Abstract
OBJECTIVE: To create a cohort for cost-effective genetic research, the Mayo Genome Consortia (MayoGC) has been assembled with participants from research studies across Mayo Clinic with high-throughput genetic data and electronic medical record (EMR) data for phenotype extraction.
PARTICIPANTS AND METHODS: Eligible participants include those who gave general research consent in the contributing studies to share high-throughput genotyping data with other investigators. Herein, we describe the design of the MayoGC, including the current participating cohorts, expansion efforts, data processing, and study management and organization. A genome-wide association study to identify genetic variants associated with total bilirubin levels was conducted to test the genetic research capability of the MayoGC.
RESULTS: Genome-wide significant results were observed on 2q37 (top single nucleotide polymorphism, rs4148325; P=5.0 × 10–62) and 12p12 (top single nucleotide polymorphism, rs4363657; P=5.1 × 10–8) corresponding to a gene cluster of uridine 5′-diphospho-glucuronosyltransferases (the UGT1A cluster) and solute carrier organic anion transporter family, member 1B1 (SLCO1B1), respectively.
CONCLUSION: Genome-wide association studies have identified genetic variants associated with numerous phenotypes but have been historically limited by inadequate sample size due to costly genotyping and phenotyping. Large consortia with harmonized genotype data have been assembled to attain sufficient statistical power, but phenotyping remains a rate-limiting factor in gene discovery research efforts. The EMR consists of an abundance of phenotype data that can be extracted in a relatively quick and systematic manner. The MayoGC provides a model of a unique collaborative effort in the environment of a common EMR for the investigation of genetic determinants of diseases.
ABI = ankle-brachial index; BORA = Biologically Oriented Repository Architectures; DUA = data use agreement; eMERGE = Electronic Medical Records and Genomics; EMR = electronic medical record; GENEVA = Gene Environment Association Studies; GWA = genome-wide association; MayoGC = Mayo Genome Consortia; PAD = peripheral arterial disease; SNP = single nucleotide polymorphism
Genome-wide association (GWA) studies have identified genetic variants associated with numerous diseases and phenotypes. This agnostic approach to discovering genetic variants important to diseases has been historically limited by inadequate sample size due to costly genotyping and phenotyping. Traditionally, GWA studies have been conducted in research cohorts designed to capture and measure outcomes and phenotypes specific to one disease domain. This has prompted the formation of large consortia comprising several research cohorts with harmonized genotype data and similar phenotype measurements to attain the statistical power required for genetic variant discovery.
Despite the high initial cost of genotyping, the static nature of germline genotypes allows such data to be mined and fitted to pursue various hypotheses, including those unrelated to the original study. As such, a collection of genotype data from existing GWA studies would be a valuable cost-saving resource for genetic research. In contrast, phenotype acquisition and standardization across different studies, which has been time-consuming, laborious, and prone to error, is a rate-limiting factor in gene discovery research efforts. The electronic medical record (EMR) consists of an abundance of phenotype data that can be extracted in a relatively quick and systematic manner. The Electronic Medical Records and Genomics (eMERGE) Network of the National Human Genome Research Institute (for more information, see http://www.gwas.org) is a national consortium formed to develop, disseminate, and apply approaches for genetic research within EMR systems.1,2 Successful use of this approach in the eMERGE Network has inspired the creation of the intramural Mayo Genome Consortia (MayoGC). The goal of the MayoGC is to assemble a large cohort of participants from research studies across Mayo Clinic with high-throughput genetic data and to use EMR for phenotype extraction for cost-effective genetic research.
Herein, we describe the design of the MayoGC, including the current participating cohorts, expansion efforts, data processing, and study management and organization. As a test of the genetic research capability of the MayoGC, we conducted a GWA study to identify genetic variants associated with total bilirubin levels. Bilirubin levels have a large variability in the population, with heritability of roughly 0.50.3 Two previous GWA studies identified variants from similar genomic locations with strong and moderate effects on bilirubin levels,4,5 making this phenotype an ideal candidate for testing. The MayoGC provides a model of a unique collaborative effort in the environment of a common EMR for the investigation of genetic determinants of diseases.
