Abstract
The pregnane X receptor (PXR) is a master regulator of xenobiotic clearance and is implicated in deleterious drug interactions (e.g., acetaminophen hepatotoxicity) and cancer drug resistance. However, small-molecule targeting of this receptor has been difficult; to date, directed synthesis of a relatively specific PXR inhibitor has remained elusive. Here we report the development and characterization of a first-in-class novel azole analog [1-(4-(4-(((2R,4S)-2-(2,4-difluorophenyl)-2-methyl-1,3-dioxolan-4-yl)methoxy)phenyl)piperazin-1-yl)ethanone (FLB-12)] that antagonizes the activated state of PXR with limited effects on other related nuclear receptors (i.e., liver X receptor, farnesoid X receptor, estrogen receptor α, peroxisome proliferator-activated receptor γ, and mouse constitutive androstane receptor). We investigated the toxicity and PXR antagonist effect of FLB-12 in vivo. Compared with ketoconazole, a prototypical PXR antagonist, FLB-12 is significantly less toxic to hepatocytes. FLB-12 significantly inhibits the PXR-activated loss of righting reflex to 2,2,2-tribromoethanol (Avertin) in vivo, abrogates PXR-mediated resistance to 7-ethyl-10-hydroxycamptothecin (SN-38) in colon cancer cells in vitro, and attenuates PXR-mediated acetaminophen hepatotoxicity in vivo. Thus, relatively selective targeting of PXR by antagonists is feasible and warrants further investigation. This class of agents is suitable for development as chemical probes of PXR function as well as potential PXR-directed therapeutics.
Introduction
The pregnane X receptor (PXR) is an adopted orphan nuclear receptor that functions as a traditional RXRα heterodimer. It controls gene expression in response to ligands as small as estradiol (268 Da) through large macrolide antibiotics [e.g., rifampicin (823 Da)]. In its role as a xenosensor, PXR regulates the expression of genes in response to potentially toxic endogenous (bile acids) and exogenous (xenobiotics) chemicals; however, ligand-activated PXR (e.g., St. John's Wort; hyperforin) has been implicated in deleterious drug interactions (e.g., reductions in serum levels of oral contraceptives and chemotherapeutics, such as camptothecins, indinavir, and cyclosporine), inconsistent drug bioavailability (e.g., digoxin), exclusion of central nervous system-acting drugs because of enhanced blood-brain barrier transporter expression (e.g., P-glycoprotein, multidrug resistance protein 2), and resistance to cancer drugs (for review, see Biswas et al., 2009). Our laboratory has demonstrated that one possible mechanism of PXR-mediated drug resistance involves the tumoral induction of cytochrome P450 and transporters (Gupta et al., 2008).
On the basis of these observations, it is evident that antagonists of PXR could be essential tools to probe PXR biology (e.g., the mechanisms underlying drug interactions mediated by PXR) and, importantly, serve as potential pharmaceuticals for a wide-based application in humans (e.g., the reversal of drug resistance and enhanced drug delivery) (for review, see Biswas et al., 2009; Harmsen et al., 2010). PXR is activated by a wide variety of structurally diverse compounds (Kortagere et al., 2010), including many anticancer drugs (Harmsen et al., 2010; Raynal et al., 2010). Although several agonists, such as rifampicin, are described in the literature, only a few weak PXR antagonists have been reported [e.g., trabectedin (ET-743), methyl 1-(5-(2-(3-benzyl-2-oxoimidazolidin-1-yl)-3,3-dimethylbutanamido)-4-hydroxy-6-phenyl-1-(4-(pyridin-2-yl)phenyl)hexan-2-ylamino)-3,3-dimethyl-1-oxobutan-2-ylcarbamate (A-792611), and sulforaphane] (for review, Biswas et al., 2009)]. Takeshita et al. (2002) originally described ketoconazole as a PXR antagonist, but its exact mechanism was elusive. Our laboratory was the first to report the likely mechanisms of ketoconazole's action by demonstrating its antagonist effect on the activation function 2 PXR coregulator interaction surface (Ekins et al., 2007; Huang et al., 2007; Wang et al., 2007). Subsequent studies demonstrated that ketoconazole exhibits distinct features pertinent to antagonists (compared with ligand pocket binding agonists), including smaller size, increased hydrophobicity and increased hydrogen bond features (Ekins et al., 2008).
The significant side effects of ketoconazole are due largely to its off-target effects (e.g., cortisol synthesis, hepatic toxicity), some of which are related to its effects as a cytochrome P450/steroid enzyme inhibitor (these effects are also dependent on the stereochemistry of the entire molecule, particularly the imidazole moiety) and others to the formation of the de-N-acetyl metabolite (Buchi et al., 1986; Rodriguez and Acosta, 1997; Rodriguez et al., 1999; Rodriguez and Buckholz, 2003). For example, in mice, the de-N-acetyl metabolite of ketoconazole is primarily responsible for hepatotoxicity. Furthermore, the down-regulation of serum amyloid A (SAA 1/2) and hepcidin are the only changes in gene expression that correlate with hepatotoxicity (Casley et al., 2007).
Therefore, we surmise that non-P450 inhibitors, as well as stereoisomers that are less toxic to hepatocytes and/or those that do not produce toxic de-N-acetyl metabolites, would be the first step in obtaining safer compounds that block PXR activation. Keeping these principles in mind, we developed several analogs of ketoconazole. One analog, 1-(4-(4-(((2R,4S)-2-(2,4-difluorophenyl)-2-methyl-1,3-dioxolan-4-yl)methoxy)phenyl)piperazin-1-yl)ethanone (FLB-12; Das et al., 2008), which lacks the imidazole group, retained PXR antagonist activity and exhibited markedly reduced CYP3A4 inhibition. Furthermore, we demonstrated that, compared with ketoconazole, FLB-12 was significantly less toxic to cells in vitro (Das et al., 2008). Here, we present data on FLB-12, which, in contrast to ketoconazole, antagonizes endogenous PXR activation but also has a “limited” antagonist profile on other related orphan and nonorphan nuclear receptors. FLB-12 is significantly less toxic to human and murine hepatocytes in vitro and in vivo. FLB-12 antagonizes PXR-mediated transcription and SN-38 drug resistance in vitro and duration of the loss of righting reflex by anesthesia in vivo. Furthermore, FLB-12 attenuates PXR-enhanced acetaminophen hepatotoxicity in mice. Together, these studies demonstrate that it is feasible to design nontoxic PXR antagonists that may be useful not only as probes for PXR biology but also as potential therapeutics in the treatment of cancer drug resistance and drug toxicity.
Materials and Methods
Materials.
LS174T cells were obtained from the American Type Culture Collection (Manassas, VA). Primary hepatocyte cultures for 4- to 6-week-old C57BL/6 mice were isolated using methods described previously with minor modifications of the two-step perfusion technique (Seglen, 1976). Human hepatocyte(s) were purchased from In Vitro Technologies, Inc. (Baltimore, MD) and maintained in InVitroGrow HI medium. All cell culture, immunochemistry, and transfection reagents were purchased from Invitrogen (Carlsbad, CA) unless otherwise indicated. Acetaminophen, pregnenolone-16α-carbonitrile (PCN), and fetal bovine serum were purchased from Sigma-Aldrich (St. Louis, MO). hPXR-LBD (S247W/S208W/C284W) triple mutant was generated using pM-Gal4-PXR-LBD as described published (Wang et al., 2008).
Cell Survival Assay.
To measure cell survival, we used the Promega MTS cell proliferation/cytotoxicity assay kit (Madison, WI) and employed a protocol previously published by our laboratory (Mani et al., 2005).
Transactivation Assays.
These assays were performed in 293T cells as described previously (Huang et al., 2007). In brief, transient transcription assays were carried out in 293T cells cotransfected with plasmids expressing hPXR (A), hPXR triple mutant (S247W/S208W/C284W) (B), mPXR (C), mCAR (D), LXRα (E), FXR (F), ERα (G), and PPARγ (H) with respective reporters, as shown in schematic diagrams (Figs. 1 and 2). After 8 h of transfection, ligands/drugs for respective receptors were added—25 μM FLB-12, and/or 10 μM rifampicin (Rif; hPXR ligand), and/or 10 μM PCN (mPXR ligand), and/or 5 μM N-(2,2,2-trifluoroethyl)-N-[4-[2,2,2-trifluoro-1-hydroxy-1-(trifluoromethyl)ethyl]phenyl]benzenesulfonamide (T0901317) (LXRα ligand), and/or 50 μM chenodeoxycholic acid (FXR ligand), and/or 20 μM estradiol (ERα ligand), and/or 10 μM rosiglitazone (PPARγ ligand), and/or 0.2 μM 1,4-bis[2-(3,5-dichloropyridyloxy)]benzene (mCAR ligand)—and incubated for a total period of 48 h. The cells were harvested in passive lysis buffer (Promega, Madison, WI), and luciferase activity was detected using the dual-luciferase reporter assay system from Promega in 20 μl of cell lysate using the 20/20n Luminometer (Promega, Madison, WI).
