Abstract
The Myc protein binds DNA and activates transcription by mechanisms that are still unclear. We used chromatin immunoprecipitation (ChIP) to evaluate Myc-dependent changes in histone acetylation at seven target loci. Upon serum stimulation of Rat1 fibroblasts, Myc associated with chromatin, histone H4 became locally hyperacetylated, and gene expression was induced. These responses were lost or severely impaired in Myc-deficient cells, but were restored by adenoviral delivery of Myc simultaneous with mitogenic stimulation. When targeted to chromatin in the absence of mitogens, Myc directly induced H4 acetylation. In addition, Myc recruited TRRAP to chromatin, consistent with a role for this cofactor in histone acetylation. Finally, unlike serum, Myc alone was very inefficient in inducing expression of most target genes. Myc therefore governs a step, most likely H4 acetylation, that is required but not sufficient for transcriptional activation. We propose that Myc acts as a permissive factor, allowing additional signals to activate target promoters.
Keywords: Chromatin, histone acetylation, Myc, TRRAP, HAT, nucleolin
The mammalian proto-oncogene c-myc is induced by mitogens and is a strong promoter of cell growth, proliferation, and death. The c-Myc protein (Myc) is a transcription factor of the basic-helix-loop-helix–leucine zipper (bHLHLZ) family. It dimerizes selectively with its bHLHLZ partner Max, binds the E-box motif CA(C/T)GTG, and activates transcription of genes linked to this motif. However, the mechanisms by which Myc regulates transcription remain unclear (Cole and McMahon 1999; Grandori et al. 2000; Amati et al. 2001).
The first level of DNA packaging in eukaryotes is its wrapping around the core histones H2A, H2B, H3, and H4 to form the nucleosome (Luger et al. 1997). In recent years, acetylation of nucleosomal histones has emerged as a key step of transcriptional control in all eukaryotic cells. Acetylation levels are controlled by a variety of histone acetyltransferases (HATs) and deacetylases (HDACs), which are recruited to promoters by sequence-specific activators and repressors, respectively, and mediate their transcriptional activities (e.g., Chen et al. 1999; Agalioti et al. 2000; Kuo et al. 2000; Reid et al. 2000; for reviews, see Kingston and Narlikar 1999; Knoepfler and Eisenman 1999; Sterner and Berger 2000; Fry and Peterson 2001).
Recent evidence suggests that Myc may modulate transcription via histone acetylation (Cole and McMahon 1999; Grandori et al. 2000; Amati et al. 2001). The N-terminal transactivation domain of Myc binds TRRAP (McMahon et al. 1998), a large protein that is part of at least two classes of multisubunit HAT complexes, GCN5/PCAF and Tip60/NuA4 (Vassilev et al. 1998; Brand et al. 1999; Ikura et al. 2000). Most likely through TRRAP, Myc also associates with the HAT subunit GCN5 in cells, and coprecipitates with HAT activity (McMahon et al. 2000; S. Taubert and B. Amati, unpubl.). Additional circumstantial evidence for a role of Myc in histone acetylation comes from its antagonists, the Mad family and Mnt. These proteins form alternative dimers with Max that bind the same E-box consensus as Myc/Max, but repress transcription by recruiting Sin3, a subunit of HDAC corepressor complexes (for reviews, see Knoepfler and Eisenman 1999; Grandori et al. 2000). Upon differentiation, myeloid cells switch from Myc/Max to Mad/Max complexes (Ayer and Eisenman 1993), accompanied by deacetylation of histones at a common target promoter, hTERT (Xu et al. 2001). However, it remains unknown whether Myc actively contributes to histone acetylation. In this work, we show that Myc mediates mitogen-induced acetylation of histone H4 at seven target loci. Consistent with observations on other transcription factors (e.g., Cosma et al. 1999; Krebs et al. 1999), we present evidence suggesting that this step is necessary but not sufficient for gene activation.
Results
Quantitative chromatin immunoprecipitation
To assess Myc-binding and histone acetylation at multiple genomic sites in vivo, we developed a quantitative chromatin immunoprecipitation (ChIP) protocol. As in published procedures (e.g., Boyd and Farnham 1997; Chen et al. 1999), live cells were cross-linked with formaldehyde, chromatin was fragmented by sonication, and protein–DNA complexes were immunoprecipitated. Recovery of specific DNA sequences was quantified by real-time PCR.
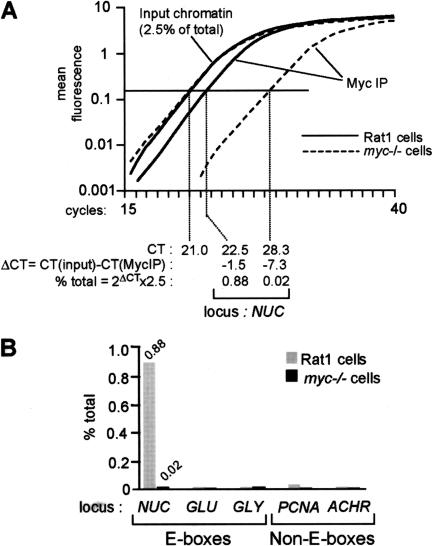
As an initial test, we analyzed binding of Myc in Rat1 cells to the E-boxes in intron 1 of nucleolin (NUC; Fig. 1, amplicon +574; Greasley et al. 2000). Representative amplification curves are shown in Figure 2A. The amount of NUC DNA coprecipitated with Myc was 0.88% of that in total input chromatin, as calculated in Figure 2A. As a control for the specificity of Myc antibodies, we also analyzed a Myc-deficient Rat1 derivative (myc−/−; Mateyak et al. 1997). The background in these cells (0.02%; Fig. 2A,B) was identical to that obtained with nonimmune precipitates from Rat1 cells (data not shown). As negative controls, we analyzed promoters containing no E-boxes, including PCNA, acetylcholine receptor (ACHR), and Topoisomerase II. None of these sequences were enriched in Myc immunoprecipitates from Rat1 cells (Fig. 2B; data not shown). In addition, Myc did not bind E-boxes in several other genes, including glucokinase (GLU), glycine methyltransferase (GLY), and socs-2 (Fig. 2B; data not shown). Myc was therefore specifically associated with the NUC E-boxes in Rat1 cells.
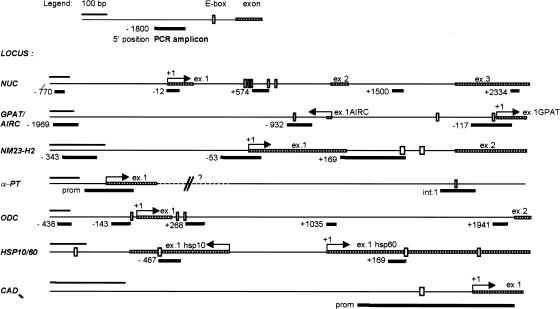
Figure 1.
Schematic representation of the genes characterized in this study and of the amplicons used in ChIP analysis. PCR amplicons are numbered with the position of their 5′ end relative to the 5′ end of exon 1 (+1), as annotated in GenBank. For the bidirectional loci AIRC/GPAT and HSP10/60, we took as reference GPAT and HSP60, respectively. The accession numbers for all genomic sequences and the sequences of the PCR primers are given in Materials and Methods.
Figure 2.
Quantification of chromatin immunoprecipitation by real-time PCR. Chromatin extracted from cross-linked Rat1 or myc−/− cells was immunoprecipitated with anti-Myc antibodies. DNA was recovered and used as a template for real-time PCR in a Taqman 5700 (Perkin Elmer). (A) Representative PCR-amplification curves, as displayed by the Taqman 5700. In this example, we used primers amplifying the NUC E-box domain (amplicon +574, Fig. 1). Calculation of the amount of immunoprecipitated NUC E-box DNA relative to that present in total input chromatin is shown at the bottom. (CT) Cycle threshold, cycle number at which each PCR reaction reaches a predetermined fluorescence threshold, set within the linear range of all reactions. (B) Graphic representation of the data for NUC and control promoters analyzed in the same experiment (GLU, GLY, PCNA, and ACHR). In this experiment, cells were cross-linked and harvested 4 h after mitogenic stimulation.
As illustrated in Figure 2A, real-time PCR allows comparison of multiple reactions in their respective linear range, even if these reactions follow very different kinetics. This is not achieved by arresting PCR reactions at a common cycle number and quantifying their products on gels, as classically performed in ChIP. For added accuracy, in every ChIP experiment shown below, each data point represents the average and standard deviation from three independent immunoprecipitations.
Association of Myc with target sites upon serum stimulation
We next examined the association of Myc with several genomic target sites in serum-stimulated Rat1cells. The sequences analyzed were E-boxes (CACGTG) located in either the promoter or the first intron of several rat genes, including NUC, CAD, NM23-H2, α-prothymosin (α-PT), ODC, and the gene pairs AIRC/GPAT and HSP10/60 (Fig. 1). Evidence for regulation of these genes by Myc was reported in the following publications: α-PT (Eilers et al. 1991; Gaubatz et al. 1994; Desbarats et al. 1996); NUC (Coller et al. 2000; Greasley et al. 2000; Boon et al. 2001); ODC (Bello-Fernandez et al. 1993; Peña et al. 1993; Wagner et al. 1993; Tavtigian et al. 1994; Shim et al. 1997; Tsuneoka et al. 1997; Coller et al. 2000; O'Hagan et al. 2000; Schuhmacher et al. 2001); HSP60 (Boon et al. 2001); HSP10, GPAT, AIRC, and NM23-H2 (Schuhmacher et al. 2001); CAD (Miltenberger et al. 1995; Boyd and Farnham 1997, 1999; Boyd et al. 1998; Schuhmacher et al. 2001).
