator and S.B. an associate of the Howard Hughes Medical Institute.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL michael.green@umassmed.edu; FAX (508) 856-5473.
Article and publication are at http://www.genesdev.org/cgi/doi/10.1101/gad.911401.
References
- Ansari AZ, Reece RJ, Ptashne M. A transcriptional activating region with two contrasting modes of protein interaction. Proc Natl Acad Sci. 1998;95:13543–13548. doi: 10.1073/pnas.95.23.13543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brachmann CB, Davies A, Cost GJ, Caputo E, Li J, Hieter P, Boeke JD. Designer deletion strains derived from Saccharomyces cerevisiae S288C: A useful set of strains and plasmids for PCR-mediated gene disruption and other applications. Yeast. 1998;14:115–132. doi: 10.1002/(SICI)1097-0061(19980130)14:2<115::AID-YEA204>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- Brown CE, Lechner T, Howe L, Workman JL. The many HATs of the transcriptional coactivators. Trends Biochem Sci. 2000;25:15–19. doi: 10.1016/s0968-0004(99)01516-9. [DOI] [PubMed] [Google Scholar]
- Carey M, Lin Y-S, Green MR, Ptashne M. A mechanism for synergistic activation of a mammalian gene by GAL4 derivatives. Nature. 1990;345:361–364. doi: 10.1038/345361a0. [DOI] [PubMed] [Google Scholar]
- Davidson I, Xiao JH, Rosales R, Staub A, Chambon P. The HeLa cell protein TEF-1 binds specifically and cooperatively to two SV40 enhancer motifs of unrelated sequence. Cell. 1988;54:931–942. doi: 10.1016/0092-8674(88)90108-0. [DOI] [PubMed] [Google Scholar]
- Dudley AM, Rougeulle C, Winston F. The Spt components of SAGA facilitate TBP binding to a promoter at a post-activator-binding step in vivo. Genes & Dev. 1999;13:2940–2945. doi: 10.1101/gad.13.22.2940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisenmann DM, Arndt KM, Ricupero SL, Rooney JW, Winston F. SPT3 interacts with TFIID to allow normal transcription in Saccharomyces cerevisiae. Genes & Dev. 1992;6:1319–1331. doi: 10.1101/gad.6.7.1319. [DOI] [PubMed] [Google Scholar]
- Eisenmann DM, Chapon C, Roberts SM, Dollard C, Winston F. The Saccharomyces cerevisiae SPT8 gene encodes a very acidic protein that is functionally related to SPT3 and TATA-binding protein. Genetics. 1994;137:647–657. doi: 10.1093/genetics/137.3.647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fromental C, Kanno M, Nomiyama H, Chambon P. Cooperativity and hierarchical levels of functional organization in the SV40 enhancer. Cell. 1988;54:943–953. doi: 10.1016/0092-8674(88)90109-2. [DOI] [PubMed] [Google Scholar]
- Grant PA, Duggan L, Cote J, Roberts SM, Brownell JE, Candau R, Ohba R, Owen-Hughes T, Allis CD, Winston F, et al. Yeast Gcn5 functions in two multisubunit complexes to acetylate nucleosomal histones: Characterization of an Ada complex and the SAGA (Spt/Ada) complex. Genes & Dev. 1997;11:1640–1650. doi: 10.1101/gad.11.13.1640. [DOI] [PubMed] [Google Scholar]
- Grant PA, Sterner DE, Duggan LJ, Workman JL, Berger SL. The SAGA unfolds: Convergence of transcription regulators in chromatin-modifying complexes. Trends Cell Biol. 1998;8:193–197. doi: 10.1016/s0962-8924(98)01263-x. [DOI] [PubMed] [Google Scholar]
- Hampsey M. A SAGA of histone acetylation and gene expression. Trends Genet. 1997;13:427–429. doi: 10.1016/s0168-9525(97)01292-4. [DOI] [PubMed] [Google Scholar]