PARTICIPANTS AND METHODS
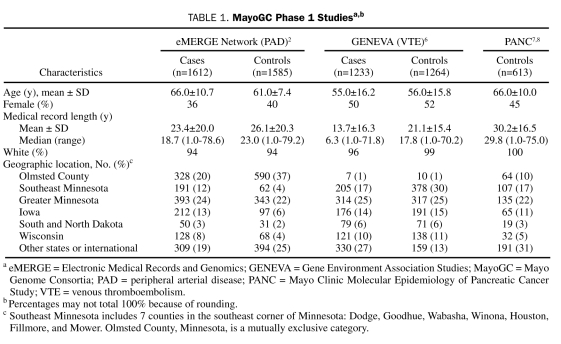
MayoGC is a large cohort of Mayo Clinic patients with EMR and genotype data. Eligible participants include those who gave general research (ie, not disease-specific) consent in the contributing studies to share high-throughput genotyping data with other investigators. This cohort is being built in 2 phases. Phase 1, which has been completed, includes participants from 3 studies funded by the National Institutes of Health, which sought to identify genetic determinants of peripheral arterial disease (PAD), venous thromboembolism, and pancreatic cancer, respectively, with a combined total sample size of 6307 unique participants (Table 1). The eMERGE study contributed genotype data for 3336 participants with PAD and control participants recruited from Mayo Clinic's noninvasive vascular and exercise stress testing laboratories, respectively.2 Peripheral arterial disease was defined by documentation of at least 1 of the following: (1) an ankle-brachial index (ABI) of 0.9 or less at rest or 1 minute after exercise, (2) the presence of poorly compressible arteries, or (3) a normal ABI but history of revascularization for PAD. Control participants had a normal ABI and no history of PAD.2
TABLE 1.
MayoGC Phase 1 Studiesa,b
The GENEVA (Gene Environment Association Studies) Study of Venous Thromboembolism of the National Human Genome Research Institute enrolled consecutive Mayo Clinic outpatients with objectively diagnosed deep venous thrombosis and/or pulmonary embolism who resided in the upper Midwest and had been referred by a Mayo Clinic physician to the Mayo Clinic Special Coagulation Laboratory or to the Mayo Clinic Thrombophilia Center.6 A deep venous thrombosis or pulmonary embolism was categorized as objectively diagnosed (1) when it was confirmed by venography or pulmonary angiography or via a pathology examination of a thrombus removed at surgery or (2) if findings on at least 1 noninvasive test (compression duplex ultrasonography, lung scan, computed tomography, magnetic resonance imaging) were positive. Persons with venous thromboembolism related to active cancer were excluded. A control group was prospectively recruited for this study. Control participants were frequency-matched to the age group (18-29 years, 30-39 years, 40-49 years, 50-59 years, 60-69 years, 70-79 years, and ≥80 years), sex, myocardial infarction or stroke status, and state of residence distribution of the cases. The study selected clinic-based controls using a database of persons undergoing general medical examinations in the Mayo Clinic Divisions of General Internal Medicine and Primary Care Internal Medicine. Additionally, persons undergoing evaluation at the Mayo Clinic Sports Medicine Center and the Department of Family Medicine were screened for inclusion as control participants. Genotype data for 2497 participants were contributed by the GENEVA study.6
The Mayo Clinic Molecular Epidemiology of Pancreatic Cancer Study contributed genotype data for 613 control participants to the MayoGC. Details of this study have been described previously.7,8 In brief, patients scheduled for a general medical examination between May 2004 and February 2007 were recruited from the Divisions of General Internal Medicine and Primary Care Internal Medicine. Control participants were frequency-matched to pancreatic cancer cases on the basis of sex, state of residence, age at recruitment, and race and ethnicity. At the time of recruitment, control participants had no personal history of cancer except nonmelanoma skin cancer.
Phase 2 is under way with the goal of expanding the MayoGC by recruiting eligible patients from other studies funded by the National Institutes of Health (Table 2). All of the MayoGC studies were approved by the Mayo Clinic Institutional Review Board, and the participants from each involved study provided written and informed consent for general research.
TABLE 2.