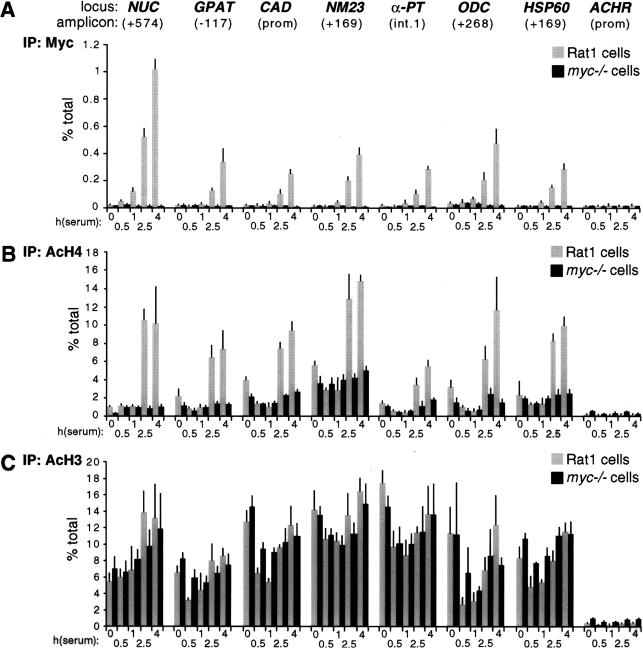
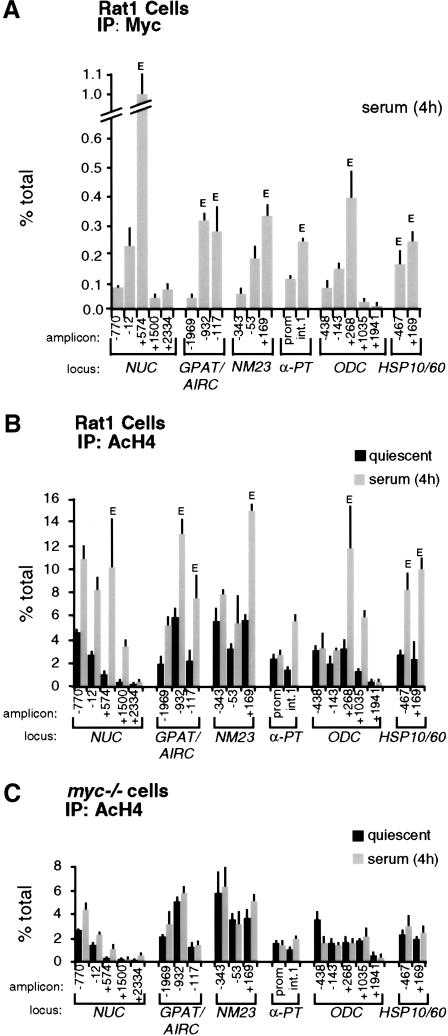
Quiescent Rat1 cells were seeded in serum-containing medium and fixed at various time points for ChIP analysis. This revealed a progressive association of Myc with genomic target sites, reaching maximal levels by 4 h (Fig. 3A, shaded bars) and decreasing thereafter (data not shown). None of these sites was enriched following precipitation of chromatin from myc−/− cells (Fig. 3A, black bars), or in nonimmune precipitates (data not shown). Binding of Myc to CAD and ODC was previously shown in NIH3T3 cells (Boyd et al. 1998; Boyd and Farnham 1999; Eberhardy et al. 2000). In conclusion, Myc is expressed following mitogenic stimulation and selectively binds E-boxes in the regulatory regions of target genes.
Figure 3.
Mitogens induce association of Myc with chromatin and acetylation of histone H4. At the indicated time points after serum stimulation, Rat1 cells (shaded bars) and myc−/− cells (black bars) were analyzed by ChIP. Antibodies against (A) Myc, (B) acetylated histone H4, and (C) acetylated histone H3 were used in parallel immunoprecipitations. Precipitated DNA samples were amplified with primers recognizing the E-box domains of Myc-target genes (see corresponding maps and amplicons in Fig. 1) or the control ACHR promoter as indicated in A. % total: as defined in Figure 2.
Mitogen-induced acetylation of histone H4 is dependent on Myc
To address whether acetylation of histones H3 and H4 at Myc-target sites is regulated by serum, we performed ChIP with antibodies specific to either acetylated histone. Serum stimulation induced a marked increase in histone H4 acetylation at all the Myc-target sites examined (Fig. 3B, shaded bars). This effect was rapid, closely paralleled binding of Myc to chromatin and, most strikingly, was lost in myc−/− cells (Fig. 3B, black bars). In contrast to H4, histone H3 acetylation was already elevated in quiescent cells and, with the possible exception of NUC, was not further increased by serum (Fig. 3C). At some loci, we observed a transient decrease in H3 acetylation upon replating, followed by a recovery to initial levels. Most importantly in the context of this work, we observed no significant differences in H3 acetylation between Rat1 and myc−/− cells (Fig. 3C). Although these data do not rule out an effect of Myc on H3 acetylation at other loci and/or in different cells, our studies here focus on histone H4.
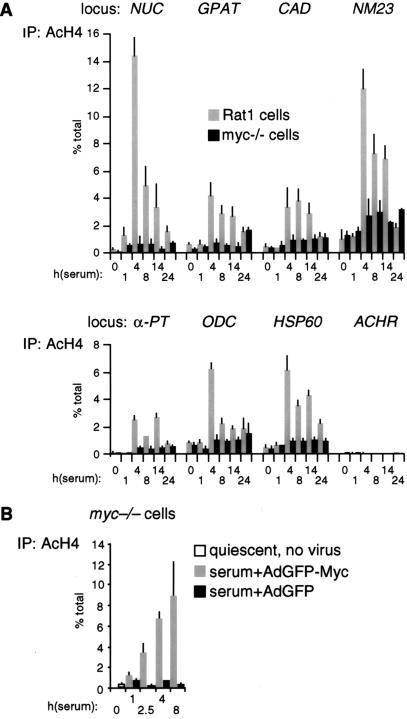
To rule out that the differences in H4 acetylation between Rat1 and myc−/− cells were an indirect consequence of slower cell-cycle progression in the latter (Mateyak et al. 1997), we analyzed H4 acetylation over a longer time course (Fig. 4A). This revealed two essential points. First, H4 acetylation in Rat1 cells peaked at 4 h and decreased thereafter (Fig. 4A, shaded bars), coinciding with the kinetics of Myc-binding to chromatin (data not shown). Second, serum induced no significant acetylation in myc−/− cells within 24 h, at which time those cells were at the G1–S boundary (Mateyak et al. 1997). To further address whether the defective response of myc−/− cells was specifically caused by lack of Myc, we performed a reconstitution experiment. Simultaneously with mitogenic stimulation, myc−/− cells were infected with an adenovirus expressing human c-myc (AdGFP-Myc), restoring binding of Myc to chromatin with kinetics comparable to those seen in Rat1 cells (data not shown). In parallel, H4 acetylation was restored by AdGFP-Myc at NUC (Fig. 4B, shaded bars) and all other loci (data not shown), but the control virus AdGFP had no effect (black bars). In summary, histone H4 at Myc-binding sites is rapidly acetylated in response to mitogens, and Myc is essential for this response.
Figure 4.
(A) Kinetics of histone H4 acetylation at Myc-binding sites. The experiment shown in Figure 3B was repeated over a longer time course. (B) Myc rescues histone H4 acetylation in myc−/− cells. Quiescent myc−/− cells (open bar) were serum-stimulated and simultaneously infected with either AdGFP (black bars) or AdGFP-Myc (shaded bars). ChIP was used to measure H4 acetylation at the NUC E-box domain (amplicon +574, Fig. 1).
In contrast with our findings, a recent study detected no changes in acetylation of either histones H4 or H3 at the CAD and ODC promoters upon mitogenic stimulation of NIH3T3 cells (Eberhardy et al. 2000). In some cells and/or conditions, acetylation levels on E-boxes may remain high at quiescence, perhaps masking mitogen-induced acetylation. This might be the case in our experiments for histone H3 (Fig. 3C). Alternatively, although both studies were based on the same antibodies, technical differences in the ChIP assay may account for the different conclusions. In particular, accurate quantitation of acetylation levels across chromatin domains critically relies on small sizes of amplicons (Fig. 1) and of sonicated chromatin fragments (bulk 100–400 bp; data not shown), as well as on the use of real-time PCR (Fig. 2).
Serum- and Myc-dependent acetylation is centered on E-boxes
If serum-induced acetylation results from a direct action of Myc on chromatin, it should localize to discrete chromatin regions centered on Myc-binding sites, as shown for other transcription factors (e.g., Krebs et al. 1999; Agalioti et al. 2000; Kuo et al. 2000; Reid et al. 2000; Vogelauer et al. 2000). We therefore mapped Myc-binding and H4 acetylation across the 5′ end of Myc target genes, using the amplicons shown in Figure 1. This revealed decreased enrichment of Myc on chromatin as a function of distance from the E-boxes (Fig. 5A, bars labeled E). In addition to those shown in Figure 3A, Myc was bound to E-boxes in intron 1 of AIRC (amplicon −932) and exon 1 of HSP10 (amplicon −467).
