Abstract
The intracellular pathogen Trypanosoma cruzi is the etiological agent of Chagas’ disease. We have isolated a full-length cDNA encoding uracil-DNA glycosylase (UDGase), a key enzyme involved in DNA repair, from this organism. The deduced protein sequence is highly conserved at the C-terminus of the molecule and shares key residues involved in binding or catalysis with most of the UDGases described so far, while the N-terminal part is highly variable. The gene is single copy and is located on a chromosome of ∼1.9 Mb. A His-tagged recombinant protein was overexpressed, purified and used to raise polyclonal antibodies. Western blot analysis revealed the existence of a single UDGase species in parasite extracts. Using a specific ethidium bromide fluorescence assay, recombinant T.cruzi UDGase was shown to specifically excise uracil from DNA. The addition of both Leishmania major AP endonuclease and exonuclease III, the major AP endonuclease from Escherichia coli, produces stimulation of UDGase activity. This activation is specific for AP endonuclease and suggests functional communication between the two enzymes.
INTRODUCTION
Uracil can arise in DNA by two mechanisms: misincorporation of deoxyuridine triphosphate during DNA synthesis and, especially, spontaneous deamination of cytosine (1,2). Repair is initiated by uracil-DNA glycosylases (UDGases), which are highly conserved and very ubiquitous and specifically remove the normal RNA base uracil from DNA. Their biological function is performed by hydrolysis of the N-glycosidic bond linking the base to the deoxyribose sugar, leaving an abasic site (AP site). The removal of AP sites is initiated by AP endonucleases (apurinic/apyrimidinic) and deoxyribophosphodiesterases. The resultant gap is filled in and sealed by the action of DNA polymerase and DNA ligase.
UDGases show high specificity for efficient excision of uracil from DNA, with weak activity against synthetic uracil analogs such as 5-fluorouracil and 5-hydroxyuracil (3,4). Excision of uracil from single-stranded DNA occurs more rapidly than from double-stranded DNA, although UDGases can utilize both substrates (5). No activity has been detected against any normal DNA base nor against uracil in RNA (6–8).
All UDGases are monomeric proteins with molecular masses ranging between 19 and 35 kDa (9) and a conserved C-terminal component. N-terminal extensions found in eukaryotic and viral enzymes are probably involved in subcellular localization and vary considerably in length and composition. The human UNG gene has been shown to encode both nuclear (UNG2) and mitochondrial (UNG1) isoforms of the enzyme. A mechanism that comprises transcription from two different promoters and alternative splicing for this enzyme has been reported (10–14).
Genes for UDGases have been cloned and sequenced from several sources (15,16), establishing that they are highly conserved proteins. There are certain organisms that contain another group of UDGases, such as the cyclin-like UDGases (17,18), dsUDGase (19) and glyceraldehyde 3-phosphate dehydrogenases (20). UDGases do not require any known cofactor or ions and are fully active in the presence of EDTA (21–24). Recently several crystal structures have been reported. The active site groove of the enzyme is highly conserved and uridine is bound in its extrahelical conformation (25–29).
Recent results indicate that human UDGase may have a function in replication (30–32) and in the repair of oxidative damage (33). Dizdaroglu et al. (33) have shown that the human enzyme can excise isodialuric acid, 5-hydroxyuracil and alloxan from DNA. These uracil derivatives are generated as major products of cytosine in DNA by hydroxyl radical attack or other oxidative processes. Finally, it has also been reported that uracil-containing DNA may play an important role in cells targeted for death. Thus, in pupating insects no detectable levels of UDGase can be found and there appears to be a correlation between cellular destruction during development and the absence of this DNA repair activity (34).
We have isolated the UDGase gene of Trypanosoma cruzi, the etiological agent of Chagas’ disease. The protein has been overexpressed as a histidine-tagged enzyme which catalyzes the removal of deoxyuracil from double-stranded DNA. The existence of stimulation of T.cruzi UDGase by Leishmania major AP endonuclease (LmAP) is shown, suggesting a functional interaction between the two enzymes during base excision repair.
MATERIALS AND METHODS
Materials and general procedures
Restriction enzymes, T4 DNA ligase, Taq polymerase, exonuclease III and the Klenow fragment of DNA polymerase were purchased from Boehringer Mannheim and used according to the instructions specified by the manufacturer. The pET expression system and His-bind resin were purchased from Novagen. Oligonucleotides were synthesized at the Analytical Services of the Instituto de Parasitología y Biomedicina ‘López Neyra’ (Granada, Spain).
Epimastigotes of T.cruzi were grown in filter-sterilized LIT medium with 10% (v/v) heat-inactivated fetal calf serum (Gibco) in tissue flasks at 28°C. Total T.cruzi genomic DNA was isolated from the Y strain by phenol extraction (35). Standard molecular biology techniques were performed as described elsewhere (36,37).
