Abstract
In abandoned coal mines, methanogenic archaea are responsible for the production of substantial amounts of methane. The present study aimed to directly unravel the active methanogens mediating methane release as well as active bacteria potentially involved in the trophic network. Therefore, the stable-isotope-labeled precursors of methane, [13C]acetate and H2-13CO2, were fed to liquid cultures from hard coal and mine timber from a coal mine in Germany. Guided by methane production rates, samples for DNA stable-isotope probing (SIP) with subsequent quantitative PCR and denaturing gradient gel electrophoretic (DGGE) analyses were taken over 6 months. Surprisingly, the formation of [13C]methane was linked to acetoclastic methanogenesis in both the [13C]acetate- and the H2-13CO2-amended cultures of coal and timber. H2-13CO2 was used mainly by acetogens related to Pelobacter acetylenicus and Clostridium species. Active methanogens, closely affiliated with Methanosarcina barkeri, utilized the readily available acetate rather than the thermodynamically more favorable hydrogen. Thus, the methanogenic microbial community appears to be highly adapted to the low-H2 conditions found in coal mines.
INTRODUCTION
Worldwide, mine gas emissions from active and abandoned coal mines release substantial amounts of methane, contributing as much as 7% of global methane formation (4). Mine gas is a hazard but is also a potential source of methane for the industry. Stable carbon and hydrogen isotopic signatures indicate that methane in mine gas has mixed thermogenic and biogenic origins (31, 33). In abandoned coal mines, thermogenic methane is a remainder of geological processes, but biogenic formation of methane is still going on (14). Besides hard coal, possible sources for methane are large amounts of mine timber used for the construction of mines and left behind after the cessation of mining.
Generally, methane is produced from either acetate, hydrogen, or methylotrophic substrates as precursors. Recently, we showed that methylotrophic methanogenesis does not have a quantitative impact on in situ processes (2). However, while hydrogen is energetically favorable, acetate is the quantitatively more available substrate (38). In previous studies we have revealed that acetate is an important intermediate of the degradation processes and the main precursor of the biogenic methane in abandoned coal mines (14). We have also shown the presence of Methanosarcina spp. as the dominating archaea (2). Methanosarcina spp. are able to use acetate as well as H2-CO2. While other investigators reported that methanogenesis in coal mines is driven mainly by H2-utilizing archaea (7, 28), our studies indicated that acetoclastic methanogenesis seems to be the main methanogenic process, at least in the abandoned coal mines we have investigated (2).
The activity of methanogens in different habitats can be studied by using stable-isotope probing (SIP) (16, 17). DNA-SIP allows the identification of specifically active members of microbial populations based on the incorporation of 13C into the DNA of cells consuming labeled substrates (18). In this technique, labeled DNA is resolved after incubation under label addition by subsequent isopycnic gradient ultracentrifugation (22). Hence, guilds of methanogens that utilize 13C-labeled methanogenic substrates such as [13C]acetate or H2-13CO2 can be recovered in 13C-enriched DNA. However, both methanogenic precursors can also be utilized by syntrophic acetate-oxidizing and/or homoacetogenic bacteria, respectively (27, 32). Until now, only a few SIP studies have been performed in coal habitats. Han et al. (10) investigated the active methanotrophic community in a Chinese coal deposit. However, no studies have revealed the activity of microorganisms directly involved in methane production in abandoned coal mines. In our present study, we wanted to identify active methanogens and to test the hypothesis that acetate is the main precursor of methane, even if hydrogen is available as an energetically more favorable electron donor.
MATERIALS AND METHODS
Sample collection and enrichment cultures.
Samples were collected in May 2007 in sealed compartments of coal mines closed in the 1960s. The mines harbor hard coal (“rock coal”) belonging to the fat coals according to the German classification (reference 33 and references therein). Briefly, large pieces of coal (2 to 15 cm in diameter) and mine timber (2 to 10 cm in diameter) were collected aseptically in glass bottles that were immediately flushed with N2 and stored at 4°C until further processing. In situ temperatures were 35 to 36°C, with 100% air humidity. Coal and mine timber samples were processed in an anaerobic chamber under a nitrogen atmosphere to prevent oxidation. Samples were homogenized and distributed into Hungate tubes containing 500 ml of sulfate-free mineral medium (36) with a salinity of 15 practical salinity units (PSU), according to in situ values. Controls were supplemented with 10 mM 2-bromoethanesulfonate (BES) to exclude abiotic degassing by inhibiting methanogenesis. Enrichment cultures were amended with 10 mM fully 13C-labeled acetate (Campro Scientific) or [13C]bicarbonate (Campro Scientific) plus H2. All incubated samples were frozen at −80°C after an incubation time of 6 months until further processing for DNA-SIP. All incubations were carried out at least in triplicate. Subsamples of all incubated samples were taken at the beginning and at month 3 of incubation. The increases in the amounts of methane and hydrogen in the headspace, as well as the stable isotopes of methane, were monitored continuously over 6 months and were analyzed by gas chromatography-mass spectrometry (GC-MS) as described previously by Krüger et al. (13). Concentrations of acetate were analyzed by high-performance liquid chromatography (Agilent Technologies) using a Zorbax Eclipse Plus C8 USP L7 column (Agilent Technologies) at 60°C. The eluent was a 5 mM H2SO4-methanol gradient at 1 ml/min. Acetate was detected by a diode array detector (DAD; Agilent Technologies).
DNA extraction, isopycnic centrifugation, and gradient fractionation.