Figure 5.
Localization of Myc and acetylated histones on Myc-target genes by ChIP. (A) Mapping of Myc-binding along its target genes, 4 h after mitogenic stimulation in Rat1 cells. (B) Distribution of acetylated histone H4 along Myc-target genes in quiescent Rat1 cells (black bars) and 4 h after serum stimulation (shaded bars). (C) Same experiment, in myc−/− cells. The identity of each amplicon is indicated below the graphs (see maps in Fig. 1). (E) E-box sites.
Similar to Myc, serum-induced acetylation of H4 was maximal at Myc-binding sites, with variable levels of spreading to adjacent domains (Fig. 5B, shaded bars). Notably, this occurred over a background of acetylation detectable in quiescent cells (black bars). For example, the NUC promoter was acetylated in quiescent cells (amplicons −770 and −12), whereas the Myc-binding sites in intron 1 displayed lower acetylation levels (amplicon +574). Serum enhanced acetylation over both domains, spreading at least 1 kb into intron 2 (amplicon +1500), but terminating within ∼2 kb (+2334). At NM23-H2, both the promoter (amplicons −343 and −53) and intron 1 (+169) showed constitutive H4 acetylation, presumably on adjacent nucleosomes, but serum-induced acetylation was localized to the Myc-binding sites in intron 1 (no data were obtained on downstream sequences). Similarly, at ODC, serum had no significant effect at the promoter (−438 and −143), but induced hyperacetylation at the E-boxes in intron 1 (+268). The effect of serum spread over at least 800 bp into intron 1 (amplicon +1035) and appeared to terminate within <2 kb (+1941). In summary, serum-induced acetylation at these loci covered discrete chromatin domains overlapping with Myc-binding sites.
To address the role of Myc in the acetylation patterns induced by serum, we determined these patterns in myc−/− cells (Fig. 5C). At quiescence, the distribution and levels of H4 acetylation were the same in Rat1 and myc−/− cells (Fig. 5B,C; black bars). Remarkably, in myc−/− cells, the effects of serum were lost across all loci examined (shaded bars). Altogether, the localized nature of serum-induced H4 acetylation, its dependence upon Myc, and its rapid kinetics strongly suggest that Myc initiates this acetylation event. However, Myc is not required for basal, mitogen-independent acetylation at the same loci.
Myc activation is sufficient for acetylation of target sites
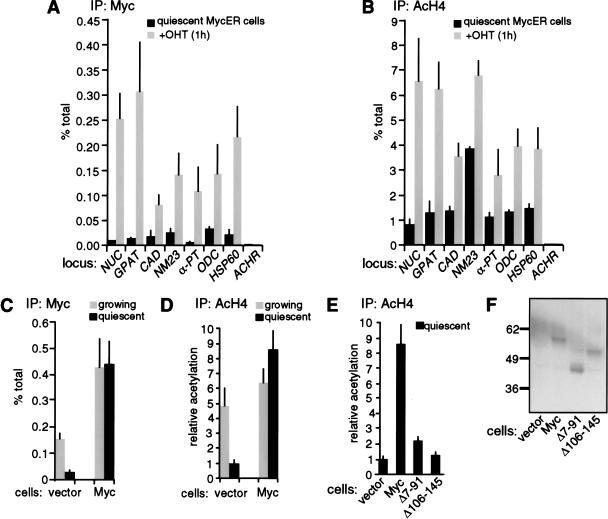
Having shown that Myc is required for mitogen-induced H4 acetylation, we addressed whether activation of Myc in the absence of mitogenic signals was sufficient to reproduce this response. Quiescent Rat1 cells expressing a 4-hydroxytamoxifen (OHT)-inducible MycER chimera (Littlewood et al. 1995) were treated with OHT and analyzed by ChIP as above. At all sites studied, binding of MycER to chromatin was detectable within 15 min, was maximal by 1 h, and remained stable 4 h after OHT treatment (Fig. 6A; data not shown). Judging from the specific recovery of E-box sequences with Myc antibodies relative to input DNA (percent total), MycER bound GPAT as efficiently as endogenous Myc in serum-stimulated Rat1 cells, but other sites were bound with somewhat lower efficiencies (cf. Figs. 3A and 6A). Binding of MycER to chromatin was accompanied by increased acetylation of histone H4 (Fig. 6B). Induction of H4 acetylation was resistant to the protein synthesis inhibitor cycloheximide, demonstrating that it was a direct effect of MycER (data not shown). Of all the loci analyzed, NUC and GPAT were most significantly acetylated by MycER, approaching the levels observed with serum (Figs. 3B and 6B, see also Fig. 5B). In addition, the pattern of MycER-induced acetylation across the NUC locus was remarkably similar to that observed with serum (data not shown). Control sites that were not bound by endogenous Myc also remained free of MycER and were not acetylated upon OHT treatment (ACHR shown in Fig. 6A,B). In conclusion, Myc directly induces H4 acetylation, and is sufficient to reproduce the effect of serum at its target loci.
Figure 6.
Targeting of Myc to chromatin in quiescent cells promotes H4 acetylation. (A,B) Quiescent Rat1 cells expressing MycER were analyzed by ChIP before (black bars) or 1 h after OHT treatment (shaded bars). Data are shown for the same E-box and control amplicons as in Figure 3. (C,D) Rat1 cells harboring a control retrovirus (vector) or a Myc-expressing virus (Myc) were analyzed by ChIP during continuous growth (shaded bars) or following withdrawal from the cell cycle (black bars), as achieved by contact inhibition and serum starvation. ChIP data are shown for the NUC E-box domain (amplicon +574, Fig. 1). ChIP was performed with antibodies against Myc (A,C) or acetylated H4 (B,D). (E) Comparison of H4 acetylation in quiescent control and Myc-expressing cells (same as in D) with cells expressing the Myc mutants Δ7-91 and Δ106-145. In D and E, all data are expressed relative to the acetylation level in quiescent control cells (vector). (F) Immunoblot analysis of wild-type Myc, Δ7-91 and Δ106-145 in the same cells used in E, with the 9E10 monoclonal antibody (BabCO).
The ability of Myc to promote H4 acetylation was confirmed in cells constitutively expressing wild-type Myc. Upon exit of Rat1 cells from the cell cycle, binding of endogenous Myc to NUC intron 1 decreased as expected (Fig. 6C, vector), whereas retrovirally expressed Myc was maintained on chromatin (Fig. 6C, Myc). In the conditions used here (contact inhibition and serum starvation), Myc-expressing cells withdrew from the cell cycle as efficiently as control cells, and did not undergo significant apoptosis within the time course of the experiment (data not shown). Simultaneously with Myc-binding, H4 acetylation decreased in control cells, but remained elevated in Myc-expressing cells (Fig. 6D). This assay was repeated with two deletion mutants, Myc Δ7-91 and Δ106-145, which lack conserved Myc-boxes I and II, respectively, and are inactive in cellular transformation (Stone et al. 1987). Figure 6E shows that Myc Δ7–91 and Δ106–145 were unable to maintain H4 acetylation in quiescent cells, although expressed at levels similar to those of wild-type Myc (Fig. 6F). ChIP analysis was inconclusive on these mutants, owing to deletions within the domain used to raise the antibody (N262; none of the other Myc antibodies tested were active in ChIP). However, Myc Δ7–91 and Δ106–145 possess an intact bHLHLZ domain, sufficient to dimerize and bind DNA. We conclude that promotion of H4 acetylation by Myc at its chromosomal sites requires domains spanning Myc-boxes I and II.
Although some increases in H3 acetylation were occasionally observed upon MycER activation, these were always minor (<1.5–2×) in comparison to H4. In addition, retrovirally expressed Myc had no effect at all on H3 acetylation (data not shown). Although our data do not rule out direct effects of Myc on H3 acetylation in other circumstances, these remain to be investigated (see Discussion).
Myc recruits TRRAP to chromatin
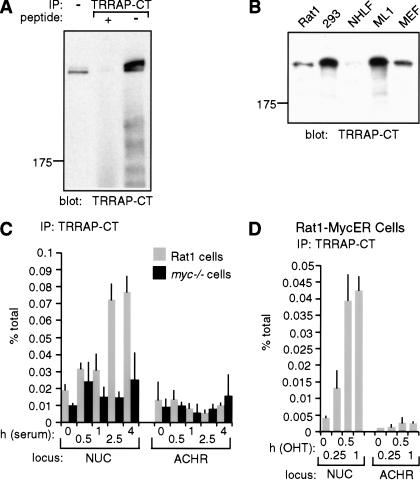
The simplest hypothesis to explain the above observations is that Myc recruits a HAT, which in turn acetylates histone H4. Myc binds TRRAP in a manner dependent on Myc-boxes I and II (McMahon et al. 1998), and TRRAP is a subunit of the HAT complexes PCAF/GCN5 and Tip60/NuA4 (Vassilev et al. 1998; Brand et al. 1999; Ikura et al. 2000). We therefore decided to investigate the recruitment of TRRAP to Myc-target sites in chromatin. To this aim, we raised a rabbit polyclonal antiserum directed against a C-terminal epitope of TRRAP (TRRAP-CT; see Materials and Methods). TRRAP-CT specifically precipitated endogenous TRRAP from lysates of 293 cells, as shown by probing immunoprecipitates with TRRAP-CT itself (Fig. 7A), or with an antibody raised against a different epitope (data not shown; L. Deleu and H. Land, unpubl.). The same band was detected by immunoblot analysis of Rat1 cells, primary human and mouse fibroblasts, human ML1 cells, and 12 additional cell lines (Fig. 7B; data not shown). ChIP analysis with TRRAP-CT in serum-stimulated Rat1 cells revealed a modest but reproducible enrichment of NUC E-box DNA (Fig. 7C, shaded bars), with a time course that paralleled Myc-binding and histone acetylation (Fig. 3A,B). This enrichment was not seen in myc−/− cells (black bars) or with preimmune serum (data not shown). Furthermore, the control ACHR promoter was not enriched. In a similar manner, MycER induced rapid recruitment of TRRAP to NUC, but not to ACHR (Fig. 7D). Although the specific enrichment of TRRAP-associated chromatin at other Myc-target loci was low, Myc-dependent increases could be seen in individual experiments (data not shown). In summary, TRRAP is recruited by Myc to chromatin. It remains to be addressed whether the effect of Myc is mediated by a TRRAP-associated HAT, such as GCN5 or Tip60.