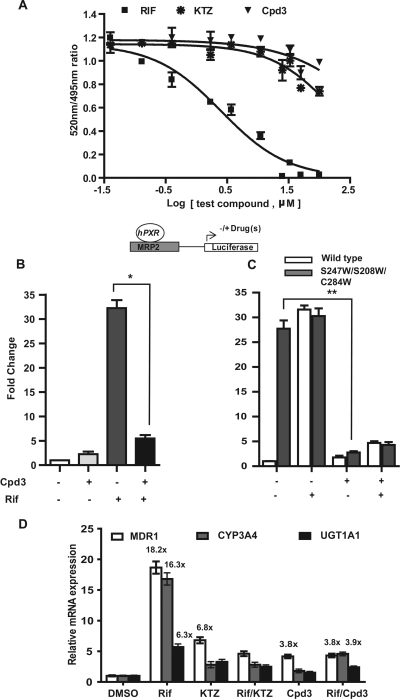
Fig. 1.
A, effect of azole compounds on the LanthaScreen TR-FRET PXR (SXR) competitive binding assay in vitro. Rifampicin was included as a positive control PXR ligand. B and C, effect of FLB-12 (Cpd3) on hPXR and hPXR triple mutant (S247W/S208W/C284W) activity. Transactivation assays were performed using plasmids encoding hPXR (B) or triple-mutant hPXR (C) with respective reporters/ligands, as shown in the schematic diagram. Transfected cells were exposed to vehicle (0.2% DMSO), 10 μM Rif, with or without 25 μM FLB-12. Columns, mean; bars, S.E.M.; *, P < 0.0048; **, P < 0.0280. D, effect of FLB-12 on PXR target gene expression. RT-qPCR for human PXR and target genes (CYP3A4, MDR1, and UGT1A1) was carried out in LS174T cells exposed to 10 μM Rif with or without 25 μM FLB-12 or KTZ or vehicle (0.2% DMSO) for 48 h. Total RNA was isolated and subjected to RT-qPCR. β-Actin served as an internal control. These experiments were performed two separate times each, assayed in triplicate. Columns, bars, S.D.
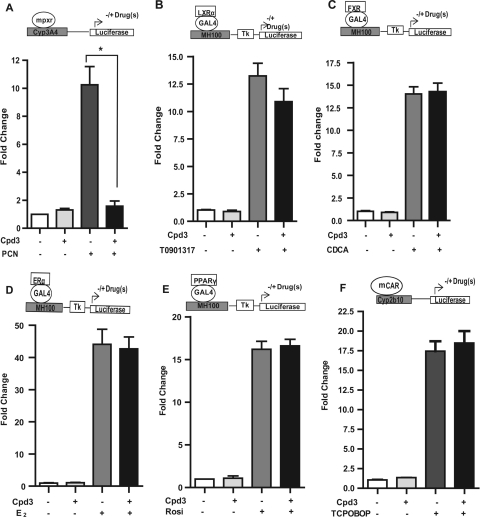
Fig. 2.
Effect of FLB-12 (Cpd3) on nuclear receptor target gene expression. Transactivation assays were performed using plasmids encoding mPXR (A), LXRα (B), FXR (C), ERα (D), PPARγ (E), and mCAR (F) with respective reporters/ligands, as shown in the schematic diagrams. Transfected cells were exposed to vehicle (0.2% DMSO), 10 μM PCN, 5 μM T0901317; 50 μM chenodeoxycholic acid (CDCA), 20 μM estradiol (E2), and 10 μM rosiglitazone (Rosi) with or without 10 μM FLB-12 or 0.2% DMSO for 48 h. These experiments were performed three times in triplicate. Columns, bars, S.E.M. *, P < 0.001. Tk, thymidine kinase; TCPOBOP, 1,4-bis[2-(3,5-dichloropyridyloxy)]benzene.
UGT Enzymatic Assay.
To assess for UGT enzyme activity, the UGT-Glo assay (Promega) was performed using UGT microsomes according to the manufacturer's protocol. The inhibitory activity of UGT1A1 was measured over a concentration range of 0 to 100 μM ketoconazole and FLB-12. Twenty-microliter UGT-Glo reactions were performed using UGT multienzyme substrate (0.4 mM) and microsomes. Two glucuronidation reactions were set up in parallel. Both reactions contained a source of UGT and proluciferin substrate, but only one contained the uridine 5′-diphosphoglucuronic acid. Reactions were incubated at 37°C for 2 h. Luciferin detection reagent plus d-cysteine was added, and reading was performed using a plate reader (luminometer). Relative light unit values were background-subtracted and then converted to percentage substrate consumed; values were plotted using Prism (GraphPad Software, San Diego, CA). Experiments were repeated three times, each in duplicate.
Time-Resolved Fluorescence Resonance Energy Transfer Assays.
A LanthaScreen time-resolved fluorescence resonance energy transfer (TR-FRET) PXR Competitive Binding Assay was conducted according to the manufacturer's protocol (Invitrogen). In brief, at first, serial dilutions of test compounds [FLB-12, ketoconazole (KTZ), or Rif; diluted in TR-FRET PXR assay buffer from Invitrogen) were dispensed into triplicate wells of a black nontreated 384-well assay plate (Corning Life Sciences, Lowell, MA). Second, 5 μl of Fluormone PXR Green was added into each well. Finally, 5 μl of a master mix containing hPXR ligand-binding domain, terbium-labeled anti-glutathione transferase (final concentration, 10 nM), and dithiothreitol (final concentration, 0.05 mM) was added into each well. The content was mixed briefly (10 s) and the plate was incubated in the dark at room temperature (22–24°C) for 1 h. TR-FRET was measured using a SpectraMax M5 Microplate Reader (Molecular Devices, Sunnyvale CA), with an excitation wavelength of 340 nm and emission wavelengths of 520 and 495 nm. TR-FRET ratio was calculated by dividing the emission signal at 520 nm by the emission signal at 495 nm. Data are expressed as a TR-FRET ratio. Error bars represent the S.E.M. of duplicate wells from two separate experiments. The curve was fit to data (TR-FRET ratio versus log test compound) using a sigmoidal dose response (variable slope) equation in Prism software.
Coactivator-Dependent Receptor Ligand Assays.
hPXR LBD [35S]methionine-labeled proteins were prepared using in vitro-transcribed and -translated protein (TnT; Promega). The GST-SRC-1 protein was expressed in Escherichia coli BL21 cells and purified using glutathione-Sepharose (GE Healthcare, Chalfont St. Giles, Buckinghamshire, UK) and pull-down experiments performed as described previously (Huang et al., 2007). In brief, purified GST fusion protein (5 μg) was incubated with 5 μl of in vitro translated 35S-labeled protein with overnight shaking at 4°C in the presence of 0.2% DMSO (vehicle), 10 μM rifampicin, 25 μM FLB-12, or the combination of rifampicin and FLB-12. GST beads were used as a negative control. The bound protein was washed three times, and the beads were collected by centrifugation at 3000 rpm for 5 min. The bound protein was eluted into SDS sample buffer and then subjected to 10% SDS-polyacrylamide gel electrophoresis, and the gel was processed has been reported previously (Huang et al., 2007). Each experiment was repeated twice, and each blot was exposed a minimum of three times.
RNA Preparation and RT-Quantitative PCR Assays.