- Horiuchi J, Silverman N, Pina B, Marcus GA, Guarente L. ADA1, a novel component of the ADA/GCN5 complex, has broader effects than GCN5, ADA2, or ADA3. Mol Cell Biol. 1997;17:3220–3228. doi: 10.1128/mcb.17.6.3220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston M. A model fungal gene regulatory mechanism: The GAL genes of Saccharomyces cerevisiae. Microbiol Rev. 1987;51:458–476. doi: 10.1128/mr.51.4.458-476.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston M, Carlson M. Regulation of carbon and phosphate utilization. In: Jones EW, et al., editors. The molecular and cellular biology of yeast Saccharomyces: Gene expression. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1992. pp. 193–281. [Google Scholar]
- Koh SS, Ansari AZ, Ptashne M, Young RA. An activator target in the RNA polymerase II holoenzyme. Mol Cell. 1998;1:895–904. doi: 10.1016/s1097-2765(00)80088-x. [DOI] [PubMed] [Google Scholar]
- Larschan E, Winston F. The S. cerevisiae SAGA complex functions in vivo as a coactivator for transcriptional activation by Gal4. Genes & Dev. 2001;15:1946–1956. doi: 10.1101/gad.911501. (this issue). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee TI, Young RA. Transcription of eukaryotic protein-coding genes. Annu Rev Genet. 2000;34:77–137. doi: 10.1146/annurev.genet.34.1.77. [DOI] [PubMed] [Google Scholar]
- Lee TI, Causton HC, Holstege FC, Shen W-C, Hannett N, Jennings EG, Winston F, Green MR, Young RA. Redundant roles for the TFIID and SAGA complexes in global transcription. Nature. 2000;405:701–704. doi: 10.1038/35015104. [DOI] [PubMed] [Google Scholar]
- Li X-Y, Virbasius A, Zhu X, Green MR. Enhancement of TBP binding by activators and general transcription factors. Nature. 1999;399:605–609. doi: 10.1038/21232. [DOI] [PubMed] [Google Scholar]
- Li X-Y, Bhaumik SR, Green MR. Distinct classes of yeast promoters revealed by differential TAF recruitment. Science. 2000;288:1242–1244. doi: 10.1126/science.288.5469.1242. [DOI] [PubMed] [Google Scholar]
- Lohr D, Venkov P, Zlatanova J. Transcriptional regulation in the yeast GAL gene family: A complex genetic network. FASEB J. 1995;9:777–787. doi: 10.1096/fasebj.9.9.7601342. [DOI] [PubMed] [Google Scholar]
- Longtine MS, McKenzie A, III, Demarini DJ, Shah NG, Wach A, Brachat A, Philippsen P, Pingle JR. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast. 1998;14:953–961. doi: 10.1002/(SICI)1097-0061(199807)14:10<953::AID-YEA293>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- Ma J, Ptashne M. Deletion analysis of GAL4 defines two transcriptional activating segments. Cell. 1987;48:847–853. doi: 10.1016/0092-8674(87)90081-x. [DOI] [PubMed] [Google Scholar]
- McKnight S, Tjian R. Transcriptional selectivity of viral genes in mammalian cells. Cell. 1986;46:795–805. doi: 10.1016/0092-8674(86)90061-9. [DOI] [PubMed] [Google Scholar]
- Melcher K, Johnston SA. GAL4 interacts with TATA-binding protein and coactivators. Mol Cell Biol. 1995;15:2839–2848. doi: 10.1128/mcb.15.5.2839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orlando V, Paro R. Mapping Polycomb-repressed domains in the bithorax complex using in vivo formaldehyde cross-linked chromatin. Cell. 1993;75:1187–1198. doi: 10.1016/0092-8674(93)90328-n. [DOI] [PubMed] [Google Scholar]
- Orphanides G, Lagrange T, Reinberg D. The general transcription factors of RNA polymerase II. Genes & Dev. 1996;10:2657–2683. doi: 10.1101/gad.10.21.2657. [DOI] [PubMed] [Google Scholar]
- Ostergaard S, Walloe KO, Gomes CSG, Olsson L, Nielsen J. The impact of GAL6, GAL80, and MIG1 on glucose control of the GAL system in Saccharomyces cerevisiae. FEMS Yeast Res. 2001;1:47–55. doi: 10.1111/j.1567-1364.2001.tb00012.x. [DOI] [PubMed] [Google Scholar]
- Ptashne M. How eukaryotic transcriptional activators work. Nature. 1988;335:683–689. doi: 10.1038/335683a0. [DOI] [PubMed] [Google Scholar]