Figure 7.
TRRAP is recruited to chromatin in a Myc-dependent manner. (A,B) Characterization of the TRRAP-CT antibody (see additional information in Materials and Methods). (A) TRRAP-CT was used in immunoprecipitation from a 293-cell lysate (500 μg), in the presence or absence of the cognate antigenic peptide, as indicated. Precipitates were analyzed by immunoblotting with TRRAP-CT itself. The first lane shows total cell lysate (50 μg). (B) TRRAP-CT was used for immunoblot analysis of the indicated cell lysates. (NHLF) normal human lung fibroblasts; (ML1) human myeloid leukemia cell line; (MEF) mouse embryonic fibroblasts. (C,D) TRRAP-binding to the NUC E-box domain (amplicon +574, Fig. 1) and control ACHR promoter was assayed by ChIP with TRRAP-CT, following (C) mitogenic stimulation of Rat1 (shaded bars) or myc−/− cells (black bars) and (D) activation of MycER in quiescent cells.
Myc is required but not sufficient for induction of most target genes
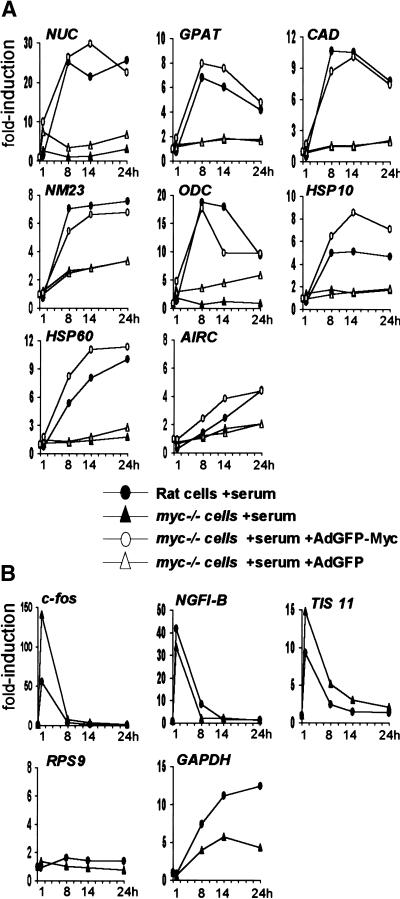
The data presented above establish a role for Myc in mitogen-induced acetylation of histone H4. To address whether this activity might be involved in gene regulation, we compared mRNA induction following serum stimulation in Rat1 and myc−/− cells. The expression of NUC, GPAT, CAD, NM23-H2, ODC, HSP10, and HSP60 was rapidly induced by serum in Rat1 cells (Fig. 8A, filled circles), but this effect was either lost or significantly diminished in myc−/− cells (filled triangles). Infection of myc−/− cells with AdGFP-Myc at the time of serum stimulation restored induction of these genes, with similar time courses as those observed in parental Rat1 cells (Fig. 8A, open circles), but the control virus AdGFP had no effect (open triangles). AIRC, the gene adjacent to GPAT (Fig. 1), was not significantly induced by serum in our experiments. However, AIRC and GPAT were induced upon Myc overexpression in B-cells (Schuhmacher et al. 2001), suggesting that both are functional targets of Myc. We were unable to reliably measure α-PT mRNA levels by real-time PCR. Other genes, such as RPS9 (Fig. 8B), were unaffected by serum in either Rat1 or myc−/− cells. Serum-responsive genes such as c-fos, NGFI-B, and TIS 11 were induced normally, or even to higher levels in myc−/− cells, showing that these cells are responsive to serum mitogens (Fig. 8B). In conclusion, at the target loci studied here, Myc is essential for both H4 acetylation and transcriptional activation in response to mitogens.
Figure 8.
Induction of Myc-target genes by mitogens is Myc-dependent. Relative mRNA levels for (A) Myc-target genes and (B) control genes, quantified by reverse transcription and real-time PCR. Rat1 cells (filled circles) and myc−/− cells (filled triangles) were analyzed at the indicated time points following mitogenic stimulation. To restore Myc expression in myc−/− cells, these cells were infected at the time of serum stimulation with the recombinant adenovirus AdGFP-Myc (open circles) or with the control virus AdGFP (open triangles). Expression of each gene is represented as the fold-induction relative to untreated quiescent cells (time 0).
It was concluded in a previous study that induction of cad by serum was lost in the same myc−/− cell line used here, whereas other target genes, including ODC, were normally induced (Bush et al. 1998). This contrasts with the defective response of ODC in myc−/− cells (Figs. 8A, 9). In addition, we tested multiple Myc-target genes without identifying any that is unaffected in myc−/− cells (M. Schroeder, S.R. Frank, and B. Amati, unpubl.). The reference gene used to normalize mRNA levels in the previous study was GAPDH. This gene, however, has shown serum- and Myc-dependent changes in different cell types (Tavtigian et al. 1994; Boon et al. 2001), and a clearly delayed serum response in myc−/− cells, as is evident in both the previous data (Bush et al. 1998) and our own (Fig. 8B). Regardless of the reasons for this defect, the use of GAPDH as a reference has the potential to mask Myc-dependent changes in gene expression.
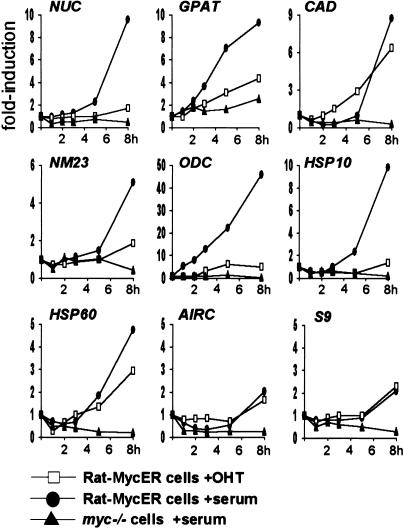
Figure 9.
Mitogenic stimuli and activation of Myc alone are not equivalent in inducing expression of Myc-target genes. Quiescent Rat1-MycER cells were stimulated with either OHT alone (open squares) or serum alone (filled circles). Serum-stimulated myc−/− cells are shown as a reference (filled triangles). mRNA expression was quantified as in Figure 8.
To examine the kinetics of gene activation relative to Myc-binding and histone acetylation, we analyzed mRNA induction by serum in an abbreviated time course (Fig. 9, filled circles). These experiments revealed that the kinetics of mRNA accumulation were slightly delayed compared to H4 acetylation at Myc-target sites. At 4 h, for example, acetylation was already maximal (Figs. 3B, 4A), while mRNA accumulation was not yet apparent for most target genes (Fig. 9). This is consistent with acetylation preceding transcription, and mRNAs accumulating steadily thereafter.
Because MycER was sufficient to induce H4 acetylation, we monitored mRNA induction by MycER (Fig. 9, open squares) and compared it to the effect of serum (filled circles). This analysis revealed that MycER induced only CAD and HSP60 comparably to serum. As noted above for the serum response, MycER-induced acetylation on CAD and HSP60 (Fig. 6B) preceded gene expression (Fig. 9): acetylation levels peaked within <1 h, whereas mRNA accumulation started at ∼3 h. Most strikingly, MycER was ineffective in activation of other target genes (Fig. 9), and analogous observations were made with longer time courses (data not shown). Interestingly, although NUC was not appreciably induced by MycER, it was efficiently acetylated (Fig. 6B). Therefore, Myc-induced acetylation does not systematically result in gene expression. In conclusion, although Myc is critical for the transcriptional activation of its target genes, it is not always sufficient to elicit a full transcriptional response.
Discussion
Myc mediates serum-induced acetylation of histone H4 and gene activation
Several immediate-early serum-responsive genes, such as c-myc, c-fos, or c-jun, encode transcription factors that are believed to drive induction of delayed-early, or secondary mitogen-responsive genes (Bravo 1990; Winkles 1998). Consistent with this view, Myc/Max dimers function as sequence-specific transcriptional activators, but their precise role and mode of action in mitogen-induced transcription have remained obscure (Cole and McMahon 1999; Grandori et al. 2000; Amati et al. 2001). In this work, we use a quantitative chromatin immunoprecipitation (ChIP) assay to demonstrate that Myc mediates serum-induced acetylation of histone H4 at seven distinct target loci. In a parallel study, Bouchard et al. (2001) report that Myc regulates cyclin D2 by a similar mechanism.
