Abstract
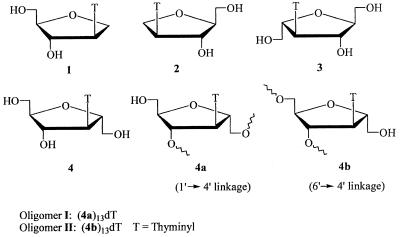
Oligonucleotides consisting of the isonucleoside repeating unit 2′,5′-anhydro-3′-deoxy-3′-(thymin-1-yl)-d-mannitol (4) were synthesized with the monomeric unit 4 incorporated into oligonucleotides as 1′→4′ linkage 4a (oligomer I) or 6′→4′ linkage 4b (oligomer II). The hybrid properties of the two oligonucleotides I and II with their complementary strands were investigated by thermal denaturation and CD spectra. Oligonucleotide I (4a) formed a stable duplex with d(A)14 with a slightly reduced Tm value of 36.6°C, relative to 38.2°C for the control duplex d(T)14/d(A)14, but oligomer II (4b) failed to hybridize with a DNA complementary single strand. The spectrum of the duplex oligomer I/d(A)14 showed a positive CD band at 217 nm and a negative CD band at 248 nm attributable to a B-like conformation. Molecular modeling showed that in the case of oligomer I the C6′ hydroxy group of each unit could be located in the groove area when hybridized to the DNA single strand, which might contribute additional hydrogen bonding to the stability of duplex formation.
INTRODUCTION
The selective inhibition of expression of specific genes by oligonucleotides via an antisense or antigene strategy provides an attractive and elegant approach to drug discovery (1). Several requirements must be fulfilled by a potential antisense oligonucleotide. Efforts to enhance the biological activity of oligonucleotides as inhibitors of gene expression have been made by improvements in their stability to nuclease digestion, in their ability to penetrate the cell membrane and in their efficient hybridization to the target RNA/DNA. Antisense oligonucleotides with phosphorothioate backbones exhibit several advantages over other forms, including relatively high nuclease resistance as well as the ability to induce degradation of the target sequence by RNase H (2,3). Currently one drug based on the antisense strategy has been approved by the FDA and clinical evaluation of many other antisense phosphorothioate ODNs and DNA–RNA oligomer chimeras is underway (4). However, phosphorothioate oligonucleotides are possibly hydrolyzed, primarily from the 3′-end, and have also been shown to block proliferation of HIV-1 in acutely infected cells in a non-sequence-specific manner (5). Another problem in the use of such modified antisense oligonucleotides is their inefficient cellular uptake. Research efforts continue in the field aimed at the construction of novel oligonucleotide analogs with improved biochemical and pharmacological properties. Oligonucleotides with various modifications of the internucleotide linkages and/or the sugar rings have been described (6,7). Most of these modifications contain a five-membered sugar ring closely resembling the natural 2-deoxyribose (8–10). Oligonucleotides consisting of hexose or cyclopentanediol moiety nucleoside analogs were reported to possess significantly increased stability towards phosphodiesterases whereby the hybridization properties are retained (11–15). Recently the locked nucleic acid (LNA) was introduced, where the backbone of the oligonucleotide consists of a conformationally restricted nucleotide with a 2′-O-4′-C-methylene bridge, which has shown unprecedented helical thermal stability when hybridized to complementary DNA and RNA (16).
