Abstract
Gene-targeted FLX titanium pyrosequencing integrated with stable isotope probing (SIP) using [13C]biphenyl substrate revealed that tidal mudflat sediments harbor novel aromatic ring hydroxylating dioxygenases (ARHD). More than 80% of the detected ARHD genes comprise four clades (0.5 distance) with 49 to 70% amino acid identity to sequences in public databases. The 16S rRNA sequences enriched in the 13C fraction were from the Betaproteobacteria, bacilli (primarily Paenibacillus-like), and unclassified phyla.
TEXT
Stable isotope probing (SIP) was used in previous studies of aerobic biphenyl metabolism to determine the active members of biphenyl-metabolizing soil microbial communities and their aromatic ring hydroxylating dioxygenase (ARHD) genes (15, 20, 21). We extended this approach to marine tidal flats since this is a very different environment: salty, high in sulfate, with regular water exchange due to tidal cycles, and often anaerobic with depth. These systems have a remarkable remineralization capacity with a high microbial diversity (3, 10, 16, 22), and some, including the one studied, receive pollutants from industrial and urban sources (12). Since biphenyl is commonly produced from oil combustion and vehicle exhaust, it would be expected to reach these tidal flats. In this study, we characterized ARHD genes and the coenriched bacterial populations from a tidal mudflat metagenome using gene-targeted amplicon FLX titanium pyrosequencing of DNA fractionated by SIP.
Sediments were sampled from a previously characterized site of the Yeochari tidal flat, Kangwha, South Korea (37°36′30″N, 126°22′58″E). The Korean tidal flats are the fifth largest in area in the world. Three replicate microcosms of 5 g each were amended with 13C-labeled biphenyl and incubated for 14 days. After incubation, DNA was extracted from the three microcosms and combined to obtain a sufficient amount of DNA for SIP. [13C]DNA was separated from [12C]DNA by CsCl density gradient ultracentrifugation, and each band was carefully extracted with a syringe needle from ethidium bromide (EtBr)-containing gradients (17). 16S rRNA genes were PCR amplified from [12C]DNA and [13C]DNA fractions by using a 27F and 518R primer set (14). ARHD genes were amplified with a published 888F and 300R primer set (11) after multiple displacement amplification (MDA) of the limited DNA recovered from the [13C]DNA fractions. Gene-targeted pyrosequencing was performed by Macrogen, Inc. (Seoul, South Korea), using a 454/Roche GS-FLX titanium instrument (Roche, Nutley, NJ). The microbial community structure was analyzed with the pyrosequencing pipeline provided by the Ribosomal Database Project (RDP) (2, 5). Operational taxonomic units (OTUs) were clustered at 97% sequence identity, and the median sequence of the cluster was used to determine taxonomy and percent identity to the nearest neighbor in Greengenes (6). Sequences from the [12C]DNA that were also found in the [13C]DNA were assumed to be contaminating and removed. The ARHD sequences were subjected to BLASTX to identify the closest sequences in the nonredundant protein sequence (nr) database. The phylogenetic tree of ARHD amino acid sequences was constructed after alignment by MUSCLE (7) with MEGA4 (13). In order to compare the conserved region with the well-known dioxygenases from Pfam, an amino acid conservation analysis was carried out using ARHD sequences (9, 23). The experimental details are described in the supplemental material.
We analyzed 3,490 and 7,699 16S rRNA gene reads produced from the [12C]DNA and [13C]DNA fractions, respectively. The retained sequences passed a quality filter that allowed up to two mismatches in the forward primer, a minimum average exponential quality score of 20, no ambiguous bases, and a required length longer than 250 bp. The Shannon index values for OTUs in the [12C]DNA and [13C]DNA were 5.56 and 3.88, respectively, suggesting a reduced bacterial diversity in [13C]DNA. The rarefaction curve (see Fig. S1 in the supplemental material) for the [13C]DNA demonstrates that the richness of the biphenyl-metabolizing populations (97% clustering) was reduced from that of the background community ([12C]DNA) such that its OTU richness was captured by using SIP-integrated deep sequencing.