LS174T cells were treated with 10 μM rifampicin, 10 μM rifampicin plus 20 μM FLB-12, or DMSO; after 48 h, total RNA was extracted from cells with an RNeasy Mini kit (QIAGEN Inc., Valencia, CA), according to the manufacturer's instructions. Wild-type C57BL/6 mice were treated with DMSO, ketoconazole, or FLB-12 (300 mg · kg−1 · day−1) for 3 days; then the animals were scarified, and total RNA was isolated from the liver using the QIAzole, Qiagen RNeasy Mini kit, according the Qiagen protocol. Two micrograms of total RNA was reverse-transcribed with random hexamer primers and SuperScript III-RT (Invitrogen). The RT-quantitative PCR for MDR1, CYP3A4, UGT1A1, hepcidins, SAA (serum amyloid A) and β-actin was performed using TaqMan universal PCR master mix and TaqMan probes, as described previously (Huang et al., 2007; Gupta et al., 2008). The following assays, available on demand, were procured from Applied Biosystems (Foster City, CA): CYP3A4, Hs00604506_m1; MDR1, Hs00184500_m1, UGT1A1, Hs02511055_s1, ACTB 4333762, hepcidin-1 Mm00519025_m1; hepcidin-2, Mm00842044_gl; SAA1, Mm00656927_gl; SAA3, Mm00441203_m1; GAPDH, Mm999999915_g1. PCR conditions for assays were 50°C for 2 min and 95°C for 10 min, followed by 40 cycles of amplification (95°C for 15 s, then 60°C for 1 min). The relative fold change in mRNA expression in treated samples compared with controls was calculated using comparative Ct method. The percentage inhibition (antagonism) was calculated using the difference in fold expression in cells treated with rifampicin versus rifampicin + FLB-12, subsequently divided by fold expression in rifampicin-treated cells (× 100%).
Animal Studies.
C57BL/6 mice (6–8 weeks old; The Jackson Laboratory, Bar Harbor, ME) were used for all animal studies. The transgenic and knockout mice have been described before. and the loss-of-righting-reflex studies were performed as described previously (Huang et al., 2007) The transgenic and knockout mice were obtained from Dr. Wen Xie (University of Pittsburgh, Pittsburgh, PA). Oral (gavage) dosing of FLB-12 (100, 200, 300, and 600 mg/kg) and ketoconazole (100, 200, 300, and 600 mg/kg) was performed using drugs solubilized in corn oil and dosed twice per day for 4 days. Mice were euthanized on day 4, and serum and liver were isolated for the estimation of liver enzyme activity, hematoxylin and eosin staining, and gene expression studies (qRT-PCR and immunoblot). For the serum transaminase studies and gene expression studies in the mouse liver, fixed doses of 300 mg/kg ketoconazole and FLB-12 were used, respectively. LORR studies were performed as described previously (Huang et al., 2007). For LORR studies, the FLB-12 dose was 300 mg · kg−1 · day−1. For studies pertaining to acetaminophen (APAP) toxicity, the dose and schedule of administration of acetaminophen was directly adopted from a study published previously (Guo et al., 2004). In these studies, FLB-12 (300 mg · kg−1 · day−1 administered in two divided doses per day) was started on day −1 before PCN dosing (75 mg/kg i.p. for 2 days) and continued through acetaminophen dosing (350 mg/kg i.p.) for a total of 4 days. For pxr(+/+), seven treatment groups (8 mice/group) were developed: vehicle control (100% corn oil), PCN, FLB-12, APAP, PCN + APAP, FLB-12 + APAP, and PCN + FLB-12 + APAP. Total blood and serum were isolated 24 h after acetaminophen dosing for the assessment of ALT levels.
Immunoblot.
Individual or pooled livers from the treated groups of FLB-12 and KTZ (six mice in each group) were homogenized on ice in Tris-HCl buffer (0.01 M, pH 7.4) containing protease inhibitor cocktail (Sigma-Aldrich). The homogenate was centrifuged at 4000g for 10 min (4°C), the pellet was discarded, and the supernatant was centrifuged at 100,000g for 30 min (4°C). The resulting pellet was resuspended in Tris-HCl buffer with protease inhibitor cocktail; expression of MDR1 (anti-P-gp-C219; Signet Labs, Dedham, MA) and Na+/K+-ATPase (loading control; Santa Cruz Biotechnology, Santa Cruz, CA) were performed as reported previously by Huang et al. (2007).
LC/MS/MS.
Blood and tissue samples were analyzed using electrospray ionization/chemical ionization (ESCi) multimode ionization (TQ Tandem Quadrupole Mass Spectrometer; Waters Corp, Parsippany, NJ). The best ionization was achieved by positive ion electrospray. The MS/MS detection was developed and optimized using IntelliStart software. Anthracene-2 was used as internal standard (Supplemental Fig. 8). The elution peak for FLB-12 occurred at 1.15 min and contained an isomer peak at 1.13 min. To demonstrate sensitivity, a linear calibration curve was created by serial dilutions of the stock standard (1–1000 ng/ml) in mouse blood plasma (data not shown; r2 = 99.9091 × 10−2). The lower limit of quantitation for this assay is 5 pg on the column, with a signal/noise ratio of 168.00. Tissue concentrations were established by normalizing to total protein content (NanoDrop 1000 spectrophotometer; Thermo Fisher Scientific, Waltham, MA) in cleared lysate (membrane-free) injected into columns.
Transaminase Enzymatic Assay.
Mouse serum was collected and prepared according to the manufacturer's instructions for the MaxDiscovery ALT and Aspartate (AST) Transaminase enzymatic assay kit (Bio-Scientific Corp, Austin, TX). A standard curve was also generated using the pyruvate control dilutions, and enzymatic activity in serum samples was reported in units per liter. Measurements were performed on a SpectraMax M5 Microplate Reader (Molecular Devices).
Manual Patch Clamp Assay.
Whole-cell patch-clamp recordings were made at 37 ± 2°C from human embryonic kidney 293 cells stably transfected with hERG gene, maintained in culture with selective agent (G148; Sigma-Aldrich). The cells were transferred to a bath mounted onto the heated stage of an inverted microscope. The bath was perfused with an external solution composed of 140 mM NaCl, 5 mM KCl, 1.8 mM CaCl2, 1 mM MgCl2, 10 mM glucose, and 10 mM HEPES, pH 7.4. The electrodes used were pulled from borosilicate glass using a micropipette puller (PMP-202; MicroData Instruments Inc., S. Plainfield, NJ) and filled with internal pipette solution containing 140 mM KCl, 1 mM MgCl2, 5 mM EGTA, 10 mM HEPES, 4 mM Mg-ATP, and 10 mM sucrose. The resistance of the electrode was 5 to 8 MΩ before touching the cell membrane. Currents were acquired at a rate of 2 Hz and filtered using a low-pass four-pole Bessel filter with cut-off rate set at 650 Hz. Baseline condition currents were recorded using the pClamp 10 acquisition suite (Molecular Devices) after achieving a gigohm seal, after a 2-min equilibration period. Currents were also recorded after 5 min of exposure to each concentration of ketoconazole (Sigma-Aldrich) or FLB-12. The cells were stimulated every 10 s with the pulse as denoted in Fig. 6A. Current analysis was performed on digitized current records using Clampfit 10. The software calculated current peak and activation, inactivation, and deactivation constants automatically. Repeat Student's t tests were used to assess statistical significance in changes from baseline conditions. Statistical significance was declared at P < 0.05.
Fig. 6.
hERG tail current density averages obtained by measuring the hERG tail peak amplitude at −7 mV in baseline conditions and in the presence of 5, 10, 20, and 50 μM ketoconazole (Table 2) or FLB-12 (Cpd3; Table 3). A, representative recordings of activating and inactivating hERG currents in baseline conditions and after exposure of the cells to 50 μM ketoconazole or 50 μM FLB-12. B, current density was measured from seven cells, averaged, normalized against baseline current density, and corrected for time and solvent effects. KTZ IC50 is 18 μM; FLB-12 IC50, not available. Statistical comparisons between current density levels after drug exposure and at baseline were made using repeat paired Student's t tests: differences were considered significant at P ≤ 0.05.
Electrochemiluminescence.
Chemiluminescence detection technology (Meso Scale Discovery, Gaithersburg, MD) was employed to measure ERK (total) and phospho-ERK in lysate of LS174T cells after cells were exposed to FLB-12 (0–25 μm), KTZ (0–25 μm), or vehicle control. Cell lysate (5 μg) was coated on 96-Well MULTI-ARRAY MSD high-bind plates according to manufacturer's instructions. The wells were incubated at room temperature for 2 h and then blocked with MSD Blocker A solution diluted in phosphate-buffered saline for 1 h at room temperature. After incubation with primary antibody (1 μg/ml) for 1 h at room temperature, the wells were washed with Blocker A solution and then incubated with the secondary antibody (1 μg/ml). After an additional hour of incubation, the wells were washed and refilled with 150 μl of MSD Read Buffer (1×) with surfactant. Readings were measured using the SECTOR Imager 2400 according to the manufacturer's instructions, and signals (light units) were plotted against the different concentration of compounds to phospho-ERK expression. Experiments were performed in duplicate and repeated twice. Anti-ERK1/2 antibody was used to normalize the data for protein loading.
Statistical Analysis.
