Abstract
The clustered regularly interspaced short palindromic repeat (CRISPR)/Cas system confers acquired heritable immunity against mobile nucleic acid elements in prokaryotes, limiting phage infection and horizontal gene transfer of plasmids. In CRISPR arrays, characteristic repeats are interspersed with similarly sized nonrepetitive spacers derived from transmissible genetic elements and acquired when the cell is challenged with foreign DNA. New spacers are added sequentially and the number and type of CRISPR units can differ among strains, providing a record of phage/plasmid exposure within a species and giving a valuable typing tool. The aim of this work was to investigate CRISPR diversity in the highly homogeneous species Erwinia amylovora, the causal agent of fire blight. A total of 18 CRISPR genotypes were defined within a collection of 37 cosmopolitan strains. Strains from Spiraeoideae plants clustered in three major groups: groups II and III were composed exclusively of bacteria originating from the United States, whereas group I generally contained strains of more recent dissemination obtained in Europe, New Zealand, and the Middle East. Strains from Rosoideae and Indian hawthorn (Rhaphiolepis indica) clustered separately and displayed a higher intrinsic diversity than that of isolates from Spiraeoideae plants. Reciprocal exclusion was generally observed between plasmid content and cognate spacer sequences, supporting the role of the CRISPR/Cas system in protecting against foreign DNA elements. However, in several group III strains, retention of plasmid pEU30 is inconsistent with a functional CRISPR/Cas system.
INTRODUCTION
Clustered regularly interspaced short palindromic repeats (CRISPRs) (23, 51, 55) are arrays of highly conserved direct DNA repeats that are interspersed by unique, similarly sized spacers. CRISPRs were first described for Escherichia coli (25), and bioinformatic analyses of sequenced genomes were able to locate them in approximately 90% of archaeal and 40% of bacterial species (30). Depending upon the species, CRISPRs range from 24 to 48 bp and CRISPR spacers range from 26 to 72 bp in length (16). Spacers matching sequences in bacteriophage genomes or transmissible plasmids (3, 23, 39) were shown to provide a defense against invasion by foreign nucleic acid elements (2, 5, 34). CRISPR arrays are flanked by a set of CRISPR-associated (cas) genes (17) that encode the Cas proteins involved in processing and binding of the pre-CRISPR RNA (pre-crRNA) transcribed from the CRISPR array (6, 18, 55). Eight different CRISPR/Cas system subtypes have been identified so far, each containing a set of subtype-specific cas genes in combination with a variable number of core cas genes (17). Each set of subtype-specific genes is associated with a definite CRISPR (30), and sequence- and structure-specific recognition between Cas proteins and CRISPRs was found to be crucial for processing of pre-crRNA (5, 20). The resulting small CRISPR RNA (crRNA)-Cas protein complexes direct immunity against foreign DNA (13, 34) or RNA (19) infection by a mechanism based on strict identity between the incorporated CRISPR spacer and the nucleic acid target (5). New spacers derived from foreign DNA elements are incorporated in a polarized manner at the 5′ end of the CRISPR array next to the leader sequence (2, 9, 56). A short contiguous sequence, the protospacer adjacent motif (PAM), directs the selection of the target sequences by the CRISPR/Cas system (24, 40). This structured integration of novel spacers provides both an evolutionary imprint and heritable acquired immunity (51). Evolutionary remnants of CRISPR spacers distinguish subsequent lineages and can be exploited for spoligotyping applications (3, 8, 10, 14, 41, 44). The unidirectional assembly mechanism of CRISPR arrays reveals genotypic divergence chronologically, with newly acquired spacers located closest to leader sequences (44), whereas strain-level variation is further provided by internal spacer deletions (9) or duplications within consecutive spacers (8, 21, 54).
Erwinia amylovora was the first phytopathogenic bacterium described and causes fire blight, the most important global disease threat to pome fruit and Rosaceae (Spiraeoideae) plants (43). Intercontinental dissemination from the United States occurred relatively recently to New Zealand (1910s), the United Kingdom and Northern Europe (1950s), and the Middle East (1960s) (4). Long-distance dispersal occurs principally via infected plant host material, limiting the number of invasion events into new regions. E. amylovora, even strains from the area of origin in North America (42), has been considered a genetically low-diversity species based on previous molecular approaches, including pulsed-field gel electrophoresis (PFGE) (27, 28), PCR fingerprinting based on PCR ribotyping (11, 37), rep-PCR (1, 46), amplified fragment length polymorphism (AFLP) analysis (47), random amplified polymorphic DNA fragment (RAPD) analysis, and amplified rRNA gene restriction analysis (ARDRA) (42). While strain patterns derived using these methods distinguish strains from Rubus (subfamily Rosoideae) and other Spiraeoideae plants (1, 37) and indicate a North American origin based on the slightly greater diversity of strains from this region (47), they provide poor discrimination of strains within local infested regions, which is required for effective implementation of phytosanitary control measures.
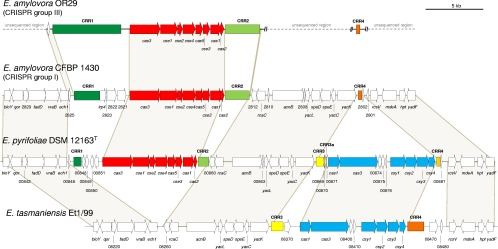
In this study, we describe a CRISPR-based approach to resolve E. amylovora strain biodiversity. Recent complete genome sequencing of E. amylovora CFBP1430 revealed the presence of three CRISPR repeat regions (CRRs), designated CRR1, CRR2, and CRR4 (renamed from CRR3 [see below]), in E. amylovora (50) (Fig. 1). We analyzed CRISPR locus organization and diversity for the potential to genotype E. amylovora strains of global origin. Additionally, we examined the potential function of the CRISPR/Cas system in protecting the fire blight pathogen against foreign nucleic acid elements by relating the results obtained with the sequences of known E. amylovora bacteriophages or with plasmid distribution in the corresponding strains.
Fig. 1.
Comparison of cas genes and associated CRRs between related Erwinia species. CRRs sharing the same repeat sequence are indicated by the same color, whereas similar colors indicate the presence of a SNP between two consensuses. In E. pyrifoliae, two independent sets of cas genes (cse and csy, corresponding to the Ecoli [red] and Ypest [blue] subtype variants, respectively) are associated with two distinct CRISPR arrays. The csy genes and one of the associated CRISPR regions show evidence of having been lost in E. amylovora, leaving only a remnant CRR4. In contrast, loss of the cse genes and both associated CRRs appears to have occurred in E. tasmaniensis. The four predicted ORFs situated between CRR1 and the cas3 gene in E. amylovora CRISPR group I and II strains (the gene arrangement of UTRJ2/pEU30 corresponds to that of CFBP1430) are remnants of the genomic island found in Rubus strains and are missing in group III strains such as OR29.
MATERIALS AND METHODS
Bacterial strains.
The 37 E. amylovora strains used throughout this study were obtained from several sources, including culture collections, generous gifts from other laboratories, and our own field samples. They originated from various areas in Europe, North America, New Zealand, and around the Mediterranean and were isolated from various cultivars of apple and pear as well as from other fire blight host plants within the Rosaceae family (Table 1). Strains were routinely grown overnight (16 to 18 h) at 28°C in Luria-Bertani (LB) broth prior to DNA extraction.
