Abstract
Liver fibrosis is a chronic disorder that is characterized by an alteration of the balance between fibrogenesis and fibrinolysis, which results in accumulation of excessive amounts of extracellular matrix (ECM) and distortion of the normal liver architecture. The activation and transformation of quiescent hepatic stellate cells (HSCs) into myofibroblast-like cells constitute a major mechanism for the increased production of ECM in the liver. The nuclear receptor farnesoid X receptor (FXR) shows potent antifibrotic activity in HSCs and protects animals in rodent models of liver fibrosis. However, the detailed mechanism remains incompletely understood. In this study, we report that treatment with 3-[2-[2-chloro-4-[[3-(2,6-dichlorophenyl)-5-(1-methylethyl)-4-isoxazolyl]methoxy]phenyl]ethenyl]benzoic acid (GW4064), a synthetic FXR ligand, led to up-regulation of microRNA-29a (miR-29a) in HSCs isolated from wild-type mice, rats, and humans but not from FXR(−/−) mice. miR-29a seems to play an inhibitory role in the regulation of ECM production because of the following: 1) transfection of HSCs with miR-29a mimic resulted in drastic down-regulation of the mRNA expression of several genes that encode ECM proteins; and 2) miR-29a significantly inhibited the expression of a reporter expression plasmid that contains the 3′-untranslated region of the corresponding ECM genes. Our results suggest that miR-29a is a FXR target gene because miR-29a promoter activity was significantly increased by pharmacologic or genetic activation of FXR. Functional analysis of human miR-29a promoter identified an imperfect inverted repeat spaced by one nucleotide DNA motif, inverted repeat-1 (5′-AGGTCAcAGACCT-3′), as a likely FXR-responsive element that is involved in miR-29a regulation. Our study uncovers a new mechanism by which FXR negatively regulates the expression of ECM in HSCs.
Introduction
Liver fibrosis is a chronic disorder that is characterized by an alteration of the balance between fibrogenesis and fibrinolysis, which results in accumulation of excessive amounts of extracellular matrix (ECM) and distortion of the normal liver architecture (Jiao, Friedman, and Aloman, 2009). The activation and transformation of quiescent hepatic stellate cells (HSCs) into myofibroblast-like cells constitute a major mechanism for the increased production of ECM in the liver (Parsons et al., 2007; Friedman, 2008). The mechanism of HSC activation is not completely understood and involves the alterations of several intracellular pathways such as TGF-β and platelet-derived growth factor signaling (Parsons et al., 2007; Friedman, 2008). Several studies have shown that nuclear receptors such as peroxisome proliferator-activated receptor γ and pregnane X receptor also play an important role in the modulation of HSC activation and their biologic functions (Miyahara et al., 2000; Galli et al., 2002; Marek et al., 2005).
Farnesoid X receptor (FXR; NR1H4) is a member of the nuclear receptor superfamily that is highly expressed in liver, kidney, adrenal glands, and intestine (Forman et al., 1995). FXR plays a key role in the homeostasis of cholesterol and bile acids by regulating the expression of genes involved in the synthesis and transport of bile acids (Chiang, 2002; Bertolotti et al., 2008). In addition to the potential of its ligands in the treatment of cholestasis (Claudel et al., 2002), FXR has been shown to be expressed in HSCs and to negatively regulate the activation of HSCs and the associated overproduction of ECM in rodent models of liver fibrosis (Fiorucci et al., 2004, 2005a,b). A study by Fiorucci et al. (2004) suggested that short heterodimer partner (SHP) played a role in the FXR-mediated antifibrotic effect. SHP is one of the FXR target genes, and it effectively blocks the AP-1 signaling that is critically involved in ECM production. However, the detailed mechanism that is involved in the antifibrotic effects of FXR remains incompletely understood.
microRNAs (miRNAs) are short noncoding RNA molecules that control gene expression by modulating the stability and/or the translational efficiency of target mRNAs (Lee et al., 1993; Ghildiyal and Zamore, 2009). miRNAs play a role in the control of a wide range of biologic functions and processes such as development, differentiation, metabolism, carcinogenesis, and immune response (Ghildiyal and Zamore, 2009). miRNAs are initially transcribed as long primary transcripts that undergo several processing steps to form the mature 22-nucleotide miRNA-miRNA* duplex. The complementary strand miRNA* is typically degraded, whereas the mature single-stranded form is incorporated into the RNA-induced silencing complex. Most miRNA binding sequences are found in the 3′-untranslated region (UTR) of the mRNAs (Ghildiyal and Zamore, 2009). However, increasing evidence suggests that miRNAs can also be targeted to both coding regions and 5′-UTR of mRNAs (Forman et al., 2008). Several studies showed that miRNAs were involved in the activation of HSCs: several miRNA species are up-regulated whereas many others are down-regulated during the process (Guo et al., 2009; Ji et al., 2009; Venugopal et al., 2010). A recent study by Roderburg et al. (2011) showed that all three members (a, b, and c) of the miR-29 family were significantly down-regulated in mouse liver in both CCl4 and common bile ligation models. Down-regulation of miR-29a, -b, and -c was also shown in activated HSCs in vitro. A probable role of miR-29 members in liver fibrosis is suggested by the following: 1) overexpression of miR-29 in mouse HSCs leads to down-regulation of the expression of several ECM genes and 2) treatment with TGF-β or lipopolysaccharide-mediated activation of NF-κB signaling leads to down-regulation of the expression of miR-29a, -b, and -c (Roderburg et al., 2011). Furthermore, the clinical significance is supported by the observation that serum levels of miR-29a in patients with cirrhosis are inversely correlated with the stages of the disease (Roderburg et al., 2011). However, it is unknown whether miRNAs are also involved in the antifibrotic effect of FXR in HSCs.
In this study, we showed that activation of FXR by a synthetic ligand, 3-[2-[2-chloro-4-[[3-(2,6-dichlorophenyl)-5-(1-methylethyl)-4-isoxazolyl]methoxy]phenyl]ethenyl]benzoic acid (GW4064), led to significant up-regulation of the expression of miR-29a in mouse, rat, and human HSCs. Functional and reporter assays suggested that miR-29a is a potent mediator that negatively regulates the expression of several ECM genes. Furthermore, a functional FXR response element (FXRE) was found in the miR-29a promoter. Our study unveils a new mechanism by which FXR negatively regulates the expression of ECM in HSCs.
Materials and Methods
Reagents and Chemicals.
