Abstract
Plants have evolved a number of adaptive responses to cope with growth in conditions of limited phosphate (Pi) supply involving biochemical, metabolic, and developmental changes. We prepared an EMS-mutagenized M2 population of an Arabidopsis thaliana transgenic line harboring a reporter gene specifically responsive to Pi starvation (AtIPS1∷GUS), and screened for mutants altered in Pi starvation regulation. One of the mutants, phr1 (phosphate starvation response 1), displayed reduced response of AtIPS1∷GUS to Pi starvation, and also had a broad range of Pi starvation responses impaired, including the responsiveness of various other Pi starvation-induced genes and metabolic responses, such as the increase in anthocyanin accumulation. PHR1 was positionally cloned and shown be related to the PHOSPHORUS STARVATION RESPONSE 1 (PSR1) gene from Chlamydomonas reinhardtii. A GFP∷PHR1 protein fusion was localized in the nucleus independently of Pi status, as is the case for PSR1. PHR1 is expressed in Pi sufficient conditions and, in contrast to PSR1, is only weakly responsive to Pi starvation. PHR1, PSR1, and other members of the protein family share a MYB domain and a predicted coiled–coil (CC) domain, defining a subtype within the MYB superfamily, the MYB–CC family. Therefore, PHR1 was found to bind as a dimer to an imperfect palindromic sequence. PHR1-binding sequences are present in the promoter of Pi starvation-responsive structural genes, indicating that this protein acts downstream in the Pi starvation signaling pathway.
Keywords: Arabidopsis thaliana, Chlamydomonas reinhardtii, coiled–coil domain, MYB domain, Pi starvation, transcription factor
Phosphorus is an essential macronutrient for growth and development of living organisms. It is a constituent of key molecules such as ATP, nucleic acids, or phospholipids, and as phosphate, pyrophosphate, ATP, ADP, or AMP, plays a crucial role in energy transfer, metabolic regulation, and protein activation (Marschner 1995). Phosphorus is one of the most limiting nutrients for plants because the form that is preferentially assimilable, phosphate (Pi), is unevenly distributed in soils and >80% is immobile and not readily available to roots (Holford 1997).
Plants have evolved adaptive responses to cope with growth under conditions of limited phosphate availability (for review, see Raghothama 1999). Biochemical and metabolic adaptations involve changes that increase the availability of endogenous and exogenous inorganic phosphate (Pi), including increased secretion of organic acids from roots, the induction of high affinity phosphate transporters, RNases, and phosphatases (Clarkson 1985; Lipton et al. 1987; Goldstein et al. 1988; Krannitz et al. 1991; Theodorou and Plaxton 1993; Bariola et al. 1994; Duff et al. 1994; Green 1994; Muchhal et al. 1996; Smith et al. 1997; C.M. Liu et al. 1998; H. Liu et al. 1998) and changes in thylakoid lipid composition, whereby a decrease in phosphatidylglycerol is accompanied by an increase in sulpholipids (Essigmann et al. 1998). Additional adaptations involve alterations in the rate of photosynthesis and photosynthate partitioning, the accumulation of the light protecting, anthocyanin pigments, and the utilization of alternative glycolytic or respiration pathways (Duff et al. 1989). These alternative glycolytic and respiratory pathways circumvent steps requiring phosphate or adenylate, contributing to the plant survival during prolonged periods of phosphate deprivation (Duff et al. 1989).
Developmental responses involve changes in root growth and architecture that enhance the exploitation of soil phosphate resources and include increases in root/shoot ratio, root hair proliferation and length, and lateral root number (Bates and Lynch 1996). Some plants further modify the soil scavenging potential of their roots by forming lateral proliferations (proteoids) or establishing symbiotic associations with mycorrhizal fungi (for review, see Harrison 1999; Watt and Evans 1999).
Several genes responsive to Pi starvation have been isolated recently from vascular plants (for review, see Raghothama 1999), and encode high-affinity Pi transporters, acid phosphatases, and RNases, among other proteins (Bariola et al. 1994; Muchhal et al. 1996; C.M. Liu et al. 1998; H. Liu et al. 1998; del Pozo et al. 1999; Haran et al. 2000). Members of the Mt4/TPSI1 gene family are also characterized by their highly specific responsiveness to Pi starvation. These genes encode RNAs with short nonconserved reading frames (Burleigh and Harrison 1997, 1999; Liu et al. 1997; Martín et al. 2000). The existence of various genes responding to Pi starvation suggests that plants are also endowed with a phosphate starvation regulon, as is the case for yeast and Escherichia coli (for reviews, see Torriani 1990; Lenburg and O'Shea 1996). In contrast to the situation with these microorganisms, very little is known about the mechanisms governing responses to phosphate starvation in vascular plants. Mutants of Arabidopsis thaliana have been isolated that are affected in phosphate accumulation, such as pho1 or pho2, or an acid phosphatase activity (Poirier et al. 1991; Delhaize and Randall 1995; Trull and Deikman 1998), but the structural or regulatory roles of the genes are not known. In addition, several Pi starvation response mutants have been identified recently by use of an elegant conditional genetic screen (Chen et al. 2000), but the corresponding genes have not yet been identified. One regulatory gene of the Pi starvation response has been identified and cloned from the unicellular algae Chlamydomonas reinhardtii, and shown to encode a member of the MYB transcription factor superfamily (Wykoff et al. 1999).
We have taken advantage of the availability of transgenic A. thaliana plants harboring a reporter gene specifically induced by Pi starvation (AtIPS1∷GUS; Martín et al. 2000) to initiate the molecular genetic dissection of Pi starvation signaling. We report on the identification and characterization of a phosphate starvation response mutant, phr1, which is impaired in various aspects of the response, such as the induction of Pi starvation-responsive genes and anthocyanin synthesis. We show that PHR1 encodes a transcription factor related to the PHOSPHORUS STARVATION RESPONSE 1 (PSR1) protein from C. reinhardtii, suggesting that the increase in complexity of the Pi starvation response during the evolution of multicellular vascular plants was achieved, at least in part, via recruitment of new functions under the control of a MYB-based regulatory system pre-existing in unicellular photosynthetic ancestors. We also show that PHR1 binds as a dimer to sequences present in the promoter of Pi starvation-responsive genes, in line with the presence in this protein of a coiled–coil domain shared with PSR1 and other members of the MYB–CC family.
Results
Isolation of the phosphate starvation response mutant phr1
The AtIPS1 gene, like other members of the Mt4/TPSI1 family, is specifically responsive to Pi starvation. A translational fusion between AtIPS1 and the coding region of the GUS gene also displays a specific response to Pi starvation in transgenic A. thaliana plants (Martín et al. 2000); transgenic plants harboring this reporter gene are therefore suitable for identifying mutants with altered Pi starvation responses. M2 seedlings of an EMS-mutagenized population were screened by use of a nondestructive GUS staining assay (Martin et al. 1997). Putative mutants were identified as follows: nine-day-old seedlings (∼25,000) grown in medium lacking Pi were stained with GUS for 6 h, during which time plates were examined every hour. Seedlings showing reduced GUS staining were selected as candidates for further analysis; 17 mutant candidates were selected, and M3 progeny was obtained from 15 of them. After preliminary phenotypic analysis (not shown), phr1-1, which was affected in the expression of several Pi starvation-inducible genes, and which did not accumulate anthocyanin during Pi starvation stress (see following section and Figs. 1 and 2, below), was selected for further analysis.
Figure 1.