DNA was extracted from 0.5 g of incubation slurry after 0, 3, and 6 months using phenol-chloroform extraction as described by Lueders et al. (18). Three parallel extractions were carried out, and extracts were pooled for each incubation treatment. DNA was checked by standard agarose gel electrophoresis and was quantified using PicoGreen staining according to the method of Wilms et al. (37).
Gradient preparation, isopycnic centrifugation, and gradient fractionation were performed as described by Lueders et al. (17) with minor modifications. Each gradient consisted of 6.3 ml of CsCl (approximately 1.72 g ml−1; Calbiochem) and ca. 1 ml of gradient buffer (100 mM Tris-HCl [pH 8.0] liter−1, 100 mM KCl liter−1, 1 mM EDTA liter−1) including 2,000 ng of DNA. Prior to centrifugation, the average density of the centrifugation medium was controlled refractometrically and was adjusted to 1.84 g cm−3. The samples were centrifuged in 6.3-ml Quick-Seal Polyallomer tubes (Beckman) in an NVT 65 near-vertical rotor (Beckman) using an LE-70 ultracentrifuge (Beckman Instruments). Centrifugation was performed at 20°C for 36 h at 44,500 rpm (184,000 × g). Gradients were fractionated as described previously by Neufeld et al. (21). Briefly, the gradients were fractionated from bottom to top into 12 equal fractions (400 μl). A precisely controlled flow rate was achieved by displacing the gradient medium with water at the top of the tube using a Graseby 3100 syringe pump at a flow rate of 1 ml min−1. The density of each collected fraction (a small aliquot of 100 μl) was measured by determining the refractory index using a digital refractometer (model AR-20; Reichert Analytical Instruments, Depew, NY). Subsequently, the DNA was precipitated using polyethylene glycol 6000 (Aldrich Chemistry). The DNA pellet was washed once with 70% ethanol and was dissolved in 25 μl of elution buffer.
Quantification of archaeal and bacterial 16S rRNA genes in density gradient fractions.
DNA was precipitated from gradient fractions and was quantified fluorometrically and by quantitative PCR (qPCR) using the Ar109f/Ar912rt and Ba27F/907R primer systems described by Lueders et al. (17). The qPCR mixtures contained 12.5 μl of the premix solution of a DyNAmo HS SYBR Green qPCR kit (New England Biolabs, Inc., Hitchin, United Kingdom), 1 μl of each primer, and 10 μl of the standard or DNA extract as a template in a final reaction mixture volume of 25 μl. The PCR was carried out in a Rotor-Gene 3000 cycler (Corbett Research, Sydney, Australia). After initial denaturation at 95°C for 15 min, 50 cycles followed. Each cycle consisted of denaturation for 30 s at 94°C, annealing for 20 s at 52°C, elongation for 30 s at 70°C, and fluorescence measurement at 70°C. To check the amplification specificity, fluorescence was also measured at the end of each cycle for 20 s at 80°C. After the last cycle, a melting curve was recorded by increasing the temperature from 50°C to 99°C (1°C every 10 s). The numbers of bacterial and archaeal 16S rRNA gene targets were calculated from the DNA concentrations according to the work of Süss et al. (30). DNA standards for quantitative (real-time) PCR were prepared as described by Wilms et al. (37) and Engelen et al. (6). Bacterial and archaeal targets were measured in at least three different dilutions of DNA extracts (1:10 to 1:1,000) and in triplicate.
PCR and DGGE analysis.
For denaturing gradient gel electrophoresis (DGGE), an 803-bp fragment of the archaeal 16S rRNA gene was amplified by using primers Ar109f (5′-AC KGC TCA GTA ACA CGT-3′) and Ar912rt (5′-GTG CTC CCC CGC CAA TTC CTT TA-3′). For the analysis of bacterial composition, primers BA27F (5′-AGA GTT TGA TCM TGG CTC AG-3′) and 907R (5′-CCG TCA ATT CCT TTG AGT TT-3′) were used to amplify an 880-bp fragment of the bacterial 16S rRNA gene. At the 5′ end of each forward primer, an additional 40-nucleotide GC-rich sequence (GC-clamp) was added to achieve a stable melting point for the DNA fragments in the DGGE according to the method of Muyzer et al. (20). PCR amplification was performed using an Eppendorf thermal cycler system (Mastercycler; Eppendorf, Hamburg, Germany) as follows: 2 μl (1 to 100 ng) of template DNA, 1 U of Taq DNA polymerase, the manufacturer's recommended buffer supplied with the polymerase enzyme, 10 mM deoxynucleoside triphosphates (dNTPs), 50 μM (each) appropriate primers, and 10 mM bovine serum albumin (BSA) were adjusted to a total volume of 50 μl with PCR-grade water (Ampuwa; Fresenius, Bad Homburg, Germany). The PCR program for archaeal and bacterial DNA included an initial denaturation step for 5 min at 94°C, followed by 30 cycles of 30 s at 94°C, 30 s at 52°C, and 1 min at 72°C. Primer extension was carried out for 5 min at 72°C. Aliquots (5 μl) of the PCR products were analyzed by agarose gel electrophoresis in 1.5% (wt/vol) agarose gels and ethidium bromide (0.8 ng ml−1) staining for 20 min on a UV transilluminator as described previously (37).