GW4064 was synthesized by following a published protocol (Maloney et al., 2000). miR-29a mimic and nonspecific control miRNA mimic were purchased from Applied Biosystems (Foster City, CA). All products for cell culture were purchased from Invitrogen (Carlsbad, CA). pCMX, pCMX-FXR, and pCMV-βgal were described previously (Umesono et al., 1991). pCMX-vpFXR (a gift from Drs. Enrique Saez and Ronald Evans, Salk Institute, San Diego, CA) was generated by fusing the VP16 activation domain from the herpes simplex virus to the N terminus of the FXR (Downes et al., 2003).
Animals and HSC Isolation.
Retired male Sprague-Dawley rats and C57BL/6 mice were obtained from Charles River Laboratories (Wilmington, MA). FXR(−/−) mice (Sinal et al., 2000) were purchased from The Jackson Laboratory (Bar Harbor, ME) and bred in the Central Animal Facility of University of Pittsburgh (Pittsburgh, PA). HSCs were isolated through in situ proteinase/collagenase perfusion followed by density gradient centrifugation, as described previously (Thirunavukkarasu et al., 2005). Primary cells were more than 95% pure. Cells were grown on standard tissue culture plastic dishes in Dulbecco's modified Eagle's medium with 10% fetal bovine serum and antibiotics. HSCs were used after activation through culturing for 7 days. All studies conform to the Guide for the Care and Use of Laboratory Animals (Institute of Laboratory Animal Resources, 1996).
LX-2, an immortalized human hepatic stellate cell line, was kindly provided by Dr. Scott L. Friedman (Mount Sinai School of Medicine, New York, NY) (Xu et al., 2005). The cells were maintained in Dulbecco's modified Eagle's medium with 10% fetal bovine serum and antibiotics.
Real-TimePolymerase Chain Reaction.
Total RNA was extracted from cells with TRIzol reagent (Invitrogen), and the first-strand cDNA was synthesized by use of SuperScript III reverse transcriptase (Invitrogen). Real-time polymerase chain reaction (PCR) analysis of rat, mouse, and human fibrosis-related genes and miR-29 precursor was performed by using SYBR Green-based assays with the ABI 7300 Real-Time PCR System (Applied Biosystems) (Li et al., 2008). Transcript abundance, normalized to β-glucuronidase expression, was expressed as fold increase over a calibrated sample.
For detection of mature miRNA, total RNA was reverse-transcribed into cDNA using miScript Reverse Transcriptase Kit (QIAGEN, Valencia, CA) according to the manufacturer's protocol. cDNA samples (2 μl) were used for real-time PCR in a total volume of 25 μl using miScript SYBR Green PCR Kit (QIAGEN) and miRNA-specific primers (QIAGEN) on a quantitative PCR machine (Applied Biosystems). The sequences of primers for all of the reverse transcription (RT)-PCR analysis were shown in Supplemental Tables 1 and 2.
Western Blot Analysis.
Protein extraction and Western blot analysis were performed as described previously (Li et al., 2008). FXR antibody and collagen 1A1 (COL1A1) antibody were purchased from Santa Cruz Biotechnology, Inc. (Santa Cruz, CA). Horseradish peroxidase-labeled goat anti-rabbit IgG and the enhanced chemiluminescence kit were purchased from GE Healthcare (Chalfont St. Giles, Buckinghamshire, UK).
Plasmid Construction.
A fragment spanning 1.98 kb of 5′-flanking sequence of the human miR-29a gene was PCR-amplified from human genomic DNA using primers 5-′cgacgcgtgtgggtaagggagagggaag-3′ and 5′-cgacgcgttgctgactgatgagaggaaa-3′ and cloned into MluI of pGL3-basic vector (Promega, Madison, WI). A mutated construct that lacks a putative IR-1 (miR-29a/IR-1) binding site was similarly generated by cloning a fragment spanning 1.87 kb of 5′-flanking sequence of the miR-29a gene that was amplified using primers 5′-cgacgcgtctggtgttcgcagctttcac-3′ and 5′-cgacgcgttgctgactgatgagaggaaa-3′. A TK-Luc construct that contains three copies of miR-29a/IR-1 sequence was generated by annealing the oligonucleotides 5′-agcttcttggaggtcacagacctcgaggtcacagacctcgaggtcacagacctcgttg-3′ and 5′-caacgaggtctgagacctcgaggtctgagacctcgag-gtctgagacctccaaga-3′ followed by ligation into HindIII/BamHI-digested TK-Luc. For functional analysis of miR-29a, 3′-UTR segments containing the miR-29a binding sequences or without binding sequences for COL1A1, collagen 3A1 (COL3A1), and elastin-1 (ELN1) genes were PCR-amplified from human genomic DNA using the corresponding forward and reverse primers (Supplemental Table 1). The PCR product was then subcloned into the SacI-MluI site downstream of the stop codon in the pMIR-REPORT firefly luciferase reporter vector (Ambion, Austin, TX).
Transfection Assays.
Normal monkey kidney fibroblast cells (CV-1 line) were grown to 60 to 70% confluence in 48-well plates. Cells were transiently transfected using Lipofectamine 2000 (Invitrogen) with pGL3-miR-29a in the presence or absence of pCMX-vpFXR or pCMX-FXR. pCMX was added to ensure identical amounts of DNA in each well. Transfection efficiency was monitored by cotransfection of pCMV-βgal plasmid. Twenty-four hours later, cells were treated with GW4064 or DMSO vehicle. Cell extracts were prepared 24 h after GW4064 treatment, the luciferase and β-galactosidase assays were performed as described previously (He et al., 2006), and luciferase activity was normalized against β-galactosidase activity. Transfection experiments were performed on at least three occasions, and in each case, experiments were done in triplicate. Data were represented as fold induction over reporter gene alone.
In a separate study, CV-1 cells were transfected with pMIR-COL1A1-3′-UTR, pMIR-COL3A1-3′-UTR, or pMIR-ELN1-3′-UTR in the presence of miR-29a mimic or nonspecific control miRNA mimic. The reporter expression was then similarly examined as described above 24 h after transfection.
Electrophoretic Mobility Shift Assay.
FXR and retinoid X receptor (RXR) proteins were generated in vitro by coupled in vitro transcription/translation (TnT system; Promega) with pCMX-FXR and pCMX-RXR plasmids. The DNA probe miR-29a/IR-1 (5′-gaagaggtcacagacctctgg-3′) was derived from a region in the human miR-29a promoter that contains a putative FXR response element (bold). It was labeled with [γ-32P]ATP by using the Klenow fragment of DNA polymerase. DNA binding reactions were carried out as follows: aliquots of in vitro translation mixture were incubated in 20 μl of binding buffer (10 mM HEPES, pH 7.9, 10 mM EGTA, 10 mM EDTA, 0.25 mM dithiothreitol, and 10% glycerol) containing 2 μg of poly(dI-dC) (Sigma-Aldrich, St. Louis, MO) and 6 to 20 × 103 cpm of DNA probe at room temperature for 20 min. For supershift assays, 0.2 μg of rabbit anti-FXR IgG was added, and the samples were incubated for another 10 min. The binding mixture was then applied onto a 5% polyacrylamide gel (0.5× Tris borate-EDTA buffer) for electrophoresis. The gels were dried and exposed at −80°C for autoradiography.