Characterization of the phr1 mutant alleles. (A) Histochemical analysis of GUS activity driven by the AtIPS1∷GUS reporter gene in response to phosphate starvation in wild type (wt; left) and in the phr1-1 mutant (right). Scale bar, 1 cm. (B) Histograms of metabolic (anthocyanin and Pi content) and developmental (root/shoot growth ratio and total weight) parameters of the wild-type (wt) and phr1-1 (1-1) and phr1-2 (1-2) mutant alleles, grown under different nutrient regimes; complete medium (+P), Pi starvation (−P), or nitrogen starvation (−N) regimes. (C) Plates containing the wild-type (bottom) and the phr1-1 (left) and phr1-2 (right) mutant alleles grown on different nutrient regimes. Scale bars, 1 cm. (D) Detail showing root hairs of wild type and phr1-1 grown under Pi starvation conditions. Scale bar, 0.5 mm. The analyses were conducted on plants grown in complete medium for 5 d, then transferred to complete medium or to medium lacking Pi or N for 7 d, except in the cases of the Pi and N starvation shown in C, in which the starvation lasted for 12 d. Data represent means of at least six independent measurements. Standard deviations are indicated by bars. Statistically significant differences using the Student's t test between wild-type and phr1 alleles were observed for anthocyanin accumulation, root-to-shoot growth ratio, and total weight for plants grown under Pi starvation conditions (P < 0.01), as well as for total Pi content for plants grown under Pi sufficient conditions (P < 0.02).
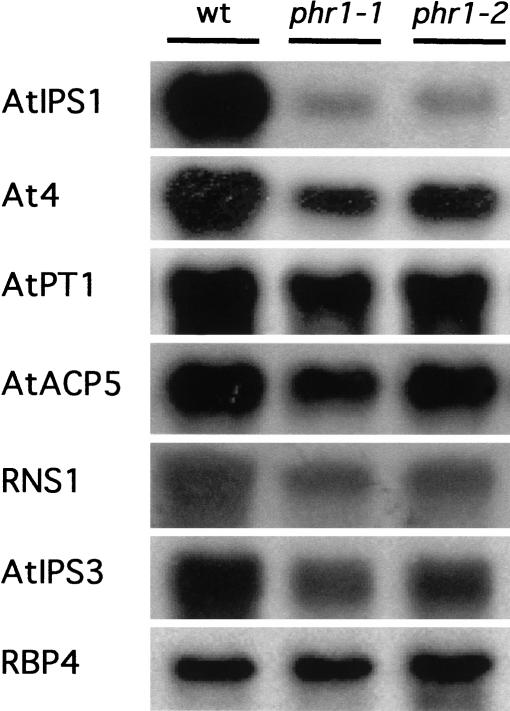
Figure 2.
Northern analysis of the effect of phr1 mutations on the expression of Pi starvation-responsive genes. Wild-type and mutant phr1-1 and phr1-2 alleles were grown for 5 d in complete medium, transferred to medium lacking Pi, and collected at 7 d. Total RNA was isolated and RNA gel blots containing 10 μg of these samples were hybridized to the AtIPS1 probe and subsequently rehybridized to probes corresponding to the related gene as follows: At4 (Burleigh and Harrison 1999); Pi transporter AtPT1 (Muchhal et al. 1996); RNS1 gene (Bariola et al. 1994); type 5 acid phosphatase AtACP5 (del Pozo et al. 1999); AtIPS3 gene, encoding a protein of unknown function (J.C. del Pozo, J. Iglesias, V. Rubio, A. Leyva, and J. Paz-Ares, unpubl.); RBP4 gene (encoding the ribosome binding protein 4; C. Konnz, unpubl.) used as loading control.
The fact that phr1-1 did not accumulate anthocyanin in response to Pi starvation suggested a simple screen for phr1 alleles. After pre-screening 100,000 seedlings grown under Pi starvation conditions for colorless cotyledons, followed by the analysis of GUS activity, we identified a single additional mutant, phr1-2, with reduced GUS staining. Results of crossing experiments (not shown) indicated that both mutants were recessive and allelic. Prior to the phenotypic analysis (detailed in the next section), the phr1-1 allele was backcrossed four times with the wild-type transgenic reporter line.
phr1 mutant alleles are impaired in different Pi starvation responses
In addition to the study of the expression of the AtIPS1∷GUS reporter gene, several metabolic and developmental traits influenced by Pi starvation, as well as the expression of six Pi starvation-responsive genes, were examined in the phr1-1 and phr1-2 mutant alleles (Figs. 1 and 2, below). The phr1 mutations resulted in reduced GUS activity driven by AtIPS1∷GUS in all parts of Pi starved plants (Fig. 1A). In addition, Pi starvation-induced increases in anthocyanin accumulation and, to a lesser, although in a statistically significant extent (P < 0.02), in the root-to-shoot growth ratio, were impaired in the plants homozygous for either of the phr1 alleles (Fig. 1B,C). The effect of phr1 mutations on these two traits was specific for Pi starvation stress, as no significant difference was observed between mutant alleles and wild type in anthocyanin accumulation or on the root/shoot growth ratio under nitrogen starvation conditions (Fig. 1B,C). Moreover, the mutants showed increased anthocyanin synthesis in response to the stress-related hormones abscisic acid and jasmonic acid similar to wild-type plants (not shown). Both mutations resulted in a statistically significant decrease in the Pi content of the plant (P < 0.01) when plants were grown under Pi sufficient conditions, and in a decrease in plant growth under Pi starvation conditions (P < 0.01; Fig. 1B). In contrast, no effect of the mutations of PHR1 was observed for the Pi starvation-induced increases in root hair length and number (Fig. 1D).
The effect of the phr1 mutations on the expression of Pi starvation-induced genes was examined by use of Northern analysis. Six Pi starvation-induced genes were analyzed, including AtIPS1 and At4, members of the Mt4/TPSI1 family (Burleigh and Harrison 1997; Martín et al. 2000); AtPT1, AtACP5, and RNS1, encoding a high-affinity Pi transporter, an acid phosphatase, and a RNase, respectively (Bariola et al. 1994; Muchhal et al. 1996; del Pozo et al. 1999); and AtIPS3, encoding a protein of unknown function (J.C. del Pozo, J. Iglesias, V. Rubio, A. Leyva, and J. Paz-Ares, unpubl.). The Pi starvation inducibility of all six genes examined was reduced in the plants homozygous for either phr1 allele, the effect being most pronounced on AtIPS1, At4, and RNS1 (Fig. 2).
PHR1 encodes a member of the MYB superfamily conserved between A. thaliana and C. reinhardtii
The PHR1 gene was cloned by a map-based chromosome walking procedure on the basis of a cross between the phr1-1 mutant (Columbia ecotype) and the Landsberg wild-type ecotype. By use of 2100 F2 seedlings showing the phr1 phenotype, the PHR1 gene was mapped to chromosome 4 (between markers RPS2 and prha) by a series of simple sequence length polymorphism markers (SSLP; Bell and Ecker 1994) and cleaved, amplified, polymorphic sequences (CAPS; Konieczny and Ausubel 1993) available in databases. The mapping was further refined by use of new SSLP markers generated from sequence information from the A. thaliana genome sequencing project (see Materials and Methods). As a result, the PHR1 locus was defined to a region of ∼120 kb between BACs F20O9 and F16A16 (Fig. 3A). Within this region, candidate genes were considered that showed homology to yeast Pi starvation signaling genes (PHO80, PHO81, PHO85, PHO2, and PHO4) (Bajwa et al. 1984; Legrain et al. 1986; Sengstag and Hinnen 1987; Madden et al. 1988; Gilliquet et al. 1990; Creasy et al. 1993) or to the recently defined PSR1 gene from the unicellular algae C. reinhardtii (Wykoff et al. 1999). Only a single gene (AT4g28610; protein CAB81449.1) in this region showed homology to any of these genes (specifically, to the C. reinhardtii PSR1 gene). Transformation of the phr1-1 mutant with a 5-kb genomic region, spanning the coding region plus sequences 2 kb upstream of the translation initiation codon and 1.3-kb sequences downstream of the termination codon, cloned in the binary vector pBIB (Becker 1990), rescued the wild-type phenotype in 12 of 19 transformants (Fig. 3B).
