Abstract
The proteomes expressed at 4°C and 18°C by the psychrophilic Antarctic bacterium Pseudoalteromonas haloplanktis were compared using two-dimensional differential in-gel electrophoresis with special reference to proteins repressed by low temperatures. Remarkably, the major cold-repressed proteins, almost undetectable at 4°C, were heat shock proteins involved in folding assistance.
TEXT
The Gram-negative bacterium Pseudoalteromonas haloplanktis is a typical representative of the Gammaproteobacteria found in cold marine environments, and strain TAC125 has been isolated from seawater sampled along the Antarctic ice shelf. Such strains thrive permanently in seawater at about −2°C to +4°C but are also anticipated to endure long-term frozen conditions when entrapped in the winter ice pack. The genome of P. haloplanktis TAC125 has been fully sequenced (11). This work has further allowed a proteomic study of its cold acclimation proteins (CAPs), i.e., proteins that are continuously overexpressed at a high level during growth at low temperatures (14). This has demonstrated that protein synthesis and protein folding are the main upregulated functions, suggesting that both cellular processes are limiting factors for bacterial development in cold environments. Here we report a proteomic survey of cold-repressed proteins at 4°C in order to complete the metabolic pattern of the bacterium's growth at low temperature.
Temperature dependence of growth.
The Antarctic bacterium maintains a doubling time of ∼4 h at 4°C in a marine broth, with an extrapolated generation time of 5 h 15 min at 0°C (Fig. 1a). When the culture temperature is raised to 20°C, the generation time decreases moderately (e.g., 1 h 40 min at 18°C) with a concomitant increase in the biomass produced at the stationary phase (Fig. 1b). At temperatures higher than 20°C, a drastic reduction in cell density at the stationary phase is recorded (Fig. 1b), indicating heat-induced stress on the cell. P. haloplanktis fails to grow above 29°C. According to this growth behavior, the temperatures of 4°C and 18°C were selected here for the differential comparison of the proteomes, as 18°C does not induce excessive stress as far as growth rate and biomass are concerned.
Fig. 1.
(a) Temperature dependence of the generation time of Pseudoalteromonas haloplanktis TAC125 grown in marine broth (solid line and circles). A typical curve for E. coli RR1 in LB broth is shown for comparison (dashed line). (b) Growth curves of P. haloplanktis at 4°C (○), 18°C (●), and 26°C (■).
Cold-induced versus cold-repressed proteins.
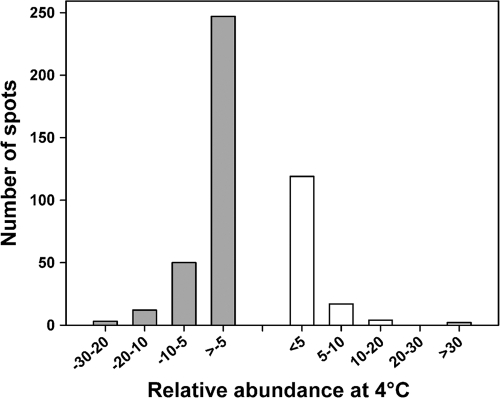
The proteomes expressed by the Antarctic bacterium at 4°C and 18°C during the logarithmic phase of growth were compared by two-dimensional (2D) differential in-gel electrophoresis as described previously (14). In a typical single 2D gel (see Fig. S1 in the supplemental material), 142 protein spots were more abundant at 4°C (CAPs), whereas 309 protein spots were less intense at 4°C than at 18°C. This unexpectedly large number of cold-repressed proteins already indicates that numerous cellular functions are downregulated during growth at low temperature. The repression factors (or induction factors for CAPs), given by relative spot abundance between 4°C and 18°C, are illustrated in Fig. 2. This distribution shows that 21% of cold-repressed proteins display a downregulation factor of between 5 and 28, revealing that some key cellular functions are severely affected. Of the 309 cold-repressed proteins, 83 were retained, which satisfied both statistical biological variation analysis and mass spectrometry identification scores. These cold-repressed proteins are listed in Table S1 in the supplemental material along with their repression factors, and their distribution in the main cellular functions is given in Table S2 in the supplemental material. It is worth mentioning that cold-repressed proteins could formally correspond to proteins that are overexpressed at 18°C. This feature should be taken into account when analyzing the proteomic results presented below.