Studies Participating in the MayoGC
MayoGC Organization
The MayoGC represents a voluntary collaboration of investigators across disciplines at Mayo Clinic. The organizational structure comprises a MayoGC Research Group and a Steering Committee. The Research Group is a collaboration of investigators with expertise in epidemiology, statistical genetics, and bioinformatics who are responsible for the creation, implementation, and maintenance of the consortia. Specifically, members of this group are responsible for study recruitment, development of policies and procedures for widespread use of MayoGC data, harmonization of genetic and phenotype data, and maintenance and storage of study data. The Steering Committee consists of a member from each of the contributing studies and members of the Research Group. This committee is responsible for approving all study policies and procedures and recommending modifications to operational policy as needed. Furthermore, the committee reviews all research proposals that use MayoGC data. Via its representative on the Steering Committee, each contributing study may formally opt out of participation in research proposals that are in conflict with its goals.
Mayo Clinic Biobank
The MayoGC is a passive collection of existing data and, as such, does not have stored biological samples on participants in the consortia. However, 6% of the participants in phase 1 were also enrolled in the Mayo Clinic Biobank. The Mayo Clinic Biobank (for more information, see http://mayoresearch.mayo.edu/biobank/) has enrolled more than 18,000 participants and has the goal of reaching 20,000 participants by the end of 2011 in an effort to support a wide array of health-related research studies throughout the institution. Study participants provide a blood sample for DNA and serum/plasma research, complete a health risk questionnaire, allow access to medical records, and consent to prospective follow-up for health outcomes. Thus, for the subset of the MayoGC participants in the Biobank, biological specimens are available. Likewise, genotype data is available through the MayoGC for users of the Biobank.
Data Use Agreement
To protect the confidentiality and privacy of the MayoGC, investigators who are granted access to MayoGC data will operate under a data use agreement (DUA). The DUA describes the terms and conditions for the following: data use and transferability, publication, termination/expiration of the data agreement, obtaining of informed consent from study participants, and compliance by data recipients with the requirements of the institutional review board of their home institution. Further, the DUA outlines the procedures for amending a current agreement and specifies how the failure to comply with the terms of the DUA will be addressed.
Bilirubin Phenotype—A GWA Application
For phase 1 of the MayoGC, all bilirubin levels clinically ordered from January 1, 1994, through August 31, 2010, were retrieved from a structured laboratory database. Extracted data included the test code and description, the date and time of the sample, the units of results, the associated reference range and indicators for low and high results, the laboratory accession number, and the results of the test in both character and numeric format. Bilirubin measurements were available for 4195 participants. Because our hypothesis focused on identifying genetic variants that affect bilirubin levels within the normal range, we excluded 726 participants who had at least 1 abnormal bilirubin level from the primary analysis. Of the 3469 participants with normal bilirubin levels, 58% had serial measurements of bilirubin. This analysis includes only the first-ever bilirubin level measured after age 18 years. To further explore the genetic effects on bilirubin levels, secondary analyses included a GWA study of all 4195 participants without exclusions and 2 subsets in which we excluded participants with any abnormal test results for alanine aminotransferase, alkaline phosphatase, aspirate aminotransferase, or γ-glutamyltransferase on the same day as the bilirubin measurement (n=2427) or those without any abnormal liver test results within 1 year before or after the bilirubin measurement (n=2191).
Genotype Harmonization
Participants in the eMERGE Network and GENEVA were genotyped using Illumina HumanHap660-Quad chips (Illumina, San Diego, CA) (Table 2). Participants from the Mayo Clinic Molecular Epidemiology of Pancreatic Cancer Study were typed on the HumanHap550 and the Human 610-Quad chips.7,8 PLINK files, in which genotypes were coded as the number of minor alleles, were provided by the individual studies. Of the participants in phase 1 of the MayoGC, 60 also participated in 2 of the contributing studies, and 1 was involved in all 3 studies. These duplicated samples were used to check genotype concordance across studies as well as to inform flipping of minor allele/strand as necessary. Single nucleotide polymorphisms (SNPs) that had more than 2 discordant genotypes among the duplicates or that were monomorphic in all phase 1 samples were excluded from the dataset. Other commonly used SNP-filtering criteria were not imposed because of their dependency on the samples used for a specific hypothesis (ie, the availability of EMR data).