Isolation of the T.cruzi uracil-DNA glycosylase gene
For this purpose and considering the presence of highly conserved sequences located at the C-terminal end of most UDGases, two degenerate oligonucleotides were synthesized. The sequences of these oligonucleotides were located in the binding pocket for uracil. The chosen sequences were 5′-IL(I)GQDPY- - - - - - - - - - VFL(M)LWG-3′. The sequences of the oligonucleotides used were designed taking into account the codon usage for this parasite: TcUNG-1, 5′-ATT(C)C(A)TT(C)(G)GGT(C)CAGGAT(C)CCT(C)(A)(G)TA-3′; TcUNG-2, 5′-GTC(G)TTT-(C)C(A)TT(C)(G)CTT(C)(G)TGGGG-3′.
PCR was carried out in a reaction mixture (100 µl) containing 100 pmol each of the two oligonucleotide primers TcUNG-1 and TcUNG-2 and 500 ng T.cruzi genomic DNA. Amplification was initiated with 2.5 U Taq polymerase. PCR parameters were 1 min at 94°C, followed by 1 min at 44°C and an extension period of 1 min at 72°C for 30 thermal cycles.
The gene was isolated from a cDNA expression library of the T.cruzi Y strain constructed using a ZAP Express cDNA Synthesis Kit (Stratagene) as described (38). The PCR product was used as hybridization probe. Hybridization and washings were conducted at 42°C. Approximately 100 000 plaques were replicated on nitrocellulose and screened following standard protocols (36,37). Phagemids were rescued from the library by co-infection of Escherichia coli XL1B with 5.2 × 105 p.f.u. of λ phage and 107 p.f.u of ExAssist helper phage in 25 ml of Luria broth. The supernatant obtained after incubation and clarification of the culture by centrifugation had a titer of 2.5 × 103 kanamycin-resistant c.f.u./ml. The isolation of plasmid DNA was performed as in standard protocols (36,37).
Contour-clamped homogeneous electric field (CHEF) electrophoresis
The chromosomal location of the UDGase gene was established by CHEF electrophoresis separation of T.cruzi chromosomes. Blocks of T.cruzi in low melting point agarose were prepared as described (39). Chromosomes were separated on a 1% (w/v) agarose gel with a CHEF system (Pharmacia). The following parameters were used: frequencies of 350 s for 24 h, 500 s for 24 h, 750 s for 24 h and 1000 s for 24 h at 84 V and 13°C. Molecular masses of the chromosomal DNA bands were assigned using DNA of Saccharomyces cerevisiae strain S13 as the standard. The resulting gel was transferred to a Hybond-N (Amersham) nylon filter and subjected to Southern blot analysis using the coding region of the ung gene as probe.
Overexpression and protein purification
Two primers were designed to amplify the coding region of the ung gene, containing NdeI and HindIII restriction sites for cloning in the pET-28a translation vector. This vector allows N-terminal fusion to a cleavable His-Tag sequence. The target plasmid, pETTcung, was established in BL21(DE3) E.coli cells. Bacterial clones were grown in Luria broth containing 50 µg/ml kanamycin. Expression of the target DNA was induced by addition of 0.5 mM isopropyl-β-d-thiogalactoside (IPTG). Cells were collected by centrifugation and frozen at –80°C when not used immediately.
Overexpressed plasmid carrying the His-Tag sequence allows purification of the protein by affinity chromatography. The protein binds to divalent cations (e.g. Ni2+) immobilized on His-Bind metal chelation resin. Unbound proteins were washed away and the target protein recovered with an imidazole gradient. Protein was eluted at 200 mM imidazole. Analysis of the purity of the samples was performed electrophoretically on SDS–polyacrylamide gels.
Aggregation state under native conditions was analyzed by chromatography in an HPLC Superdex 75 HR10/30 column using phosphate buffer (50 mM PO4H2K, 150 mM NaCl, pH 7).
Assay of UDGase activity
UDGase activity was determined by measuring the enhanced fluorescence of ethidium bromide when intercalated into double-stranded DNA at pH 12 (40,41). Removal of uracil by UDGase results in the appearance of AP sites in DNA. Since the phosphodiester backbone of DNA is not broken after uracil elimination, the level of fluorescence or intercalated ethidium bromide remains unaltered. Heat treatment at pH 12 results in hydrolysis of AP sites and irreversible denaturation of substrate DNA molecules containing AP sites. Unreacted substrate consisting of covalent closed circular (ccc)DNA molecules rapidly reanneals and regains most of the initial ethidium bromide fluorescence. A plasmid containing uracil was used as a substrate and prepared by isolating phagemid pBluescript II KS from E.coli strain CJ236, which is deficient in dUTPase (dut–) and UDGase (ung–) (42). Control DNA was derived from the same plasmid isolated from E.coli XL1B cells. Assay mixtures and reactions where carried out as described elsewhere (41) using 1.5 µg plasmid DNA (10 nM) and 0.5 µg enzyme (168 nM) in a volume of 70 µl. Samples were measured in a Perkin-Elmer LS-5 Luminescence Spectrometer. Loss of fluorescence due to non-specific nicking was monitored by adding 2 vol DMSO to the samples prior to dilution into pH 12 buffer, which causes denaturation of nicked open circular DNA but has no effect upon DNA containing AP sites. EDTA (20 mM) added to the reaction mixture inhibited almost all non-specific nuclease activities.