Fig. 2.
Distribution of the relative abundance of cold-repressed proteins (dashed, negative values, 18°C/4°C spot volume ratio) and of cold acclimation proteins (positive values, 4°C/18°C spot volume ratio) in the proteome of P. haloplanktis grown at 4°C.
HSPs and protein folding.
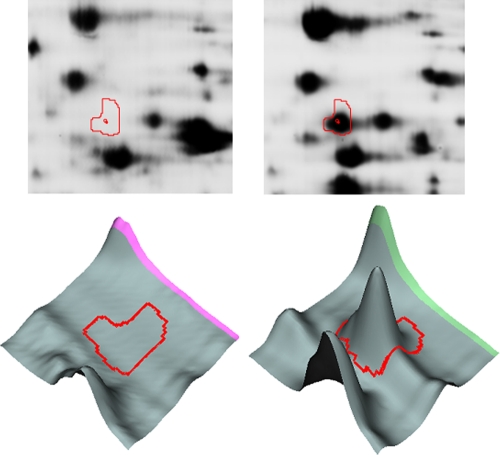
The major heat shock proteins (HSPs) such as the chaperone DnaK, the chaperonin GroEL, and the chaperone Hsp90, as well as the so-called small HSPs (IbpA and -B), were identified here as strongly cold-repressed proteins in the proteome of P. haloplanktis. The overexpression of bacterial HSPs at elevated temperatures is well recognized as being indicative of a heat-induced cellular stress (5, 16), and this is obviously relevant for the Antarctic bacterium grown at 18°C. In Escherichia coli, it has been shown that synthesis of HSPs is repressed during growth at low temperatures but also that these HSPs are harmful to cells at 4°C, as their induced expression reduces cell viability at this temperature (9). Analysis of the P. haloplanktis HSP spots (Fig. 3; see Fig. S2 in the supplemental material) shows that these HSPs are present in only trace amounts at 4°C and are therefore true cold-repressed proteins in the Antarctic strain. However, downregulation of this protein group in the Antarctic bacterium severely impairs an essential cellular function, as these HSPs are chaperones assisting co- or posttranslational protein folding (7). Such a detrimental situation reinforces our previous suggestion that overexpression of the trigger factor (37-times upregulation; see Fig. S2 in the supplemental material) by P. haloplanktis TAC125 at 4°C may rescue the chaperone function at low temperatures (14). Indeed, the trigger factor is the first chaperone interacting with virtually all newly synthesized polypeptides on the ribosome (12) but it is also a cold shock protein in E. coli (9). Low temperature slows down the folding reaction and is well known to reduce misfolding and aggregation (10), possibly contributing to a limiting of the detrimental effects of HSP repression. Nevertheless, it is now clear that regulation of the expression of these proteins involved in thermal stress is a primary adaptation to bacterial growth at low temperatures.
Fig. 3.
Comparative analysis of spots containing the chaperone DnaK from Pseudoalteromonas haloplanktis grown at 4°C (left panels) and 18°C (right panels). Spot view on a 2D gel seen in fluorescence (upper panels) and three-dimensional images (lower panels) obtained with DeCyder software.
Metabolism depression at low temperatures.
Nearly half of the proteins downregulated at 4°C are related to functions involved in general bacterial metabolism (see Table S2 in the supplemental material). This includes the degradation or biosynthesis of compounds and the production of energy. Most of these proteins are involved in oxidative metabolism, in particular to glycolysis, the pentose phosphate pathway, the Krebs cycle, and electron chain transporters. Accordingly, the Antarctic bacterium depresses its general metabolism when grown at low temperature. This is in agreement with the reduced biomass produced at 4°C (Fig. 1b). It is worth mentioning that ancient bacterial survival has been reported in frozen samples up to half a million years old and such viability has been correlated with minimal cellular metabolic activity and the capacity to slowly repair DNA (8). We have previously shown that protein synthesis and folding are limiting factors in the growth of P. haloplanktis at cold temperatures (14). The present proteomic data indicate that when these limitations are alleviated at 18°C, the bacterium proliferates by the activation of its general metabolism and therefore divides actively and produces more biomass. This can also be regarded as an adaptive strategy to increase the viable population during short warmer periods. From an ecological point of view, and in the context of possible global warming, a rise in environmental temperature would result mainly in the proliferation of bacteria such as P. haloplanktis.