Isonucleosides represent a novel class of carbohydrate-modified nucleosides in which the nucleobase is linked to various positions of ribose other than C1′. The torsion angles in the sugar–phosphate backbones of such oligonucleotides consisting of isonucleosides exhibit profound changes compared to regular oligonucleotides. These alternations in torsion angles might affect the recognition of such oligomers by nucleases. It could also be anticipated that the bases in the modified oligonucleotides retain their hybridization properties with complementary sequences. This prompted us to study the hybridization properties and enzymatic stability of oligonucleotides bearing such isonucleosides (17–20). Taktakishvili et al. reported the recognition and inhibition of HIV integrase by some novel dinucleotides consisting of isonucleoside (21). We have recently reported the synthesis and duplex stabilization of oligonucleotides consisting of isonucleosides. Interestingly it was found that oligonucleotides consisting of 1′,4′-anhydro-2′-deoxy-2′-(thymin-1-yl)-d-arabinitol (1) could form a duplex with complementary d(A)14; the Tm value showed a slight decrease compared with that of the parent duplex d(A)14:d(T)14. An oligonucleotide consisting of 1′,4′-anhydro-2′-deoxy-2′-(thymin-1-yl)-l-arabinitol (2), the enantiomer of isonucleoside 1, could not form regular base pairing. Oligonucleotides consisting of 2′,5′-anhydro-3′-deoxy-3′-(thymin-1-yl)-l-mannitol (3) (6′→4′ linkage), containing an extra hydroxymethyl group in 2, leads to a common type of duplex structure (22). This shows that the additional hydroxymethyl group in the sugar moiety can increase duplex stability.
In the context of our study on antisense oligonucleotides we initiated a study of the synthesis and properties of oligonucleotides consisting of the repeating unit 2′,5′-anhydro-3′-deoxy-3′-(thymin-1-yl)-d-mannitol (4), the enantiomer of isonucleoside 3, containing an extra C-hydroxymethyl group at the C1′ position of isonucleoside 1. This distinct structural feature and hybrid property should provide information for further understanding the recognition of modified oligonucleotide:DNA/RNA hybrids. In this report monomeric unit 4 was incorporated into oligonucleotides in two distinct structural forms, namely as a (1′→4′) linkage (4a) or (6′→4′) linkage (4b), respectively. Oligonucleotides I and II were designed such that the different conformers in these phosphodiester linkages lie on one face of the sugar rings and the nucleobases are located on the other face (Fig. 1). In order to understand whether oligomer I or II with different linkage modes could influence the interaction with complementary DNA, computer-assisted molecular dynamics simulations of the two duplexes were performed to mimic the solution structures.
Figure 1.
Structures of isonucleosides and oligomers mentioned in this paper.
MATERIALS AND METHODS
General
All solvents were dried and distilled prior to use. Thin layer chromatography (TLC) was performed using silica gel GF-254 (Qing-Dao Chemical Co., China) plates with detection by UV. Column chromatography was performed on silica gel (200–300 mesh; Qing-Dao Chemical Co.). UV spectra were recorded with a Pharmacia LKB Biochrom 4060 spectrophotometer. Optical rotations were recorded on a Perkin-Elmer 243B polarimeter. ZAB-HS, KYKY-ZHP-5 and LSI1700 (Linear Scientific Inc.) instruments were used for mass spectra and MALDI-TOF mass spectra. NMR spectra were recorded with a Varian VXR-300 or Bruker DPX-400 instrument with TMS as internal standard. Evaporations were carried out under reduced pressure with a bath temperature <45°C.
2′,5′-Anhydro-6′-O-(dimethoxytrityl)-1′-O-benzoyl-3′-deoxy-3′-(thymin-1-yl)-d-mannitol (11)
To a solution of 2′,5′-anhydro-1′,6′-O-dibenzoyl-3′-deoxy-3′-(thymin-1-yl)-d-mannitol (8) (1.2 g, 2.5 mmol) in anhydrous methanol (20 ml) was added saturated ammonia/anhydrous methanol (10 ml). The reaction mixture was stirred at room temperature for ∼1 h and carefully monitored by TLC. Once the fully deprotected product 4 was detected, the reaction was stopped by removing the solvent under reduced pressure. The resulting residue was applied to a silica gel column, eluting with 5% methanol in methylene chloride, to afford the partially deprotected products 2′,5′-anhydro-1′-O-benzoyl-3′-deoxy-3′-(thymin-1-yl)-d-mannitol (9) (40%) and 2′,5′-anhydro-6′-O-benzoyl-3′-deoxy-3′-(thymin-1-yl)-d-mannitol (10) (35%) and some starting material (8).