DGGE was performed using an INGENYphorU-2 system (Ingeny, Goes, Netherlands). PCR products and loading buffer (40% [wt/vol] glycerol, 60% [wt/vol] 1× Tris-acetate-EDTA [TAE], and bromphenol blue) were mixed in a 1:2 ratio. The PCR amplicons were applied directly to 6% (wt/vol) polyacrylamide gels with a linear gradient of 30 to 80% denaturant for archaeal PCR products and 50 to 70% denaturant for bacterial PCR products (100% denaturant corresponds to 7 M urea and 40% [vol/vol] formamide). Electrophoresis was carried out in 1× TAE buffer (40 mM Tris-acetate [pH 7.4], 20 mM sodium acetate, 1 mM Na2 EDTA) at a constant voltage of 100 V and a temperature of 60°C for 20 h. After electrophoresis, the gels were stained for 2 h in 1× SYBR Gold solution (Molecular Probes, Eugene, OR) in 1× TAE and were washed for 20 min with distilled water. The gel was digitized using a digital imaging system (BioDocAnalyze; Biometra, Göttingen, Germany) with UV transillumination (302 nm).
Reamplification and sequencing of DGGE bands.
DGGE bands were excised for sequencing using a sterile scalpel and were treated as described by Del Panno et al. (3). Briefly, the bands were transferred into 50 μl of PCR-grade water and were incubated for 48 h at 4°C. For reamplification, 2 μl of the supernatant was taken as a template using the reaction mixture described above with a final volume of 50 μl. The PCR protocol adjusted for the reamplification was the same as that described above, with the following minor modifications: the total number of cycles was 25, and the final elongation was carried out at 72°C for 10 min. PCR products were purified by using the QIAquick PCR purification kit (Qiagen GmbH) and were eluted in 30 μl of PCR-grade water. DNA yields were estimated fluorometrically in a microtiter plate reader (FLUOstar Optima; BMG Labtechnologies, Offenburg, Germany) using a 1:200-diluted PicoGreen reagent according to a modified manufacturer′s protocol (Molecular Probes, Eugene, OR) as described in detail by Wilms et al. (37). Sequence analyses were performed by GATC Biotech AG (Konstanz, Germany). Sequences were compared to those in GenBank using the BLAST tool of the National Center for Biotechnology Information server (1).
Nucleotide sequence accession numbers.
The sequences determined in this study have been deposited in the GenBank nucleotide sequence database under accession no. FR838951 to FR838999.
RESULTS
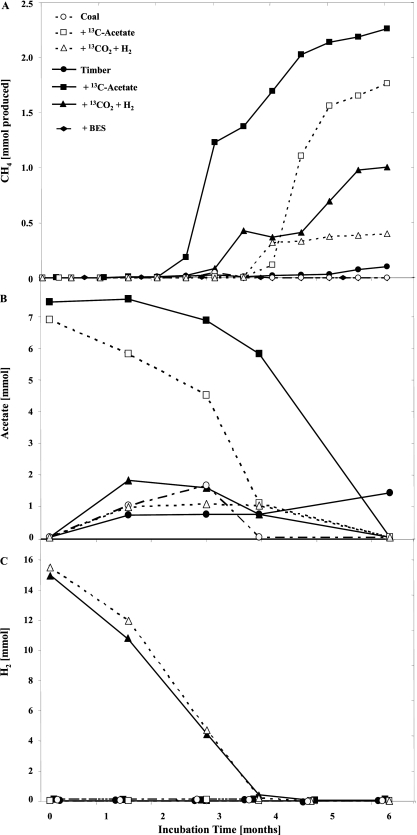
For stable-isotope probing (SIP), microcosms with coal or timber samples were amended with [13C]acetate or H2-13CO2 and were incubated under in situ conditions. The accumulation of CH4 was followed over a period of 6 months. To identify active community members, samples for DNA extraction were taken at the beginning of incubation and after 3 and 6 months (Fig. 1A). The day zero incubation samples served as SIP controls.
Fig. 1.
Long-term incubation of weathered hard coal and mine timber amended with 13C-labeled acetate or H2-CO2. Shown are microbial methane formation (A) and acetate (B) and hydrogen (C) depletion/formation. Measurements were carried out in 5 replicates.
Acetate is the precursor of methane.
CH4 production was observed in all incubated samples (Fig. 1A), with higher activities in timber than in coal enrichment cultures. The highest CH4 formation rates (0.13 μmol per g [wet weight] and day) were detected in the [13C]acetate enrichment cultures between months 3 and 6 of incubation. The addition of H2-13CO2 resulted in less stimulation of methanogenesis (maximum rate, 0.05 μmol per g [wet weight] and day). The isotopic signature of CH4 indicated that methane was formed from the labeled substrates added (data not shown). No methane formation was observed in control samples incubated with the methanogenesis inhibitor 2-bromoethanesulfonate (BES). Therefore, the possibility of abiotic degassing of adsorbed methane from the incubated samples can be excluded.
While H2 was largely used up after 3 months, acetate was completely depleted after 6 months (Fig. 1B and C). Interestingly, acetate formation was observed in the H2-13CO2 cultures, while there was no H2 formation in the samples incubated with acetate. The fact that acetate formation was also detected in the unamended incubated samples indicates the central role of acetate in the process of methanogenesis. The isotopic signature of the acetate formed showed strong labeling, indicating its formation from 13CO2 (data not shown).
DNA-SIP reveals that methanogenesis is mediated by Methanosarcina spp.