Student's t test (two-tailed; α,β = 0.05) was used to analyze differences between two groups. Trends analyses between three or more groups were performed using analysis of variance. All the results were expressed as means ± S.E.M. P < 0.05 was considered significant. All analyses were performed using Prism software.
Results
FLB-12 Antagonizes PXR Activation In Vitro.
FLB-12 has been shown to antagonize PXR activation in transformed hepatocyte cell lines (Das et al., 2008). Furthermore, this compound is significantly less toxic to cells than KTZ (Das et al., 2008). However, its effect on endogenous PXR activation is unknown. First, using the TR-FRET assay, we demonstrated that the FLB-12 does not quench fluorescence, suggesting low affinities for binding and competing with strong ligands within the ligand binding pocket (Fig. 1A). However, FLB-12 (25 μM) was found to significantly antagonize 10 μM rifampicin-mediated activation of human PXR in a PXR-transactivation assay (Fig. 1B). Because full-length PXR cannot be purified from mammalian systems, we employed a genetic approach toward determining whether FLB-12 might bind outside the ligand binding pocket, as suggested by experiments in Fig. 1, A and B. We used a PXR ligand-binding pocket triple mutant plasmid (S247W/S208W/C284W) that was constructed by mutating three amino acids to more bulky amino acid (tryptophan) at positions Ser247/Ser208/Cys284, resulting in ligand binding occlusion and, by serendipity, ligand-independent constitutive activation (Chrencik et al., 2005; Wang et al., 2008). We found that FLB-12 (25 μM) significantly antagonized hPXR-LBD triple mutant in the PXR-transactivation assay (Fig. 1C), which implies that PXR is likely to be antagonized by FLB-12 at a site distinct from the ligand-binding pocket. Indeed, FLB-12 is able to disrupt the interaction between PXR and steroid receptor co-activator 1 protein on a protein pull-down assay (Supplemental Fig. 1A).
To determine whether endogenous PXR is antagonized by FLB-12, LS174T cells were exposed to 10 μM rifampicin, 25 μM ketoconazole, and/or 25 μM FLB-12. After 48 h of exposure to drug(s), total RNA was isolated from treated cells and subjected to RT-qPCR of the PXR target genes CYP3A4, MDR1, and UGT1A1. The human PXR ligand rifampicin induced a 16.3-, 18.2-, and 6.3-fold mean change in CYP3A4, MDR1, and UGT1A1 mRNA abundance, respectively (Fig. 1D). FLB-12 alone induces the expression of MDR1 (3.8-fold), but has negligible effect on CYP3A4 (1.8-fold) and UGT1A1 (1.7-fold). It is noteworthy that there is no apparent induction of MDR1 protein by FLB-12 in mouse liver, suggesting lack of significant induction of MDR1 in vivo (Supplemental Fig. 1B). However, in the presence of rifampicin, FLB-12 inhibits rifampicin-mediated CYP3A4, MDR1, and UGT1A1 transcription by ∼76, 79, and 60%, respectively.
FLB-12 Effects on Orphan Receptor Mediated Transcription In Vitro.
We have shown previously that ketoconazole antagonizes the ligand-mediated activation of mPXR, CAR, LXR, and FXR (receptors closely related to PXR); however, there was no effect on ERα- or PPARγ-mediated activation (Huang et al., 2007). FLB-12 (25 μM) was found to inhibit the 10 μM PCN-mediated activation of mPXR (Fig. 2A), whereas it showed no significant inhibitory effects on the ligand-mediated activation of LXR (5 μM T0901315; Fig. 2B), FXR (50 μM chenodeoxycholic acid; Fig. 2C), ERα (20 μM estradiol; Fig. 2D), PPARγ (10 μM rosiglitazone; Fig. 2E), and mCAR (0.2 μM 1,4-bis[2-(3,5-dichloropyridyloxy)]benzene; Fig. 2F) in our reporter assays. It is noteworthy that FLB-12 did not significantly activate these nuclear receptors (Fig. 2B–F).
LC/MS/MS of FLB-12 in the Liver, Blood, and Intestine.
The LC and MS-MS spectrum for FLB-12 is shown in Supplemental Fig. 2. FLB-12 is orally absorbed in mice. A single oral dose of FLB-12 (200 mg/kg) yields an AUC(0-t) values of ∼121.7, 254.9, and 537.7 μM · h−1 in blood, liver, and proximal small bowel, respectively (Supplemental Fig. 3). This suggests that there is either accumulation and/or persistence of drug in these tissue compartments. At 2 h after dosing, the drug concentration in blood and tissues ranges from ∼25.4 to 38.1 μM, respectively. At 48 h, there is virtually no drug in the blood compartment (less than lower limit of quantitation); however, the concentrations in the tissues range from 0.5 to 1.9 μM. The approximate half-life of FLB-12 is ∼0.53, 0.77, and 8.59 h in blood, liver, and proximal small bowel, respectively. This implies that oral dosing [multiple times per day (e.g., twice per day)] would be necessary to maintain tissue levels in the micromolar range (one exponential phase, r2 ∼ 0.99; Prism software; Supplemental Fig. 3). These data also suggest that there are tissue-specific residence times for the drug, and perhaps highly metabolic tissues such as blood and liver clear the drug faster than other tissues. Therefore, we made the assumption that if we increased the dose of FLB-12 and administered the compound at least three times per day, after 2 or more days of continuous dosing, steady-state trough levels should approximate ≳10 μM. Accordingly, we chose to perform a second experiment in which we dosed mice at 300 mg/kg three times per day by gavage. However, after 1 day we reduced the frequency to two times per day because of significant throat irritation in >90% of mice. Eight doses were delivered before sacrificing the mice. The mice were sacrificed 10 h after the last delivered dose by gavage. Because steady states for most drugs are usually reached after ∼5 · t1/2, we surmised that, at this point, all mice would exhibit steady-state blood and tissue concentrations of FLB-12. The blood and tissue concentrations after eight doses of FLB-12 are shown in Table 1.
TABLE 1.
FLB-12 concentrations in mouse and human tumor xenograft tissues
Six- to eight-week-old C57BL/6 female and SCID/NOD mice xenografted with LS174T cells (group average flank tumor volume ∼0.45 mm3) were injected with rifampicin (50 mg · kg−1 · day−1 i.p.) and/or orally gavaged FLB-12 (300 mg · kg−1 · day−1) for 4 days. All tissues were harvested on day 4 for assessment of FLB-12 concentrations, as described under Materials and Methods.
| Samples & Treatment Groups | Concentration |
|---|---|
| μM | |
| Plasma | |
| FLB-12 | 28.0 ± 3.5 |
| Rifampicin/FLB-12 | 33.31 ± 1.69 |
| Liver | |
| FLB-12 | 23.45 ± 1.55 |
| Rifampicin/FLB-12 | 23.1 ± 4.4 |
| Tumor (LS174T xenografts) | |
| FLB-12 | 24.5 ± 2.1 |
| Rifampicin/FLB-12 | 23.475 ± 3.325 |
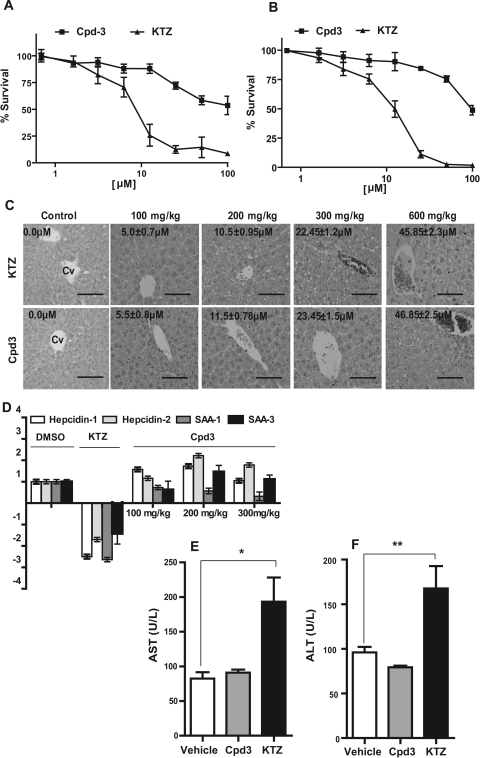
FLB-12, Unlike Ketoconazole, Is Significantly Less Toxic to Hepatocytes In Vitro and In Vivo.