Upon serum stimulation of quiescent Rat1 fibroblasts, we observed binding of Myc in vivo to E-boxes in the following target genes: NUC, CAD, NM23-H2, α-PT, ODC, and the linked gene pairs GPAT/AIRC and HSP10/60 (see maps in Fig. 1). Simultaneously with Myc-binding, serum enhanced histone H4 acetylation at these sites. Enhancement was maximal over E-boxes and spread at variable, discrete distances on either side. The effects of serum on H4 acetylation were lost in a c-myc-deficient Rat1 cell line (myc−/−; Mateyak et al. 1997), but were restored by adenoviral delivery of Myc. Altogether, the strict dependence of serum-induced acetylation upon Myc and its localized nature strongly suggested that acetylation was directly initiated by Myc. Indeed, activation of a MycER chimera (Littlewood et al. 1995) in quiescent cells rapidly induced H4 acetylation, with distribution patterns resembling those induced by serum. Reciprocally, ectopic expression of wild-type Myc prevented deacetylation of histone H4 upon cell-cycle exit. In summary, upon binding to chromatin, Myc induces localized acetylation of histone H4 and, as such, is a key component in translating mitogenic stimuli into chromatin modifications.
Besides H4 acetylation, Myc also mediates gene activation. Myc-target genes were rapidly induced by serum in Rat1 cells but not in myc−/− cells, whereas reconstitution of myc−/− cells with AdGFP-Myc restored expression, showing that Myc is essential for this response. Time-course experiments showed that mRNA accumulation closely followed H4 acetylation. Altogether, these observations strongly suggest that acetylation is an essential preliminary step in transcriptional activation of Myc-target genes.
In spite of its essential role, Myc was not always sufficient for a full transcriptional response. Indeed, activation of MycER alone in quiescent cells induced most target genes only poorly in comparison to serum stimulation. This might be for multiple reasons. For example, some Myc-target sites in chromatin remained poorly accessible in quiescent cells, and may require additional chromatin-remodeling events prior to Myc-binding. Indeed, in our experiments, MycER bound only one target site (GPAT) with the same efficiency as endogenous Myc in serum-stimulated cells. Alternatively, as we observed on NUC and GPAT, Myc alone may induce acetylation without being sufficient to induce gene expression. Together with previous observations in other systems (e.g., Krebs et al. 1999), these data suggest that H4 acetylation is only one step in gene activation. We propose that Myc-induced acetylation plays an essential permissive role, allowing gene activation by additional signals, normally triggered by mitogens. These signals may be multiple, and may differ among Myc-target genes and/or cell types, explaining why some genes are responsive to Myc alone in a given cell line (e.g., CAD; Fig. 9), but most others are not.
In previous studies, MycER activation in quiescent cells has been used as a hallmark in defining Myc-target genes (Cole and McMahon 1999; Amati et al. 2001). Recent screens based on cDNA microarrays made use of this system (Coller et al. 2000; O'Hagan et al. 2000). These conditions undoubtedly allowed identification of bona fide Myc-target genes, but revealed only few genes and, as illustrated by our data, very suboptimal transcriptional responses. Although activation of Myc (or MycER) alone can induce cellular responses such as cell cycle entry and apoptosis (e.g., Eilers et al. 1991; Evan et al. 1992), these effects are sometimes dependent on other signals (e.g., Leone et al. 1997). We assume that the gene-expression programs governed by Myc in different cell types will be fully revealed only upon combinatorial activation of signaling pathways, as achieved by complex mitogenic stimuli or multiple oncogenic lesions.
Myc-mediated acetylation: which HAT?
We have demonstrated here that Myc recruits TRRAP to chromatin. We have also shown that Myc-induced acetylation of histone H4 requires two domains in the Myc N terminus, which span Myc-boxes I and II, required for association with TRRAP (McMahon et al. 1998). TRRAP is not only a Myc-binding protein, but is also a subunit of at least two classes of HAT complexes (Vassilev et al. 1998; Brand et al. 1999; Ikura et al. 2000). The first class includes the related complexes PCAF, GCN5, STAGA, and TFTC (the last three likely being isolates of the same complex), which contain either one of the homologous HAT subunits PCAF or GCN5 (Martinez et al. 1998; Ogryzko et al. 1998; Brand et al. 1999). Consistent with its interaction with TRRAP, Myc also associates with GCN5 in cells (McMahon et al. 2000). The second TRRAP-containing complex includes the HAT subunit Tip60 (Ikura et al. 2000). Two other subunits of this complex, the ATPases Tip48 and Tip49, are also associated with Myc in cells (Wood et al. 2000). These observations suggest that Myc may be able to interact with both the Tip60 and GCN5 complexes, and that one of these is likely to be involved in Myc-induced acetylation.
One salient feature of the acetylation events reported here is their selectivity toward histone H4, whereas no significant effects of serum and/or Myc were seen on H3. Many in vitro experiments indicate that GCN5/PCAF and Tip60/NuA4 acetylate nucleosomal histones with tight selectivities toward H3 and H4, respectively (Smith et al. 1998; Xu et al. 1998; Allard et al. 1999; Brand et al. 1999; Ikura et al. 2000; for review, see Sterner and Berger 2000). Consistent with these in vitro specificities, loss of GCN5 and ESA1 (the Tip60 homolog) in yeast led to selective defects in acetylation of H3 and H4 (Kuo et al. 2000; Reid et al. 2000; Vogelauer et al. 2000). Therefore, although our observations do not rule out a role for GCN5 in Myc-mediated H4 acetylation in vivo, they raise the possibility that Tip60 might be involved instead. It remains equally possible that in other cell types and physiological circumstances, or at different target loci, Myc may regulate H3 acetylation through the recruitment of GCN5.
Finally, Myc may contribute mechanisms other than H4 acetylation to activate transcription. In particular, Myc binds INI1/SNF5 (Cheng et al. 1999), a subunit of the chromatin remodeling complex SWI/SNF (Wang et al. 1996). In transient transfection assays, Myc-activated transcription was suppressed by a dominant-negative mutant of the SWI/SNF catalytic subunit BRG1 (Cheng et al. 1999). There is a wealth of evidence indicating that SWI/SNF and HAT complexes have interdependent functions in gene regulation (e.g., Hassan et al. 2001; for reviews, see Kingston and Narlikar 1999; Fry and Peterson 2001). For example, at the yeast HO promoter, the transcription factor Swi5 is required for the sequential recruitment of SWI/SNF and SAGA complexes, prior to DNA-binding by another factor (SBF) and transcriptional activation (Cosma et al. 1999; Krebs et al. 1999). Based on these observations, we previously speculated that Myc controls chromatin modifications by both SWI/SNF and HAT complexes (Amati et al. 2001). We demonstrate here that Myc mediates acetylation of histone H4. It remains to be seen whether it recruits the SWI/SNF complex and induces chromatin remodeling, in concert with the recruitment of TRRAP and a yet unidentified HAT.
Materials and methods
Cells and adenoviruses
Rat1 and myc−/− cells (Mateyak et al. 1997), and Rat1 cells expressing the chimeric protein MycER (Littlewood et al. 1995) were cultured in D-MEM medium supplemented with 10% fetal-calf serum (FCS). Cells were rendered quiescent by growth to confluent density followed by incubation for 2 d in serum-free medium. To induce entry into the cell cycle, cells were harvested by trypsinization and reseeded onto plates containing D-MEM/10% serum, at densities of either 1:3 (for ChIP) or 1:6 (for mRNA analysis). For analysis of Myc by ChIP, cells from 2–3 confluent 150-mm dishes, or the equivalent amount of cells following splitting, were used per time point. For ChIP with antibodies to acetylated histones, cells from 1 confluent 150-mm dish (or equivalent) were used per time point.
The recombinant adenovirus AdGFP-Myc was constructed by ligation of a human c-myc cDNA into the plasmid vector AdTrack-CMV; followed by recombination in Escherichia coli, viral production, and amplification in 293T cells; and purification of recombinant adenoviruses as previously described by He et al. (1998). To improve gene delivery, adenoviral infections were performed using a modification of a previously described procedure (Fasbender et al. 1997): to infect cells from a single confluent 150-mm dish, 4 × 1010 viral particles were diluted in 2.8 mL of serum-free medium containing 2.3 μL of Superfect (QIAGEN). The mixture was incubated for 10 min to allow virus/lipid complex formation, and was added to cells at the time of replating.
Generation of TRRAP-specific antibodies
The antiserum TRRAP-CT was raised against a chemically synthesized, KLH-conjugated peptide comprising the C-terminal amino acids of human TRRAP (KVNTLVAAANSLDNLCRM DPAWHPWL). Antibodies were affinity purified by binding to peptide-conjugated Sulfolink gel (Pierce). The corresponding preimmune serum was purified by affinity on a Protein A column. TRRAP-CT specifically recognized a transfected C-terminal fragment of TRRAP (McMahon et al. 1998) as well as the cellular TRRAP protein, but immunoprecipitation of endogenous TRRAP was dependent on prior denaturation with SDS, analogous to the conditions used in our ChIP assay (Fig. 7A; data not shown). The identity of the immunoprecipitated protein as TRRAP was confirmed by immunoblotting with an antibody directed against a different TRRAP peptide (L. Deleu and H. Land, unpubl.).