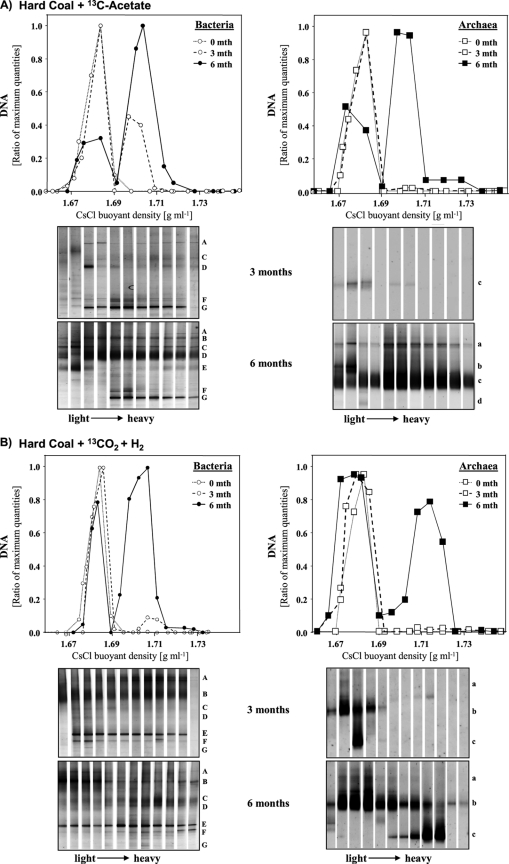
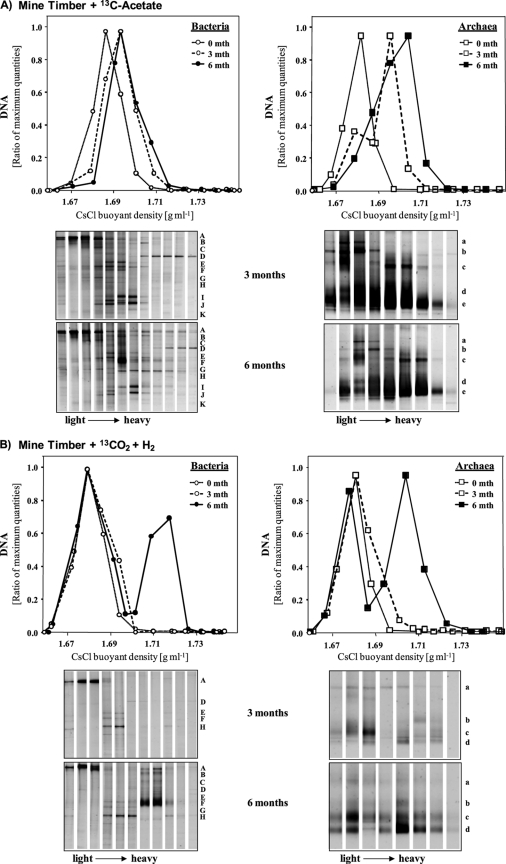
In order to identify active community members, samples from the 13C-enriched incubated samples were analyzed by SIP. Density-resolved archaeal DNA was detected in gradient fractions using archaeal qPCR (Fig. 2 and 3). The amount of archaeal DNA detected in the heavy fractions increased over the incubation time between months 3 and 6. Only the timber cultures showed 13C-labeled archaeal DNA after 3 months, with maximum quantities in DNA gradients from acetate enrichment cultures. After 6 months, “light” and “heavy” DNA fractions substantiated a clear labeling of archaeal DNA in timber and coal cultures amended with [13C]acetate or 13CO2 (Fig. 2 and 3).
Fig. 2.
Quantitative PCR distribution and DGGE community profiles of density-resolved bacterial and archaeal DNA in SIP centrifugation gradients after 3 and 6 months of incubation of hard coal with [13C]acetate (A) or H2-13CO2 (B). Each letter on the right of a DGGE profile indicates the same band throughout the gradient. Band letters correspond to those in Table 1. The same band letter indicates the same organism.
Fig. 3.
Quantitative PCR distribution and DGGE community profiles of density-resolved bacterial and archaeal DNA in SIP centrifugation gradients after 3 and 6 months of incubation of mine timber with [13C]acetate (A) or H2-13CO2 (B). Each letter on the right of a DGGE profile indicates the same band throughout the gradient. Band letters correspond to those in Table 2. The same band letter indicates the same organism.
Next, the archaeal community members detected in “light” and “heavy” gradient fractions were analyzed by denaturing gradient gel electrophoresis (DGGE) and subsequent band sequencing (Tables 1 and 2). In general, the compositions of the total archaeal communities in the [13C]acetate and H2-13CO2 cultures were similar, and these communities were dominated by relatives of Methanosarcina spp., Methanosaeta spp., and uncultured Crenarchaeota (Tables 1 and 2). However, clear 13C labeling was evident mainly for relatives of Methanosarcina spp. Increased band intensities reflected larger amounts of 13C-labeled DNA in acetate cultures (Fig. 2 and 3). To a minor extent, labeled DNA was also observed for a DGGE band related to Methanosaeta spp. in the acetate enrichment cultures. Members of the Crenarchaeota were abundant in the original coal and timber samples but showed no incorporation of 13C-labeled substrates.
Table 1.