Figure 3.
Positional cloning and structure of the PHR1 gene. (A) PHR1 was first mapped between CAPS markers RPS2 and phra, and finally narrowed the physical localization between BACs F20O9 and F16A16. Within this region, a homolog to the PSR1 gene from C. reinhardtii (Wykoff et al. 1999) was identified (At4g28610). Sequencing of the region corresponding to the PSR1 homolog in the two alleles, phr1-1 and phr1-2 revealed that each had a mutation in this gene. The mutation in phr1-1 was a C-to-T transition, causing the introduction of a premature stop codon. The mutation in phr1-2 was also a G-to-A substitution, which impaired a GT splicing donor site. Nucleotides in the intron are shown in italics. The exon structure derived from comparison of the genomic and cDNA sequences is highlighted with boxes (empty, noncoding exons, or parts; full, coding exons, or parts). (B) Complementation of the phr1-1 mutant with plasmid pBIB∷PHR1, harboring the PHR1-coding region plus 2 kb upstream and 1.3 kb downstream sequences. T2 progeny of a transgenic plant harboring a copy of the pBIB∷PHR1 T-DNA (middle), displays a 3:1 segregation of the colored phenotype when germinated directly in Pi starvation medium. Control progeny from wild-type and phr1-1 homozygous plants are shown at left and right, respectively. Scale bar, 0.5 cm. (C) Nucleotide and deduced amino acid sequence from the PHR1 cDNA. The two regions conserved between PHR1 and the C. reinhardtii PSR1 protein, corresponding to the MYB domain and to a predicted coiled–coil domain, are highlighted in reverse contrast and gray, respectively. A putative nuclear localization signal is shown (underlined).
To define experimentally the structure of the PHR1 gene, overlapping cDNA fragments were isolated following the Marathon RACE protocol (Clontech) by use of oligonucleotides derived from the gene sequence (gene AT4g28610, EMBL accession no. AL161573; see Materials and Methods). The cDNA sequence (Fig. 3C) showed that the predicted intron/exon structure in gene AT4g28610 is correct except for the sixth exon, which uses an AG acceptor site 39 nucleotides downstream of that predicted (position 1195 instead of 1156, in which the A of the start codon is defined as nucleotide 1). In addition, the first 91 nucleotides of the cDNA sequence shown in Figure 3C correspond to a 5′ untranslated exon. Sequencing of the transcribed region of the two alleles revealed that each contained a point mutation. In the case of the phr1-1 allele, the mutation was a C-to-T transition at nucleotide 663 of the genomic sequence (in which the A of the predicted start codon is defined as nucleotide 1), which results in the replacement of Gln 192 with an ochre stop codon. The phr1-2 mutation is a G-to-A transition at nucleotide 838, which results in the loss of a splice donor site in the third intron, and probably generates a truncated protein.
The presence of a single repeat MYB domain classifies PHR1, as well as PSR1, as members of the MYB superfamily of DNA-binding proteins. A singular characteristic of the PHR1 and PSR1 proteins is that they share a second motif predicted to adopt a coiled–coil conformation (using COILS at www.ch.embnet.org/software/COILS_form.html), which is a potential dimerization motif. Searches in the SPTREMBL or EMBL databanks allowed us to identify 18 proteins (15 in A. thaliana) containing both domains (Fig. 4A). A phylogram of all of these proteins was constructed using the neighbor-joining method (Saitou and Nei 1987) of the CLUSTAL program (Higgins et al. 1996). Two subgroups can be distinguished, with PHR1 and the C. reinhardtii PSR1 protein belonging to the same subgroup (I) (Fig. 4B).
Figure 4.
Sequence comparison among PHR1 and related proteins in databanks. (A) Alignment of the MYB (top) and predicted coiled–coil (bottom) conserved domains constructed by use of the CLUSTAL program (Higgins et al. 1996). The protein accession number given in the SPTREMBL or EMBL databanks is preceded by a species identifier as follows: A. thaliana (At), Nicotiana tabacum (Nt), Mesembryanthemum crystallinum (Mc), and C. reinhardtii (Cr). Arrowhead indicates the position of an insertion of 8 and 16 amino acid residues in the Q9SVP8 and Q9LRN5 sequences, respectively. Alignment was colored according to the average BLOSUM62 score (0.5–1.49, light gray; 1.5–2.9, gray; ≥3.0, black). (B) Phylogram of proteins described in A sharing the MYB and predicted coiled–coil conserved domains, constructed by use of the CLUSTAL (Higgins et al. 1996) program and the neighbor-joining method (Saitou and Nei 1987). The bootstrap (Felsenstein 1992) value of each node is indicated (of 1000 samples). Scale bar, 0.05 substitutions/site. To construct the alignment and the tree, only the two conserved regions were considered.
Sequence-specific binding of PHR1 in the promoter of phosphate starvation-responsive genes
To test the sequence-specific DNA-binding properties of PHR1, we performed electrophoretic mobility shift assays (EMSA) with in vitro-translated PHR1 protein and DNA fragments from the 5′ region (upstream of the first ATG) of AtIPS1, a gene whose Pi starvation induction is severely impaired in phr1-1 and phr1-2 (Fig. 2). Five overlapping fragments encompassing 707 bp upstream of the first ATG were amplified and radiolabeled by PCR and incubated with the in vitro-translated PHR1 protein. A slower migrating band was observed when either of two overlapping fragments (a and b) were incubated with PHR1 (Fig. 5A). To confirm that the shifted band contained PHR1 and to examine which part of the PHR1 protein was responsible for the binding activity observed, several carboxy- or amino-terminally truncated derivatives were generated and subjected to binding and EMSA. As a probe, the 45-bp fragment representing the overlap between fragments a and b (extending from −612 to −568) was used. In all cases in which the retarded band was observed, the mobility shift correlated with the size of the truncated protein, confirming the presence of PHR1 in the protein–DNA complex (Fig 5B). At least 207 amino-terminal amino acids (Δ207Nt) close to the start of the MYB domain could be deleted without compromising DNA binding. In contrast, deletion of 78 amino acids in the carboxy-terminal region abolished DNA binding of the truncated protein. This deletion does not affect the MYB domain, but the second domain conserved between PHR1 and PSR1 from C. reinhardtii, in agreement with the idea that the coiled–coil domain of these proteins is also necessary for correct DNA binding.
Figure 5.