Table 1.
Origins and relevant characteristics of strains used in this study
| Strain/plasmida | Geographic origin | Host (subfamily)b | Yr of isolationc | Source or referenced |
|---|---|---|---|---|
| CRISPR group I strains | ||||
| 01SFR-BO | Ravenna, Italy | Sorbus sp. (S) | 1991 | K. Geider |
| ACW56400/pEI70 | Fribourg, Switzerland | Pyrus communis (pear) (S) | 2007 | ACW |
| ACW63230 | Lucerne, Switzerland | Cydonia oblonga (quince) (S) | 2008 | ACW |
| ACW63579 | St. Gallen, Switzerland | Malus domestica (apple) (S) | 2008 | ACW |
| ACW63889 | Lucerne, Switzerland | Cotoneaster dammeri (S) | 2008 | ACW |
| ACW64132 | St. Gallen, Switzerland | M. domestica (S) | 2008 | ACW |
| CFBP1232T | United Kingdom | P. communis (S) | 1959 | CFBP |
| CFBP1430 | Lille, France | Crataegus sp. (hawthorn) (S) | 1972 | CFBP |
| CFBP2301 | Haute Savoie, France | Pyracantha sp. (firethorn) (S) | 1981 | CFBP |
| CFBP3020 | Netherlands | P. communis cv. Clapp's Favorite (S) | 1981 | CFBP |
| CFBP3025 | New Zealand | M. domestica (S) | 1953 | CFBP |
| CFBP3049 | Ontario, Canada | M. domestica cv. Jonathan (S) | 1981 | CFBP |
| CFBP3098 | Masluc, Israel | M. domestica cv. Ein-Shemer (S) | 1987 | CFBP |
| Ea110R | Michigan | M. domestica cv. Jonathan (S) | 1975 | V. Stockwell |
| Ea153 | Oregon | M. domestica (S) | 1989 | V. Stockwell |
| Ea209/pEL60 | Israel | P. communis (S) | 1997 | S. Manulis |
| Ea263 | Israel | C. oblonga (S) | 1997 | S. Manulis |
| Ea273/pEA72 | New York | M. domestica (S) | 1971 | K. Geider |
| Ea4/82 | Egypt | P. communis (S) | 1982 | K. Geider |
| Ea7/74 | Northern Germany | Cotoneaster sp. (S) | 1974 | K. Geider |
| Ea02 | Vollèges, Switzerland | Cotoneaster sp. (S) | 2002 | ACW |
| LebA3/pEL60 | Lebanon | M. domestica (S) | 1998–1999 | G. Sundin |
| LebB66/pEL60 | Lebanon | C. oblonga (S) | 1998–1999 | G. Sundin |
| UPN527 (ΔpEA29) | Navarra, Spain | M. domestica (S) | 1997 | M. Lopez |
| CRISPR group II strains | ||||
| CFBP3792 | Idaho | Prunus salicina (plum) (S) | 1996 | CFBP |
| UTFer2/pEU30 | Utah | M. domestica (S) | NA | G. Sundin |
| UTRJ2/pEU30 | Utah | M. domestica (S) | NA | G. Sundin |
| CRISPR group III strains | ||||
| JL1185 | Washington, USA | P. communis (S) | 1988 | V. Stockwell |
| JL1189/pEU30 | Washington, USA | P. communis (S) | 1988 | G. Sundin |
| LA092/pEU30 | Washington, USA | P. communis (S) | 1988 | G. Sundin |
| LA476 | Oregon, USA | P. communis (S) | 2002 | V. Stockwell |
| OR25/pEU30 | Oregon, USA | P. communis (S) | NA | G. Sundin |
| OR29/pEU30 | Oregon, USA | P. communis (S) | NA | G. Sundin |
| Other strains | ||||
| Ea6-96r*/pEA2.8 | Canada | Rubus sp. (raspberry) (R) | 1996 | 35 |
| Ea7-96r* | Canada | Rubus sp. (R) | 1996 | 35 |
| IH3-1/pEA1.7 | Louisiana | Rhaphiolepis indica (Indian hawthorn) (S) | 1998 | G. Sundin |
| IL-5/pEA2.8 | Illinois | Rubus sp. (R) | 1977 | G. Sundin |
Names of detected plasmids are indicated after the strain names. With the exception of UPN527, all strains were positive for plasmid pEA29. Strains carrying a variant of pEA29 similar to the one observed in the Rubus strain MR1 are marked by an asterisk.
R, Rosoideae; S, Spiraeoideae.
NA, not available.
ACW, Agroscope Changins-Wädenswil (our collection); CFBP, Collection Française de Bactéries Phytopathogènes.
PCR amplification and DNA sequencing of CRISPR arrays and cas genes.
Chromosomal DNAs were isolated with a Wizard genomic DNA purification kit (Promega, Dübendorf, Switzerland) from 1.5-ml aliquots of overnight cultures grown at 28°C in LB. All primers used for DNA amplification and sequencing in this work are provided in Table S1 in the supplemental material. The three CRRs were amplified by PCR, using primer pairs C1f01/C1r01, C2f01/C2r01, and C4f01/C4r01. Amplification of the cas operon was performed for strains UTRJ2 and OR29, using three primer pairs (cas3-f1/cse1-r1, cas3-f4/cas5-r1, and cse4-f1/C2r04 [UTRJ2] or cse4-f1/C2r05 [OR29]), to obtain an equivalent number of amplicons of about 3 kb covering the complete 8.5-kb region containing the eight cas genes. Each PCR was performed with 10 ng of genomic DNA in a total volume of 40 μl, using a 0.4 mM concentration of each primer in a final concentration of 1× master mix from a HotStarTaq master mix kit (Qiagen, Basel, Switzerland). The PCR program consisted of denaturation and activation of the HotStarTaq enzyme for 15 min at 95°C, followed by 35 cycles of denaturation for 30 s at 95°C, annealing for 30 s at 60°C, and extension for 3 min at 72°C. Positive PCR amplification was verified by loading 5 μl of each reaction mixture into a 1.2% agarose gel. All PCR products were purified using a MultiScreen PCR plate (Millipore, Molsheim, France) and directly sequenced using an ABI Prism BigDye Terminator v1.1 cycle sequencing kit (Applied Biosystems, Foster City, CA) with the same primers used for amplification. Except for CRR4, a primer walking strategy using primers designed from the nonrepetitive spacers of the CRISPR arrays (or internal primers in the case of cas genes) was needed to complete sequencing of the PCR products.
Spacer crawling.
Several strains that contained plasmid pEU30 did not produce any CRR1 amplicons. We found that a larger CRR1 sequence (i.e., containing a larger number of spacers and repeats) was present, creating problems for amplifying the region with a single conventional PCR. Thus, we developed a “spacer crawling” strategy using primers designed from both flanking regions of CRR1 (echF and cas3R) in combination with primers targeting the CRR1 repeat on the cDNA strand (C1repR and C1repF, respectively). The PCR was performed using a slightly modified protocol with respect to that described above (i.e., with annealing for 30 s at 57°C and extension for 7 min at 72°C). Reactions produced a number of amplicons of increasing length, each differing by 61 bp (i.e., one spacer and one repeat) from the next-smaller amplicon, resulting in a stuttering pattern on the gel. After purification on a MultiScreen PCR plate, the PCR product combinations could be utilized as templates for subsequent sequencing reactions using respective primers located on the flanking regions. Once the beginning of the repeat region was reached, further primers based on the spacers of CRR1 were designed until sequencing of the original PCR products was possible. As necessary, the entire procedure was reiterated starting from a further location in the sequence until the left and right contigs could be joined together over at least three spacers.