Bacterial and archaeal diversity in incubated hard-coal samples amended with [13C]acetate or H2-13CO2a
| Labeled substrate and closest cultured relative | Similarity (%) | Band letter in Fig. 2 | GenBank accession no. |
|---|---|---|---|
| [13C]acetate | |||
| Bacteria | |||
| Burkholderia cepacia | 100 | A | FR838981 |
| Pseudoalteromonas sp. | 100 | B | FR838963 |
| Clostridium sp. | 99 | C | FR838991 |
| Pseudomonas stutzeri | 99 | D | FR838972 |
| Desulfovibrio africanus | 100 | E | FR838976 |
| Pelobacter acetylenicus | 95 | F | FR838952 |
| Uncultured Geobacteraceae | 99 | G | FR838962 |
| Archaea | |||
| Methanosaeta sp. | 98 | a | FR838958 |
| Uncultured Crenarchaeota | 98 | b | FR838970 |
| Methanosarcina barkeri | 99 | c | FR838992 |
| Methanosarcina sp. | 99 | d | FR838971 |
| H2-13CO2 | |||
| Bacteria | |||
| Burkholderia cepacia | 100 | A | FR838986 |
| Desulfovibrio alkaliphilus | 92 | B | FR838983 |
| Clostridium sp. | 99 | C | FR838998 |
| Desulfovibrio africanus | 100 | D | FR838979 |
| Pelobacter acetylenicus | 95 | E | FR838951 |
| Uncultured Geobacteraceae | 98 | F | FR838960 |
| Xanthomonas sp. | 100 | G | FR838954 |
| Archaea | |||
| Methanosaeta sp. | 98 | a | FR838957 |
| Uncultured Crenarchaeota | 99 | b | FR838966 |
| Methanosarcina barkeri | 93 | c | FR838997 |
Detected by denaturing gradient gel electrophoresis of density-resolved DNA gradient fractions. The phylogenetic affiliations of the 16S rRNA genes from microbes that incorporated 13C-labeled substrates are shaded.
Table 2.
Bacterial and archaeal diversity in incubated mine timber samples amended with [13C]acetate and H2-13CO2a
| Labeled substrate and closest cultured relative | Similarity (%) | Band letter in Fig. 3 | GenBank accession no. |
|---|---|---|---|
| [13C]acetate | |||
| Bacteria | |||
| Acholeplasma sp. | 95 | A | FR838980 |
| Uncultured Bacteroidetes | 99 | B | FR838984 |
| Clostridium sp. | 99 | C | FR838974 |
| Burkholderia cepacia | 100 | D | FR838994 |
| Hyphomicrobium sp. | 99 | E | FR838953 |
| Hydrogenophaga sp. | 94 | F | FR838955 |
| Desulfovibrio sp. | 95 | G | FR838961 |
| Desulfovibrio africanus | 100 | H | FR838982 |
| Halochromatium sp. | 92 | I | FR838989 |
| Desulfuromonas acetexigens | 95 | J | FR838965 |
| Uncultured Geobacteraceae | 96 | K | FR838964 |
| Archaea | |||
| Uncultured Crenarchaeota | 98 | a | FR838968 |
| Uncultured Crenarchaeota | 98 | b | FR838969 |
| Methanosaeta sp. | 98 | c | FR838959 |
| Methanosarcina barkeri | 99 | d | FR838995 |
| Methanosarcina barkeri | 93 | e | FR838996 |
| H2-13CO2 | |||
| Bacteria | |||
| Acholeplasma sp. | 95 | A | FR838990 |
| Uncultured Bacteroidetes | 99 | B | FR838988 |
| Clostridium sp. | 99 | C | FR838975 |
| Burkholderia cepacia | 99 | D | FR838987 |
| Hyphomicrobium sp. | 99 | E | FR838956 |
| Hydrogenophaga sp. | 94 | F | FR838985 |
| Desulfovibrio sp. | 95 | G | FR838973 |
| Desulfovibrio africanus | 95 | H | FR838999 |
| Archaea | |||
| Uncultured Crenarchaeota | 98 | a | FR838967 |
| Methanosarcina sp. | 100 | b | FR838977 |
| Methanosarcina sp. | 100 | c | FR838978 |
| Methanosarcina barkeri | 99 | d | FR838993 |
Detected by denaturing gradient gel electrophoresis of density-resolved DNA gradient fractions. The phylogenetic affiliations of the 16S rRNA genes from microbes that incorporated 13C-labeled substrates are shaded.
Identification of active Bacteria.
Quantities of labeled bacterial 16S rRNA genes increased over the incubation time (Fig. 2 and 3), as found for the Archaea. While strongly labeled bacterial DNA was detected especially in the [13C]acetate-amended coal samples after 3 months, H2-13CO2 cultures showed substantial quantities of 13C-labeled DNA only after 6 months of incubation.
In the acetate-amended coal samples, bacterial DNA labeled after 3 months was affiliated with a lineage of uncultured Geobacteraceae and Pelobacter spp., and bacterial DNA labeled after 6 months was additionally affiliated with Pseudoalteromonas and Clostridium spp. Most surprisingly, however, abundant unlabeled and labeled DNA was detected for a relative of a Pseudomonas sp. after 6 months only. In H2-13CO2-amended coal samples, clearly labeled DNA was evident from gradient fractions in DGGE analysis after 6 months only and was affiliated with Clostridium, Desulfovibrio, and Pelobacter spp., as well as, again, with uncultured Geobacteraceae.