Sequence-specific DNA-binding properties of PHR1. (A) EMSA of in vitro translated PHR1 protein binding to overlapping DNA fragments from the 5′ region of AtIPS1. Mock-translated reticulocyte lysate was used in the indicated lanes. The DNA fragments used in the experiment are represented diagrammatically at top, and span 707 bp upstream of the first ATG of AtIPS1 (a, from −726 to −568; b, from −612 to −440; c, from −484 to −343; d, from −384 to −245; e, from −285 to −130; f, from −152 to −19). Arrows show PHR1-bound or free DNA (B or F, respectively). Other bands detected in the autoradiograph are shared for each fragment between the lanes corresponding to the mock-translated and to the in vitro-translated PHR1 protein, and thus represent interactions between the fragment and proteins of the reticulocyte lysate. (B) EMSA of amino-terminal and carboxy-terminal deletion derivatives binding to the overlapping region between fragments a and b (from −612 to −568). A diagrammatic representation of the PHR1 protein is shown at top. The black and gray boxes represent the MYB and the predicted coiled–coil domains, respectively. Arrows show the amino terminus and the carboxyl terminus of the amino- and carboxy-terminally truncated derivatives, respectively. (C) Scan mutagenesis of the 10 bp sequence containing the PHR1-binding site (P1BS). Wild-type P1BS is shown with the imperfect palindromic repeats indicated by arrows. Base changes in the mutants tested are indicated in the lines below. Broken lines indicate positions with the same base as in the wild-type P1BS. (D) EMSA of PHR1 to the P1BS-related sequence present in the AtIPS3 promoter (see Table 1). (E) PHR1 homodimerization. EMSA was conducted with the full-size PHR1 protein and with the Δ207Nt deletion derivative binding to the sequence used in B. Proteins were translated in vitro, alone, or in combination.
To further delimit the PHR1-binding site, DNA-binding assays and EMSAs were performed by use of three overlapping deletion derivatives of the 45-bp fragment (spanning positions; −612 to −593, −599 to −575, and −586 to −568), As a result, only one (−599 to −575) was bound by PHR1 (not shown). Scanning mutagenesis, consisting of consecutive 4-bp substitutions throughout this 25-bp fragment, was performed, and the substitution derivatives were subjected to binding and EMSA with PHR1 (not shown). As a result, a 10-bp sequence (Fig. 5C) was found to be sufficient for PHR1 binding. This sequence included an imperfectly palindromic sequence GCATATTC. Analysis of the effect of mutations on the 10-bp sequence further highlighted the relevance of the imperfect-palindromic sequence for binding by PHR1. Mutations in the sequence of the imperfect palindrome did not greatly affect binding by PHR1. The same was true for mutations in positions 2 and 7 (C and T, respectively), the positions that do not conform to the rules of a canonical palindromic sequence.
Given that mutations at PHR1 affected the activity of all Pi starvation-induced genes tested, we asked whether they contained the PHR1-binding sequence (GNATATNC, P1BS) in their promoter region. All Pi starvation-induced genes examined contained a P1BS-related sequence (Table 1). The P1BS present in the AtIPS3 gene is bound by PHR1 (Fig. 5D).
Table 1.
Sequences related to the PHR1-binding site found at the upstream region of phosphate starvation-responsive genes from several plant species
| Gene
|
Species
|
Sequence
|
Position
|
Reference
|
|---|---|---|---|---|
| AtlPS1 | Arabidopsis thaliana | GCATATTC | −598 | Martin et al. 2000) |
| AtlPS3 | Arabidopsis thaliana | GAATATGC GAATATGC | −570 −745 | (J.C. del Pozo, J. Iglesia, V. Rubio, A. Leyva, and J. Paz-Ares, unpubl.) |
| AtACP5 | Arabidopsis thaliana | GAATATCC | −290 | (del Pozo et al. 1999) |
| AtPT1 | Arabidopsis thaliana | GTATATCC | −200 | (Muchhal et al. 1996) |
| At4 | Arabidopsis thaliana | GCATATTC GTATATGC | −245 −782 | (Burleigh and Harrison 1999) |
| RNS1 | Arabidopsis thaliana | GTATATAC | −188 | (Bariola et al. 1994) |
| PAP1 | Arabidopsis thaliana | GGATATAC | −149 | (Haran et al. 2000) |
| TPSI1 | Lycopersicum esculentum | GCATATCC | −551 | (Liu et al. 1997) |
| Mt4 | Medicago truncatula | GCATATCC | −230 | (Burleigh and Harrison 1997) |
The position is given for the most 5′-upstream nucleotide with respect to the first ATG in the transcribed region.
The imperfect-palindromic nature of the PHR1 binding sequence (P1BS) raised the possibility that PHR1 would bind its target as a dimer. To address this question, the strategy of Hope and Struhl (1987) was followed. The full-length and the ΔNt207 derivative were translated in vitro, alone or in combination, and the resulting products were tested in DNA binding and EMSA with P1BS. In addition to the band corresponding to the full-length or the deletion derivative, a band of intermediate mobility appeared when the cotranslation products were used (Fig. 5E). This intermediate mobility band corresponds to the mobility expected for a heterodimer, indicating that PHR1 recognizes its target as a dimer.
PHR1 transcription and nuclear localization of a GFP∷PHR1 fusion protein is found independent of Pi status
To evaluate whether control of PHR1 activity occurs pretranslationally, we examined transcript levels in plants grown under different conditions. As shown in Figure 6, PHR1 RNA is detected independently of the Pi status of the plant, but in contrast to PSR1 in C. reinhardtii counterpart, is only moderately responsive to Pi starvation (twofold induction for PHR1 vs. 13-fold induction for PSR1; Wykoff et al. 1999).
Figure 6.
Northern analysis of PHR1 gene expression. A. thaliana plants were grown in complete medium for 5 d, then transferred to medium containing Pi (+P) for 7 d or lacking Pi (−P) for 2 or 7 d. Poly A+-enriched RNA was isolated from these samples and RNA gel blots containing 0.5 μg of these samples were hybridized to the PHR1 probe and subsequently rehybridized to a probe corresponding to the RBP4 gene used as loading control.
A common mechanism to regulate nutrient starvation responses is to control the subcellular localization of a transcription factor (O'Neill et al. 1996; Beck and Hall 1999). To test whether this could be the case for PHR1, we prepared a 35S∷GFP∷PHR1 chimeric gene in which the 35S promoter drives the expression of the coding region of the green fluorescence protein 5 (Siemering et al. 1996) fused to the full-size PHR1 ORF. The vectors containing the 35S∷GFP∷PHR1 fusion gene or the 35S∷GFP gene alone were used to transform wild-type and phr1 mutant plants. Analysis of the phr1 plants transformed with the fusion protein showed that it could phenotypically complement the Pi starvation response defect, indicating that the GFP∷PHR1 fusion protein is functional (data not shown). Microscopic analysis showed that, whereas in the plants harboring the control GFP construct, fluorescence was distributed throughout the cell, in the plants harboring the GFP∷PHR1 construct, fluorescence was found in the nucleus, both in the wild-type and in the mutant phr1 grown under any Pi regimen. This is shown in Figure 7 for the case of wild-type transformed plants, and indicates that the subcellular localization to the nucleus is independent of the Pi status. A similar Pi status-independent nuclear localization was found for the C. reinhardtii PSR1 protein (Wykoff et al. 1999).
Figure 7.
Subcellular localization of a GFP∷PHR1 fusion protein in wild-type plants grown under Pi sufficient and Pi starvation conditions. Microscopic images of root cells from transgenic A. thaliana Col-0 plants harboring a control gene 35S∷GFP (top) or a 35S∷GFP∷PHR1 fusion gene (bottom). Plants were grown in complete medium for 5 d, then transferred to medium containing Pi (+P, left) or lacking Pi (−P, right ) for 7 d before analysis. Scale bar, 20 μm.
Discussion
The mechanisms underlying Pi starvation signaling are well understood in bacteria and yeast (for reviews, see Torriani 1990; Lenburg and O'Shea 1996). However, little is known about this process in vascular plants. In this study, we have isolated and characterized a Pi starvation response mutant and cloned the corresponding gene PHR1, the first gene involved in the control of Pi responses in vascular plants to be characterized at the molecular level.