Bioinformatic analysis.
The obtained DNA sequences were analyzed, edited, and assembled using Sequencher, version 4.9 (Gene Codes Corporation, Ann Arbor, MI). CRISPR spacers and repeats were identified using CRISPRfinder (16). The similarity of each spacer to sequences deposited in GenBank was analyzed by BLASTN, using an E value cutoff of 0.1. Only sequences pointing to mobile genetic elements and viruses were considered legitimate hits, whereas those pointing to implausible targets for the CRISPR/Cas system were discarded as random matches (moreover, the similarity to these sequences was usually well below 100%, except in the case of mammalian genomic libraries, where the hits were often located within the plasmid vectors used to construct the libraries). Sequences of CRRs and associated cas genes in the complete genomes of the closely related strains Erwinia pyrifoliae DSM12163T (49) and Erwinia tasmaniensis Et1/99 (29), which contain four (CRR1 to CRR4) and two (CRR3 and CRR4) CRRs, respectively, were included in our analysis for comparison (Fig. 1). The online software WebLogo (7) was used to visualize PAMs (40) in the sequence of E. amylovora plasmid pEU30 (12). The secondary structure of RNA derived from direct repeat motifs of each single CRR was predicted using RNAfold (22), a software product that calculates the stability of each possible secondary structure.
Cluster analysis.
An unweighted-pair group method using average linkages (UPGMA)-based tree that included all E. amylovora strains used in this study was generated using MEGA, version 4.0 (53), with distance calculations applied on the binary-coded data as generated based upon the presence or absence of each individual spacer in the concatenated CRR arrays. Throughout this work and for the purpose of comparative analysis between strains unbiased by single DNA mutations, similar spacers diverging from each other by no more than three single-nucleotide polymorphisms (SNPs) were considered equivalent if they were found in the same succession on the same CRR. The rationale for this was to avoid misleading discrimination between spacers simply due to unrelated acquisition of SNPs within spacers.
Plasmid content.
All strains were tested by PCR for the presence of known E. amylovora plasmids pEU30, pEL60, pEA29, pEI70, pEA1.7, and pEA2.8. Specific primers were designed in our study or were retrieved as available from the literature (see Table S1 in the supplemental material). When necessary, the amplicons obtained were sequenced to confirm that they did correspond to a specific plasmid.
RESULTS
Sequence analysis of CRISPRs and organization of cas genes.
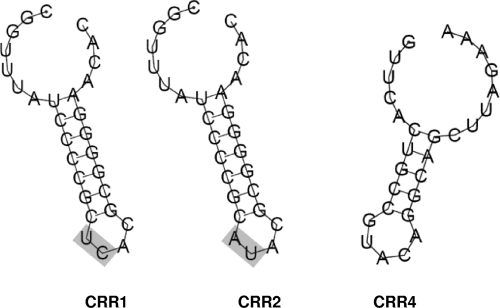
Direct repeats of both CRR1 and CRR2 were 29 bp long and differed in only two adjacent base pairs in the central region of their sequences that are expected to be situated in the loop region of the hypothesized RNA secondary structure, while the CRR4 repeat was 28 bp long and showed no sequence similarity with the two previous sequences (Fig. 2). As observed previously in other organisms (26), the last repeat at the 3′ end of each CRR was degenerate to some extent. The predicted correspondence between cas gene subtypes (17) and repeat clusters (30) was corroborated herein, confirming that Ecoli-subtype genes appear exclusively in the proximity of structured repeat cluster 2, while Ypest-subtype genes correspond strictly to cluster 4 (Fig. 1). Comparison with CRRs of related Erwinia species showed that sequences of CRR1 and CRR2 repeats (belonging to Kunin's cluster 2) in E. amylovora are identical to the equivalent regions in E. pyrifoliae, whereas the repeat of CRR4 (Kunin's cluster 4) perfectly matches the one found in CRR4 of E. pyrifoliae (5′-GTTCACTGCCGTACAGGCAGCTTAGAAA-3′), diverging by 1 bp (in bold) from the CRR4 repeat (5′-GTTCACTGCCGTACAGGCAGCTTAGAAG-3′) of E. tasmaniensis Et1/99. The CRR3 repeats of E. pyrifoliae and E. tasmaniensis are identical to each other but differ by 1 bp (5′-GTTCACTGCCGCACAGGCAGCTTAGAAA-3′) from the CRR4 repeat of E. amylovora and E. pyrifoliae DSM12163T, hence the decision to rename the third CRR found in E. amylovora (50) CRR4. In E. pyrifoliae, two independent sets of cas genes (cse and csy, corresponding to the Ecoli and Ypest subtype variants, respectively) are associated with two distinct CRRs (49). The csy genes and one of the associated CRRs were apparently lost in E. amylovora, leaving only CRR4 as a relic. This fact, along with the relative invariability of the CRR4 array in E. amylovora (see below), may be an indication that this region is no longer evolutionarily active in this species. Meanwhile, loss of the cse genes as well as both related CRRs seems to have occurred in E. tasmaniensis, where a single slightly degenerate repeat was found in the intergenic region between ech1 and ETA_08260 (Fig. 1).
Fig. 2.
Sequences and predicted secondary structures of Erwinia amylovora CRISPRs, obtained using RNAfold. Sequence comparison shows that both CRR1 and CRR2 belong to cluster 2, while CRR4 belongs to cluster 4 of the proposed repeat-based classification of CRISPR arrays (30). The gray boxes highlight the differences in repeat consensus sequence between CRR1 and CRR2.
In all three Rubus strains, the four genes that in E. amylovora CFBP1430 are found between CRR1 and cas3 are interleaved in an ∼20-kb genomic island which is flanked by an IS630 family transposase and embraces several other genes, including a type II nonribosomal peptide synthase/polyketide synthase (NRPS/PKS) gene (45).
Analysis of CRISPR spacers.
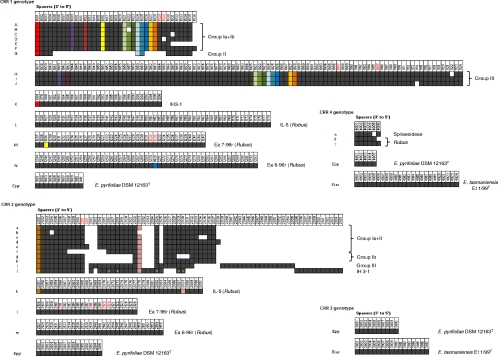
A total of 454 distinctive CRISPR spacers were identified among the 37 E. amylovora strains (Fig. 3; see Table S2A to D in the supplemental material) and were numbered progressively (strain by strain, in order of discovery) in 3′-to-5′ orientation with a four-digit code, whereby the first digit designates affiliation with the equivalent CRR. Over 95% of the spacers were 32 bp long, with the remaining spacers ranging mostly from 30 to 34 bp. Notable exceptions were a 10-bp spacer in CRR1 of strain IL-5 (spacer 1144), consisting of a (GT)5 repeat; a 42-bp spacer in the CRR2 of Ea6-96r (spacer 2143) displaying a duplication of its last 10 bp; and a 63-bp spacer (spacer 1043) found in CRR1 of group III strains (for strain groupings, see below). The first 32 bp of the last spacer were identical with those of spacer 1012, suggesting that spacer 1043 may have been generated by the fusion of two contiguous spacers after the loss of the middle repeat (Fig. 3).
