Abstract
Multiple immune factors control host responses to Mycobacterium tuberculosis (Mtb) infection, including the formation of granulomas, aggregates of immune cells whose function may reflect success or failure of the host to contain infection. One such factor is tumor necrosis factor-α (TNF). TNF has been experimentally characterized to have the following activities in Mtb infection: macrophage activation, apoptosis, chemokine and cytokine production. Availability of TNF within a granuloma has been proposed to play a critical role in immunity to Mtb. However, in vivo measurement of a TNF concentration gradient and activities within a granuloma are not experimentally feasible. Further, processes that control TNF concentration and activities in a granuloma remain unknown. We developed a multi-scale computational model that includes molecular, cellular and tissue scale events that occur during granuloma formation and maintenance in lung. We use our model to identify processes that regulate TNF concentration and cellular behaviors and thus influence the outcome of infection within a granuloma. Our model predicts that TNF receptor 1 internalization kinetics play a critical role in infection control within a granuloma, controlling whether there is clearance of bacteria, excessive inflammation, containment of bacteria within a stable granuloma, or uncontrolled growth of bacteria. Our results suggest that there is an inter-play between TNF and bacterial levels in a granuloma that is controlled by the combined effects of both molecular and cellular scale processes. Finally, our model elucidates processes involved in immunity to Mtb that may be new targets for therapy.
INTRODUCTION
Tuberculosis (TB), a disease caused by the intracellular pathogen Mycobacterium tuberculosis (Mtb), is responsible for 2–3 million deaths per year. In the presence of an effective immune response, only 5–10% of infected people develop clinical signs of active TB (known as primary TB). However, immunological testing provides evidence of a state of latent infection, with no clinical symptoms, in one third of the world population (1). Latent TB represents a state of equilibrium in which the host controls the infection but is unable to clear it, allowing bacteria to survive at relatively constant but low levels (2). Latent infection may reactivate to active disease (reactivation TB) with an average 10% per lifetime frequency, as a result of, for example, age, impaired immunity (as in the case of HIV co-infection), malnutrition, or anti-inflammatory drug administration that interferes with host immunity (3).
The key pathological feature of TB that arises as a result of the immune response is the formation of aggregates of bacteria and immune cells within the lung called granulomas. TB granulomas, especially in humans, form as organized spherical structures composed of bacteria, a macrophage-rich core including resting, infected and activated macrophages, and a surrounding mantle of lymphocytes. Granulomas act to immunologically restrain and physically contain Mtb infection (4–10). Latent and active TB in humans comprise a heterogeneous mixture of granulomas in both lung and lymph nodes that provide a range of physiological microenvironments associated with bacterial replication, persistence and killing. Characterization of different types of granulomas will provide a framework for understanding the immunobiology of TB that can lead to the development of new strategies for control and therapy (11, 12).
In addition to cellular components, studies in animal models and humans have identified a variety of cytokines involved in granuloma formation and function, including tumor necrosis factor-α (TNF) and interferon-γ (IFN-γ) (reviewed in (13)). These molecules are secreted from cellular sources (macrophages and T cells) as a result of Mtb infection, interact with receptors on target cells, trigger intracellular signaling pathways, and induce cell responses that ultimately contribute to formation of granulomas and immunologic control of Mtb infection (13–15). One can hypothesize that molecular scale processes that lie between the availability of particular extracellular cytokines and the final cytokine-mediated response may influence the outcome of Mtb infection. Receptor/ligand binding and trafficking (defined here to include synthesis, internalization, recycling and degradation of the ligand and receptors) are a group of molecular scale processes that take place under physiological conditions and are believed to play a major role in receptor-mediated cell responses (16). However, the significance of trafficking processes in controlling the effect of cytokines on the host immune response (TB immune response in particular) has never been studied. Hence, a multi-scale approach that considers events at the molecular, cellular and tissue scales is required for comprehensive analysis of the role of cytokines in the complex immune response to Mtb.
Our study is focused on TNF interactions with immune cells that form a granuloma. The pleiotropic cytokine TNF is produced by a variety of immune cells, especially infected and activated macrophages and pro-inflammatory T cells (17, 18), and functions as part of the immune response to Mtb infection via several mechanisms. TNF (in conjunction with the cytokine IFN-γ) induces macrophage activation (19–21), enhances immune cell recruitment to the site of infection (22), and augments chemokine expression by macrophages through activation of the NF-κB signaling pathway (23). TNF can also mediate cell death via inducing the caspase-mediated apoptotic pathway (24, 25). Data identifying the roles of TNF include: TNF knockout/neutralization experiments in mice and monkeys (17, 26–8), TNF receptor 1 (TNFR1) knockout experiments in mice (17), and mathematical modeling studies (29, 30). Despite this wealth of information on the critical role of TNF in immunity to Mtb, many fundamental questions remain unanswered regarding the mechanisms that regulate TNF activity at different biological scales. For example, it is not known how the dynamics of molecular events such as TNF/TNFR binding and trafficking influence a granuloma's ability to control Mtb infection. We have recently suggested via mathematical modeling that organization of immune cells as well as the processes of TNF/TNFR binding and trafficking control steady state TNF availability within an existing granuloma. This results from a TNF concentration gradient that is created with the highest concentration at the core of granuloma (31). However, important unanswered questions remain: What factors control such a gradient during a long-term immune response to Mtb infection that includes formation and maintenance of granulomas? How does this gradient regulate TNF-associated processes and ultimately translate to the outcome of Mtb infection? Is there an inter-play between TNF availability and bacterial load in a granuloma? Are there TNF-level processes that, if targeted, could present new strategies for disease therapy?
These questions invoke multiple biological scales (in length and time) are currently difficult to address experimentally. Hence, a systems biology approach that incorporates computational modeling to generate and test hypotheses, run virtual experiments, and make experimentally testable predictions is uniquely suited to address these questions. We develop a multi-scale computational model that describes the immune response to Mtb in the lung over three biological length scales: molecular, cellular and tissue. We use the model to track formation and maintenance of a granuloma in space and time. The model captures the dynamics of TNF/TNFR interactions that occur on the second to minute time scales within the long-term immune response to Mtb infection, a complex process that lasts for months to years. We identify TNF-associated processes that influence infection outcome at the granuloma scale as well as predicting cellular scale processes that influence TNF availability. Finally, we identify processes that regulate TNF concentration and cellular behaviors and thus influence the outcome of infection within a granuloma.
METHODS
An overview of the multi-scale granuloma model
To build a multi-scale model necessary to address the questions herein regarding TNF-regulated immune responses to Mtb infection in the lung, we need to first have working models at both the cellular/tissue scale and the molecular/single-cell scale. We briefly describe these models below and then describe our approach for linking them. Cellular and tissue scale dynamics are captured via a set of well-described interactions between immune cells and Mtb at the site of infection using stochastic simulations in the form of an agent-based model (ABM). Single-cell molecular scale processes that control TNF/TNFR binding and trafficking for each individual cell are captured by a set of nonlinear ordinary differential equations (ODEs). Figure 1 indicates how these models exist separately and how they are linked into a single multi-scale granuloma model. The linkage is achieved via TNF-induced cell responses (i.e. apoptosis and NF-κB activation) that are modeled as Poisson processes with rate parameters computed as functions of molecular concentrations from the ODE model. Further details of the rules, equations and parameters of the multi-scale model are described below and in Supporting Text and Supplementary Tables S1–5 at http://malthus.micro.med.umich.edu/lab/movies/Multiscale/GranSim/.
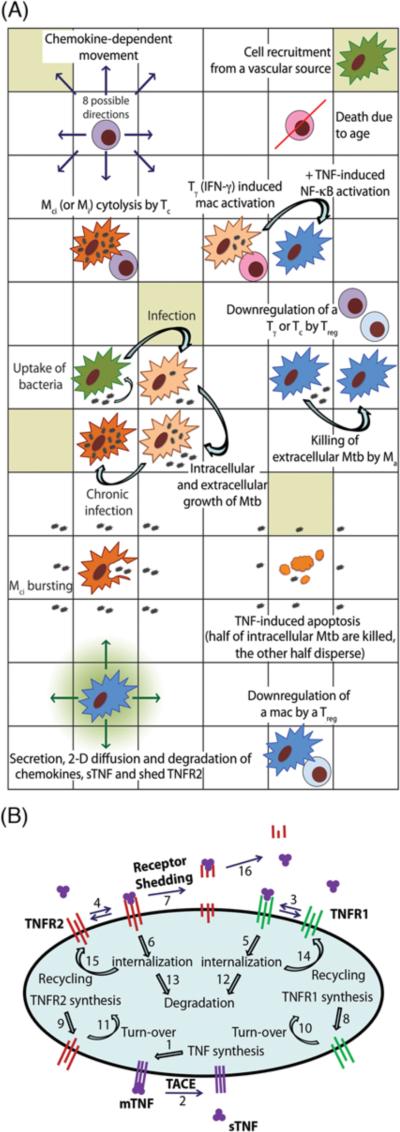
Figure 1.
Schematic representation of the multi-scale model of the immune response to Mtb infection in the lung. (A) An overview of selected cell-level ABM rules based on known immunological activities and interactions. (B) Schematic representation of binding interactions and reactions controlling TNF/TNFR dynamics at the single-cell level with numbers that represent model processes as listed in Supplementary Table S3.