Compound 9 (0.26 g, 0.69 mmol) was dissolved in pyridine (10 ml), and dimethoxytrityl chloride (0.45 g) was added. The solution was stirred at room temperature for 24 h. After evaporation the mixture was purified by silica gel chromatography, eluting with petroleum ether/CHCl2/EtOAc (8:1:1–0:1:1) (0.5% Et3N added), to afford compound 11 as a colorless foam in 79% yield.
1H-NMR (DMSO-d6): δ 1.70 (s, 3H, 5-CH3), 3.14 (m, 2H, H-6′), 3.70 (s, 6H, DMT-OCH3), 4.01 (m, 2H, H-1′), 4.26 (m, 1H), 4.38 (m, 3H), 4.82 (m, 1H, H-3′), 5.64 (d, J = 5.4 Hz, D2O exchangeable, 1H, 4′-OH), 6.86 (m, 4H), 7.19–7.29 (m, 7H), 7.27 (s, 1H, H-6), 7.41 (m, 2H), 7.53 (m, 2H), 7.71 (m, 2H), 7.99 (d, J = 7.5 Hz, 2H), 11.32 (s, 1H, D2O exchangeable, NH). FAB-MS m/z: 679 [M+H]+. Calculated for C39H38N2O9: C, 69.03; H, 5.60; N, 4.13. Found: C, 68.69; H, 6.09; N, 4.60.
2′,5′-Anhydro-1′-O-(dimethoxytrityl)-6′-O-benzoyl-3′-deoxy-3′-(thymin-1-yl)-d-mannitol (12)
Analogous to the preparation of 11 from 9, compound 12 was obtained from 10 in 61% yield as a colorless foam.
1H-NMR (300 MHz, DMSO-d6): δ 1.76 (s, 3H, 5-CH3), 3.09 (d, J = 4.5 Hz, 2H, H-1′), 3.70 (s, 6H, DMT-OCH3), 4.13 (m, 2H, H-6′), 4.38–4.50 (m, 3H), 4.81 (t, J = 8.4 Hz, 1H, H-3′), 5.78 (d, J = 5.4 Hz, D2O exchangeable, 1H, 4′-OH), 6.80 (m, 4H), 7.20 (s, 1H, H-6), 7.15–7.45 (m, 7H), 7.55–7.95 (m, 4H), 8.01 (d, J = 7.5 Hz, 2H), 11.32 (s, 1H, D2O exchangeable, NH). FAB-MS m/z: 679 [M+H]+. Calculated for C39H38N2O9: C, 69.03; H, 5.60; N, 4.13. Found: C, 60.31; H, 6.00; N, 4.44.
2′,5′-Anhydro-6′-O-(dimethoxytrityl)-1′-O-benzoyl-3′-deoxy-3′-(thymin-1-yl)-d-mannitol-4′-(2-cyanoethyl-N,N-diisopropylphosphoramidite) (13)
Compound 11 (0.250 g, 0.37 mmol) was dried by repeated co-evaporation with MeCN (3 × 2 ml) in vacuo and dissolved in anhydrous THF (3.5 ml) under argon. To this solution was added diisopropylethylamine (DIPEA, 0.26 ml, 1.48 mmol) and 2-cyanoethyl-N,N-diisopropyl-chlorophosphoramidite (0.165 ml, 0.74 mmol). The mixture was stirred at 0°C for 10 min, then at room temperature for 30 min. The reaction mixture was quenched by addition of MeOH (1 ml). After stirring for 10 min, EtOAc (25 ml) was added and the organic layer was washed with 5% aqueous NaHCO3 (2 × 7.0 ml), followed by H2O (1 × 7.0 ml). The solution was dried (Na2SO4), then evaporated under reduced pressure to an oily residue and the residue purified by silica gel column chromatography eluting with petroleum ether/CHCl2/EtOAc (6:1:1–2:1:1, 0.5% Et3N) to afford compound 13 as a colorless foam (0.265 g, 81%).
31P-NMR (CDCl3): δ 152.6, 153.9.