At concentrations >10 μM, FLB-12 is significantly less toxic to primary mouse (Fig. 3A) and human (Fig. 3B) hepatocytes. Indeed, vacuolization and cellular ballooning is a hallmark of drug-induced toxicity (Lee, 1995) and ketoconazole hepatotoxicity (Lewis et al., 1984). On hematoxylin and eosin-stained sections of mice livers exposed to multiple doses of FLB-12 or ketoconazole, it is evident that there is a profound increase in vacuolization and cellular ballooning at ketoconazole doses ≥200 mg/kg (≳10 μM); however, these effects are only beginning to be seen at concentrations of FLB-12 ≳23 μM. At the highest dose delivered, 600 mg/kg ketoconazole plus FLB-12, there was significant cellular ballooning in the ketoconazole-exposed mice. There was a less dramatic effect on livers exposed to FLB-12 (Fig. 3C). At this dose, the mouse livers and sera were isolated, and tissue was extracted for gene expression and enzymatic assays, respectively. FLB-12, unlike ketoconazole, does not inhibit expression of hepcidin-1 and -2 and SAA-1 and -3 (Fig. 3D). In addition, high serum levels of ALT and AST were statistically significant only in the ketoconazole-treated mice. The FLB-12-exposed mice had AST (Fig. 3E) and ALT (Fig. 3F) levels similar to those of control mice.
Fig. 3.
Effect of FLB-12 (Cpd3) on liver cells in vivo. Primary murine (A) and human (B) hepatocytes were exposed to ketoconazole (1–100 μM), FLB-12 (1–100 μM), or vehicle (0.2% DMSO) for 48 h. Cells were then harvested and subjected to MTS assay (Materials and Methods). Experiments were repeated three separate times each in triplicate. Points, mean; bars, S.D. C, mice were treated for 3 days with 0 to 600 mg · kg−1 · day−1 p.o. ketoconazole or FLB-12; after animals were euthanized, liver tissues were stained with hematoxylin and eosin. Representative sections from control and drug-treated mice are shown. For orientation, the first (control) section denotes the central vein (Cv). There is striking ballooning of hepatocytes in ketoconazole-treated mice (liver concentration, ∼45.9 μM). Drug concentrations in the liver, analyzed as described under Materials and Methods, are illustrated in black. Hematoxylin and eosin staining; magnification, 20×; scale bar, 50 μm. D, effect of ketoconazole and FLB-12 on the expression of hepcidin-1 and -2 and SAA-1 and -3. Mice were treated for 3 days with 100 (n = 3), 200 (n = 3), or 300 mg · kg−1 · day−1 (n = 2) FLB-12 or 100 (n = 3) mg · kg−1 · day−1 of ketoconazole, by oral administration. On day 3 after euthanasia, for each dose group, the livers were harvested and pooled, and total RNA was isolated from the pooled fraction and subjected to RT-qPCR, as described under Materials and Methods. The same RNA was assayed in three separate RT-qPCR experiments, each performed in duplicate. Columns, bars, S.E.M. Transaminase enzymatic assays were performed in pooled liver samples obtained from mice treated at 300 mg · kg−1 · day−1 for AST (E) and ALT (F) as described under Materials and Methods. The assay was repeated two times each, performed in triplicate. Columns, bars, S.E.M. *, P < 0.001; **, P < 0.001.
FLB-12 Inhibits PXR-Mediated SN-38 Drug Resistance in Colon Cancer cells In Vitro.
Previously published data have demonstrated that PXR ligands such as rifampicin can induce increased resistance to SN-38-mediated cytotoxicity, in part through enhanced glucuronidation via UGT1A1 (Gupta et al., 2008; Raynal et al., 2010). We exposed the LS174T colon cancer cells (high endogenous PXR; transduced with scrambled shRNA or PXR shRNA; Supplemental Figs. 4 and 5) to vehicle, SN-38, 10 μM rifampicin, and/or 25 μM FLB-12 (Fig. 4, A and B). The data show that FLB-12 significantly inhibited rifampicin-mediated effects on the cytotoxicity of SN-38 in scrambled shRNA cells (SN-38/Rif versus SN-38/Rif/FLB-12; one-way ANOVA P < 0.00001) (Fig. 4A). FLB-12 did not alter rifampicin induced SN-38 cytotoxicity in PXR shRNA cells (Fig. 4B). In this context, because azole compounds inhibit CYP450 and UGT enzymes, which could independently affect SN-38 pharmacodynamics in cells, it has been noted previously that ketoconazole inhibits UGT1A1 and UGT1A9 (Yong et al., 2005); however, FLB-12 has no significant effects on UGT1A1 activity (a major determinant of SN-38 detoxification in cells) (Supplemental Fig. 6).
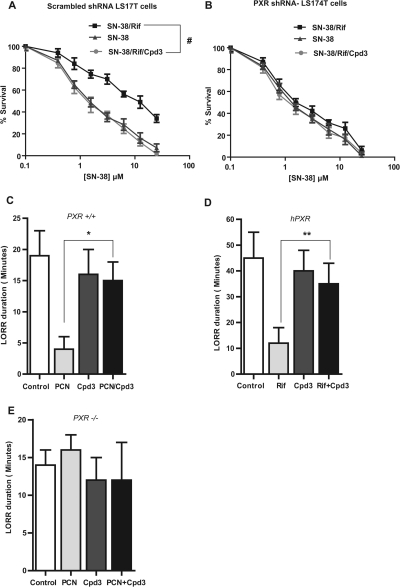
Fig. 4.
The effects of FLB-12 (Cpd3) on PXR-induced chemoresistance in vitro and anesthetic (2,2,2-tribromoethanolamine) metabolism in vivo. A and B, cell cytotoxicity from the anticancer drug SN-38 in LS174T (transduced with scrambled or PXR shRNA) colon cancer cells pretreated with Rif or vehicle (0.2% DMSO) and/or FLB-12. LS174T cells were pretreated (for 24 h) with 15 μM Rif or vehicle and/or 10 μM FLB-12 and then exposed to SN-38 (0.0–50 μM) for another 24 h. Each experiment was performed three times in triplicate. Points or columns, mean; bars, S.D. #, P < 0.0001. FLB-12 modulates anesthetic (2,2,2-tribromoethanolamine) metabolism in vivo. Loss of righting reflex (LORR) duration was performed as described under Materials and Methods (Animal Studies) to determine the effect of FLB-12 in pxr(+/+) (C), hPXR (D), and pxr(−/−) mice (E). Columns, bars, S.E.M. *, P < 0.001; **, P < 0.05.
FLB-12 Antagonizes PXR Activation In Vivo.
Previously published data have established that ketoconazole reverses the duration of the loss of righting reflex mediated by PXR ligands in pxr(+/+) and humanized (h) PXR mice, but not in pxr(−/−) mice (Huang et al., 2007). In this assay, the consequences of activating PXR can be studied using mice challenged with 2,2,2-tribromoethanol (Avertin) anesthesia, where the drug-induced change in the duration of the loss of righting reflex (LORR) acts as a phenotypic measure of PXR target gene activity and xenobiotic metabolism (Huang et al., 2007). We verified that the effects of ketoconazole were pxr-specific by comparing wild-type [pxr (+/+)] and pxr-null [pxr(−/−)] mice. We used the same assay to study the effects of FLB-12 on this pxr-mediated phenotype in vivo. Because FLB-12 inhibits both human and mouse pxr (when ligand tethered), we performed LORR studies in all three genotypes: pxr(+/+), hPXR, and pxr (−/−). The data indicate that FLB-12 reverses the duration of LORR mediated by a pxr/PXR ligand only in pxr(+/+) (Fig. 4C) and hPXR (Fig. 4D) but not in pxr(−/−) (Fig. 4D). These results indicate that 1) FLB-12 antagonizes pxr/PXR function in vivo and 2) FLB-12 has pxr/PXR-specific effects in vivo.
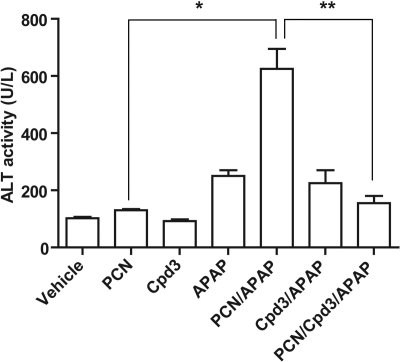
A second model used to study the in vivo activation effects of PXR in the liver shows a significant enhancement of acetaminophen-induced liver toxicity in mice (Guo et al., 2004) (Wolf et al., 2005; Cheng et al., 2009). In this model, pxr(+/+) were pretreated with the pxr activator PCN for 2 days before administration of one APAP intraperitoneal dose dissolved in alkaline solution. FLB-12 was administered 1 day before administration of PCN and continued for a total of 4 days. The mice were sacrificed 24 h after APAP administration, and blood was immediately aspirated from a cardiac puncture for determination of ALT levels (see Materials and Methods). In pxr(+/+) mice, PCN significantly augments APAP-induced ALT levels, signifying hepatotoxicity (Fig. 5; *, P < 0.05). These data also indicate that FLB-12 abrogates PCN-mediated APAP hepatotoxicity (Fig. 5; **, P < 0.05).