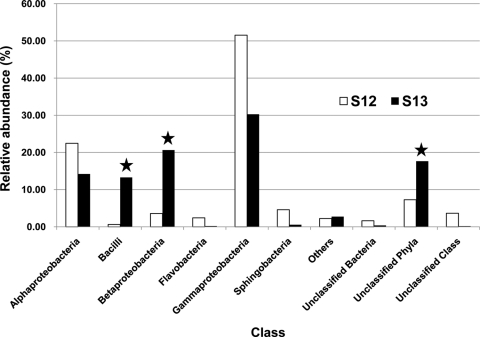
Bacteria that were significantly enriched in the [13C]DNA, compared to the [12C]DNA, belonged to Betaproteobacteria, bacilli, and unclassified phyla (P = 0.0097) (Fig. 1). A relatively high abundance of the unclassified group with the [13C]DNA indicated the possibility of novel biphenyl-degrading bacteria in this mudflat community. The dominant enriched members were affiliated with the genera Paenibacillus (28.6% of the total [13C]DNA), Pusillimonas (14.1%), and Alcaligenes (4.4%) (Table 1). Polychlorinated biphenyl (PCB) degradation capabilities have previously been reported for Paenibacillus and Alcaligenes isolated from PCB-contaminated soil (1, 18), whereas Pusillimonas isolated from polycyclic aromatic hydrocarbon (PAH)-contaminated soil has been reported to degrade pyrene (8). Notably, the dominant biphenyl-metabolizing populations found in this study were different from those in rhizosphere soils (15, 21) or river sediment (20) where the biphenyl-SIP approach was also used. Only Variovorax and Pseudomonas, detected by [13C]DNA with low abundances (0.56% and 0.10%, respectively), were found in those previous studies. These data suggest that the biphenyl-metabolizing bacteria from the tidal mudflat may contain a PCB degradative capability with a phylogenetic signature distinctly different from that of the terrestrial biphenyl-metabolizing populations.
Fig. 1.
Relative abundances of [12C]DNA (S12) and [13C]DNA (S13) in microbial community members. RDP Classifier was used at the class level with a 50% bootstrap threshold (4). Solid stars indicate the class members that exhibited significant enrichment in S13 compared to S12.
Table 1.
Phylogenetic classification of 16S rRNA genes detected only in the [13C]DNA (S13) fractiona
| S13 (%) | Closest relative by Greengenes | Accession no. | Class | Identity (%) |
|---|---|---|---|---|
| 6.25 | Pusillimonas terrae | DQ466075 | Betaproteobacteria | 98.74 |
| 5.65 | Paenibacillus sp. strain KO_CM21 | GQ497919 | Bacilli | 93.75 |
| 5.12 | Pusillimonas terrae | DQ466075 | Betaproteobacteria | 96.14 |
| 4.99 | Paenibacillus sp. Eur1 9.9 | DQ444978 | Bacilli | 87.93 |
| 4.92 | Paenibacillus sp. KCTC 13564 | GQ303568 | Bacilli | 88.10 |
| 1.92 | Pusillimonas terrae | DQ466075 | Betaproteobacteria | 97.54 |
| 1.90 | Paenibacillus favisporus strain GMP03 | AY308758 | Bacilli | 85.85 |
| 1.68 | Alcaligenes sp. strain L6 | X92415 | Betaproteobacteria | 97.03 |
| 1.68 | Paenibacillus sp. strain P-3 | AM411970 | Bacilli | 87.52 |
| 1.58 | Paenibacillus sp. strain KBC101 | AB186915 | Bacilli | 87.16 |