2′,5′-Anhydro-1′-O-(dimethoxytrityl)-6′-O-benzoyl-3′-deoxy-3′-(thymin-1-yl)-d-mannitol-4′-(2-cyanoethyl-N,N-diisopropylphosphoramidite) (14)
Analogous to the preparation of 13, compound 14 was prepared from 12 as a white foam (0.251 g, 77.5%).
31P-NMR (CDCl3): δ 152.8, 153.5.
Solid phase synthesis of oligonucleotides I and II
Oligonucleotide syntheses were carried out on the 1 µmol scale on a DNA synthesizer model 391A (Applied Biosystems) applying regular phosphoramidite chemistry. Cleavage and deprotection of the oligomers were performed in concentrated aqueous ammonia solution at 50°C for 24 h. The oligomers were purified using oligonucleotide purification cartridges (Perkin Elmer, Applied Biosystems). The pure oligonucleotides were lyophilized and stored at –20°C.
Enzymatic stability of the oligomers
The oligonucleotides (0.2 OD) in 1.0 ml of buffer solution (0.1 M NaCl, 14 mM MgCl2, 0.1 M Tris–HCl, pH 8.6) were digested with 1.2 U snake venom phosphodiesterase (SVPDE) at 37°C. During digestion the increase in absorbance at 260 nm was followed. The absorption versus time curve of the digestion was plotted and the hyperchromicity was evaluated.
UV melting experiments
UV melting experiments were recorded with a Pharmacia LKB Biochrom 4060 spectrophotomer. Samples were dissolved in a buffer solution containing 0.14 M NaCl, 0.01 M Na2HPO4, 1 mM EDTA, pH 7.2. The solution, containing each ODN and the complementary d(A)14, was heated at 80°C for 5 min, then cooled gradually to 4°C and used for the thermal denaturation studies. Thermally induced transitions of each mixture were monitored at 260 nm. Sample temperature was increased at 0.5°C/min between 15 and 75°C. In all experiments the concentration of each oligonucleotide strand was 2 µM.
CD spectra
CD spectra were measured at 5°C with a J 720 polarized spectrophotometer (JAC) in thermostatically controlled 1 cm cuvettes. The oligomers were dissolved and analyzed in buffer containing 10 mM Na2HPO4, 0.14 M NaCl, 1.0 mM EDTA, pH 7.2, and at a concentration of 4 µM each strand.
Molecular modeling
All molecular modeling processes were performed on a SGI Indy workstation. The structures of the duplexes were constructed using the Biopolymer module in INSIGHT II 95.0 (Biosym).
RESULTS AND DISCUSSION
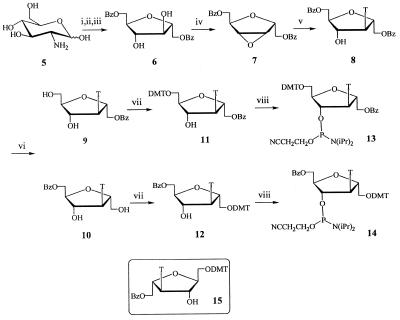
Building blocks 13 and 14, suitable for solid phase oligonucleotide synthesis, were obtained starting from 2′,5′-anhydro-1′,6′-O-dibenzoyl-3′-deoxy-3′-(thymin-1-yl)-d-mannitol (8), which was prepared earlier in our laboratory from d-glucosamine in five steps (23; Fig. 2). When compound 8 was debenzoylated with 5% NH3/CH3OH at room temperature the reaction was found to be sequential and time-dependent. The rate of hydrolysis of the two benzoyl groups in 8 is almost the same. This useful process led to the partially debenzoylated products 9 and 10 in one step. Thus, by carefully monitoring the deprotection process the desirable intermediates 9 and 10 were afforded in 40 and 35% yields, respectively. After protection with a dimethoxytrityl group and phosphitylation by the standard procedure, 9 and 10 gave phosphoramidites 13 and 14, respectively, in good yields.
Figure 2.