Chromatin Immunoprecipitation Assay.
Chromatin immunoprecipitation (ChIP) assay was performed by using a ChIP assay kit (Millipore Corporation, Billerica, MA) according to the manufacturer's instructions. Soluble chromatin was prepared from LX-2 cells treated with 1 μM GW4064 for 24 h. Chromatin was immunoprecipitated with antibodies (2 μg) directed against FXR. Final DNA extractions were PCR amplified using primer pairs that cover an IR-1 consensus sequence in the miRNA-29a promoter as follows: forward, 5′-GTGGGTAAGGGAGAGGGAAG-3′; antisense, 5′-ACATTGCCTTCTCCCCAAAG-3′.
Statistical Analysis.
All data are expressed as means ± S.E.M. unless otherwise stated. Comparisons between two groups were made with unpaired Student's t test. Comparisons among three or more groups were made with analysis of variance followed by Tukey-Kramer post hoc analysis. In all cases, P < 0.05 was considered statistically significant.
Results
Treatment of Rat HSCs with GW4064 Led to Significant Inhibition of the mRNA Expression of Several ECM Genes.
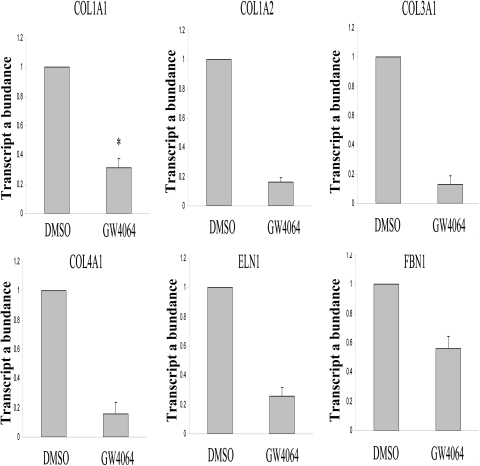
Using 6α-ethyl-chenodeoxycholic acid as a specific ligand, Fiorucci et al. (2004) have previously shown that activation of FXR leads to a significant inhibition of COL1A1 expression in both primary rat HSCs and an immortalized human hepatic stellate cell line HSC-T6. In this experiment, we examined whether GW4064 could similarly inhibit the expression of COL1A1 in rat HSCs. GW4064 is also a synthetic ligand that is highly specific for FXR and has been widely used in studying FXR-mediated gene regulation in vitro and in vivo (Maloney et al., 2000; Li et al., 2009). Figure 1 shows that GW4064 treatment resulted in a significant down-regulation of the expression of COL1A1 mRNA in rat HSCs. GW4064 also significantly inhibited the expression of several other fibrosis-related genes including COL1A2, COL3A1, COL4A1, COL5A1, ELN1, and fibrillin 1 (FBN1) (Fig. 1). Inhibition of ECM protein expression by GW4064 was also confirmed as shown in Western analysis of COL1A1 expression (Supplemental Fig. 1).
Fig. 1.
GW4064 treatment led to down-regulation of the mRNA expression of ECM genes in rat HSCs. Rat HSCs were isolated as described under Materials and Methods and cultured for 7 days to allow transactivation. HSCs were then treated with GW4064 (1 μM) or DMSO vehicle. The mRNA expression levels of several ECM genes were determined by real-time RT-PCR 24 h after the treatment. n = 3; *, P < 0.05 (versus DMSO).
GW4064 Treatment Led to Up-Regulation of miR-29a in Rat and Mouse HSCs.
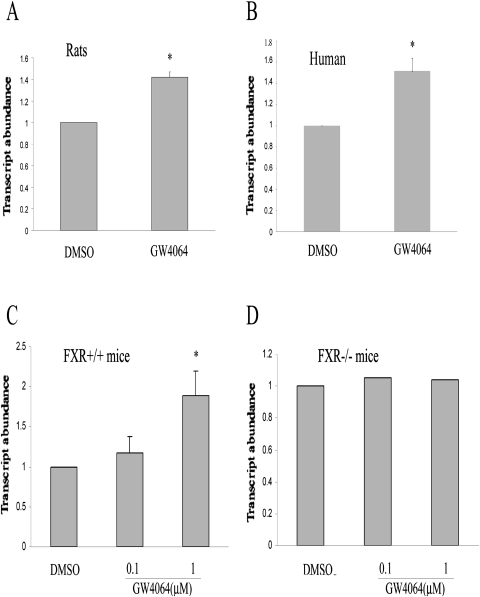
After the demonstration of the inhibition of the mRNA expression of several ECM genes by GW4064, we went on to explore the potential mechanism involved. We hypothesized that a miRNA might be involved because a cluster of ECM-related genes was affected by GW4064 treatment. Multiple algorithms were used to screen for miRNAs that may be involved in the regulation of ECM including MicroCosm Targets (http://www.ebi.ac.uk/enright-srv/microcosm/cgi-bin/targets/v5/search.pl), TargetScan (http://www.targetscan.org/), and Probability of Interaction by Target Accessibility (PITA; Lewis et al., 2003; Xin et al., 2009; Dong et al., 2010). Members of miR-29 family, including miR-29a, miR-29b, and miR-29c, were identified by all three programs to be the best candidates as ECM-targeting miRNAs (Supplemental Table 4). As an initial step to study a potential role of miR-29a in GW4064-mediated effects, we examined whether the expression of miR-29a is regulated by GW4064 in HSCs. Figure 2A shows that GW4064 treatment resulted in a significant increase in the expression of miR-29a gene as assessed by real-time RT-PCR analysis of miR-29a precursor. A similar induction of miR-29a expression by GW4064 was also observed in LX-2 cells (an immortalized human hepatic stellate cell line) (Fig. 2B) and HSCs isolated from wild-type mice (Fig. 2C). However, no induction of miR-29a was observed in HSCs prepared from FXR(−/−) mice (Fig. 2D), which suggests that induction of miR-29a by GW4064 was mediated by FXR. It is interesting to note that GW4064 treatment had no effect on the expression of miR-29b or miR-29c in both rat and mouse HSCs (Supplemental Fig. 2). A similar result was obtained when miR-29a expression was examined by RT-PCR analysis of mature miR-29a (data not shown). Induction of miR-29a in mouse liver was also observed after oral delivery of GW4064 (Supplemental Fig. 3).
Fig. 2.