Modulation of UDGase activity by exonuclease III (from E.coli; Boehringer Mannheim) or LmAP was measured in the same mixture after adding from 40 to 175 U exonuclease III or a molar excess of up to 100-fold AP endonuclease. Pure preparations of His-tagged AP endonuclease from L.major were obtained as previously described (43).
Preparation of antibodies and western blot analysis
Polyclonal anti-TcUDGase antibodies were obtained by immunization of rabbits with 300 µg purified recombinant protein dissolved in 1 ml of complete Freund’s adjuvant (Bacto). Antiserum was collected 8 weeks after initial injection. Epimastigotes of T.cruzi were lysed by sonication and cell supernatants were prepared by centrifugation at 12 000 g for 30 min. For western blot analysis protein samples were transferred from SDS–PAGE gels to Immobilon-P membranes (Millipore). Bound antibodies were detected using horseradish peroxidase-conjugated anti-rabbit IgG as the secondary antibody. Visualization was performed with the ECL western blotting reagents (Amersham).
RESULTS
Isolation and characterization of the ung gene
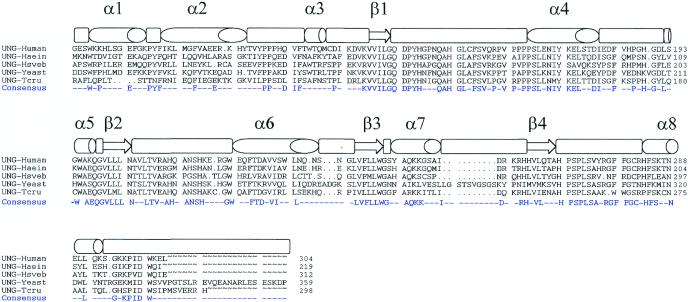
Using a PCR-based strategy we have isolated a cDNA from T.cruzi encoding UDGase. The sequence amplified by PCR corresponds to a highly conserved region of the uracil-binding site and was used as a probe to screen a cDNA library from T.cruzi constructed in λ-ZAP Express. After analysis of several positive clones, we found one phage containing a non-truncated cDNA for UDGase of T.cruzi (all the other positive clones were identical cDNAs truncated at the 5′-end). Thus, the insert of 1045 bp contained 23 bp of the spliced leader (a 35 nt sequence added by trans-splicing) at the 5′-end and a polyadenylated tail was evident at the 3′-end. The largest ORF identified contained two ATG codons and the first one after the spliced leader was considered as the probable site of translation initiation based on codon usage studies and characteristics of the 5′-end of other protozoan genes. According to this, the coding sequence had 897 bp and 298 amino acids. The predicted amino acid sequence from the ung gene of T.cruzi was aligned with sequences from different eukaryotic and prokaryotic counterparts (Fig. 1). The major secondary structure elements are indicated above the sequence alignment. The protozoan enzyme shows an identity of 51.6% with UDGase of Haemophilus influenzae, although this latter enzyme is 80 amino acids shorter than the T.cruzi protein. With human UDGase the identity was 48.6% and both enzymes are of similar size. The N-terminus varied significantly with regard to most of the UDGase proteins described so far. The T.cruzi sequence shows the highly conserved amino acids that are involved in binding and catalysis, such as the conserved motifs GQDPYH and HPSPLS. The selected ORF presented a codon usage that strongly corresponds to the codon frequency described for other T.cruzi genes.
Figure 1.
Structure-based alignment of UDGase sequences (Swiss-Prot accession nos in parentheses): human (P13051), H.influenzae (P43731), equine herpes virus type I (P28866), S.cerevisiae (P12887) and T.cruzi. The GenBank accession no. for the T.cruzi UDGase cDNA sequence is AF152347. The secondary structure motifs identified in the crystal structure of human UDGase are indicated above the sequence alignment (26,35). The consensus sequence is also shown.
Chromosomal localization and genomic organization of the ung gene
Genomic DNA from a clone of T.cruzi Y strain was digested with different endonucleases, blotted and probed with a fragment that encompasses the entire coding region of the UDGase gene. The number of bands observed in the digests suggests that the gene is single copy (data not shown).
Chromosomes of T.cruzi range in size between 450 kb (or even smaller) and 2.5 Mb (44) and can be resolved by CHEF electrophoresis. The ung gene was labeled by random priming and hybridized to a filter replicate of the T.cruzi chromosomes. Under the CHEF conditions used in this study UDGase sequences were located on a chromosome with a molecular weight between 2.1 and 1.5 Mb (data not shown).
Overexpression, purification and western blot analysis
To enhance the level of expression, the entire coding region of the ung gene was amplified by PCR. The gene was cloned in the expression vector pET-28a, which allows N-terminal fusion to a cleavable His-Tag sequence; the target protein is expressed with an extension of 20 amino acids containing six consecutive histidine residues at the N-terminal end. This construct was used to transform BL21(DE3) cells. Lysates of cells induced with IPTG displayed a band of ∼37 kDa not present in uninduced cells.