Synthesis protocol of isonucleosides and isonucleoside phosphoramidites. Reagents and conditions: (i) NaNO2, concentrated HCl, H2O, 0°C; (ii) NaBH4, H2O, room temperature; (iii) BzCl, pyridine, CH2Cl2 (1:2 v/v), room temperature; (iv) PPh3, diethyl azodicarboxylate, 1,4-dioxane, 70°C; (v) thymine, DBU, DMF, 90–100°C; (vi) 5% NH3/CH3OH, room temperature; (vii) DMTrCl, pyridine, room temperature; (viii) chloro-(2-cyanoethoxy)-(diisopropylamine)phosphine, Et(i-Pr)2N, THF, room temperature. DMTr, 4,4′-dimethoxytrityl; Bz, benzoyl; T, thyminyl.
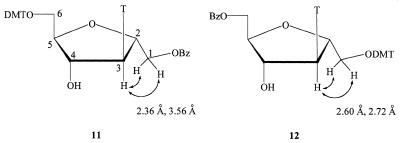
We have reported the synthesis of 2′,5′-anhydro-1′-O-benzoyl-3′-deoxy-3′-(thymin-1-yl)-5′-O-(dimethoxytrityl)-l-mannitol (15) (22). The 1H-NMR spectra of 11 and 15 were identical, suggesting that they are enantiomers of each other. This structural assignment was supported by the NOESY spectra of 11 and 12, which clearly showed a NOE correlation between H1′ and H3′ in compounds 11 and 12. The chemical shift of H1′ in 11 was downfield compared to that in 12, because the benzoyl group is a stronger electron acceptor than the DMT group (Fig. 3).
Figure 3.
NOE in compounds 11 and 12.
Oligonucleotides I and II, fully constructed with 4 in two different structural forms, were assembled on an automated DNA synthesizer on the 1 µmol scale. For convenience the synthesis was started with commercially available thymidine-loaded controlled pore glass. Synthesis followed the standard protocol except for a prolonged coupling time of 200 s to ensure adequate coupling yields. The coupling efficiency was determined by release of DMT (13, 80%; 14, 84%). The lower coupling yields are probably due to steric hindrance caused by the benzoyl group-protected side chain in building blocks 13 and 14. After cleavage from the solid phase support and concomitant removal of the protecting groups with concentrated aqueous ammonia (24 h at 50°C) the crude DMT-protected oligomers were purified and detritylated on an oligonucleotide purification cartridge and the compositions of oligomers I and II confirmed by MALDI-TOF mass spectrometry (Table 1).
Table 1. Sequence data, incorporation yield and mass spectrometric analysis of oligomers I and II.
| Oligomer | Sequence | Incorporation | MALDI-TOF-MS | |
| |
|
(%) |
Theoretical |
Observed |
| I | (4a)13dT (1′→4′) | 80 | 4584.8 | 4582.0 |
| II | (4b)13dT (6′→4′) | 84 | 4584.8 | 4583.6 |
The enzymatic stabilities of oligonucleotides I and II were analyzed by monitoring the hyperchromic effect upon addition of SVPDE (3′-exonuclease) to a buffer solution of the oligomers. The results indicated that no significant change in UV absorbance occurred within 30 min when oligomer I or II was incubated with SVPDE, while regular d(T)14 gave a time-dependent increase in absorbance (Table 2).
Table 2. Enzymatic stabilitya and Tm datab of oligomers I and II.
| Entry |
Duplex |
t1/2 (min) |
Henzym.c (%) |
Tm (°C) |
ΔTm/mod. (°C) |
Hdenat.d (%) |
| 1 | I/d(A)14 | >30.0 | n.d.e | 36.6 | –0.12 | 7.5 |
| 2 | II/d(A)14 | >30.0 | n.d. | <20.0 | n.d. | |
| 3 | d(T)14/d(A)14 | 2.5 | 23.0 | 38.2 | 18.7 |
aMeasured in 0.1 M Tris–HCl, pH 8.6, 0.1 M NaCl, 14 mM MgCl2. [c]oligomer 1 µM.
bMeasured in 10 mM Na2HPO4, pH 7.2, 0.14 M NaCl, 1.0 mM EDTA. [c]tol 4 µM. Absorbance detected at 260 nm.
cEnzymatic hyperchromicity.
dDenaturation hyperchromicity.
en.d., not detected.