Cellular/tissue scale model
The two-dimensional (2-D) ABM used in this study is an updated version of our previous model that captures cellular scale interactions leading to a tissue-level readout, namely granuloma formation in response to Mtb infection in primates (30). Because multi-scale analysis of the long-term immune response to Mtb is the aim of this study, an extensive sensitivity analysis is required. Therefore, choosing a 2-D model that can be used to run a large number of simulations in a reasonable timeframe is key for our study. A comparison with a simple three-dimensional version has shown that our 2-D model can capture important dynamics of Mtb infection (32). Figure 1A depicts a schematic overview of selected immunological interactions tracked at the cellular scale. A full description of all ABM rules that reflect known biological activities is provided in Supporting Text and significant updates to the original model are highlighted. Briefly, rule events include: chemotactic movement and recruitment of immune cells to site of infection, intracellular and extracellular growth of Mtb, phagocytosis of bacteria by macrophages, cell death and apoptosis, macrophage/T-cell interactions such as cytolytic functions of cytotoxic T cells (Tc) and IFN-γ based activation of macrophages by pro-inflammatory T cells (Tγ), down-regulation of immune cells by regulatory T cells (Treg), secretion of chemokines, and caseation.
One important simplification in our model is the choice to include only cell types with well-characterized roles in Mtb granulomas (macrophages, Tc, Tγ and Treg cells). Cell types that may have important roles but are not sufficiently characterized at this point to include in mechanistic ways in the model include neutrophils (with protective roles in early infection that may be immunomodulatory in nature (33–35)), multi-nucleate giant cells (may modulate chemokine production (36, 37)), dendritic cells (for optimal antigenic stimulation of T cells (38, 39)), and foamy cells (possible nutrient source for bacteria (40, 41)). Future work can easily incorporate them into the model when more mechanistic data become available.
Molecular/single-cell scale TNF/TNFR model
The kinetic processes of TNF/TNFR binding and trafficking (synthesis, internalization, recycling and degradation of ligand and receptors) occurring in individual cells within a granuloma can be described by ODEs (31). As schematically shown in Figure 1B, TNF is first synthesized by TNF-producing cells, including infected macrophages, chronically infected macrophages, NF-κB-activated resting macrophages, activated macrophages and T cells as a membrane-bound precursor form (mTNF) that can then be processed and released as a soluble form (sTNF) into extracellular spaces. Two types of TNF receptors (TNFR1 and TNFR2) are synthesized and expressed on the cell membrane. The equations describing TNF/TNFR processes for an individual cell are detailed in Supporting Text and Supplementary Tables S3, S4.
Linking the individual models via sTNF-induced cell responses
Activation of the two major TNF-induced signaling pathways, the caspase-mediated apoptotic pathway and the NF-κB pathway, are both controlled at the level of sTNF/TNFR1 interactions and thus serve as the link between the molecular/single-cell scale TNF/TNFR kinetic model and the cellular/tissue scale model. The NF-κB signaling pathway is initiated by sTNF-bound cell surface TNFR1 and apoptosis depends on the internalized sTNF/TNFR1 complexes (42–44). As reported in the literature, NF-κB activation of macrophages is a necessary but not sufficient factor in successful immune response to mycobacterial infection. Its role includes induction of a variety of inflammatory-related genes such as TNF and chemokines as well as controlling phagolysosome fusion-mediated killing of mycobacteria by activated macrophages (20, 45). TNF-induced apoptosis of macrophages kills intracellular bacteria and is associated with a better outcome of infection (3). In addition to sTNF, mTNF has also been shown to contribute in part to control of Mtb infection in mice (46–48). However, experimental data regarding molecular and cellular-level details of mTNF-mediated signaling and reverse signaling in Mtb immune responses are limited. Thus, at this time we only consider sTNF/TNFR1-mediated signaling in our model.
A recent study has shown that TNF-induced NF-κB activation is a process with a discrete nature at the single-cell level, with fewer cells responding at lower doses (49). Accordingly, we describe TNF-induced NF-κB activation for each individual macrophage as a Poisson process with a probability determined within each time-step (Δt), based on a Poisson rate parameter that is a function of the NF-κB activation rate constant (kNF-κB), the concentration of cell surface sTNF/TNFR1 complexes [sTNF/TNFR1], and the concentration threshold for cell surface sTNF/TNFR1 (τNF-κB):
| (1) |
Similarly, we model TNF-induced apoptosis for each individual cell (macrophage and T cell) by
| (2) |
We use a Poisson process with a probability computed as a function of the apoptosis rate constant (kapopt), the concentration of internalized sTNF/TNFR1 complexes [sTNF/TNFR1i], and the concentration threshold for internalized sTNF/TNFR1 (τapopt). Scale-linking parameters, or simply linking parameters, i.e. TNF-response parameters (defined here to include parameters introduced in Equations 1 and 2), are listed in Supplementary Table S5.
To analyze how TNF affects infected versus resting macrophages in a granuloma, we define infected:resting cell ratios, Rapoptosis and RNF-κB, as follows. Rapoptosis is defined as the ratio of the number of infected macrophages that undergo TNF-mediated apoptosis to the number of resting macrophages that undergo TNF-mediated apoptosis during a 200-day period post-infection. RNF-κB is similarly defined as the number of infected macrophages that undergo TNF-mediated NF-κB activation to the number of resting macrophages that undergo TNF-mediated NF-κB activation during a 200-day period post-infection.
Parameter estimation and control experiments
We estimate ABM parameter values from literature data as described in detail by Ray et al (30). When data are not available, we use uncertainty analysis to explore the entire parameter space of possible model outcomes as described in (50). Cell-specific TNFR densities and rate constants for TNF/TNFR processes are estimated based on experimental data from our group (31) and other groups as indicated in Supplementary Table S2. Values of parameters used to describe TNF-induced cell responses, including NF-κB activation and apoptosis (i.e. linking parameters) are estimated via uncertainty analysis by varying parameter values in ranges that are qualitatively consistent with experimental and modeling data on time-scales and thresholds for TNF-induced cell responses (49, 51, 52).
Using the above methods, we specify a baseline set of parameter values that robustly leads to control of infection in granulomas with organized structures as reported for humans and non-human primates (Supplementary Tables S1, S2 and S5). We then explore parameter changes that shift infection outcome to clearance or uncontrolled growth of Mtb. To further test the ability of the model to predict different infection outcomes under pathological conditions compatible with both experimental and previous modeling data on granuloma formation, we simulate gene knockouts of previously identified essential components of the Mtb immune response (e.g. TNF, IFN-γ and T cell knockouts). To do this, we set relevant probabilities or rate constants to zero from the beginning of simulations.
Sensitivity analysis
When computational models include parameters describing a large number of known biological processes, it is critical to understand the role that each of these parameters plays in determining output. Sensitivity analysis is a technique to identify critical parameters of a model and quantify how input uncertainty impacts model outputs. Latin hypercube sampling (LHS) is an algorithm that allows multiple parameters to be varied and sampled simultaneously in a computationally efficient manner (53). We use LHS sensitivity analysis as described for application to ABMs (50) to analyze the impact of TNF/TNFR trafficking and TNF response (linking) parameter values as well as TNF-independent and cellular scale parameter values, as listed in Supplementary Tables S1, S2 and S5, on model outputs. For clarity, these outputs are grouped, as TNF function-related outputs (total number of TNF-induced events, including NF-κB activation and apoptosis in different types of cells), cellular-level outputs (total bacteria, macrophage and T cell numbers), tissue-level outputs (granuloma size and caseation area) and average tissue concentrations of TNF and chemokines. The correlation of model outputs with each parameter is quantified via calculation of a partial rank correlation coefficient (PRCC). PRCC values vary between −1 (perfect negative correlation) and +1 (perfect positive correlation) and can be differentiated based on p-values derived from Student's t test. LHS simulations sampled each parameter 250 times. Each sampled parameter set was run 4 times and averages of the outputs were used to calculate PRCC values. The choice of the number of simulations is determined by the desired significance level for the PRCC (50, 53). Here, 250 runs imply that PRCC values above +0.24 or below −0.24 are significantly different from zero (p < 0.001). To study how processes at different scales interact with each other, we analyze the effect of parameters associated with each scale on the outputs of the same scale (intra-scale sensitivity analysis) as well as on the outputs of the other scale (inter-scale sensitivity analysis).
Computer simulations and visualization
The model was implemented in C++. We use Qt, a C++ framework for developing cross-platform applications with a graphical user interface (GUI), to visualize and track different aspects of the granuloma, including the structure and molecular concentration gradients, as it forms and is maintained. Simulations can be run with or without graphical visualization. Simulations were run on Linux and Mac operating systems.
RESULTS
Prediction of different infection outcomes at the granuloma level
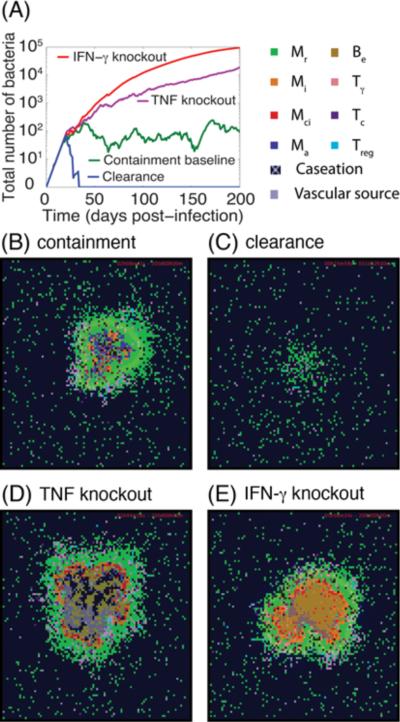
We first tested whether our multi-scale computational model could capture key features of granuloma formation and maintenance. Using a combination of parameter estimation and uncertainty analysis as described in Methods, we identified a set of baseline values for model parameters, including cellular/tissue scale parameters (Supplementary Table S1), molecular/single-cell scale TNF/TNFR parameters (Supplementary Table S2) and parameters that link the two scales in the model (Supplementary Table S5). This set of parameter values leads to containment: control of Mtb infection within a well-circumscribed granuloma containing stable bacterial levels (<103 total bacteria) at 200 days post-infection (Figures 2A, B, Movie 1 at http://malthus.micro.med.umich.edu/lab/movies/Multiscale/GranSim/). This recapitulates a state that has been described as an equilibrium between the host and Mtb at the level of single granuloma and is referred to as a solid granuloma with caseous center (Mtb containment) (3, 54). As observed in Movie 1, simulated granulomas form as organized immune structures predominantly composed of uninfected macrophages surrounding a core of bacteria and infected and activated macrophages with T cells localized at the periphery (4–10). As reported for most animal models of TB, bacterial growth increases logarithmically until reaching a plateau coincident with T cell response initiation (55).