GW4064 treatment led to up-regulation of miR-29a expression in HSCs. Rat HSCs (A) or LX2 cells (B) were treated with GW4064 or DMSO vehicle. The expression level of miR-29a was determined by real-time RT-PCR 24 h after the treatment. The effect of GW4064 on miR-29a expression was similarly examined in HSCs isolated from wild-type mice (C) or FXR(−/−) mice (D). n = 3; *, P < 0.05 (versus DMSO).
miR-29a Negatively Regulated the Expression of ECM in HSCs.
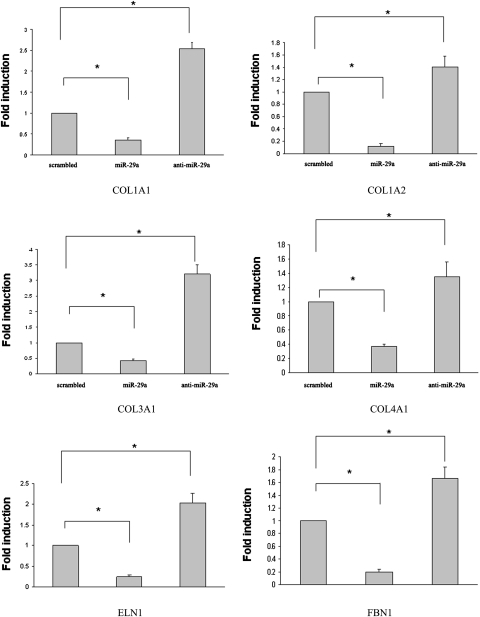
After demonstration of induction of miR-29a by GW4064, we then examined the effect of miR-29a overexpression on the expression of several ECM genes in HSCs to also establish a role of miR-29a in the GW4064/FXR-mediated antifibrotic effect. HSCs were transfected with a miR-29a mimic, a control sequence, or a miR-29a inhibitor, and the mRNA expression of several ECM genes was examined 24 h later. As shown in Fig. 3, overexpression of miR-29a mimic in HSCs resulted in a significant inhibition of the mRNA expression of all six ECM genes examined. Higher levels of ECM mRNAs were observed in HSCs treated with miR-29a inhibitor compared with cells treated with control sequence. This result might be due to the inhibition of endogenous miR-29a that is involved in the control of the basal levels of ECM expression. The effect of miR-29a appears to be target sequence-specific because it showed no effect on the expression of FXR and SHP expression at both mRNA and protein level over a wide range of concentrations (Supplemental Figs. 4 and 5). miR-29a treatment had no effect either on GW4064-mediated induction of SHP expression, which suggests that the function of FXR was well retained in miR-29a-treated HSCs (Supplemental Fig. 6). In contrast, miR-29a inhibited the expression of COL1A1 at both mRNA and protein levels in a dose-dependent manner (Supplemental Figs. 4 and 5).
Fig. 3.
Overexpression of miR-29a resulted in down-regulation of the mRNA expression of ECM genes in rat HSCs. HSCs were transfected with miR-29a mimic, nonspecific control miRNA mimic, or miR-29a inhibitor (25 pmol/well in 6-well plates by 10 μl of Lipofectamine) for 24 h. The mRNA expression levels of several ECM genes were then determined by real-time RT-PCR. n = 3; *, P < 0.05 (versus control miRNA).
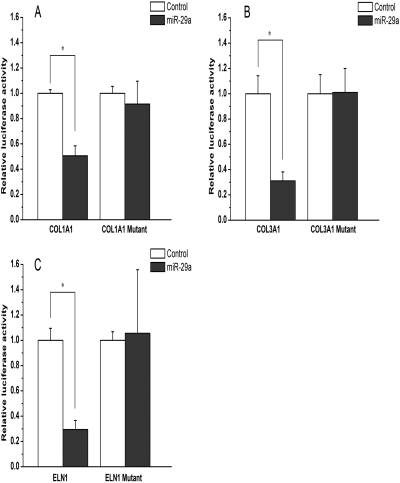
To also establish a role of miR-29a in the regulation of ECM expression, we examined the effect of miR-29a on the expression of several reporter constructs that contain the respective 3′-UTRs from COL1A1, COL3A1, and ELN1 genes. All three 3′-UTRs contain a putative miR-29a target sequence as analyzed by TargetScan algorithm. As shown in Fig. 4A, transfection of cells with miR-29a mimic significantly inhibited the expression of the reporter construct with an intact COL1A1 3′-UTR. Such inhibitory effect was completely lost for a mutant reporter construct lacking the miR-29a target sequence. These results suggest that the presence of the miRNA target site in the COL1A1 3′-UTR of the reporter construct is necessary for the inhibition by miR-29a. Similar results were observed with the construct with a 3′-UTR from either COL3A1 or ELN1 gene (Figs. 4, B and C).
Fig. 4.
miR-29a regulates the expression of ECM genes through targeting at the 3′-UTR of their mRNAs. CV-1 cells were transfected with a luciferase construct with Col1A1-3′-UTR (A), Col3A1-3′-UTR (B) or ELN-3′-UTR (C) in the presence of miR-29a mimic or nonspecific control miRNA mimic. Luciferase assay was performed 24 h after the transfection. Data shown in the panels represent mean (S.D.) from triplicate assays. n = 3; *, P < 0.05 (versus control miRNA).
Human miR-29a Is a Probable Target Gene of FXR.
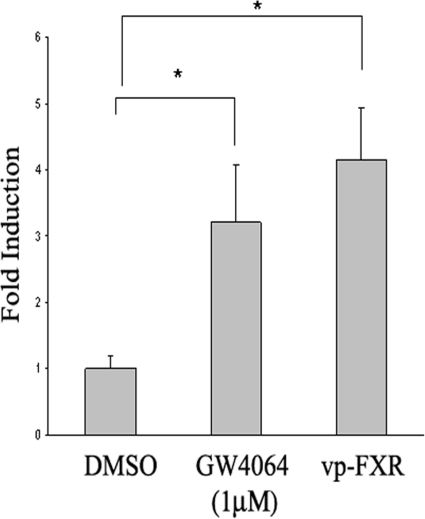
Up-regulation of miR-29a expression by GW4064 suggests that activation of FXR modulates miR-29a expression at the transcriptional level. We then hypothesized that activation of FXR enhances miR-29a expression by exerting its stimulatory activity on miR-29a promoter. To test this hypothesis, we constructed a luciferase reporter expression plasmid (pGL3-miR-29a) that is driven by a 1.98-kb sequence of human miR-29a promoter. To examine promoter activation, CV-1 cells were cotransfected with pGL3-miR-29a and pCMX-FXR expression vector, followed by GW4064 treatment. CV-1 cells instead of HSCs were used for transfection because of low transfection efficiency in the HSCs. As shown in Fig. 5, treatment with GW4064 resulted in a significant increase in the transcriptional activity of the miR-29a promoter. To elucidate a role of FXR in regulating miR-29a promoter activity, we then cotransfected pGL3-miR-29a with an expression plasmid encoding a constitutively activated FXR, vpFXR. vpFXR was generated by fusing the VP16 activation domain to the N terminus of FXR cDNA (Downes et al., 2003). Figure 5 shows that coexpression of vpFXR in CV-1 cells significantly enhanced the miR-29a promoter activity, clearly demonstrating that a genetic activation of FXR enhances miR-29a promoter activity.