The hybrid properties of oligonucleotides I and II with complementary d(A)14 were investigated by thermal denaturation studies. Oligomer II (with a 6′→4′ linkage) failed to hybridize with d(A)14, nevertheless, a stable duplex of oligomer I (with a 1′→4′ linkage) with d(A)14 was formed with a slightly reduced Tm value of 36.6°C and a ΔTm of –0.12°C per modification, relative to the Tm of 38.2°C for the control duplex d(T)14/d(A)14. Another aspect in the thermal denaturation experiments was that the hyperchromicity value for duplex I/d(A)14 was much less than that for the control duplex d(T)14/d(A)14, which reflected poor base stacking within duplex I/d(A)14 (Table 2).
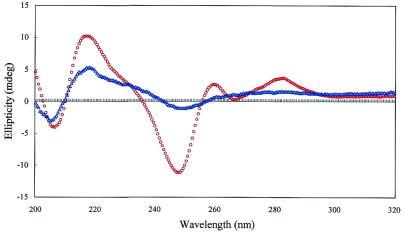
To study the global conformation of the duplex the CD spectrum of a duplex composed of oligomer I and the complementary d(A)14 was measured. The spectrum of the control duplex d(T)14/d(A)14 showed a positive CD band at 217 nm and a negative CD band at 248 nm attributable to a B-like DNA conformation. The shape of the spectrum of duplex I/d(A)14 was very similar to that of the control duplex. However, the intensity of the negative and positive bands in the spectrum of the duplex was much reduced compared with those for the control duplex. These results suggested that the duplex formed by oligonucleotide I with a complementary single strand may adopt a B-like DNA conformation and that the formation of Watson–Crick hydrogen bonding was perturbed by the torsion of the backbone in such a duplex, which caused poor base stacking (Fig. 4).
Figure 4.
CD spectra of ODN–DNA duplexes at 5°C (Co = 4 µM, d = 1 cm, in 10 mM Na2HPO4, 0.14 M NaCl, 1.0 mM EDTA, pH 7.2). Blue triangles, I/d(A)14; red circles, II/d(A)14; black squares, d(T)14/d(A)14
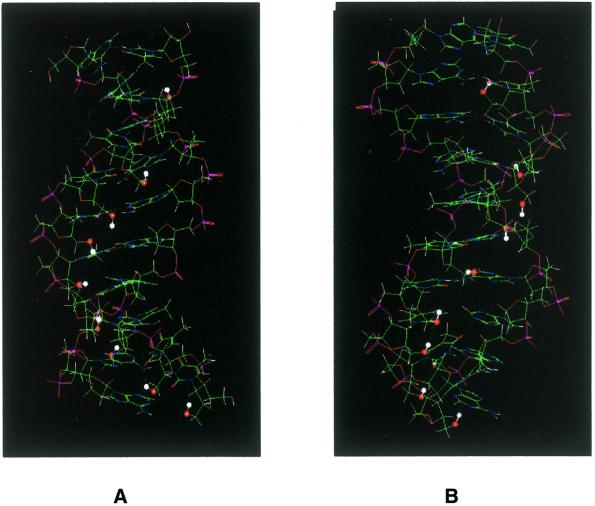
As described previously, oligonucleotides consisting of isonucleoside 3 can form duplexes with complementary single strands with a higher stability due to the additional hydroxymethyl group in building block 3 (22). However, while oligonucleotides I and II both consist of isonucleoside 4, they show different hybrid properties. This indicates that the location of the hydroxymethyl group in oligomers I and II may play a key role in the formation of duplexes. For a further understanding of the recognition of oligonucleotides I and II by DNA single strands we investigated duplex formation by molecular modeling. In I/d(A)10 the C6′ hydroxy group of each unit is located in the groove area when hybridized to the complementary strand, where it can form hydrogen bonds with water in the medium and could contribute to the stability of formation of the duplex, while in II/d(A)10 most of the C1′ hydroxy groups are directed to the inside of the duplex (Fig. 5).
Figure 5.