Figure 2.
Simulation results for the Mtb dynamics and granuloma structures at 200 days post-infection under different pathological conditions. (A) Changes of total number of Mtb (intracellular and extracellular bacteria, i.e. Bint + Bext) with time for simulation of containment baseline, a scenario of Mtb clearance, a TNF (or TNFR1) knockout scenario and a IFN-γ knockout scenario. Granuloma snapshots for: (B) a scenario of containment, (C) clearance of Mtb infection in less than five weeks as a result of an efficient immune response, (D) a TNF (or TNFR1) knockout scenario, and (E) an IFN-γ knockout scenario. Cell types and status are shown by different color squares, as indicated in the upper right corner of the figure (Mr: resting macrophage, Mi: infected macrophage, Mci: chronically infected macrophage, Ma: activated macrophage, Be: extracellular bacteria, Tγ: pro-inflammatory IFN-γ producing T cell, Tc: cytotoxic T cell, Treg: regulatory T cell). Caseation and vascular sources are also indicated.
As observed in non-human primate models as well as in humans, several types of granuloma are observed in Mtb infection (56). Our multi-scale model is also able to recapitulate different granuloma types with different abilities to control infection as we vary specific parameters identified as important via sensitivity analysis from their baseline values (see Supplementary Tables S1, S2 and S5). Cellular scale processes identified to significantly control bacterial numbers, caseation and granuloma size at 200 days post-infection are bacterial growth, Tγ-induced STAT-1 activation of macrophages and T cell movement and recruitment (Tγ cells in particular). These results are highlighted in Table I and are consistent with available experimental data reviewed in (3, 57, 58) and our previous modeling studies (30, 59). Greater intracellular Mtb growth rates, in agreement with published data (60), lead to higher bacterial loads and larger granulomas with larger caseation areas. STAT-1 activation of macrophages by IFN-γ producing Tγ cells is required for activation of macrophages and killing of intracellular and extracellular Mtb. Recruitment of IFN-γ producing pro-inflammatory T cells (Tγ cells) to site of infection is a critical component of immunity to Mtb as a smaller TNF/chemokine concentration threshold for Tγ recruitment leads to more efficient responses. Further, the ability of T cells to migrate through a dense uninfected macrophage network surrounding bacteria and infected macrophages at the core of a granuloma helps determine the efficiency of the T cell-mediated immune response to Mtb.
Table I.
Model parameters significantly correlated with outputs of interest, bacterial numbers, granuloma size, caseation area and TNF concentration at day 200 post-infection. Detailed sensitivity analysis results are presented in Supplementary Tables S6, S7.
| Selected model outputs | Important TNF-independent and cellular scale parameters *† | Important TNF/TNFR-associated molecular and linking parameters *‡ |
|---|---|---|
| Total number of bacteria | αBi (++) | ksynthMac (−−) |
| PSTAT1 (−−) | δTNF (++) | |
| TmoveM (−−) | Kd1 (+) | |
| Trecr (−−) | kint1 (+) | |
| τrecTgam (++) | TNFR1mac (−) | |
| Dchem (−−) | kNFκB (−−) | |
| δchem (++) | τNFκB (++) | |
|
| ||
| Granuloma size | αBi (++) | ksynthMac (−−) |
| PSTAT1 (−) | δTNF (++) | |
| Mrecr (++) | Kd1 (++) | |
| TmoveM (−−) | kint1 (+) | |
| Trecr (−−) | TNFR1mac (−−) | |
| Papop/Fas (−) | kNFκB (−−) | |
| τrecTgam (++) | τNFκB (++) | |
|
| ||
| Caseation | αBi (++) | ksynthMac (−−) |
| PSTAT1 (−) | δTNF (++) | |
| Mrecr (++) | Kd1 (++) | |
| TmoveM (−−) | kint1 (+) | |
| Trecr (−−) | Krec1 (++) | |
| τrecTgam (++) | TNFR1mac (−−) | |
| δchem (++) | kapop (−) | |
| τchem (+) | kNFκB (++) | |
| τNFκB (++) | ||
|
| ||
| Average tissue concentration of sTNF | αBi (++) | ksynthMac (−−) |
| PSTAT1 (−) | Kd1 (+) | |
| TmoveM (−−) | TNFR1mac (−) | |
| Trecr (−−) | kNFκB (−−) | |
| τrecTgam (++) | τNFκB (+) | |
| δchem (++) | ||
| τchem (+) | ||
Only parameters with significant PRCC values are indicated. Significant positive and negative correlations are shown using + and − as follows: −/+: 0.001 < p-value < 0.01, −−/++: p-value < 0.001.
TNF-independent and cellular scale parameter descriptions are as follows: αBi intracellular Mtb growth rate, PSTAT1 probability of STAT-1 activation in Mr or Mi, TmoveM probability of T cell moving to a macrophage-containing location, Trecr probability of T cell recruitment, Mrecr probability of Mr recruitment, τrecTgam TNF/chemokine concentration threshold for Tγ recruitment, Papop/Fas probability of Fas/FasL apoptosis by Tγ, Dchem diffusion coefficient of chemokines, δchem chemokine degradation rate constant. τchem minimum chemokine concentration detection threshold.
TNF/TNFR associated parameter descriptions are as follows: ksynthMac mTNF synthesis rate for macrophages, δTNF sTNF degradation rate constant, Kd1 equilibrium dissociation constant of sTNF/TNFR1, kint1 TNFR1 internalization rate constant, krec1 TNFR1 recycling rate constant, TNFR1mac TNFR1 density on the surface of macrophages, kapop rate constant for TNF-induced apoptosis in all cell types, kNFκB rate constant for TNF-induced NF-κB activation in macrophages, τNFκB cell surface sTNF/TNFR1 threshold for TNF-induced NF-κB activation.
In addition to containment, we can reproduce other possible outcomes of Mtb infection, including clearance and uncontrolled growth of bacteria, by manipulating values of important model parameters. For example, an increase in the ability of T cells to penetrate into the site of infection at the core of granuloma by increasing the value of model parameter Tmove, the probability of a T cell moving onto a macrophage-containing micro-compartment, significantly increases the efficiency of the T cell-mediated response and thus favors Mtb clearance (Figure 2A, C, Movie 2 at http://malthus.micro.med.umich.edu/lab/movies/Multiscale/GranSim/). On the other hand, simulations of gene knockouts of essential components of the Mtb immune response such as TNF or TNFR1 knockouts (ksynthMac = ksynthTcell = 0 or TNFR1mac = TNFR1Tcell = 0) and IFN-γ knockout (PSTAT1 = 0) lead to uncontrolled growth of Mtb. This is consistent with a variety of data on the crucial role of these cytokines in immunity to Mtb (3, 13, 61). In this case, granulomas that form are greater in size, irregular in structure and include very high numbers of extracellular Mtb, large numbers of infected macrophages and wide-spread caseation (dead tissue caused by multiple deaths of macrophages in tissue usually within the core of the granuloma) (Figure 2A, D, E, Movies 3, 4 at http://malthus.micro.med.umich.edu/lab/movies/Multiscale/GranSim/). Overall, our multi-scale model that includes molecular (TNF-associated), cellular, and tissue scale events predicts dynamics of Mtb infection for different infection scenarios, including containment, clearance and uncontrolled growth of bacteria as well as a variety of structural and functional outcomes that are expected to occur under different pathological conditions. Our results for these conditions are in agreement with our previous study using a model without molecular (TNF-associated) events (30) and a variety of experimental data. We now turn our analysis to the important role that TNF plays, and the factors that affect the ability of TNF to play that role, during the immune response to Mtb.
Cellular scale processes control TNF concentration by affecting bacterial load
We know from experimental studies that artificial manipulation of TNF concentration via anti-TNF treatments negatively affects infection outcome in mice, humans and non-human primates (26, 27, 62, 63). Are there physiological processes within the granuloma that also affect TNF concentration, and does alteration of those processes similarly affect infection outcome? To answer this question, we used sensitivity analysis to identify critical TNF-independent and cellular scale parameters that influence TNF concentration. Interestingly, processes highlighted in the previous section to be important determinants of bacterial numbers, caseation and granuloma size also significantly impact TNF concentration and thus the number of TNF-induced NF-κB activation and apoptosis events (see Table I and Supplementary Table S6 at http://malthus.micro.med.umich.edu/lab/movies/Multiscale/GranSim/). This is because TNF production within the granuloma strongly depends on the level of infection. For example, reducing the probability of T cell recruitment (Trecr) decreases the level of T cell-mediated macrophage activation, a process that is necessary for limiting Mtb growth in infected macrophages. Therefore, intracellular Mtb can grow, disperse in the tissue after bursting of chronically infected macrophages, and infect more resting macrophages. This ultimately increases the number of extracellular Mtb as well as numbers of infected macrophages that act as further TNF sources in the tissue. TNF also enhances activation and recruitment of immune cells, leading to larger granulomas. This is in agreement with data from animal models that show increased bacterial numbers result in increased inflammation and more immune cell recruitment (64).
TNF/TNFR molecular processes control TNF actions and thus ability of a granuloma to control infection
The analysis above indicates that physiological processes that affect bacterial load indirectly affect TNF concentration. Are there other processes that more directly act on TNF, and would manipulation of those processes alter infection outcome? To answer this question, we performed sensitivity analysis to identify TNF/TNFR-associated molecular scale and linking parameters that influence model outcomes. Both signaling and trafficking processes are identified to significantly influence granuloma function, as indicated in Table I and Supplementary Table S7 at http://malthus.micro.med.umich.edu/lab/movies/Multiscale/GranSim/. TNF-induced NF-κB activation of resting and infected macrophages strongly correlates with bacterial numbers. Increasing the rate constant for TNF-induced NF-κB activation or reducing its cell surface sTNF/TNFR1 concentration threshold leads to faster macrophage NF-κB activation responses by smaller concentrations of available sTNF. Thus, these parameters can significantly influence the outcome of infection. This is consistent with the published data on the role of NF-κB activation of macrophages in killing mycobacteria (20).