In [13C]acetate-amended timber cultures, members of chemoautotrophic bacteria (Hydrogenophaga and Hyphomicrobium spp.) as well as sulfate and sulfur reducers (Desulfovibrio and Desulfuromonas spp.) were detected primarily in “intermediate” and “heavy” gradient fractions. Surprisingly, the DGGE band dominating the highest-density DNA gradient fractions was related to Burkholderia spp. This could be a methodological bias due to the high-GC DNA content of Burkholderia spp. In H2-13CO2-amended timber microcosms, Hydrogenophaga spp. clearly dominated “heavy” DNA after 6 months, while a relative of Desulfovibrio spp. was detected in “intermediate” gradient fractions after 3 months.
DISCUSSION
In the present study, we have identified active microbes responsible for methane formation in samples taken from abandoned coal mines. We demonstrated that acetate is the main precursor of methane and identified the microbes involved in the processes leading to methane formation.
Within the past few years, methane release has also been observed and studied in other coal mines worldwide. Several of the community members we have detected in our samples have been found previously in other coal mine deposits (especially Pelobacter acetylenicus, Clostridium spp., Pseudomonas spp., uncultured Geobacteraceae, and Methanosarcina spp.), indicating their potential role in the process of methane release (8, 12, 25, 26). A predominance of acetoclastic Methanosarcinales was already shown in two other coal seams investigated recently (9, 34) and in comparable habitats, i.e., hydrocarbon-contaminated aquifers (5). However, in coal mine water and drainage liquids, hydrogenotrophic methanogens were prevalent (7, 29, 35). Hydrogenotrophic methanogenesis dominated in the drainage water of coal reservoirs. In the floating systems, easily degradable substrates that lead to intermediate hydrogen formation might be released.
Methane release via acetoclastic methanogenesis.
The fact that Methanosarcina spp. were responsible for methane production in our enrichment cultures is in agreement with our earlier studies showing that the Methanosarcinales are most abundant in in situ coal and timber samples (2). Although Methanosarcina spp. are known to use either hydrogen or acetate, those identified here seem to be strictly adapted to the conditions in this habitat. In the mines studied, acetate seems to be quantitatively more available for the Methanosarcinales. Hydrogen might hardly be formed at low metabolic rates and therefore might not be available for methanogens. Even after an incubation time of 6 months with an adequate supply of hydrogen, coal mine methanogens did not make direct use of hydrogen for methane production. Hydrogen appeared rather to be used by acetogens producing acetate, which then, in turn, was utilized by the Methanosarcinales.
Active Geobacteraceae predominate in coal.
Acetotrophic members of the Geobacteraceae were found to be labeled in the coal enrichment cultures. Their abundance could be increased by amendment with acetate. However, they were also active in the samples incubated with hydrogen. This could be explained by secondary cross-feeding processes, since labeled acetate was formed (15). In our cultures, it is not clear which electron acceptor is used by the Geobacteraceae for acetate oxidation. One possibility could be the utilization of electron acceptors, such as sulfur, directly from the coal, since members of the family Geobacteraceae are reported to be elemental-sulfur reducers (11). Jones et al. (12) also obtained high numbers of Geobacter species from coal, but none of the known electron acceptors was present, suggesting that Geobacter might be capable of coupling the degradation of organics to an electron- or H2-accepting partner. This could also be proposed in our case. However, in contrast to our strictly anoxic enrichment cultures, coal mines showed low concentrations of oxygen in layers of coal near the surface, and Geobacteraceae constituted the bulk of the overall bacterial community in the original coal samples (F. Gründger, unpublished data).
Coal and timber: two substrates, two different active bacterial communities.
The active bacterial communities in coal and timber differ. Amendment with acetate or H2-CO2 did not have a strong effect on the composition of the active community. Besides the Geobacteraceae, the active community in the coal samples comprised Pelobacter acetylenicus and members of Clostridium and Pseudomonas species. In earlier studies, Pseudomonas stutzeri was isolated from coal samples; this species is potentially able to utilize polycyclic aromatic hydrocarbons (PAHs) (24). This suggestion can be supported by the fact that the first event of coal fragmentation is exoenzymatic hydrolysis into small PAHs (29). Moreover, Pseudomonas stutzeri is even more active when acetate is present as a second electron donor (19), and that could be one reason for its predominance in the coal enrichment cultures amended with acetate. However, oxygen and nitrate were not available as electron acceptors.
Accumulation of acetate was detected in both supplemented and nonsupplemented cultures. The presence of isotopically heavy acetate in samples incubated with H2-13CO2 indicated its new formation from CO2. Relatives of Pelobacter and Clostridium species might be involved in this process. For Pelobacter acetylenicus, acetate formation from acetylene has been described as well (23). Which other coal and timber compounds could be feasible substrates for acetogenesis is not known. However, acetate formation appeared to continue, since the levels do not approach zero in most cases.
In the timber enrichment cultures, active bacteria similar to Hydrogenophaga and Clostridium species predominated, suggesting that they might use timber compounds or secondary fermentative products for metabolism. Only the H2-13CO2-amended coal cultures showed distinct labeling of bacteria after 6 months. The slight heavy shift in the acetate-amended coal cultures might not be an indication of bacterial activity, but the result of an increase in the amount of bacterial DNA with a high GC content, such as Burkholderia species (Fig. 3; Table 2). Like the coal cultures, the H2-13CO2-enriched timber cultures also showed acetate formation. Obviously, timber provides acetogenic substrates other than those from coal. However, Hydrogenophaga as well as Clostridium species were recently also detected in coal samples from another mine (12).
Acetate is the main precursor of methane.