| 1.40 | Paenibacillus ruminocola strain CA8 | DQ085278 | Bacilli | 82.20 |
| 1.27 | Paenibacillus sp. strain KO_CM21 | GQ497919 | Bacilli | 92.25 |
| 0.82 | Paenibacillus sp. KCTC 13564 | GQ303568 | Bacilli | 88.70 |
| 0.78 | Paenibacillus validus strain SB 3263 | GU191921 | Bacilli | 99.74 |
| 0.70 | Paenibacillus koreensis strain YC300 | AF130254 | Bacilli | 88.16 |
| 0.68 | Alcaligenes sp. strain L6 | X92415 | Betaproteobacteria | 96.01 |
| 0.64 | Alcaligenes sp. strain L6 | X92415 | Betaproteobacteria | 97.16 |
| 0.64 | Paenibacillus koreensis strain YC300 | AF130254 | Bacilli | 87.02 |
| 0.56 | Variovorax paradoxus strain S110 | CP001635 | Betaproteobacteria | 100.00 |
| 0.55 | Bacillus mucilaginosus strain HSCC 1605T | AB045091 | Bacilli | 93.65 |
| 0.53 | Alcaligenes sp. strain L6 | X92415 | Betaproteobacteria | 95.24 |
| 0.53 | Paenibacillus ehimensis strain IFO 15659 | AB021184 | Bacilli | 86.05 |
| 0.51 | Pusillimonas terrae | DQ466075 | Betaproteobacteria | 96.80 |
Only taxa of >0.5% abundance are shown.
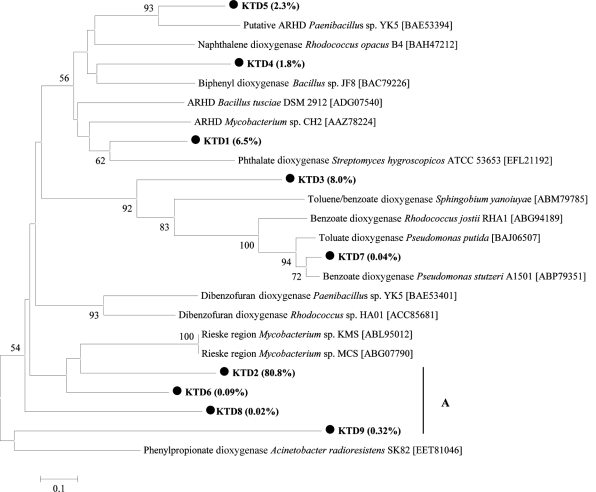
We also PCR amplified from the [13C]DNA and pyrosequenced ARHD gene fragments that contain coding regions for the Rieske-type [2Fe-2S] cluster. Using a strict quality filtering procedure described in the supplemental material, 4,924 ARHD gene reads (60.3%) were retained from a total of 8,169 reads. Most of these sequences (99.8%) coded for the conserved Rieske domain, which is essential for function and indicates that the retained sequences were for the correct protein. A BLASTX search against the nr database revealed best matches to other ARHD genes but that the obtained genes were diverse and different from those known. More than 80% of the detected ARHD genes were in clade A and had only 49 to 70% amino acid identity to known ARHD genes (Fig. 2). The clade represented by KTD2, which had 80.8% of the sequences, also contains considerable amino acid diversity (40 to 60% amino acid identity) since the clade was clustered at a distance of 0.5. A few of the sequences matched to a putative ring hydroxylating dioxygenase (2.3% of 4,664 reads) of Paenibacillus, which was a dominant genus in the 16S rRNA analysis. This suggests that Paenibacillus-like bacteria are capable of biphenyl metabolism in the tidal mudflat community.
Fig. 2.
Neighbor-joining phylogenetic tree (1,000 bootstraps) based on approximately 100-amino-acid sequences of ARHD genes. The Korean tidal flat dioxygenase (KTD) sequences are marked by dark circles. The number in the parentheses after each KTD sequence indicates the percentage of sequences in that cluster.