Receptor and ligand trafficking processes that strongly influence infection outcome are: mTNF synthesis by infected and activated macrophages, sTNF degradation, TNFR1 affinity for sTNF, TNFR1 density on the membrane of macrophages, and sTNF-induced internalization of TNFR1 (Tables I, S7). The rate of mTNF synthesis by infected and activated macrophages positively correlates with two key TNF functions (NF-κB activation and apoptosis) and negatively correlates with bacterial load, numbers of infected and chronically infected macrophages as well as granuloma and caseation size. This is consistent with human and animal model data and our previous studies on the crucial role of TNF in controlling Mtb infection (26, 29, 30, 62, 65). Similarly, the rate of sTNF degradation negatively influences TNF activities and thus positively correlates with bacterial numbers within a granuloma. TNFR1 density on macrophage membranes has a negative impact on bacterial numbers. This is consistent with experimental data on the importance of TNFR1 in controlling Mtb infection (17). A greater TNFR1 affinity for binding to sTNF (smaller Kd1) also enhances the level of TNF-induced NF-κB activation and apoptosis and reduces total bacterial numbers. Internalization of TNFR1 occurs as a result of sTNF binding to TNFR1 on cell membranes and is not only required for TNF-induced apoptosis (42–44), but also reduces sTNF in a granuloma (31). Thus TNFR1 internalization enhances apoptosis in TNF-secreting infected macrophages but reduces levels of NF-κB activation in non-TNF-secreting resting macrophages by limiting TNF concentrations near these cells (see Supplementary Table S7). Overall, our sensitivity analysis predicts that sTNF-induced TNFR1 internalization increases bacterial levels within a granuloma. We focus our next analysis on potential effects of manipulation in TNFR1 internalization.
TNF/TNFR trafficking dynamics balance inflammation and bacterial killing
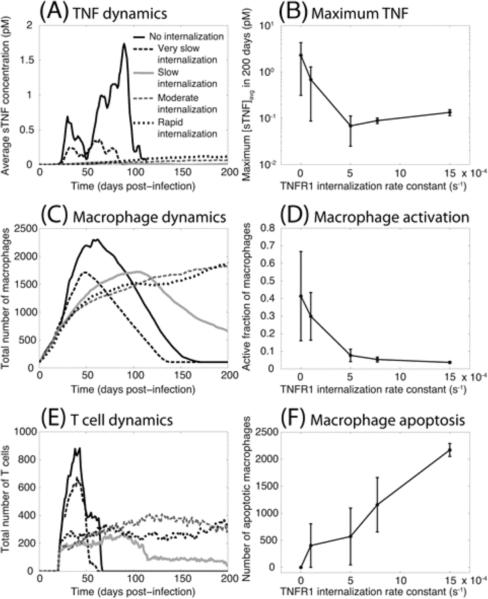
The effect of changing the rate constant for TNFR1 internalization on sTNF concentration, recruitment of immune cells, macrophage activation and apoptosis is shown in Figure 3. As described above, sTNF-induced TNFR1 internalization, the key process in TNF/TNFR trafficking, has a significant impact on responses at molecular, cellular and tissue scales. The value of the rate constant for TNFR1 internalization (kint1) controls sTNF concentration dynamics during the immune response to Mtb (Figure 3A). The physiological rate of TNFR1 internalization (half-time of ~15 min (66, 67), kint1 = 7.7×10−4 s−1) leads to much less extracellular sTNF in the tissue compared with the scenario in which sTNF/TNFR1 complex on the cell membrane is not at all (kint1 = 0) or is very slowly internalized (half-time of ~115 min, kint1 = 1.0×10−4 s−1) (Figure 3B). Although TNF is required for control of Mtb infection and the protective granulomatous response, high concentrations of TNF may lead to excessive inflammation and cause immunopathology (61, 68). Therefore, we predict that TNF/TNFR trafficking plays an important role in preventing excessive inflammation. Indeed, the rate of sTNF-induced internalization of TNFR1 controls the concentration of available TNF in tissue and regulates cell infiltration by affecting the extent and dynamics of TNF-dependent recruitment and activation of immune cells as well as mediating TNF-induced apoptosis (Figure 3C–F). Thus, a hyper-inflammatory state may occur in the absence of a sufficiently rapid sTNF-induced TNFR1 internalization, leading to early and extensive recruitment of macrophages and T cells as well as uncontrolled activation of a large fraction of macrophages that are unable to efficiently undergo apoptosis. Interestingly, increasing TNFR1 internalization rate constant to kint1 = 1.5×10−3 s−1 (corresponding to a half-time of ~7.7 min) does not have a large effect on either sTNF concentration or immune cell population dynamics but does significantly enhance the number of apoptotic macrophages. However, further analysis reveals that other model outputs may be significantly affected by increasing TNFR1 internalization rate constant.
Figure 3.
TNFR1 internalization dynamics control sTNF concentration as well as macrophage and T cell recruitment and behavior. Simulation results show: (A) sTNF concentration dynamics, (B) maximum simulated sTNF concentration as a function of TNFR1 internalization rate constant (kint1), (C) macrophage recruitment dynamics, (D) maximum fraction of macrophages that become activated following Mtb infection, (E) T cell recruitment dynamics, (F) TNF-induced macrophage apoptosis within a 200 day period after Mtb infection (No internalization: kint1 = 0, very slow internalization: kint1 = 1.0×10−4 s−1, slow internalization: kint1 = 5.0×10−4 s−1, moderate internalization: kint1 = 7.7×10−4 s−1, and rapid internalization: kint1 = 1.5×10−3 s−1).
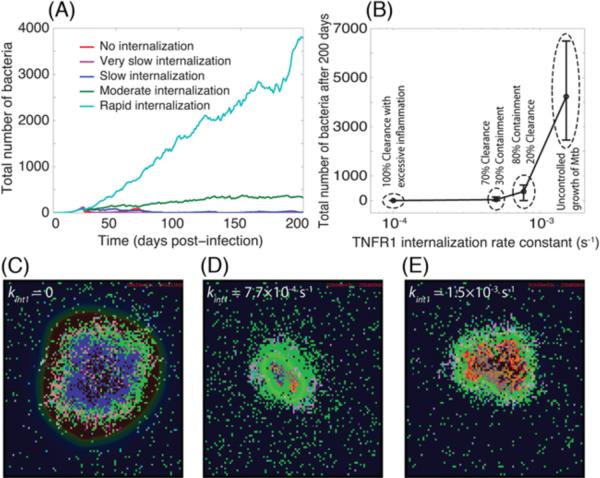
In addition to an impact on inflammation, TNF/TNFR trafficking dynamics are capable of exerting a dramatic effect on the bacterial outcome of Mtb infection in a granuloma (Figure 4). Zero to slow rates of sTNF-induced TNFR1 internalization (half-time of ≥23 min, kint1 ≤ 5.0×10−4 s−1) favor clearance of bacteria; a moderate (physiological) rate (half-time of ~15 min, kint1 = 7.7×10−4 s−1) leads to containment of bacteria, and a rapid rate of TNFR1 internalization (half-time of ~7.7 min. kint1 = 1.5×10−3 s−1) results in uncontrolled growth of Mtb within the 200-day period of infection (Figure 4, Movies 5–8 at http://malthus.micro.med.umich.edu/lab/movies/Multiscale/GranSim/). However, zero or very slow rates of TNFR1 internalization (e.g. half-time of ~115 min, kint1 = 1.0×10−4 s−1) result in clearance of infection at the expense of extensive inflammation. Thus, our model suggests that there may exist an optimum rate of sTNF-induced TNFR1 internalization that balances the impacts that TNF has on control of Mtb infection and inflammation in tissue. We now investigate mechanisms underlying this balance.
Figure 4.
TNFR1 internalization dynamics control bacterial load during Mtb infection. Simulation results show: (A) Mtb dynamics within a 200 day period after Mtb infection, (B) granuloma outcomes and bacterial load 200 days post-infection, (C) granuloma snapshot at the time of Mtb clearance (day 45) in the absence of TNFR1 internalization (kint1 = 0), (D)–(E) granuloma snapshots 200 days after Mtb infection for moderate (kint1 = 7.7×10−4 s−1) and rapid (kint1 = 1.5×10−3 s−1) rates of TNFR1 internalization. The colors representing cells of different type and status in granuloma snapshots are the same as shown in Figure 2.
Do high rates of TNFR1 internalization and slow rates of TNF synthesis have the same effects?