Molecular modeling results. The hydroxy groups of the hydroxymethyl moiety are highlighted, with red balls representing oxygen atoms and white balls hydrogen atoms. (A) (4a)9dT/d(A)10 duplex. (B) (4b)9dT/d(A)10.
The stability of nucleic acid structures in solution depends on the formation of hydrogen bonds and base stacking. In our case the base stacking in duplex I/dA14 is unfavorable owing to the torsion of a backbone composed of isonucleoside 4, but the additional hydrogen bonding formed by the hydroxy group located in the groove area with water in the medium could be advantageous for duplex formation.
In summary, two novel classes of oligonucleotide analogs consisting of isonucleoside unit 4 were synthesized. Both of them were highly resistant to SVPDE (3′-exonuclease). Oligomer I, composed of isonucleoside unit 4a with a 1′→4′ linkage, exhibited nearly the same hybridization potential as unmodified oligonucleotides towards complementary DNA. However, oligomer II, consisting of unit 4b with a 6′→4′ linkage, failed to form a duplex with complementary DNA. The modified oligomer I reported here contains a supplementary hydroxyl group which, when located in the groove area in a duplex, could provide additional hydrogen bonds with water in the medium and might contribute to the stability of duplex formation.
Acknowledgments
ACKNOWLEDGEMENTS
This work was supported by the National Natural Science Foundation of China and by research grant G1998051103 awarded by The Ministry of Science and Technology, People’s Republic of China.
References
- 1.Zamecnik P.C. and Stephenson,M.L. (1978) Inhibition of Rous sarcoma virus replication and cell transformation by a specific oligonucleotide. Proc. Natl Acad. Sci. USA, 75, 280–284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Stein C.A. and Cohen,J.S. (1988) Oligodeoxynucleotides as inhibitors of gene expression: a review. Cancer Res., 48, 2659–2668. [PubMed] [Google Scholar]
- 3.Uhlmann E. and Peyman,A. (1990) Antisense oligonucleotides: a new therapeutic principle. Chem. Rev., 90, 543–584. [Google Scholar]
- 4.Nemunaitis J., Holmlund,J.T., Kraynak,M., Richards,D., Bruce,J., Ognoskie,N., Kwoh,T.J., Geary,R., Dorr,A., Von Hoff,D. and Eckhardt,S.G. (1999) Phase I evaluation of ISIS 3521, an antisense oligonucleotide to protein kinase Cα, in patients with advanced cancer. J. Clin. Oncol., 17, 3586–3595. [DOI] [PubMed] [Google Scholar]
- 5.Matsukura M., Shinozuka,K., Zon,G., Mitsuya,H., Reitz,M., Cohen,J.C. and Broder,S. (1987) Phosphorothiate analogs of replication and cytopathic effects of human immunodeficiency virus. Proc. Natl Acad. Sci. USA, 84, 7706–7710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.De Mesmaeker A., Haner,R., Martin,P. and Moser,H.E. (1995) Antisense oligonucleotides. Acc. Chem. Res., 28, 366–374. [Google Scholar]
- 7.Wenzel T. and Nair,V. (1998) Self-complementary oligodeoxyribonucleotides incorporating l-related isonucleosides: synthesis, physical characterization, enzymology and CD studies. Bioconjugate Chem., 9, 683–690. [DOI] [PubMed] [Google Scholar]