The enzyme from T.cruzi was purified using an affinity Ni2+ column. This method allows purification under native conditions, maintaining the activity of the protein. After washing out all the unbound proteins the target protein is recovered by elution with a gradient of 0–500 mM imidazole. In this way we have obtained a homogeneous purified protein, as judged by SDS–PAGE and Coomassie blue staining (Fig. 2). Immunoblotting analysis performed using antibodies raised against purified recombinant protein showed that UDGase present in cellular extracts of epimastigote forms of the parasite has the same mobility on SDS–PAGE gels as the recombinant purified protein expressed from the deduced start codon (Fig. 3), which further corroborates the correct designation of the initiator methionine.
Figure 2.
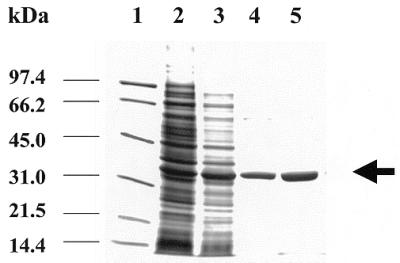
SDS–PAGE of purified UDGase. Lane 1, positions of molecular mass standards; lane 2, soluble BL21(DE3)/pETTcung cell extracts without IPTG induction; lane 3, soluble BL21(DE3)/ pETTcung extracts after 2 h induction with IPTG; lanes 4 and 5, fractions of purified UDGase after affinity column elution.
Figure 3.

Western blot analysis of T.cruzi UDGase. Lane 1, purified recombinant UDGase; lane 2, sonicated soluble extracts of the T.cruzi Y strain. Samples were made to react with the polyclonal antibodies and detection was performed as described in Materials and Methods.
In order to determine the aggregation state of the protein under native conditions we used a Superdex 75 HR10/30 HPLC column. Trypanosoma cruzi UDGase elutes between ovalbumin (42 998 Da) and chymotrypsinogen (25 666 Da) under native conditions (data not shown), with a calculated molecular mass of ∼42 000 Da, which suggests that it is a monomeric protein since the theoretical molecular mass of the His-tagged protein based on the amino acid sequence is 35 347.6 Da. This is the aggregation state of all the UDGases described to date (9).
Measurement of UDGase activity
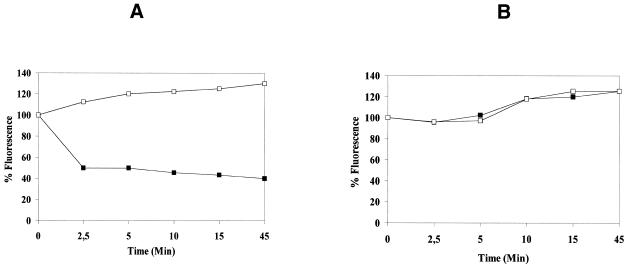
UDGase activity was determined using uracil-enriched DNA as substrate. Incubations of up to 45 min allowed for measurable UDGase uracil excision activity (Fig. 4A). Thus, there is a quantitative loss of substrate cccDNA, reflected in a loss of fluorescence due to breakage of the DNA backbone at the heat-sensitive AP site. Due to the characteristics of the buffer in which the fluorescence was measured (low salt buffer, pH 12), after uracil excision the denatured and separated strands of the substrate DNA cannot reanneal. Substrate DNA treated with DMSO does not undergo strand denaturation and therefore no decrease in percentage fluorescence is observed. Changes in the level of fluorescence do not occur after heating or DMSO treatment of control cccDNA (Fig. 4B).
Figure 4.
Assay of T.cruzi UDGase. Closed squares correspond to ethidium bromide fluorescence of cccDNA after heating the samples at 95°C for 8 min. Open squares correspond to ethidium bromide fluorescence after treating the samples with 2 vol DMSO. The data at time 0 were normalized to 100%. (A) Assay using uracil-containing cccDNA. (B) Assay using control cccDNA.
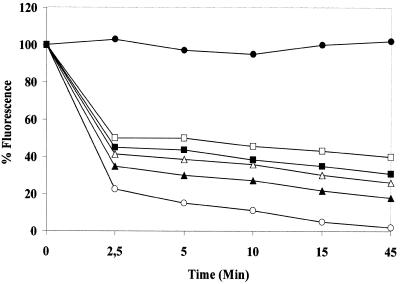
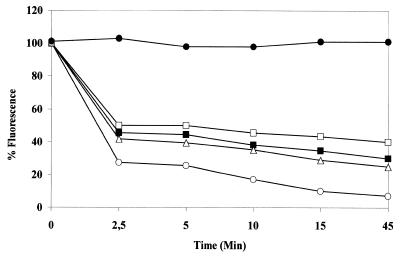
It has been reported (13,45) that the activity of human UDGase is stimulated in the presence of AP endonuclease and that this effect requires a molar excess of enzyme. In the presence of the AP endonuclease of L.major (another protozoan belonging to the Trypanosomatidae family) (43), activity was stimulated with an increasing molar excess of AP enzyme. Stimulation varied from 23 to 96% with a molar excess between 2 and 100 (data not shown). At short incubation times (5 min), before a significant number of AP sites had been removed, 2 and 10 molar excesses of LmAP enhanced T.cruzi activity, while LmAP alone did not produce a decrease in the level of fluorescence (Fig. 5). We have also tested the effect of exonuclease III from E.coli (Fig. 6), which stimulated T.cruzi UDGase activity by 25–83% with 40–175 U enzyme. It has been hypothesized that this interaction may be of functional importance in protecting the cells from toxic AP site intermediates (13,45).