Fig. 3.
CRR genotypes found in a collection of 37 E. amylovora strains. CRISPR spacers are represented by boxes, with the spacer positions numbered at the top of the columns. For each single spacer, the DNA sequence and results of BLAST similarity searches are available in Table S2A to D in the supplemental material. The arrays are oriented in the 3′-to-5′ direction, meaning that the spacers next to the leader sequence are on the right side of the picture and that within the same block, the numbering generally increases chronologically from the most ancient to the more recent spacers. Identical spacers within the same block are vertically aligned and colored gray, whereas identical spacers between different blocks are represented by the same color. A white box indicates the absence of the corresponding spacer in a given genotype. A yellow “x” inside a box points to the presence of SNPs in one spacer with respect to the consensus. Duplicated spacers within the same array are characterized by the same number at the spacer position (highlighted in red). Strains or CRISPR groups encompassing one or more CRR genotypes are indicated to the right of the array.
Sequence diversity in CRISPR arrays.
The numbers of spacers in the first two CRRs showed large variations among the different E. amylovora strains. CRR1 was found to contain between 12 (strains UTRJ2 and UTFer2) and 98 (strains OR29, LA092, and JL1189) spacers, whereas between 23 (strain Ea110R) and 49 (strains in group III) spacers were counted in CRR2. On the other hand, the number of spacers in CRR4 was almost invariant and included the same five spacers in all but three strains. Those three exceptions were isolated from Rubus host plants, and they had only minor changes in the identity of the last one or two spacers at the 5′ end of the array.
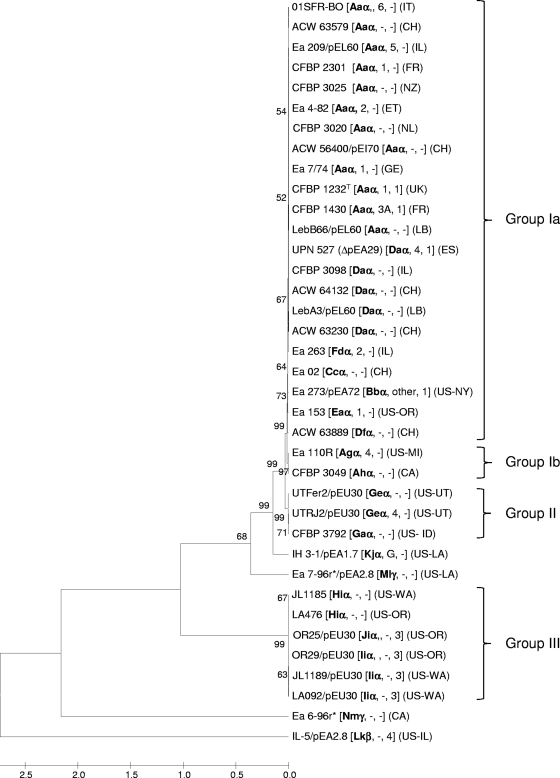
Across the 37 E. amylovora strains analyzed in detail, 14 and 13 distinct genotypes were resolved for CRR1 and CRR2, respectively, and 3 were resolved for the CRR4 region (Fig. 3). When sequence results for all three CRISPR regions were combined, the 37 strains could be assigned to 18 distinct genotypes (Fig. 4). A high degree of genetic homogeneity among E. amylovora strains from Spiraeoideae plants is reflected herein. Of the 19 European and Mediterranean strains examined, 84% belonged to CRR genotype Aaα or Daα, irrespective of country origin. In several cases, differences between strains were obtained due to mere deletion, addition, or duplication of single spacers, while in others entire regions were deleted or at some point divergent evolution occurred at the 5′ end of the array (Fig. 3), leading to the formation of three major genotypic groups following cluster analysis (Fig. 4). Since the construction of the unrooted UPGMA tree was based upon total numbers of differences between pairwise strain comparisons, its topology (but not its clustering) is biased by the relative size of the different CRISPR region in each strain. Thus, the early branching of group III probably does not reflect its real phylogenetic position with respect to other E. amylovora strains but may rather be the result of rapid divergence caused by the increased accumulation of spacers in CRR1 (Fig. 3; see Fig. S1 in the supplemental material).
Fig. 4.
Clustering of E. amylovora strains based on cumulative spacer patterns of CRR1, CRR2, and CRR4. The codes in brackets indicate the CRR genotype obtained in this work (shown in bold in Fig. 3), the PFGE type (27, 57), and the PCR ribotype (11, 37). An absence of data is indicated by a dash. The geographic origin of each strain is indicated in parentheses as follows: CA, Canada; CH, Switzerland; DE, Germany; ES, Spain; ET, Egypt; FR, France; IL, Israel; IT, Italy; LB, Lebanon; NL, Netherlands; NZ, New Zealand; UK, United Kingdom; and US, United States. Results of CRISPR typing were converted in a binary array according to the presence or absence of each spacer, and taxonomy was inferred using the UPGMA method (1,000 bootstrap replications; only values of >50% are shown; there were 461 positions in the final data set).
Group I can be divided into subgroups IA and IB, with the latter characterized by deletion of spacers 2014 to 2021 in CRR2, and contains strains of diverse geographical origins, i.e., Europe, the Mediterranean, New Zealand, and North America. Among these strains, Ea273 was the one most related to non-North American isolates, suggesting that an isolate with a similar genotype was responsible for the introduction of the disease outside North America. Both group II and group III comprised exclusively strains originating from the United States, many of them carrying plasmid pEU30. Compared to strains from group I, strains in group II showed deletion of over 20 spacers in CRR1 (spacers 1005 to 1023 and 1026 to 1029), one of which (1022) matches plasmid pEU30 (see below). The same array of strains of CRISPR group III was much larger and contained up to 98 spacers, and only 10 and 2 were shared with group I and II strains, respectively (Fig. 3). Furthermore, group III strains presented a peculiar set of 17 spacers (2038 to 2055) at the 5′ end of CRR2, suggesting that these strains diverged from the other two groups before their encounter with plasmid pEU30 (signaled by spacers 2042 to 2047 [see Table S2B in the supplemental material]). In Indian hawthorn strain IH3-1, the more ancient 3′-end region of CRR2 matched to some extent the spacers in the corresponding array of E. amylovora strains from Malus or Pyrus, even if it displayed the displacement of a few spacers and some sequence variation (SNPs) within the spacers themselves. The CRR1 region of strain IH3-1, in contrast, shared only the most ancient 3′-terminal spacer with other strains isolated from Spiraeoideae plants.
Apart from the modest variation described above for CRR4, the other two CRRs of Rubus-derived strains Ea696r, Ea796r, and IL-5 showed little commonality with each other or with Spiraeoideae-derived strains. Only the 3′-terminal spacer of CRR2 in strain IL-5 matched the corresponding sequence in Spiraeoideae strains. Comparison with spacers found in the related Erwinia strains E. pyrifoliae DSM12163T and E. tasmaniensis Et1/99 showed no similarity with those in E. amylovora strains for any CRR (Fig. 3).