Fig. 5.
FXR enhances the transcriptional activity of the miR-29a gene promoter. A luciferase reporter driven by a human miR-29a promoter of 1984 base pairs was used to study the miR-29a promoter activity. CV-1 cells were transiently transfected with pGL3-miR-29a in the presence or absence of pCMX-FXR. Five hours later, the transfection medium was replaced with complete medium and cells were incubated for 20 h. Cells were then cultured in the presence of GW4064 or vehicle DMSO for 24 h. Luciferase assay was then performed. Data shown in the panels represent mean (S.D.) from triplicate assays. To examine the effect of genetic activation of FXR on miR-29a promoter activity, CV-1 cells were transfected with pGL3-miR-29a in the presence or absence of pCMX-vpFXR. Luciferase assays were then examined as described above. n = 3; *, P < 0.05 (versus DMSO).
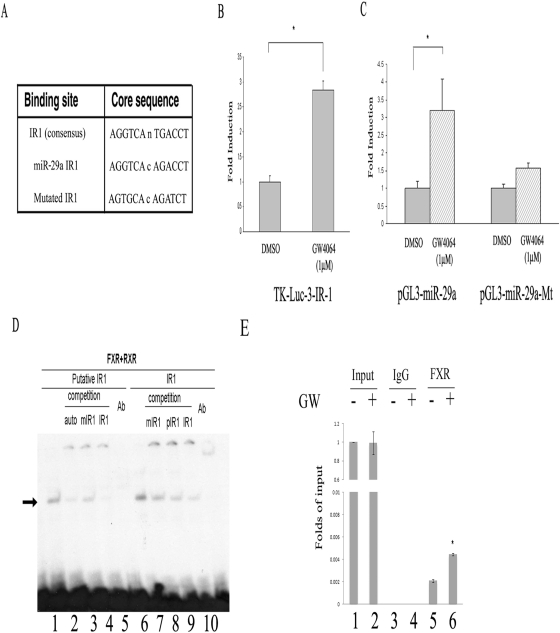
To search for FXREs that may mediate miR-29a induction by GW4064, the 2-kb miR-29a promoter sequence was subjected to in silico analysis with a Web-based algorithm (NUBIScan, http://www.nubiscan.unibas.ch/). One imperfect inverted repeat spaced by one nucleotide (IR-1) site was identified, and its sequences and location are shown in Fig. 6A. To determine whether miR-29a/IR-1 is necessary and sufficient in mediating FXR transactivation, a heterologous thymidine kinase-luciferase reporter gene that contains three copies of miR-29a/IR-1 element was generated and tested for FXR transactivation in CV-1 cells. As shown in Fig. 6B, the synthetic thymidine kinase reporter gene was activated by FXR in the presence of its agonist GW4064. To determine whether this IR-1 sequence is responsible for FXR-mediated transactivation of miR-29a promoter, we also generated a mutant pGL3-miR-29a in which this putative FXR binding site was eliminated. The wild-type and mutated plasmids were then similarly transfected into the CV-1 cells, and their transfection efficiency was compared. Figure 6C shows that the FXR-mediated activation of miR-29a promoter was substantially diminished when the miR-29a/IR-1 site was eliminated. The above studies strongly suggested a probable role of miR-29a/IR-1 element in FXR-mediated transactivation of human miR-29a promoter.
Fig. 6.
Analysis of putative FXREs in human miR-29a promoter. A, identification of a putative FXRE in human miR-29a promoter through an in silico analysis with a Web-based algorithm (NUBIScan). B, miR-29a/IR-1 mediates FXR transactivation of a heterologous tk-luciferase reporter gene. n = 3; *, P < 0.05 (versus DMSO). C, mutation of miR-29a/IR-1 on the miR-29a promoter eliminates activation by FXR. n = 3; *, P < 0.05 (versus DMSO). D, electrophoretic mobility shift assay analysis of the binding of FXR/RXR to miR-29a/IR-1 in human miR-29a promoter. Double-stranded oligonucleotides (−1924/−1936) were end-labeled with [γ-32P]ATP using T4 polynucleotide kinase. The labeled probe was incubated with in vitro-translated RXR/FXR for 20 min. The reactions were analyzed by electrophoresis in a nondenaturing 5% polyacrylamide gel followed by autoradiography. In some studies, the samples were preincubated with anti-FXR antibody before gel electrophoresis. E, ChIP analysis of the binding of FXR to miR-29a/IR-1 in human miR-29a promoter in LX-2 cells. Soluble chromatin was prepared from LX2 cells treated with 1 μM GW4064 or DMSO for 24 h. Chromatin was immunoprecipitated with antibodies directed against FXR. The extracted DNA was PCR amplified using primer pairs that cover an IR-1 consensus sequence in the miRNA-29a promoter.
Figure 6D shows the result of an electrophoretic mobility shift assay with a 20-base-pair oligonucleotide that contained the putative FXRE (Fig. 6D, lanes 1∼5). A typical IR-1/FXRE oligonucleotide was also included as a positive control (Fig. 6D, lanes 6∼10) because FXR is known to bind to IR-1 with high specificity and affinity. Interaction of the oligonucleotide with in vitro-translated FXR/RXR yielded a DNA/protein band of expected mobility (Fig. 6D, lane 1). This binding was specific, because it was inhibited by addition of excess unlabeled miR-29a/IR-1 (Fig. 6D, lane 2) or IR-1/FXRE (Fig. 6D, lane 4) but not a mutated miR-29a/IR-1 (Fig. 6D, lane 3). Addition of antibody against FXR to the reaction mixture resulted in the disappearance of the radiolabeled band (Fig. 6D, lane 5). This supershifting confirms the identity of the protein that interacts with the DNA as being FXR.
To also demonstrate the binding of FXR to miR-29a/IR-1, ChIP assay was then performed. In this experiment, LX-2 cells were treated with vehicle (DMSO) or GW4064 for 24 h before ChIP analysis using an anti-FXR antibody or the control mouse IgG. As shown in Fig. 6E, treatment of LX-2 cells with GW4064 led to a significant increase in the recruitment of FXR to miR-29a/IR-1. Taken together, these results suggested that miR-29a is a direct transcriptional target of FXR.