Chromatin immunoprecipitation (ChIP) assay
Rat1 cells and their derivatives were cultured as above. Formaldehyde (Fisher) was added to the culture medium to a final concentration of 1%. Cross-linking was allowed to proceed for 10 min at room temperature and stopped by addition of glycine at a final concentration of 0.125 M, followed by an additional incubation for 5 min. Fixed cells were washed twice with TBS (20 mM Tris at pH 7.4, 150 mM NaCl) and harvested in SDS Buffer (50 mM Tris at pH 8.1, 0.5% SDS, 100 mM NaCl, 5 mM EDTA, and protease inhibitors). Cells were pelleted by centrifugation, and suspended in 4 mL of IP Buffer (100 mM Tris at pH 8.6, 0.3% SDS, 1.7% Triton X-100, and 5 mM EDTA). Cells were disrupted by sonication with a 1/4-in.-diameter tapered probe for 20 sec in a Branson 250 sonicator, at a power setting of 3 and 100% duty cycle, yielding genomic DNA fragments with a bulk size of 100–400 bp. The lysate was then diluted with IP buffer to a final volume of 9 mL. For each immunoprecipitation, 1 mL of diluted lysate was precleared by addition of 30 μL of blocked protein A beads (50% slurry protein A-Sepharose, Amersham; 0.5 mg/mL fatty acid-free BSA, Sigma; and 0.2 mg/mL salmon sperm DNA in TE). Samples were immunoprecipitated overnight at 4°C with polyclonal antibodies specific for either c-Myc (2 μg N262, Santa Cruz, Cat. no. SC764), acetylated H3 (3 μg, Upstate Biotech, Cat. no. 06-599), acetylated H4 (3 μL, Upstate Biotech, Cat. no. 06-866), or TRRAP (affinity-purified TRRAP-CT, 2 μg). Immune complexes were recovered by adding 30 μL of blocked protein A beads and incubated for 2 h at 4°C. Beads were washed and eluted, and cross links were reversed as described by Aparicio (1999). This included successive washes in 1 mL of Mixed Micelle Buffer (20 mM Tris at pH 8.1, 150 mM NaCl, 5 mM EDTA, 5% w/v sucrose, 1% Triton X-100, and 0.2% SDS), Buffer 500 (50 mM HEPES at pH 7.5, 0.1% w/v deoxycholic acid, 1% Triton X-100, 500 mM NaCl, and 1 mM EDTA), LiCl Detergent Wash Buffer (10 mM Tris at pH 8.0, 0.5% deoxycholic acid, 0.5% NP-40, 250 mM LiCl, and 1 mM EDTA), and TE (pH 7.5). The eluted material was phenol/chloroform-extracted and ethanol-precipitated. DNA was resuspended in 300–600 μL of water. PCR was performed with 9 μL of DNA and 800 nM primers diluted to a final volume of 30 μL in SYBR Green Reaction Mix (Perkin Elmer). Accumulation of fluorescent products was monitored by real-time PCR using a GeneAmp 5700 Sequence Detector (Perkin Elmer). Each PCR reaction generated only the expected specific amplicon, as shown by the melting-temperature profiles of final products (dissociation curve, automatically measured by the Taqman 5700) and by gel electrophoresis of test PCR reactions. No PCR products were observed in the absence of template.
Primers for ChIP analysis
The following primers were used to amplify rat genomic sequences at Myc-target loci, including the amplicons shown in Figure 1. NUC (GenBank no. M55015), amplicon (−770): CAG GCAGGCCGAGTACTTCT and TCCAGGTTCAGAGAGG TGACTTC; (−12): CAAGCTCAGTCTTTTGCCTCAGA and GGATCGGCGGGTAATGAAG; (+574): CGCGTCCGAGGCA GTG and TCCATCTACCGTCACGGTCAG; (+1500): CCAC GACAAATGACAAGTTTAGTGA and AAGCTGAGACCCT GATTCCATCT; (+2334): GGTAGTAAAAGGAGTGAAACC AGCA and GAGGCAGGAGCAGCAGGAG. AIRC/GPAT (GenBank no. D37978), amplicon (−1969): GAGCAGCAAAC AGCCACAAG and ACCTTAGCAGTTTCACTCTTTCAGC; (−932): GTTTATCGCACGTCTCCAAGC and CGTTTGGAT CTTAGCCCTTCTG; (−117): AAGGAGGAGGCAGTCTGTA GCTC and AGAAGCGATGCACCCGAA. NM23-H2 (GenBank no. D89068), amplicon (−343): GTAGTGTTGGCTGAAG ACAGACTTG and TATGTCTTCCTCATCCAAAATGACA; (−53): CTCCGTACAAAGCCCTGCA and TGATCTTGCCAG TCGTCTCTGT; (+169): ACCAGCTTTCGGTAAGGCTTG and GGACACGTGGCTTTCCAGAT. α-PT (GenBank no. S70441 and S69916), promoter: TCCCTGTACTCAACCAA TAGCGT and GCGTTGGAGGAGACGGTG; intron 1 E-box: CACCGCTCACCCAGAGAACT and GGCAAACTCGCTCA CCTAGATG. ODC (GenBank no. 07944), amplicon (−438): TTCCAGGCCAGGGCTCA and TGGTCGTCCGTCTTGCA AC; (−143): TCGGGAACCGGGTTGG and GTCGTCATG GAGACGCACC; (+268): AGGCTGGTGACCTTGCGA and TGACACGATGCGGGCTC; (+1035): GCCAAAGCCCATTC ATCAGT and CGCCAGCCAACTCAGTTGTAT; (+1941): CGTAGTTCTGACCTCACAGTTGATC and TCTTACCATT AAAGGTTACACACTGCA. HSP10/60 (GenBank no. U68562), amplicon (−467): CGTGAAAAGACAGCGCAGC and CCGCC TTCTCTCACGCTAAG; (+169): AGGCTCGGTTCACTTGT TCAG and TGATGAAAGCAGTATGGCCCTAC. CAD promoter: GCCGTCGCAGTCGTGCT and ACCGACCCGTCC TCCAA. In the absence of the rat sequence, mouse CAD is shown in Figure 1. The 5′ primer is conserved in hamster (M31621) and mouse (AF053338). The 3′ primer lies shortly after the start codon and is conserved in hamster, mouse, and human (NM_004341). The E-box in conserved in all species. The cap site shown in Figure 1 was deduced from NM_004341.
The following primers were used to amplify E-boxes in promoters not bound by Myc (Fig. 2B). GLU (GenBank no. X94615): CAGTGTTCTGTCATCCTGTCTCATAG and ATACCCTGC CTAGTGTCACAAGG. GLY (GenBank no. X07833): CAAGC GCCGTCAGGATG and CAGTACGGCTGCGGGTG. The following primers were used to amplify control promoters without E-boxes (Fig. 2B). PCNA (GenBank no. X67329): TCAAACCAC GGATACGATTGG and CCCGCAACCGTCTAATGC. ACHR (GenBank no. L22646): TGCCTCGGGTGAACTAAGATG and GCCTCATTCGTCTTGGGAACT.
mRNA analysis
Total RNA was extracted with an RNAeasy Minikit (QIAGEN), including a DNase treatment before elution from the column. Five μg of RNA was reverse-transcribed for 50 min at 42 °C in a 100-μL reaction containing 2.5 μg of Oligo-dT, 250 ng of random hexamers (Boehringer), 1× first strand buffer (GIBCO BRL), 10 mM DTT, 0.5 mM deoxynucleotides (A, G, C, T), 80 units of RNAsin (Roche), and 500 units of Superscript II RT (GIBCO BRL). Real-time PCR was performed in a GeneAmp 5700 Sequence Detector (Perkin Elmer). Each PCR reaction contained 10 ng of cDNA template and primers at a concentration of 400 nM in a final volume of 20 μL of SYBR Green Reaction Mix (Perkin Elmer).
The following primers were used to amplify rat cDNA following reverse transcription. NUC (GenBank no. AH002217): CTCCAGCAAAGAAGGTGGTTGT and CAGGTGTTGGAA CTACTTTGGCT. AIRC (GenBank no. D37979): GAGTCAT GCCACACAAGC and GCCATAGTCTCCAGGAATC. GPAT (GenBank no. D10853): ATGTAGGAAGAACCTTCATTC and TGTTTATTCCCATGAAGCAC. NM23-H2 (GenBank no. X58965, human): TGGTGGGCGAGATCATCAA and TCAG GTCAATGTAGTGCTGCTTC. ODC (GenBank no. M16982): ACTGAAATACAGTTGGTGC and AATCATCAGTGGCA ATCC. HSP10 (GenBank no. U68562): AAAGGTGGCAT TATGCTTCC and TCCAACTTTCACACTGACAG. HSP60 (GenBank no. U68562): ACAAGTGATGTTGAAGTGAATG and ATTGCAGGAATTTTAAGTGCTC. CAD (GenBank no. AB007768): TGCGCCGTGTGGTCTTG and GGCCACTTCC GTACATCTTGTC. GAPDH (GenBank no. NM_017008): CC ATGCCATCACTGCCAC and GGGTAGGAACACGGAAGG. RPS9 (GenBank no. NM_031108): GGGATGTTCACCACCTG and GCAAGATGAAGCTGGATTAC. NGFI-B (or NR4A, GenBank no. U17254): GCTTGGGTGTTGATGTTCC and AG CAGACGTGACAGGCAG. TIS 11 (or Zfp36, Nup475; GenBank no. X63369): TCACCCTCACCTACTTCG and AGTTCCGT TTTGTACTTGG. c-fos (GenBank no. X06769): CTTTGATGA CTTCTTGTTTCCG and GCTCCAGCTCTGTGACCA. For most genes, the expression patterns shown in Figures 8 and 9 were confirmed with a different primer pair.