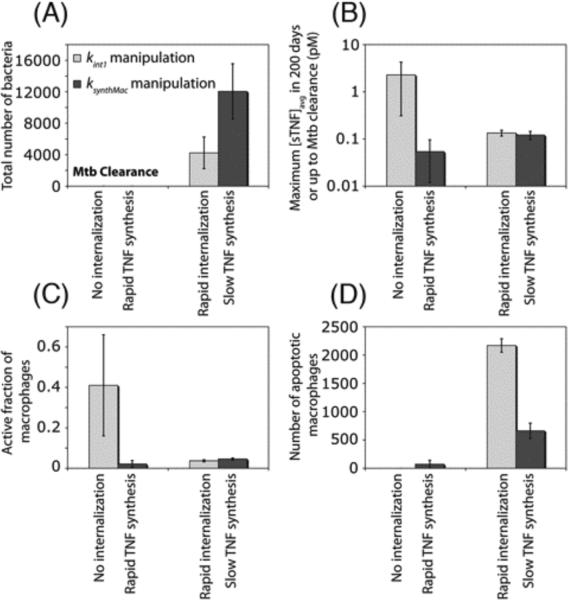
In the previous section, we showed that the rate of sTNF-induced TNFR1 internalization significantly affects the immune response to Mtb; a small value of TNFR1 internalization rate constant favors Mtb clearance, while increasing the rate of TNFR1 internalization leads to uncontrolled growth of Mtb. One might argue that such an effect is simply attributable to the role of TNFR1 internalization in reducing sTNF concentration in the granuloma, and that therefore an increase or a decrease in the rate of TNF synthesis may compensate for the effects of increasing or decreasing the rate of TNFR1 internalization. To test this hypothesis, we compare the effects of manipulating rates of TNFR1 internalization and mTNF synthesis by macrophages on model outputs (such as bacterial numbers and inflammation). As indicated in Figure 5A, a zero rate of TNFR1 internalization and a high rate of TNF synthesis both result in Mtb clearance. However, high rates of TNF synthesis, in contrast to very slow or zero rates of TNFR1 internalization, do not lead to dramatic increases in sTNF concentration and macrophage activation (Figures 5B, C). This is because impairing TNFR1 internalization has a negative effect on rates of TNF-induced apoptosis (Figure 5D), a process that has been suggested to be important for controlling the level of inflammation (30). However, high rates of TNF synthesis favor apoptosis of macrophages and thus do not lead to extensive inflammation.
Figure 5.
Manipulations in the rate constants for TNFR1 internalization (kint1) and mTNF synthesis (ksynthMac) lead to different effects on model outputs. Simulation results show the effect of manipulations in kint1 and ksynthMac on (A) bacterial levels 200 days after Mtb infection, (B) maximum sTNF concentration, (C) maximum fraction of macrophages that become activated following Mtb infection and (D) TNF-induced macrophage apoptosis within a 200-day period after Mtb infection (No internalization: kint1 = 0, rapid TNF synthesis: ksynthMac = 1 #/cell.s, rapid internalization: kint1 = 1.5×10−3 s−1, slow TNF synthesis: ksynthMac = 0.1 #/cell.s).
On the other hand, a rapid rate of TNFR1 internalization and a small rate of TNF synthesis both result in uncontrolled growth of Mtb, although to different extents (Figure 5A). This difference can be explained by high levels of TNF-induced apoptosis in macrophages (and thus infected macrophages) for high values of TNFR1 internalization rate constant, whereas a small rate of TNF synthesis leads to lower levels of apoptosis (Figure 5D). Apoptosis of infected macrophages can help with reducing intracellular bacterial burden (25). Thus, our results suggest that the impact of TNF/TNFR trafficking on Mtb infection is more complex than simply changing the TNF concentration in the granuloma.
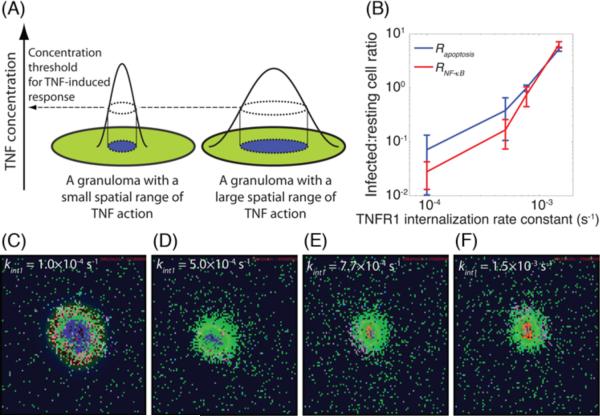
TNFR1 internalization controls the spatial range of TNF action within a granuloma
As demonstrated above, sTNF-induced TNFR1 internalization controls both Mtb infection and inflammation in tissue. How does this process play such a key role? Here, we explore the possibility that the spatial range of TNF action underlies the important effects of the rate of TNFR1 internalization on granuloma function. By spatial range of TNF action, we mean the area surrounding the center of granuloma, as indicated in Figure 6A, within which macrophages become activated or undergo apoptosis via autocrine or paracrine stimulation pathways (19, 25, 69). As infected macrophages are located in the core of granuloma surrounded by resting macrophages, the spatial range of TNF action is correlated with the infection status of macrophages affected by TNF. Thus, motivated by our sensitivity analysis (Supplementary Table S7), we explore here the possibility that TNF/TNFR trafficking leads to differential effects on TNF-mediated responses in cells of different infection status. We analyzed the infection status of macrophages affected by TNF-induced events (either NF-κB activation or apoptosis) following Mtb infection by computing infected:resting cell ratios, RNF-κB and Rapoptosis, as defined in Methods (Figure 6B). These ratios compare TNF effects on infected macrophages versus resting macrophages during the Mtb immune response. Our model predicts a very significant effect of TNFR1 internalization on both Rapoptosis and RNF-κB (Figure 6B). At very slow rates of sTNF-induced TNFR1 internalization (half-time of ~115 min, kint1 = 1.0×10−4 s−1) resting macrophages are the main cells affected by both TNF-mediated apoptosis and NF-κB signaling pathways (Rapoptosis and RNF-κB << 1). However, with a dramatic increase in the rate of TNFR1 internalization (to a half-time of ~7.7 min, kint1 = 1.5×10−3 s−1), infected macrophages become the main responders to TNF-induced activities (Rapoptosis and RNF-κB >> 1). Figures 6C–F display how granulomas are affected by the rate at which sTNF/TNFR1 complexes become internalized; these snapshots are taken early after T cell recruitment to the site of infection. While a significant fraction of resting macrophages surrounding the infected core of granuloma become activated as a result of slow rates of TNFR1 internalization (i.e. there is a greater spatial range of TNF action, as seen in Figure 6C), only a small number of infected macrophages in the core may become activated with a rapid rate of TNFR1 internalization (Figure 6F). Thus, we suggest that the spatial range of TNF action within a granuloma is an important factor that controls the effect of TNFR1 internalization on the bacterial outcome of Mtb infection as well as the level of inflammation in the tissue.
Figure 6.
TNFR1 internalization dynamics control the spatial range of TNF action within a granuloma. (A) Schematic definition of spatial range of TNF action in a granuloma, (B) The effect of kint1 on Rapoptosis and RNF-κB, the ratios of total number of TNF-induced (apoptotic and NF-κB activated) infected macrophages to the number of TNF-induced resting macrophages within a 200 day period after Mtb infection, (C)–(F) Granuloma snapshots early after recruitment of T cells for very slow, slow, medium and rapid rates of sTNF-induced TNFR1 internalization. Simulated granuloma snapshots are shown at 5 weeks after Mtb infection, right before clearance of bacteria in (C) and at 8 weeks after Mtb infection in (D–F). The colors representing cells of different type and status in granuloma snapshots are the same as shown in Figure 2.
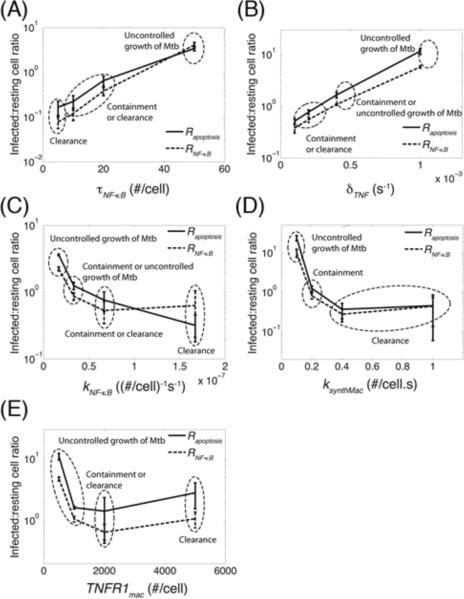
A robust metric for assessing TNF impact on granuloma function
In the previous section, we demonstrated that the impact of the rate of TNFR1 internalization on bacterial levels in a granuloma is significantly correlated with infection status of macrophages that undergo TNF-mediated apoptosis or NF-κB activation (Rapoptosis and RNF-κB). Here, we explore the possibility that such a correlation between TNF activities and infection outcome also exists for other processes. We analyzed the effect of varying values of important TNF-associated molecular scale and linking parameters on Rapoptosis and RNF-κB during a 200-day period post-infection. A significant correlation was observed between bacterial levels and infected:resting cell ratios, Rapoptosis and RNF-κB. As indicated in Figure 7, an increase of one order of magnitude in cell surface sTNF/TNFR1 concentration threshold for NF-κB activation and TNF degradation rate constant, or a decrease of one order of magnitude in NF-κB activation rate constant and the rate of mTNF synthesis by macrophages around baseline parameter values led to significant increases in both Rapoptosis and RNF-κB as well as bacterial levels. Outcomes of uncontrolled growth of Mtb generally occur at Rapoptosis and RNF-κB values of 1–10 or greater, while the chance of achieving clearance is greater for smaller values of these ratios. However, as indicated in Figure 7E, when macrophage TNFR1 density is varied, the correlation between these ratios and bacterial levels (in clearance and containment cases in particular) does not appear very significant. This is probably because TNFR1 density has contradictory effects on TNF functions; although greater TNFR1 densities lead to more sensitive responses to smaller TNF concentrations, at the same time such larger densities enhance TNF uptake by macrophages limiting TNF availability in a granuloma. Overall, we suggest that infected:resting cell ratios we introduced here to compare TNF effects on infected versus resting macrophages, Rapoptosis and RNF-κB,translate the effects of a variety of TNF-associated processes to granuloma outcomes.
Figure 7.