Discussion
In this study, we have demonstrated that miR-29a was significantly up-regulated in HSCs after treatment with GW4064, a synthetic FXR-specific agonist. Overexpression of miR-29a in HSCs resulted in a significant inhibition of the mRNA expression of several ECM genes, including collagens, elastin, and fibrillin. miR-29a also significantly inhibited the expression of a reporter expression plasmid that contains a full-length 3′-UTR from COL1A1, COL3A1, or ELN1 genes. These results are consistent with the notion that miR-29a may be critically involved in the FXR-mediated inhibition of ECM expression in HSCs.
Induction of miR-29a in HSCs is likely to be mediated by FXR because of the following: 1) GW4064 is highly specific for FXR and is often used as a “chemical tool” to show that bile acids target genes are regulated in a FXR-specific manner (Maloney et al., 2000); and 2) the induction of miR-29a was abolished in FXR(−/−) mouse HSCs. We have also identified an imperfect IR-1 as a FXRE that appears to be involved in GW4064-mediated up-regulation of miR-29a in HSCs. IR-1 is a typical FXRE that has be implicated in the regulation of several FXR target genes. Thus, miR-29a is a likely target gene of FXR.
Various biological functions have been reported for members of miR-29 family including miR-29a, b, and c. These include modulations of self-renewal in hematopoietic progenitor cells (Han et al., 2010), regulation of Wnt signaling in human osteoblasts (Kapinas et al., 2010), control of host HIV-1 interactions (Nathans et al., 2009), and regulation of the expression of ECM in fibroblasts and stellate cells (Jiang et al., 2010) etc. It is clear that the biological functions of miRNAs are complex and may be tissue- or cell type-specific. A study from van Rooij et al. (2008) has shown that dysregulation of miR-29 plays an important role in cardiac fibrosis after myocardial infarction. All three members of the miR-29 family were down-regulated in the region of the heart adjacent to the infarct. Systemic delivery of a cholesterol-conjugated miR-29b inhibitor led to increased expression of collagen in liver, kidney, and heart, which suggests a role of miR-29 in regulating the expression of ECM in vivo (van Rooij et al., 2008). Down-regulation of miR-29 has also been shown in rodent models of liver fibrosis and in liver biopsies from patients with hepatitis C virus infection (Kwiecinski et al., 2009; Pogribny et al., 2010). The role of miR-29 members in liver fibrosis was also demonstrated in a study by Roderburg et al. (2011) in which all members of miR-29 were significantly down-regulated in mouse liver in both CCl4 and common bile ligation models. They have also shown that TGF-β or NF-κB signaling negatively regulated the expression of miR-29, which suggests TGF-β↑→miR-29↓ or NF-κB↑→miR-29↓ as an important mechanism in the development of liver fibrosis (Roderburg et al., 2011). A similar role of TGF-β↑→miR-29↓ was also demonstrated in a mouse model of lung fibrosis (Cushing et al., 2010). These data, together with our observation that activation of FXR led to up-regulation of miR-29a, strongly support an important role of miR-29 in the control of ECM expression under both physiological and pathophysiologic conditions. They also suggest the potential of miR-29 as a novel therapeutic for the treatment of various fibrotic diseases, including liver fibrosis.
It is likely that other mechanisms are involved in the antifibrotic effect of FXR/FXR ligands in addition to a direct activation of the miR-29a gene. Several studies, including our work, have suggested that many nuclear receptors, such as FXR and SHP, inhibit various types of inflammatory signaling, including NF-κB, AP-1, and TGF-β, through competition for essential cofactors or recruitment of corepressors (He et al., 2006; Zhang et al., 2010). Therefore, treatment of HSCs with FXR ligands at quiescent stage may prevent the NF-κB- and/or TGF-β-mediated down-regulation of miR-29 expression during their transactivation process. Studies are currently ongoing in our laboratory to test this hypothesis. Alternatively, FXR and/or SHP may directly inhibit the expression of fibrosis-related genes through the interference of AP-1 and/or TGF-β signaling, as suggested by Fiorucci et al. (2004). It is clear that more studies are required in the future to better understand the contribution of each potential mechanism to the overall antifibrotic effect of FXR.
It should be noted that, despite a similar function in regulation of ECM expression, miR-29a, -b, and -c may be differentially up- or down-regulated under different physiologic or pathophysiologic conditions. miRNA-29a and miR-29b1 are clustered together and are located in chromosome 7 in humans, whereas the miR-29b2/miR-29c cluster is found on chromosome 1. Pogribny et al. (2010) showed that miR-29c was down-regulated in a mouse model of dietary nonalcoholic steatohepatitis. In contrast, in a rat model of common bile duct ligation and in liver biopsies from patients with hepatitis C virus, prominent down-regulation of miR-29a and moderate down-regulation of miR-29b were observed (Kwiecinski et al., 2009). We observed a significant up-regulation of miR-29a in both mouse and rat HSCs only after GW4064 treatment, which is probably due to lack of functional FXR response element in the miR-29b2/miR-29c transcriptional unit. This is in contrast to down-regulation of all members of miR-29 family in mouse models of liver fibrosis reported by Roderburg et al. (2011), which may be due to the fact that the expression of both miRNA-29a/miR-29b1 and miR-29b2/miR-29c is negatively regulated by NF-κB and/or TGF-β signaling. This may reflect a complex mechanism in the regulation of miRNA expression.
In summary, we have shown for the first time that activation of FXR led to expression of miR-29a in HSCs, which may play an important role in FXR-mediated antifibrotic effect. Our study provides new insight into the mechanism by which FXR/FXR ligands control the expression of ECM in HSCs. It also suggests a miR-29a-based new therapy for the treatment of liver fibrosis.
Supplementary Material
Acknowledgments
We thank Dr. Scott L. Friedman (Mount Sinai School of Medicine, New York, NY) for generously providing the LX-2 cell line.
The online version of this article (available at http://molpharm.aspetjournals.org) contains supplemental material.
This work was supported by the National Institutes of Health National Heart, Lung and Blood Institute [Grants HL68688, HL091828-01]; and in part by a grant from University of Pittsburgh Central Research Development Fund.
Article, publication date, and citation information can be found at http://molpharm.aspetjournals.org.
doi:10.1124/mol.110.068247.