Downregulation of iron metabolism at low temperatures.
Iron uptake and iron-related proteins are clearly downregulated at 4°C. Two transport systems of this essential element were found to be downregulated at 4°C: the ABC transporter FbpA and a TonB-dependent receptor. The first is involved in the uptake of weakly soluble ferric ion (Fe3+) directly from the environment, and the second is required for the transport of heme complexes and ferric siderophores through the cell membrane (3). The reduced need of P. haloplanktis for iron at 4°C can be partly explained by the downregulation of the Krebs cycle and the respiratory chain (and their iron-containing complexes such as SdhB), by the repression of HmgA, which requires Fe2+ to degrade cyclic amino acids, or by the strong downregulation of catalase (which is made up of four heme groups). Hemes are tetrapyrroles that have porphobilinogen as a precursor: this is in agreement with the downregulation of both GltX (glutamyl-tRNA synthetase) and HemB (5-aminolevulinate dehydratase), which are responsible for porphobilinogen synthesis.
Various metallic ions are essential for cell metabolism, and therefore, the fact that proteomic data only point to cold repression of iron-related proteins is puzzling. Iron in a redox-active form (Fe2+) is potentially deleterious, as it is able to induce oxidative cell damage by the Fenton reaction, for instance (17). It can be tentatively proposed that, as a result of the improved stability of ROS (reactive oxygen species) at low temperatures, the downregulation of iron-related proteins could contribute to an avoidance of such detrimental iron-based reactions. In this respect, it should be mentioned that the genome of P. haloplanktis entirely lacks the ubiquitous ROS-producing molybdopterin metabolism (11). This suggests that the Antarctic bacterium tends to avoid ROS production involving metallic ions.
Oxidative stress-related proteins.
The second group of proteins that displays the highest repression factors at 4°C is represented by the oxidative stress-related proteins catalase, glutathione reductase, and peroxiredoxin (see Table S1 in the supplemental material). At first sight, this may be regarded as a conflicting result because conclusive findings have indicated that psychrophiles, including P. haloplanktis, are exposed to permanent oxidative stress at low temperatures, which originates from improved oxygen solubility and increased ROS stability (1, 2, 4, 11, 13–15). However, it should be recalled that the general aerobic metabolism of the Antarctic bacterium is stimulated at 18°C, also resulting in ROS production. Accordingly, the identified oxidative stress-related proteins would be better regarded as being induced at 18°C rather than repressed at 4°C.
The upregulation of catalase and peroxiredoxin at 18°C shows that the bacterium needs to be protected against ROS like H2O2 as both enzymes catalyze its decomposition into O2 and H2O. Under oxidative stress, the NADPH supply for reduced glutathione regeneration is also dependent on glucose-6-phosphate dehydrogenase (Zwf) in the first step of the pentose phosphate pathway, which is upregulated at 18°C. Glutathione reductase (Gor) plays a central role in the reoxidation of NADPH from the pentose phosphate pathway, allowing the formation of reduced glutathione, an important cellular antioxidant. The upregulated DNA-binding DPS protein DpsB plays a major role in the protection of bacterial DNA from damage by ROS and is induced under stress conditions (6). There is obviously a finely tuned balance between the cellular mechanisms protecting against oxidative stresses generated by low temperatures (resulting from ROS stability and oxygen solubility) and those generated by high temperatures (resulting from stimulated metabolic activity) which deserves further investigation.
Supplementary Material
Acknowledgments
We thank E. De Pauw for access to the GIGA mass spectrometry facilities. The Institut Polaire Français Paul Emile Victor is also acknowledged for support at early stages of this work.
This work was supported by F.R.S.-FNRS (National Fund for Scientific Research), Belgium (FRFC grants to G.F., an FRSM grant to P.L., and a Crédits aux Chercheurs grant to S.D. and G.F.). F.P. was a FRIA research fellow and S.D. was an F.R.S.-FNRS postdoctoral researcher during this work. G.M. is an F.R.S.-FNRS logistics collaborator, and P.L. is an F.R.S.-FNRS research associate. F.P. was supported by the European Space Agency (Exanam-Prodex Experiment Arrangement).