Plasmid distributions in individual E. amylovora strains.
The investigated strains were found to carry a range of the currently described E. amylovora plasmids (Fig. 4; Table 1). Previous observations that have demonstrated the near ubiquity of pEA29 (NCBI GenBank accession number NC_013957) were confirmed here, with the plasmid being absent only in strain UPN527 (32). For Rubus-derived strains Ea6-96r and Ea7-96r, but not for IL-5, the amplicon produced by primers AJ484 and AJ485, targeting the msrA gene of pEA29, was shorter than that obtained from E. amylovora strains obtained from Spiraeoideae plants, suggesting that both strains may carry a variant of pEA29 that is similar or identical to the one observed in the Rubus-derived strain MR1, where a 1,264-bp sequence was found to replace a 1,890-bp region typically conserved in all other strains (35).
Many group II and group III strains selected for this study were shown to carry pEU30 (GenBank accession no. NC_005247), with the exception of CFBP3792, JL1185, and LA476. Conversely, pEU30 was not detected in any group I strain. The distribution of other plasmids proved numerically less important. The presence of plasmid pEL60 (NC_005246) was restricted to a number of strains originating from the Middle East, specifically from Israel (Ea209) and Lebanon (LebA3 and LebB66); plasmids pEA72 (NC_013973) and pEI70 (31) were each found only once within our collection of strains (i.e., in Ea273 and ACW56400, respectively); and plasmid pEA1.7 (NC_004940) was detected only in the Indian hawthorn strain IH3-1. Positive amplification was obtained using primer pair pEA2.8-fw/pEA2.8-rev, targeting ColE1 plasmid pEA2.8 (NC_004446), with Rubus strain Ea7-96r. The size of the amplicon and part of its sequence were different from those reported previously for strain IL-5 (36), but the region around the ColE1 origin was well conserved.
Sequences of CRISPR spacers and correlation with plasmid content.
Only a fraction of spacers (21.8%) showed significant similarity to any sequences in current databases (39), and most of them were homologous to fragments of plasmid (15.9%) or viral (1.3%) genes rather than to chromosomal sequences. Noteworthy exceptions pointed to mobile elements integrated into the bacterial chromosome, such as prophages (4.0%) (e.g., spacers 1005, 1180, and 2106) or transposon (spacer 1275)-related sequences (see Table S2A to D in the supplemental material). This observation supports the current theory that the CRISPR/Cas system acts as a defense against foreign mobile genetic elements via an RNA-mediated interference mechanism (23, 33). Nevertheless, no spacer matched the sequence of any of the four fully sequenced phages published thus far for Erwinia species (i.e., Felix01 [GenBank accession no. AF320576], ΦEa21-4 [NC_011811], Era103 [NC_009014], and ΦEa1 [48]).
In the collection of strains investigated, no CRISPR spacer was found to be directed against the nearly ubiquitous plasmid pEA29, which is absent only in strain UPN527, or against plasmids pEI70 and pEA1.7, found in strains ACW56400 and IH3-1, respectively. Spacer 2008, presenting three mismatches to a region of ColE1-type plasmid pEA2.8, was shared by all strains under investigation, with the exception of Rubus-derived strains IL-5 and Ea7-96r. Another spacer, 2133, presented an additional inserted base compared to a corresponding sequence found on the same plasmid in strain Ea6-96r (see Table S2B in the supplemental material). In agreement with the current model about the immunity function of the CRISPR/Cas system, plasmid pEA2.8 was detected in only two of the strains (i.e., IL-5 and Ea7-96r) that did not display any matching spacers in their CRISPR arrays, although the number of nucleotide substitutions between protospacer and spacer and the location of the latter near the 3′ end of CRR2 indicate that the encounter of E. amylovora with this ColE1-type plasmid was not recent.
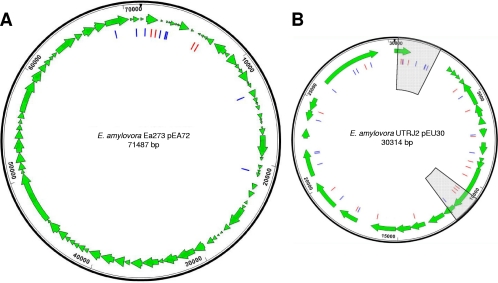
The presence of CRISPR spacers against pEA72 in strains corresponded to the absence of this plasmid. All strains from Spiraeoideae plants included at least one spacer perfectly matching some region of pEA72, with the exception of the apple strain Ea273. In Ea273, this plasmid was present, and compared to other group I and II strains, CRR2 spacer 2004 had been lost (Fig. 3). It is noteworthy that on pEA72, sequences matching actual CRISPR spacers were not distributed stochastically but were concentrated primarily in or around the repA gene, encoding the putative replication protein, whereas an extensive, >40-kb region of the plasmid was completely devoid of protospacers (Fig. 5A). This may be ascribed to the mosaic-like nature of the plasmid itself, which appears to share part of its sequence with pEU30 and pET46 (see Fig. S2 in the supplemental material), and it may suggest that pEA72 originated via the insertion of a wide region of foreign DNA from a related ancestor plasmid after the latter had challenged E. amylovora for the first time.
Fig. 5.
Distribution of protospacers on plasmids pEA72 (A) and pEU30 (B). Each bar indicates the position of a sequence integrated as a spacer in the CRISPR region of at least one of the E. amylovora strains considered in this work. Blue and red bars represent protospacers in forward and reverse orientations, respectively. Sections of pEU30 which were sequenced in group III strain OR29 to evaluate the presence of potential mutations in the 12 associated protospacers are represented by the shaded area.
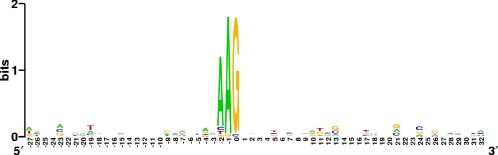
Also in accordance with the currently accepted model (13), spacer 1022 against pEU30 was located in a section of CRR1 in group I isolates that was missing in group II isolates (Fig. 3), therefore allowing the presence of the plasmid in the latter strains. However, in group III strains, six consecutive spacers at the 5′ end of CRR2, as well as up to 31 spacers in the CRR1 region, were shown to correspond to sequences present in plasmid pEU30, an observation that for four strains (JL1189, LA092, OR25, and OR29) is in obvious disagreement with the model of mutual exclusion between a foreign genetic element and CRISPR spacers matching its sequence. For group III strain OR29, Sanger sequencing of two separate sections of pEU30, encompassing part of the repA gene along with its downstream region (1,866 bp) or spanning virB4 and virB5 (1,489 bp) and covering a total of 12 protospacers (Fig. 5B), showed perfect identity to the corresponding sequence of the plasmid in group II strain UTRJ2 (12), indicating that in these regions the plasmid did not undergo any mutation in either the protospacer itself or the PAM to overcome the CRISPR/Cas system. Sites on pEU30 that corresponded to sequences of CRISPR spacers in group III strains were evenly distributed around the whole plasmid in a way that most of its genes could be related to at least one spacer (Fig. 5B). Analysis of the PAM in pEU30 revealed that a three-nucleotide consensus sequence (AAG) situated immediately upstream of the target region is likely accountable for the selection of spacers to be incorporated into CRRs (Fig. 6).