- ECM
- extracellular matrix
- TGF
- transforming growth factor
- HSC
- hepatic stellate cell
- FXR
- farnesoid X receptor
- SHP
- short heterodimer partner
- miRNA
- microRNA
- UTR
- untranslated region
- AP-1
- activator protein 1
- NF-κB
- nuclear factor-κB
- GW4064
- 3-[2-[2-chloro-4-[[3-(2,6-dichlorophenyl)-5-(1-methylethyl)-4-isoxazolyl]methoxy]phenyl]ethenyl]benzoic acid
- FXRE
- FXR response element
- RT
- reverse transcription
- PCR
- polymerase chain reaction
- COL1A1
- collagen 1A1
- kb
- kilobase(s)
- IR-1
- inverted repeat spaced by one nucleotide
- COL3A1
- collagen 3A1
- ELN1
- elastin-1
- DMSO
- dimethyl sulfoxide
- RXR
- retinoid X receptor
- ChIP
- chromatin immunoprecipitation.
Authorship Contributions
Participated in research design: J. Li and S. Li.
Conducted experiments: J. Li, Zhang, and Gao.
Contributed new reagents or analytic tools: Kuruba, Gao, Gandhi, and Xie. .
Performed data analysis: J. Li, Zhang, and S. Li.
Wrote or contributed to the writing of the manuscript: J. Li, Zhang, and S. Li.
Other: S. Li acquired funding for the research.
References
- Bertolotti M, Gabbi C, Anzivino C, Carulli L, Loria P, Carulli N. (2008) Nuclear receptors as potential molecular targets in cholesterol accumulation conditions: insights from evidence on hepatic cholesterol degradation and gallstone disease in humans. Curr Med Chem 15:2271–2284 [DOI] [PubMed] [Google Scholar]
- Chiang JY. (2002) Bile acid regulation of gene expression: roles of nuclear hormone receptors. Endocr Rev 23:443–463 [DOI] [PubMed] [Google Scholar]
- Claudel T, Sturm E, Duez H, Torra IP, Sirvent A, Kosykh V, Fruchart JC, Dallongeville J, Hum DW, Kuipers F, et al. (2002) Bile acid-activated nuclear receptor FXR suppresses apolipoprotein A-I transcription via a negative FXR response element. J Clin Invest 109:961–971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cushing L, Kuang PP, Qian J, Shao F, Wu J, Little F, Thannickal VJ, Cardoso WV, Lu J. (2010) Mir-29 is a major regulator of genes associated with pulmonary fibrosis. Am J Respir Cell Mol Biol doi:10.1165/rcmb.2010-0323OC [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong H, Paquette M, Williams A, Zoeller RT, Wade M, Yauk C. (2010) Thyroid hormone may regulate mRNA abundance in liver by acting on microRNAs. PLoS One 5:e12136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Downes M, Verdecia MA, Roecker AJ, Hughes R, Hogenesch JB, Kast-Woelbern HR, Bowman ME, Ferrer JL, Anisfeld AM, Edwards PA, et al. (2003) A chemical, genetic, and structural analysis of the nuclear bile acid receptor FXR. Mol Cell 11:1079–1092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiorucci S, Antonelli E, Rizzo G, Renga B, Mencarelli A, Riccardi L, Orlandi S, Pellicciari R, Morelli A. (2004) The nuclear receptor SHP mediates inhibition of hepatic stellate cells by FXR and protects against liver fibrosis. Gastroenterology 127:1497–1512 [DOI] [PubMed] [Google Scholar]
- Fiorucci S, Rizzo G, Antonelli E, Renga B, Mencarelli A, Riccardi L, Morelli A, Pruzanski M, Pellicciari R. (2005a) Cross-talk between farnesoid-X-receptor (FXR) and peroxisome proliferator-activated receptor gamma contributes to the antifibrotic activity of FXR ligands in rodent models of liver cirrhosis. J Pharmacol Exp Ther 315:58–68 [DOI] [PubMed] [Google Scholar]
- Fiorucci S, Rizzo G, Antonelli E, Renga B, Mencarelli A, Riccardi L, Orlandi S, Pruzanski M, Morelli A, Pellicciari R. (2005b) A farnesoid X receptor-small heterodimer partner regulatory cascade modulates tissue metalloproteinase inhibitor-1 and matrix metalloprotease expression in hepatic stellate cells and promotes resolution of liver fibrosis. J Pharmacol Exp Ther 314:584–595 [DOI] [PubMed] [Google Scholar]
- Forman BM, Goode E, Chen J, Oro AE, Bradley DJ, Perlmann T, Noonan DJ, Burka LT, McMorris T, Lamph WW, et al. (1995) Identification of a nuclear receptor that is activated by farnesol metabolites. Cell 81:687–693 [DOI] [PubMed] [Google Scholar]
- Forman JJ, Legesse-Miller A, Coller HA. (2008) A search for conserved sequences in coding regions reveals that the let-7 microRNA targets Dicer within its coding sequence. Proc Natl Acad Sci USA 105:14879–14884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedman SL. (2008) Hepatic stellate cells: protean, multifunctional, and enigmatic cells of the liver. Physiol Rev 88:125–172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galli A, Crabb DW, Ceni E, Salzano R, Mello T, Svegliati-Baroni G, Ridolfi F, Trozzi L, Surrenti C, Casini A. (2002) Antidiabetic thiazolidinediones inhibit collagen synthesis and hepatic stellate cell activation in vivo and in vitro. Gastroenterology 122:1924–1940 [DOI] [PubMed] [Google Scholar]
- Ghildiyal M, Zamore PD. (2009) Small silencing RNAs: an expanding universe. Nat Rev Genet 10:94–108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo CJ, Pan Q, Li DG, Sun H, Liu BW. (2009) miR-15b and miR-16 are implicated in activation of the rat hepatic stellate cell: an essential role for apoptosis. J Hepatol 50:766–778 [DOI] [PubMed] [Google Scholar]
- Han YC, Park CY, Bhagat G, Zhang J, Wang Y, Fan JB, Liu M, Zou Y, Weissman IL, Gu H. (2010) microRNA-29a induces aberrant self-renewal capacity in hematopoietic progenitors, biased myeloid development, and acute myeloid leukemia. J Exp Med 207:475–489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He F, Li J, Mu Y, Kuruba R, Ma Z, Wilson A, Alber S, Jiang Y, Stevens T, Watkins S, et al. (2006) Downregulation of endothelin-1 by farnesoid X receptor in vascular endothelial cells. Circ Res 98:192–199 [DOI] [PubMed] [Google Scholar]
- Institute of Laboratory Animal Resources (1996) Guide for the Care and Use of Laboratory Animals 7th ed. Institute of Laboratory Animal Resources, Commission on Life Sciences, National Research Council, Washington DC [Google Scholar]