We retained 1 duplicated sample with more nonmissing genotypes from each of the 61 study participants. PLINK was then used for sample-wise quality control on the remaining data. Relatedness was determined on the basis of identity-by-descent estimates generated from the “--Zgenome” option in PLINK. Genotype data of trios from the Centre d'Etude du Polymorphisme Humain collected in Utah, USA, with ancestry from northern and western Europe were used to define an identity-by-descent threshold for relatedness. No cryptic relatedness was detected. Because the patterns of missingness differed somewhat across studies, we excluded samples on the basis of the notion of within-study outlier. This approach eliminated 12 samples with a comparably high missing genotype rate. On the basis of the genotype data, a large majority of the participants in phase 1 of MayoGC were of European ancestry. The 48 samples in which population admixture was evident were filtered out. Cross-checking of the sex of study participants identified and removed 5 with a mismatch between the reported and deduced sex. These quality controls left 6307 study participants in whom 583,129 SNPs were available for analysis. No SNP-wise quality control filtering based on Hardy-Weinberg equilibrium or minor allele frequency was completed at this stage because these measures may change depending on the sample with the phenotype of interest.
Data Management and Data Security
The Bioinformatics Core at Mayo Clinic has developed a system for the storage of processed genomics data. The Biologically Oriented Repository Architectures (BORA) system includes components to facilitate the processing and analysis of data produced by high-throughput genomics platforms. BORA features fully deployed methods for data security and access control, including user authentication and user authorization. Authentication involves validation of the institution's user ID and password and verification that the user ID is included in the BORA list of authorized users. Authorization of data use is defined on a user and user group level. A user or user group is provided access to explicitly defined samples or sample sets. Designation of specific user or user group authorization to access the MayoGC data is approved by the MayoGC Steering Committee. The BORA system stores and provides access to MayoGC genomics data that have been quality controlled and imputed following the described procedures.
Statistical Analyses
Genotype association with bilirubin levels was performed using a linear regression model implemented in PLINK. Age at the time of first bilirubin measurement in adulthood and sex were accommodated in the model as covariates. Latent variables indicative of study of origin were also included to avoid study-induced bias. Bilirubin values were log transformed to accommodate skewness. P<10–6 is deemed to have genome-wide significance.
RESULTS
The MayoGC phase 1 population is 44% female, has a mean age of 60.3 years, and is predominately white (Table 1). As a whole, the cohort is a rich source of phenotypes, with a mean medical record length of 22.4 years.
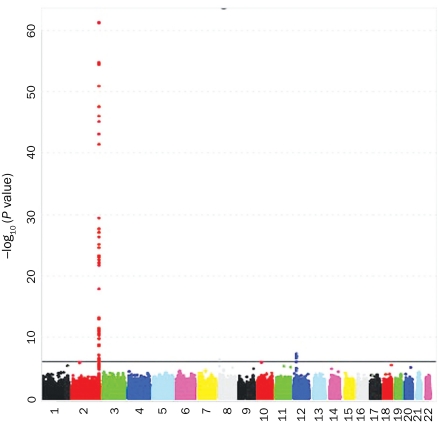
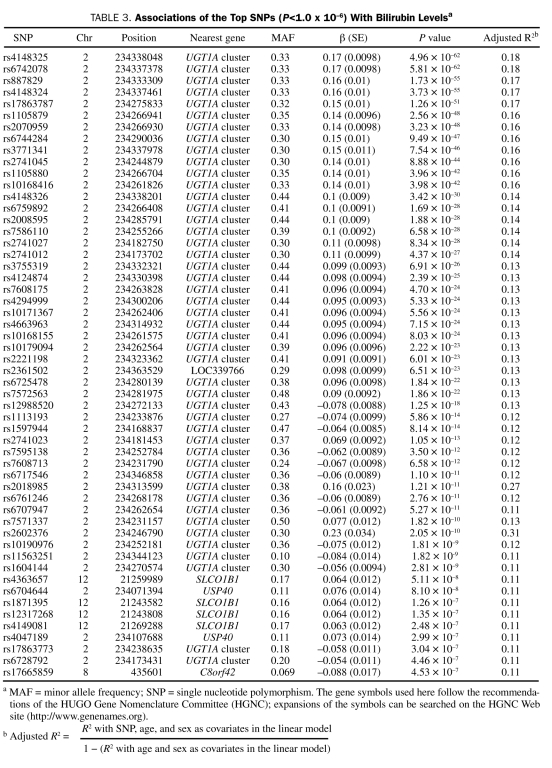
Bilirubin levels (0.1-1.1 mg/dL [to convert to μmol/L, multiply by 17.104]) were available for 3469 participants: 1726 in the eMERGE Network, 1316 in GENEVA, and 427 with pancreatic cancer. Of participants in this group, 47% were women, the mean ± SD age at the time of first bilirubin measurement in adulthood was 57±12.3 years, and the mean ± SD bilirubin level was 0.56±0.20 mg/dL. All studies showed significantly higher bilirubin levels in men (P<.001). The top GWA results were similar with and without adjustment for study; thus, only age- and sex-adjusted results are reported (Figure 1). Genome-wide significant