Fig. 5.
Determination of ALT activities in serum of PXR wild-type mice after PCN, APAP, and FLB-12 (Cpd3) combinatorial treatment groups. PXR wild-type mice (n = 8/groups) were pretreated with corn oil (vehicle) or FLB-12 on day −1 before PCN dosing and continued through acetaminophen dosing for a total of 4 days. Blood was collected and serum was separated as described under Materials and Methods. ALT levels were measured according to the manufacturer's instructions. Columns, bars, S.E.M. *, P < 0.05; **, P < 0.05.
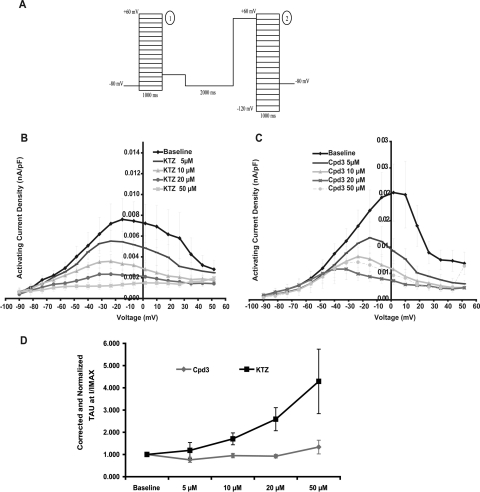
Effect of FLB-12 and Ketoconazole on hERG Current Amplitude.
hERG currents activate in an upward curve after depolarization and deactivate/inactivate when the cells' holding potential is brought back to −90 mV (Fig. 6A). Ketoconazole and FLB-12 caused concentration-dependent decreases in hERG activating current density. A shift in voltage for the activation curve was observed for both compounds, which suggests that both ketoconazole and FLB-12 were more potent hERG inhibitors at more depolarized holding potentials. In addition, ketoconazole and FLB-12 produced a concentration-dependent inhibition of hERG tail current density. At the peak of the current/voltage curve (measured immediately after the repolarization of the cells), ketoconazole inhibited 69% of the current, whereas FLB-12 inhibited 42% of the hERG current, at approximately +7 mV. The IC50 for ketoconazole was 18 μM. but IC50 for FLB-12 was not available (Fig. 6B; Tables 2 and 3). The reversal potential for the current was extrapolated to −98 mV, close to the theoretical reversal potential, calculated to be −85 mV. It did not vary after exposure to either ketoconazole or FLB-12, suggesting that neither molecule alters the selectivity of the channels.
TABLE 2.
Normalized hERG tail current values at −7 mV when exposed to ketoconazole and corrected for the DMSO effect
| Conditions | Corrected Normalized Current Density | S.E.M. | P | n |
|---|---|---|---|---|
| Baseline | 1.000 | 0.000 | N.A. | 7 |
| Ketoconazole | ||||
| 5 μM | 0.706 | 0.086 | 0.0064 | 7 |
| 10 μM | 0.608 | 0.084 | 0.0007 | 7 |
| 20 μM | 0.474 | 0.078 | 0.0001 | 7 |
| 50 μM | 0.302 | 0.038 | 0.0000 | 7 |
N.A., not available.
TABLE 3.
Normalized hERG tail current values at −7 mV when exposed to FLB-12 and corrected for the DMSO effect
| Conditions | Corrected Normalized Current Density | S.E.M. | P | n |
|---|---|---|---|---|
| Baseline | 1.000 | 0.000 | N.A. | 7 |
| FLB-12 | ||||
| 5 μM | 0.822 | 0.067 | 0.0126 | 7 |
| 10 μM | 0.732 | 0.072 | 0.0013 | 7 |
| 20 μM | 0.644 | 0.125 | 0.0058 | 7 |
| 50 μM | 0.576 | 0.123 | 0.0026 | 7 |
N.A., not available.
Effect of FLB-12 and Ketoconazole on hERG Activation, Deactivation, and Inactivation Kinetics.
Ketoconazole caused a concentration-dependent deceleration in hERG activation; such an effect was not observed with FLB-12. This translated into a hERG current that activated more slowly and reached a lower level in the presence of ketoconazole. In contrast, the activating current measured in the presence of FLB-12 was lowe, but activated at the same rate regardless of the concentration of FLB-12 present in the bath (Fig. 7).
Fig. 7.
Effects of ketoconazole and FLB-12 (Cpd3) on the activation phase of the hERG current. Voltage-pulse protocol used. A, pulse 1 was used to determine channel activation kinetics and steady-state current-voltage relationship, whereas pulse 2 was used to determine the channel inactivation and current rectification. B and C, current/voltage relationship for current measured at the end of depolarizing step from pulse 1 when exposed to ketoconazole (B) and to FLB-12 (C). D, changes in hERG activation time constant induced by ketoconazole and FLB-12: activation constants were corrected for the effect of DMSO on activation kinetics.
Neither ketoconazole nor FLB-12 had any effect on the deactivation and inactivation kinetics of the current. Inactivation kinetics decelerated with voltage, but varying concentrations of either ketoconazole or FLB-12 did not enhance the inactivation kinetics of the currents. It is a coincidence that the currents that inactivate faster were used to test FLB-12, leading to inactivation constant values (τ) that were lower for FLB-12 currents (i.e., faster inactivation). Regardless of the rate of inactivation, statistical tests confirmed that the current kinetics exhibited pharmacological sensitivity identical to that of ketoconazole and FLB-12 (data not shown).
Discussion
Inappropriate PXR activation has some undesired and important pathophysiologic consequences (e.g., promoting drug interactions that alter drug pharmacokinetics, efficacy, and toxicity and cancer drug resistance). In addition, there is a lack of nontoxic PXR antagonists that would allow researchers to chemically reduce the activity of PXR in different biologic systems, so that its molecular consequences may be fully characterized. Hence, from a biological and clinical perspective, developing PXR-specific antagonists is an important but daunting task. Based on some well known observations about the chemical moieties of ketoconazole, we explored the possibility of first developing less toxic analogs of ketoconazole that still served as PXR antagonists. We demonstrated that imidazole moiety-deleted analogs of ketoconazole are less toxic to cells. Further modifications of the halogen on the phenyl group led to the discovery of compounds that could inhibit PXR but not CYP3A4 (Das et al., 2008).
One lead compound, FLB-12, was demonstrated to abrogate endogenous PXR activation in vitro and in vivo and was less toxic to liver cells in vivo compared with ketoconazole. Indeed, markers of azole-mediated hepatotoxicity (e.g., liver enzymes, hepcidin and SAA expression profile) suggested that FLB-12 was likely to have significantly less toxic effect on the liver than ketoconazole. It is noteworthy that FLB-12 did not inhibit mCAR, LXR, and FXR to the same extent that ketoconazole did (Huang et al., 2007), which suggests that there are analogs that could be made more PXR-specific without compromising their inhibitory potency.
Ketoconazole has off-target effects on many enzymes and protein channels (e.g., CYP450, UGTs, 11β-hydroxylases, BK channels, hERG) (Loose et al., 1983; Yong et al., 2005; Power et al., 2006; Yoshida and Niwa, 2006; Nardi and Olesen, 2008). To begin to explore some of the off-target properties of FLB-12, compared with ketoconazole, we evaluated the potency of ketoconazole and FLB-12 in modifying hERG current density and current kinetics. The data presented herein suggest that FLB-12 is significantly less potent than ketoconazole at inhibiting the hERG tail current. Both ketoconazole and FLB-12 exhibit voltage-dependent hERG tail current inhibition, with a more pronounced effect at more depolarized potentials. This effect may translate into a decreased repolarization reserve, because the demand for hERG current in vivo is greatest at the end of phase 0 of the action potential, when the cardiac cells are maximally depolarized. Unfortunately, this is also a window of membrane potentials where the cardiac cells in the ventricles spend a significant amount of time (approximately 350 ms per heartbeat). The loss of repolarization from ketoconazole exposure is expected to be greater than from FLB-12; ketoconazole is twice as potent as FLB-12 at decreasing current peak, and the activation of the current is decreased for ketoconazole-treated samples (but not FLB-12). This is likely to lead to a greater decrease in charge transport (the area under the activating current curve) for ketoconazole. The kinetics of deactivation/inactivation was not affected by the compounds tested.