Figure 5.
Enhancement of UDGase activity by LmAP. Decrease in fluorescence observed in the presence of different concentrations of LmAP. Open squares, UDGase; closed squares, UDGase + 2LmAP; open triangles, UDGase + 10 molar excess LmAP; closed triangles, UDGase + 50 molar excess LmAP; open circles, UDGase + 100 molar excess LmAP; closed circles, LmAP.
Figure 6.
Enhancement of UDGase activity by exonuclease III. Decrease in fluorescence observed in the presence of different concentrations of exonuclease III. Open squares, UDGase; closed squares, UDGase + 40 U; triangles, UDGase +80 U; open circles, UDGase +175 U; closed circles, LmAP.
DISCUSSION
Trypanosoma cruzi is the hemoflagellate protozoan that causes Chagas’ disease, one of the most important public health problems in South America (46,47). A vaccine to prevent transmission of T.cruzi is not available, neither is there an effective chemotherapeutic treatment. There is little knowledge of many aspects of the biology of this parasite that could be used as a tool against the organism. Details about the enzymatic mechanisms involved in DNA repair or the elimination of modified bases from the genome of this parasite are unknown. Uracil metabolism has an additional interest since it has been reported (48) that the modified base β-d-glucosyl-hydroxymethyluracil is a normal constituent of DNA in kinetoplastida and its presence seems to be related to regulation of gene expression. Recently, Otterlei et al. (31) have demonstrated that in humans UDGase (UNG2) is responsible for removal of incorporated dUMP, since neutralizing antibodies to UNG2 efficiently protected uracil from excision. UDGase may also be crucial for the integrity of the DNA of this intracellular pathogen, since T.cruzi is exposed to oxidative stress and probably undergoes cytosine deamination due to the unfavorable habitat of the host cell where it multiplies.
In humans two different isoenzymes have been reported with mitochondrial and nuclear localizations. The unique 44 N-terminal amino acids present in UNG2 are required, although not sufficient, for sorting to nuclei, while the motif 17RKR19 located in this sequence appears to be essential. It has been shown that residues 13RK14 and 26LSRL29 present in the N-terminal part of UNG1 are essential for mitochondrial targeting (14). Mitochondrial targeting sequences have been reported for proteins from early branching eukaryotes, such as Crithidia, Trypanosoma and Trichomonas (49). In this regard, predictive analysis of the N-terminal region of T.cruzi UDGase reveals a possible dual localization, with both mitochondrial and nuclear targeting sequences. Thus, the prediction indicates a cleavage site for mitochondrial localization located at motif QRT/LL and the potential existence of a nuclear or mitochondrial signal sequence at motif RKRR.
The pET system allowed production of high levels of soluble recombinant protein which behaved as a monomer in gel filtration, similar to all UDGases characterized so far (9). We have used an assay that is dependent on enhanced fluorescence of ethidium bromide when intercalated into double-stranded DNA at pH 12. Although it has been reported that a number of DNA glycosylases, such as T4 endonuclease V and E.coli endonuclease III, have AP endonuclease activity (50), this is not the case for T.cruzi UDGase; in fact no UDGase has yet been reported to possess AP endonuclease function.
Stimulation of T.cruzi UDGase by AP endonuclease from L.major increased with an increasing molar excess of this enzyme. The enzyme activity in T.cruzi also showed stimulation in the presence of the E.coli AP endonuclease exonuclease III and, as in the case of LmAP, increasing amounts of this enzyme further enhanced its effects, suggesting a functional conservation between AP endonucleases from different organisms. Trypanosoma cruzi UDGase does not break the deoxyribose–phosphate backbone under the assay conditions since heat-sensitive AP sites remain after incubation with the enzyme. A similar activation has been shown for human UDGase and HAP1 endonuclease (45). Apparently it is not due to AP site elimination since early in incubation, before a significant number of AP sites have been removed, a low molar excess of LmAP is sufficient for activation. This stimulatory effect was not due to non-specific protein interactions since bovine serum albumin does not induce activity. Based on these observations, a model of DNA base excision repair initiation has been proposed (45). When UDGase finds a uracil-containing nucleotide this is flipped out of the DNA base stack into the active site, the glycosidic bond is cleaved and UDGase remains bound to the products of the reaction: free uracil and an abasic site. When the AP enzyme finds an AP site bound to UDGase it induces the latter enzyme to dissociate from DNA. This may be an important role of UDGase which acts to protect the cells from toxic AP intermediates. The required molar excess for stimulation supports a functional rather than a protein–protein interaction between UDGase and the AP enzymes, as previously reported (13,45).