results were observed on 2q37 (top SNP, rs4148325; P=5.0 × 10–62) and 12p12 (top SNP, rs4363657; P=5.1 × 10–8), corresponding to a gene cluster of uridine 5′-diphospho-glucuronosyltransferases (the UGT1A cluster) and solute carrier organic anion transporter family, member 1B1 (SLCO1B1), respectively. Table 3 contains a complete list of significant results, and Figures 2 and 3 show the regional association plots for chromosomes 2 and 12. Figures S1 through S3 (see Supporting Online Material, a link to which is provided at the end of this article) illustrate that the signal on chromosome 2 was robust to different exclusion criteria because it was highly significant both for all study participants with bilirubin measurements (N=4195) and for the 2 subsets restricted to participants with normal liver enzymes either measured on the same day as the bilirubin (n=2427) or within 1 year before or after bilirubin measurement (n=2191).
FIGURE 1.
Genome-wide –log10 P-value plot for total serum bilirubin levels. Solid horizontal line represents significant level of P=10–6.
TABLE 3.
Associations of the Top SNPs (P<1.0 x 10–6) With Bilirubin Levelsa
FIGURE 2.
Regional view of UGT1A cluster for associations with total serum bilirubin levels. The gene symbols used here follow the recommendations of the HUGO Gene Nomenclature Committee (HGNC); expansions of the symbols can be searched on the HGNC Web site (http://www.genenames.org).
FIGURE 3.
Regional view of SLCO1B1 region for associations with total serum bilirubin levels. The gene symbols used here follow the recommendations of the HUGO Gene Nomenclature Committee (HGNC); expansions of the symbols can be searched on the HGNC Web site (http://www.genenames.org).
DISCUSSION
Inspired by the National Center for Biotechnology Information's Database of Genotypes and Phenotypes, the MayoGC represents a model for genetic research that capitalizes on existing genetic data in the environment of a common EMR. Furthermore, this cost-effective approach expands the use of genetic data, derived from available data within the medical record, to phenotypes unrelated to the original study. The utility of the MayoGC is demonstrated by genome-wide significant results for clinically ordered serum bilirubin levels that replicate findings from several cohort studies. In addition to classic gene discovery, other applications of the MayoGC resource include replication of results from other studies, association of SNPs with multiple phenotypes, and pilot studies for rare diseases.
Recruitment is under way for phase 2 of MayoGC, which is intended to expand the cohort and to develop the infrastructure and processes needed for wider and long-term use of the resource. Table 2 lists the cohorts that have agreed to participate. Phase 2 is expected to be completed by the end of 2011 and will include more than 10,000 participants with harmonized and imputed high-throughput genetic data. This effort requires processing of a greater variety of platforms. Thus, a fully automated workflow for forward strand mapping and imputation of genotyping data will be implemented. The workflow will standardize the genotyping data stored in BORA to enable integrated analysis of genotypes from different platforms and from multiple studies collected at Mayo Clinic. In addition, current genotyping-focused quality control procedures will be extended with generalized procedures for population structure validation as well as patient relationship cross-checking to monitor for duplicates and relatedness. BORA is also being interfaced with the Enterprise Data Trust, a central data warehouse in which minable clinical information on Mayo Clinic's patients will be stored and can be accessed. The interface between the 2 systems will greatly facilitate phenotype-genotype association and replication studies.