Another important effect of ketoconazole is its inhibition of cytochrome P450s (e.g., CYP3A4) and UGTs (e.g., UGT1A1). We have extended our findings to show that FLB-12 does not significantly inhibit UGT1A1 (Supplemental Fig. 6). Hence, it is possible to change the activity of analogs to several nonreceptor protein targets without affecting their potency toward the target protein of interest, PXR. Indeed, FLB-12 increased the cytotoxicity of SN-38/Rif in colon cancer because of its effects on PXR rather than azole-mediated (e.g., ketoconazole-mediated) inhibition of CYP3A4 (Das et al., 2008) and/or UGT1A1 (Fig S6) (Yong et al., 2005).
On the other hand, some properties relating to ketoconazole's chemical structure have potential pharmacological benefits in human pathophysiology (e.g., BK channels are pharmacological targets for stroke treatment). For example, some structural variants of ketoconazole block or potentiate BK channel (Maxi-K or slo-1) activity. This property is dependent on phenyl and phenoxy moieties, and the imidazole moiety is not important (Power et al., 2006; Nardi and Olesen, 2008). Hence, FLB-12, which lacks the imidazole moiety but has preserved its phenyl group, may still have an effect on these channels. In addition, there are several other enzyme/protein systems/networks that are affected by ketoconazole (Loose et al., 1983; Gergely et al., 1984; Rodriguez et al., 1999; Rodriguez and Buckholz, 2003; Kinobe et al., 2006; Yoshida and Niwa, 2006), but current evidence suggests that the loss of the imidazole moiety might not adversely affect the pharmacological properties of FLB-12 in human applications. However, further studies are needed to profile the effects of ketoconazole and FLB-12 on all human NRs and special channel proteins such as BK and Kv1.5. In this context, it is important to note that FLB-12, unlike ketoconazole, has negligible (inductive) effect on phosphorylation of ERK, a common downstream effector of GPCRs and other relevant receptor-targeted pathways (Supplemental Fig. 7) (Chong et al., 2004; Osmond et al., 2005; Leroy et al., 2007). Finally, it would be important to study the metabolism of radiolabeled FLB-12 to determine the production of de-N-acetyl metabolites in vitro and in vivo.
In summary, our findings validate the feasibility of developing nontoxic inhibitors of the adopted orphan nuclear receptor PXR. It is also feasible to design analogs that have fewer off-target effects on cross-talking nuclear receptors. These drugs will not only serve as valuable chemical tools for probing PXR action in vitro and in vivo but will also be important adjuncts for novel targeted approaches against cancer drug resistance and other drug-related toxicities (e.g., acetaminophen hepatotoxicity). Because environmental xenogens (e.g., bisphenol A) are commonly found in humans at concentrations that could potentially activate PXR, the notion that some of the PXR-mediated pathophysiological processes could have clinical utility now seems justified (Takeshita et al., 2001; Cobellis et al., 2009; Creusot et al., 2010).
Supplementary Material
Acknowledgments
We thank Dr. Ronald Evans, Dr. Wen Xie (for pxr knockout and humanized PXR mice), and Dr. Prem Reddy for reagents.
The online version of this article (available at http://molpharm.aspetjournals.org) contains supplemental material.
This work was supported by the National Institutes of Health National Cancer Institute [Grant CA12723101]; the Damon Runyon Foundation [Clinical Investigator Award CI 1502]; and Phase I Program, Albert Einstein College of Medicine.
The current ketoconazole analogs are under patent protection (WO/2009/110955), by Albert Einstein College of Medicine, Bronx, NY 10461.
Article, publication date, and citation information can be found at http://molpharm.aspetjournals.org.
doi:10.1124/mol.111.071787.
- PXR
- pregnane X receptor
- ET-743
- trabectedin
- A-792611
- methyl 1-(5-(2-(3-benzyl-2-oxoimidazolidin-1-yl)-3,3-dimethylbutanamido)-4-hydroxy-6-phenyl-1-(4-(pyridin-2-yl)phenyl)hexan-2-ylamino)-3,3-dimethyl-1-oxobutan-2-ylcarbamate
- SAA
- serum amyloid A
- PCN
- pregnenolone-16α-carbonitrile
- LBD
- ligand binding domain
- T0901317
- N-(2,2,2-trifluoroethyl)-N-[4-[2,2,2-trifluoro-1-hydroxy-1-(trifluoromethyl)ethyl]phenyl]benzenesulfonamide
- Rif
- rifampicin
- h
- human
- m
- mouse
- mCAR
- mouse constitutive androstane receptor
- LXR
- liver X receptor
- FXR
- farnesoid X receptor
- ERα
- estrogen receptor α
- UGT
- UDP glucuronosyltransferase
- TR-FRET
- time-resolved fluorescence resonance energy transfer
- DMSO
- dimethyl sulfoxide
- PCR
- polymerase chain reaction
- RT
- reverse transcription
- APAP
- acetaminophen
- ALT
- alanine aminotransferase
- KTZ
- ketoconazole
- LC
- liquid chromatography
- MS/MS
- tandem mass spectrometry
- AST
- aspartate aminotransferase
- hERG
- human ether-a-go-go-related gene
- ERK
- extracellular signal-regulated kinase
- qPCR
- quantitative polymerase chain reaction
- MDR1
- multidrug resistance protein 1
- PPAR
- peroxisome proliferator-activated receptor
- shRNA
- short hairpin RNA
- LORR
- loss of righting reflex.
Authorship Contributions
Participated in research design: Venkatesh, Cayer, Salvail, and Mani.
Conducted experiments: Venkatesh, Wang, Cayer, and Leroux.
Contributed new reagents or analytic tools: Das.
Performed data analysis: Venkatesh, Cayer, Salvail, and Mani.
Wrote or contributed to the writing of the manuscript: Venkatesh, Cayer, Salvail, Wrobel, and Mani.
Other: Mani acquired funding for the research.
References
- Biswas A, Mani S, Redinbo MR, Krasowski MD, Li H, Ekins S. (2009) Elucidating the ‘Jekyll and Hyde’ nature of PXR: the case for discovering antagonists or allosteric antagonists. Pharm Res 26:1807–1815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchi KN, Gray PD, Tolman KG. (1986) Ketoconazole hepatotoxicity: an in vitro model. Biochem Pharmacol 35:2845–2847 [DOI] [PubMed] [Google Scholar]
- Casley WL, Ogrodowczyk C, Larocque L, Jaentschke B, LeBlanc-Westwood C, Menzies JA, Whitehouse L, Hefford MA, Aubin RA, Thorn CF, Whitehead AS, Li X. (2007) Cytotoxic doses of ketoconazole affect expression of a subset of hepatic genes. J Toxicol Environ Health A 70:1946–1955 [DOI] [PubMed] [Google Scholar]
- Cheng J, Ma X, Krausz KW, Idle JR, Gonzalez FJ. (2009) Rifampicin-activated human pregnane X receptor and CYP3A4 induction enhance acetaminophen-induced toxicity. Drug Metab Dispos 37:1611–1621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chong MP, Barritt GJ, Crouch MF. (2004) Insulin potentiates EGFR activation and signaling in fibroblasts. Biochem Biophys Res Commun 322:535–541 [DOI] [PubMed] [Google Scholar]
- Chrencik JE, Orans J, Moore LB, Xue Y, Peng L, Collins JL, Wisely GB, Lambert MH, Kliewer SA, Redinbo MR. (2005) Structural disorder in the complex of human pregnane X receptor and the macrolide antibiotic rifampicin. Mol Endocrinol 19:1125–1134 [DOI] [PubMed] [Google Scholar]
- Cobellis L, Colacurci N, Trabucco E, Carpentiero C, Grumetto L. (2009) Measurement of bisphenol A and bisphenol B levels in human blood sera from healthy and endometriotic women. Biomed Chromatogr 23:1186–1190 [DOI] [PubMed] [Google Scholar]
- Creusot N, Kinani S, Balaguer P, Tapie N, LeMenach K, Maillot-Maréchal E, Porcher JM, Budzinski H, Aït-Aïssa S. (2010) Evaluation of an hPXR reporter gene assay for the detection of aquatic emerging pollutants: screening of chemicals and application to water samples. Anal Bioanal Chem 396:569–583 [DOI] [PubMed] [Google Scholar]