Acknowledgments
We are grateful to Steen Hansen, Emma Lees, and David Parry for thoughtful discussions and comments on the manuscript. We thank Sandra Zurawski and Sylvia Lo for the amplification and purification of adenoviruses, Stephen Hurst and Tom Morrison for their guidance with real-time PCR, and Konstantinos Alevizopoulos for his initial help in generating the TRRAP-CT antibody. We also thank Gerard Evan, David Hancock, and Trevor Littlewood for Rat1-MycER cells, John Sedivy for the myc−/− cell line, Michael Cole and Steven McMahon for TRRAP-encoding plasmids, Hartmut Land and Laurent Deleu for providing anti-TRRAP peptide sera, Bert Vogelstein for adenoviral vectors, and Martin Eilers and Bernhard Lüscher for communicating their unpublished data. S.T. was supported by an H.F.S.P.O. grant to B.A. DNAX Research Institute is supported by Schering-Plough.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL bruno.amati@dnax.org; FAX (650) 496-1200.
Article and publication are at http://www.genesdev.org/cgi/doi/10.1101/gad.906601.
References
- Agalioti T, Lomvardas S, Parekh B, Yie J, Maniatis T, Thanos D. Ordered recruitment of chromatin modifying and general transcription factors to the IFN-β promoter. Cell. 2000;103:667–678. doi: 10.1016/s0092-8674(00)00169-0. [DOI] [PubMed] [Google Scholar]
- Allard S, Utley RT, Savard J, Clarke A, Grant P, Brandl CJ, Pillus L, Workman JL, Cote J. NuA4, an essential transcription adaptor/histone H4 acetyltransferase complex containing Esa1p and the ATM-related cofactor Tra1p. EMBO J. 1999;18:5108–5119. doi: 10.1093/emboj/18.18.5108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amati B, Frank SR, Donjerkovic D, Taubert S. Function of the c-Myc oncoprotein in chromatin remodeling and transcription. Biochim Biophys Acta. 2001;1471:M135–M145. doi: 10.1016/s0304-419x(01)00020-8. [DOI] [PubMed] [Google Scholar]
- Aparicio OM. Characterization of proteins bound to chromatin by immunoprecipitation from whole-cell extracts. In: Ausubel FM, Brent R, Kingston RE, Moore DM, Seidman JG, Smith JA, Struhl K, editors. Current protocols in molecular biology. New York: John Wiley; 1999. pp. 21.23.21–21.23.12. [Google Scholar]
- Ayer DE, Eisenman RN. A switch from Myc:Max to Mad:Max heterocomplexes accompanies monocyte/macrophage differentiation. Genes & Dev. 1993;7:2110–2119. doi: 10.1101/gad.7.11.2110. [DOI] [PubMed] [Google Scholar]
- Bello-Fernandez C, Packham G, Cleveland JL. The ornithine decarboxylase gene is a transcriptional target of c-Myc. Proc Natl Acad Sci. 1993;90:7804–7808. doi: 10.1073/pnas.90.16.7804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boon K, Caron HN, van Asperen R, Valentijn L, Hermus MC, van Sluis P, Roobeek I, Weis I, Voute PA, Schwab M, et al. N-myc enhances the expression of a large set of genes functioning in ribosome biogenesis and protein synthesis. EMBO J. 2001;20:1383–1393. doi: 10.1093/emboj/20.6.1383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouchard C, Dittrich O, Kiermaier A, Dohmann K, Menkel A, Eilers M, Lüscher B. Regulation of cyclin D2 gene expression by the Myc/Max/Mad network: Myc-dependent TRRAP recruitment and histone acetylation at the cyclin D2 promoter. Genes & Dev. 2001;15:2042–2047. doi: 10.1101/gad.907901. (this issue). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyd KE, Farnham PJ. Myc versus USF: Discrimination at the cad gene is determined by core promoter elements. Mol Cell Biol. 1997;17:2529–2537. doi: 10.1128/mcb.17.5.2529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ————— Coexamination of site-specific transcription factor binding and promoter activity in living cells. Mol Cell Biol. 1999;19:8393–8399. doi: 10.1128/mcb.19.12.8393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyd KE, Wells J, Gutman J, Bartley SM, Farnham PJ. c-Myc target gene specificity is determined by a post-DNA binding mechanism. Proc Natl Acad Sci. 1998;95:13887–13892. doi: 10.1073/pnas.95.23.13887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brand M, Yamamoto K, Staub A, Tora L. Identification of TATA-binding protein-free TAFII-containing complex subunits suggests a role in nucleosome acetylation and signal transduction. J Biol Chem. 1999;274:18285–18289. doi: 10.1074/jbc.274.26.18285. [DOI] [PubMed] [Google Scholar]
- Bravo R. Growth factor-responsive genes in fibroblasts. Cell Growth Differentiation. 1990;1:305–309. [PubMed] [Google Scholar]
- Bush A, Mateyak M, Dugan K, Obaya A, Adachi S, Sedivy J, Cole M. c-myc null cells misregulate cad and gadd45 but not other proposed c-Myc targets. Genes & Dev. 1998;12:3797–3802. doi: 10.1101/gad.12.24.3797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H, Lin RJ, Xie W, Wilpitz D, Evans RM. Regulation of hormone-induced histone hyperacetylation and gene activation via acetylation of an acetylase. Cell. 1999;98:675–686. doi: 10.1016/s0092-8674(00)80054-9. [DOI] [PubMed] [Google Scholar]
- Cheng SW, Davies KP, Yung E, Beltran RJ, Yu J, Kalpana GV. c-MYC interacts with INI1/hSNF5 and requires the SWI/SNF complex for transactivation function. Nat Genet. 1999;22:102–105. doi: 10.1038/8811. [DOI] [PubMed] [Google Scholar]
- Cole MD, McMahon SB. The Myc oncoprotein: A critical evaluation of transactivation and target gene regulation. Oncogene. 1999;18:2916–2924. doi: 10.1038/sj.onc.1202748. [DOI] [PubMed] [Google Scholar]
- Coller HA, Grandori C, Tamayo P, Colbert T, Lander ES, Eisenman RN, Golub TR. Expression analysis with oligonucleotide microarrays reveals that MYC regulates genes involved in growth, cell cycle, signaling, and adhesion. Proc Natl Acad Sci. 2000;97:3260–3265. doi: 10.1073/pnas.97.7.3260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cosma MP, Tanaka T, Nasmyth K. Ordered recruitment of transcription and chromatin remodeling factors to a cell cycle- and developmentally regulated promoter. Cell. 1999;97:299–311. doi: 10.1016/s0092-8674(00)80740-0. [DOI] [PubMed] [Google Scholar]
- Desbarats L, Gaubatz S, Eilers M. Discrimination between different E-box-binding proteins at an endogenous target gene of c-myc. Genes & Dev. 1996;10:447–460. doi: 10.1101/gad.10.4.447. [DOI] [PubMed] [Google Scholar]
- Eberhardy SR, D'Cunha CA, Farnham PJ. Direct examination of histone acetylation on myc target genes using chromatin immunoprecipitation. J Biol Chem. 2000;275:33798–33805. doi: 10.1074/jbc.M005154200. [DOI] [PubMed] [Google Scholar]
- Eilers M, Schirm S, Bishop JM. The MYC protein activates transcription of the α-prothymosin gene. EMBO J. 1991;10:133–141. doi: 10.1002/j.1460-2075.1991.tb07929.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evan GI, Wyllie AH, Gilbert CS, Littlewood TD, Land H, Brooks M, Waters CM, Penn LZ, Hancock DC. Induction of apoptosis in fibroblasts by c-Myc protein. Cell. 1992;69:119–128. doi: 10.1016/0092-8674(92)90123-t. [DOI] [PubMed] [Google Scholar]
- Fasbender A, Zabner J, Chillon M, Moninger TO, Puga AP, Davidson BL, Welsh MJ. Complexes of adenovirus with polycationic polymers and cationic lipids increase the efficiency of gene transfer in vitro and in vivo. J Biol Chem. 1997;272:6479–6489. doi: 10.1074/jbc.272.10.6479. [DOI] [PubMed] [Google Scholar]
- Fry CJ, Peterson CL. Chromatin remodeling enzymes: Who's on first? Curr Biol. 2001;11:R185–R197. doi: 10.1016/s0960-9822(01)00090-2. [DOI] [PubMed] [Google Scholar]
- Gaubatz S, Meichle A, Eilers M. An E-box element localized in the first intron mediates regulation of the prothymosin α gene by c-myc. Mol Cell Biol. 1994;14:3853–3862. doi: 10.1128/mcb.14.6.3853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grandori C, Cowley SM, James LP, Eisenman RN. The Myc/Max/Mad network and the transcriptional control of cell behavior. Annu Rev Cell Dev Biol. 2000;16:653–699. doi: 10.1146/annurev.cellbio.16.1.653. [DOI] [PubMed] [Google Scholar]
- Greasley PJ, Bonnard C, Amati B. Myc induces the nucleolin and BN51 genes: Possible implications in ribosome biogenesis. Nucleic Acids Res. 2000;28:446–453. doi: 10.1093/nar/28.2.446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hassan AH, Neely KE, Workman JL. Histone acetyltransferase complexes stabilize swi/snf binding to promoter nucleosomes. Cell. 2001;104:817–827. doi: 10.1016/s0092-8674(01)00279-3. [DOI] [PubMed] [Google Scholar]