In connecting the results from our earlier investigations (2, 14) with our new findings in the present study, we present the following arguments supporting the notion that acetoclastic methanogenesis is the main route for methane formation. First, the natural isotopic signatures of methane indicated an acetoclastic origin, supported by the isotopic signatures of acetate that was formed from 13CO2 (14). Second, the highest methane formation rates were observed in the acetate-amended enrichment cultures of coal and timber, while H2 gave lower activities. Third, acetate was depleted in the acetate cultures but accumulated in the H2-CO2- and BES-treated enrichment cultures. Finally, DNA-SIP revealed that relatives of Methanosarcina spp. were responsible for methane production in both [13C]acetate- and H2-13CO2-amended cultures, and in the latter, their activity was coupled to the activity of bacteria related to acetogens.
ACKNOWLEDGMENTS
We thank Daniela Zoch (Federal Institute for Geosciences and Natural Resources [BGR], Hannover, Germany) and Katrin Hörmann (Helmholtz Zentrum, Munich, Germany) for technical assistance.
Financial support was provided by the BGR. Funding for M.K. was partially provided by the Deutsche Forschungsgemeinschaft (DFG) (grants KR 3311/5-1 and 6-1) and the Bundesministerium für Bildung und Forschung (grant 03G0697A). F.V.N. was financed by a grant from the DFG to T.L. within priority program SPP 1319 (grant LU 1188/4-1).
Footnotes
Published ahead of print on 1 April 2011.
REFERENCES
- 1. Altschul S. F., et al. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389–3402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Beckmann S., Krüger M., Engelen B., Gorbushina A. A., Cypionka H. 2011. Role of bacteria, archaea and fungi involved in methane release in abandoned coal mines. Geomicrobiol. J. 28(4):UGMB-2010-0055.R1 [Google Scholar]
- 3. Del Panno M., Morelli I., Engelen B., Berthe-Corti L. 2005. Effect of petrochemical sludge concentration on microbial communities during a soil bioremediation process. FEMS Microbiol. Ecol. 53:305–316 [DOI] [PubMed] [Google Scholar]
- 4. Denman K., et al. 2007. Couplings between changes in the climate system and biogeochemistry, chapter 7. In Solomon S., Qin D., Manning M., Chen Z., Marquis M., Averyt K. B., Tignor M., Miller H. L. (ed.), Climate change 2007: the physical science basis. Contribution of Working Group I to the Fourth Assessment Report of the Intergovernmental Panel on Climate Change Cambridge University Press, Cambridge, England [Google Scholar]
- 5. Dojka M. A., Hugenholtz P., Haack S. K., Pace N. R. 1998. Microbial diversity in a hydrocarbon- and chlorinated-solvent-contaminated aquifer undergoing intrinsic bioremediation. Appl. Environ. Microbiol. 64:3869–3877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Engelen B., et al. 2008. Fluids from the oceanic crust support microbial activities within the deep biosphere. Geomicrobiol. J. 25:56–66 [Google Scholar]
- 7. Flores R. M. 2008. Microbes, methanogenesis, and microbial gas in coal. Int. J. Coal Geol. 76:1–2 [Google Scholar]
- 8. Fry C. J., et al. 2009. Prokaryotic populations and activities in an interbedded coal deposit, including a previously deeply buried section (1.6-2.3 km) above ∼150 Ma basement rock. Geomicrobiol. J. 26:163–178 [Google Scholar]
- 9. Green M. S., Flanegan K. C., Gilcrease P. C. 2008. Characterization of a methanogenic consortium enriched from a coalbed methane well in the Powder River Basin, U.S.A. Int. J. Coal Geol. 76:34–45 [Google Scholar]
- 10. Han B., et al. 2009. Diversity and activity of methanotrophs in alkaline soil from a Chinese coal mine. FEMS Microbiol. Ecol. 70:40–51 [DOI] [PubMed] [Google Scholar]
- 11. Holmes D. E., Kelly P. N., Lovley D. R. 2004. Comparison of 16S rRNA, nifD, recA, gyrB, rpoB, and fusA genes within the family Geobacteraceae fam. nov. Int. J. Syst. Evol. Microbiol. 54:1591–1599 [DOI] [PubMed] [Google Scholar]
- 12. Jones E. J. P., Voytek M. A., Corum M. D., Orem W. H. 2010. Stimulation of methane generation from nonproductive coal by addition of nutrients or a microbial consortium. Appl. Environ. Microbiol. 76:7013–7022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Krüger M., Frenzel P., Conrad R. 2001. Microbial processes influencing methane emission from rice fields. Global Change Biol. 7:49–63 [Google Scholar]
- 14. Krüger M., et al. 2008. Microbial methane formation from hard coal and timber in an abandoned coal mine. Geomicrobiol. J. 25:315–321 [Google Scholar]
- 15. Lanoil B. D., Han S.-K. 2003. Identification of microbes responsible for acetate consumption in soils under different wetting regimes. Kearney Foundation of Soil Science: Soil Carbon and California′s Terrestrial Ecosystems, 2001-2006 Mission. Final Report: 2001028, 1/1/2002-12/31/2003. Kearney Foundation, Division of Agriculture and Natural Resources, University of California, Davis, CA [Google Scholar]