The phr1 mutations isolated in this work affect a broad spectrum of Pi starvation responses, including some responses shared with other stress responses. For example, they affect anthocyanin accumulation, which is also increased in response to nitrogen starvation and to a large number of environmental stresses, including cold and UV radiation (Dixon and Paiva 1995; Mol et al. 1996). This defect in anthocyanin accumulation in the phr1 mutants is specific for phosphate starvation, however, as no alteration in its accumulation is observed in these mutants in response to nitrogen starvation (Fig. 1B,D) or to the stress-related hormones tested, abscisic and jasmonic acids (data not shown). These observations indicate that responses common to several different stresses may be controlled by signaling pathways specific to each stress type. In addition, they suggest that anthocyanin accumulation is a primary response to Pi starvation, rather than a secondary nonspecific side effect controlled by a common mechanism as suspected previously (Trull et al. 1997), as the phr1 mutant is stressed with respect to Pi when grown under Pi starvation conditions (Fig. 1B).
PHR1 was cloned by chromosome walking assisted by the choice of gene candidates and was shown to be related to the PSR1 gene from C. reinhardtii. This finding further underlines the importance of C. reinhardtii as a model system for photosynthetic eukaryotes. In addition, it shows that evolution of the Pi starvation rescue system in vascular plants involved the recruitment of new responses, such as anthocyanin accumulation, to a conserved regulatory system preexisting in the unicellular ancestors. The PHR1 protein is 409 amino acids in size and contains two domains shared with PSR1, a MYB-related domain, characteristic of DNA-binding proteins, and a predicted coiled–coil domain, potentially involved in protein–protein interactions. In line with the presence of these two domains, DNA-binding assays showed that PHR1 binds to DNA as a dimer in a sequence-specific manner. Thus, the MYB domain is likely to be involved in sequence-specific recognition, and the coiled–coil domain may be the dimerization domain. In agreement with this interpretation, a deletion derivative of PHR1 lacking part of the coiled–coil domain showed impaired high-affinity sequence-specific DNA binding.
The combination of a characteristic MYB and a coiled–coil domain is shared by several other plant proteins (15 in A. thaliana) defining a family that we term the MYB–CC family. Phylogenetic analysis within this family reveals two subgroups, I and II. PHR1 and PSR1 from C. reinhardtii belong to subgroup I, which contains several other members from A. thaliana more closely related to PHR1 than to PSR1. This raises the possibility of functional heterodimeric interactions between MYB–CC proteins through their coiled–coil domains, as well as the possibility of partial redundancy between members of this family. In line with this hypothesis, the two phr1 mutants studied, which probably carry loss-of-function mutations, only showed partial impairment of most of the molecular/physiological responses studied, in contrast to the case of the psr1 mutations in C. reinhardtii.
Scanning mutagenesis of the DNA target site for PHR1 allowed us to determine the sequence requirements for interaction. PHR1 binds to an imperfect palindrome of 8 bp. Interestingly, this sequence is present in all Pi starvation-induced genes examined except PHR1 itself, including those acting in the scavenging, mobilization, and uptake of Pi. We conclude that PHR1 (or its highly related counterparts) acts downstream in the Pi starvation signal-transduction pathway, and does not appear to be self regulated transcriptionally.
It remains to be shown whether other members of the MYB–CC family control other aspects of the Pi starvation response. It is well known that there are two types of response to Pi deprivation in plants, one which is systemically controlled by whole-plant Pi status and the other governed by local Pi status (Drew 1975; Drew and Saker 1984; Burleigh and Harrison 1999). Notably, of the various responses studied, the only ones that were unaffected in the phr1 alleles were root hair length and number, which are the only ones known to be controlled by local Pi status (Bates and Lynch 1996).
Expression of PHR1, like that of PSR1, is detected independent of the Pi status. In both cases, there is increased RNA accumulation in response to Pi starvation, although to a lesser extent in the case of PHR1. Similarly, both PHR1 and PSR1 proteins are localized in the nucleus under any Pi regime. The presumed presence of PHR1 in the nucleus of plants grown in Pi sufficient conditions raises the questions as to its role under these conditions and to its regulation. In this regard, it is noticeable that the Pi content of the mutant is lower than that of the wild type when grown under a Pi sufficient regime, suggesting that PHR1 participates in the control of the plant Pi status under any Pi regime. On the other hand, PHR1 is either post-translationally modified or the protein is a constitutively active, specific component of Pi starvation signaling and a second component of the signaling cascade responds to the signal.
The relevance of the Pi starvation rescue system in the context of phosphorus nutrition efficiency is indicated by the fact that the phr1 mutants display reduced growth under Pi starvation conditions (Fig. 1B). The identification and cloning of PHR1 provides a potentially useful tool toward engineering plants that require less phosphate fertilizer.
Materials and methods
Strains and growth conditions
All A. thaliana plants used in this study, including mutants and transgenic plants, were on the Columbia (Col-0) or the Landsberg erecta background. Plants were grown in complete medium (+P) as described by Bates and Lynch (1996), using one-strength nutrient salts (Johnson et al. 1957). In the Pi deficient medium (−P), KH2PO4 was replaced by equimolar amounts of KCl2. In nitrogen deficient medium (−N), Ca(NO3)2 and KNO3 were replaced by equimolar amounts of CaCl2 and K2SO4, respectively. Growth chamber conditions were 22°C, 60% humidity, and a 16-h light/8-h dark photoperiod with 100 μE/m2 per sec of white light.
Isolation of mutants
A. thaliana seeds (50,000) of a homozygous line harboring the phosphate starvation responsive AtIPS1∷GUS reporter gene (Martín et al. 2000) were mutagenized with ethyl EMS by treating hydrated seeds (soaked overnight in distilled water) with 0.3% EMS for 13 h, then sowed directly onto soil in 56 pots. Seeds were harvested separately as M1 families. M2 seeds (450) from each M1 family were plated directly on low-phosphorus medium and, after 9 d, screened for GUS expression using a nondestructive assay (Martin et al. 1997). Putative mutants showing impaired phosphate starvation response were recovered on fresh complete medium and transferred to soil. M3 seeds were retested for inheritance of the observed phenotype.
A search for phr1-1 alleles was performed by taking advantage of the colorless phenotype of phr1 plants grown under Pi starvation conditions. M2 seedlings (100,000) were grown under Pi starvation conditions for 12 d, and plants showing colorless cotyledons were subsequently subjected to analysis of GUS activity, and the one showing reduced GUS activity was selected. Prior to the phenotypic analysis, the phr1-1 allele was backcrossed four times with the wild-type transgenic reporter line.
Physiological measurements
Anthocyanin was extracted from rosettes of plants grown on complete medium (+P) for 5 d, then transferred to +P, −P, or −N medium for 7 d. Anthocyanin content was measured as described previously (Swain and Hillis 1959). The method of Ames (1966) was used to determine the cellular phosphorus content of seedlings grown on complete medium (+P) for 5 d, and then transferred to +P or −P medium for 7 d. Mean values were compared by use of a Student's t test.