Fig. 6.
WebLogo plot of the PAM consensus created utilizing the entire pool of 37 protospacers found on plasmid pEU30. All entries are oriented in the same way as the cognate sequences in CRR1 and CRR2 of E. amylovora and are aligned relative to the 5′ end of the protospacer (base 1). Sequences include 28 nucleotides upstream of the first base of the protospacer (containing the PAM) and the protospacer itself (positive numbers).
Sequencing of cas genes in group III E. amylovora strains.
The large number of spacers of some group III strains that perfectly match a sequence on plasmid pEU30 is hardly compatible with the currently accepted mode of action for the CRISPR/Cas system. The option of CRISPR autoimmunity (52) was discarded, as no spacers were found to match the sequence of any of the cas genes or of the CRISPR array itself. We therefore explored the possibility that one or more defective cas genes may prevent the CRISPR machinery from fulfilling its role in group III strains by sequencing the section of the genome between CRR1 and CRR2 in pEU30-containing strains UTRJ2 and OR29 (CRISPR groups II and III, respectively). Comparison with complete genome sequences of E. amylovora strains CFBP1430 and Ea273 (50) revealed perfect identity among both group I strains and UTRJ2 over the entire cas gene cluster, including the leader sequence of CRR1. In contrast, five SNPs were detected in strain OR29, all of which were located at position 3 of the predicted codon. Four of these account for silent mutations in three different genes, specifically two in cas1 and one each in cas2 and cse2. A fifth one (60G→C) is predicted to cause the substitution of a glutamine for a histidine (Q20H) in Cas protein Cse1. Another peculiarity of OR29 was the absence of four predicted open reading frames (ORFs) situated between CRR1 and the cas3 gene in strains belonging to groups I and II (e.g., CFBP1430, UTRJ2, and Ea273) (Fig. 1). However, none of these genes appeared to have a function which could obviously be related to the CRISPR machinery or was found to be associated physically with the cas gene cluster in other bacteria. The presence of these ORFs inside the ∼20-kb genomic island inserted in the same spot of the genome in Rubus-derived strains suggests instead that they may be remnants of this region in group I and II strains (45).
DISCUSSION
All E. amylovora strains from Spiraeoideae or Prunus plants fit within one of three main CRISPR groups, while strains obtained from Rosoideae (Rubus spp.) plants and Indian hawthorn strain IH3-1 clustered independently. Similarity between Prunus and Spiraeoideae strains of E. amylovora was reported previously (38). Spacers of E. tasmaniensis and E. pyrifoliae show no commonality with those of E. amylovora, while Rubus and Spiraeoideae strains are highly similar regarding CRR4, confirming the placement of both as E. amylovora. CRISPR groups II and III are composed of strains from the United States, whereas CRISPR group I contains primarily strains from Europe and the Mediterranean. Groupings based on CRISPR diversity matched PCR ribotyping results. Ribotype 1 has been reported to be the dominant pattern found in E. amylovora strains from eastern North America and New Zealand (37), and it was the sole profile recovered among European strains as well as for Ea273 (11). Ribotype 3 is prevalent in strains from the western United States (37). Thus, PCR ribotypes 1 and 3 appear to correspond with CRISPR groups I and III, although CRISPR analysis further resolves these two ribotypes into nine and three distinct genotypes, respectively. Conversely, CRISPR genotyping did not completely overlap with PFGE-derived Pt grouping used to outline geographic distribution patterns for E. amylovora (27) but was able to discriminate similar strains within individual PFGE Pt groups, thus demonstrating deeper resolution power than that of PFGE, in particular within populations of similar geographic origin. For instance, a comparatively high level of diversity was revealed for five Swiss strains of similar origin (years 2007 to 2008; ACW56400, ACW63230, ACW63579, ACW63889, and ACW64132) that fell into three different CRISPR genotypes. While this may also reflect an increased diversity of more recent strains than of older and more clonal populations, no new spacer was acquired at the 5′ terminus in any of these strains, and diversity was mainly the result of duplications or deletions within CRRs. From this perspective, E. amylovora Ea263 appears to be the most evolutionarily recent strain in CRISPR group I, carrying an additional spacer compared to the other strains at the 5′ ends of both CRR1 and CRR2. Both spacers match sequences on plasmid SL483 of Salmonella enterica subsp. enterica (GenBank accession no. CP001137) and were possibly acquired from a related promiscuous enterobacterial plasmid.
Analysis of the different CRRs supports ideas about the origin of fire blight on domesticated Malus and Pyrus species. Global biodiversity among E. amylovora strains isolated from Spiraeoideae plants is very low in comparison to that among strains isolated from Rubus spp. We hypothesize that the Spiraeoideae genotype of E. amylovora was selectively enriched from the broader genetic pool present on wild host plants in North America with the introduction of European apple and pear. These new hosts may have selected pathogen genotypes highly virulent for domestic Malus and Pyrus. The primarily vegetative propagation of apple and pear trees employed in horticulture (i.e., grafting) would have resulted in clonal host populations unable to circumvent the disease, thus eliminating the need for diversification in the pathogen. This could explain the relatively small number of type III secretion system effectors, which reflect prolonged coevolutionary adaptation to diverse hosts, in E. amylovora (50) compared to other plant pathogens (e.g., Pseudomonas syringae and Xanthomonas) (15).
Based on CRISPR data, the most probable genotype of the earliest causal agent of fire blight in apple and pear was similar to that of group IA strains, with spacers 2012 and 2013 probably lost after the separation of group III strains from the common ancestor. In fact, neither group IB nor group II strains can be ancestral, because compared to group IA strains they display extensive deletions in the central regions of CRR2 and CRR1, respectively. Likewise, the 5′ end of CRR2 of group III strains shows a recent incorporation of new spacers possibly resulting from the encounter with plasmid pEU30. Accumulation of anti-pEU30 spacers is also evident in CRR1, which experienced major rearrangements leaving only a few spacers that share commonality with group I and II strains. The intermediate genotype observed for E. amylovora IH3-1 from Indian hawthorn warrants further analysis of strains from this ornamental, which was introduced into the United States from China, as a possible selective host for advanced divergence from Rubus or other Spiraeoideae genotypes. Together, our data support the hypothesis that fire blight and E. amylovora originated in the eastern United States and suggest a slow evolution during westward migration across North America. The genetic similarity observed within strains of the cosmopolitan CRISPR group IA also supports historical evidence of recent global dissemination of a founder strain. As with any biogeographic study, refinement based on deeper analysis of individual populations is needed, and this can be facilitated using the CRISPR tools described here (27).