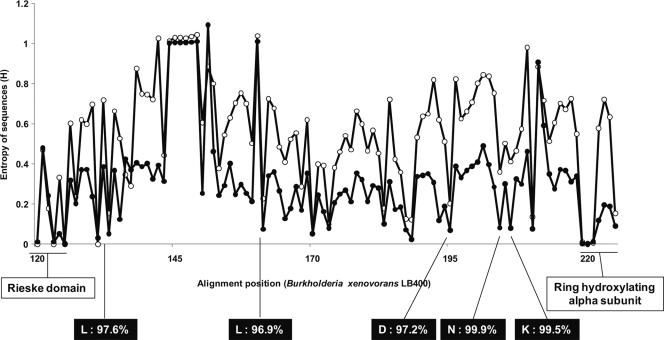
ARHD sequences were compared with 467 representative sequences (see the supplemental material) from the Pfam protein family database by using the amino acid conservation analysis method of Iwai et al. (9). Conservation analysis identified several conserved amino acids in the ARHD genes beyond those known to be important for function. They were located between the end of the Rieske [2Fe-2S] domain and the beginning of the ring hydroxylating alpha subunit (catalytic domain) (Fig. 3). Because even a single amino acid substitution in a functionally important, conserved region can enhance the rate of the aromatic oxygenation by biphenyl dioxygenase (19), these findings suggest that the host strains may exhibit different degradation kinetics and/or PCB congener specificities compared to the known ARHD.
Fig. 3.
Shannon entropy (H′) at each alignment position and conserved residues among the obtained and reference ARHD sequences. Open circles indicate the entropy of reference sequences, and solid circles indicate the entropy of the SIP-enriched (obtained) sequences. Known domains, amino acids, and their percentages of the total obtained sequences are shown in boxes.
Using the SIP-pyrosequencing method, we discovered apparently novel biphenyl-metabolizing bacteria and ARHD genes from a tidal mudflat. Bacteria similar to Paenibacillus, which are known to be a terrestrial PCB-degrading population (18), were identified as one of the dominant biphenyl-utilizing populations at this marine site.
Nucleotide sequence accession numbers.
All sequences have been deposited in the Short Read Archive database at NCBI (accession no. SRA028415, SRA028416, and SRA028424).
Supplementary Material
Acknowledgments
This research was supported by the WCU (World Class University) Program of the National Research Foundation of Korea, funded by the Ministry of Education, Science and Technology (R33-10076), and by NIEHS project P42-ES004911-20 under the Superfund Basic Science Program.
Footnotes
Supplemental material for this article may be found at http://aem.asm.org/.
Published ahead of print on 15 April 2011.
REFERENCES
- 1. Bedard D. L., Wagner R. E., Brennan M. J., Hanerl M. L., Brown J. F., Jr 1987. Extensive degradation of Aroclors and environmentally transformed polychlorinated biphenyls by Alcaligenes eutrophus H850. Appl. Envion. Microbiol. 53:1094–1102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Cardenas E., Cole J. R., Tiedje J. M., Park J. 2009. Microbial community analysis using RDP-II (Ribosomal Database Project II) methods, tools and new advances. Environ. Eng. Res. 14:3–9 [Google Scholar]
- 3. Carling P. A. 1982. Temporal and spatial variation in intertidal sedimentation rates. Sedimentology 29:17–23 [Google Scholar]
- 4. Claesson M. J., et al. 2009. Comparative analysis of pyrosequencing and a phylogenetic microarray for exploring microbial community structures in the human distal intestine. PLoS One 4:e6669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Cole J. R., et al. 2008. The Ribosomal Database Project: improved alignments and new tools for rRNA analysis. Nucleic Acids Res. 37:141–145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. DeSantis T. Z., et al. 2006. Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Appl. Environ. Microbiol. 72:5069–5072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Edgar R. C. 2004. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32:1732–1797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Hilyard E. J., Jones-Meehan J. M., Spargo B. J., Hill R. T. 2008. Enrichment, isolation, and phylogenetic identification of polycyclic aromatic hydrocarbon-degrading bacteria from Elizabeth River sediments. Appl. Environ. Microbiol. 