The impact of TNF/TNFR-associated processes on granuloma outcome is correlated with infection status of macrophages that undergo TNF-mediated apoptosis or NF-κB activation. Simulation results show the effect of: (A) cell surface sTNF/TNFR1 concentration threshold for NF-κB activation (τNF-κB), (B) sTNF degradation rate constant (δTNF), (C) NF-κB activation rate constant (kNF-κB), (D) rate of mTNF synthesis by macrophages (ksynthMac), and (E) macrophage TNFR1 density (TNFR1mac) on infected:resting cell ratios Rapoptosis and RNF-κB within a 200-day period after Mtb infection. Also indicated is granuloma outcome (clearance, containment, or uncontrolled growth of Mtb).
DISCUSSION
TNF was long suggested, based on experimental data from mice, to be essential for formation of granulomas in response to Mtb (17, 26, 70). However, recent TNFR1 knockout and TNF neutralization experiments in zebrafish and non-human primate models have shown that TNF, although not required for the formation of a granuloma, is important to restrict mycobacterial growth in a granuloma (27, 71). This suggests that TNF activities within a granuloma determine our ability to control Mtb infection. The important questions are, then, how TNF activities influence granuloma function, and what mechanisms control TNF activities in a granuloma during a long-term immune response to Mtb? To answer these questions, we need information about the spatial and temporal dynamics of TNF concentration during granuloma development in vivo. These experiments are not at present feasible, and thus, theses questions have remained unanswered. In this study, we use computational modeling/systems biology to address these questions. Our novel hypothesis is that events at different biological scales (molecular, cellular and tissue scales) may influence TNF activities in a granuloma, ultimately determining a granuloma's ability to control infection and inflammation. To address this hypothesis, our model was developed to link the dynamics of molecular scale TNF/TNFR interactions that occur on the second to minute time scales to cellular/tissue scale events that control the long-term immune response to Mtb. One of our interesting findings is that both TNF-independent cellular/tissue scale events (e.g. T cell recruitment or chemotactic movement of immune cells) and TNF-associated molecular scale processes (e.g. mTNF synthesis or TNFR1 internalization) influence TNF availability and activity in the granuloma, but in different ways. TNF-independent cellular scale processes influence bacterial numbers and that controls TNF availability. However, TNF-associated molecular scale processes directly affect TNF availability and activities that control both the level of inflammation and bacterial numbers. Thus, there is an inter-play between TNF availability and bacterial population at the site of infection that is controlled by the combined effects of molecular and cellular scale processes. An equilibrium state in this inter-play leads to control of infection within a granuloma.
Our model reveals, for the first time, the importance of TNF-associated molecular processes (TNFR1 internalization in particular) in immunity to Mtb. We found that TNFR1 internalization regulates a balance between paracrine and autocrine TNF-induced responses, including NF-κB activation and apoptosis, in resting versus infected macrophages. Because resting macrophages do not express TNF, they become activated by TNF-producing cells only in a paracrine manner. However, infected macrophages express and release TNF to the extracellular space. Hence, they can become activated under the effect of TNF via both autocrine and paracrine pathways. Our results show that TNFR1 internalization favors activation of infected macrophages in an autocrine manner by restricting the diffusion of TNF from TNF-producing cells. TNF-induced activation of resting macrophages in addition to infected macrophages is necessary for controlling Mtb infection. Uncontrolled activation of resting macrophages, on the other hand, may result in excessive inflammation. Thus, a balance between the autocrine and paracrine TNF-induced responses is required for an efficient granuloma response to Mtb and an optimum rate of TNFR1 internalization can provide this balance. This finding can be considered in future studies examining approaches to control and therapy of TB or inhibition of TB reactivation as several ways have already been proposed to influence the rate of TNFR1 internalization in vitro (42, 72, 73).
Another novel hypothesis from this study is that the efficacy of TNF in controlling Mtb infection is strongly affected by whether or not macrophages induced by TNF-mediated signaling pathways (NF-κB activation and apoptosis) are infected. Bacterial numbers are positively correlated with the ratio of infected macrophages to (uninfected) resting macrophages that become activated by TNF. Thus, we suggest that this ratio is a critical factor that controls the outcome of Mtb infection at the granuloma level. This might be of particular interest in the case of TB reactivation as a result of using TNF-neutralizing drugs (e.g. for treatment of inflammatory diseases such as rheumatoid arthritis and Crohn's disease). As drug penetrates into a granuloma, resting macrophages, compared with infected macrophages in the granuloma core, are exposed to smaller concentrations of TNF and are affected by higher concentrations of the drug. This can potentially impair TNF function, leading to TB reactivation.
Finally, our findings may predict new therapies for control of TB as they suggest novel host targets (e.g. TNFR1 internalization and NF-κB activation) that play key roles in control of Mtb immune response. Further modeling studies including molecular detail of additional processes, such as those involving other cytokines (e.g. IL-10, IL-6 and IL-12) and chemokines, and using a similar approach to that identified here may also identify other important targets for therapy. Our multi-scale computational model also provides a platform at the level of single granuloma to identify and compare therapeutic strategies as well as to investigate mechanisms by which TNF-neutralizing drugs (used to treat inflammatory diseases) or other drugs that diffuse in TB lesions may interfere with immune response to Mtb and reactivate TB.
Supplementary Material
ACKNOWLEDGEMENT
We thank Joe Waliga for management of Movies and Supplementary Materials at http://malthus.micro.med.umich.edu/lab/movies/Multiscale/GranSim/. We also thank JoAnne Flynn for helpful discussions.
Grant Support This work was supported by National Institute of Health (NIH) grants R33HL092844, R33HL092853, and N01 AI50018.
Abbreviations used in this paper
- TB
tuberculosis
- Mtb
Mycobacterium tuberculosis
- TNFR
TNF receptor
- ABM
agent-based model
- ODE
ordinary differential equation
- 2-D
two-dimensional
- Tc
cytotoxic T cell
- Tγ
pro-inflammatory T cell
- Treg
regulatory T cell
- mTNF
membrane-bound TNF
- sTNF
soluble TNF
- LHS
Latin hypercube sampling
- PRCC
partial rank correlation coefficient
- GUI
graphical user interface
- kNF-κB
NF-κB activation rate constant
- τNF-κB
concentration threshold of cell surface sTNF/TNFR1 complexes for NF-κB activation
- kapopt
apoptosis rate constant
- τapopt
concentration threshold of internalized sTNF/TNFR1 complexes for apoptosis
- Tmove
probability of a T cell moving onto a macrophage-containing micro-compartment
- ksynthMac
rate of mTNF synthesis by macrophages
- ksynthTcell
rate of mTNF synthesis by a T cell
- TNFR1mac
TNFR1 density on a macrophage
- TNFR1Tcell
TNFR1 density on a T cell
- PSTAT1
probability of STAT-1 activation of a macrophage
- Trecr
probability of T cell recruitment
- Kd1
equilibrium dissociation constant for TNF/TNFR binding
- kint1
TNFR1 internalization rate constant
REFERENCES
- 1.Mortellaro A, Robinson L, Ricciardi-Castagnoli P. Spotlight on mycobacteria and dendritic cells: will novel targets to fight tuberculosis emerge? EMBO Mol. Med. 2009;1:19–29. doi: 10.1002/emmm.200900008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Russell DG, Barry CE, 3rd, Flynn JL. Tuberculosis: what we don't know can, and does, hurt us. Science. 2010;328:852–856. doi: 10.1126/science.1184784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lin PL, Flynn JL. Understanding latent tuberculosis: a moving target. J. Immunol. 2010;185:15–22. doi: 10.4049/jimmunol.0903856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Algood HM, Chan J, Flynn JL. Chemokines and tuberculosis. Cytokine Growth Factor Rev. 2003;14:467–477. doi: 10.1016/s1359-6101(03)00054-6. [DOI] [PubMed] [Google Scholar]
- 5.Morel PA, Ta'asan S, Morel BF, Kirschner DE, Flynn JL. New insights into mathematical modeling of the immune system. Immunol. Res. 2006;36:157–165. doi: 10.1385/IR:36:1:157. [DOI] [PubMed] [Google Scholar]
- 6.Davis JM, Ramakrishnan L. “The very pulse of the machine”: the tuberculous granuloma in motion. Immunity. 2008;28:146–148. doi: 10.1016/j.immuni.2008.01.002. [DOI] [PubMed] [Google Scholar]