Compatible with the established mode of action of the CRISPR/Cas system, there was generally a good correlation between the presence of spacers and the absence of target plasmids bearing cognate sequences. However, in group III strains such as OR29, pEU30 should theoretically be repelled by the accumulation of spacers perfectly matching several regions of the plasmid, and its retention may result from a partially defective CRISPR/Cas system. It is apparent that of the three distinct stages that shape the CRISPR/Cas defense mechanism (55), adaptation of the array via recognition of alien DNA and integration of new spacers is unaffected, while transcription analysis reveals that both cas gene and pre-crRNA are expressed at comparable levels in all three major CRISPR strain groups (our unpublished preliminary data). Thus, further work will be needed to assess whether the point mutation causing an amino acid substitution in Cse1 of group III strains is sufficient to affect subsequent processing of pre-crRNA or to disturb the interference mechanism itself, as the functions of individual cas gene products have not been elucidated completely. The four predicted ORFs between CRR1 and cas3 are not obviously involved in the CRISPR/Cas system, and their absence in group III strains is most likely not accountable for any malfunction thereof. However, their presence in group I and II strains as remnants of the genomic island found in Rubus strains further emphasizes the fact that group III strains are not ancestral in apple and pear trees.
Although CRISPR analysis applied to E. amylovora has a lower resolution than that of spoligotyping for Yersinia (44), Streptococcus (3), and E. coli (10), it offers greater strain discrimination than previously possible and provides a foundation for developing inoculum source-tracking methods. The chronological succession of spacers within CRRs also offers insight into fire blight spread, phage resistance, and plasmid content/evolution.
Supplementary Material
ACKNOWLEDGMENTS
We thank G. Sundin and G. McGhee for generously sharing strains with pEU30 and pEL60.
This work was supported by the Swiss Federal Office of Agriculture and conducted within the research network COST 864.
Footnotes
Supplemental material for this article may be found at http://aem.asm.org/.
Published ahead of print on 1 April 2011.
REFERENCES
- 1. Barionovi D., Giorgi S., Stoeger A. R., Ruppitsch W., Scortichini M. 2006. Characterization of Erwinia amylovora strains from different host plants using repetitive-sequences PCR analysis, and restriction fragment length polymorphism and short-sequence DNA repeats of plasmid pEA29. J. Appl. Microbiol. 100:1084–1094 [DOI] [PubMed] [Google Scholar]
- 2. Barrangou R., et al. 2007. CRISPR provides acquired resistance against viruses in prokaryotes. Science 315:1709–1712 [DOI] [PubMed] [Google Scholar]
- 3. Bolotin A., Quinquis B., Sorokin A., Ehrlich S. D. 2005. Clustered regularly interspaced short palindrome repeats (CRISPRs) have spacers of extrachromosomal origin. Microbiology 151:2551–2561 [DOI] [PubMed] [Google Scholar]
- 4. Bonn W. G., van der Zwet T. 2000. Distribution and economic importance of fire blight, p. 37–53 In Vanneste J. L. (ed.), Fire blight: the disease and its causative agent, Erwinia amylovora. CAB International, Wallingford, United Kingdom [Google Scholar]
- 5. Brouns S. J. J., et al. 2008. Small CRISPR RNAs guide antiviral defense in prokaryotes. Science 321:960–964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Carte J., Wang R., Li H., Terns R. M., Terns M. P. 2008. Cas6 is an endoribonuclease that generates guide RNAs for invader defense in prokaryotes. Genes Dev. 22:3489–3496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Crooks G. E., Hon G., Chandonia J.-M., Brenner S. E. 2004. WebLogo: a sequence logo generator. Genome Res. 14:1188–1190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Cui Y., et al. 2008. Insight into microevolution of Yersinia pestis by clustered regularly interspaced short palindromic repeats. PLoS One 3:e2652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Deveau H., et al. 2008. Phage response to CRISPR-encoded resistance in Streptococcus thermophilus. J. Bacteriol. 190:1390–1400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Díez-Villaseñor C., Almendros C., García-Martínez J., Mojica F. J. M. 2010. Diversity of CRISPR loci in Escherichia coli. Microbiology 156:1351–1361 [DOI] [PubMed] [Google Scholar]
- 11. Donat V., Biosca E. G., Peñalver J., López M. M. 2007. Exploring diversity among Spanish strains of Erwinia amylovora and possible infection sources. J. Appl. Microbiol. 103:1639–1649 [DOI] [PubMed] [Google Scholar]
- 12. Foster G. C., McGhee G. C., Jones A. L., Sundin G. W. 2004. Nucleotide sequences, genetic organization, and distribution of pEU30 and pEL60 from Erwinia amylovora. Appl. Environ. Microbiol. 70:7539–7544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Garneau J. E., et al. 2010. The CRISPR/Cas bacterial immune system cleaves bacteriophage and plasmid DNA. Nature 468:67–71 [DOI] [PubMed] [Google Scholar]
- 14. Goyal M., Saunders N. A., van Embden J. D., Young D. B., Shaw R. J. 1997. Differentiation of Mycobacterium tuberculosis isolates by spoligotyping and IS6110 restriction fragment length polymorphism. J. Clin. Microbiol. 35:647–651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Grant S. R., Fisher E. J., Chang J. H., Mole B. M., Dangl J. L. 2006. Subterfuge and manipulation: type III effector proteins of phytopathogenic bacteria. Annu. Rev. Microbiol. 60:425–449 [DOI] [PubMed] [Google Scholar]
- 16. Grissa I., Vergnaud G., Pourcel C. 2007. CRISPRFinder: a web tool to identify clustered regularly interspaced short palindromic repeats. Nucleic Acids Res. 35:W52–W57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Haft D. H., Selengut J., Mongodin E. F., Nelson K. E. 2005. A guild of 45 CRISPR-associated (Cas) protein families and multiple CRISPR/Cas subtypes exist in prokaryotic genomes. PLoS Comput. Biol. 1:e60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Hale C., Kleppe K., Terns R. M., Terns M. P. 2008. Prokaryotic silencing (psi)RNAs in Pyrococcus furiosus. RNA 14:2572–2579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Hale C. R., et al. 2009. RNA-guided RNA cleavage by a CRISPR RNA-Cas protein complex. Cell 139:945–956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Haurwitz R. E., Jinek M., Wiedenheft B., Zhou K., Doudna J. A. 2010. Sequence- and structure-specific RNA processing by a CRISPR endonuclease. Science 329:1355–1358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Held N. L., Herrera A., Cadillo-Quiroz H., Whitaker R. J. 2010. CRISPR associated diversity within a population of Sulfolobus islandicus. PLoS One 5:e12988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Hofacker I. L., et al. 1994. Fast folding and comparison of RNA secondary structures. Monatshefte Chemie 125:167–188 [Google Scholar]
- 23. Horvath P., Barrangou R. 2010. CRISPR/Cas, the immune system of bacteria and archaea. Science 327:167–170 [DOI] [PubMed] [Google Scholar]