Molecular procedures
Routine molecular work was performed as described previously, except where indicated (Sambrook et al. 1989; del Pozo et al. 1999). Genomic DNA was isolated as described by Doyle and Doyle (1990). A full-length cDNA of PHR1 was isolated by use of the Marathon RACE Kit as described by the manufacturer (Clontech), Expand High Fidelity polymerase (Boehringer), the AP1 primer (from the Marathon RACE Kit) and the PHR1 primers 5′-GATTCGTCTTCCTTGGTACCGGATTGCTTC-3′ and 5′-GGGATTGAAGAAGTGCGTGTGAGG-3′. These PCR fragments were cloned into SmaI-digested pBluescript SK plasmids (Stratagene) and sequenced.
Genetic analysis and positional cloning of PHR1
phr1-1 mutant plants were backcrossed four times to wild-type plants (Col-0 ecotype) to test the linkage of the different phenotypes observed to a single recessive mutation. For mapping purposes, phr1-1 plants (Col-0 ecotype) were crossed to wild-type plants of the Landsberg erecta ecotype (Ler). Twelve-day-old F2 seedlings that displayed the anthocyaninless phenotype under phosphorus starvation were isolated. DNA from these plants was prepared and used to analyze linkage of the phr1-1 mutation to previously described SSLP (Bell and Ecker 1994) and CAPS (Konieczny and Ausubel 1993). PHR1 was mapped to chromosome 4 between RPS2 and prha CAPS markers. To identify the mutant gene, we generated molecular markers (Table 2).
Table 2.
New markers generated for positional cloning of PHR1
| Markera
|
Size Col (bp)/Ler
|
Primers
|
|---|---|---|
| T15N24 | Col (177) > Ler | 5′-GAAGTTTCCACAGACGAGG-3′ 5′-CAGCTAAGGGTTCTGCATCC-3′ |
| F27G19 | Col (179) > Ler | 5′-CCCACTTCAACACAATTCTTCC-3′ 5′-CAATTGCAGCTACAGATCGC-3′ |
| F20O9 | Col (135) < Ler | 5′-CTCATAATGCATTGAACCC-3′ 5′-GTTACAATACCATGCACG-3′ |
| F16A16 | Col (167) > Ler | 5′-GCATATCAAACTCAGAGG-3′ 5′-CACATATCTAAATACAGACC-3′ |
| F19B15 | Col (181) > Ler | 5′-GTATGTTTTTCAATGTATTTGG-3′ 5′-GACAACTACTTTTGTGGATG-3′ |
All were named for the BAC from which they are derived.
Plant transformation
Two types of constructs were used to transform A. thaliana plants by use of the vacuum infiltration method (Bechtold et al. 1993). One corresponded to the genomic DNA containing the PHR1 gene, which was used to complement the mutant phr1-1. The genomic fragment was obtained by PCR amplification from wild-type plants using as primers: 5′-CAACGAAGATTAC GAAGCTCGAAAGTACG-3′ (positions −2063 to −2035 relative to the start of translation) and 5′-CATCGAAGGCT GAGATACTGCTGGAGGTCGG-3′ (positions 2580 to 2550). The PCR fragment was digested with HindIII and cloned in the binary vector pBIB plasmid (Becker 1990), which confers hygromycin resistance in planta.
The second construct corresponded to a GFP∷PHR1 translational fusion used to examine the subcellular localization of PHR1. To do this, a PHR1 fragment of 1327 bp was amplified by PCR using the PHR1 primers 5′-AAAAAAAGATCTATTC GTCTTCCTTGGTCCTGGATTGC-3′ and 5′- GAAGAACCT CAAGATAAGAGCTCG-3′. This fragment was digested with BglII and fused translationally to the 3′ end of the GFP ORF contained in the pAVA393 plasmid, which encodes GFP version 5 (Siemering et al. 1996). This construct and the pAVA393 (as a positive control) were used to transform both the wild type and the phr1 mutant.
Visualization of GFP and derivatives
GFP fluorescence was visualized by use of a Leica DM-R microscope (Leica Microsystem, Wetzlar GmBH, Germany). GFP excitation was performed with standard FITC filters. Images of roots were taken through FITC filters with an Apogee KX85 CCD camera (Apogee Instruments), splitting the emission signal into two channels, one for GFP emission (green channel) and one for autofluorescence (red channel, not shown). To visualize nuclei, roots were submerged in a DAPI solution (1 μg/mL DAPI in 100 mM phosphate buffer, 0.5% Triton X-100) for 1 h, and the nuclear-specific dye DAPI was visualized with light microscopy to certify the identity of nuclei (data not shown).
Protein synthesis, DNA-binding reactions, and EMSA
Full-length PHR1 and the PHR1 deletion derivatives were generated by in vitro translation (or cotranslation in the dimerization experiments) using the flexi-rabbit reticulocyte system (Promega) as described (Solano et al. 1997). PCR and labeling of promoter fragments and oligonucleotides, DNA-binding reactions, and EMSA were performed as described (Solano et al. 1995). The 45-bp promoter fragment of the AtIPS1 gene was obtained by PCR amplification using the following primers: 5′-CAATTTTGGTAACGCGCATATTCC-3′ and 5′-GAGAATT TTGGATCACCGATG-3′.
The deletion derivatives of the 45-bp promoter fragment were obtained by annealing and end filling by use of the Klenow fragment of DNA polymerase I and the following sets of two overlapping primers: AtIPS1AF, 5′-CAATTTTGGTAACGCG CATA-3′; AtIPS1AR, 5′-TATGCGCGTTACCAAAATTG-3′; AtIPS1BF, 5′-GCGCATATTCCATCGGATGA-3′; AtIPS1BR, 5′-TTGGATCATCCGATGGA-3′; AtIPS1CF, 5′-CGGATGAT CCAAAATTCTC-3′; AtIPS1CR, 5′-GAGAATTTTGGATCAT CCG-3′.
Mutant versions of the phosphate starvation response box were obtained by consecutive 4-bp substitutions or by point mutation of the AtIPS1B primers.
The promoter fragment of the AtIPS3 gene was obtained by end filling the following overlapping primers: AtIPS3F, 5′-CCAAATATGGGCTAAGACCAACG-3′; AtIPS3R, 5′-CACA TTTCATAAGAATGAATATGC-3′.
Computer programs for protein and nucleic acid analysis
Databank searches (Swissprot, EMBL, and NCBI) were performed by use of the FASTA and BLAST programs (Pearson and Lipman 1988; Altschul et al. 1990). Alignment, tree construction by the neighbor-joining method, and its bootstrapping (1000 samples) were performed as described previously (Saitou and Nei 1987; Felsenstein 1992; Higgins et al. 1996).
Accession number of PHR1 cDNA sequence
The PHR1 cDNA sequence has been deposited in the EMBL databank under accession number AJ310799.
Acknowledgments
We thank Professors Cathie Martin and Francesco Salamini for critical reading of the manuscript, and Catherine Mark for editorial assistance. We also thank Luis Sanchez Pulido for help with the tree construction. The excellent technical assistance of Maria Jesus Benito is also acknowledged. A.C.M. was recipient of a postdoctoral fellowship from the Comunidad de Madrid. The research was funded by the EU (programs PCP and REGIA, contract numbers BIO4-CT96-0770 and QLG-CT1999-00876) and the Spanish CICYT (contract number BIO99-0229).
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL jpazares@cnb.uam.es; FAX 34-91-585-4506.
Article and publication are at http://www.genesdev.org/cgi/doi/10.1101/gad.204401.