Significant associations were observed on chromosome 2 corresponding to the UGT1A cluster (P=5.0 × 10–62) and chromosome 12 in the SLCO1B1 gene (P=5.1 × 10–8), confirming previous results in GWA and linkage studies.4,5,9,10 The UGT1A cluster encodes 9 transferase genes (UGT1A* 1, 3, 4, 5, 6, 7, 8, 9, 10), of which 8 are known active gene transcripts. All of the UGT1A proteins are identical in the amino acid sequence encoded by exons 2 through 5 with unique alternate first exons.11 The glucuronidation of bilirubin by UGT1A and specifically by UGT1A1 is essential for the elimination of bilirubin from the body.12 Three heritable forms of hyperbilirubinemia (ie, Crigler-Najjar syndrome types 1 and 2 and Gilbert syndrome) all result from mutations in UGT1A1, and candidate gene studies in multiple ethnic populations have shown that UGT1A1 is the major gene influencing total bilirubin levels.13-17 The SLCO1B1 locus encodes a protein that mediates sodium-dependent uptake of bilirubin. Although not as influential as UGT1A1, SLCO1B1 is a major contributor to total bilirubin levels and variations in it have been associated with hyperbilirubinemia.18-20
The MayoGC is a collaborative effort for data sharing and, as such, has no stored DNA for the study participants for de novo genotyping. Likewise, phenotyping is restricted to data available within the medical record. For a subset of MayoGC participants, biological specimens are available through the Mayo Clinic Biobank. Biological specimens may also be available in the contributing studies; however, large-scale de novo measurements are likely not practical. The ability to contact and consent participants for additional measurements is possible for MayoGC participants whose contributing study consent did not prohibit recontact or for those who are enrolled in the Mayo Clinic Biobank. Despite these limitations, which are common to genetic consortia, the MayoGC leverages the success of EMR-derived phenotyping with high-throughput genetic data obtained at considerable research costs to enhance genetic research. Furthermore, the relatively stable population in Olmsted County, Minnesota, the county in which Mayo Clinic in Rochester is located, and surrounding counties and the ability to search EMR data spanning decades are unique features of Mayo Clinic that enhance the utility of the MayoGC. However, the MayoGC can serve as a template for other institutions with EMR-phenotyping capability.
CONCLUSION
Genome-wide association studies have identified genetic variants associated with numerous phenotypes but have been limited historically by inadequate sample size due to costly genotyping and phenotyping. Large consortia with harmonized genotype data have been assembled to attain sufficient statistical power, but phenotyping remains a rate-limiting factor in gene discovery research efforts. The EMR offers an abundance of phenotype data that can be extracted in a relatively quick and systematic manner. The utility of the MayoGC was successfully demonstrated by the replication of GWA results for bilirubin levels obtained from the EMR. Thus, MayoGC provides a model of a unique collaborative effort in the environment of a common EMR for the investigation of genetic determinants of diseases.
Supplementary Material
Footnotes
Funding for this research was provided by the Electronic Medical Records and Genomics (eMERGE) Network of the National Human Genome Research Institute (HG05499), Mayo Clinic Genome-wide Association Study of Venous Thromboembolism (HG04735) from the National Human Genome Research Institute (NHGRI; Gene Environment Association Studies [GENEVA] consortium), Mayo Clinic Specialized Programs of Research Excellence (SPORE) in Pancreatic Cancer (P50CA102701) from the National Cancer Institute, and Mayo Clinic Cancer Center (Genetic Epidemiology and Risk Assessment [GERA] Program). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. Dr Limburg served as a consultant for Genomic Health, Inc from August 12, 2008, through April 19, 2010. Mayo Clinic has licensed Dr Limburg's intellectual property to Exact Sciences, and he and Mayo Clinic have contractual rights to receive royalties through this agreement.
An earlier version of this article appeared Online First.