- 24. Horvath P., et al. 2008. Diversity, activity, and evolution of CRISPR loci in Streptococcus thermophilus. J. Bacteriol. 190:1401–1412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Ishino Y., Shinagawa H., Makino K., Amemura M., Nakata A. 1987. Nucleotide sequence of the iap gene, responsible for alkaline phosphatase isozyme conversion in Escherichia coli, and identification of the gene product. J. Bacteriol. 169:5429–5433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Jansen R., van Embden J. D. A., Gaastra W., Schouls L. M. 2002. Identification of genes that are associated with DNA repeats in prokaryotes. Mol. Microbiol. 43:1565–1575 [DOI] [PubMed] [Google Scholar]
- 27. Jock S., Donat V., López M. M., Bazzi C., Geider K. 2002. Following spread of fire blight in Western, Central and Southern Europe by molecular differentiation of Erwinia amylovora strains with PFGE analysis. Environ. Microbiol. 4:106–114 [DOI] [PubMed] [Google Scholar]
- 28. Jock S., Geider K. 2004. Molecular differentiation of Erwinia amylovora strains from North America and of two Asian pear pathogens by analyses of PFGE patterns and hrpN genes. Environ. Microbiol. 6:480–490 [DOI] [PubMed] [Google Scholar]
- 29. Kube M., et al. 2008. The genome of Erwinia tasmaniensis strain Et1/99, a non-pathogenic bacterium in the genus Erwinia. Environ. Microbiol. 10:2211–2222 [DOI] [PubMed] [Google Scholar]
- 30. Kunin V., Sorek R., Hugenholtz P. 2007. Evolutionary conservation of sequence and secondary structures in CRISPR repeats. Genome Biol. 8:R61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Llop P., et al. 2006. The new plasmid pEI70 is present in Erwinia amylovora European strains. Acta Hort. 793:131–136 [Google Scholar]
- 32. Llop P., López M. M., Cabrefiga J., Ruz L., Montesinos E. 2008. Study of the virulence in wild-type strains of Erwinia amylovora devoid of the plasmid pEa29. Acta Hort. 793:145–148 [Google Scholar]
- 33. Makarova K. S., Grishin N. V., Shabalina S. A., Wolf Y. I., Koonin E. V. 2006. A putative RNA-interference-based immune system in prokaryotes: computational analysis of the predicted enzymatic machinery, functional analogies with eukaryotic RNAi, and hypothetical mechanisms of action. Biol. Direct 1:7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Marraffini L. A., Sontheimer E. J. 2008. CRISPR interference limits horizontal gene transfer in staphylococci by targeting DNA. Science 322:1843–1845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. McGhee G. C., Jones A. L. 2000. Complete nucleotide sequence of ubiquitous plasmid pEA29 from Erwinia amylovora strain Ea88: gene organization and intraspecies variation. Appl. Environ. Microbiol. 66:4897–4907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. McGhee G. C., et al. 2002. Relatedness of chromosomal and plasmid DNAs of Erwinia pyrifoliae and Erwinia amylovora. Appl. Environ. Microbiol. 68:6182–6192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. McManus P. S., Jones A. L. 1995. Genetic fingerprinting of Erwinia amylovora strains isolated from tree-fruit crops and Rubus spp. Phytopathology 85:1547–1553 [Google Scholar]
- 38. Mohan S. K., Thompson S. V. 1996. An outbreak of fire blight in plums. Acta Hort. 411:73–76 [Google Scholar]
- 39. Mojica F. J., Diez-Villasenor C., Garcia-Martinez J., Soria E. 2005. Intervening sequences of regularly spaced prokaryotic repeats derive from foreign genetic elements. J. Mol. Evol. 60:174–182 [DOI] [PubMed] [Google Scholar]
- 40. Mojica F. J. M., Díez-Villaseñor C., García-Martínez J., Almendros C. 2009. Short motif sequences determine the targets of the prokaryotic CRISPR defence system. Microbiology 155:733–740 [DOI] [PubMed] [Google Scholar]
- 41. Mokrousov I., et al. 2009. Novel macroarray-based method of Corynebacterium diphtheriae genotyping: evaluation in a field study in Belarus. Eur. J. Clin. Microbiol. Infect. Dis. 28:701–703 [DOI] [PubMed] [Google Scholar]
- 42. Momol M. T., Aldwinckle H. S. 2000. Genetic diversity and host range of Erwinia amylovora, p. 55–72 In Vanneste J. L. (ed.), Fire blight: the disease and its causative agent, Erwinia amylovora. CABI Publishing, Wallingford, United Kingdom [Google Scholar]
- 43. Potter D., et al. 2007. Phylogeny and classification of Rosaceae. Plant Syst. Evol. 266:5–43 [Google Scholar]
- 44. Pourcel C., Salvignol G., Vergnaud G. 2005. CRISPR elements in Yersinia pestis acquire new repeats by preferential uptake of bacteriophage DNA, and provide additional tools for evolutionary studies. Microbiology 151:653–663 [DOI] [PubMed] [Google Scholar]
- 45. Powney R., et al. 2011. Genome sequence of an Erwinia amylovora strain with restricted pathogenicity to Rubus plants. J. Bacteriol. 193:785–786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Rico A., Führer M. E., Ortiz-Barredo A., Murillo J. 2008. Polymerase chain reaction fingerprinting of Erwinia amylovora has a limited phylogenetic value but allows the design of highly specific molecular markers. Phytopathology 98:260–269 [DOI] [PubMed] [Google Scholar]
- 47. Rico A., Ortiz-Barredo A., Ritter E., Murillo J. 2004. Genetic characterization of Erwinia amylovora strains by amplified fragment length polymorphism. J. Appl. Microbiol. 96:302–310 [DOI] [PubMed] [Google Scholar]
- 48. Schnabel E. L., Jones A. L. 2001. Isolation and characterization of five Erwinia amylovora bacteriophages and assessment of phage resistance in strains of Erwinia amylovora. Appl. Environ. Microbiol. 67:59–64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Smits T. H. M., et al. 2010. Complete genome sequence of the fire blight pathogen Erwinia pyrifoliae DSM 12163T and comparative genomic insights into plant pathogenicity. BMC Genomics 11:2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Smits T. H. M., et al. 2010. Complete genome sequence of the fire blight pathogen Erwinia amylovora CFBP 1430 and comparison to other Erwinia species. Mol. Plant Microbe Interact. 23:384–393 [DOI] [PubMed] [Google Scholar]
- 51. Sorek R., Kunin V., Hugenholtz P. 2008. CRISPR—a widespread system that provides acquired resistance against phages in bacteria and archaea. Nat. Rev. Microbiol. 6:181–186 [DOI] [PubMed] [Google Scholar]
- 52. Stern A., Keren L., Wurtzel O., Amitai G., Sorek R. 2010. Self-targeting by CRISPR: gene regulation or autoimmunity? Trends Genet. 26:335–340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Tamura K., Dudley J., Nei M., Kumar S. 2007. MEGA4: molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol. Biol. Evol. 24:1596–1599 [DOI] [PubMed] [Google Scholar]
- 54. Tyson G. W., Banfield J. F. 2008. Rapidly evolving CRISPRs implicated in acquired resistance of microorganisms to viruses. Environ. Microbiol. 10:200–207 [DOI] [PubMed] [Google Scholar]
- 55. van der Oost J., Jore M. M., Westra E. R., Lundgren M., Brouns S. J. J. 2009. CRISPR-based adaptive and heritable immunity in prokaryotes. Trends Biochem. Sci. 34:401–407 [DOI] [PubMed] [Google Scholar]
- 56. van der Ploeg J. R. 2009. Analysis of CRISPR in Streptococcus mutans suggests frequent occurrence of acquired immunity against infection by M102-like bacteriophages. Microbiology 155:1966–1976 [DOI] [PubMed] [Google Scholar]
- 57. Zhang Y., Merighi M., Bazzi C., Geider K. 1998. Genomic analysis by pulsed-field gel electrophoresis of Erwinia amylovora strains from the Mediterranean region including Italy. J. Plant Pathol. 80:225–232 [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.