- 7.Tsai MC, Chakravarty S, Zhu G, Xu J, Tanaka K, Koch C, Tufariello J, Flynn J, Chan J. Characterization of the tuberculous granuloma in murine and human lungs: cellular composition and relative tissue oxygen tension. Cell. Microbiol. 2006;8:218–232. doi: 10.1111/j.1462-5822.2005.00612.x. [DOI] [PubMed] [Google Scholar]
- 8.Ulrichs T, Kosmiadi GA, Trusov V, Jorg S, Pradl L, Titukhina M, Mishenko V, Gushina N, Kaufmann SH. Human tuberculous granulomas induce peripheral lymphoid follicle-like structures to orchestrate local host defence in the lung. J. Pathol. 2004;204:217–228. doi: 10.1002/path.1628. [DOI] [PubMed] [Google Scholar]
- 9.Lin PL, Pawar S, Myers A, Pegu A, Fuhrman C, Reinhart TA, Capuano SV, Klein E, Flynn JL. Early events in Mycobacterium tuberculosis infection in cynomolgus macaques. Infect. Immun. 2006;74:3790–3803. doi: 10.1128/IAI.00064-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Turner OC, Basaraba RJ, Frank AA, Orme IM. Granuloma formation in mouse and guinea pig models of experimental tuberculosis. In: Boros DL, editor. Granulomatous infections and inflammations: cellular and molecular mechanisms. 1st ed. ASM Press; Washington, D.C.: 2003. pp. 65–84. [Google Scholar]
- 11.Barry CE, 3rd, Boshoff HI, Dartois V, Dick T, Ehrt S, Flynn J, Schnappinger D, Wilkinson RJ, Young D. The spectrum of latent tuberculosis: rethinking the biology and intervention strategies. Nat. Rev. Microbiol. 2009;7:845–855. doi: 10.1038/nrmicro2236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kirschner DE, Young D, Flynn JL. Tuberculosis: global approaches to a global disease. Curr. Opin. Biotechnol. 2010;21:524–531. doi: 10.1016/j.copbio.2010.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Flynn JL. Immunology of tuberculosis and implications in vaccine development. Tuberculosis (Edinb) 2004;84:93–101. doi: 10.1016/j.tube.2003.08.010. [DOI] [PubMed] [Google Scholar]
- 14.Cooper AM. Cell-mediated immune responses in tuberculosis. Annu. Rev. Immunol. 2009;27:393–422. doi: 10.1146/annurev.immunol.021908.132703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hanlon AM, Jang S, Salgame P. Signaling from cytokine receptors that affect Th1 responses. Front. Biosci. 2002;7:d1247–54. doi: 10.2741/hanlon. [DOI] [PubMed] [Google Scholar]
- 16.Lauffenburger D, Linderman JJ. Receptors: models for binding, trafficking, and signaling. Oxford University Press; New York: 1993. [Google Scholar]
- 17.Flynn JL, Goldstein MM, Chan J, Triebold KJ, Pfeffer K, Lowenstein CJ, Schreiber R, Mak TW, Bloom BR. Tumor necrosis factor-alpha is required in the protective immune response against Mycobacterium tuberculosis in mice. Immunity. 1995;2:561–572. doi: 10.1016/1074-7613(95)90001-2. [DOI] [PubMed] [Google Scholar]
- 18.Saunders BM, Briscoe H, Britton WJ. T cell-derived tumour necrosis factor is essential, but not sufficient, for protection against Mycobacterium tuberculosis infection. Clin. Exp. Immunol. 2004;137:279–287. doi: 10.1111/j.1365-2249.2004.02518.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Harris J, Hope JC, Keane J. Tumor necrosis factor blockers influence macrophage responses to Mycobacterium tuberculosis. J. Infect. Dis. 2008;198:1842–1850. doi: 10.1086/593174. [DOI] [PubMed] [Google Scholar]
- 20.Gutierrez MG, Mishra BB, Jordao L, Elliott E, Anes E, Griffiths G. NF-kappa B activation controls phagolysosome fusion-mediated killing of mycobacteria by macrophages. J. Immunol. 2008;181:2651–2663. doi: 10.4049/jimmunol.181.4.2651. [DOI] [PubMed] [Google Scholar]
- 21.Mosser DM, Edwards JP. Exploring the full spectrum of macrophage activation. Nat. Rev. Immunol. 2008;8:958–969. doi: 10.1038/nri2448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhou Z, Connell MC, MacEwan DJ. TNFR1-induced NF-kappaB, but not ERK, p38MAPK or JNK activation, mediates TNF-induced ICAM-1 and VCAM-1 expression on endothelial cells. Cell. Signal. 2007;19:1238–1248. doi: 10.1016/j.cellsig.2006.12.013. [DOI] [PubMed] [Google Scholar]
- 23.Algood HM, Lin PL, Yankura D, Jones A, Chan J, Flynn JL. TNF influences chemokine expression of macrophages in vitro and that of CD11b+ cells in vivo during Mycobacterium tuberculosis infection. J. Immunol. 2004;172:6846–6857. doi: 10.4049/jimmunol.172.11.6846. [DOI] [PubMed] [Google Scholar]
- 24.Keane J, Shurtleff B, Kornfeld H. TNF-dependent BALB/c murine macrophage apoptosis following Mycobacterium tuberculosis infection inhibits bacillary growth in an IFN-gamma independent manner. Tuberculosis (Edinb) 2002;82:55–61. doi: 10.1054/tube.2002.0322. [DOI] [PubMed] [Google Scholar]
- 25.Keane J, Balcewicz-Sablinska MK, Remold HG, Chupp GL, Meek BB, Fenton MJ, Kornfeld H. Infection by Mycobacterium tuberculosis promotes human alveolar macrophage apoptosis. Infect. Immun. 1997;65:298–304. doi: 10.1128/iai.65.1.298-304.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chakravarty SD, Zhu G, Tsai MC, Mohan VP, Marino S, Kirschner DE, Huang L, Flynn J, Chan J. Tumor necrosis factor blockade in chronic murine tuberculosis enhances granulomatous inflammation and disorganizes granulomas in the lungs. Infect. Immun. 2008;76:916–926. doi: 10.1128/IAI.01011-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lin PL, Myers A, Smith L, Bigbee C, Bigbee M, Fuhrman C, Grieser H, Chiosea I, Voitenok NN, Capuano SV, Klein E, Flynn JL. Tumor necrosis factor neutralization results in disseminated disease in acute and latent Mycobacterium tuberculosis infection with normal granuloma structure in a cynomolgus macaque model. Arthritis Rheum. 2010;62:340–350. doi: 10.1002/art.27271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Roach DR, Bean AG, Demangel C, France MP, Briscoe H, Britton WJ. TNF regulates chemokine induction essential for cell recruitment, granuloma formation, and clearance of mycobacterial infection. J. Immunol. 2002;168:4620–4627. doi: 10.4049/jimmunol.168.9.4620. [DOI] [PubMed] [Google Scholar]
- 29.Marino S, Sud D, Plessner H, Lin PL, Chan J, Flynn JL, Kirschner DE. Differences in reactivation of tuberculosis induced from anti-TNF treatments are based on bioavailability in granulomatous tissue. PLoS Comput. Biol. 2007;3:1909–1924. doi: 10.1371/journal.pcbi.0030194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ray JC, Flynn JL, Kirschner DE. Synergy between individual TNF-dependent functions determines granuloma performance for controlling Mycobacterium tuberculosis infection. J. Immunol. 2009;182:3706–3717. doi: 10.4049/jimmunol.0802297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fallahi-Sichani M, Schaller MA, Kirschner DE, Kunkel SL, Linderman JJ. Identification of key processes that control tumor necrosis factor availability in a tuberculosis granuloma. PLoS Comput. Biol. 2010;6:e1000778. doi: 10.1371/journal.pcbi.1000778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Warrender C, Forrest S, Koster F. Modeling intercellular interactions in early Mycobacterium infection. Bull. Math. Biol. 2006;68:2233–2261. doi: 10.1007/s11538-006-9103-y. [DOI] [PubMed] [Google Scholar]
- 33.Pedrosa J, Saunders BM, Appelberg R, Orme IM, Silva MT, Cooper AM. Neutrophils play a protective nonphagocytic role in systemic Mycobacterium tuberculosis infection of mice. Infect. Immun. 2000;68:577–583. doi: 10.1128/iai.68.2.577-583.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Seiler P, Aichele P, Bandermann S, Hauser AE, Lu B, Gerard NP, Gerard C, Ehlers S, Mollenkopf HJ, Kaufmann SH. Early granuloma formation after aerosol Mycobacterium tuberculosis infection is regulated by neutrophils via CXCR3-signaling chemokines. Eur. J. Immunol. 2003;33:2676–2686. doi: 10.1002/eji.200323956. [DOI] [PubMed] [Google Scholar]
- 35.Zhang X, Majlessi L, Deriaud E, Leclerc C, Lo-Man R. Coactivation of Syk kinase and MyD88 adaptor protein pathways by bacteria promotes regulatory properties of neutrophils. Immunity. 2009;31:761–771. doi: 10.1016/j.immuni.2009.09.016. [DOI] [PubMed] [Google Scholar]
- 36.Zhu XW, Friedland JS. Multinucleate giant cells and the control of chemokine secretion in response to Mycobacterium tuberculosis. Clin. Immunol. 2006;120:10–20. doi: 10.1016/j.clim.2006.01.009. [DOI] [PubMed] [Google Scholar]
- 37.Lay G, Poquet Y, Salek-Peyron P, Puissegur MP, Botanch C, Bon H, Levillain F, Duteyrat JL, Emile JF, Altare F. Langhans giant cells from M. tuberculosis-induced human granulomas cannot mediate mycobacterial uptake. J. Pathol. 2007;211:76–85. doi: 10.1002/path.2092. [DOI] [PubMed] [Google Scholar]
- 38.Uehira K, Amakawa R, Ito T, Tajima K, Naitoh S, Ozaki Y, Shimizu T, Yamaguchi K, Uemura Y, Kitajima H, Yonezu S, Fukuhara S. Dendritic cells are decreased in blood and accumulated in granuloma in tuberculosis. Clin. Immunol. 2002;105:296–303. doi: 10.1006/clim.2002.5287. [DOI] [PubMed] [Google Scholar]
- 39.Giri PK, Schorey JS. Exosomes derived from M. Bovis BCG infected macrophages activate antigen-specific CD4+ and CD8+ T cells in vitro and in vivo. PLoS One. 2008;3:e2461. doi: 10.1371/journal.pone.0002461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ordway D, Henao-Tamayo M, Orme IM, Gonzalez-Juarrero M. Foamy macrophages within lung granulomas of mice infected with Mycobacterium tuberculosis express molecules characteristic of dendritic cells and antiapoptotic markers of the TNF receptor-associated factor family. J. Immunol. 2005;175:3873–3881. doi: 10.4049/jimmunol.175.6.3873. [DOI] [PubMed] [Google Scholar]