For editorial comment, see page 597
Supporting Online Material
www.mayoclinicproceedings.com/content/86/7/606/suppl/DC1 Figures S1-S3
REFERENCES
- 1. McCarty CA, Chisholm RL, Chute CG, et al. ; eMerge Team The eMERGE Network: a consortium of biorepositories linked to electronic medical records data for conducting genomic studies. BMC Med Genomics. 2011;4(1):13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Kullo IJ, Fan J, Pathak J, Savova GK, Ali Z, Chute CG. Leveraging informatics for genetic studies: use of the electronic medical record to enable a genome-wide association study of peripheral arterial disease. J Am Med Inform Assoc. 2010;17(5):568-574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Bathum L, Petersen HC, Rosholm JU, Hyltoft Petersen P, Vaupel J, Christensen K. Evidence for a substantial genetic influence on biochemical liver function tests: results from a population-based Danish twin study. Clin Chem. 2001;47(1):81-87 [PubMed] [Google Scholar]
- 4. Johnson AD, Kavousi M, Smith AV, et al. Genome-wide association meta-analysis for total serum bilirubin levels. Hum Mol Genet. 2009;18:2700-2710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Sanna S, Busonero F, Maschio A, et al. Common variants in the SLCO1B3 locus are associated with bilirubin levels and unconjugated hyperbilirubinemia. Hum Mol Genet. 2009;18:2711-2718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Cornelis MC, Agrawal A, Cole JW, et al. The Gene, Environment Association Studies consortium (GENEVA): maximizing the knowledge obtained from GWAS by collaboration across studies of multiple conditions. Genet Epidemiol. 2010;34(4):364-372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Amundadottir L, Kraft P, Stolzenberg-Solomon RZ, et al. Genome-wide association study identifies variants in the ABO locus associated with susceptibility to pancreatic cancer. Nat Genet. 2009;41(9):986-990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Petersen GM, Amundadottir L, Fuchs CS, et al. A genome-wide association study identifies pancreatic cancer susceptibility loci on chromosomes 13q22.1, 1q32.1 and 5p15.33. Nat Genet. 2010;42(3):224-228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Kang TW, Kim HJ, Ju H, et al. Genome-wide association of serum bilirubin levels in Korean population. Hum Mol Genet. 2010;19(18):3672-3678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Melton PE, Haack K, Göring HH, et al. Genetic influences on serum bilirubin in American Indians: the Strong Heart Family Study. Am J Hum Biol. 2011;23(1):118-125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Tukey RH, Strassburg CP. Human UDP-glucuronosyltransferases: metabolism, expression, and disease. Annu Rev Pharmacol Toxicol. 2000;40:581-616 [DOI] [PubMed] [Google Scholar]
- 12. Ghosh SS, Lu Y, Lee SW, et al. Role of cysteine residues in the function of human UDP glucuronosyltransferase isoform 1A1 (UGT1A1). Biochem J. 2005;392(3):685-692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Hong AL, Huo D, Kim HJ, et al. UDP-glucuronosyltransferase 1A1 gene polymorphisms and total bilirubin levels in an ethnically diverse cohort of women. Drug Metab Dispos. 2007;35(8):1254-1261 [DOI] [PubMed] [Google Scholar]
- 14. Lin JP, O'Donnell CJ, Schwaiger JP, et al. Association between the UG-T1A1*28 allele, bilirubin levels, and coronary heart disease in the Framingham Heart Study. Circulation. 2006;114(14):1476-1481 [DOI] [PubMed] [Google Scholar]
- 15. Lin R, Wang Y, Wang Y, et al. Common variants of four bilirubin metabolism genes and their association with serum bilirubin and coronary artery disease in Chinese Han population. Pharmacogenet Genomics. 2009;19(4):310-318 [DOI] [PubMed] [Google Scholar]
- 16. Lingenhel A, Kollerits B, Schwaiger JP, et al. Serum bilirubin levels, UGT1A1 polymorphisms and risk for coronary artery disease. Exp Gerontol. 2008;43(12):1102-1107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Borucki K, Weikert C, Fisher E, et al. Haplotypes in the UGT1A1 gene and their role as genetic determinants of bilirubin concentration in healthy German volunteers. Clin Biochem. 2009;42(16-17):1635-1641 [DOI] [PubMed] [Google Scholar]
- 18. Huang CS, Huang MJ, Lin MS, Yang SS, Teng HC, Tang KS. Genetic factors related to unconjugated hyperbilirubinemia amongst adults. Pharmacogenet Genomics. 2005;15(1):43-50 [DOI] [PubMed] [Google Scholar]
- 19. Ieiri I, Suzuki H, Kimura M, et al. Influence of common variants in the pharmacokinetic genes (OATP-C, UGT1A1, and MRP2) on serum bilirubin levels in healthy subjects. Hepatol Res. 2004;30(2):91-95 [DOI] [PubMed] [Google Scholar]
- 20. Zhang W, He YJ, Gan Z, et al. OATP1B1 polymorphism is a major determinant of serum bilirubin level but not associated with rifampicin-mediated bilirubin elevation. Clin Exp Pharmacol Physiol. 2007;34(12):1240-1244 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.