74:1176–1182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Iwai S., et al. 2010. Gene-targeted-metagenomics reveals extensive diversity of aromatic dioxygenase genes in the environment. ISME J. 4:279–285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Kim B. S., et al. 2008. Rapid phylogenetic dissection of prokaryotic community structure in tidal flat using pyrosequencing. J. Microbiol. 46:357–363 [DOI] [PubMed] [Google Scholar]
- 11. Kitagawa W., Suzuki A., Hoaki T., Masai E., Fukuda M. 2001. Multiplicity of aromatic ring hydroxylation dioxygenase genes in a strong PCB degrader, Rhodococcus sp. strain RHA1 demonstrated by denaturing gradient gel electrophoresis. Biosci. Biotechnol. Biochem. 65:1907–1911 [DOI] [PubMed] [Google Scholar]
- 12. Koh C. 2001. The Korean tidal flat: environment, biology and human. Seoul National University Press, Seoul, Republic of Korea [Google Scholar]
- 13. Kumar S., Nei M., Dudley J., Tamura K. 2008. MEGA: a biologist-centric software for evolutionary analysis of DNA and protein sequences. Brief. Bioinform. 9:299–306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Lee T. K., et al. 2010. Discovery of commonly existing anode biofilm microbes in two different wastewater treatment MFCs using FLX titanium pyrosequencing. Appl. Microbiol. Biotechnol. 87:2335–2343 [DOI] [PubMed] [Google Scholar]
- 15. Leigh M. B., et al. 2007. Biphenyl-utilizing bacteria and their functional genes in a pine root zone contaminated with polychlorinated biphenyls (PCBs). ISME J. 1:134–148 [DOI] [PubMed] [Google Scholar]
- 16. Mussmann M., Ishii K., Rabus R., Amann R. 2005. Diversity and vertical distribution of cultured and uncultured deltaproteobacteria in an intertidal mud flat of the Wadden Sea. Environ. Microbiol. 7:405–418 [DOI] [PubMed] [Google Scholar]
- 17. Neufeld J. D., et al. 2007. DNA stable-isotope probing. Nat. Protocol 2:860–866 [DOI] [PubMed] [Google Scholar]
- 18. Sakai M., Ezaki S., Suzuki N., Kurane R. 2005. Isolation and characterization of a novel polychlorinated biphenyl-degrading bacterium, Paenibacillus sp. KBC101. Appl. Microbiol. Biotechnol. 68:111–116 [DOI] [PubMed] [Google Scholar]
- 19. Suenaga H., Mitsuoka M., Ura Y., Watanabe T., Furukawa K. 2001. Directed evolution of biphenyl dioxygenase: emergence of enhanced degradation capacity for benzene, toluene, and alkylbenzenes. J. Bacteriol. 183:5441–5444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Sul W. J., et al. 2009. DNA-stable isotope probing integrated with metagenomics for retrieval of biphenyl dioxygenase genes from polychlorinated biphenyl-contaminated river sediment. Appl. Environ. Microbiol. 75:5501–5506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Uhlik O., et al. 2009. Biphenyl-metabolizing bacteria in the rhizosphere of horseradish and bulk soil contaminated by polychlorinated biphenyls as revealed by stable isotope probing. Appl. Environ. Microbiol. 75:6471–6477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Wilms R., et al. 2006. Specific bacterial, archaeal, and eukaryotic communities in tidal-flat sediments along a vertical profile of several meters. Appl. Environ. Microbiol. 72:2756–2764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Zhang S. W., Zhang Y. L., Pan Q., Cheng Y. M., Chou K. C. 2008. Estimating residue evolutionary conservation by introducing von Neumann entropy and a novel gap-treating approach. Amino Acids 35:495–501 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.