Figure 2 shows a structure-based sequence alignment of several UDGases indicating the classical α–β–α structure with a small β-sheet sandwiched between the helices at the core of the molecule. Three-dimensional structures combined with structure-based sequence alignments have allowed the identification of some UDGase motifs having a key role in binding and catalysis in humans (45). These motifs are totally conserved in T.cruzi. Modeling studies performed using the Swiss Model suite of programs showed a very well conserved tertiary structure when compared with human UDGase. Slight differences include two additional β-sheet strands in T.cruzi UDGase. One is comprised of residues 28KVIL31 located between two α-helices and the other encompasses residues 121VEA123 located between a β-sheet and an α-helix.
The existence of a gene encoding UDGase in addition to the recently described AP endonuclease genes in both L.major and T.cruzi (43) provides molecular evidence for the existence of a base excision repair pathway in these organisms. These observations, together with the description of an enzyme with dUTPase activity in Leishmania (51), bestow information which will help to establish details concerning the removal of uracil from DNA in the Trypanosomatidae family.
Acknowledgments
ACKNOWLEDGEMENTS
These studies were supported by grants from the Spanish Programa Nacional de Biotecnología (BIO97-0659) and the Plan Andaluz de Investigación (Cod. CVI-199). C.G. is a fellow of the Ramon Areces Foundation.
References
- 1.Kubota Y., Nash,R.A., Klungland,A., Schar,P., Barnes,D.E. and Lindahl,T. (1996) Reconstitution of DNA base excision-repair with purified human proteins: interaction between DNA polymerase β and the XRCC1 protein. EMBO J., 15, 6662–6670. [PMC free article] [PubMed] [Google Scholar]
- 2.Nicholl I.D., Nealon,K. and Kenny,M.K. (1997) Reconstitution of human base excision repair with purified proteins. Biochemistry, 36, 7557–7566. [DOI] [PubMed] [Google Scholar]
- 3.Mauro D.J., De Reil,J.K., Tallarida,R.J. and Sirover,M.A. (1993) Mechanisms of excision of 5-fluoracil by uracil DNA glycosylase in normal human cells. Mol. Pharmacol., 43, 854–857. [PubMed] [Google Scholar]
- 4.Hatahet Z., Kow,Y.W., Purmal,A.A., Cunningham,R.P. and Wallace,S.S. (1994) New substrates for old enzymes. 5-Hydroxy-2′-deoxycytidine and 5-hydroxy-2′-deoxyuridine are substrates for Escherichia coli endonuclease III and formamidopyrimidine DNA. J. Biol. Chem., 269, 18814–18820. [PubMed] [Google Scholar]
- 5.Eftedal I., Guddal,P.H., Sluppaug,G., Volden,G. and Krokan,H.E. (1993) Consensus sequences for good and poor removal of uracil from double stranded DNA by uracil-DNA glycosylase. Nucleic Acids Res., 21, 2095–2101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cone R., Duncan,J., Hamilton,L. and Friedberg,E.C. (1977) Partial purification and characterization of a uracil DNA-N glycosylase from Bacillus subtilis. Biochemistry, 16, 3194–3201. [DOI] [PubMed] [Google Scholar]
- 7.Lindhal T., Ljungquist,S., Siegert,W., Niberg,B. and Sperens,B. (1977) DNA N-glycosylases. Properties of uracil-DNA glycosylases from E. coli. J. Biol. Chem., 252, 3286–3294. [PubMed] [Google Scholar]
- 8.Krokan H. and Wittwer,C.U. (1981) Uracil-DNA-glycosylases from HeLa cells: general properties, substrate specificity and effect of uracil analogs. Nucleic Acids Res., 9, 2599–2613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Friedberg E.C., Walker,G.C. and Siede,W. (1995) DNA Repair and Mutagenesis. ASM Press, Washington, DC.
- 10.Slupphaug G., Markussen,F.H., Olsen,L.C., Aasland,R., Aarsaether,N., Bakke,O., Krokan,H.E. and Helland,D.E. (1993) Nuclear and mitochondrial forms of human uracil-DNA glycosylase are encoded by the same gene. Nucleic Acids Res., 21, 2579–2584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nilsen H., Otterlei,M., Haug,T., Solum,K., Nagelhus,T.A., Skorpen,F. and Krokan,H. (1997) Nuclear and mitochondrial uracil-DNA glycosylases are generated by alternative splicing and transcription from different positions in the UNG gene. Nucleic Acids Res., 25, 750–755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Haug T., Skorpen,F., Aas,P.A., Malm,V., Skjelbred,C. and Krokan,H.E. (1998) Regulation of expression of nuclear and mitochondrial forms of human uracil-DNA glycosylase. Nucleic Acids Res., 26, 1449–1457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bharati S., Krokan,H.E., Kristiansen,L., Otterlei,M. and Slupphaug,G. (1998) Human mitochondrial uracil-DNA glycosylase preform (UNG1) is processed to two forms one of which is resistant to inhibition by AP-sites. Nucleic Acids Res., 26, 4953–4959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Otterlei M., Haug,T., Nagelhus,T.A., Slupphaug,G., Lindmo,T. and Krokan,H.E. (1998) Nuclear and mitochondrial splice forms of human uracil-DNA glycosylase contain a complex nuclear localisation signal and a strong classical mitochondrial localisation signal respectively. Nucleic Acids Res., 26, 4611–4617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Percival K.J., Klein,M.B. and Burgers,P.M.J. (1989) Molecular cloning and primary structure of the uracil-DNA-glycosylase gene from Saccharomyces cerevisiae. J. Biol. Chem., 264, 2593–2598. [PubMed] [Google Scholar]