- Ji J, Zhang J, Huang G, Qian J, Wang X, Mei S. (2009) Over-expressed microRNA-27a and 27b influence fat accumulation and cell proliferation during rat hepatic stellate cell activation. FEBS Lett 583:759–766 [DOI] [PubMed] [Google Scholar]
- Jiang X, Tsitsiou E, Herrick SE, Lindsay MA. (2010) MicroRNAs and the regulation of fibrosis. FEBS J 277:2015–2021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiao J, Friedman SL, Aloman C. (2009) Hepatic fibrosis. Current Opinion in Gastroenterology 25:223–229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapinas K, Kessler C, Ricks T, Gronowicz G, Delany AM. (2010) miR-29 modulates Wnt signaling in human osteoblasts through a positive feedback loop. J Biol Chem 285:25221–25231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwiecinski M, Strack I, Noetel A, Schievenbusch S, Scheffler M, Elfimova N, Dienes HP, Odenthal M. (2009) MicroRNA-29: a novel antifibrogenic mediator in liver fibrogenesis. J Hepatol 50 (Suppl 1):S110 [Google Scholar]
- Laffitte BA, Kast HR, Nguyen CM, Zavacki AM, Moore DD, Edwards PA. (2000) Identification of the DNA binding specificity and potential target genes for the farnesoid X-activated receptor. J Biol Chem 275:10638–10647 [DOI] [PubMed] [Google Scholar]
- Lee RC, Feinbaum RL, Ambros V. (1993) The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell 75:843–854 [DOI] [PubMed] [Google Scholar]
- Lewis BP, Shih IH, Jones-Rhoades MW, Bartel DP, Burge CB. (2003) Prediction of mammalian microRNA targets. Cell 115:787–798 [DOI] [PubMed] [Google Scholar]
- Li J, Wilson A, Gao X, Kuruba R, Liu Y, Poloyac S, Pitt B, Xie W, Li S. (2009) Coordinated regulation of dimethylarginine dimethylaminohydrolase-1 and cationic amino acid transporter-1 by farnesoid X receptor in mouse liver and kidney and its implication in the control of blood levels of asymmetric dimethylarginine. J Pharmacol Exp Ther 331:234–243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Wilson A, Kuruba R, Zhang Q, Gao X, He F, Zhang LM, Pitt BR, Xie W, Li S. (2008) FXR-mediated regulation of eNOS expression in vascular endothelial cells. Cardiovasc Res 77:169–177 [DOI] [PubMed] [Google Scholar]
- Maloney PR, Parks DJ, Haffner CD, Fivush AM, Chandra G, Plunket KD, Creech KL, Moore LB, Wilson JG, Lewis MC, et al. (2000) Identification of a chemical tool for the orphan nuclear receptor FXR. J Med Chem 43:2971–2974 [DOI] [PubMed] [Google Scholar]
- Marek CJ, Tucker SJ, Konstantinou DK, Elrick LJ, Haefner D, Sigalas C, Murray GI, Goodwin B, Wright MC. (2005) Pregnenolone-16alpha-carbonitrile inhibits rodent liver fibrogenesis via PXR (pregnane X receptor)-dependent and PXR-independent mechanisms. Biochem J 387:601–608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyahara T, Schrum L, Rippe R, Xiong S, Yee HF, Jr, Motomura K, Anania FA, Willson TM, Tsukamoto H. (2000) Peroxisome proliferator-activated receptors and hepatic stellate cell activation. J Biol Chem 275:35715–35722 [DOI] [PubMed] [Google Scholar]
- Nathans R, Chu CY, Serquina AK, Lu CC, Cao H, Rana TM. (2009) Cellular microRNA and P bodies modulate host-HIV-1 interactions. Mol Cell 34:696–709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parsons CJ, Takashima M, Rippe RA. (2007) Molecular mechanisms of hepatic fibrogenesis. J Gastroenterol Hepatol 22:S79–S84 [DOI] [PubMed] [Google Scholar]
- Pogribny IP, Starlard-Davenport A, Tryndyak VP, Han T, Ross SA, Rusyn I, Beland FA. (2010) Difference in expression of hepatic microRNAs miR-29c, miR-34a, miR-155, and miR-200b is associated with strain-specific susceptibility to dietary nonalcoholic steatohepatitis in mice. Lab Invest 90:1437–1446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roderburg C, Urban GW, Bettermann K, Vucur M, Zimmermann H, Schmidt S, Janssen J, Koppe C, Knolle P, Castoldi M, et al. (2011) Micro-RNA profiling reveals a role for miR-29 in human and murine liver fibrosis. Hepatology 53:209–218 [DOI] [PubMed] [Google Scholar]
- Sinal CJ, Tohkin M, Miyata M, Ward JM, Lambert G, Gonzalez FJ. (2000) Targeted disruption of the nuclear receptor FXR/BAR impairs bile acid and lipid homeostasis. Cell 102:731–744 [DOI] [PubMed] [Google Scholar]
- Thirunavukkarasu C, Uemura T, Wang LF, Watkins SC, Gandhi CR. (2005) Normal rat hepatic stellate cells respond to endotoxin in LBP-independent manner to produce inhibitor(s) of DNA synthesis in hepatocytes. J Cell Physiol 204:654–665 [DOI] [PubMed] [Google Scholar]
- Umesono K, Murakami KK, Thompson CC, Evans RM. (1991) Direct repeats as selective response elements for the thyroid hormone, retinoic acid, and vitamin D3 receptors. Cell 65:1255–1266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Rooij E, Sutherland LB, Thatcher JE, DiMaio JM, Naseem RH, Marshall WS, Hill JA, Olson EN. (2008) Dysregulation of microRNAs after myocardial infarction reveals a role of miR-29 in cardiac fibrosis. Proc Natl Acad Sci USA 105:13027–13032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venugopal SK, Jiang J, Kim TH, Li Y, Wang SS, Torok NJ, Wu J, Zern MA. (2010) Liver fibrosis causes downregulation of miRNA-150 and miRNA-194 in hepatic stellate cells, and their overexpression causes decreased stellate cell activation. Am J Physiol Gastrointest Liver Physiol 298:G101–G106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xin F, Li M, Balch C, Thomson M, Fan M, Liu Y, Hammond SM, Kim S, Nephew KP. (2009) Computational analysis of microRNA profiles and their target genes suggests significant involvement in breast cancer antiestrogen resistance. Bioinformatics 25:430–434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu L, Hui AY, Albanis E, Arthur MJ, O'Byrne SM, Blaner WS, Mukherjee P, Friedman SL, Eng FJ. (2005) Human hepatic stellate cell lines, LX-1 and LX-2: new tools for analysis of hepatic fibrosis. Gut 54:142–151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Hagedorn CH, Wang L. (2010) Role of nuclear receptor SHP in metabolism and cancer. Biochim Biophys Acta doi:10.1016/j.bbadis.2010.10.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.