References
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Ames BN. Assay of inorganic phosphate, total phosphate and phosphatases. Methods Enzymol. 1966;8:115–118. [Google Scholar]
- Bajwa W, Meyhack B, Rudolph H, Schweingruber AM, Hinnen A. Structural analysis of the two tandemly repeated acid phosphatase genes in yeast. Nucleic Acids Res. 1984;12:7721–7739. doi: 10.1093/nar/12.20.7721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bariola PA, Howard CJ, Taylor CB, Verburg MT, Jaglan VD, Green PJ. The Arabidopsis ribonuclease gene RSN1 is tightly controlled in response to phosphate limitation. Plant J. 1994;6:673–685. doi: 10.1046/j.1365-313x.1994.6050673.x. [DOI] [PubMed] [Google Scholar]
- Bates TR, Lynch JP. Stimulation of root hair elongation in Arabidopsis thaliana by low phosphorus availability. Plant Cell Environ. 1996;19:529–538. [Google Scholar]
- Bechtold N, Ellis J, Pelletier G. In Planta Agrobacterium mediated gene transfer by infiltration of adult Arabidopsis thaliana plants. C R Acad Sci Paris Life Sci. 1993;316:15–18. [Google Scholar]
- Beck T, Hall MN. The TOR signalling pathway controls nuclear localization of nutrient-regulated transcription factors. Nature. 1999;402:689–692. doi: 10.1038/45287. [DOI] [PubMed] [Google Scholar]
- Becker D. Binary vectors which allow the exchange of plant selectable markers and reporter genes. Nucleic Acids Res. 1990;18:203. doi: 10.1093/nar/18.1.203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell CJ, Ecker JR. Assignment of 30 microsatellite loci to the linkage map of Arabidopsis. Genomics. 1994;19:137–144. doi: 10.1006/geno.1994.1023. [DOI] [PubMed] [Google Scholar]
- Burleigh SH, Harrison MJ. A novel gene whose expression in Medicago truncatula roots is suppressed in response to colonization by vesicular-arbuscular mycorrhizal (VAM) fungi and to phosphate nutrition. Plant Mol Biol. 1997;34:199–208. doi: 10.1023/a:1005841119665. [DOI] [PubMed] [Google Scholar]
- ————— The down-regulation of Mt4-like genes by phosphate fertilization occurs systemically and involves phosphate translocation to the shoots. Plant Physiol. 1999;119:241–248. doi: 10.1104/pp.119.1.241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen DL, Delatorre CA, Bakker A, Abel S. Conditional identification of phosphate-starvation-response mutants in Arabidopsis thaliana. Planta. 2000;211:13–22. doi: 10.1007/s004250000271. [DOI] [PubMed] [Google Scholar]
- Clarkson DT. Factors affecting mineral nutrient acquisition by plants. Annu Rev Plant Physiol. 1985;36:77–115. [Google Scholar]
- Creasy CL, Madden SL, Bergman LW. Molecular analysis of the PHO81 gene of Saccharomyces cerevisiae. Nucleic Acids Res. 1993;21:1975–1982. doi: 10.1093/nar/21.8.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- del Pozo JC, Allona I, Rubio V, Leyva A, de la Peña A, Aragoncillo C, Paz-Ares J. A type 5 acid phosphatase gene from Arabidopsis thaliana is induced by phosphate starvation and by some other types of phosphate mobilising/oxidative stress conditions. Plant J. 1999;19:579–589. doi: 10.1046/j.1365-313x.1999.00562.x. [DOI] [PubMed] [Google Scholar]
- Delhaize E, Randall PJ. Characterization of a phosphate-accumulator mutant of Arabidopsis thaliana. Plant Physiol. 1995;107:207–213. doi: 10.1104/pp.107.1.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixon RA, Paiva NL. Stress-induced phenylpropanoid metabolism. Plant Cell. 1995;7:1085–1097. doi: 10.1105/tpc.7.7.1085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doyle JJ, Doyle JL. Isolation of plant DNA from fresh tissue. Focus. 1990;12:13–15. [Google Scholar]
- Drew MC. Comparison of the effects of a localised supply of phosphate, nitrate, ammonium and potassium. New Phytol. 1975;75:479–490. [Google Scholar]
- Drew MC, Saker LR. Uptake and long-distance transport of phosphate, potassium and chloride in relation to internal ion concentrations in barley: Evidence of non-allosteric regulation. Planta. 1984;160:500–507. doi: 10.1007/BF00411137. [DOI] [PubMed] [Google Scholar]
- Duff SMG, Moorhead GBG, Lefebvre DD, Plaxton WC. Phoshate starvation inducible “bypasses” of adenylate and phosphate dependent glycolytic enzymes in Brassica nigra suspension cells. Plant Physiol. 1989;90:1275–1278. doi: 10.1104/pp.90.4.1275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duff SMG, Gautam S, Plaxton WC. The role of acid phosphatases in plant phosphorus metabolism. Physiol Plant. 1994;90:791–800. [Google Scholar]
- Essigmann B, Guler S, Narang RA, Linke D, Benning C. Phosphate availability affects the thylakoid lipid composition and the expression of SQD1, a gene required for sulfolipid biosynthesis in Arabidopsis thaliana. Proc Natl Acad Sci. 1998;95:1950–1955. doi: 10.1073/pnas.95.4.1950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felsenstein J. Estimating effective population size from samples of sequences: A bootstrap Monte Carlo integration method. Genet Res. 1992;60:209–220. doi: 10.1017/s0016672300030962. [DOI] [PubMed] [Google Scholar]
- Gilliquet V, Legrain M, Berben G, Hilger F. Negative regulatory elements of the Saccharomyces cerevisiae PHO system: Interaction between PHO80 and PHO85 proteins. Gene. 1990;96:181–188. doi: 10.1016/0378-1119(90)90251-l. [DOI] [PubMed] [Google Scholar]
- Goldstein AH, Baertlein DA, McDaniel RG. Phosphate starvation inducible metabolism in Licopersicum esculentum I. Excretion of acid phosphatase by tomato plants and suspension-cultured cells. Plant Physiol. 1988;87:711–715. doi: 10.1104/pp.87.3.711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green PJ. The ribonuclease of higher plants. Annu Rev Plant Physiol Plant Mol Biol. 1994;45:421–445. [Google Scholar]
- Haran S, Logendra S, Seskar M, Bratanova M, Raskin I. Characterization of Arabidopsis acid phosphatase promoter and regulation of acid phosphatase expression. Plant Physiol. 2000;124:615–626. doi: 10.1104/pp.124.2.615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison MJ. Molecular and cellular aspects of the arbuscular mycorrhizal symbiosis. Annu Rev Plant Physiol Plant Mol Biol. 1999;50:361–389. doi: 10.1146/annurev.arplant.50.1.361. [DOI] [PubMed] [Google Scholar]
- Higgins DG, Thompson JD, Gibson TJ. Using CLUSTAL for multiple sequence alignments. Methods Enzymol. 1996;266:383–402. doi: 10.1016/s0076-6879(96)66024-8. [DOI] [PubMed] [Google Scholar]
- Holford ICR. Soil phosphorus: Its measurement, and its uptake by plants. Aust J Soil Res. 1997;35:227–239. [Google Scholar]
- Hope IA, Struhl K. GCN4, a eukaryotic transcriptional activator protein, binds as a dimer to target DNA. EMBO J. 1987;6:2781–2784. doi: 10.1002/j.1460-2075.1987.tb02573.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson CM, Stout PR, Broyer TC, Carlton AB. Comparative chlorine requirements of different plants species. Plant Soil. 1957;8:337–353. [Google Scholar]