- 16.Olsen L.C., Aasland,R., Wittwer,C.U., Krokan,H.E. and Hellard,D.E. (1989) Molecular cloning of human uracil-DNA glycosylase. EMBO J., 8, 3121–3125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Muller-Weeks S.J. and Caradonna,S. (1996) Specific association of cyclin-like uracil-DNA glycosylase with the proliferating cell nuclear antigen. Exp. Cell Res., 226, 346–355. [DOI] [PubMed] [Google Scholar]
- 18.Muller S.J. and Caradonna,S. (1991) Isolation and characterization of a human cDNA encoding uracil-DNA glycosylase. Biochim. Biophys. Acta, 1088, 197–207. [DOI] [PubMed] [Google Scholar]
- 19.Gallinari P. and Jirincy,J. (1996) A new class of uracil-DNA glycosylases related to human thymine-DNA glycosylase. Nature, 383, 735–738. [DOI] [PubMed] [Google Scholar]
- 20.Meyer-Siegler K., Mauro,D.J., Seal,G., Wurzer,J., DeRiel,J.K. and Sirover,M.A. (1991) A human nuclear uracil DNA glycosylase is the 37-kDa subunit of glyceraldehyde-3-phosphate dehydrogenase. Proc. Natl Acad. Sci. USA, 88, 8460–8464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Duncan B.K. (1981) In Boyer,P.E. (ed.), The Enzymes. Academic Press, Orlando, FL.
- 22.Krokan H.E., Standal,R. and Slupphaug,G. (1997) DNA glycosylases in base excision repair of DNA. Biochem. J., 325, 1–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mosbaugh D.W. and Bennett,S.E. (1994) Uracil-excision DNA repair. Prog. Nucleic Acid Res. Mol. Biol., 48, 315–370. [DOI] [PubMed] [Google Scholar]
- 24.Lindahl T. (1982) DNA repair enzymes. Annu. Rev. Biochem., 51, 61–87. [DOI] [PubMed] [Google Scholar]
- 25.Savva R., McAuley-Hecht,K., Brown,T. and Pearl,L. (1995) The structural basis of specific base-excision repair by uracil-DNA glycosylase. Nature, 373, 487–493. [DOI] [PubMed] [Google Scholar]
- 26.Mol C.D., Arvai,A.S., Slupphaug,G., Kavli,B., Alseth,I., Krokan,H.E. and Tainer,J.A. (1995) Crystal structure and mutational analysis of human uracil-DNA glycosylase: structural basis for specificity and catalysis. Cell, 80, 869–878. [DOI] [PubMed] [Google Scholar]
- 27.Mol C.D., Arvai,A.S., Sanderson,R.J., Slupphaug,G., Kavli,B., Krokan,H.E., Mosbaugh,D.W. and Tainer,J.A. (1995) Crystal structure of human uracil-DNA glycosylase in complex with a protein inhibitor: protein mimicry of DNA. Cell, 82, 701–708. [DOI] [PubMed] [Google Scholar]
- 28.Savva R. and Pearl,L.H. (1995) Nucleotide mimicry in the crystal structure of the uracil-DNA glycosylase–uracil glycosylase inhibitor protein complex. Nature Struct. Biol., 2, 752–757. [DOI] [PubMed] [Google Scholar]
- 29.Slupphaug G., Mol,C.D., Kavli,B., Arvai,A.S., Krokan,H.E. and Tainer,J.A. (1996) A nucleotide-flipping mechanism from the structure of human uracil-DNA glycosylase bound to DNA. Nature, 384, 87–92. [DOI] [PubMed] [Google Scholar]
- 30.Nagelhus T.A., Haug,T., Singh,K.K., Keshav,K.F., Skorpen,F., Otterlei,M., Bharati,S., Lindmo,T., Benichou,S., Benarous,R. and Krokan,H.E. (1997) A sequence in the N-terminal region of human uracil-DNA glycosylase with homology to XPA interacts with the C-terminal part of the 34-kDa subunit of replication protein A. J. Biol. Chem., 272, 6561–6566. [DOI] [PubMed] [Google Scholar]
- 31.Otterlei M., Warbrick,E., Nagelhus,T.A., Haug,T., Slupphaug,G., Akbari,M., Aas,P.A., Steinsbekk,K., Bakke,O. and Krokan,H.E. (1999) Post-replicative base excision repair in replication foci. EMBO J., 18, 3834–3844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Muller-Weeks S., Mastran,B. and Caradonna,S. (1998) The nuclear isoform of the highly conserved human uracil-DNA glycosylase is an Mr 36,000 phosphoprotein. J. Biol. Chem., 273, 21909–21917. [DOI] [PubMed] [Google Scholar]
- 33.Dizdaroglu M., Karabaya,A., Jaruga,P., Sluppaug,G. and Krokan,H.E. (1996) Novel activities of human uracil DNA N-glycosylase for cytosine-derived products of oxidative DNA damage. Nucleic Acids Res., 24, 418–422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dudley B., Hammond,A. and Deutsch,W.A. (1992) The presence of uracil-DNA glycosylase in insects is dependent upon developmental complexity. J. Biol. Chem., 267, 11964–11967. [PubMed] [Google Scholar]
- 35.Coderre J.A., Beverley,S.M., Schimke,R.T. and Santi,D.V. (1983) Overproduction of a bifunctional thymidylate synthetase-dihydrofolate reductase and DNA amplification in methotrexate-resistant Leishmania tropica. Proc. Natl Acad. Sci. USA, 80, 2132–2136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ausubel F.M., Brent,R., Kinsgton,R.E., Moore,D.D., Seidman,J.G., Smith,J.A. and Struhl,K. (1987) Current Protocols in Molecular Biology. John Wiley & Sons, New York, NY.