- 16. Lu Y., Lueders T., Friedrich M. W., Conrad R. 2005. Detecting active methanogenic populations on rice roots using stable isotope probing. Environ. Microbiol. 7:326–336 [DOI] [PubMed] [Google Scholar]
- 17. Lueders T., Wagner B., Claus P., Friedrich M. W. 2004. Stable isotope probing of rRNA and DNA reveals a dynamic methylotroph community and trophic interactions with fungi and protozoa in oxic rice field soil. Environ. Microbiol. 6:60–72 [DOI] [PubMed] [Google Scholar]
- 18. Lueders T., Manefield M., Friedrich M. W. 2004. Enhanced sensitivity of DNA- and rRNA-based stable isotope probing by fractionation and quantitative analysis of isopycnic centrifugation gradients. Environ. Microbiol. 6:73–78 [DOI] [PubMed] [Google Scholar]
- 19. Mittal M., Rockne K. J. 2008. Indole production by Pseudomonas stutzeri strain NAP-3 during anaerobic naphthalene biodegradation in the presence of dimethyl formamide. J. Environ. Sci. Health A Tox. Hazard. Subst. Environ. Eng. 43:1027–1034 [DOI] [PubMed] [Google Scholar]
- 20. Muyzer G., de Waal E. C., Uitterlinden A. G. 1993. Profiling of complex microbial populations by denaturing gradient gel electrophoresis analysis of polymerase chain reaction-amplified genes coding for 16S rRNA. Appl. Environ. Microbiol. 59:695–700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Neufeld J. D., et al. 2007. DNA stable-isotope probing. Nat. Protoc. 2:860–866 [DOI] [PubMed] [Google Scholar]
- 22. Radajewski S., Ineson P., Parekh N. R., Murell J. C. 2000. Stable-isotope probing as a tool in microbial ecology. Nature 403:646–649 [DOI] [PubMed] [Google Scholar]
- 23. Schink B. 1985. Fermentation of acetylene by an obligate anaerobe, Pelobacter acetylenicus sp. nov. Arch. Microbiol. 142:295–301 [Google Scholar]
- 24. Schreiber A. F., Winkler U. K. 1983. Transformation of tetralin by whole cells of Pseudomonas stutzeri AS39*. Eur. J. Appl. Microbiol. Biotechnol. 18:6–10 [Google Scholar]
- 25. Shimizu S., et al. 2007. Molecular characterization of microbial communities in deep coal seam groundwater of northern Japan. Geobiology 5:423–433 [Google Scholar]
- 26. Shin S. G., Lee C. S., Hwang K., Ahn J. H., Hwang S. 2008. Use of order-specific primers to investigate the methanogenic diversity in acetate enrichment system. J. Ind. Microbiol. Biotechnol. 35:1345–1352 [DOI] [PubMed] [Google Scholar]
- 27. Stams A. J. M. 1994. Metabolic interactions between anaerobic bacteria in methanogenic environments. Antonie Van Leeuwenhoek 66:271–294 [DOI] [PubMed] [Google Scholar]
- 28. Strapoc D., Mastalerz M., Schimmelmann A., Drobniak A., Hedges S. 2008. Variability of geochemical properties in a microbially dominated gas system from the eastern margin of the Illinois Basin, U. S. A. Int. J. Coal Geol. 76:98–110 [Google Scholar]
- 29. Strapoc D., et al. 2008. Methane-producing microbial community in a coal bed of the Illinois Basin. Appl. Environ. Microbiol. 74:2424–2432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Süss J., et al. 2006. Widespread distribution and high abundance of Rhizobium radiobacter within Mediterranean subsurface sediments. Environ. Microbiol. 8:1753–1763 [DOI] [PubMed] [Google Scholar]
- 31. Tao M. X., et al. 2007. Secondary biological coalbed gas in the Xinji area, Anhui province, China: evidence from the geochemical features and secondary changes. Int. J. Coal Geol. 71:358–370 [Google Scholar]
- 32. Thauer R. K., Möller-Zinkhan D., Spormann A. M. 1989. Biochemistry of acetate catabolism in anaerobic chemotrophic bacteria. Annu. Rev. Microbiol. 43:43–67 [DOI] [PubMed] [Google Scholar]
- 33. Thielemann T., Cramer B., Schippers A. 2004. Coalbed methane in the Ruhr Basins, Germany: a renewable energy resource? Org. Geochem. 35:1537–1549 [Google Scholar]
- 34. Ulrich G., Bower S. 2008. Active methanogenesis and acetate utilization in Powder River Basin coals, United States. Int. J. Coal Geol. 76:25–33 [Google Scholar]
- 35. Warwick P. D., Breland F. C., Hackley P. C. 2008. Biogenic origin of coalbed gas in the northern Gulf of Mexico Coastal Plain, U.S.A. Int. J. Coal Geol. 76:119–137 [Google Scholar]
- 36. Widdel F., Bak F. 1992. Gram-negative mesophilic sulfate-reducing bacteria, p. 3352–3378 In Balows A., Dworkin M., Harder W., Schleifer K. H. (ed.), The prokaryotes, 2nd ed Springer-Verlag, New York, NY [Google Scholar]
- 37. Wilms R., Sass H., Köpke B., Cypionka H., Engelen B. 2007. Methane and sulfate profiles within the subsurface of a tidal flat are reflected by the distribution of sulfate-reducing bacteria and methanogenic archaea. FEMS Microbiol. Ecol. 59:611–621 [DOI] [PubMed] [Google Scholar]
- 38. Zinder S. H. 1993. Physiological ecology of methanogens, p. 128–206 In Ferry J. G. (ed.), Methanogenesis: ecology, physiology, biochemistry and genetics. Chapman & Hall, New York, NY [Google Scholar]