- He TC, Zhou S, da Costa LT, Yu J, Kinzler KW, Vogelstein B. A simplified system for generating recombinant adenoviruses. Proc Natl Acad Sci. 1998;95:2509–2514. doi: 10.1073/pnas.95.5.2509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikura T, Ogryzko VV, Grigoriev M, Groisman R, Wang J, Horikoshi M, Scully R, Qin J, Nakatani Y. Involvement of the TIP60 histone acetylase complex in DNA repair and apoptosis. Cell. 2000;102:463–473. doi: 10.1016/s0092-8674(00)00051-9. [DOI] [PubMed] [Google Scholar]
- Kingston RE, Narlikar GJ. ATP-dependent remodeling and acetylation as regulators of chromatin fluidity. Genes & Dev. 1999;13:2339–2352. doi: 10.1101/gad.13.18.2339. [DOI] [PubMed] [Google Scholar]
- Knoepfler PS, Eisenman RN. Sin meets NuRD and other tails of repression. Cell. 1999;99:447–450. doi: 10.1016/s0092-8674(00)81531-7. [DOI] [PubMed] [Google Scholar]
- Krebs JE, Kuo MH, Allis CD, Peterson CL. Cell cycle-regulated histone acetylation required for expression of the yeast HO gene. Genes & Dev. 1999;13:1412–1421. doi: 10.1101/gad.13.11.1412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuo MH, vom Baur E, Struhl K, Allis CD. Gcn4 activator targets Gcn5 histone acetyltransferase to specific promoters independently of transcription. Mol Cell. 2000;6:1309–1320. doi: 10.1016/s1097-2765(00)00129-5. [DOI] [PubMed] [Google Scholar]
- Leone G, DeGregori J, Sears R, Jakoi L, Nevins JR. Myc and Ras collaborate in inducing accumulation of active cyclin E/Cdk2 and E2F. Nature. 1997;387:422–426. doi: 10.1038/387422a0. [DOI] [PubMed] [Google Scholar]
- Littlewood TD, Hancock DC, Danielian PS, Parker MG, Evan GI. A modified oestrogen receptor ligand-binding domain as an improved switch for the regulation of heterologous proteins. Nucleic Acids Res. 1995;23:1686–1690. doi: 10.1093/nar/23.10.1686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luger K, Mader AW, Richmond RK, Sargent DF, Richmond TJ. Crystal structure of the nucleosome core particle at 2.8 Å resolution. Nature. 1997;389:251–260. doi: 10.1038/38444. [DOI] [PubMed] [Google Scholar]
- Martinez E, Kundu TK, Fu J, Roeder RG. A human SPT3–TAFII31–GCN5-L acetylase complex distinct from transcription factor IID. J Biol Chem. 1998;273:23781–23785. doi: 10.1074/jbc.273.37.23781. [DOI] [PubMed] [Google Scholar]
- Mateyak MK, Obaya AJ, Adachi S, Sedivy JM. Phenotypes of c-myc-deficient rat fibroblasts isolated by targeted homologous recombination. Cell Growth Differ. 1997;8:1039–1048. [PubMed] [Google Scholar]
- McMahon SB, Van Buskirk HA, Dugan KA, Copeland TD, Cole MD. The novel ATM-related protein TRRAP is an essential cofactor for the c-Myc and E2F oncoproteins. Cell. 1998;94:363–374. doi: 10.1016/s0092-8674(00)81479-8. [DOI] [PubMed] [Google Scholar]
- McMahon SB, Wood MA, Cole MD. The essential cofactor TRRAP recruits the histone acetyltransferase hGCN5 to c-Myc. Mol Cell Biol. 2000;20:556–562. doi: 10.1128/mcb.20.2.556-562.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miltenberger RJ, Sukow KA, Farnham PJ. An E-box-mediated increase in cad transcription at the G1/S-phase boundary is suppressed by inhibitory c-Myc mutants. Mol Cell Biol. 1995;15:2527–2535. doi: 10.1128/mcb.15.5.2527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogryzko VV, Kotani T, Zhang X, Schlitz RL, Howard T, Yang XJ, Howard BH, Qin J, Nakatani Y. Histone-like TAFs within the PCAF histone acetylase complex. Cell. 1998;94:35–44. doi: 10.1016/s0092-8674(00)81219-2. [DOI] [PubMed] [Google Scholar]
- O'Hagan RC, Schreiber-Agus N, Chen K, David G, Engelman JA, Schwab R, Alland L, Thomson C, Ronning DR, Sacchettini JC, et al. Gene-target recognition among members of the myc superfamily and implications for oncogenesis. Nat Genet. 2000;24:113–119. doi: 10.1038/72761. [DOI] [PubMed] [Google Scholar]
- Peña A, Reddy CD, Wu S, Hickok NJ, Reddy EP, Yumet G, Soprano DR, Soprano KJ. Regulation of human ornithine decarboxylase expression by the c-Myc/Max protein complex. J Biol Chem. 1993;268:27277–27285. [PubMed] [Google Scholar]
- Reid JL, Iyer VR, Brown PO, Struhl K. Coordinate regulation of yeast ribosomal protein genes is associated with targeted recruitment of Esa1 histone acetylase. Mol Cell. 2000;6:1297–1307. doi: 10.1016/s1097-2765(00)00128-3. [DOI] [PubMed] [Google Scholar]
- Schuhmacher M, Kohlhuber F, Holzel M, Kaiser C, Burtscher H, Jarsch M, Bornkamm GW, Laux G, Polack A, Weidle UH, et al. The transcriptional program of a human B cell line in response to Myc. Nucleic Acids Res. 2001;29:397–406. doi: 10.1093/nar/29.2.397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shim H, Dolde C, Lewis BC, Wu CS, Dang G, Jungmann RA, Dalla-Favera R, Dang CV. c-Myc transactivation of LDH-A: Implications for tumor metabolism and growth. Proc Natl Acad Sci. 1997;94:6658–6663. doi: 10.1073/pnas.94.13.6658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith ER, Eisen A, Gu W, Sattah M, Pannuti A, Zhou J, Cook RG, Lucchesi JC, Allis CD. ESA1 is a histone acetyltransferase that is essential for growth in yeast. Proc Natl Acad Sci. 1998;95:3561–3565. doi: 10.1073/pnas.95.7.3561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sterner DE, Berger SL. Acetylation of histones and transcription-related factors. Microbiol Mol Biol Rev. 2000;64:435–459. doi: 10.1128/mmbr.64.2.435-459.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stone J, de Lange T, Ramsay G, Jakobvits E, Bishop JM, Varmus H, Lee W. Definition of regions in human c-myc that are involved in transformation and nuclear localization. Mol Cell Biol. 1987;7:1697–1709. doi: 10.1128/mcb.7.5.1697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tavtigian SV, Zabludoff SD, Wold BJ. Cloning of mid-G1 serum response genes and identification of a subset regulated by conditional myc expression. Mol Biol Cell. 1994;5:375–388. doi: 10.1091/mbc.5.3.375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuneoka M, Nakano F, Ohgusu H, Mekada E. c-myc activates RCC1 gene expression through E-box elements. Oncogene. 1997;14:2301–2311. doi: 10.1038/sj.onc.1201067. [DOI] [PubMed] [Google Scholar]
- Vassilev A, Yamauchi J, Kotani T, Prives C, Avantaggiati ML, Qin J, Nakatani Y. The 400 kDa subunit of the PCAF histone acetylase complex belongs to the ATM superfamily. Mol Cell. 1998;2:869–875. doi: 10.1016/s1097-2765(00)80301-9. [DOI] [PubMed] [Google Scholar]
- Vogelauer M, Wu J, Suka N, Grunstein M. Global histone acetylation and deacetylation in yeast. Nature. 2000;408:495–498. doi: 10.1038/35044127. [DOI] [PubMed] [Google Scholar]
- Wagner AJ, Meyers C, Laimins LA, Hay N. c-Myc induces the expression and activity of ornithine decarboxylase. Cell Growth Diff. 1993;4:879–883. [PubMed] [Google Scholar]
- Wang W, Xue Y, Zhou S, Kuo A, Cairns BR, Crabtree GR. Diversity and specialization of mammalian SWI/SNF complexes. Genes & Dev. 1996;10:2117–2130. doi: 10.1101/gad.10.17.2117. [DOI] [PubMed] [Google Scholar]
- Winkles JA. Serum- and polypeptide growth factor-inducible gene expression in mouse fibroblasts. Prog Nucleic Acid Res Mol Biol. 1998;58:41–78. doi: 10.1016/s0079-6603(08)60033-1. [DOI] [PubMed] [Google Scholar]
- Wood MA, McMahon SB, Cole MD. An ATPase/helicase complex is an essential cofactor for oncogenic transformation by c-Myc. Mol Cell. 2000;5:321–330. doi: 10.1016/s1097-2765(00)80427-x. [DOI] [PubMed] [Google Scholar]
- Xu D, Popov N, Hou M, Wang Q, Bjorkholm M, Gruber A, Menkel AR, Henriksson M. Switch from Myc/Max to Mad1/Max binding and decrease in histone acetylation at the telomerase reverse transcriptase promoter during differentiation of HL60 cells. Proc Natl Acad Sci. 2001;98:3826–3831. doi: 10.1073/pnas.071043198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu W, Edmondson DG, Roth SY. Mammalian GCN5 and P/CAF acetyltransferases have homologous amino-terminal domains important for recognition of nucleosomal substrates. Mol Cell Biol. 1998;18:5659–5669. doi: 10.1128/mcb.18.10.5659. [DOI] [PMC free article] [PubMed] [Google Scholar]