- 8.Debart F., Rayner,B., Degols,G. and Imbach,J.-L. (1992) Synthesis and base-paring properties of the nuclease-resistant α-[r(UCUUAACCCACA)]. Nucleic Acids Res., 20, 1193–1200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sproat B.S., Beijer,B. and Iribarren,A. (1990) New synthetic routes to protected purine 2′-O-methylriboside-3′-O-phosphoramidites using a novel alkylation procedure. Nucleic Acids Res., 18, 41–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sagi J., Szemzo,A., Szecsi,J. and Otvos,L. (1990) Biochemical properties of oligo[(+)-carbocyclicthymidylates] and their complexes. Nucleic Acids Res., 18, 2133–2140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Augustyns K., Vandendriessche,F., Aerschot,A.V., Busson,R., Urbanke,C. and Herdewijn,P. (1992) Incorporation of hexose nucleoside analogues into oligonucleotides: synthesis, base-paring properties and enzymatic stability. Nucleic Acids Res., 20, 4711–4716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Marangoni M., Aerschot,A.V., Augustyns,P., Rozenski,J. and Herdewijn,P. (1997) Synthesis and hybridization properties of inverse oligonucleotides. Nucleic Acids Res., 25, 3034–3041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pitsch S., Wendeborn,S., Jaun,B. and Eschmemoser,A. (1993) Why pentose- and not hexose-nucleic acids? Part II, pyranosyl-RNA (’p-RNA). Helv. Chim. Acta, 76, 2161–2183. [Google Scholar]
- 14.Pitsch S., Krishnamurthy,R., Bolli,M., Wendebom,S., Holzner,A., Minton,M., Lesueur,C., Schlonvogt,I., Jaun,B. and Eschenmoser,A. (1995) Pyranosyl-RNA (’p-RNA): base-pairing selectivity and potential to replicate. Helv. Chim. Acta, 78, 1621–1635. [Google Scholar]
- 15.Beier M., Reck,F., Wagner,T., Kirshnamurthy,R. and Eschenmoser,A. (1999) Chemical etiology of nucleic acid structure: comparing pentopyranosyl-(2′→4′) oligonucleotides with RNA. Science, 283, 699–703. [DOI] [PubMed] [Google Scholar]
- 16.Wengel J., Koshkin,A., Singh,K.S., Nielson,P., Meldgaard,M., Rajwanshi,V.K., Kumar,R., Skouv,J., Nielsen,C.B., Jacobsen,J.P., Jacobsen,N. and Olsen,C.E. (1999) LNA (locked nucleic acids). Nucl. Nucl., 18, 1365–1370. [Google Scholar]
- 17.Yu H.-W., Zhang,L.-R., Ma,L.-T. and Zhang,L.-H. (1996) Studies on the synthesis and biological activities of 4′-(R)-hydroxy-5′-(S)-hydroxymethyl-tetrahydrofuranyl purines and pyrimidines. Bioorg. Med. Chem., 4, 609–614. [DOI] [PubMed] [Google Scholar]
- 18.Zhang H.-Y., Yu,H.-W., Ma,L.-T., Min,J.-M. and Zhang,L.-H. (1998) Synthesis of 2-C-(4-aminocarbonyl-2-thiazoyl)-1,4-anhydro-l-xylitols and their fluoro derivatives. Tetrahedron Asymmetry, 9, 141–149. [Google Scholar]
- 19.Yang Z.-J., Yu,H.-W., Min,J.-M., Ma,L.-T. and Zhang,L.-H. (1997) Stereoselective synthesis of 4-deoxy-4-nucleobase-2,5-anhydro-l-mannitol derivatives. Tetrahedron Asymmetry, 8, 2739–2347. [Google Scholar]
- 20.Yu H.-W., Zhang,H.-Y., Yang,Z.-J., Min,J.-M., Ma,L.-T. and Zhang,L.-H. (1998) Studies on the synthesis and biological activities of isonucleosides. Pure Appl. Chem., 70, 435–438. [Google Scholar]
- 21.Taktakishvili M., Neamati,N., Pommier,Y., Pal,S. and Nair,V. (2000) Recognition and inhibition of HIV integrase by novel dinucleotides. J. Am. Chem. Soc., 122, 5671–5677. [DOI] [PubMed] [Google Scholar]
- 22.Yang Z., Zhang,H., Min,J., Ma,L. and Zhang,L. (1999) Synthesis and duplex stabilization of oligonucleotides consisting of isonucleosides. Helv. Chim. Acta, 82, 2037–2043. [Google Scholar]
- 23.Lei Z., Min,J.M. and Zhang,L.H. (2000) Synthesis of 3-deoxy-3-nucleobase-2,5-anhydro-d-mannitol: a novel class of hydroxymethy-branched isonucleosides, Tetrahedron Asymmetry, 11, 2899–2906. [Google Scholar]