- Ptashne M, Gann A. Transcriptional activation by recruitment. Nature. 1997;386:569–577. doi: 10.1038/386569a0. [DOI] [PubMed] [Google Scholar]
- Roberts SM, Winston F. SPT20/ADA5 encodes a novel protein functionally related to the TATA-binding protein and important for transcription in Saccharomyces cerevisiae. Mol Cell Biol. 1996;16:3206–3213. doi: 10.1128/mcb.16.6.3206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ————— Essential functional interactions of SAGA, a Saccharomyces cerevisiae complex of Spt, Ada, and Gcn5 proteins, with the Snf/Swi and Srb/mediator complexes. Genetics. 1997;147:451–465. doi: 10.1093/genetics/147.2.451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roeder RG. The role of general initiation factors in transcription by RNA polymerase II. Trends Biochem Sci. 1996;21:327–335. [PubMed] [Google Scholar]
- Roth SY, Denu JM, Allis CD. Histone acetyltransferases. Annu Rev Biochem. 2001;70:81–120. doi: 10.1146/annurev.biochem.70.1.81. [DOI] [PubMed] [Google Scholar]
- Shen W-C, Green MR. Yeast TAFII145 functions as a core promoter selectivity factor, not a general coactivator. Cell. 1997;90:615–624. doi: 10.1016/s0092-8674(00)80523-1. [DOI] [PubMed] [Google Scholar]
- Stargell LA, Struhl K. Mechanisms of transcriptional activation in vivo: Two steps forward. Trends Genet. 1996;12:311–315. doi: 10.1016/0168-9525(96)10028-7. [DOI] [PubMed] [Google Scholar]
- Sterner DE, Berger SL. Acetylation of histones and transcription-related factors. Microbiol Mol Biol Rev. 2000;64:435–459. doi: 10.1128/mmbr.64.2.435-459.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sterner DE, Grant PA, Roberts SM, Duggan LJ, Belotserkovkaya R, Pacella LA, Winston F, Workman JL, Berger SL. Functional organization of the yeast SAGA complex: Distinct components involved in structural integrity, nucleosome acetylation, and TATA-binding protein interaction. Mol Cell Biol. 1999;19:86–98. doi: 10.1128/mcb.19.1.86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strahl-Bolsinger S, Hecht A, Luo K, Grunstein M. SIR2 and SIR4 interactions differ in core and extended telomeric heterochromatin in yeast. Genes & Dev. 1997;11:83–93. doi: 10.1101/gad.11.1.83. [DOI] [PubMed] [Google Scholar]
- Struhl K. Chromatin structure and RNA polymerase II connection: Implications for gene regulation. Cell. 1996;84:179–182. doi: 10.1016/s0092-8674(00)80970-8. [DOI] [PubMed] [Google Scholar]
- Woontner M, Wade PA, Bonner J, Jaehning JA. Transcriptional activation in an improved whole-cell extract from Saccharomyces cerevisiae. Mol Cell Biol. 1991;11:4555–4560. doi: 10.1128/mcb.11.9.4555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Y, Reece RJ, Ptashne M. Quantitation of putative activator-target affinities predicts transcriptional activating potentials. EMBO J. 1996;15:3951–3963. [PMC free article] [PubMed] [Google Scholar]
- Xie Y, Denison C, Yang S-H, Fancy DA, Kodadek T. Biochemical characterization of the TATA-binding protein–Gal4 activation domain complex. J Biol Chem. 2000a;275:31914–31920. doi: 10.1074/jbc.M003760200. [DOI] [PubMed] [Google Scholar]
- Xie Y, Sun L, Kodadek T. TATA-binding protein and the Gal4 transactivator do not bind to promoters cooperatively. J Biol Chem. 2000b;275:40797–40803. doi: 10.1074/jbc.M007019200. [DOI] [PubMed] [Google Scholar]
- Zaman Z, Ansari AZ, Gaudreau L, Nevado J, Ptashne M. Gene transcription by recruitment. Cold Spring Harb Symp Quant Biol. 1998;63:167–171. doi: 10.1101/sqb.1998.63.167. [DOI] [PubMed] [Google Scholar]