- Konieczny A, Ausubel FM. A procedure for mapping Arabidopsis mutations using co-dominant ecotype-specific PCR-based markers. Plant J. 1993;4:403–410. doi: 10.1046/j.1365-313x.1993.04020403.x. [DOI] [PubMed] [Google Scholar]
- Krannitz PG, Aarssen LW, Lefebvre DD. Relationships between physiological and morphological attributes related to phosphate uptake in 25 genotypes of Arabidopsis thaliana. Plant Soil. 1991;133:169–175. [Google Scholar]
- Legrain M, De Wilde M, Hilger F. Isolation, physical characterization and expression analysis of the Saccharomyces cerevisiae positive regulatory gene PHO4. Nucleic Acids Res. 1986;14:3059–3073. doi: 10.1093/nar/14.7.3059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lenburg ME, O'Shea EK. Signaling phosphate starvation. Trends Biochem Sci. 1996;21:383–387. [PubMed] [Google Scholar]
- Lipton DS, Blanchar RW, Blevins DG. Citrate, malate and succinate concentration in exudates from P-sufficient and P-stressed Medicago sativa L. seedlings. Plant Physiol. 1987;85:315–317. doi: 10.1104/pp.85.2.315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu CM, Muchhal US, Raghothama KG. Differential expression of TPS11, a phosphate starvation-induced gene in tomato. Plant Mol Biol. 1997;33:867–874. doi: 10.1023/a:1005729309569. [DOI] [PubMed] [Google Scholar]
- Liu CM, Muchhal US, Mukatira U, Kononowicz AK, Raghothama KG. Tomato phosphate transporter genes are differentially regulated in plant tissues by phosphorus. Plant Physiol. 1998;116:91–99. doi: 10.1104/pp.116.1.91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H, Trieu AT, Blaylock LA, Harrison MJ. Cloning and characterization of two phosphate transporters from Medicago truncatula roots: Regulation in response to phosphate and to colonization by arbuscular mycorrhizal (AM) fungi. Mol Plant Microbe Interaction. 1998;11:14–22. doi: 10.1094/MPMI.1998.11.1.14. [DOI] [PubMed] [Google Scholar]
- Madden SL, Creasy CL, Srinivas V, Fawcett W, Bergman LW. Structure and expression of the PHO80 gene of Saccharomyces cerevisiae. Nucleic Acids Res. 1988;16:2625–2637. doi: 10.1093/nar/16.6.2625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marschner H. Mineral nutrition of higher plants. San Diego, CA: Academic press; 1995. [Google Scholar]
- Martín AC, del Pozo JC, Iglesias J, Rubio V, Solano R, De La Peña A, Leyva A, Paz-Ares J. Influence of cytokinins on the expression of phosphate starvation responsive genes in Arabidopsis. Plant J. 2000;24:1–11. doi: 10.1046/j.1365-313x.2000.00893.x. [DOI] [PubMed] [Google Scholar]
- Martin T, Hellmann H, Schmidt R, Willmitzer L, Frommer WB. Identification of mutants in metabolically regulated gene expression. Plant J. 1997;11:53–62. doi: 10.1046/j.1365-313x.1997.11010053.x. [DOI] [PubMed] [Google Scholar]
- Mol J, Jenkins G, Schäfer E, Weiss D. Signal perception, transduction, and gene expression involved in anthocyanin biosynthesis. Crit Rev Plant Sci. 1996;15:525–557. [Google Scholar]
- Muchhal US, Pardo JM, Raghathama KG. Phosphate transporter from higher plant Arabidopsis thaliana. Proc Natl Acad Sci. 1996;93:10519–10523. doi: 10.1073/pnas.93.19.10519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Neill EM, Kaffman A, Jolly ER, O'Shea EK. Regulation of PHO4 nuclear localization by the PHO80–PHO85 cyclin–CDK complex. Science. 1996;271:209–212. doi: 10.1126/science.271.5246.209. [DOI] [PubMed] [Google Scholar]
- Pearson WR, Lipman DJ. Improved tools for biological sequence comparison. Proc Natl Acad Sci. 1988;85:2444–2448. doi: 10.1073/pnas.85.8.2444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poirier Y, Thoma S, Somerville C, Schiefelbein J. A mutant of Arabidopsis deficient in xylem loading of phosphate. Plant Physiol. 1991;97:1087–1093. doi: 10.1104/pp.97.3.1087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raghothama KG. Phosphate acquisition. Annu Rev Plant Physiol. 1999;50:665–693. doi: 10.1146/annurev.arplant.50.1.665. [DOI] [PubMed] [Google Scholar]
- Saitou N, Nei M. The neighbor-joining method: A new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T. Molecular cloning: A Laboratory Manual. 2nd ed. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- Sengstag C, Hinnen A. The sequence of the Saccharomyces cerevisiae gene PHO2 codes for a regulatory protein with unusual amino acid composition. Nucleic Acids Res. 1987;15:233–246. doi: 10.1093/nar/15.1.233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siemering KR, Golbik R, Sever R, Haseloff J. Mutations that suppress the thermosensitivity of green fluorescent protein. Curr Biol. 1996;6:1653–1663. doi: 10.1016/s0960-9822(02)70789-6. [DOI] [PubMed] [Google Scholar]
- Smith FW, Ealing PM, Dong B, Delhaize E. The cloning of two Arabidopsis genes belonging to a phosphate transporter family. Plant J. 1997;11:83–92. doi: 10.1046/j.1365-313x.1997.11010083.x. [DOI] [PubMed] [Google Scholar]
- Solano R, Nieto C, Avila J, Canas L, Diaz I, Paz-Ares J. Dual DNA binding specificity of a petal epidermis-specific MYB transcription factor (MYB.Ph3) from Petunia hybrida. EMBO J. 1995;14:1773–1784. doi: 10.1002/j.1460-2075.1995.tb07166.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solano R, Fuertes A, Sanchez-Pulido L, Valencia A, Paz-Ares J. A single residue substitution causes a switch from the dual DNA binding specificity of plant transcription factor MYB.Ph3 to the animal c-MYB specificity. J Biol Chem. 1997;272:2889–2895. doi: 10.1074/jbc.272.5.2889. [DOI] [PubMed] [Google Scholar]
- Swain T, Hillis HE. Phenolic constituents of Prunus domestica. I. Quantitative analysis of phenolic constituents. J Sci Food Agr. 1959;10:63–68. [Google Scholar]
- Theodorou ME, Plaxton WC. Metabolic adaptations of plant respiration to nutricional phosphate deprivation. Plant Physiol. 1993;101:339–344. doi: 10.1104/pp.101.2.339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torriani A. From cell membrane to nucleotides: The phosphate regulon in Escherichia coli. BioEssays. 1990;12:371–376. doi: 10.1002/bies.950120804. [DOI] [PubMed] [Google Scholar]
- Trull MC, Deikman J. An Arabidopsis mutant missing one acid phosphatase isoform. Planta. 1998;206:544–550. doi: 10.1007/s004250050431. [DOI] [PubMed] [Google Scholar]
- Trull MC, Guiltinan MJ, Lynch JP, Deikman J. The responses of wilde-type and ABA mutant Arabidopsis thaliana plants to phosphorus starvation. Plant, Cell & Environ. 1997;20:85–92. [Google Scholar]
- Watt M, Evans JR. Proteoid roots. Physiology and development . Plant Physiol. 1999;121:317–324. doi: 10.1104/pp.121.2.317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wykoff DD, Grossman AR, Weeks DP, Usuda H, Shimogawara K. Psr1, a nuclear localized protein that regulates phosphorus metabolism in Chlamydomonas. Proc Natl Acad Sci. 1999;96:15336–15341. doi: 10.1073/pnas.96.26.15336. [DOI] [PMC free article] [PubMed] [Google Scholar]