- 41.Peyron P, Vaubourgeix J, Poquet Y, Levillain F, Botanch C, Bardou F, Daffe M, Emile JF, Marchou B, Cardona PJ, de Chastellier C, Altare F. Foamy macrophages from tuberculous patients' granulomas constitute a nutrient-rich reservoir for M. tuberculosis persistence. PLoS Pathog. 2008;4:e1000204. doi: 10.1371/journal.ppat.1000204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Schutze S, Machleidt T, Adam D, Schwandner R, Wiegmann K, Kruse ML, Heinrich M, Wickel M, Kronke M. Inhibition of receptor internalization by monodansylcadaverine selectively blocks p55 tumor necrosis factor receptor death domain signaling. J. Biol. Chem. 1999;274:10203–10212. doi: 10.1074/jbc.274.15.10203. [DOI] [PubMed] [Google Scholar]
- 43.Schneider-Brachert W, Tchikov V, Neumeyer J, Jakob M, Winoto-Morbach S, Held-Feindt J, Heinrich M, Merkel O, Ehrenschwender M, Adam D, Mentlein R, Kabelitz D, Schutze S. Compartmentalization of TNF receptor 1 signaling: internalized TNF receptosomes as death signaling vesicles. Immunity. 2004;21:415–428. doi: 10.1016/j.immuni.2004.08.017. [DOI] [PubMed] [Google Scholar]
- 44.Schutze S, Tchikov V, Schneider-Brachert W. Regulation of TNFR1 and CD95 signalling by receptor compartmentalization. Nat. Rev. Mol. Cell Biol. 2008;9:655–662. doi: 10.1038/nrm2430. [DOI] [PubMed] [Google Scholar]
- 45.Gupta S. A decision between life and death during TNF-alpha-induced signaling. J. Clin. Immunol. 2002;22:185–194. doi: 10.1023/a:1016089607548. [DOI] [PubMed] [Google Scholar]
- 46.Saunders BM, Tran S, Ruuls S, Sedgwick JD, Briscoe H, Britton WJ. Transmembrane TNF is sufficient to initiate cell migration and granuloma formation and provide acute, but not long-term, control of Mycobacterium tuberculosis infection. J. Immunol. 2005;174:4852–4859. doi: 10.4049/jimmunol.174.8.4852. [DOI] [PubMed] [Google Scholar]
- 47.Fremond C, Allie N, Dambuza I, Grivennikov SI, Yeremeev V, Quesniaux VF, Jacobs M, Ryffel B. Membrane TNF confers protection to acute mycobacterial infection. Respir. Res. 2005;6:136. doi: 10.1186/1465-9921-6-136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Olleros ML, Guler R, Vesin D, Parapanov R, Marchal G, Martinez-Soria E, Corazza N, Pache JC, Mueller C, Garcia I. Contribution of transmembrane tumor necrosis factor to host defense against Mycobacterium bovis bacillus Calmetteguerin and Mycobacterium tuberculosis infections. Am. J. Pathol. 2005;166:1109–1120. doi: 10.1016/S0002-9440(10)62331-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tay S, Hughey JJ, Lee TK, Lipniacki T, Quake SR, Covert MW. Single-cell NF-kappaB dynamics reveal digital activation and analogue information processing. Nature. 2010;466:267–271. doi: 10.1038/nature09145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Marino S, Hogue IB, Ray CJ, Kirschner DE. A methodology for performing global uncertainty and sensitivity analysis in systems biology. J. Theor. Biol. 2008;254:178–196. doi: 10.1016/j.jtbi.2008.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Fotin-Mleczek M, Henkler F, Samel D, Reichwein M, Hausser A, Parmryd I, Scheurich P, Schmid JA, Wajant H. Apoptotic crosstalk of TNF receptors: TNF-R2-induces depletion of TRAF2 and IAP proteins and accelerates TNF-R1-dependent activation of caspase-8. J. Cell. Sci. 2002;115:2757–2770. doi: 10.1242/jcs.115.13.2757. [DOI] [PubMed] [Google Scholar]
- 52.Rangamani P, Sirovich L. Survival and apoptotic pathways initiated by TNF-alpha: modeling and predictions. Biotechnol. Bioeng. 2007;97:1216–1229. doi: 10.1002/bit.21307. [DOI] [PubMed] [Google Scholar]
- 53.Blower SM, Dowlatabadi H. Sensitivity and uncertainty analysis of complex models of disease transmission: an HIV model, as an example. Int. Stat. Rev. 1994;62:229–243. [Google Scholar]
- 54.Bold TD, Ernst JD. Who benefits from granulomas, mycobacteria or host? Cell. 2009;136:17–19. doi: 10.1016/j.cell.2008.12.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lazarevic V, Nolt D, Flynn JL. Long-term control of Mycobacterium tuberculosis infection is mediated by dynamic immune responses. J. Immunol. 2005;175:1107–1117. doi: 10.4049/jimmunol.175.2.1107. [DOI] [PubMed] [Google Scholar]
- 56.Lin PL, Rodgers M, Smith L, Bigbee M, Myers A, Bigbee C, Chiosea I, Capuano SV, Fuhrman C, Klein E, Flynn JL. Quantitative comparison of active and latent tuberculosis in the cynomolgus macaque model. Infect. Immun. 2009;77:4631–4642. doi: 10.1128/IAI.00592-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Flynn JL, Chan J. Immunology of tuberculosis. Annu. Rev. Immunol. 2001;19:93–129. doi: 10.1146/annurev.immunol.19.1.93. [DOI] [PubMed] [Google Scholar]
- 58.Sada-Ovalle I, Chiba A, Gonzales A, Brenner MB, Behar SM. Innate invariant NKT cells recognize Mycobacterium tuberculosis-infected macrophages, produce interferon-gamma, and kill intracellular bacteria. PLoS Pathog. 2008;4:e1000239. doi: 10.1371/journal.ppat.1000239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Segovia-Juarez JL, Ganguli S, Kirschner D. Identifying control mechanisms of granuloma formation during M. tuberculosis infection using an agent-based model. J. Theor. Biol. 2004;231:357–376. doi: 10.1016/j.jtbi.2004.06.031. [DOI] [PubMed] [Google Scholar]
- 60.Theus S, Eisenach K, Fomukong N, Silver RF, Cave MD. Beijing family Mycobacterium tuberculosis strains differ in their intracellular growth in THP-1 macrophages. Int. J. Tuberc. Lung Dis. 2007;11:1087–1093. [PubMed] [Google Scholar]
- 61.Lin PL, Plessner HL, Voitenok NN, Flynn JL. Tumor necrosis factor and tuberculosis. J. Investig. Dermatol. Symp. Proc. 2007;12:22–25. doi: 10.1038/sj.jidsymp.5650027. [DOI] [PubMed] [Google Scholar]
- 62.Keane J, Gershon S, Wise RP, Mirabile-Levens E, Kasznica J, Schwieterman WD, Siegel JN, Braun MM. Tuberculosis associated with infliximab, a tumor necrosis factor alpha-neutralizing agent. N. Engl. J. Med. 2001;345:1098–1104. doi: 10.1056/NEJMoa011110. [DOI] [PubMed] [Google Scholar]
- 63.Winthrop KL. Risk and prevention of tuberculosis and other serious opportunistic infections associated with the inhibition of tumor necrosis factor. Nat. Clin. Pract. Rheumatol. 2006;2:602–610. doi: 10.1038/ncprheum0336. [DOI] [PubMed] [Google Scholar]
- 64.Flynn JL. Lessons from experimental Mycobacterium tuberculosis infections. Microbes Infect. 2006;8:1179–1188. doi: 10.1016/j.micinf.2005.10.033. [DOI] [PubMed] [Google Scholar]
- 65.Bean AG, Roach DR, Briscoe H, France MP, Korner H, Sedgwick JD, Britton WJ. Structural deficiencies in granuloma formation in TNF gene-targeted mice underlie the heightened susceptibility to aerosol Mycobacterium tuberculosis infection, which is not compensated for by lymphotoxin. J. Immunol. 1999;162:3504–3511. [PubMed] [Google Scholar]
- 66.Grell M, Wajant H, Zimmermann G, Scheurich P. The type 1 receptor (CD120a) is the high-affinity receptor for soluble tumor necrosis factor. Proc. Natl. Acad. Sci. U. S. A. 1998;95:570–575. doi: 10.1073/pnas.95.2.570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Higuchi M, Aggarwal BB. TNF induces internalization of the p60 receptor and shedding of the p80 receptor. J. Immunol. 1994;152:3550–3558. [PubMed] [Google Scholar]
- 68.Bekker LG, Moreira AL, Bergtold A, Freeman S, Ryffel B, Kaplan G. Immunopathologic effects of tumor necrosis factor alpha in murine mycobacterial infection are dose dependent. Infect. Immun. 2000;68:6954–6961. doi: 10.1128/iai.68.12.6954-6961.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Engele M, Stossel E, Castiglione K, Schwerdtner N, Wagner M, Bolcskei P, Rollinghoff M, Stenger S. Induction of TNF in human alveolar macrophages as a potential evasion mechanism of virulent Mycobacterium tuberculosis. J. Immunol. 2002;168:1328–1337. doi: 10.4049/jimmunol.168.3.1328. [DOI] [PubMed] [Google Scholar]
- 70.Algood HM, Lin PL, Flynn JL. Tumor necrosis factor and chemokine interactions in the formation and maintenance of granulomas in tuberculosis. Clin. Infect. Dis. 2005;41(Suppl 3):S189–93. doi: 10.1086/429994. [DOI] [PubMed] [Google Scholar]
- 71.Clay H, Volkman HE, Ramakrishnan L. Tumor necrosis factor signaling mediates resistance to mycobacteria by inhibiting bacterial growth and macrophage death. Immunity. 2008;29:283–294. doi: 10.1016/j.immuni.2008.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Schneider-Brachert W, Tchikov V, Merkel O, Jakob M, Hallas C, Kruse ML, Groitl P, Lehn A, Hildt E, Held-Feindt J, Dobner T, Kabelitz D, Kronke M, Schutze S. Inhibition of TNF receptor 1 internalization by adenovirus 14.7K as a novel immune escape mechanism. J. Clin. Invest. 2006;116:2901–2913. doi: 10.1172/JCI23771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Neumeyer J, Hallas C, Merkel O, Winoto-Morbach S, Jakob M, Thon L, Adam D, Schneider-Brachert W, Schutze S. TNF-receptor I defective in internalization allows for cell death through activation of neutral sphingomyelinase. Exp. Cell Res. 2006;312:2142–2153. doi: 10.1016/j.yexcr.2006.03.014. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.