- Das BC, Madhukumar AV, Anguiano J, Kim S, Sinz M, Zvyaga TA, Power EC, Ganellin CR, Mani S. (2008) Synthesis of novel ketoconazole derivatives as inhibitors of the human pregnane X receptor (PXR; NR1I2; also termed SXR, PAR). Bioorg Med Chem Lett 18:3974–3977 [DOI] [PubMed] [Google Scholar]
- Ekins S, Chang C, Mani S, Krasowski MD, Reschly EJ, Iyer M, Kholodovych V, Ai N, Welsh WJ, Sinz M, et al. (2007) Human pregnane X receptor antagonists and agonists define molecular requirements for different binding sites. Mol Pharmacol 72:592–603 [DOI] [PubMed] [Google Scholar]
- Ekins S, Kholodovych V, Ai N, Sinz M, Gal J, Gera L, Welsh WJ, Bachmann K, Mani S. (2008) Computational discovery of novel low micromolar human pregnane X receptor antagonists. Mol Pharmacol 74:662–672 [DOI] [PubMed] [Google Scholar]
- Gergely P, Nékám K, Láng I, Kalmár L, González-Cabello R, Perl A. (1984) Ketoconazole in vitro inhibits mitogen-induced blastogenesis, antibody-dependent cellular cytotoxicity, natural killer activity and random migration of human leukocytes. Immunopharmacology 7:167–170 [DOI] [PubMed] [Google Scholar]
- Guo GL, Moffit JS, Nicol CJ, Ward JM, Aleksunes LA, Slitt AL, Kliewer SA, Manautou JE, Gonzalez FJ. (2004) Enhanced acetaminophen toxicity by activation of the pregnane X receptor. Toxicol Sci 82:374–380 [DOI] [PubMed] [Google Scholar]
- Gupta D, Venkatesh M, Wang H, Kim S, Sinz M, Goldberg GL, Whitney K, Longley C, Mani S. (2008) Expanding the roles for pregnane X receptor in cancer: proliferation and drug resistance in ovarian cancer. Clin Cancer Res 14:5332–5340 [DOI] [PubMed] [Google Scholar]
- Harmsen S, Meijerman I, Febus CL, Maas-Bakker RF, Beijnen JH, Schellens JH. (2009) PXR-mediated induction of P-glycoprotein by anticancer drugs in a human colon adenocarcinoma-derived cell line. Cancer Chemother Pharmacol 66:765–771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang H, Wang H, Sinz M, Zoeckler M, Staudinger J, Redinbo MR, Teotico DG, Locker J, Kalpana GV, Mani S. (2007) Inhibition of drug metabolism by blocking the activation of nuclear receptors by ketoconazole. Oncogene 26:258–268 [DOI] [PubMed] [Google Scholar]
- Kinobe RT, Dercho RA, Vlahakis JZ, Brien JF, Szarek WA, Nakatsu K. (2006) Inhibition of the enzymatic activity of heme oxygenases by azole-based antifungal drugs. J Pharmacol Exp Ther 319:277–284 [DOI] [PubMed] [Google Scholar]
- Kortagere S, Krasowski MD, Reschly EJ, Venkatesh M, Mani S, Ekins S. (2010) Evaluation of computational docking to identify pregnane X receptor agonists in the ToxCast database. Environ Health Perspect 118:1412–1417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee WM. (1995) Drug-induced hepatotoxicity. N Engl J Med 333:1118–1127 [DOI] [PubMed] [Google Scholar]
- Leroy D, Missotten M, Waltzinger C, Martin T, Scheer A. (2007) G protein-coupled receptor-mediated ERK1/2 phosphorylation: towards a generic sensor of GPCR activation. J Recept Signal Transduct Res 27:83–97 [DOI] [PubMed] [Google Scholar]
- Lewis JH, Zimmerman HJ, Benson GD, Ishak KG. (1984) Hepatic injury associated with ketoconazole therapy. Analysis of 33 cases. Gastroenterology 86:503–513 [PubMed] [Google Scholar]
- Loose DS, Stover EP, Feldman D. (1983) Ketoconazole binds to glucocorticoid receptors and exhibits glucocorticoid antagonist activity in cultured cells. J Clin Invest 72:404–408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mani S, Huang H, Sundarababu S, Liu W, Kalpana G, Smith AB, Horwitz SB. (2005) Activation of the steroid and xenobiotic receptor (human pregnane X receptor) by nontaxane microtubule-stabilizing agents. Clin Cancer Res 11:6359–6369 [DOI] [PubMed] [Google Scholar]
- Nardi A, Olesen SP. (2008) BK channel modulators: a comprehensive overview. Curr Med Chem 15:1126–1146 [DOI] [PubMed] [Google Scholar]
- Osmond RI, Sheehan A, Borowicz R, Barnett E, Harvey G, Turner C, Brown A, Crouch MF, Dyer AR. (2005) GPCR screening via ERK 1/2: a novel platform for screening G protein-coupled receptors. J Biomol Screen 10:730–737 [DOI] [PubMed] [Google Scholar]
- Power EC, Ganellin CR, Benton DC. (2006) Partial structures of ketoconazole as modulators of the large conductance calcium-activated potassium channel (BK(Ca)). Bioorg Med Chem Lett 16:887–890 [DOI] [PubMed] [Google Scholar]
- Raynal C, Pascussi JM, Leguelinel G, Breuker C, Kantar J, Lallemant B, Poujol S, Bonnans C, Joubert D, Hollande F, et al. (2010) A Pregnane X Receptor (PXR) expression in colorectal cancer cells restricts irinotecan chemosensitivity through enhanced SN-38 glucuronidation. Mol Cancer 9:46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez RJ, Acosta D., Jr (1997) N-deacetyl ketoconazole-induced hepatotoxicity in a primary culture system of rat hepatocytes. Toxicology 117:123–131 [DOI] [PubMed] [Google Scholar]
- Rodriguez RJ, Buckholz CJ. (2003) Hepatotoxicity of ketoconazole in Sprague-Dawley rats: glutathione depletion, flavin-containing monooxygenases-mediated bioactivation and hepatic covalent binding. Xenobiotica 33:429–441 [DOI] [PubMed] [Google Scholar]
- Rodriguez RJ, Proteau PJ, Marquez BL, Hetherington CL, Buckholz CJ, O'Connell KL. (1999) Flavin-containing monooxygenase-mediated metabolism of N-deacetyl ketoconazole by rat hepatic microsomes. Drug Metab Dispos 27:880–886 [PubMed] [Google Scholar]
- Seglen PO. (1976) Preparation of isolated rat liver cells. Methods Cell Biol 13:29–83 [DOI] [PubMed] [Google Scholar]
- Takeshita A, Koibuchi N, Oka J, Taguchi M, Shishiba Y, Ozawa Y. (2001) Bisphenol-A, an environmental estrogen, activates the human orphan nuclear receptor, steroid and xenobiotic receptor-mediated transcription. Eur J Endocrinol 145:513–517 [DOI] [PubMed] [Google Scholar]
- Takeshita A, Taguchi M, Koibuchi N, Ozawa Y. (2002) Putative role of the orphan nuclear receptor SXR (steroid and xenobiotic receptor) in the mechanism of CYP3A4 inhibition by xenobiotics. J Biol Chem 277:32453–32458 [DOI] [PubMed] [Google Scholar]
- Wang H, Huang H, Li H, Teotico DG, Sinz M, Baker SD, Staudinger J, Kalpana G, Redinbo MR, Mani S. (2007) Activated pregnenolone X-receptor is a target for ketoconazole and its analogs. Clin Cancer Res 13:2488–2495 [DOI] [PubMed] [Google Scholar]
- Wang H, Li H, Moore LB, Johnson MD, Maglich JM, Goodwin B, Ittoop OR, Wisely B, Creech K, Parks DJ, et al. (2008) The phytoestrogen coumestrol is a naturally occurring antagonist of the human pregnane X receptor. Mol Endocrinol 22:838–857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf KK, Wood SG, Hunt JA, Walton-Strong BW, Yasuda K, Lan L, Duan SX, Hao Q, Wrighton SA, Jeffery EH, Evans RM, Szakacs JG, von Moltke LL, Greenblatt DJ, Court MH, Schuetz EG, Sinclair PR, Sinclair JF. (2005) Role of the nuclear receptor pregnane X receptor in acetaminophen hepatotoxicity. Drug Metab Dispos 33:1827–1836 [DOI] [PubMed] [Google Scholar]
- Yong WP, Ramirez J, Innocenti F, Ratain MJ. (2005) Effects of ketoconazole on glucuronidation by UDP-glucuronosyltransferase enzymes. Clin Cancer Res 11:6699–6704 [DOI] [PubMed] [Google Scholar]
- Yoshida K, Niwa T. (2006) Quantitative structure-activity relationship studies on inhibition of HERG potassium channels. J Chem Inf Model 46:1371–1378 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.