Footnotes
Supplemental material for this article may be found at http://aem.asm.org/.
Published ahead of print on 8 April 2011.
REFERENCES
- 1. Ayub N. D., Tribelli P. M., Lopez N. I. 2009. Polyhydroxyalkanoates are essential for maintenance of redox state in the Antarctic bacterium Pseudomonas sp. 14-3 during low temperature adaptation. Extremophiles 13:59–66 [DOI] [PubMed] [Google Scholar]
- 2. Bakermans C., et al. 2007. Proteomic analysis of Psychrobacter cryohalolentis K5 during growth at subzero temperatures. Extremophiles 11:343–354 [DOI] [PubMed] [Google Scholar]
- 3. Clarke T. E., Tari L. W., Vogel H. J. 2001. Structural biology of bacterial iron uptake systems. Curr. Top. Med. Chem. 1:7–30 [DOI] [PubMed] [Google Scholar]
- 4. Duchaud E., et al. 2007. Complete genome sequence of the fish pathogen Flavobacterium psychrophilum. Nat. Biotechnol. 25:763–769 [DOI] [PubMed] [Google Scholar]
- 5. Goodchild A., Raftery M., Saunders N. F., Guilhaus M., Cavicchioli R. 2005. Cold adaptation of the Antarctic archaeon, Methanococcoides burtonii assessed by proteomics using ICAT. J. Proteome Res. 4:473–480 [DOI] [PubMed] [Google Scholar]
- 6. Haikarainen T., Papageorgiou A. C. 2010. Dps-like proteins: structural and functional insights into a versatile protein family. Cell. Mol. Life Sci. 67:341–351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Hartl F. U., Hayer-Hartl M. 2009. Converging concepts of protein folding in vitro and in vivo. Nat. Struct. Mol. Biol. 16:574–581 [DOI] [PubMed] [Google Scholar]
- 8. Johnson S. S., et al. 2007. Ancient bacteria show evidence of DNA repair. Proc. Natl. Acad. Sci. U. S. A. 104:14401–14405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Kandror O., Goldberg A. L. 1997. Trigger factor is induced upon cold shock and enhances viability of Escherichia coli at low temperatures. Proc. Natl. Acad. Sci. U. S. A. 94:4978–4981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. King J., Haase-Pettingell C., Robinson A. S., Speed M., Mitraki A. 1996. Thermolabile folding intermediates: inclusion body precursors and chaperonin substrates. FASEB J. 10:57–66 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Médigue C., et al. 2005. Coping with cold: the genome of the versatile marine Antarctica bacterium Pseudoalteromonas haloplanktis TAC125. Genome Res. 15:1325–1335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Merz F., et al. 2008. Molecular mechanism and structure of Trigger Factor bound to the translating ribosome. EMBO J. 27:1622–1632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Methé B. A., et al. 2005. The psychrophilic lifestyle as revealed by the genome sequence of Colwellia psychrerythraea 34H through genomic and proteomic analyses. Proc. Natl. Acad. Sci. U. S. A. 102:10913–10918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Piette F., et al. 2010. Proteomics of life at low temperatures: trigger factor is the primary chaperone in the Antarctic bacterium Pseudoalteromonas haloplanktis TAC125. Mol. Microbiol. 76:120–132 [DOI] [PubMed] [Google Scholar]
- 15. Rabus R., et al. 2004. The genome of Desulfotalea psychrophila, a sulfate-reducing bacterium from permanently cold Arctic sediments. Environ. Microbiol. 6:887–902 [DOI] [PubMed] [Google Scholar]
- 16. Rosen R., Ron E. Z. 2002. Proteome analysis in the study of the bacterial heat-shock response. Mass Spectrom. Rev. 21:244–265 [DOI] [PubMed] [Google Scholar]
- 17. Valko M., Morris H., Cronin M. T. 2005. Metals, toxicity and oxidative stress. Curr. Med. Chem. 12:1161–1208 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.