- 37.Sambrook J., Fritsch,E.F. and Maniatis,T. (1989) Molecular Cloning: A Laboratory Manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 38.Peña-Diaz J., Montalvetti,A., Camacho,A., Gallego,C., Ruiz-Perez,L.M. and Gonzalez-Pacanowska,D. (1997) A soluble eukaryotic HMGCoA reductase in the protozoan Trypanosoma cruzi. Biochem. J., 324, 619–626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Garvey E.P. and Santi,D.V. (1986) Stable amplified DNA in drug-resistant Leishmania exists as extrachromosomal circles. Science, 233, 535–540. [DOI] [PubMed] [Google Scholar]
- 40.Morgan A.R. and Chlebek,J. (1988) Quantitative assays for uracil-DNA glycosylase of high sensitivity. Biochem. Cell Biol., 66, 157–160. [DOI] [PubMed] [Google Scholar]
- 41.Upton C., Stuart,D. and McFadden,G. (1993) Identification of a poxvirus gene encoding a uracil-DNA-glycosylase. Proc. Natl Acad. Sci. USA, 90, 4518–4522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kunkel T.A., Roberts,J.D. and Zakour,R.A. (1987). Rapid and efficient site-specific mutagenesis without phenotypic selection. Methods Enzymol., 154, 367–382. [DOI] [PubMed] [Google Scholar]
- 43.Perez J., Gallego,C., Bernier-Villamor,V., Camacho,A., Gonzalez-Pacanowska,D. and Ruiz-Perez,L.M. (1999) Apurinic/apyrimidinic endouclease genes from the trypanosomatidae Leishmania major and Trypanosoma cruzi confer resistance to oxidizing agents in DNA repair-deficient Escherichia coli. Nucleic Acids Res., 27, 771–777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Henriksson J., Ashmd,L., Macina,R.A., Franke de Cazzulo,B.M., Cazzulo,J.J., Frasch,A.C.C. and Petersson,U. (1990) Chromosomal localisation of seven cloned antigen genes provides evidence of diploidy and further demonstration of karyotype variability in Trypanosoma cruzi. Mol. Biochem. Parasitol., 42, 213–223. [DOI] [PubMed] [Google Scholar]
- 45.Parikh S., Mol,C.D., Slupphaug,G., Bharati,S., Krokan,H.E. and Tainer,J. (1998) Base excision repair initiation revealed by crystal structures and binding kinetics of human uracil-DNA-glycosylase with DNA. EMBO J., 17, 5214–5226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Amorin D.S., Mello de Oliveira,J.A., Meira de Oliveira,J.S., Manco,J.C., Gallo,L,.Jr, Marin Neto,J.A. and Koberle,F. (1976) Heart block in Chagas’ disease. Arq. Bras. Cardiol., 28, 79–82. [PubMed] [Google Scholar]
- 47.Tanowitz H.B., Kirchhoff,L.V., Simon,D., Morris,S.A., Weiss,L.M. and Wittner,M. (1992) Chagas’ disease. Clin. Microbiol. Rev., 5, 400–419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Borst P. and Leeuwen,F.V. (1997) β-d-Glucosyl-hydroxymethyluracil, a novel base in African trypanosomes and other Kinetoplastida. Mol. Biochem. Parasitol., 90, 1–8. [DOI] [PubMed] [Google Scholar]
- 49.Häusler T., Stierhof,Y.D., Blattner,J. and Clayton,C. (1997) Conservation of mitochondrial targeting sequence function in mitochondrial and hydrogenosomal proteins from the early-branching eukaryotes Crithidia, Trypanosoma and Trichomonas. Eur. J. Cell Biol., 73, 240–251. [PubMed] [Google Scholar]
- 50.Doetsch P.W. and Cunningham,R.P. (1990) The enzymology of apurinic/apyrimidinic endonucleases. Mutat. Res., 236, 173–201. [DOI] [PubMed] [Google Scholar]
- 51.Camacho A., Arrebola,R., Peña-Díaz,J., Ruiz-Perez,L.M. and González-Pacanowska,D. (1997) Description of a novel eukaryotic deoxyuridine 5′-triphosphate nucleotidohydrolase in Leishmania major. Biochem. J., 325, 441–447. [DOI] [PMC free article] [PubMed] [Google Scholar]